SUMMARY
Localization of a variety of RNAs to non-membrane-bound cellular compartments such as nucleoli and Cajal bodies is critical for their stability and function. The molecular mechanisms that underly the recruitment and exclusion of RNAs from these phase-separated organelles is incompletely understood. Telomerase is a ribonucleoprotein composed of the reverse transcriptase protein telomerase reverse transcriptase (TERT), the telomerase RNA (TR), and several auxiliary proteins, including TCAB1. Here we show that in the absence of TCAB1, a large fraction of TR is tightly bound to the nucleolus, while TERT is largely excluded from the nucleolus, reducing telomerase assembly. This suggests that nuclear compartmentalization by the non-membrane-bound nucleolus counteracts telomerase assembly, and TCAB1 is required to retain TR in the nucleoplasm. Our work provides insight into the mechanism and functional consequences of RNA recruitment to organelles formed by phase separation and demonstrates that TCAB1 plays an important role in telomerase assembly.
Graphical Abstract
In brief
Klump et al. show that TCAB1, a telomerase cofactor, facilitates telomerase assembly. The results indicate that TCAB1 is not required for telomerase catalysis and that TCAB1 association with the telomerase RNA prevents its entry into the nucleolus to facilitate its interaction with the telomerase reverse transcriptase.
INTRODUCTION
Human cells contain several non-membrane-bound organelles that carry out critical cellular functions. For instance, nucleoli and Cajal bodies are phase-separated nuclear organelles that play important roles in the biogenesis and maturation of cellular RNAs.1,2 Nucleoli and Cajal bodies contain small nucleolar RNAs (snoRNAs) and small Cajal-body-specific RNAs (scaRNAs). A subset of these snoRNAs and scaRNAs are bound by the H/ACA complex, which contains NOP10, NHP2, GAR1, and the dyskerin.3 A key difference between snoRNAs and scaRNAs is the presence of the Cajal-body box (CAB-box) motif in scaRNAs, which directly associates with the telomerase Cajal-body protein 1 (TCAB1).4-6 TCAB1 is required for the recruitment of scaRNAs to Cajal bodies, and scaRNAs localize to the nucleolus when the CAB-box is mutated.6,7 Therefore, TCAB1 controls which phase-separated nuclear organelle scaRNAs associate with.
The telomerase RNA (TR) is an scaRNA and its association with nucleoli and Cajal bodies is controlled by TCAB1.4 Telomere maintenance by telomerase is essential for continuous proliferation of stem cell populations in the human body, and most cancers require telomerase activity for their survival.8 To compensate for the incomplete replication of chromosome ends, telomerase appends TTAGGG repeats to the telomeric single-stranded overhang.4 Telomerase-mediated telomere maintenance requires three critical steps: telomerase assembly, telomerase recruitment to telomeres, and telomeric repeat synthesis.4 Mutations in several genes that cause deficiencies in one of these critical steps lead to a variety of diseases known as telomere syndromes.9 In addition, telomerase is inappropriately activated in >90% of cancers.8 While telomerase recruitment to telomeres10 and telomerase catalysis11 have been studied extensively, much less is known about telomerase assembly. Importantly, telomerase assembly could be targeted to reduce telomerase activity in cancer cells or to increase telomerase function in patients affected by genetically defined telomerase deficiency syndromes.12,13
Telomerase is a complex ribonucleoprotein (RNP). The core components of telomerase are the telomerase reverse transcriptase (TERT) protein, TR, the H/ACA complex, and TCAB1.4 The primary function of the H/ACA complex is to stabilize TR by preventing its exonucleolytic degradation.14,15 The 3′-end formation of TR is tightly regulated by the competing activities of the poly(A) polymerase PAPD5 and the nuclease PARN.16,17 Loss of TCAB1 function leads to telomere attrition in a variety of cell lines.6,18-20 In addition, mutations in TCAB1 cause misfolding of TCAB1 and lead to dyskeratosis congenita, a telomere syndrome.18,21 While these observations highlight that TCAB1 is necessary for telomere synthesis, the underlying molecular mechanism is unclear. Initially, it was proposed that TCAB1 is required for telomerase recruitment to telomeres.6,22 A more recent study suggested that TCAB1 is required for the correct folding of TR and that its absence results in a reduction in telomerase activity.20 All previous studies claim that TCAB1 does not mediate telomerase assembly.
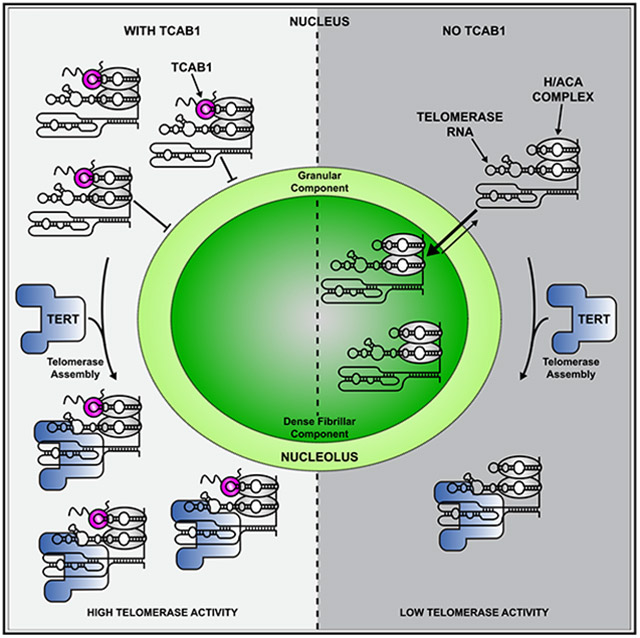
Here, we analyze telomerase assembly in intact cells and by purification of the telomerase RNP and demonstrate that TCAB1 facilitates telomerase assembly in cancer cells. In the absence of TCAB1, a large fraction of TR is tightly associated with the nucleolus while TERT is largely excluded from the nucleolus. This spatial separation of TERT and TR is incompatible with proper telomerase assembly. Furthermore, we show that the limited amount of telomerase that can assemble in the absence of TCAB1 is fully active, suggesting that TCAB1 is not necessary for the enzymatic function of telomerase. Finally, analysis of the sub-cellular dynamics of TCAB1 indicates that it rarely enters the nucleolus, suggesting that TCAB1 associates with TR in the nucleoplasm and prevents its entry into the nucleolus. Collectively, our results support a model in which TCAB1 facilitates telomerase assembly by promoting nucleoplasmic accumulation of TR to increase encounters with TERT, which is also localized in the nucleoplasm. Furthermore, our observations suggest that the nucleolar phase separation constitutes a barrier for telomerase RNP assembly and that incompletely assembled telomerase RNPs are associated with the nucleolus and do not readily enter the nucleoplasm. Thus, it raises the possibility that cellular compartmentalization by phase-separated organelles, such as the nucleolus, can directly regulate RNP function in human cells.
RESULTS
Loss of TCAB1 leads to nucleolar accumulation of TR
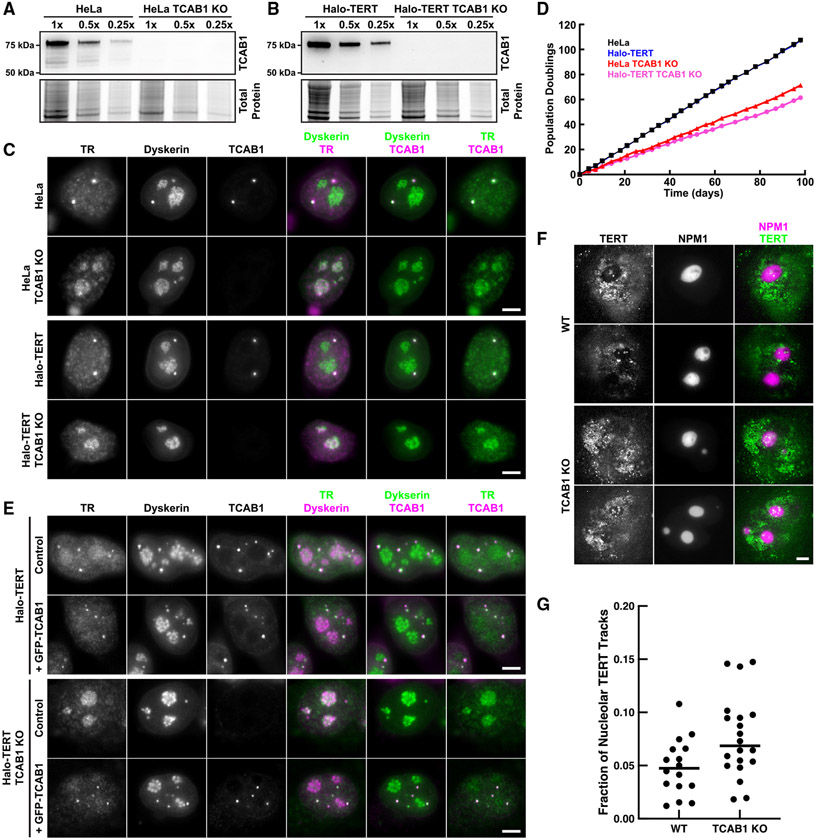
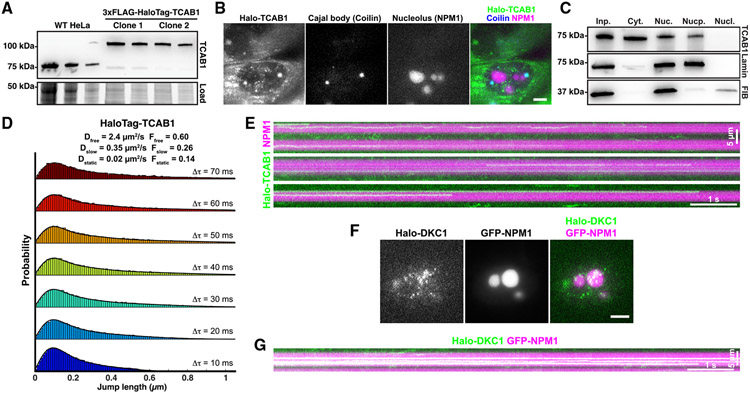
To assess whether TR is sequestered in the nucleolus in the absence of TCAB1, we knocked out TCAB1 in HeLa cells and in HeLa cells expressing 3xFLAG-HaloTag-TERT (Halo-TERT; Figure S1A). TCAB1 knockout was validated by Southern blot, PCR, western blot, and immunofluorescence (IF) imaging (Figures 1A-1C and S1B-S1D). Both HeLa and Halo-TERT TCAB1 knockout cell lines continuously grew at approximately 60% of the rate of their parental cell lines (Figure 1D). Telomeres in cells lacking TCAB1 were stable over time at a shorter length than telomeres in control cells, as previously described (Figure S1E).19 Fluorescence in situ hybridization (FISH) demonstrated that TR accumulated in the nucleolus in cells that lack TCAB1 (Figure 1C). To confirm that dyskerin signals mark nucleoli, we transiently expressed HaloTag-dyskerin and GFP-NPM1, a marker of the granular component (GC) of the nucleolus, in control and TCAB1 knockout cells. Dyskerin was enriched in nucleolar regions with low NPM1 intensity in the presence and absence of TCAB1 (Figure S1F), consistent with its localization to the dense fibrillar component (DFC) of the nucleolus.23 Importantly, expression of GFP-TCAB1 in TCAB1 knockout cells rescued TR localization to Cajal bodies (Figure 1E), confirming that the mislocalization of TR to nucleoli is caused by absence of TCAB1. These observations indicate that TCAB1 is required to prevent TR accumulation in nucleoli.
Figure 1. TR is localized to nucleoli in TCAB1 knockout cells.
(A and B) Western blot probed with TCAB1 antibody of TCAB1 knockouts from (A) HeLa and (B) Halo-TERT cells.
(C) IF with anti-dyskerin and anti-TCAB1 antibodies and FISH using TR probes (scale bar, 5 μm).
(D) Growth rate of parental and TCAB1 knockout cell lines.
(E) IF with anti-dyskerin and anti-TCAB1 antibodies and FISH with TR probes (scale bar, 5 μm).
(F) Maximum intensity projection (1,000 frames) of HaloTag-TERT (JFX650) in control and TCAB1 knockout cells expressing GFP-NPM1 (scale bar, 5 μm).
(G) Quantification of the fraction of TERT trajectories that overlap with the nucleolus in control and TCAB1 knockout cells (n = 16 and 20 cells, mean).
