Abstract
Synthetic biology enables the creative combination of engineering and molecular biology for exploration of fundamental aspects of biological phenomena. However, there are limited resources available for such applications in the educational context, where straightforward setup, easily measurable phenotypes and extensibility are of particular importance. We developed unigems, a set of ten plasmids that enable classroom-based investigation of gene-expression control and biological logic gates to facilitate teaching synthetic biology and genetic engineering. It is built on a high-copy plasmid backbone and is easily extensible thanks to a common primer set that facilitates Gibson assembly of PCR-generated or synthesized DNA parts into the target vector. It includes two reporter genes with either two constitutive (high- or low-level) or two inducible (lactose- or arabinose-) promoters, as well as a single-plasmid implementation of an AND logic gate. The set can readily be employed in undergraduate teaching settings, during outreach events and for training of iGEM teams. All plasmids have been deposited in Addgene.
Keywords: synthetic biology, Gibson assembly, iGEM, plasmids, education
Data Summary
The plasmids have been deposited at Addgene (https://www.addgene.org/Jaroslaw_Bryk/). An accompanying Figshare repository (https://figshare.com/projects/Unigems_paper/114069) contains SnapGene sequences of all constructs as well as EPS and Adobe Illustrator files with the plasmid maps to allow educators create their own high-quality figures of their assemblies.
Impact Statement.
There is a dearth of practical synthetic biology resources available – plasmids, DNA parts and ways to generate and manipulate them – that could easily be implemented in teaching, outreach events or as foundational constructs for more complex designs. We designed a set of ten plasmids that can be broken down to five functional sections (reporter gene and promoter, origin of replication, antibiotic resistance and repressor) using the same set of overlapping PCR primers, therefore making each section compatible with any plasmid in the set. Each of the sections can also be replaced with synthetic DNA parts and/or via PCR with Gibson assembly. We have been using the system in introductory undergraduate synthetic biology laboratory classes, in student research projects, in iGEM teams training and in outreach events and expect they will become a valuable tool for teachers, university instructors and educators. All plasmids and corresponding source files and illustrations are available through Addgene and Figshare.
Introduction
The development and standardization of biological parts (DNA or RNA fragments) to reliably achieve a predictable output in living organisms has been an often-emphasized aspect of synthetic biology [1]. Remarkable progress has been achieved in characterization of bacterial promoters [2], transcriptional terminators [3], insulators [4], translation optimization [5] and promoter inducibility [6], however advances on a higher level of systems’ complexity have not been as forthcoming [7–9] and major synthetic biology applications (e.g. [10–14]) were achieved by a host of sophisticated and custom-developed genetic engineering [15, 16].
Nevertheless, standardized biological parts are particularly well suited to use in an educational context, as they emphasize utility and remove complexities associated with protocol development [17–19]. These advantages are best illustrated by the BioBuilder lessons, which provide detailed protocols that combine lab-based experiments with engineering tasks to demonstrate fundamental principles of biological and electronic circuits [20] (https://biobuilder.org/), and the International Genetically Engineered Machines competition (iGEM), where thousands of students worldwide create synthetic biology designs and constructs based on and inspired by a set of DNA parts provided by the iGEM Foundation [21]. Notably, practical synthetic biology exemplified by building novel biological constructs from compatible genetic parts, favour active learning approaches in teaching, which were shown to improve students’ academic attainment [22, 23].
To expand the availability of education-friendly resources for synthetic biology we developed unigems, a set of plasmids with simple features such as strong and weak constitutive promoters or single- and dual-inducible promoters and an olfactory construct that can be used directly in laboratory classes. In addition, each plasmid can be split into functional parts that can be exchanged either between the plasmids or replaced with novel parts with PCR and Gibson assembly.
Methods
Plasmids
All the plasmids in the unigems set have been assembled using Gibson assembly [24] on the backbone of the pJ401 high-copy plasmid obtained from Atum (Newark, CA, USA). The assembled parts were synthesized by IDT DNA (Coralville, IO, USA) and Life Technologies (now Thermo Fisher Scientific, Waltham, MA, USA).
Transformation
All transformations were performed using the Sub-cloning Efficiency DH5α™ Competent cells (Invitrogen, now Thermo Fisher Scientific, Waltham, USA) as per the manufacturer’s protocol.
Growth and storage conditions
All cells were cultured using LB Lennox media and agar plates, with incubation at 37◦C and agitation at 180 r.p.m. for liquid cultures. Volumes of cultures were 1 or 5 ml, depending on context (smaller for plating transformed cells, sequencing and fluorescence analysis, larger for plasmid isolation). For selective media, both kanamycin and ampicillin (Sigma-Aldrich, now Merck, Darmstadt, Germany) were used at concentration of 50 µg ml−1. Plates and cultures were stored at 4◦C or preserved for long-term storage at −80◦C with 500 µl of overnight culture suspended in 500 µl of 50 % glycerol.
