Abstract
Lactobacilli and Acetobacter sp. are commercially important bacteria that often form communities in natural fermentations, including food preparations, spoilage, and in the digestive tract of the fruit fly Drosophila melanogaster. Communities of these bacteria are widespread and prolific, despite numerous strain-specific auxotrophies, suggesting they have evolved nutrient interdependencies that regulate their growth. The use of a chemically-defined medium (CDM) supporting the growth of both groups of bacteria would facilitate the identification of the molecular mechanisms for the metabolic interactions between them. While numerous CDMs have been developed that support specific strains of lactobacilli or Acetobacter, there has not been a medium formulated to support both genera. We developed such a medium, based on a previous CDM designed for growth of lactobacilli, by modifying the nutrient abundances to improve growth yield. We further simplified the medium by substituting casamino acids in place of individual amino acids and the standard Wolfe’s vitamins and mineral stocks in place of individual vitamins and minerals, resulting in a reduction from 40 to 8 stock solutions. These stock solutions can be used to prepare several CDM formulations that support robust growth of numerous lactobacilli and Acetobacters. Here, we provide the composition and several examples of its use, which is important for tractability in dissecting the genetic and metabolic basis of natural bacterial species interactions.
Introduction
Lactic Acid Bacteria (LAB) and Acetic Acid Bacteria (AAB) coexist in nature in a wide variety of environments [1]. These include food fermentations such as wine [1], beer [2], kefir [3, 4], sauerkraut [5], kimchi [6], bread [7], and cacao beans [8]. They also have been found together in agricultural feed, such as silage [9]. LAB and AAB can coexist as well in mammalian and insect gastrointestinal tracts where they have probiotic benefits [10]. This includes the intestine of the genetic model animal Drosophila melanogaster, where lactobacilli and Acetobacter sp. are the core types of LAB and AAB, respectively [11]. We note that due to the recent reclassifications within the former Lactobacillus genus [12], we refer to the LABs used in this study by their newly given formal scientific names, Lactiplanibacillus plantarum (Lp. plantarum) and Levilactobacillus brevis (Ll. brevis) or collectively by their common name, lactobacillus (plural lactobacilli).
Why LAB and AAB co-occur so prevalently is the subject of ongoing investigations. A synergistic metabolism between lactobacilli and Acetobacters has been demonstrated based on sharing of organic short chain fatty acids (SCFAs), including lactic acid and acetic acid. These compounds are generally known to mutually promote growth through cross-feeding, such that lactate stimulates Acetobacter growth and acetate stimulates lactobacilli [13, 14]. There is biomedical relevance of SCFA as their production by the gut microbiome plays a key role in gut epithelial cell metabolism and immune homeostasis [15, 16] potentially underlying the role of LABs as probiotics [10]. Furthermore, in the mammalian colonic crypts, microbial assemblages include lactobacilli and other Lactobacillales such as Streptococcus, and α-Proteobacteria besides Acetobacter, including Sphingomonas and Paracoccus [17, 18], suggesting there may be a broader phylogenetic pattern of co-existence for these groups. Studying these relationships in synthetic microbial communities could provide insights into their roles in more complex environments with multiple trophic levels such as the digestive tract. For instance, cross-feeding may extend beyond SFCAs to other metabolites, namely B-vitamins, which are known to impact host health [19, 20]. Cross-fed nutrients could also influence secondary metabolite production [21]. Thus, there exists a pressing need to study the molecular mechanisms of metabolic interactions in natural gut microbial communities, and the lactobacilli-Acetobacter communities in the Drosophila gut are a promising model.
Identifying the nutrient requirements for organismal growth [22] as well as the metabolites produced [23] during fermentation is greatly improved with the use of chemically-defined growth media (CDM). Various CDM recipes have been developed to support growth of lactobacilli [24–30], yet no efforts have been made to develop a CDM with the express purpose of supporting lactobacillus-Acetobacter communities for microbiology experiments. As a resource to investigate the mechanisms of lactobacillus-Acetobacter nutritional cross-feeding, we developed a chemically-defined medium (CDM) that independently supports growth of several Lp. plantarum strains. By altering the nutrient composition, we show that the CDM can enable growth of both lactobacilli and Acetobacter sp. isolated from the fruit fly gut microbiome and other sources in high throughput. Therefore, this CDM fulfills the purpose of growing lactobacilli and Acetobacter sp. under chemically-defined conditions, which will enable experiments to determine the molecular and genetic mechanisms of their nutrient exchange.
We based this medium on the formulation of [29]. Our medium may be modified to optimize the growth of either lactobacilli or Acetobacters to a density of ∼ 109 cells/mL (OD600 > 1) or to support co-cultures. In this short report, we provide the chemical composition of the medium, some guides on its preparation, an approach to circumvent strain-specific auxotrophies, and some known issues that can result from chemical impurities. We focus our results on the lactobacilli and Acetobacter sp. from the Drosophila gut microbiome.
Materials and methods
Chemicals
A complete list of all chemicals used in the various CDM formulations is provided in S1 Table in S1 File.