TERT is not enriched in nucleoli in the absence of TCAB1
Our previous observations suggested that TERT does not enter nucleoli in human cancer cells.24 To test whether TERT, like TR, is enriched in nucleoli in the absence of TCAB1, we performed single-molecule imaging of Halo-TERT in living HeLa and TCAB1 knockout cells expressing GFP-NPM1 to mark nucleoli. Halo-TERT was largely excluded from the nucleolus in the presence and absence of TCAB1 with a small number of TERT molecules localizing to nucleoli (Figure 1F; Videos S1 and S2). Single-particle tracking demonstrated that 4.9% and 7.7% of TERT trajectories overlapped with the nucleolus in control and TCAB1 knockout cells, respectively (Figure 1G). To exclude the possibility that nucleolar exclusion is a consequence of the 3xFLAG-HaloTag fused to TERT, we expressed the 3xFLAG-HaloTag fused to a nuclear localization sequence (NLS) in HeLa cells and showed that the nuclear 3xFLAG-HaloTag signals overlapped with nucleoli (Figures S1G and S1H; Video S3). These results demonstrate that Halo-TERT is largely excluded from the nucleolus in the presence and absence of TCAB1 and that this exclusion is an intrinsic property of TERT. In contrast, TR accumulates in the nucleolus in the absence of TCAB1, which demonstrates that the vast majority of TERT does not co-localize with TR in cells lacking TCAB1, suggesting that TERT may not be assembled with TR.
TCAB1 facilitates telomerase RNP assembly
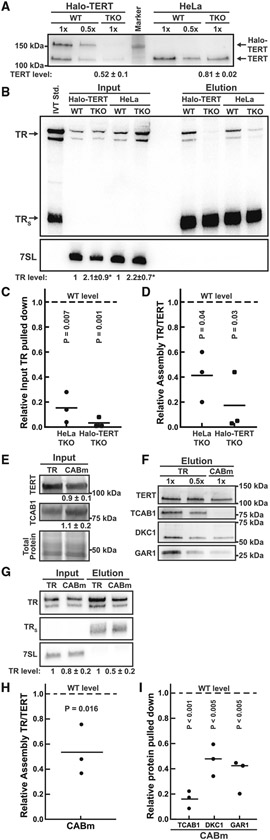
Previous studies have claimed that telomerase assembly is unaffected by the absence of TCAB1, and whether TCAB1 is required for telomerase activity is controversial.6,19,20 To quantitatively analyze the role of TCAB1 in telomerase assembly, we immuno-purified endogenous telomerase using a well-established TERT antibody.25 The amount of TERT purified from TCAB1 knockout cells was reduced to 80% and 50% for the two independent TCAB1 knockout clones compared with control cells (Figure 2A), indicating a lower expression level of TERT in cells lacking TCAB1. In contrast, TR levels were increased ~2-fold (p < 0.02) in TCAB1 knockout cells compared with controls (Figure 2B). To quantify telomerase assembly, we measured TERT levels by western blot and quantified co-purified TR using northern blot (Figures 2A and 2B). The amount of TR associated with TERT relative to total cellular TR was reduced to <20% in TCAB1 knockout cells compared with controls (Figures 2B and 2C). In addition, the ratio of TR relative to TERT, which is a direct measure of telomerase assembly, was reduced to 20%–40% in cells lacking TCAB1 relative to controls (Figures 2A, 2B, and 2D). This excludes the possibility that the lower amount of TR co-purified with TERT from TCAB1 knockout cells is a consequence of the reduction of total TERT purified from these cells (Figures 2A and 2D). These observations strongly suggest that telomerase assembly is defective in cells lacking TCAB1.
Figure 2. Telomerase assembly is reduced in the absence of TCAB1.
(A) Western blots analyzing endogenous TERT immunoprecipitation (IP) probed with a rabbit anti-TERT antibody. TERT level normalized to wild type (WT) (n = 6, mean, SEM).
(B) Northern blot probed for TR and 7SL (loading control) RNA from input and purified endogenous TERT samples. Standards are TR and truncated TRS. Input TR levels relative toWTcontrol normalized to 7SL RNA (n = 3, mean, SD, t test, *p value < 0.05).
(C and D) Quantification of the amount of (C) TR purified relative to input RNA levels, and (D) the ratio of TR relative to endogenous TERT in telomerase purified from TCAB1 knockout cells compared with controls that were normalized to 1 (n = 3, mean, t test).
(E–I) TERT IP from HeLa cells overexpressing TERT and TR (WT) or TRG414C (CABm). (E) Western blots to analyze TERT and TCAB1 levels in cell lysates normalized to total protein levels (n = 3, mean, SD).
(F) Western blots of purified TERT probed with anti-TERT, TCAB1, DKC1, and GAR1 antibodies.
(G) Northern blot probed for TR and 7SL (loading control) of RNA from input and purified endogenous TERT samples. Standards are TR and truncated TRS. Input TR levels relative to WT control normalized to 7SL RNA (n = 3, SD).
(H and I) Quantification of the amount of (H) TR relative to TERT (n = 3, mean, t test), (I) the ratio of TCAB1, DKC1, and GAR1 (n = 3, mean, t test) co-purified with TERT from cells expressing TRG414C (CABm) relative to WT controls normalized to 1.
To verify that the reduction in telomerase assembly observed in TCAB1 knockout cells is a consequence of the inability of TR to interact with TCAB1, we overexpressed TERT and a previously described CAB-box mutant (G414C) of TR that disrupts the TCAB1 interaction site in parental HeLa cells.5,26 TRG414C was enriched in nucleoli (Figure S2A), consistent with previous findings.5 Compared with wild-type TR, telomerase assembly was significantly reduced to about 54% in HeLa cells expressing TRG414C (Figures 2E-2I). In addition, we assessed telomerase assembly by measuring the amount of dyskerin and GAR1 associated with TERT via TR, which showed a similar reduction (~50%) in cells expressing TRG414C compared with wild-type TR (Figures 2F and 2I). We also confirmed that TCAB1 did not associate with TERT purified from cells expressing TRG414C (Figures 2F and 2I).
To ensure that reduction of telomerase assembly observed in the absence of TCAB1 was not a result of altered TR and TERT expression in the TCAB1 knockout cells (Figures 2A and 2B), we overexpressed TERT and TR in parental and TCAB1 knockout cells and purified TERT (Figures S2B-S2L). Consistent with a reduction in telomerase assembly, both TR and dyskerin associated with TERT purified from TCAB1 knockout cells were significantly reduced to ~20%–30% compared with controls (Figures S2B-S2L).
Finally, to ensure that the reduction in telomerase assembly observed in TCAB1 knockout cells is not a consequence of an altered cell-cycle distribution of cells that lacked TCAB1, we purified telomerase from cells synchronized in various stages of the cell cycle. We synchronized cells using thymidine, released and harvested cells immediately (early S-phase), 4 h (mid S-phase), and 8 h (G2/M-phase) after release. Cell-cycle synchronization was confirmed by DNA content analysis (Figure S2M). Telomerase assembly was reduced in TCAB1 knockout cells in all cell-cycle stages analyzed (Figure S2N-S2P). This demonstrates that the reduction of telomerase assembly observed in cells lacking TCAB1 is not a consequence of an aberrant cell-cycle distribution of TCAB1 knockout cells. Altogether, these results demonstrate that telomerase assembly is significantly reduced in the absence of TCAB1 or when the TR-TCAB1 interaction is disrupted by a mutation in the CAB-box.
TCAB1 is not required for telomerase catalytic activity
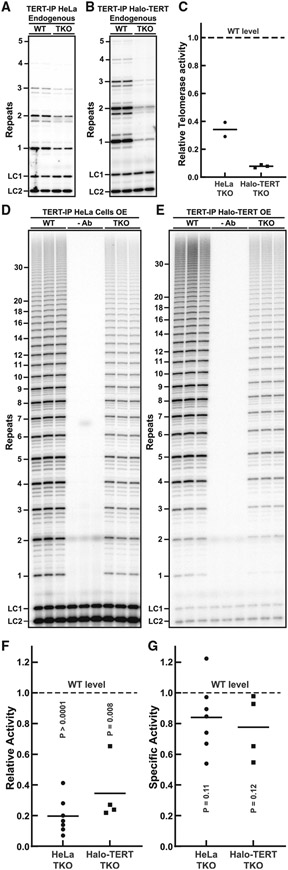
To assess whether TCAB1 plays a role in telomerase catalysis, we analyzed the enzymatic activity of endogenous telomerase purified from TCAB1 knockout cells using the direct telomerase extension assays (Figures 3A and 3B). Consistent with previous results,20 telomerase activity was reduced in the absence of TCAB1 (Figure 3C). To address whether this reduction in telomerase activity was a consequence of the defect in telomerase assembly observed in TCAB1 knockout cells, we determined the specific activity of telomerase by dividing the measured activity by the amount of TR present in the respective telomerase sample. Due to the small amount of TR detected in endogenous telomerase samples (Figures 2B and 2C), the specific activity of endogenous telomerase was highly variable in TCAB1 knockout cells but did not appear to be reduced compared with telomerase purified from control cells (Figure S3A). To overcome this limitation, we determined the specific activity of telomerase purified from cells overexpressing TERT and TR. Similar to endogenous telomerase, activity of overexpressed telomerase purified from HeLa and Halo-TERT cells lacking TCAB1 was significantly reduced to 24% and 34% compared with controls, respectively (Figures 3D-3F and S3B). The specific activity of overexpressed telomerase purified from HeLa and Halo-TERT cells lacking TCAB1 was slightly reduced (84% and 77% relative to control, respectively), but this reduction was not statistically significant (Figure 3G). Together, these observations demonstrate that net cellular telomerase activity is reduced in the absence of TCAB1, while specific enzymatic activity is not. The reduction in cellular telomerase activity closely corresponds to the reduction of telomerase assembly observed in TCAB1 knockout cells, suggesting that the smaller number of telomerase RNPs that form in the absence of TCAB1 are fully active.
Figure 3. The specific activity of telomerase is unchanged in the absence of TCAB1.
(A and B) Direct telomerase extension assay (300 mM KCl) of endogenous telomerase purified from parental (WT) and TCAB1 knockout (A) HeLa and (B) Halo-TERT cell lines (LC1 and LC2 loading controls, n = 2 and 3, mean).
(C) Quantification of endogenous telomerase activity in samples from TCAB1 knockout cells relative to parental controls (n = 3, mean). TKO, TCAB1 knockout.
(D and E) Direct telomerase extension assay (150 mM KCl) of telomerase purified from parental (WT) and TCAB1 knockout overexpressing TERT and TR (–Ab is a no-TERT antibody control). (D) HeLa and (E) Halo-TERT cell lines (LC1 and LC2 loading controls). TKO, TCAB1 knockout.
(F) Quantification of telomerase activity in samples from TCAB1 knockout cells relative to parental controls (n = 4–7, mean, t test).
(G) Specific activity (total activity/TR in sample; see Figure 3F) of overexpressed telomerase purified from TCAB1 knockout cells relative to parental controls normalized to 1 (n = 4–7, mean, t test).