Gibson assembly
Fragments to be inserted were designed with 20–40 bp overlaps of the vector primer binding sites using SnapGene and synthesized by Integrated DNA Technologies. Platinum SuperFi Green PCR Master Mix (Invitrogen) or Q5 High Fidelity Polymerase (New England Biolabs, Ipswich, MA, USA) were used for generation of backbones from p005 and p006 vectors following the manufacturers’ protocols. PCR products were purified with GeneJET PCR purification kit (Thermo Fisher Scientific) and concentration measured using NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific). Homemade Gibson Assembly master mix was prepared and assembly carried out as outlined by [25], using 2-3 : 1 of molar ratio of the donor parts to target vectors.
Plasmid verification
Following transformation, colony PCR was performed using DreamTaq Green PCR Master Mix (2X) (Thermo Fisher Scientific) as described at https://openwetware.org/wiki/Endy:Colony_PCR. Colony PCR products were verified on 1 % agarose gels stained with RedSafe (ChemBio, Oxford, UK) and then purified with GeneJET PCR purification kit (Thermo Fisher Scientific) and sequenced by SourceBioscience (Nottingham, UK).
Analysis of fluorescence
Horiba FluoroMax−4 spectrofluorometer (Horiba, Kyoto, Japan) was used to obtain emission and excitation spectra for green and red fluorescent proteins. We used 1 ml of overnight cultures resuspended in phosphate buffered saline (PBS, Sigma-Aldrich) to an OD600 below 0.120. Guava easyCyte 5HT system flow cytometer (Merck) was used to determine fluorescence of 10 000 events of each triplicate culture. A signal from RFP cultures, indicating protein expression, was only detected after a 24 h period of incubation.
Olfactory measurements
The banana smell was identified using the protocol by Dixon and Kuldell [17], following overnight culture at 37◦C at 180 r.p.m. to reach stationary growth phase.
Results
Assembled plasmids
The unigems' set includes ten plasmids shown in Table 1.
Table 1.
Unigems plasmids
|
Plasmidname |
AddgeneID |
Antibiotic resistance |
Reporter gene |
Function |
|---|---|---|---|---|
|
p006-strongGFP |
108 313 |
kanR |
GFP |
pFAB4026 strong constitutive |
|
p006-weakGFP |
108 314 |
kanR |
GFP |
pFAB4282 weak constitutive |
|
p005-strongRFP |
108 317 |
kanR |
RFP |
pFAB4005 strong constitutive |
|
p005-weakRFP |
108 316 |
kanR |
RFP |
pFAB4024 weak constitutive |
|
p006-pBADGFP |
108 315 |
kanR |
GFP |
pBAD |
|
p006kanGFP |
58 534 |
kanR |
GFP |
T5-pLacO |
|
p005kanRFP |
58 533 |
kanR |
RFP |
T5-pLacO |
|
p007ampGFP |
58 535 |
ampR |
GFP |
T5-pLacO |
|
p006-ANDGFP |
112 237 |
kanR |
GFP |
pLac +pBAD AND logic gate |
|
p006-Banana-Late |
112 251 |
kanR |
ATFI |
osmY |
The promoters and terminators are derived from the BIOFAB collection: pFAB4026, pFAB4282, pFAB4005, pFAB4024 [26] and BBa B0062-R [3], respectively. The pBAD-araC and osmY-ATF1 parts come from IGEM repository (BBa K808000, positions 1 : 1200 bp) and BBa J45250, while the AND logic gate is based on the D61 clone from [27]. The RFP reporter gene is a synthetic gene (ID 97752) generated by Atum (Newark, CA, USA) by random assembly, but it has a 76 % sequence identity to an RFP from a strawberry coral Corynactis californica. GFP is a standard reporter gene from Aquorea victoria. All plasmids also include the lacI repressor under control of a weak constitutive Amp promoter, even when they do not have T5-pLacO promoter themselves.
The unigems system
The unigems system is built upon a set of six pairs of overlapping PCR primer binding sites (Table 2) that split each plasmid into functional sections that can be replaced or exchanged. Thanks to this arrangement, each plasmid can be a source of parts for other recipient plasmids and new parts can be generated by direct synthesis or PCR with overhanging primers matching the primer binding sites on the unigems plasmids (Fig. 1).
Table 2.