Media preparation
Stock solutions were individually prepared in ultra-pure water or in appropriate solvents as indicated in Table 1. All stock solutions were passed through 0.22 μm filter and kept in dark at 4°C except nucleotides, which were stored at -20°C. FeSO4 ⋅ 7H2O was freshly prepared. Medium was initially made with 30% less water to allow customized additions. Final pH of CDM was adjusted to 6.5 and the medium was passed through a 0.22 μm filter. CDM was stored at 4°C and used within 2 days.
Table 1. Composition of the chemically-defined growth medium.
| Compound | Concentration | Units | Supplier | Part Number | Stock |
|---|---|---|---|---|---|
| Base components | |||||
| MOPS | 40 | mM | Millipore | 475898 | 10x in H2O |
| K2HPO4 | 5 | mM | Fisher | P288 | 10x in H2O |
| NaCl | 0.2 | mM | Fisher | S271 | 100x in H2O |
| NH4Cl | 20 | mM | Fisher | A649 | 100x in H2O |
| K2SO4 | 10 | mM | Sigma | 746363 | 50x in H2O |
| MgCl2.6H2O | 1 | mM | Fisher | BP214 | 100x in H2O |
| MnCl2.4H2O | 0.05 | mM | Sigma | M3634 | 100x in H2O |
| FeSO4.7H2O | 0.05 | mM | Sigma | F8633 | 100x in H2O |
| Amino acids | |||||
| L-Alanine | 14 | mM | Sigma | A7627 | 40x in H2O |
| L-Arginine | 0.36 | mM | Sigma | A5131 | 200x in H2O |
| Glycine | 3.41 | mM | Sigma | G7126 | 200x in H2O |
| L-Lysine | 3.59 | mM | Sigma | L5626 | 200x in H2O |
| L-Proline | 1.737 | mM | Sigma | P0380 | 200x in H2O |
| L-Histidine | 2.664 | mM | Sigma | H8125 | 200x in H2O |
| L-Serine | 7.374 | mM | Sigma | S4500 | 200x in H2O |
| L-Threonine | 2.098 | mM | Sigma | T8625 | 200x in H2O |
| L-Aspartic acid | 0.083 | mM | Sigma | A9256 | 200x in 1 M HCl |
| L-Asparagine | 4.162 | mM | Sigma | A0884 | 200x in 1 M HCl |
| L-Tyrosine | 1.1035 | mM | Sigma | T3754 | 200x in 1 M NaOH |
| L-Cysteine-HCl | 4.758 | mM | Sigma | C1276 | 200x in H2O |
| L-Valine | 4.268 | mM | Sigma | V0500 | 200x in 1 M NaOH |
| L-Glutamic acid | 1.417 | mM | Sigma | G1251 | 200x in 1 M HCl |
| L-Tryptophane | 1.371 | mM | Sigma | T0254 | 200x in 1 M NaOH |
| L-Phenylalanine | 1.513 | mM | Sigma | P2126 | 200x in 1 M HCl |
| L-Glutamine | 3.267 | mM | Sigma | G3126 | 200x in 1 M NaOH |
| L-Leucine | 1.905 | mM | Sigma | L8000 | 2000x in 0.5 M HCl |
| L-Isoleucine | 1.905 | mM | Sigma | I2752 | 2000x in 0.5 M HCl |
| L-Methionine | 0.67 | mM | Sigma | M9625 | 200x in 0.1 M HCl |
| Nucleotides | |||||
| Guanine | 0.033 | mM | Sigma | G6779 | 200x in 0.1 M NaOH |
| Uracil | 0.0445 | mM | Sigma | U1128 | 200x in 0.1 M NaOH |
| Xanthine | 0.0325 | mM | Sigma | X4002 | 200x in 0.1 M NaOH |
| Adenine | 0.037 | mM | Sigma | A2786 | 200x in 0.1 M HCl |
| Carbon | |||||
| Glucose | 125 | mM | Sigma | G8270 | 50x in H2O |
| Acetate | 10 | mM | Fisher | S210 | 100x in H2O |
| D,L-Lactate | 0.6 | mM | Sigma | 69785 | 100x in H2O |
| Optional | |||||
| Ascorbic acid | 1.4 | mM | Fisher | AA3623714 | 100x in H2O |
| Lipoic acid | 1 | mM | Sigma | T1395 | direct |
| Fructose | 62.5 or 125 | mM | Sigma | F3510 | 50x in H2O |
To make a carbon-free CDM, the following components were combined in the following order in a final volume of 17.5 mL: 8.375 mL ultra-pure water, 2.5 mL MOPS buffer, 0.25 mL K2HPO4, 0.25 mL NaCl, 0.25 mL NH4Cl, 0.5 mL K2SO4, 0.625 mL L-Alanine, 0.125 mL L-Arginine, 0.125 mL Glycine, 0.125 mL L-Lysine, 0.5 mL L-Proline, 0.125 mL L-Histidine, 0.125 mL L-Serine, 0.125 mL L-Threonine, 0.125 mL L-Aspartic acid, 20 uL of 10 N NaOH for pH correction, 0.125 mL L-Asparagine, 10 uL of 10 N NaOH for pH correction, 0.125 mL L-Tyrosine, 0.125 mL L-Cysteine-HCl, 0.125 mL L-Valine, 0.125 mL L-Glutamic acid, 0.125 mL L-Tryptophane, 0.125 mL L-Phenylalanine, 0.125 mL L-Glutamine, 0.125 mL L-Leucine, 0.125 mL L-Isoleucine, 0.125 mL L-Methionine, 0.125 mL Ca-D-(+)-pantothenate, 0.125 mL Lipoic acid, 0.125 mL Nicotinic acid, 0.125 mL para-Aminobenzoic acid, 0.125 mL Pyridoxine-HCl, 0.125 mL Thiamine-HCl, 0.125 mL Biotin, 0.125 mL Ascorbic acid, 0.125 mL Folic acid, 0.125 mL Guanine, 0.125 mL Uracil, 0.125 mL Xanthine, 0.125 mL Adenine, 0.25 mL MgCl2 ⋅ 6H2O, 0.25 mL MnCl2 ⋅ 4H2O, 0.25 mL FeSO4 ⋅ 7H2O.