TCAB1 mediates telomerase assembly in living cells
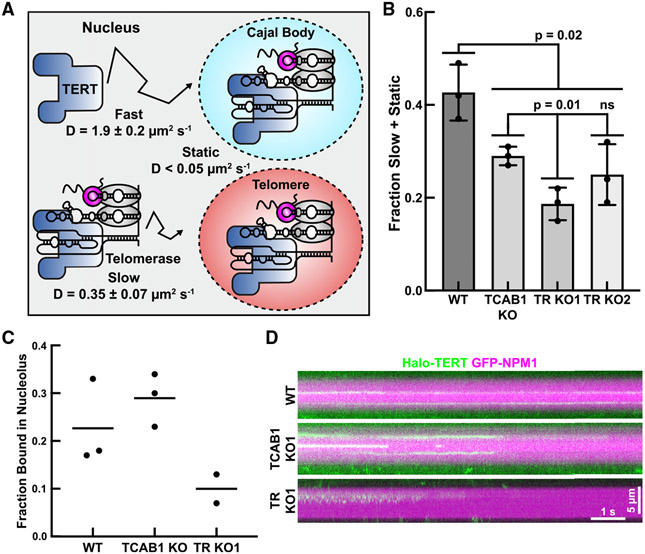
The experiments presented thus far demonstrate that telomerase assembly is reduced in the absence of TCAB1 but were carried out in fixed cells or cell lysates. To analyze telomerase assembly in intact cells, we carried out live-cell single-molecule imaging of Halo-TERT (Videos S4 and S5). We have previously demonstrated that there are three distinct TERT populations in the nuclei of human cancer cells24: a static population (assembled telomerase RNPs bound to telomeres, Cajal bodies, or other cellular structures), a slowly diffusing population, and a rapidly diffusing population (Figure 4A). The slowly diffusing population likely includes assembled telomerase RNPs, while the rapidly diffusing particles represents TERT molecules, which are not assembled with TR (Figures 4A and 4B). To determine the diffusion properties of TERT when telomerase assembly cannot occur, we knocked out TR. TR knockout was confirmed by PCR and Sanger sequencing, FISH, and qPCR (Figures S4A-S4C). To define the diffusion rate of free TERT molecules, we determined the diffusion coefficient of rapidly diffusing Halo-TERT molecules in TR knockout cells using the Spot-On tool (Figures 4A, 4B, and S4D).27 Similarly, we defined the rate of diffusion of assembled telomerase RNPs by measuring the diffusion coefficient of slowly moving Halo-TERT molecules in control cells (Figures 4A, 4B, and S4D). These measurements are the most accurate estimations possible for the diffusion coefficient of free TERT (Dfast = 1.9 ± 0.2 μm2/s) and TERT that is part of a telomerase RNP (Dslow = 0.35 ± 0.07 μm2/s), respectively. Using these diffusion coefficients, we fit the step-size distributions of TERT trajectories in control, TR knockout, and TCAB1 knockout cells to determine the fraction of TERT molecules in the fast, slow, and static TERT populations. Our key assumption is that rapidly diffusing TERT particles represent free TERT, while the much larger telomerase RNP is part of the slowly moving and static TERT populations. Consistent with this assumption, the fraction of TERT particles in the slowly diffusing and static populations was significantly reduced in TR knockout cells (19% ± 4%, 25% ± 7%) compared with control cells (43% ± 6%) (Figure 4B). It is important to note that, even in TR knockout cells, 19%–25% of TERT particles are slowly diffusing or static (Figure 4B). Because TR is absent in these cells, the slowly diffusing and static TERT molecules must be the result of interactions of TERT with cellular components other than Cajal bodies or telomeres. Importantly, we also observed a significant reduction in the fraction of slowly diffusing and static TERT particles in TCAB1 knockout cells (29% ± 2%) compared with control cells (43% ± 6%) (Figure 4B), consistent with a defect in telomerase assembly when TCAB1 is absent. The fraction of slowly diffusing and static TERT particles in TCAB1 knockout cells (29% ± 2%) was higher than in TR knockout cells (19% ± 4%, 25% ± 7%), suggesting that telomerase assembly is strongly reduced but not completely lost in the absence of TCAB1. In addition, in the absence of TCAB1, the reduction of telomerase assembly was only detected for TERT particles that localized to the nucleoplasm (i.e., the non-nucleolar region of the nucleus) but not the nucleolus (Figures 4C, 4D, and S4E).
Figure 4. Telomerase assembly is reduced in live TCAB1 knockout cells.
(A) Diagramof distinct populations of TERT particles detected in control cells (three replicates of >15 cells, mean ± SD).
(B) Fraction of slow plus static 3xFLAG-HaloTag-TERT particles in control, TCAB1 knockout, and TR knockout cells (three replicates of >15 cells, mean ± SD, t test).
(C) Quantification of the fraction of TERT particles statically bound to the nucleolus in control (WT), TCAB1 knockout, and TR knockout cells (two or three replicates of >18 cells, mean).
(D) Kymographs of HaloTag-TERT particles that co-localized with nucleoli (GFP-NPM1) in control (WT), TCAB1 knockout, and TR knockout cells.
Because stable binding of telomerase to the telomere requires base pairing of TR with the single-stranded telomeric overhang,28 we analyzed the interaction of TERT with telomeres. To assess TERT association with telomeres, we plotted the step size of TERT trajectories vs. the distance from the closest telomere for each step of these trajectories (Figure S5A). In control cells, we observed an enrichment of TERT particles in close proximity to telomeres that do not move (i.e., have small step sizes), consistent with TERT interactions with the chromosome end (Figure S5A, red boxes). In contrast, TERT trajectories in TCAB1 and TR knockout cells lacked this enrichment, indicating that interactions between TERT and telomeres are reduced (Figure S5A). Next, we filtered out TERT trajectories that came into proximity with telomeres marked by mEOS3.2-TRF2, as previously described.24 Diffusion analysis of these TERT trajectories using Spot-On demonstrated that the fraction of static TERT particles at telomeres was reduced from 12% in control cells to 4%–5% in TCAB1 and TR knockout cells (Figure S5B). These observations indicate that, in the absence of either TCAB1 or TR, stable interactions of telomerase with telomeres occur at a lower frequency because they require TR to stably bind to the chromosome end.28 Together, these single-molecule imaging experiments indicate that, in living cells, telomerase assembly is significantly reduced in the absence of TCAB1.
TERT is not retained in the nucleolus in the absence of TR
The experiments presented thus far do not address the sub-cellular localization of telomerase assembly. It was previously suggested that telomerase is assembled in the nucleolus and active telomerase is enriched in the nucleolus in the absence of TCAB1.29 Alternatively, TERT could associate with TR outside of the nucleolus, and a small fraction of the telomerase RNP could localize to the nucleolus after assembly. Importantly, telomerase assembly within the nucleolus would require TERT to localize to the nucleolus independently of TR. To address whether TERT can enter the nucleolus in the absence of TR, we analyzed TERT trafficking at the single-molecule level in TR knockout cells synchronized in S-phase of the cell cycle. Nucleoli were marked by transient expression of GFP-NPM1 and TERT trajectories were split up into tracks that overlapped with the location of the nucleolus (Figure S4E, colored tracks), and nucleoplasmic trajectories that did not overlap with the nucleolus (Figure S4E, black tracks). A small fraction of TERT particles was detected within the nucleolus in control and TCAB1 knockout cells (Figure S4E, colored tracks). Analysis of the diffusion properties of TERT particles co-localized with GFP-NPM1 demonstrated that the fraction of static TERT molecules was slightly increased in TCAB1 knockout cells (29%) compared with controls (23%, Figures 4C, 4D, S4E, and S4F; Video S6). In contrast, static TERT molecules that co-localized with the nucleolus were less frequently detected in TR knockout cells (10%; Figures 4C, 4D, S4E, and S4F; Video S6). Furthermore, we observed very rare TERT molecules in TR knockout cells that were mobile but constrained within a sub-region of the nucleolus, which, based on our other observations, likely corresponds to TERT within the DFC that is not bound to TR (Figure 4D; Video S6). In total, these results demonstrate that, although rare, TERT can enter the nucleolus independently of TR but only transitions into a stably bound state in the presence of TR.
TCAB1 is excluded from the nucleolus
Our experiments demonstrate that the loss of TCAB1 leads to an accumulation of TR in the nucleolus (Figure 1). TCAB1 could lead to the depletion of TR from the nucleolus by binding to TR in the nucleoplasm and preventing it from entering the nucleolus. Alternatively, TCAB1 could bind to TR within the nucleolus and accelerate the export of TR from the nucleolus, which requires TCAB1 entry into the nucleolus. To analyze the sub-nuclear localization of TCAB1, we introduced a 3xFLAG-HaloTag at the endogenous TCAB1 locus and generated single-cell clones that exclusively expressed 3xFLAG-HaloTag-TCAB1 (Figure 5A). HaloTag-TCAB1 strongly accumulated at Cajal bodies (marked by BFP-coilin) and was excluded from the nucleolus (Figure 5B; Video S7). Additionally, a large fraction of HaloTag-TCAB1 localized to the cytoplasm, suggesting that its import into the nucleus is restricted (Figure 5B). To ensure that the cytoplasmic localization of HaloTag-TCAB1 was not an artifact of fusing TCAB1 to the HaloTag, we carried out cell fractionation experiments, which demonstrated that untagged endogenous TCAB1 was also detected in the cytoplasm and nucleus but not in the nucleolus (Figure 5C), consistent with our imaging experiments. Single-particle tracking of HaloTag-TCAB1 within the nucleus showed that the diffusion dynamics of TCAB1 were similar to those of TERT (Figures 4B and 5D). The step-size distribution of HaloTag-TCAB1 particles fitted well to a three-state model (Figure 5D). The freely diffusing state of TCAB1 had a higher diffusion coefficient than TERT (Dfast = 2.4 μm2/s vs. 1.9 μm2/s). Strikingly, the diffusion coefficients of the slow-diffusing state for TCAB1 and TERT were identical (Dslow = 0.35 μm2/s), consistent with TCAB1 and TERT being part of assembled RNPs. On rare occasions, we observed HaloTag-TCAB1 molecules that associated with the nucleolus (Figure 5E; Videos S7 and S8). In contrast, HaloTag-dyskerin was readily detected stably bound to the nucleolus (Figures 5F and 5G). In total, these results demonstrate that TCAB1 is largely excluded from the nucleolus and that TCAB1 molecules diffuse through the nucleoplasm at a rate similar to assembled telomerase RNPs.
Figure 5. TCAB1 is excluded from the nucleolus.
(A) Western blot probed for TCAB1 of parental and genome-edited HeLa cells expressing HaloTag-TCAB1.
(B) Maximum intensity projection of a single-molecule imaging movie of HaloTag-TCAB1 (JFX650) in cells expressing BFP-coilin and GFP-NPM1 to mark Cajal bodies and nucleoli, respectively (scale bar, 5 μm).
(C) Western blots probed with TCAB1, fibrillarin, and lamin B1 antibodies of samples of cellular fractionation experiments (left to right: input, cytoplasm, nucleus, nucleoplasm, nucleolus) from HeLa cells.
(D) Probability density functions of the step sizes derived from nuclear HaloTag-TCAB1 trajectories and the corresponding three-state model fit using the Spot-On tool (pooled data from two replicates of two independent HaloTag-TCAB1 clones, >10 cells per replicate).
(E) Kymograph of nucleolar HaloTag-TCAB1 particles over time.
(F) Live-cell fluorescence images of single 3xFLAG-HaloTag-dyskerin particles, nucleoli marked by GFP-NPM1 (scale bar, 5 μm).
(G) Kymograph of nucleolar 3xFLAG-HaloTag-dyskerin particles over time.
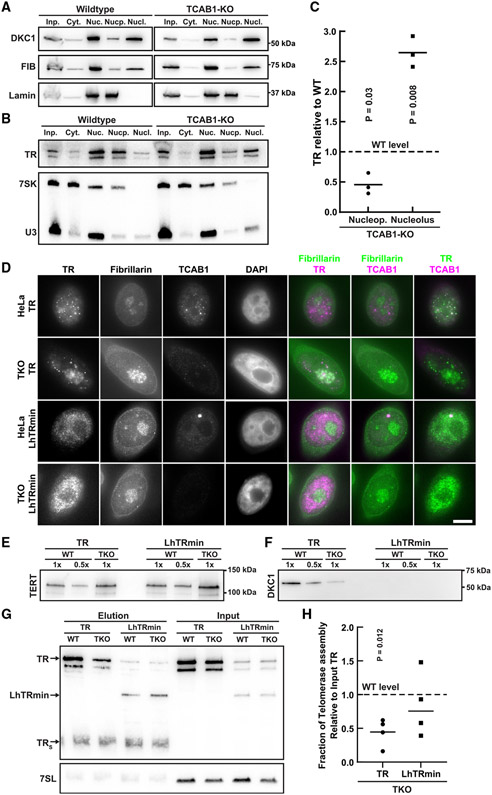
The majority of TR is tightly associated with the nucleolus in absence of TCAB1
Single-molecule imaging of TERT indicated that <10% of TERT molecules localize to the nucleolus in control and TCAB1 knockout cells (Figure 1G). To determine the fraction of TR that associates with the nucleolus in the absence of TCAB1, we carried out cellular fractionations to isolate nucleoli.30 Isolated nucleoli were enriched with the nucleolar protein fibrillarin and U3 snoRNA while being depleted of lamin B1 and the 7SK RNA (Figures 6A and 6B), which served as nucleoplasmic markers. To determine the amount of TR found in the nucleolus and the nucleoplasm, we quantified the level of TR relative to that of the U3 or the 7SK RNA, respectively. In control cells, the majority of TR was found in the nucleoplasmic fraction, and a small amount of TR was detected in nucleoli (Figure 6B), consistent with previous work that analyzed TR localization by live-cell imaging.31 In TCAB1 knockout cells, TR was depleted from the nucleoplasm and enriched in the nucleolus (Figures 6B and 6C). To assess the impact of high salt concentrations20 on nucleolar integrity, we supplemented ruptured nuclei with potassium chloride, prior to isolating nucleoli. After exposure to a high salt concentration, fibrillarin and TR were found in the nucleoplasmic fraction instead of the nucleolar pellet (Figure S6), demonstrating that nucleoli are disrupted and TR is released under these conditions. These observations confirm that >50% of TR is sequestered in the nucleolus in the absence of TCAB1 and strongly suggest that the majority of TR is tightly associated with the nucleolus.