Primers for the unigems plasmids
|
Primer name |
Primer sequence (5’ → 3’) |
|---|---|
|
A-REV |
CTCGAAAATAATAAAGGGAAAATCAG |
|
AGFP-FWD |
TTCTCCCTCTCCACTGACAG |
|
ARFP-FWD |
TACGGTTTGCCTGTACCTTC |
|
B-FWD |
CTCAGAAGTGAAACGCCGTA |
|
BGFP-REV |
GGGCACAAATTTTCTGTCAG |
|
BRFP-REV |
GTACAGGCAAACCGTATGAG |
|
C-FWD |
TCACCACCCTGAATTGACTC |
|
C-REV |
ACTACCATCGGCGCTACG |
|
d-FWD |
CTCACGTTAAGGGATTTTGG |
|
d-REV |
CGCCCGGAAGAGAGTC |
|
E-FWD |
ACTCAAAGGCGGTAATACGG |
|
E-REV |
CCAAAATCCCTTAACGTGAG |
|
F-FWD |
CTGATTTTCCCTTTATTATTTTCGAGA |
|
F-REV |
CCTGATTCTGTGGATAACCG |
Fig. 1.
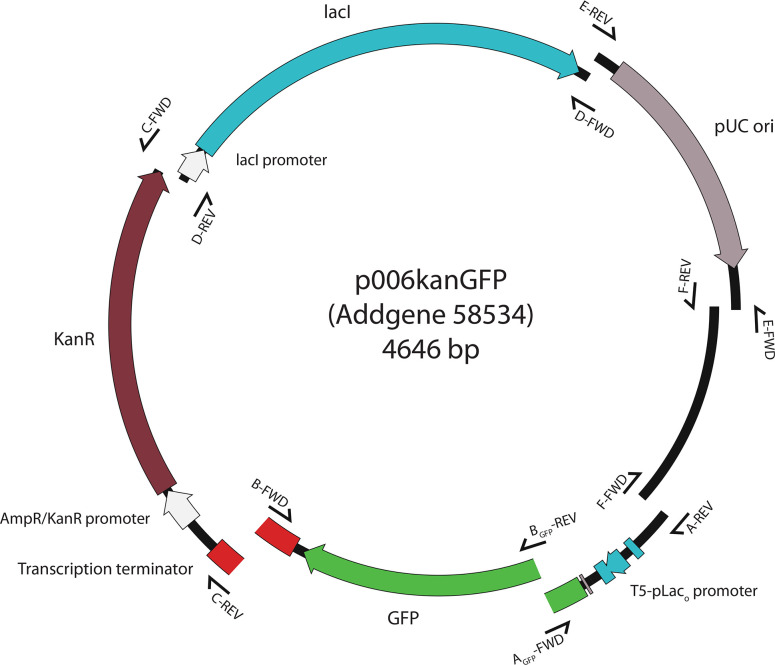
Building sections of the standard unigems plasmid, with the location of each pair of overlapping primer binding sites: primers F-FWD and A-REV, AGFP-FWD and BGFP-REV, B-FWD and C-REV, C-FWD and d-REV, d-FWD and E-REV, E-FWD and F-REV overlap such that PCR products made with them can be directly used in Gibson assembly. Note that all primer pairs except AGFP-FWD and AGFP-REV are identical in all plasmids.