Appropriate carbon sources were provided in CDM to grow specific isolates. A final concentration of 1% glucose and 0.1% acetate were added for the growth of Lp. plantarum. A final concentration of 1% glucose, 0.5% fructose and 0.1% acetate supported limited growth of Ll. brevis. A final concentration of 1% fructose was used for the growth of A. pasteurianus, and 1% glucose was used for the growth of A. tropicalis.
Bacterial strains and growth
Bacterial strains used in this study are listed in S2 Table in S1 File. Prior to CDM experiments, strains were initially cultured in rich media to maximize growth rate. These include MRS medium for lactobacilli and Mannitol-Yeast Extract-Peptone medium supplemented with 1% DL-Lactic acid (MYPL) for Acetobacters. Rich medium streak plates were routinely prepared from glycerol stocks and incubated at 30°C for 48 hours and used only once. From these plates, 2–3 mL rich media were inoculated with single colonies and grown at 30°C for 16 to 18 h. To facilitate efficient growth in CDM for growth kinetics experiments, cultures were passaged once in a 1:1 mixture of rich medium and CDM followed by 1–3 passages in CDM. Turbidity was then adjusted to OD600 = 1 and cultures were diluted 1:100 into 200 μL CDM in optically clear 96-well microplates. Microplates were incubated at 30°C in a temperature-controlled plate reader either statically or with double orbital shaking at a speed of 425 CPM and an orbital radius of 3 mm. For static cultures, plates were subjected to linear shaking at 425 CPM for 10 seconds prior to each measurement to resuspend bacteria. OD600 measurements were taken every 60 minutes in static cultures and every 5–15 minutes in shaken cultures for 48 hours. Static culture plates were sealed with a low-evaporation plastic lid. Shaken culture plates were sealed with a Breath-Easy film, and off-center holes were poked over each well out of the way of the detector with a 16 gauge needle to ensure optimal aeration for Acetobacter sp. M1000 (Tecan), Magellan, and Epoch 2 (Biotek) plate readers were used. OD600 measurements were background-subtracted using the OD600 measurement of medium-only controls. Each experiment was performed for 12 experimental replicates from three biological replicates.
Statistical analysis
Growth curves were smoothed with a cubic spline and standard error of the mean was calculated on a five time point window rolling average across the 12 replicates. All analysis was performed in MATLAB R2021b.
Results and discussion
Formulation of the CDM for lactobacillus growth
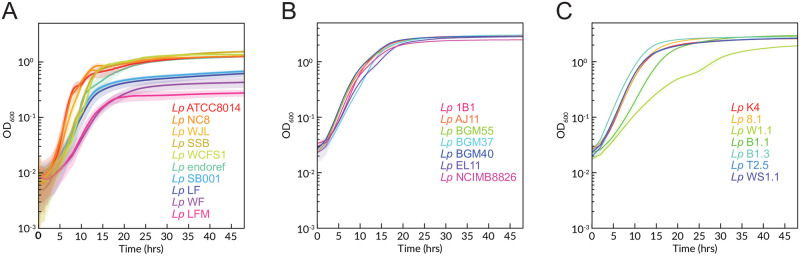
With the goal of formulating a CDM that supports growth of the lactobacilli and Acetobacter isolated from the fruit fly gut (see S2 Table in S1 File), we first tested a previous chemically-defined medium that was developed for lactobacilli [29]. This medium contains glucose, all amino acids, and nucleotides. It supported limited growth of lactobacilli to an OD600 of only approximately 0.2, and it precipitated upon storage. We eliminated precipitation by decreasing the amino acid concentrations to the levels described by Palmer et el. 2007 [31] and added 40 mM MOPS buffer (3-morpholinopropane-1-sulfonic acid). This modified medium supported growth of Lp. plantarum LFM, a D. melanogaster isolate, to an OD600 of ≈ 0.2 (S3 Table in S1 File), corresponding to ∼ 108 cells/mL. This low value is ∼ 10-fold lower density than reported for the Lp. plantarum strain analyzed in the previous study [29]. As strain-specific differences in media preferences are widely reported in lactobacilli [32], we further refined the composition by individually adding amino acids, vitamins and nucleotides at 10-fold higher concentrations into the original CDM (S4 Table in S1 File). Higher amounts of tyrosine and cysteine increased media precipitation, causing a higher OD600 reading, which would interfere with high throughput growth measurements. Improved growth yield with excess tryptophan was reproducible, thus we increased the concentrations in the CDM to 1.4 mM, (Table 1). We also experimented with increasing or decreasing the total amino acids, vitamins, and nucleic acids (S3 Table in S1 File), which showed we could double these components to increase Lp. plantarum growth without substantially affecting other lactobacilli and Acetobacter species. We next simplified the medium to reduce the total number of stock solutions. We first replaced the vitamins and minerals with Wolfe’s vitamins and Wolfe’s minerals stocks. We then replaced the individual amino acids with casamino acids, which are a mixture of amino acids derived from acid hydrolysis of casein, having a representative abundance of every individual amino acid except tryptophan. This variant of the medium, named CDML still required additional alanine (14 mM) and cysteine (12 mM) (S1 Fig in S1 File; Table 2) but supported rapid growth of a variety of Lp. plantarum strains from different sources (Fig 1, S2 Table in S1 File) to an OD600 > 1, corresponding to ∼ 109 cells/mL under aerobic (Fig 1A) and anaerobic conditions (Fig 1B and 1C).