Figure 6. Localization of TR to the nucleoplasm rescues telomerase assembly in the absence of TCAB1.
(A) Western blots probed with TCAB1, fibrillarin, and lamin B1 antibodies of samples of cellular fractionation experiments (left to right: input, cytoplasm, nucleus, nucleoplasm, nucleolus) from control and TCAB1 knockout cells.
(B) Northern blots probed for TR, U3 (nucleolar marker), and 7SK (nucleoplasmic marker) of samples of cellular fractionation experiments (left to right: input, cytoplasm, nucleus, nucleoplasm, nucleolus) from control and TCAB1 knockout cells.
(C) Quantification of the nucleoplasmic and nucleolar abundance of TR in TCAB1 knockout cells relative to control cells (n = 3, mean, t test).
(D) IF-FISH of control and TCAB1 knockout cells overexpressing TERT and full-length TR or LhTRmin probed with antibodies for fibrillarin and TCAB1 and TR-FISH probes (scale bar, 5 μm).
(E and F) Western blots of TERT purified from control and TCAB1 knockout cells overexpressing TERT and full-length TR or LhTRmin probed with antibodies for (E) TERT and (F) dyskerin.
(G) Northern blot probed for TR and 7SL (loading control) of RNA extracted from input and TERT samples purified from control and TCAB1 knockout cells overexpressing TERT and full-length TR or LhTRmin. Standards are TR and truncated TRS.
(H) Quantification of TR or LhTRmin associated with TERT relative to input RNA of TERT samples purified from TCAB1 knockout cells compared with control cells (n = 4, mean, t test).
Localization of TR to the nucleoplasm rescues telomerase assembly in the absence of TCAB1
Our data demonstrate that stable association of TERT with the nucleolus requires its interaction with TR (Figures 4C-D and S4). Importantly, we cannot distinguish whether static TERT molecules within the nucleolus represent telomerase RNPs that have assembled in the nucleolus or the nucleoplasm. To address whether the telomerase RNP can assemble in the nucleoplasm, we expressed a truncate variant of TR (LhTRmin) that lacks the H/ACA domain19 and therefore does not bind to dyskerin and TCAB1. As a consequence, LhTRmin does not localize to the nucleolus or Cajal bodies (Figure 6D).19 Because LhTRmin lacks the H/ACA region of TR, it does not accumulate to the same level as wild-type TR (Figure 6D).19 While TERT assembly with full-length TR and dyskerin was significantly reduced in the absence of TCAB1, TERT association with LhTRmin was not significantly different in TCAB1 knockout cells compared with control cells expressing LhTRmin (Figures 6E-6H). Importantly, no dyskerin was associated with TERT purified from cells expressing LhTRmin (Figure 6F). These results demonstrate that forcing TR localization to the nucleoplasm suppresses the telomerase assembly defect observed in TCAB1 knockout cells and indicate that the telomerase RNP can form outside of the nucleolus. Importantly, these observations suggest that the sequestration of TR in the nucleolus causes the reduction in telomerase assembly observed in the absence of TCAB1.
Analysis of nucleolar TR and snoRNP dynamics
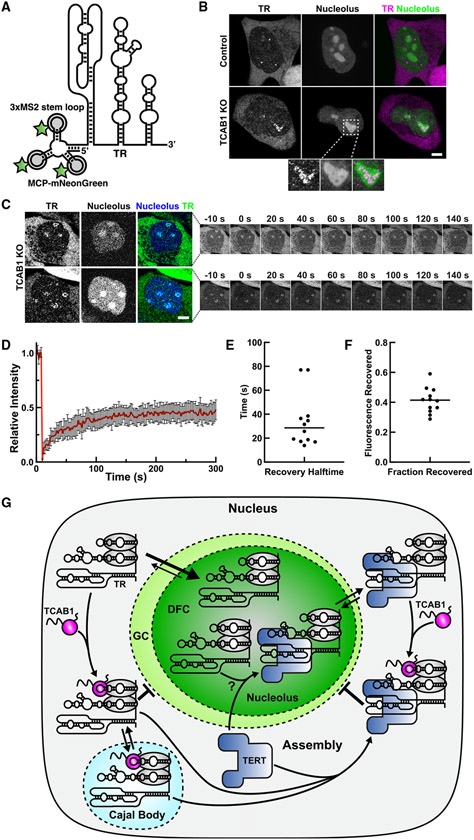
The results presented thus far have demonstrated that TR accumulates in nucleoli in the absence of TCAB1 by cell fractionation and in fixed cells, but we have not addressed the dynamic localization of TR in living cells. We first used dyskerin as a surrogate for TR and H/ACA snoRNPs in general. We transiently expressed HaloTagged dyskerin in parental HeLa and TCAB1 knockout cells and analyzed dyskerin binding to the nucleolus using fluorescence recovery after photo bleaching (FRAP;, Figure S7A; Videos S9 and S10). The dyskerin signal recovered rapidly (t1/2 = 28 s), but only ~65% of the signal was recovered after >4 min (Figures S7B-S7D; Videos S9 and S10). This indicates that there are at least two distinct populations of dyskerin molecules in the nucleolus: a rapidly exchanging population and a static population that does not dissociate from the nucleolus over the time course of this experiment (Figure S7D). The presence of a mobile dyskerin population was confirmed by analysis of unbleached nucleoli, which lost fluorescence signal with similar kinetics (Figures S7A, S7E, and S7F). Importantly, no significant difference in dyskerin dynamics was observed in TCAB1 knockout cells compared with parental controls (Figure S7A-S7F), which suggests that TCAB1 is not required to extract dyskerin-containing scaRNPs from nucleoli.
To directly analyze the dynamics of TR binding to the nucleolus, we introduced three MS2 stem loops at the 5′ end of the endogenous TR gene (Figures 7A, S7G, and S7H), as previously described.31 MS2-TR can be detected in living cells using the MS2 coat protein (MCP) fused to the mNeonGreen fluorescent protein (Figure 7A). Similar to endogenous TR (Figure 1), MS2-TR localized to nucleoplasmic foci in control cells (likely Cajal bodies) and to nucleoli in absence of TCAB1 (Figures 7B and S7I). Like dyskerin (Figure S1F), MS2-TR was specifically enriched in nucleolar regions with reduced GC marker (Figure S1F) intensity in the absence of TCAB1 (Figure 7B, zoomed panels), which corresponds to the DFC of the nucleolus. To measure the dynamics of TR association with the nucleolus in TCAB1 knockout cells, we carried out FRAP of MS2-TR clearly localized within DFC of the nucleolus (Figure 7C; Videos S11 and S12). After bleaching, ~40% of the MS2-TR signal recovered with a half-time of recovery of ~30 s (Figures 7D-7F). Importantly, no MS2-TR accumulation in nucleoli was observed in control cells, precluding its analysis by FRAP (Video S13). This demonstrates that, in the absence of TCAB1, the majority of TR is tightly associated with the nucleolus and the remaining TR molecules exchange with the nucleoplasm with kinetics comparable with dyskerin. In total, these results strongly suggest that the reduction of telomerase assembly, observed in the absence of TCAB1, is a consequence of the sequestration of TR in the nucleolus, where it infrequently encounters TERT, which is largely localized to the nucleoplasm (Figure 7G).
Figure 7. Analysis of the nucleolar binding dynamics of dyskerin and TR.
(A) Model of 3xMS2-TR bound to MCP-mNeon-Green.
(B) Live-cell microscopy images of control and TCAB1 knockout cells expressing 3xMS2-TR, MCP-mNeonGreen, and BFP-NLS to mark the nucleolus (scale bar, 5 μm).
(C) FRAP of 3xMS2-TR within the DFC of the nucleolus marked by BFP-NLS (scale bar, 5 μm).
(D) FRAP of MS2-TR within nucleoli of TCAB1 knockout cells (n = 12 cells, mean and 95% confidence interval).
(E) Quantification of the recovery half-time from single-exponential fits of individual recovery curves (n = 12, mean).
(F) Quantification of the fraction of fluorescence recovered from single-exponential fits of individual recovery curves (n = 12 cells, mean).
(G) Model for TCAB1-mediated retention of TR in the nucleoplasm which facilitates TR assembly with TERT.
DISCUSSION
TCAB1 facilitates telomerase assembly
The importance of TCAB1 for telomere synthesis is undisputed.6,20 Knockout or depletion of TCAB1 results in telomere shortening,6,19,20 and mutations in TCAB1 lead to dyskeratosis congenita.18 Prior studies concluded that TCAB1 is not required for telomerase assembly but instead plays a role in telomerase trafficking to Cajal bodies and telomeres or is required for telomerase catalysis.6,18-20,22 The work presented here demonstrates that, in the absence of TCAB1, telomerase assembly is reduced. This finding is supported by two completely distinct and complementary approaches: telomerase purification and live-cell single-molecule imaging. The purification experiments indicate that telomerase assembly is reduced to 20%–40% in the absence of TCAB1 compared with control cells. A similar reduction is observed in the live-cell single-molecule imaging experiments. When telomerase assembly is impossible (in TR knockout cells), the vast majority (~75%–80%) of TERT molecules rapidly diffuse through the nucleus. In contrast, in the presence of TR, only ~55% of TERT molecules rapidly diffuse through the nucleus, because the association of TERT with TR leads to the formation of a large RNP that moves through the nucleus at a slower rate. In the absence of TCAB1, the fraction of rapidly moving TERT molecules is significantly increased to ~70%, consistent with a reduction in telomerase assembly. Assuming a maximal change of 25% in the fraction of rapidly diffusing TERT molecules (from 55% in wild type to 80% in TR knockout cells), the 15% increase, observed in the absence of TCAB1 compared with wild type, corresponds to a 60% reduction (15 out of 25) in telomerase assembly. In addition, we demonstrate that the TERT less frequently forms stable interactions with telomeres in the absence of TCAB1, consistent with fewer TERT molecules being bound to TR. The reduction in telomerase assembly in cells lacking TCAB1 leads to a lower number of telomerase RNPs per cell and, in turn, telomere shortening. Importantly, telomerase assembly is reduced but not completely abolished when TCAB1 is absent, which is sufficient to support continuous cell proliferation with a short telomere length set point.
TCAB1 is not required for telomerase catalysis
Previous work by others has reported conflicting results regarding the role of TCAB1 in telomerase catalysis, ranging from full enzymatic activity to substantial activity defects.6,19,20 Chen et al. proposed that TCAB1 is required for proper folding of the CR4/CR5 region of TR, which directly associates with TERT, without affecting telomerase assembly.20 Due to the misfolding of TR, telomerase was suggested to adopt a low-activity state in the absence of TCAB1. Experimentally, such a low-activity state would manifest itself as a reduction in the specific activity of telomerase. Our experiments strongly suggest that, while telomerase assembly is reduced in the absence of TCAB1, the limited amount of telomerase that can assemble is close to fully active (i.e., does not have reduced specific activity). One possible explanation for the discrepancies between the work by Chen et al. and our study is the methodology used to generate cell lysates. Our results demonstrate that the high salt concentration used by Chen et al. to generate nuclear extracts dissolves nucleoli and releases TR (Figure S6) and could override the localization of TR and TERT to distinct sub-cellular compartments. Importantly, our single-molecule imaging of TERT dynamics is consistent with a reduction of telomerase assembly in intact cells. Altogether, our enzymatic analysis does not support a role of TCAB1 in telomerase catalysis but is fully consistent with TCAB1 facilitating telomerase assembly.