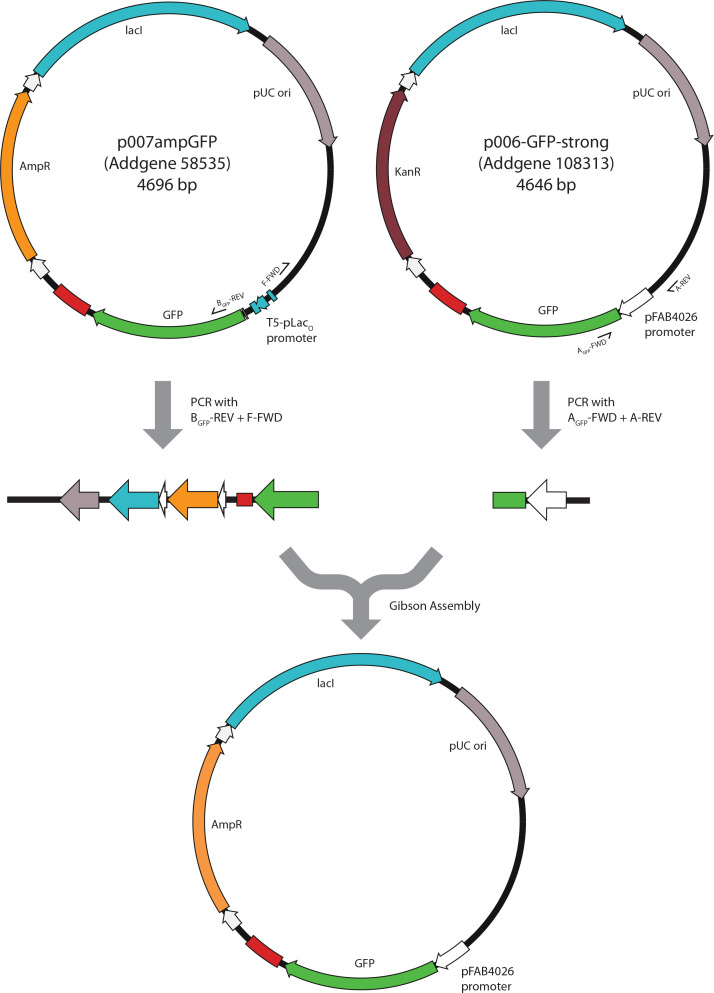
This principle can be illustrated with the following example (Fig. 2). Let our starting point be the p007ampGFP plasmid (Addgene 58535), where a reporter gene (GFP) is under control of the lactose-inducible promoter (T5-pLacO) and the plasmid contains an ampicilin-resistance gene. To replace the lactose-inducible promoter with a constitutive one (for example, a BIOFAB promoter pFAB4026, available in p006-GFP-strong (Addgene 108313)), we would run two PCR reactions: one to generate the recipient vector and one to generate a donor part (p005ampGFP and p006-GFP-strong, respectively).
Fig. 2.
Example of novel plasmid assembly using existing unigems plasmids as sources of vector and insert parts with PCR and Gibson assembly. See text for details.
We would use primers BGFP-REV and F-FWD to generate linearized recipient plasmid and primers AGFP-FWD and A-REV to generate the donor part. Because the primers BGFP-REV and AGFP-FWD, as well as F-FWD and A-REV are overlapping, both of their PCR products are directly usable for Gibson assembly. After purification and quantification, they can be mixed in appropriate molar proportions to assemble a new plasmid.
The same principle can be used to exchange or remove the entire reporter gene, antibiotic resistance gene, origin of replication or the repressor gene between the plasmids or with a newly generated or synthesized part. The combination of different promoters, reporters and antibiotic resistances ensures identification of a successful assembly. In our example, the colonies would be grown on ampicillin-containing media with no lactose or IPTG to identify GFP-fluorescent bacteria, indicating correctly assembled plasmids.
Because primers AGFP-FWD and BGFP-REV are located inside the ORFs of the reporter genes, they are specific to these genes. However, change of the promoter could also be achieved by inserting the entire ORF of the reporter gene combined with the promoter and part of the terminator. In this case, primers A-REV and B-FWD would be used and they would be identical for every reporter-promoter combination to be exchanged in any of the unigems plasmids.
Following assembly, we experimentally verified the characteristics of the plasmids.
Constitutive promoters
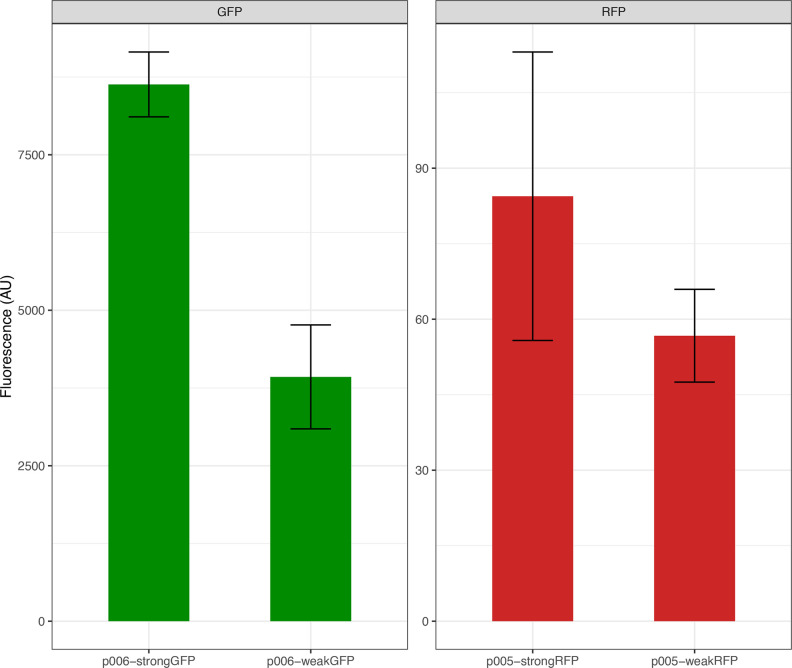
GFP and RFP reporter genes, placed under the control of either a strong or a weak promoter (pFAB4026 and pFAB4282 for GFP and pFAB4005 and pFAB4024 for RFP, respectively) (BIOFAB collection [26]) were analysed using a flow cytometer on three replicate samples each (separate colonies from the same transformation event). To characterize the properties of the fluorescent proteins, we used a spectrofluorometer to test a range of excitation wavelengths for both GFP and RFP. We found that the GFP can be excited between 350 to 420 nm (with a peak at 395 nm) for emission at 506 nm. The RFP can be excited between 480 and 505 nm (with a peak at 505 nm) for emission at 560 nm. The fluorescence levels of the reporter genes were measured using the Guava easyCyte 5HT, with cells grown in liquid culture (Fig. 3).