Table 2. Composition of CDML.
| Stock solution # | Chemical | Final concentration (1x) |
|---|---|---|
| Buffer and salts | ||
| 1 | MOPS | 40 mM |
| 1 | K2HPO4 | 5 mM |
| 1 | NH4Cl | 20 mM |
| 1 | Na2SO4 | 10 mM |
| Metals | ||
| 2 | MgCl2 ⋅ 6 H2O | 1 mM |
| 2 | MnCl2 ⋅ 4 H2O | 0.05 mM |
| 2 | FeSO4 ⋅ 7 H2O | 0.05 mM |
| Carbon source | ||
| 3 | Glucose | 125 mM |
| 3 | Ammonnium acetate | 10 mM |
| Amino acids | ||
| 4 | Casamino acids | 3 g/L |
| 5 | Cysteine-HCl ⋅ H2O | 0.145 g/L |
| 6 | Tryptophan | 0.05 g/L |
| 7 | Wolfe’s vitamins | 1x |
| 8 | Wolfe’s minerals | 1x |
10x Wolfe’s vitamins contains 10 mg/L Ca-(D)-(+)-pantothenate (Sigma C8731), 10 mg/L Nicotinic acid (Sigma N4126), 10 mg/L para-Aminobenzoic acid (Nutr. Biochem. R-238), 20 mg/L Pyridoxine-HCl (Alfa aesar A12041), 10 mg/L Thiamine HCl (EMD Millipore 5871), 4 mg/L Biotin (Sigma B4639) in 0.1 M NaOH, 4 mg/L Folic acid (Sigma F8758) in 0.1 M NaOH, 0.2 mg/L Vitamin B12 (TCI C0449). 10x Wolfe’s minerals contains 3 g/L Nitrilotriacetic Acid (Sigma N9877), 6 g/L MgSO4 ⋅ 7 H2O (EMD Millipore MX0070–1), 1 g/L MnSO4 ⋅ H2O (Fisher M113), 2 g/L NaCl (Fisher S271), 0.2 g/L CaCl2 (Mallinckrodt 4160), 0.2 g/L FeSO4 ⋅ 7 H2O (Sigma F8633), 0.2 g/L CoCl2 ⋅ 6 H2O (Sigma 202185), 0.2 g/L ZnSO4 ⋅ 7 H2O (Fisher Z68), 20 mg/L CuSO4 ⋅ 5 H2O (VWR 330), 20 mg/L AlK(SO)4 ⋅ 12 H2O (Sigma 237086), 20 mg/L H3BO3 (Fisher A78), 20 mg/L Na2MoO4 ⋅ 2 H2O (Acros Organics 446360250).
Fig 1. Growth of Lactiplantibacillus plantarum isolates in CDML.
Growth of Lactiplantibacillus plantarum isolates from different sources grown with variations of CDML in different laboratories (S2 Table in S1 File). (A) 10 isolates predominantly from Drosophila guts grown aerobically with continuous shaking in the CDML with a starting inoculum of 0.01 OD600. Note Lp WCFS1 is a human mouth isolate. (B) Seven isolates of L. plantarum from food fermentations and mammals grown anaerobically in CDML, shaken intermittently before each OD reading. Note Lp WCFS1 in panel A is a single colony derivative of Lp NCIMB8826 in panel B. Both are from the same human mouth isolate. (C) Seven isolates of L. plantarum from food fermenatations grown anaerobically in a rich formulation of CDML with 10x L-cysteine. Solid lines represent means of 12 technical replicates. In panel A, Time point OD600 readings were taken every 5 minutes, and shaded areas represent standard error on a 5-point rolling average. for panels B and C, time point OD600 readings were taken every every 1 hour for panels B and C, and shaded areas represent standard error at each time point (note the error in B and C is very small except at early time points).
We note several technical issues that arose when constructing and testing the various CDMs. First, as mentioned previously, using high concentrations of some amino acids in the CDM often leads to formation of a precipitate (likely an amino acid metal salt complex) when stored for >48 hours. We found that this problem is exacerbated when preparing Wolfe’s minerals with metals sourced from different suppliers. Re-filtering the media to remove the precipitate did not inhibit growth of the lactobacilli tested (not shown). While this suggests that lower metal concentrations are tolerable for some strains, this may impede the growth of lactobacilli that exhibit more stringent mineral requirements.
