Abstract
Brain cells are arranged in laminar, nuclear, or columnar structures, spanning a range of scales. Here, we construct a reliable cell census in the frontal lobe of human cerebral cortex at micrometer resolution in a magnetic resonance imaging (MRI)–referenced system using innovative imaging and analysis methodologies. MRI establishes a macroscopic reference coordinate system of laminar and cytoarchitectural boundaries. Cell counting is obtained with a digital stereological approach on the 3D reconstruction at cellular resolution from a custom-made inverted confocal light-sheet fluorescence microscope (LSFM). Mesoscale optical coherence tomography enables the registration of the distorted histological cell typing obtained with LSFM to the MRI-based atlas coordinate system. The outcome is an integrated high-resolution cellular census of Broca’s area in a human postmortem specimen, within a whole-brain reference space atlas.
Advancements in imaging and analysis enable the construction of a cellular atlas of the human cerebral cortex.
INTRODUCTION
The human brain is a complex organ organized across an astonishing range of spatial scales. To understand its properties and functionality, it is critical to produce a comprehensive characterization of the neuronal cell types that is one of its major constituents and to visualize their distributions through the whole brain volume. Although notable technological advances (1) have made it possible to obtain a comprehensive cell census in animal models such as mouse and marmoset monkey (2), no current imaging technology can directly visualize the defining microscopic features in the whole human brain without substantial distortion. Small mammals’ brain can be acquired in their entirety using high-resolution optical method such as light-sheet fluorescence microscopy (LSFM) allowing an easy registration (3) to standard atlas such as the Allen Mouse Brain Common Coordinate Framework. On the other hand, cytoarchitectural parcellations of the human brain (4, 5) are limited by unavoidable distortions introduced by the process of slice-to-slice sectioning, clearing, staining, and mounting involved in current histochemistry protocols. This results in artifacts that prevent accurate visualization of the tissue’s morphomolecular properties such as individual cells across different regions or laminar and cytoarchitectural boundaries that form the natural coordinate system for a cell census of the human brain.
Several important steps have been made toward building three-dimensional (3D) models of the human brain with cellular resolution through the use of magnetic resonance imaging (MRI) (6, 7), standard histology (5, 8), and polarized light imaging (9). For example, the Big Brain project (4) required 5 years and 1000 person-hours to obtain a comprehensive dataset of one human brain with a nominal resolution of 20 μm. In parallel, data analysis and atlasing methods have been proposed to manage the very large datasets generated during the reconstruction and mapping of the volumes to standardized templates (10). While these technologies represent remarkable advances, they still do not produce the undistorted 3D images of the cytoarchitecture of the human brain that are needed to build accurate models with nuclear and laminar resolution, a critical component of any cellular atlas.
Here, we overcome the inherent limitations of each individual methodology using different innovative imaging and analysis techniques. We propose to bridge microscopic volumetric histological imaging of single slabs acquired with the LSFM and macroscopic MRI reconstruction of the whole-hemisphere using mesoscopic optical coherence tomography (OCT) as an intermediate modality to enable registration of the distorted histological volumes containing cell type information to an MRI-based atlas coordinate system. OCT is an optical imaging technique that measures the backscattered photons from the sample to provide high-resolution cross-sectional images and volumetric reconstruction up to several hundred micrometers in depth in fixed ex vivo biological tissues, in a contact-free, staining-free, and noninvasive manner (11). Integrated with a mechanical sectioning device, serial sectioning OCT can further image tissue blocks up to several centimeters in each dimension. As imaging is performed before slicing, the 3D information is preserved, providing an accurate registration target to the MRI-based coordinate system. Moreover, the slices sectioned during OCT acquisition can be further collected to perform histological staining. In this study, the area of interest can be blocked from the hemisphere and acquired prior the preparation for LSFM imaging with serial-sectioning OCT. For 3D histological analysis with specific high subcellular resolution, confocal LSFM (12) is used to image the large volumes of human brain tissue slabs collected from serial sectioning OCT acquisition. LSFM enables fast optical reconstruction of the sample in 3D by using a specific configuration where the illumination axis is orthogonal to the acquisition axis (3, 12). Several advantages of LSFM are nondestructive optical sectioning, fast acquisition times, micrometric resolution, high penetration depths, and 3D tomography. However, LSFM requires the sample to be transparent to the light; therefore, a clearing step is necessary. Biological tissues are opaque because of light scattering. The principle of tissue clearing relies on the homogenization of the refractive index inside and outside the sample. Here, LSFM is coupled with a dedicated tissue transformation technique, SWITCH—H2O2—antigen Retrieval—TDE (SHORT) (13), to specifically label neuron subtypes and clear the tissue. Last, a stereological assessment is performed on the LSFM reconstructions to obtain a cell type–specific quantitative census of the neurons.