Fig. 3.
GFP fluorescence of three replicates of each p006-strongGFP and p006-weakGFP and RFP yellow fluorescence of p005-strongRFP and p005-weakRFP. Note the difference in scale in GFP vs RFP: the lack of differences between strong and weak RFP fluorescence is due to a mismatch between its optimal excitation wavelength (550 nm) and the 488 nm excitation laser in the flow cytometer (see Discussion).
Inducible promoters and logic gate
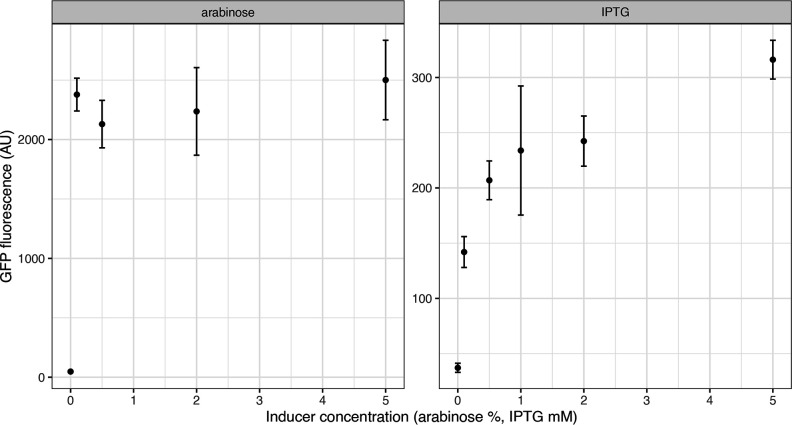
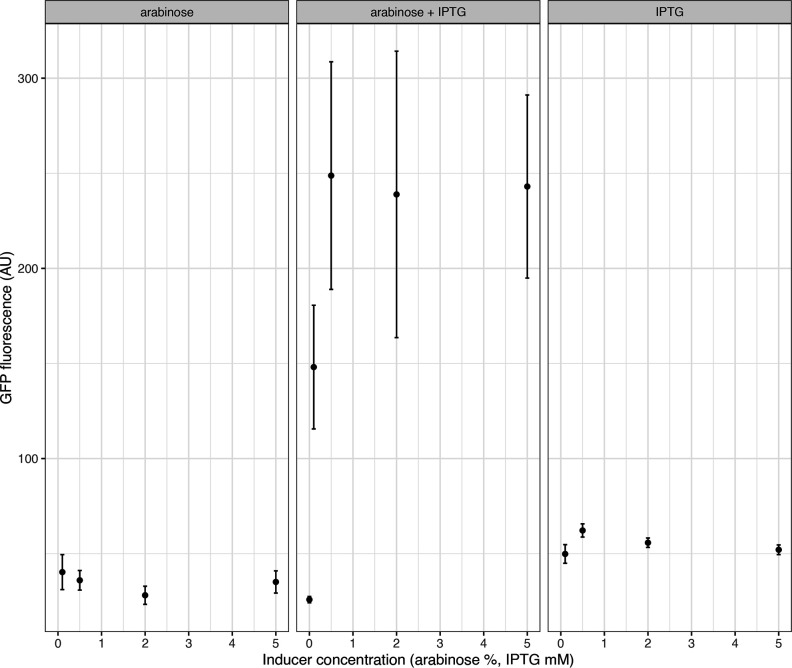
To demonstrate the inducibility of the pBAD and T5-pLacO promoters, we measured fluorescence of GFP with an increasing concentration of inducers, arabinose (0–5 %) and IPTG (0–5 mM). We observed clear activation depending on the inducer. pBAD exhibits a binary-like induction, where already at 0.1 % concentration of arabinose it produces 90 % of fluorescence observed at 5 % arabinose. In contrast, T5-pLacO produces a more dose-dependent pattern of induction (Fig. 4).