Location and molecular mechanism of telomerase assembly
The sub-cellular location and order in which telomerase RNP components associate with TR in human cells have largely been unknown. It has been suggested that telomerase assembly occurs in the DFC of the nucleolus.29 While this study by Lee et al. clearly demonstrated that catalytically active telomerase can localize to the DFC of the nucleolus,29 it failed to show that telomerase is assembled in the nucleolus. Results by others have demonstrated that eliminating Cajal bodies does not affect telomerase activity or telomere maintenance, suggesting that Cajal bodies are not necessary for telomerase assembly.19,20,32 Our single-molecule live-cell imaging of TERT showed that TERT is almost exclusively localized to the nucleoplasm. Since Cajal bodies are dispensable for telomerase assembly, and TERT is largely excluded from the nucleolus, we believe telomerase assembly occurs in the nucleoplasm. The available data do not rule out that telomerase can also assemble in Cajal bodies, but it is certainly not obligatory. Our model is further supported by our observation that forcing TR localization to the nucleoplasm by expression of LhTRmin rescues the telomerase assembly defect observed in the absence of TCAB1. However, it is important to point out that we do observe a very small number of TERT molecules that localize to nucleoli in control and TCAB1 knockout cells. These static nucleolar TERT molecules require the presence of TR and therefore likely represent TERT associated with TR. Unfortunately, we are unable to determine whether this assembly of TERT and TR occurred in the nucleolus or prior to import of an assembled telomerase RNP into the nucleolus.
Altogether, our observations support our model that TCAB1 facilitates telomerase assembly by counteracting TR accumulation in the nucleolus to enable its assembly with TERT in the nucleoplasm (Figure 7G). Our analysis of the dynamics of nucleolar TR suggests that the majority of TR is tightly associated with the DFC when it is not bound by TCAB1. Since TCAB1 rarely enters the nucleolus, we believe the primary role of TCAB1 is to prevent the entry of TR into the nucleolus. In the absence of TCAB1, the equilibrium of TR localization is shifted toward the nucleolus, because re-entry is not inhibited by TCAB1 and the dissociation of TR from the DFC is very slow. The resulting nucleolar accumulation of TR effectively reduces the amount of TR available to assemble with TERT. Importantly, a very small number of TERT molecules bound to TR can be imported and trapped in the nucleolus, which would further decrease the amount of telomerase available to elongate telomeres.
Regulation of RNP assembly by nucleolar phase separation
In addition to the mechanistic insight into the role of TCAB1 in telomerase function, our results also demonstrate that nucleolar phase separation can regulate telomerase RNP assembly in the nucleus of human cells (Figure 7G). How RNA molecules are specifically recruited into or excluded from non-membrane-bound organelles is a key unanswered question. One model suggests that gradual replacement of non-specific, multivalent interactions of pre-ribosomal RNAs with nucleolar proteins, such as NPM1 and fibrillarin, with specific, high-affinity interactions with ribosomal proteins leads to the ejection of mature ribosomal subunits from the nucleolus.33 In this model, a key driving force for the retention of RNA in the nucleolus is regions of RNA that are not yet bound by ribosomal proteins and that are available to interact with nucleolar proteins.33 By analogy, this model would explain why TR bound by the H/ACA complex but not associated with TERT would be sequestered in the nucleolus. Our experiments demonstrate that 60% of nucleolar TR is tightly associated with the nucleolus while the remaining 40% can exchange with the nucleoplasm, which closely corresponds to fraction of telomerase assembly observed in TCAB1 knockout cells. We believe that TR bound to TERT can exchange with the nucleoplasm, but TR not associated with TERT remains trapped in the nucleolus. In addition to the interactions formed by the H/ACA complex with nucleolar proteins and RNA, the regions of TR that are bound by TERT would be available to form non-specific, multivalent interactions with nucleolar proteins to strengthen the association of TR with the nucleolus and prevent its release when it is not associated with TERT (Figure 7G).
The mechanism by which TCAB1 binding leads to the exclusion of TR and other scaRNA from the nucleolus remains a key unanswered question. As outlined above, we believe that TCAB1 prevents localization of scaRNAs to the nucleolus, rather than extracting scaRNAs that are already localized to the DFC. One potential explanation is that TCAB1 counteracts scaRNA recruitment to the nucleolus by inhibiting the nucleolar localization signals within dyskerin.34 Dissecting the molecular mechanism by which TCAB1 leads to exclusion of TR from the nucleolus in future studies will undoubtably shed light on the fundamental principles of RNP recruitment to non-membrane-bound organelles and its physiological role in cell biology.
Limitations of the study
Our work demonstrates that TCAB1 facilitates telomerase assembly in human cancer cells. We cannot rule out that there are differences between cancer cells and stem cells in which TCAB1 also plays a critical role, although the fundamental molecular mechanism of telomerase assembly is likely the same in all human cells. In addition, the analysis of telomerase assembly could be confounded by the methodology used to generate cell lysates. Finally, while we have generated multiple independent TCAB1 knockout cell lines and re-expression of TCAB1 rescues telomerase localization to Cajal bodies, and TRG414C that also revealed reduced assembly, it is possible that these cell lines contain additional mutations that could affect the observed phenotypes.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Any additional information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jens Schmidt (schmi706@msu.edu).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact with a completed material transfer agreement.
Data and code availability
All unprocessed data is available upon request from the lead contact. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Cell cultures
All cell lines were based on HeLa-EM2-11ht43 and were cultured in Dulbecco’s Modified Eagle Medium including L-glutamine (Gibco) supplemented with 10% fetal bovine serum, 100 units/ml penicillin and 100 μg/mL streptomycin at 37°C with 5% CO2. Live cell imaging was carried out using CO2 independent media supplemented with 2 mM GlutaMAX (Life Technologies), 10% fetal bovine serum, 100 units/ml penicillin and 100 μg/mL streptomycin at 37°C with 5% CO2. For single-molecule imaging of HaloTag-TERT cell were cultured in homemade imaging dishes made by gluing 22 × 22 mm Nexterion coverslips (170 ± 5 μm, Schott) onto the bottom of plastic 3.5 × 1.0 cm cell culture dishes with a hole in the middle using an epoxy adhesive. Prior to chamber assembly the coverslips were washed with 1 M KOH and 100% for 30 min each in a sonicating water bath. To enrich for cells in S-phase for live cell imaging experiments, cultures were treated with complete media including 2 mM thymidine for a minimum of 16 h. Cells were released 2 h prior to imaging by replacing the thymidine containing media with fresh media without thymidine. Puromycin selection was carried out at a concentration of 1 μg/mL.
METHOD DETAILS
Plasmid construction and genome editing
All plasmids were generated by Gibson assembly (NEB) using standard protocols or by inverse PCR. All plasmids will be made available on Addgene. All Cas9 and sgRNA expression plasmids were based on pX330.37 The homologous recombination donor for the TR knock-out was generated by assembling the genomic sequences immediately upstream and downstream (~500 bp each, gBlocks IDT) of the TR sequence flanking a puromycin resistance cassette into HpaI linearized pFastBac. The homologous recombination donor for the insertion of the 3xMS2-tag at the endogenous locus was generated by cloning a left homology arm (418 bp upstream of the TERC gene, gBlock IDT) followed by 3xMS2-TR and 100 bp of the genomic sequence downstream of TR, followed by an SV40-promoter driven puromycin resistance marker, and finally a right homology arm (473 bp, 101–573 bp downstream of the TERC gene, gBlock IDT) into pFastBac. The sgRNA were designed to cut at the junction of the 3xMS2-TR insert junctions and the PAM sites were mutated to not be present in the homologous recombination donor plasmid, to assure the recombined allele was not cut by Cas9. The lentiviral plasmids for expression of MCP-NeonGreen was generated by replacing the sfGFP sequence in the pHAGE-UBC-MCP-sfGFP plasmid (a kind gift from Agnel Sfeir) with the coding sequence for mNeonGreen. The lentivirus and cell lines expressing MCP-mNeonGreen were generated as previously described.44 The homologous recombination donor for the insertion of the 3xFLAG-HaloTag into the TCAB1 gene was generated by cloning the 3xFLAG-HaloTag sequence including an SV40-promoter driven puromycin resistance marker flanked by LoxP-sites in between two homology arms (500 bp up and downstream of the start codon, gBlocks IDT). The 3xFLAG-HaloTag-NLS plasmids was generated by adding a 3xFLAG-tag to a previously described HaloTag-NLS plasmid (a kind gift from X. Darzacq and A. Hansen).27 The 3xFLAG-HaloTag-dyskerin plasmid was generated by replacing TERT in our previously described 3xFLAG-HaloTag-TERT expression plasmid with the dyskerin coding sequence.24 The mCherry-dyskerin plasmid was generated by replacing TERT in our previously described mCherry-TERT expression plasmid with the dyskerin coding sequence.38 Unless otherwise stated, transfections were carried out with Lipofectamine 2000 (Invitrogen) using the manufacturer’s instructions. For FRAP analysis of dyskerin 1x106 HeLa cells were transfected with 1 μg of 3xFLAG-HaloTag-dyskerin plasmid using the Lonza 4D-Nucleofector with the SE Cell Line 4D-Nucleofector X kit (Cat. V4XC-1012) and program CN-114. For single-molecule imaging of dyskerin 1 μg of a GFP-NPM1 plasmid was included in addition to the 1 μg of 3xFLAG-HaloTag-dyskerin plasmid. GFP-NPM1 WT was a gift from Xin Wang (Addgene plasmid #17578; http://n2t.net/addgene:17578; RRID:Addgene_17578).41 For single-molecule imaging of 3xFLAG-HaloTag-NLS, 1 μg of a GFP-Nucleolin plasmid was included in addition to the 1 μg of 3xFLAG-HaloTag-NLS plasmid. The GFP-Nucleolin plasmid was a gift from Michael Kastan (Addgene plasmid #28176; http://n2t.net/addgene:28176; RRID: Addgene_28176).40 For FRAP analysis of nucleolar 3xMS2-TR 1 μg of mTagBFP-Nucleus-7 was transfected into HeLa cells expressing 3xMS2-TR and MCP-mNeonGreen. mTagBFP-Nucleus-7 was a gift from Michael Davidson (Addgene plasmid # 55265; http://n2t.net/addgene:55265; RRID:Addgene_55265). TCAB1 was knocked-out using two separate sgRNA and Cas9 encoding plasmids that were transfected alongside a GFP-expressing plasmid. 24 h after transfection single-cell clones were sorted using the GFP signal. TCAB1 knock-out clones were screened by PCR and confirmed by Western blot, Southern Blotting of the TCAB1 locus and immunofluorescence imaging. TR was knocked out by transfecting two sgRNA plasmids and a homologous recombination donor plasmid. 48 h after transfection puromycin selection was initiated and 1 week after the initiation of selection single-cell clones were generated by dilution into 96-well plates. TR knock-out was confirmed using PCR and Sanger sequencing, fluorescence in situ hybridization, and RT-qPCR. TRG414C plasmid was generated by inverse PCR using the vector of the WT TR plasmid.
Immunofluorescence and fluorescence in situ hybridization imaging
Fixed cell immunofluorescence imaging and fluorescence in situ hybridization was carried out as previously described.38 Briefly, cells grown on coverslips were fixed in PBS supplemented with 4% formaldehyde. When using the HaloTag for fluorescence detection cells were incubated with 100 nM of JF646 HaloTag-ligand for 30 min prior to fixation. Unincorporated ligand was removed by 3 washes with complete media followed by placing the cells back in the incubator for 10 min to let additional dye leak out of the cells. mEOS3.2-TRF2 was detected using the intrinsic fluorescence of green form of mEOS3.2. After removing the fixation solution using 2 PBS washes, coverslips were transferred into aluminum foil covered humidity chambers with a parafilm layer and rinsed with 1 mL of PBS with 0.2% Triton X-100. Cells were than incubated in blocking buffer (PBS, 0.2% Triton X-100, 3% BSA) for 30 min, followed by incubation with primary antibodies diluted in blocking buffer for 1 h. All primary antibodies were used at a concentration of 1 μg/mL. After three washes with PBS +0.2% Triton X-100, coverslips were incubated with secondary antibodies diluted in PBS +0.2% Triton X-100 for 1 h. All secondary antibodies were used at a concentration of 4 μg/mL. Cells were washed three times PBS +0.2% Triton X-100 prior to a second fixation with PBS +4% formaldehyde. In cases where nuclear staining was used, the first of the three washing steps also included 0.1 μg/mL HOECHST. After the second fixation steps, coverslips were dehydrated in three steps with ethanol (70%, 95%, 100%), re-hydrated in 2xSSC +50% formamide, blocked for 1 h in hybridization buffer (100 mg/mL dextran sulfate, 0.125 mg/ml E. coli tRNA, 1 mg/mL nuclease free BSA, 0.5 mg/mL salmon sperm DNA, 1 mM vanadyl ribonucleoside complexes, 50% formamide, 2xSSC) at 37°C, before incubating the coverslips in hybridization buffer supplemented with three TR probes (30 ng per coverslip,/5Cy5/GCTGACATTTTTTGTTTGCTCTAGAATGAACGGTGGAAGGCGGCAGGCCGAGGCTT,/5Cy5/CTCCGTTCCTCTTCCTGCGGCCTGAAAGGCCTGAACCTCGCCCTCGCCCCCGAGAG,/ATGTGTGAGCCGAGTCCTGGGTGCACGTCCCACAGCTCAGGGAATCGCGCCGCGCGC) overnight at 37°C. Probe sequences were previously described.45 After hybridization coverslips were washed twice for 30 min in 2xSSC +50% formamide and then mounted on slides using ProLong Antifade Diamond mounting media (Life Technologies). Microscopy was carried out using a DeltaVision Elite microscope using a 60x PlanApo objective (1.42 NA) and a pco.edge sCMOS camera. We acquired 20 Z sections spaced by 0.2 μm, followed by image deconvolution and maximum intensity projection of the sections using the DeltaVision Softworx software.