Second, while decreasing amino acid concentration inhibits precipitation, this could present challenges when culturing lactobacilli that are deficient in amino acid synthesis. Indeed, some strains grown on nutrient rich agar plates such as De Man, Rogosa and Sharpe (MRS) agar do not grow well when inoculated directly into CDML. We found that conditioning the strains by passaging them through a mixture of 50:50 MRS:CDML before 100% CDML greatly improved growth rate. For weak growers, we found that up to three passages in 100% CDML were required to facilitate robust growth.
Optimization of the CDM for Acetobacter growth and co-culturing
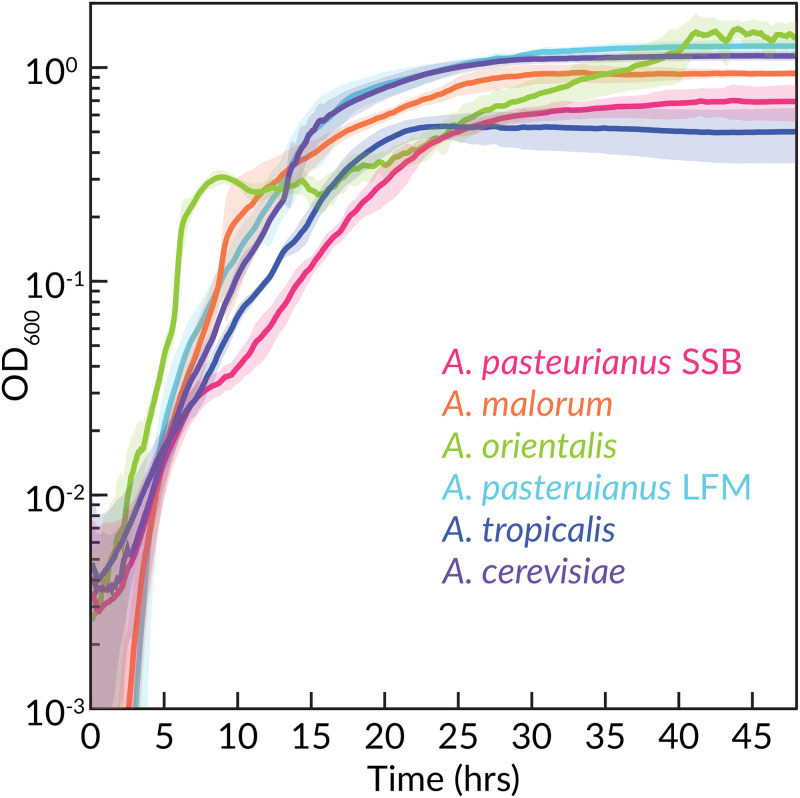
To test the CDM’s ability to support growth of AAB, we incubated A. pasteurianus and A. tropicalis in CDML, which produced extremely limited growth (S5 Table in S1 File). In an attempt to improve growth, we repeated the 10-fold additions experiment for Lp growth and found only ascorbate provided growth improvement (S4 Table in S1 File). Because ascorbate reacts with Wolfe’s minerals as evidenced by discoloration of the media after 48 h at room temperature, we use it optionally. Through a series of trials, we found that supplementing CDML with 50 mM D,L-lactic acid was sufficient to enable growth. This formulation, CDMA, supports an OD600 of ∼ 1.0 (∼ 109 cells/mL) for the six Acetobacters tested (Fig 2). We note that it was critical to supply adequate aeration to Acetobacters by shaking and poking a 0.5 mm hole in the breathable sealing film over the plate. Increased shaking speed increased the clumping of the cultures, which appears as increased variance in the growth curves between replicates (Fig 2). Reducing the shaking speed reduced clumping but also reduced the growth rate. We also found that mannitol can serve as a carbon source for Acetobacters.
Fig 2. Growth of Acetobacter sp. isolates.
Acetobacter sp. from several different sources (S2 Table in S1 File) were grown with CDMA under aerobic conditions with continuous shaking. Solid lines represent means of 12 technical replicates. Shaded areas represent standard error on a 5-point rolling average. Time point OD600 readings were taken every 5 minutes.