Fig. 4.
GFP fluorescence of three replicates of p006-strongGFP under arabinose-inducible p006-pBADGFP or IPTG-inducible p006kanGFP.
We also used combined lactose- and arabinose-inducible promoter based on design by Cox and colleagues [27]. This promoter acts as a biological AND gate, where both inputs (IPTG and arabinose) are necessary to activate the output – expression of the GFP. We tested the performance of the logic gate with arabinose only, IPTG only and both, at increasing concentrations. GFP fluorescence does not increase significantly in the presence of either IPTG or arabinose alone, but is clearly inducible in the presence of both inducers (Fig. 5).
Fig. 5.
GFP fluorescence of three replicates of p006-GFP-Logic-AND induced by individual inducers (arabinose or IPTG) or by both inducers.
Olfactory construct
The p006-Banana-Late construct is identical to the Eau de Smell described by Dixon and colleagues [17] and produces ATFI enzyme (alcohol acetyltransferase I) that converts isoamyl alcohol to isoamyl acetate, which has a strong banana odour. ATFI production is controlled by the osmY stationary phase promoter, therefore the banana odour can only be detected once the cells reach the stationary growth phase. The construct was tested in broth culture.
Discussion
We have assembled and characterized a set of plasmids that enable out-of-the-box investigations of gene expression control and straightforward extensibility with PCR and Gibson assembly. The phenotypes presented are clearly distinguishable and interpretable in an educational context rather than being designed and tested to produce precise quantitative output. For instance, Mutalik and colleagues reported (2013) that the relative difference in GFP fluorescence driven by BIOFAB promoters pFAB4026 and pFAB4282 is sevenfold vs threefold in our characterization (Fig. 3), the discrepancy that can reasonably be attributed to differences in bacterial chassis, fluorescent marker sequence and culture conditions. The minimal signal from RFP fluorescence shown on Fig. 3 is due to a mismatch between its optimal excitation wavelength (550 nm) and the 488 nm excitation laser in the flow cytometer. The GFP’s excitation and emission reported here are similar to those reported by Heim and Tsien [28] for an unmodified protein [28]. The RFP’s excitation and emission spectra behave similarly to that quantified by Baird et al. [29]. Overall, the performance of the plasmids in the K12 E. coli strain DH5alpha is suitable for demonstrating the quantitative differences between various promoters and inducers, including those in the bi-inducible promoter.
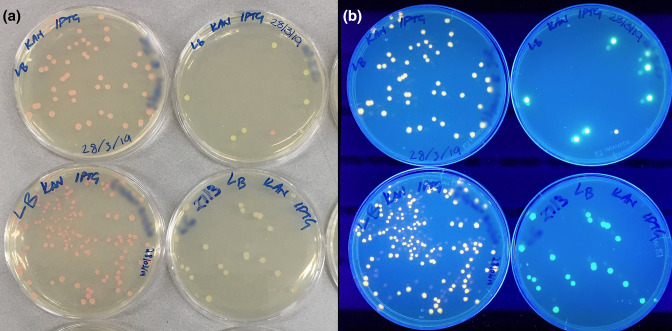
A standard benchtop UV transilluminator or a keyring UV torch, with excitation at ~400 nm, is the only equipment required to verify the expression of the reporter genes. Expression of RFP is also visible in daylight without any equipment (Fig. 6). Fluorometer or flow cytometer are only needed to quantify the expression level of GFP or RFP.
Fig. 6.
Images of p005-kan-RFP (left column) and p006-kan-GFP (right column) in daylight (a) and on a benchtop UV transilluminator (excitation wavelength 395 nm) (b). Transformed DH5α cells were grown with 50 µg ml−1 of kanamycin and induced with 100 mM IPTG. The single RFP-expressing colony on the top right plate is due to a mix of plasmids used by the students in this transformation. Photographs were taken with a mobile phone by JB.
We (AS, JB, JS, DM) have been using the unigems plasmids in a variety of contexts, ranging from outreach events, where participants transform E. coli with p006kanGFP or p005kanRFP using a 15 min protocol to observe a spectacular fluorescence after an overnight incubation to undergraduate research projects, where students have to design a new compatible part (e.g. a NOT gate, or a quorum-sensing system) and characterize its functions following a successful assembly. The plasmids themselves were first tested in two synthetic biology workshops run for prospective members of iGEM teams. Students’ feedback from this course is included in the report available in the Figshare repository (https://figshare.com/articles/media/Unigems_report_from_a_synthetic_biology_workshop_for_undergraduates/14627211). Since deposition in Addgene, Unigems plasmids have enjoyed a stable stream of requests.