Single-molecule live cell imaging
Live cell single-molecule imaging was carried out on an Olympus IX83 inverted microscope equipped with a 4-line cellTIRF illuminator (405 nm, 488 nm, 561 nm, 640 nm lasers), an Excelitas X-Cite TURBO LED light source, a Olympus UAPO 100x TIRF objective (1.49 NA), a CAIRN TwinCam beamsplitter, 2 Andor iXon 897 Ultra EMCCD cameras or 2 Hamamatsu Orca BT Fusion cameras, a cellFRAP with a 100 mW 405 nm laser, and a blacked-out environmental control enclosure. The microscope was operated using the Olympus cellSense software. 3xFLAG-HaloTag-TERT was labeled for 2 min in complete media supplemented with 100 nM JF646-HaloTag ligand.35 After removing the HaloTag-ligand with three washes in complete media, cells were placed back in the incubator for 10 min to allow unincorporated dye to leak out of the cells. Cells were then transferred into CO2 independent media and put on the microscope which was heated to 37°C. Single-molecule imaging was carried out at 50 or 100 frames per second using highly inclined laminated optical sheet illumination.46 Moviewere typically 20 s in length (2000 frames) and were followed by a transmitted light acquisition to visualize overall cell morphology. For single-molecule imaging of 3xFLAG-HaloTag-Dyskerin, cells were labeled with 100 pM of JFX650-HaloTag Ligand47 for 1 min. Imaging was carried out at 100 frames per second and images of GFP-NPM1 were taken before and after single-molecule Movieof dyskerin to assure the position of the nucleolus had not shifted.
RT-qPCR
RNA samples for RT-qPCR analysis were generated by using RNeasy Mini kits (Qiagen) using ~2 million cells as starting material. Reverse transcription was carried out using random hexamer primers and SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. qPCR was carried out using the Maxima SYBR Green qPCR master mix (Thermo Scientific) using primers for GAPDH and TR according to the manufacturer’s instructions. All qPCR reactions were carried out in triplicates and three independent biological replicates were analyzed.
Southern blotting
Southern blotting was carried out using standard protocols.48 Briefly, genomic DNA generated by phenol-chloroform extraction after cell lysis using TE supplemented with 0.5% SDS and 0.1 mg/mL Proteinase K, was digested with BamHI (generating a 1394 bp fragment spanning exons 1–3 of the TCAB1 locus) and separated on a 0.8% agarose gel. The DNA was then transferred on a Hybond-N+ nylon membrane using capillary transfer. The TCAB1 locus was detected using radioactive probes (alpha-49 P-dCTP) generated by randomly primed DNA synthesis using an 800 bp PCR product overlapping with the 1394 bp restriction fragment as a template and Klenow polymerase (NEB). Telomeric restriction fragment analysis was carried out as previously described.50
Western blotting
Mini-PROTEAN TGX stain-free gels (Bio-Rad) were used for SDS-PAGE. Total protein was detected using a ChemiDoc MP (Bio-Rad) after a 45 s UV activation. Western transfer was carried out using the Trans-Blot Turbo transfer system (Bio-Rad) according to the manufacturer’s instructions using the mixed molecular weight transfer setting. Immuno-blotting was carried out using standard protocols. The C-terminal TCAB1 antibody (Proteintech, 14761-1-AP) was used at a 1:2000 dilution, the N-terminal TCAB1 antibody (Novus Biologicals, NB100-68252) was used at a 1:1000 dilution, the TERT antibody (Abcam, ab32020) was used at a 1:4000 dilution, the dyskerin antibody (Santa Cruz Biotech, sc-373956) was used at a 1:200 dilution, the GAR1 antibody was used at a 1:2000 dilution (Proteintech, 11711-1-AP), the fibrillarin antibody was used at a 1:2000 dilution (Novus Biologicals, NB300-269), and the lamin B1 antibody was used a 1:2000 dilution. Secondary antibodies were used at a 1:5000 dilution.
Northern blotting
RNA was extracted from cell lysates, cellular fractions, and purified telomerase samples using the RNeasy Mini kit (Qiagen) and eluted in 30 μl of RNase free water. Purified telomerase samples were supplemented with 10 ng of a loading and recovery control prior to RNA extraction (in vitro transcribed TR 34–328). 15 μl of eluted RNA was mixed with 15 μl of 2x formamide loading buffer (0.1XTBE, 25 mM EDTA, 0.1% bromophenol blue, 0.1% xylene cyanol, 93% formamide) and heated to 60°C for 5 min. Samples were separated on a 6% TBE, 7M Urea, polyacrylamide gel (Life Technologies), and transferred to a Hybond N+ membrane (Cytiva) using a wet-blotting apparatus in 1x TBE for 2 h at 0.5 A of constant current in the cold room. After transfer, membranes were UV-cross-linked, and pre-hybridized in Church buffer for 2 h at 50°C. Three DNA oligos complementary to TR (GACTCGCTCCGTTCCTCTTC, GCTCTAGAATGAACGGTGGAA, CCTGAAAGGCCTGAACCTC, CGCCTACGCCCTTCTCAGT, ATGTGTGAGCCGAGTCCTG), 7SL (GCGGACACCCGATCGGCATAGC), U3 (GCCGGCTTCACGCTCAGGAGAAAACGCTACCTCTCTTCCTCGTGG), and 7SK (GTGTCTGGAGTCTTGGAAGC) were radioactively labeled using T4 PNK (NEB) and ~10x106 cpm of each probe were added to the membrane. Hybridization was carried out at 50°C overnight. Membranes were washed three times with 2xSSC, 0.1% SDS prior to exposure to a storage phosphorescence screen (Cytiva) which was then imaged on an Amersham Typhoon IP phosphoimager (Cytiva).
Telomerase expression and purification
Cell lines were transfected in 15-cm tissue culture plates at ~90% confluency (~25-30x106 cells) using 7.5 μg of TERT plasmid, 30 μg of TR plasmid and 75 μL of Lipofectamine 2000 in 1875 μl of Opti-MEM.39 The same amounts were used in transfecting TRG414C, LhTRmin19 or pBS U3-hTR-500 (Addgene plasmid # 28170; http://n2t.net/addgene:28170; RRID:Addgene_28170)42 plasmids, which were generously gifted by Kathleen Collins. Transfected cells were split to three 15 cm dishes 24 h after transfection. 48 h after transfection cells were counted, harvested, and snap frozen in liquid nitrogen. Cells were lysed in 1 mL of CHAPS lysis buffer supplemented with 5 μL of RiboLock RNase inhibitor (10 mM TRIS pH 7.5, 1 mM MgCl2, 1 mM EGTA pH 8.0, 0.5% CHAPS, 10% glycerol) per 100x106 cells and rotated at 4°C for 30 min. Lysates were cleared in a table-top centrifuge at 21,000xg for 15 min at 4°C. Identical cell equivalents were used for all samples. 45 μg of anti-TERT antibody was added per ml of cleared lysate and samples were rotated for 1 h at 4°C. Lysates were then added to 100 μL of protein G agarose and rotated for 1 h at 4°C. The resin was spun down at 1000xg and washed four times with 1 mL of Buffer W (20 mM HEPES pH 7.9, 300 mM KCl, 2 mM MgCl2, mM MgCl2, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, 0.1% Triton X-100, 10% glycerol). TERT was eluted in 100 μL of Buffer W supplemented with 5 μL of 1 mM TERT peptide by rotating for 30 min at room temperature.
Telomerase activity assays
Telomerase assays were carried out in 20 μL of reaction buffer (50 mM TRIS pH 8.0, 150 mM KCl, 1 mM MgCl2, 2 mM DTT, 100 nM TTAGGGTTAGGGTTAGG oligo, 10 μM dATP, 10 μM dGTP, 10 μM dTTP, 0.165 μM dGTP [α-32P] 3000 Ci/mmol) including 2 μL of purified telomerase for 1 h at 30°C. Telomerase was incubated with the substrate oligo for 15 min at room temperature, prior to initiating the reaction by addition of dNTPs. Reactions were stopped by adding 100 μL of 3.6 M of ammonium acetate supplemented with 20 μg of glycogen and 32P 5′-end labeled loading control oligos (TTAGGGTTAGGGTTAGGG, TTAGGGTTAGGGTTAG). Reaction products were precipitated using 500 μL of ice-cold ethanol and stored at −20°C over-night. Reaction products were spun down in a table-top centrifuge at max speed for 30 min at 4°C, washed with 500 μL of 70% ethanol, and spun down again speed for 30 min at 4°C. The 70% ethanol was decanted, and the reaction products were dried in an Eppendorf vacuum concentrator at 45°C. Reaction products were resuspended in 20 μL of loading buffer (0.05XTBE, 25 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol, 46.5% formamide) and incubated at 95°C for 5 min 10 μL of each sample was separated on a 12% polyacrylamide, 7 M urea sequencing gel pre-run for 45 min at 90W. Gels were dried and exposed to a storage phosphorescence screen (Cytiva) and imaged on an Amersham Typhoon IP phosphoimager (Cytiva).
Nucleolar isolation
Cellular fractionation was carried out using a previously described method.30 All procedures were carried out on ice and centrifugations at 4°C. Approximately 1x106 million cells were harvested by trypsinization, washed with PBS, followed by incubation in a hypotonic buffer (10 mM HEPES pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT) to swell the cells. A small fraction of the swollen cells was collected as input sample. Swollen cells were then ruptured using pre-cooled Dounce homogenizer and the tight pestle (VWR Cat. 62400-595). The ruptured cells were centrifuged at 218xg for 5 min to pellet nuclei. Nuclei where then resuspended in buffer S1 (0.25 M sucrose, 10 mM MgCl2), layered on top of buffer S2 (0.35 M sucrose, 0.5 mM MgCl2) in a 15 mL conical tube, and centrifuged at 1430xg in a swinging bucket rotor for 5 min to further purify nuclei. Nuclei were resuspended in buffer S2 and sonicated on ice for 10 s at 30% power (Fisherbrand Model 505, 500W). The sonicated nuclei were then layered on top of buffer S3 (0.88 M sucrose, 0.5 mM MgCl2) and centrifuged at 3000xg in a swinging bucket rotor for 5 min to further purify nucleoli. The nucleolar pellet was suspended in buffer S2 and centrifuged a final time at 1430xg to yield a highly purified nucleolar pellet, which was resuspended in buffer S2. Equal fractions of input, cytoplasm, nuclei, nucleoplasm, and nucleoli samples were collected and analyzed by western and northern blots. To test the impact of salt concentration on the integrity of nucleoli, nuclei ruptured by sonication were mixed 1:1 with buffer S2 containing 40 mM HEPES pH7.9 with and without 715 mM KCl, prior layering the solution on top of buffer S3.