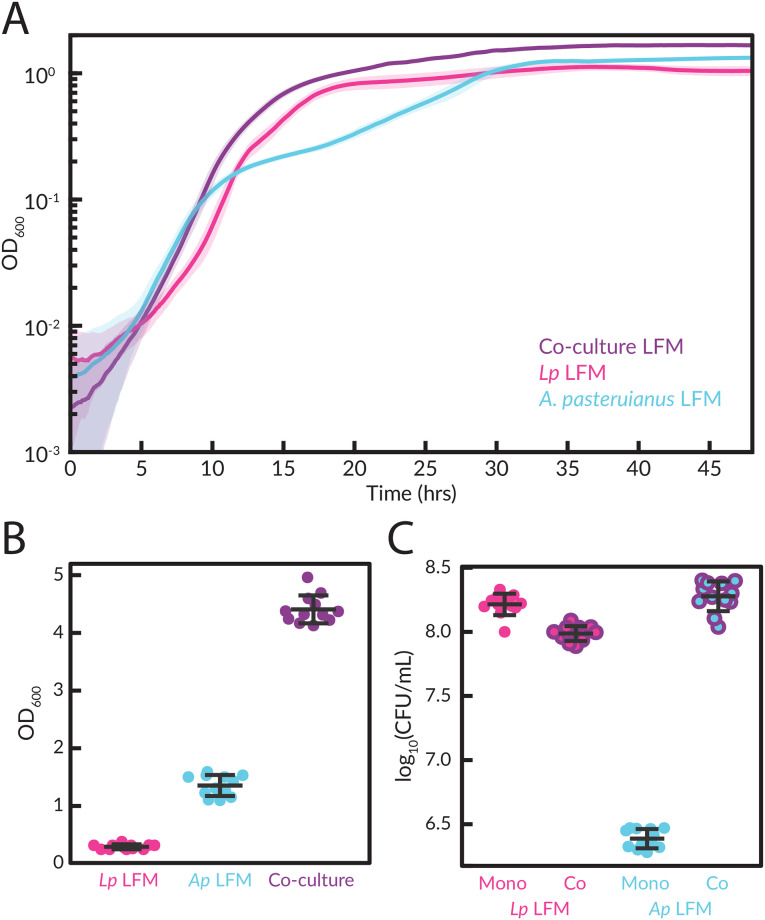
For co-cultures with Lp. plantarum and A. pasteurianus, we add glucose and potassium acetate to CDMA (Table 3). In these co-cultures, the OD600 approximated the combined OD600 for the two strains grown separately (Fig 3A) but reached an overall higher final yield of ≈ 4 versus ≈ 1 for A. pasteurianus and < 1 for Lp. plantarum (Fig 3B). By plating the co-culture on selective media (MRS for Lp. plantarum versus MYPL for A. pasteurianus), we found that A. pasteurianus increased growth yield from ∼ 106 CFU/mL to ∼ 108 CFU/mL while Lp. plantarum yield remained roughly constant at ∼ 108 CFU/mL (Fig 3C), indicating that growth with Lp. plantarum provides an advantage to A. pasteurianus in these specific co-culture conditions.
Table 3. Composition of CDMA.
| Stock solution # | Chemical | Final concentration (1x) |
|---|---|---|
| Buffer and salts | ||
| 1 | MOPS | 40 mM (1x) |
| 1 | K2HPO4 | 5 mM (1x) |
| 1 | NH4Cl | 20 mM (1x) |
| 1 | Na2SO4 | 10 mM (1x) |
| Metals | ||
| 2 | MgCl2 ⋅ 6 H2O | 1 mM (1x) |
| 2 | MnCl2 ⋅ 4 H2O | 0.05 mM (1x) |
| 2 | FeSO4 ⋅ 7 H2O | 0.05 mM (1x) |
| Carbon source | ||
| 3 | DL-Lactic acid | 0.6 mM (1x) |
| 3 | Glucose or mannitol (optional) | 125 mM (1x) |
| 3 | Ammonium acetate (optional) | 10 mM |
| Amino acids | ||
| 4 | Casamino acids | 3 g/L |
| 5 | Cysteine-HCl | 0.145 g/L |
| 6 | Tryptophan | 0.05 g/L |
| 7 | Wolfe’s vitamins | 1x |
| 8 | Wolfe’s minerals | 1x |
Fig 3. Co-culture growth of Lp. plantarum and A. pasteurianus.
Lp. plantarum and A. pasteurianus were grown in co-culture in CDMA with glucose and potassium acetate added. (A) Growth curves (solid lines) display means of 12 experimental replicates. Shaded areas represent standard error on a 5-point rolling average. Time point OD600 readings were taken every 5 minutes. (B) OD measurements were taken at the end of the growth curve after resuspension of the culture supernatant for each of the 12 replicates. Mean and standard deviation are shown. We note that the apparently 4-fold lower OD of Lp. plantarum in the resupsended culture versus the time course in A is likely due to settling of the cells and adherence to the plate during the time course. The CFU/mL counts are in C. (C) CFU/ml was counted by plating the resuspended cultures on MRS and MYPL for each of the 12 replicates. Mean and standard deviation shown.
Conclusions
The CDM formulated provides a tractable growth medium for the investigation of lactobacillus-Acetobacter community growth and metabolism under defined conditions so that genes and metabolites can be causally linked through experiments.
The CDM may also provide a starting point for investigation of more complex LAB and AAB communities, which are often observed in nature. We have investigated many Lp. plantarum strains and species of Acetobacter and found consistent growth. Due to the known species- and strain-dependence of lactobacilli growth, investigators of other LAB may find this CDM a useful starting point for devising new CDMs that support growth of other LAB species and genera. For instance, initial tests with Ll. brevis indicated weak but consistent growth when certain additives were supplied (S4 Table in S1 File).
The complex metabolic interactions in natural microbial communities have important impacts on their environments, including soils, oceans, and the guts of animals. Studying the fundamental biology of these interactions is facilitated by the use of CDMs and naturally low diversity microbial communities. For instance, with an optically clear CDM such as this one, experiments to combinatorically altering the chemical components are possible by running growth experiments in 96-well plates. We hope that this CDM for the Drosophila gut microbiome provides a foundation for the molecular characterization of the complex interactions in this naturally low diversity community.