The availability and use of GMOs is regulated in almost all jurisdictions [30–32]. In the UK, at the time of writing, the use of the unigems plasmids needs to comply with the UK’s Health and Safety Authority’s Genetically Modified Organisms (Contained Use) Regulations 2014 (https://www.hse.gov.uk/pubns/books/l29.htm). These guidelines allow for exemptions if the constructs and parts have a long history of safe use, but they are still restrictive compared to the US regulations, which only require that constructs do not pose an unreasonable risk (Toxic Substances Control Act, Environmental Protection Agency, https://www.epa.gov/laws-regulations/summary-toxic-substances-control-act). Therefore, use of the unigems system within high school classrooms would be possible in some jurisdictions, but it would pose an administrative burden in most. On the other hand, as most undergraduate biology teaching facilities already comply with GMO containment requirements, unigems can be straightforwardly employed at higher education institutions.
Funding information
This work was funded by the European Commission (FP7 People: Marie-Curie Actions, Grant number - 300038) and Wellcome Trust Society Award (WT103054MA).
Acknowledgements
The authors wish to thank all members of the University of Reading 2013 and 2014 iGEM teams and University of Huddersfield 2015–2019 project students for help in testing the parts and plasmids. JB would like to dedicate this manuscript to Dr. Dean R. Madden, Director of the National Centre for Biotechnology Education, University of Reading.
Author contributions
A.S., A.A.W. investigation, A.S. formal analysis, writing – original draft, writing – review & editing, J.S. formal analysis (digital illustration), J.A.D. formal analysis (regulation), writing – review & editing, D.M., JSchollar. resources, D.M., JSchollar, J.B. conceptualization, funding acquisition, methodology, supervision, project administration, J.B. investigation, formal analysis, validation, writing – original draft, writing – review & editing.
Conflicts of interest
The author(s) declare that there are no conflicts of interest.
Footnotes
Abbreviations: GFP, green fluorescent protein; IGEM, International Genetically Engineered Machines; RFP, red fluorescent protein.
References
- 1.Endy D. Foundations for engineering biology. Nature. 2005;438:449–453. doi: 10.1038/nature04342. [DOI] [PubMed] [Google Scholar]
- 2.Mutalik VK, Guimaraes JC, Cambray G, Mai QA, Christoffersen MJ, et al. Quantitative estimation of activity and quality for collections of functional genetic elements. Nat Methods. 2013;10:347–353. doi: 10.1038/nmeth.2403. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y-J, Liu P, Nielsen AAK, Brophy JAN, Clancy K, et al. Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat Methods. 2013;10:659–664. doi: 10.1038/nmeth.2515. [DOI] [PubMed] [Google Scholar]
- 4.Lou C, Stanton B, Chen YJ, Munsky B, Voigt CA. Ribozyme-based insulator parts buffer synthetic circuits from genetic context. Nat Biotechnol. 2012;30:1137–1142. doi: 10.1038/nbt.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reeve B, Hargest T, Gilbert C, Ellis T. Predicting translation initiation rates for designing synthetic biology. Front Bioeng Biotechnol. 2014;2:1. doi: 10.3389/fbioe.2014.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyer AJ, Segall-Shapiro TH, Glassey E, Zhang J, Voigt CA. Escherichia coli “Marionette” strains with 12 highly optimized small-molecule sensors. Nat Chem Biol. 2019;15:196–204. doi: 10.1038/s41589-018-0168-3. [DOI] [PubMed] [Google Scholar]
- 7.Freemont PS. Synthetic biology industry: data-driven design is creating new opportunities in biotechnology. Emerg Top Life Sci. 2019;3:651–657. doi: 10.1042/ETLS20190040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelwick R, MacDonald JT, Webb AJ, Freemont P. Developments in the tools and methodologies of synthetic biology. Front Bioeng Biotechnol. 2014;2:60. doi: 10.3389/fbioe.2014.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young R, Haines M, Storch M, Freemont PS. Combinatorial metabolic pathway assembly approaches and toolkits for modular assembly. Metab Eng. 2021;63:81–101. doi: 10.1016/j.ymben.2020.12.001. [DOI] [PubMed] [Google Scholar]