Cell synchronization and flow cytometry
Cell lines in 15-cm tissue culture plates at a 70% confluency were blocked with 2 mM thymidine then released from thymidine after 24 h and harvested at four time points post release: 0, 4, 8 h. Along with each synchronization, asynchronous population was also harvested. Cells were harvested using PBS supplemented with 5 mM EDTA then fixed with ethanol, stained with propidium iodide for DNA content, and finally filtered to generate a single cell suspension. Stained samples were run through BD Accuri C6 cytometer and cell-cycle distributions were analyzed on OriginLab.
Single-particle tracking
Single-particle tracking was carried out in MATLAB 2019a using a batch parallel-processing version of SLIMfast modified to allow the input of TIFF files (kindly provided by Xavier Darzacq and Anders Hansen),27 an implementation of the Multiple-Target-Tracing algorithm,51 with the following settings: Exposure Time = 10 ms, NA = 1.49, Pixel Size = 0.16 μm, Emission Wavelength = 664 nm, Dmax = 5 μm2/s, Number of gaps allowed = 2, Localization Error = 10−5, Deflation Loops = 0. Diffusion coefficients and the fraction of molecules in each distinct particle population were determined using the MATLAB version of the Spot-On tool (kindly provided by Xavier Darzacq and Anders Hansen)27 with the following settings: TimeGap = 10 ms or 20 ms, dZ = 0.700 μm, GapsAllowed = 2, TimePoints = 8, JumpsToConsider = 4, BinWidth = 0.01 μm, PDF-fitting, D_Free1_3State = [1 25], D_Free2_3State = [0.1 1], D_Bound_3State = [0.0001 0.1]. For all experiments we carried out 3 independent biological replicates with at least 15 cells for each cell line. The statistical significance of differences in particle fractions and diffusion coefficients were assessed using a two-tailed T Test.
For the analysis of dyskerin trajectories a mask of the nucleolus was generated manually using the threshold function in FIJI. Dyskerin trajectories whose coordinates overlapped with the nucleolar mask for a single frame were designated as nucleolar trajectories. The remaining trajectories were designated nuclear trajectories. All data sets were analyzed using Spot-On as described above.
Fluorescence recovery after photo bleaching
Fluorescence recovery experiments (FRAP) of Halo-DKC1 were carried out using the same Olympus microscope used for single-molecule imaging. Cells were stained for 10 min with 100 nM JFX650-HaloTag ligand in complete media. After removing the HaloTag-ligand with three washes in complete media, cells were placed back in the incubator for 10 min to allow unincorporated dye to leak out of the cells. Cells were then transferred into CO2 independent media and put on the microscope which was heated to 37°C. We identified cells with two clearly visible nucleoli and bleached one of them by placing three diffraction limited bleach spots within the nucleolar 3xFLAG-HaloTag-dyskerin signal. Each spot was bleached for 100 ms at 50% laser power, which lead to complete loss of the fluorescence within the nucleolus. Cells were imaged prior to and after bleaching at 1 frame per second using the Excelitas X-cite TURBO LED light source and the 100x objective. Photobleaching due to LED exposure was negligible. To quantify FRAP we first drift corrected the movie using NanoJ,52 we then placed a region of interest (ROI) within the nucleolus and quantified mean intensity within the ROI over time. Background signal was determined in an area of the field of view that was not covered by a cell and subtracted from the nucleolar ROI. In addition, the mean fluorescence after the bleaching pulse was divided by the fraction of total cellular fluorescence remaining after the bleaching pulse. Because the laser pulse bleaches a significant amount of total cellular fluorescence (typically 20–40%), this normalization is critical to determine the maximal amount of fluorescence recovery possible. For example, if 30% of total cellular fluorescence is lost due to the bleaching pulse, the maximal fraction of pre-bleach fluorescence than can theoretically be recovered is 70%. The recovery data was then fit using a single exponential function (1-A*e−kt+C), where k corresponds to the rate constant and C to the fraction of the initial signal that is not recovered (i.e. the static fraction).
For the FRAP analysis of nucleolar 3xMS2-TR bound by MCP-mNeonGreen cells were imaged on a Nikon A1R confocal microscope in the MSU Confocal Laser Scanning Microscopy core. 3xMS2 TR clearly localized to the DFC of the nucleolus marked by mTagBFP-Nucleus-7 was bleached using the 488 nm laser line and recovery of the MCP-mNeonGreen was imaged every 2 s using the 488 nm laser line. Data analysis was varied out as described above.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification of RT-qPCR data
RT-qPCR experiments were carried out in triplicate and the TR Ct value was normalized to the GAPDG Ct value. The mean ΔCt (Ct of TR – Ct of GAPDH) value from three independent experiments and the corresponding standard deviation were plotted.
Quantification of western blots, northern blots, and telomerase activity assays
Gel images from western Blots, northern Blots, and telomerase activity assays were analyzed using ImageQuant TL 8.2. To quantify TR levels in Northern blots the TR band intensity was normalized to the loading and recovery control signal added to the RNA sample prior to RNA purification. To quantify telomerase activity, the whole lane intensity starting at repeat 1 was determined and divided by the sum of the loading control signals. Telomerase processivity was calculated by dividing product intensity >7 repeats by the total signal in the respective lane. The statistical significance of the observed differences was calculated using a two-tailed T test using a minimum of three biological replicates. For telomerase activity assays each biological replicate (independent telomerase expression and purification) was analyzed in technical triplicate.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-TCAB1 rabbit polyclonal antibody | Proteintech | 14761-1-AP |
| anti-TCAB1 rabbit polyclonal antibody | Abcam | ab224444 |
| anti-TCAB1 rabbit polyclonal antibody | Novus Biologicals | NB100-68252 |
| anti-GAR1 rabbit polyclonal antibody | Proteintech | 11711-1-AP |
| anti-Coilin mouse monoclonal antibody | Abcam | ab87913 |
| anti-Dyskerin mouse monoclonal antibody | Santa Cruz Biotech. | sc-373956 |
| anti-TERT recombinant rabbit monoclonal antibody | Abcam | ab30202 |
| anti-TERT sheep polyclonal antibody | Scott Cohen | Abx120550 |
| anti-Fibrillarin mouse monoclonal antibody | Novus Biologicals | NB300-269 |
| anti-Lamin B1 rabbit polyclonal antibody | Proteintech | 12987-1-AP |
| anti-Rabbit Cy3 secondary antibody | Invitrogen | A-10520 |
| anti-Mouse Alexa 647 secondary antibody | Invitrogen | A-21235 |
| anti-Mouse Alexa 405 secondary antibody | Invitrogen | A-31553 |
| anti-Mouse HRP | Invitrogen | A-31430 |
| anti-Rabbit HRP | Invitrogen | A-31460 |
| Chemicals, peptides, and recombinant proteins | ||
| JF646 HaloTag Ligand | Grimm et al.35 | N/A |
| Prolong Diamond Antifade Mountant | Life Technologies | P36970 |
| Thymidine | Sigma | T1895 |
| Puromycin 10 mg/mL | Gibco | A1113803 |
| Lipofectamine 2000 | Invitrogen | 11668019 |
| dGTP [α-32P] 3000 Ci/mmol 10 mCi/mL | Perkin Elmer | BLU514H250UC |
| TERT peptide ARPAEEATSLEGALSGTRH | Cohen et al.36 | N/A |
| Dextrane sulfate | Sigma | D8906-10G |
| E. coli tRNA | Sigma | 10109541001 |
| Salmon Sperm DNA | Invitrogen | 15632011 |
| Vanadyl Ribonucleoside Complexes | Sigma | R3380-5ML |
| CHAPS | Fisher BioReagents | 211911 |
| SuperSignal West Femto Maximum Sensitivity Substrate | Thermo Scientific | 34096 |
| Clarity Western ECL substrate | Bio-Rad | 1705061 |
| Experimental models: Cell lines | ||
| HeLa EM2-11ht, TCAB1 KO | This paper | N/A |
| HeLa EM2-11ht, FLAG-HaloTag-TERT WT, HA-mEOS3.2-TRF2 | Schmidt et al.24 | N/A |
| HeLa EM2-11ht, FLAG-HaloTag-TERT WT, HA-mEOS3.2-TRF2, TCAB1 KO | This paper | N/A |
| HeLa EM2-11ht, FLAG-HaloTag-TERT WT, HA-mEOS3.2-TRF2, TR KO | This paper | N/A |
| HeLa EM2-11ht, FLAG-HaloTag-TERT WT, HA-mEOS3.2-TRF2, 3xMS2-TR | This paper | N/A |
| HeLa EM2-11ht, FLAG-HaloTag-TERT WT, HA-mEOS3.2-TRF2, 3xMS2-TR, TCAB1 KO | This paper | N/A |
| Oligonucleotides | ||
| All Oligonucleotides used in this study can be found in Table S1 | ||
| Recombinant DNA | ||
| pHTN HaloTag® CMV-neo | Promega | G7721 |
| pFastBac1 | Life Technologies | 10360014 |
| pX330 | Cong et al.37 | Addgene: 42230 |
| pmaxGFP | Lonza | Control from Kits |
| mCherry-TERT | Schmidt et al.38 | N/A |
| TERT (pVAN107) | Cristofari and Lingner39 | N/A |
| mCherry-Dyskerin | This paper | N/A |
| 3xFLAG-HaloTag-dyskerin | This paper | N/A |
| GFP-Nucleolin | Takagi et al.40 | Addgene: 28176 |
| GFP-NPM1 | Wang et al.41 | Addgene: 17578 |
| mTagBFP-Nucleus-7 (BFP-NLS) | Addgene | Addgene: 55265 |
| BFP-coilin | Schmidt et al.24 | N/A |
| pSUPER TR | Cristofari and Lingner39 | N/A |
| LhTRmin | Vogan et al.19 | N/A |
| pBS U3-hTR-500 | Fu and Collins42 | Addgene: 28170 |
| TR knockout HRD | This paper | N/A |
| pHAGE-UBC-MCP-mNeonGreen | This paper | N/A |
| 3xMS-TR-100-PURO HRD | This paper | N/A |
| Software and algorithms | ||
| MATLAB 2019a | Mathworks Inc., USA | http://mathworks.com |
| ParallelProcess_fastSPT_JF646.m | Hansen et al.27 | https://github.com/elifesciences-publications/SPT_LocAndTrack |
| ImageQuant TL 8.2 | Cytiva | cytivalifesciences.com |
| Spot-On | Hansen et al.27 | https://github.com/elifesciences-publications/spot-on-matlab |
| Other | ||
| SuperScript III | Invitrogen | 18080044 |
| Maxima SYBR Green qPCR Master Mix | Thermo Scientific | K0251 |
| Random Hexamers | Invitrogen | N8080127 |
| RiboLock RNase Inhibitor | Thermo Scientific | EO0382 |
| HOECHST 33342 | Invitrogen | 62249 |
| Protein G Agarose | Millipore Sigma | 11243233001 |
| 4-15% Mini-PROTEAN TGX precast protein gels | Bio-Rad | 4561083 |
| Trans-Blot Turbo RTA Mini 0.2 μm Nitrocellulose Kit | Bio-Rad | 1704270 |
| FxCycle Propidium iodide/RNase staining solution | Thermo Scientific | F10797 |
| RNeasy Mini Kit | Qiagen | 74106 |
Highlights.