Supporting information
(PDF)
Acknowledgments
The authors acknowledge members of the Ludington, Marco, and Taga labs who read and gave feedback on the manuscript.
Data Availability
All relevant data are within the paper and its Supporting information files.
Funding Statement
W.B.L. acknowledges National Institutes of Health (www.nih.gov) grants DP5OD017851 and R01DK128454, National Science Foundation (www.nsf.gov) Integrative Organismal Systems grants 2032985 and 2144342, and the Carnegie Institution for Science Endowment (carnegiescience.edu). M.L.M. acknowledges United States Department of Agriculture National Institute of Food and Agriculture grant no. 2015-67017-23116 and California Department of Food and Agriculture (www.usda.gov) grant no. 19-0001-050-SF. M.E.T. acknowledges National Institutes of Health grant DP2AI117984. E.T.S. acknowledges National Science Foundation Graduate Research Fellowship Program grant 1650042. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Reese V. Some effects of association and competition on Acetobacter. Journal of Bacteriology. 1938;36(4):357–367. doi: 10.1128/jb.36.4.357-367.1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dysvik A, Leanti La Rosa S, De Rouck G, Rukke EO, Westereng B, Wicklund T. Microbial Dynamics in Traditional and Modern Sour Beer. Applied and Environmental Microbiology. 2020;86(14):1–14. doi: 10.1128/AEM.00566-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. da Cruz Pedrozo Miguel MG, Gomes Cardoso P, de Assis Lago L, Freitas Schwan R. Diversity of bacteria present in milk kefir grains using culture-dependent and culture-independent methods. Food Research International. 2010;43(5):1523–1528. doi: 10.1016/j.foodres.2010.04.031 [DOI] [Google Scholar]
- 4. Gulitz A, Stadie J, Wenning M, Ehrmann MA, Vogel RF. The microbial diversity of water kefir. International Journal of Food Microbiology. 2011;151(3):284–288. doi: 10.1016/j.ijfoodmicro.2011.09.016 [DOI] [PubMed] [Google Scholar]
- 5. Wang Z, Shao Y. Effects of microbial diversity on nitrite concentration in pao cai, a naturally fermented cabbage product from China. Food Microbiology. 2018;72:185–192. doi: 10.1016/j.fm.2017.12.003 [DOI] [PubMed] [Google Scholar]
- 6. Wang ZM, Lu ZM, Shi JS, Xu ZH. Exploring flavour-producing core microbiota in multispecies solid-state fermentation of traditional Chinese vinegar. Scientific Reports. 2016;6(March):1–10. doi: 10.1038/srep26818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li H, Fu J, Hu S, Li Z, Qu J, Wu Z, et al. Comparison of the effects of acetic acid bacteria and lactic acid bacteria on the microbial diversity of and the functional pathways in dough as revealed by high-throughput metagenomics sequencing. International Journal of Food Microbiology. 2021;346(March):109168. doi: 10.1016/j.ijfoodmicro.2021.109168 [DOI] [PubMed] [Google Scholar]
- 8. Ho VTT, Fleet GH, Zhao J. Unravelling the contribution of lactic acid bacteria and acetic acid bacteria to cocoa fermentation using inoculated organisms. International journal of food microbiology. 2018;279:43–56. doi: 10.1016/j.ijfoodmicro.2018.04.040 [DOI] [PubMed] [Google Scholar]
- 9. Guan H, Yan Y, Li X, Li X, Shuai Y, Feng G, et al. Microbial communities and natural fermentation of corn silages prepared with farm bunker-silo in Southwest China. Bioresource Technology. 2018;265(April):282–290. doi: 10.1016/j.biortech.2018.06.018 [DOI] [PubMed] [Google Scholar]
- 10. Viladomiu M, Hontecillas R, Yuan L, Lu P, Bassaganya-Riera J. Nutritional protective mechanisms against gut inflammation. The Journal of Nutritional Biochemistry. 2013;. doi: 10.1016/j.jnutbio.2013.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wong CNA, Ng P, Douglas AE. Low-diversity bacterial community in the gut of the fruitfly Drosophila melanogaster. Environmental Microbiology. 2011;13(7):1889–1900. doi: 10.1111/j.1462-2920.2011.02511.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zheng J, Wittouck S, Salvetti E, Franz CMAP, Harris HMB, Mattarelli P, et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. International Journal of Systematic and Evolutionary Microbiology. 2020;70(4):2782–2858. doi: 10.1099/ijsem.0.004107 [DOI] [PubMed] [Google Scholar]