- 10.Gibson DG, Glass JI, Lartigue C, Noskov VN, Chuang R-Y, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 11.Gleizer S, Ben-Nissan R, Bar-On YM, Antonovsky N, Noor E, et al. Conversion of Escherichia coli to generate all biomass carbon from CO2 . Cell. 2019;179:1255–1263. doi: 10.1016/j.cell.2019.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoshika S, Leal NA, Kim MJ, Kim MS, Karalkar NB, et al. Hachimoji DNA and RNA: a genetic system with eight building blocks. Science. 2019;363:884–887. doi: 10.1126/science.aat0971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 14.Schuler ML, Mantegazza O, Weber APM. Engineering C4 photosynthesis into C3 chassis in the synthetic biology age. Plant J. 2016;87:51–65. doi: 10.1111/tpj.13155. [DOI] [PubMed] [Google Scholar]
- 15.Canton B, Labno A, Endy D. Refinement and standardization of synthetic biological parts and devices. Nat Biotechnol. 2008;26:787–793. doi: 10.1038/nbt1413. [DOI] [PubMed] [Google Scholar]
- 16.de Lorenzo V, Schmidt M. Biological standards for the knowledge-based bioeconomy: what is at stake. N Biotechnol. 2018;40:170–180. doi: 10.1016/j.nbt.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Dixon J, Kuldell N. Methods of Enzymology. Vol. 497. Elsevier; 2011. BioBuilding; pp. 255–271. vol. [DOI] [PubMed] [Google Scholar]
- 18.Penumetcha P, Lau K, Zhu X, Davis K, Eckdahl TT, et al. Improving the lac system for synthetic biology. Bios. 2010;81:7–15. doi: 10.1893/011.081.0104. [DOI] [Google Scholar]
- 19.Wolyniak MJ. Improved student linkage of Mendelian and molecular genetic concepts through a yeast-based laboratory module. Biochem Mol Biol Educ. 2013;41:163–172. doi: 10.1002/bmb.20679. [DOI] [PubMed] [Google Scholar]
- 20.Bernstein R, Ingram K, Hart KM. BioBuilder: Synthetic Biology in the Lab. O’Reilly Media, Inc; 2015. [Google Scholar]
- 21.Mitchell R, Dori YJ, Kuldell NH. Experiential engineering hrough iGEM—An undergraduate summer competition in synthetic biology. J Sci Educ Technol. 2011;20:156–160. doi: 10.1007/s10956-010-9242-7. [DOI] [Google Scholar]
- 22.Freeman S, Eddy SL, McDonough M, Smith MK, Okoroafor N, et al. Active learning increases student performance in science, engineering, and mathematics. Proc Natl Acad Sci. 2014;111:8410–8415. doi: 10.1073/pnas.1319030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Theobald EJ, Hill MJ, Tran E, Agrawal S, Arroyo EN, et al. Active learning narrows achievement gaps for underrepresented students in undergraduate science, technology, engineering, and math. Proc Natl Acad Sci. 2020;117:6476–6483. doi: 10.1073/pnas.1916903117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibson D. One-step enzymatic assembly of DNA molecules up to several hundred kilobases in size. Protoc Exch. 2009 doi: 10.1038/nprot.2009.77. [DOI] [PubMed] [Google Scholar]
- 25.Gibson DG. Enzymatic assembly of overlapping DNA fragments. Methods Enzymol. 2011;498:349–361. doi: 10.1016/B978-0-12-385120-8.00015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mutalik VK, Guimaraes JC, Cambray G, Lam C, Christoffersen MJ, et al. Precise and reliable gene expression via standard transcription and translation initiation elements. Nat Methods. 2013;10:354–360. doi: 10.1038/nmeth.2404. [DOI] [PubMed] [Google Scholar]
- 27.Cox RS, Surette MG, Elowitz MB. Programming gene expression with combinatorial promoters. Mol Syst Biol. 2007;3:145. doi: 10.1038/msb4100187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heim R, Tsien RY. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- 29.Baird GS, Zacharias DA, Tsien RY. Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci. 2000;97:11984–11989. doi: 10.1073/pnas.97.22.11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buhk HJ. Synthetic biology and its regulation in the European Union. N Biotechnol. 2014;31:528–531. doi: 10.1016/j.nbt.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 31.Hansson SO. How to be cautious but open to learning: time to update biotechnology and GMO legislation. Risk Anal. 2016;36:1513–1517. doi: 10.1111/risa.12647. [DOI] [PubMed] [Google Scholar]
- 32.Reeves RG, Denton JA, Santucci F, Bryk J, Reed FA. Scientific standards and the regulation of genetically modified insects. PLoS Negl Trop Dis. 2012;6:e1502. doi: 10.1371/journal.pntd.0001502. [DOI] [PMC free article] [PubMed] [Google Scholar]