TCAB1 facilitates telomerase assembly in cancer cells
In the absence of TCAB1, the telomerase RNA is sequestered in the nucleolus
TCAB1 is not required for telomerase catalysis
TCAB1 is excluded from nucleoli and prevents entry of the telomerase RNA into the nucleolus
ACKNOWLEDGMENTS
We would like to thank members of the Schmidt lab and J. Nandakumar for discussions and critical reading of the manuscript and Luke Lavis for providing HaloTag dyes. This work was supported by grants from the NIH (R00 GM120386, R01GM141354) to J.C.S. J.C.S. was a Damon Runyon Dale F. Frey Scientist supported (in part) by the Damon Runyon Cancer Research Foundation (DFS-24-17). S.B.C. acknowledges sustained support from the Ernest & Piroska Major Foundation. We acknowledge the Flow Cytometry core (Research Technology Support Facility, Michigan State University) and the Confocal Laser Scanning Microscopy core (Center for Advanced Microscopy, Michigan State University) for supporting our work.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112577.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Hyman AA, Weber CA, and Jülicher F (2014). Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol 30, 39–58. 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- 2.Mitrea DM, and Kriwacki RW (2016). Phase separation in biology; functional organization of a higher order. Cell Commun. Signal 14, 1. 10.1186/s12964-015-0125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angrisani A, Vicidomini R, Turano M, and Furia M (2014). Human dyskerin: beyond telomeres. Biol. Chem 395, 593–610. 10.1515/hsz-2013-0287. [DOI] [PubMed] [Google Scholar]
- 4.Schmidt JC, and Cech TR (2015). Human telomerase: biogenesis, trafficking, recruitment, and activation. Gene Dev. 29, 1095–1105. 10.1101/gad.263863.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jády BE, Bertrand E, and Kiss T (2004). Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body–specific localization signal. J. Cell Biol 164, 647–652. 10.1083/jcb.200310138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, Veenstra TD, Terns MP, and Artandi SE (2009). A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science 323, 644–648. 10.1126/science.1165357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tycowski KT, Shu M-D, Kukoyi A, and Steitz JA (2009). A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol. Cell 34, 47–57. 10.1016/j.molcel.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stewart SA, and Weinberg RA (2006). Telomeres: cancer to human aging. Annu. Rev. Cell Dev. Biol 22, 531–557. 10.1146/annurev.cellbio.22.010305.104518. [DOI] [PubMed] [Google Scholar]
- 9.Armanios M, and Blackburn EH (2012). The telomere syndromes. Nat. Rev. Genet 13, 693–704. 10.1038/nrg3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nandakumar J, and Cech TR (2013). Finding the end: recruitment of telomerase to telomeres. Nat. Rev. Mol. Cell Biol 14, 69–82. 10.1038/nrm3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu RA, Upton HE, Vogan JM, and Collins K (2017). Telomerase mechanism of telomere synthesis. Annu. Rev. Biochem 86, 439–460. 10.1146/annurev-biochem-061516-045019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagpal N, Wang J, Zeng J, Lo E, Moon DH, Luk K, Braun RO, Burroughs LM, Keel SB, Reilly C, et al. (2020). Small-molecule PAPD5 inhibitors restore telomerase activity in patient stem cells. Cell Stem Cell 26, 896–909.e8. 10.1016/j.stem.2020.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shukla S, Jeong H-C, Sturgeon CM, Parker R, and Batista LFZ (2020). Chemical inhibition of PAPD5/7 rescues telomerase function and hematopoiesis in dyskeratosis congenita. Blood Adv. 4, 2717–2722. 10.1182/bloodadvances.2020001848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stuart BD, Choi J, Zaidi S, Xing C, Holohan B, Chen R, Choi M, Dharwadkar P, Torres F, Girod CE, et al. (2015). Exome sequencing links mutations in PARN and RTEL1 with familial pulmonary fibrosis and telomere shortening. Nat. Genet 47, 512–517. 10.1038/ng.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tummala H, Walne A, Collopy L, Cardoso S, de la Fuente J, Lawson S, Powell J, Cooper N, Foster A, Mohammed S, et al. (2015). Poly(A)-specific ribonuclease deficiency impacts telomere biology and causes dyskeratosis congenita. J. Clin. Invest 125, 2151–2160. 10.1172/jci78963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tseng C-K, Wang H-F, Burns AM, Schroeder MR, Gaspari M, and Baumann P (2015). Human telomerase RNA processing and quality control. Cell Rep. 13, 2232–2243. 10.1016/j.celrep.2015.10.075. [DOI] [PubMed] [Google Scholar]
- 17.Shukla S, Schmidt JC, Goldfarb KC, Cech TR, and Parker R (2016). Inhibition of telomerase RNA decay rescues telomerase deficiency caused by dyskerin or PARN defects. Nat. Struct. Mol. Biol 23, 286–292. 10.1038/nsmb.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong F, Savage SA, Shkreli M, Giri N, Jessop L, Myers T, Chen R, Alter BP, and Artandi SE (2011). Disruption of telomerase trafficking by TCAB1 mutation causes dyskeratosis congenita. Gene Dev. 25, 11–16. 10.1101/gad.2006411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vogan JM, Zhang X, Youmans DT, Regalado SG, Johnson JZ, Hockemeyer D, and Collins K (2016). Minimized human telomerase maintains telomeres and resolves endogenous roles of H/ACA proteins, TCAB1, and Cajal bodies. Elife 5, e18221. 10.7554/elife.18221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen L, Roake CM, Freund A, Batista PJ, Tian S, Yin YA, Gajera CR, Lin S, Lee B, Pech MF, et al. (2018). An activity switch in human telomerase based on RNA conformation and shaped by TCAB1. Cell 174, 218–230.e13. 10.1016/j.cell.2018.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freund A, Zhong FL, Venteicher AS, Meng Z, Veenstra TD, Frydman J, and Artandi SE (2014). Proteostatic control of telomerase function through TRiC-mediated folding of TCAB1. Cell 159, 1389–1403. 10.1016/j.cell.2014.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stern JL, Zyner KG, Pickett HA, Cohen SB, and Bryan TM (2012). Telomerase recruitment requires both TCAB1 and cajal bodies independently. Mol. Cell Biol 32, 2384–2395. 10.1128/mcb.00379-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao R-W, Xu G, Wang Y, Shan L, Luan P-F, Wang Y, Wu M, Yang L-Z, Xing Y-H, Yang L, and Chen LL (2019). Nascent pre-rRNA sorting via phase separation drives the assembly of dense fibrillar components in the human nucleolus. Mol. Cell 76, 767–783.e11. 10.1016/j.molcel.2019.08.014. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt JC, Zaug AJ, and Cech TR (2016). Live cell imaging reveals the dynamics of telomerase recruitment to telomeres. Cell 166, 1188–1197.e9. 10.1016/j.cell.2016.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen SB, Graham ME, Lovrecz GO, Bache N, Robinson PJ, and Reddel RR (2007). Protein composition of catalytically active human telomerase from immortal cells. Science 315, 1850–1853. 10.1126/science.1138596. [DOI] [PubMed] [Google Scholar]
- 26.Egan ED, and Collins K (2010). Specificity and stoichiometry of subunit interactions in the human telomerase holoenzyme assembled in vivo. Mol. Cell Biol 30, 2775–2786. 10.1128/mcb.00151-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hansen AS, Woringer M, Grimm JB, Lavis LD, Tjian R, and Darzacq X (2018). Robust model-based analysis of single-particle tracking experiments with Spot-On. Elife 7, e33125. 10.7554/elife.33125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt JC, Zaug AJ, Kufer R, and Cech TR (2018). Dynamics of human telomerase recruitment depend on template-telomere base pairing. Mol. Biol. Cell 29, 869–880. 10.1091/mbc.e17-11-0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JH, Lee YS, Jeong SA, Khadka P, Roth J, and Chung IK (2014). Catalytically active telomerase holoenzyme is assembled in the dense fibrillar component of the nucleolus during S phase. Histochem. Cell Biol 141, 137–152. 10.1007/s00418-013-1166-x. [DOI] [PubMed] [Google Scholar]
- 30.Lam YW, and Lamond AI (2006). Cell biology. In Part Organelles Cell Struct Sect 1isolation Plasma Membr Organelles Cell Struct Sect 1 Isol Plasma Membr Organelles Cell Struct, Third Edition, pp. 103–107. 10.1016/b978-012164730-8/50087-3. [DOI] [Google Scholar]
- 31.Laprade H, Querido E, Smith MJ, Guérit D, Crimmins H, Conomos D, Pourret E, Chartrand P, and Sfeir A (2020). Single-molecule imaging of telomerase RNA reveals a recruitment-retention model for telomere elongation. Mol. Cell 79, 115–126.e6. 10.1016/j.molcel.2020.05.005. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y, Deng Z, Jiang S, Hu Q, Liu H, Songyang Z, Ma W, Chen S, and Zhao Y (2015). Human cells lacking coilin and Cajal bodies are proficient in telomerase assembly, trafficking and telomere maintenance. Nucleic Acids Res. 43, 385–395. 10.1093/nar/gku1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riback JA, Zhu L, Ferrolino MC, Tolbert M, Mitrea DM, Sanders DW, Wei M-T, Kriwacki RW, and Brangwynne CP (2020). Composition-dependent thermodynamics of intracellular phase separation. Nature 581, 209–214. 10.1038/s41586-020-2256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heiss NS, Girod A, Salowsky R, Wiemann S, Pepperkok R, and Poustka A (1999). Dyskerin localizes to the nucleolus and its mislocalization is unlikely to play a role in the pathogenesis of dyskeratosis congenita. Hum. Mol. Genet 8, 2515–2524. 10.1093/hmg/8.13.2515. [DOI] [PubMed] [Google Scholar]
- 35.Grimm JB, English BP, Chen J, Slaughter JP, Zhang Z, Revyakin A, Patel R, Macklin JJ, Normanno D, Singer RH, et al. (2015). A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods 12, 244–250, 3 p following 250 p following 250. 10.1038/nmeth.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cohen SB, and Reddel RR (2008). A sensitive direct human telomerase activity assay. Nat Methods 5, 355–360. 10.1038/nmeth.f.209. [DOI] [PubMed] [Google Scholar]
- 37.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, and Zhang F (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt JC, Dalby AB, and Cech TR (2014). Identification of human TERT elements necessary for telomerase recruitment to telomeres. Elife 3, e03563. 10.7554/elife.03563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cristofari G, and Lingner J (2006). Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 25, 565–574. 10.1038/sj.emboj.7600952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takagi M, Absalon MJ, McLure KG, and Kastan MB (2005). Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell 123, 49–63. 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 41.Wang W, Budhu A, Forgues M, and Wang XW (2005). Temporal and spatial control of nucleophosmin by the Ran–Crm1 complex in centrosome duplication. Nat. Cell Biol 7, 823–830. 10.1038/ncb1282. [DOI] [PubMed] [Google Scholar]
- 42.Fu D, and Collins K (2003). Distinct biogenesis pathways for human telomerase RNA and H/ACA small nucleolar RNAs. Mol. Cell 11, 1361–1372. 10.1016/s1097-2765(03)00196-5. [DOI] [PubMed] [Google Scholar]
- 43.Weidenfeld I, Gossen M, Löw R, Kentner D, Berger S, Görlich D, Bartsch D, Bujard H, and Schönig K (2009). Inducible expression of coding and inhibitory RNAs from retargetable genomic loci. Nucleic Acids Res. 37, e50. 10.1093/nar/gkp108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Querido E, Sfeir A, and Chartrand P (2020). Imaging of telomerase RNA by single-molecule inexpensive FISH combined with immunofluorescence. STAR Protoc. 1, 100104. 10.1016/j.xpro.2020.100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomlinson RL, Ziegler TD, Supakorndej T, Terns RM, and Terns MP (2006). Cell cycle-regulated trafficking of human telomerase to telomeres. Mol. Biol. Cell 17, 955–965. 10.1091/mbc.e05-09-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tokunaga M, Imamoto N, and Sakata-Sogawa K (2008). Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Methods 5, 159–161. 10.1038/nmeth1171. [DOI] [PubMed] [Google Scholar]
- 47.Grimm JB, Xie L, Casler JC, Patel R, Tkachuk AN, Choi H, Lippincott-Schwartz J, Brown TA, Glick BS, Liu Z, et al. (2020). Deuteration improves small-molecule fluorophores. Preprint at bioRxiv. 10.1101/2020.08.17.250027. [DOI] [Google Scholar]
- 48.Southern E. (2006). Southern blotting. Nat. Protoc 1, 518–525. 10.1038/nprot.2006.73. [DOI] [PubMed] [Google Scholar]
- 49.Ghanim GE, Fountain AJ, van Roon A-MM, Rangan R, Das R, Collins K, and Nguyen THD (2021). Structure of human telomerase holoenzyme with bound telomeric DNA. Nature, 1–5. 10.1038/s41586-021-03415-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nandakumar J, Bell CF, Weidenfeld I, Zaug AJ, Leinwand LA, and Cech TR (2012). The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature 492, 285–289. 10.1038/nature11648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sergé A, Bertaux N, Rigneault H, and Marguet D (2008). Dynamic multiple-target tracing to probe spatiotemporal cartography of cell membranes. Nat. Methods 5, 687–694. 10.1038/nmeth.1233. [DOI] [PubMed] [Google Scholar]
- 52.Laine RF, Tosheva KL, Gustafsson N, Gray RDM, Almada P, Albrecht D, Risa GT, Hurtig F, Lindås AC, Baum B, et al. (2019). NanoJ: a high-performance open-source super-resolution microscopy toolbox. J. Phys. D Appl. Phys 52, 163001. 10.1088/1361-6463/ab0261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All unprocessed data is available upon request from the lead contact. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.