- 13. Consuegra J, Grenier T, Akherraz H, Rahioui I, Gervais H, da Silva P, et al. Metabolic cooperation among commensal bacteria supports Drosophila juvenile growth under nutritional stress. iScience. 2020; p. 101232. doi: 10.1016/j.isci.2020.101232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Henriques SF, Dhakan DB, Serra L, Francisco AP, Carvalho-Santos Z, Baltazar C, et al. Metabolic cross-feeding in imbalanced diets allows gut microbes to improve reproduction and alter host behaviour. Nature Communications. 2020;11(1):4236. doi: 10.1038/s41467-020-18049-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim G, Huang JH, McMullen JG, Newell PD, Douglas AE. Physiological responses of insects to microbial fermentation products: Insights from the interactions between Drosophila and acetic acid. Journal of Insect Physiology. 2018;106(May):13–19. doi: 10.1016/j.jinsphys.2017.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rivera-Chávez F, Zhang LF, Faber F, Lopez CA, Byndloss MX, Olsan EE, et al. Depletion of Butyrate-Producing Clostridia from the Gut Microbiota Drives an Aerobic Luminal Expansion of Salmonella. CHOM. 2016;19(4):443–454. doi: 10.1016/j.chom.2016.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pédron T, Mulet C, Dauga C, Frangeul L, Chervaux C, Grompone G, et al. A Crypt-Specific Core Microbiota Resides in the Mouse Colon. mBio. 2012;3(3):262–267. doi: 10.1128/mBio.00116-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Saffarian A, Mulet C, Regnault B, Amiot A, Tran-Van-Nhieu J, Ravel J, et al. Crypt- and Mucosa-Associated Core Microbiotas in Humans and Their Alteration in Colon Cancer Patients. mBio. 2019;10(4):G351–20. doi: 10.1128/mBio.01315-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Degnan PH, Taga ME, Goodman AL. Vitamin B12 as a Modulator of Gut Microbial Ecology. Cell metabolism. 2014;20(5):769–778. doi: 10.1016/j.cmet.2014.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sannino DR, Dobson AJ, Edwards K, Angert ER, Buchon N. The Drosophila melanogaster Gut Microbiota Provisions Thiamine to Its Host. mBio. 2018;9(2). doi: 10.1128/mBio.00155-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. San Roman M, Wagner A. An enormous potential for niche construction through bacterial cross-feeding in a homogeneous environment. PLoS Computational Biology. 2018;14(7):1–29. doi: 10.1371/journal.pcbi.1006340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Elli M, Zink R, Rytz A, Reniero R, Morelli L. Iron requirement of Lactobacillus spp. in completely chemically defined growth media. Journal of Applied Microbiology. 2000;88(4):695–703. doi: 10.1046/j.1365-2672.2000.01013.x [DOI] [PubMed] [Google Scholar]
- 23. Ponomarova O, Gabrielli N, Sévin DC, Mülleder M, Zirngibl K, Bulyha K, et al. Yeast Creates a Niche for Symbiotic Lactic Acid Bacteria through Nitrogen Overflow. Cell Systems. 2017;5(4):345–357.e6. doi: 10.1016/j.cels.2017.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chervaux C, Ehrlich SD, Maguin E. Physiological Study of Lactobacillus delbrueckii subsp. bulgaricus Strains in a Novel Chemically Defined Medium. Applied and Environmental Microbiology. 2000;66(12):5306–5311. doi: 10.1128/aem.66.12.5306-5311.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grobben GJ, Sikkema J, Smith MR, de Bont JAM. Production of extracellular polysaccharides by Lactobacillus delbrueckii ssp. bulgaricus NCFB 2772 grown in a chemically defined medium. Journal of Applied Bacteriology. 1995;79(1):103–107. doi: 10.1111/j.1365-2672.1995.tb03130.x [DOI] [Google Scholar]
- 26. Petry S, Furlan S, Crepeau MJ, Cerning J, Desmazeaud M. Factors affecting exocellular polysaccharide production by Lactobacillus delbrueckii subsp bulgaricus grown in a chemically defined medium. Applied and Environmental Microbiology. 2000;66(8):3427–3431. doi: 10.1128/aem.66.8.3427-3431.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ricciardi A, Ianniello RG, Parente E, Zotta T. Modified chemically defined medium for enhanced respiratory growth of Lactobacillus caseiand Lactobacillus plantarumgroups. Journal of Applied Microbiology. 2015;119(3):776–785. doi: 10.1111/jam.12894 [DOI] [PubMed] [Google Scholar]
- 28. Saguir FM, de Nadra MCM. Improvement of a Chemically Defined Medium for the Sustained Growth of Lactobacillus plantarum: Nutritional Requirements. Current microbiology. 2007;54(6):414–418. doi: 10.1007/s00284-006-0456-0 [DOI] [PubMed] [Google Scholar]
- 29. Savijoki K, Suokko A, Palva A, Varmanen P. New convenient defined media for [35S]methionine labelling and proteomic analyses of probiotic lactobacilli. Letters in Applied Microbiology. 2006;42(3):202–209. doi: 10.1111/j.1472-765X.2005.01853.x [DOI] [PubMed] [Google Scholar]
- 30. Wegkamp A, van Oorschot W, de Vos WM, Smid EJ. Characterization of the role of para-aminobenzoic acid biosynthesis in folate production by Lactococcus lactis. Applied and Environmental Microbiology. 2007;73(8):2673–2681. doi: 10.1128/AEM.02174-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Palmer KL, Aye LM, Whiteley M. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. Journal of Bacteriology. 2007;189(22):8079–8087. doi: 10.1128/JB.01138-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hayek SA, Ibrahim SA. Current limitations and challenges with lactic acid bacteria: a review. Food and Nutrition Sciences. 2013;4(November):73–87. doi: 10.4236/fns.2013.411A010 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting information files.