This multimodal imaging infrastructure was developed to bridge the resolution gap between macroscopic and microscopic techniques, resulting in a platform that integrates cellular anatomical information within a whole-brain reference space. The approach successfully allowed us to collect the cell census of different neuronal types in the Broca’s area within the whole-hemisphere MRI reference system of a postmortem human brain.
The work represents a large multi-institutional collaborative effort across the Massachusetts General Hospital, Boston University, the European Laboratory for Non-Linear Spectroscopy, and the Icahn School of Medicine at Mount Sinai.
RESULTS
Imaging and analysis overview
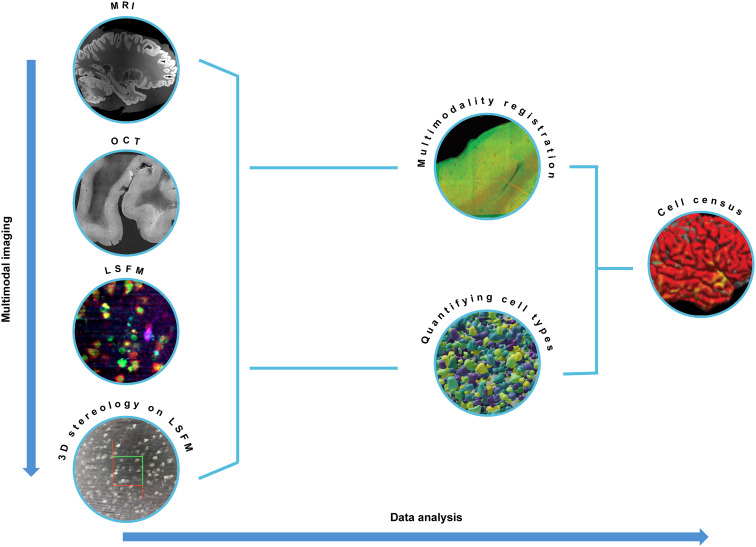
In this work, we generated a cell census of the human cerebral cortex centered on a Broca’s area (Brodmann’s areas 44/45) blocked from a whole human hemisphere. The postmortem specimen included in this project was obtained from a subject who had no neurologic or psychiatric illnesses. Figure 1 provides an overview of the multimodal imaging performed on the sample (left, from top to bottom) and the subsequent data analysis (left to right).
Fig. 1. Imaging and analysis overview.
From top to bottom: Multimodal imaging. MRI, OCT, LSFM, and 3D stereology on LSFM images are performed on the same sample. Left to right: Data analysis. Registration between MRI, OCT, LSFM, and 3D stereology data is performed to align all the datasets back to the MRI coordinate system. Cell counting on LSFM images with digital stereology reliable quantification of cell types. Multimodal registration between the images and the counting is needed to lastly obtain the cell census of the neurons in an MRI-based atlas coordinate system.
The whole hemisphere was first imaged with MRI before the sample was blocked around Broca’s area and imaged with serial sectioning OCT. 3D histological fluorescence imaging on the acquired slices was then performed with LSFM. A stereological evaluation was performed digitally on the LSFM data with a laminar level of resolution. An atlas of the data was obtained through nonlinear registration of the three modalities: LSFM to OCT and OCT to MRI. Cell counts and manually labeled features were also registered along with the volumes on which they were generated, thereby mapping the results of our analysis to a within-subject coordinate system. The registration to whole-hemisphere MRI makes transferring the data to in vivo coordinate systems straightforward. All the data collected are made available on the Distributed Archives for Neurophysiology Data Integration (DANDI) repository (14). In the following sections, details are provided on each step, and the cellular resolution atlas of the area is provided.
Whole-hemisphere MRI and analysis for reference coordinate system establishment
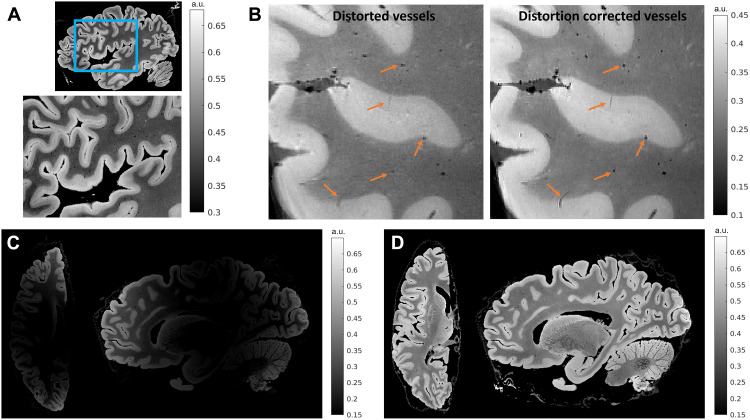
As the first step in the pipeline, a reference coordinate system for the cellular atlas was established with ex vivo MRI of the whole hemisphere. Structural images with 150-μm isotropic resolution were acquired using a multiflip angle, multi-echo fast low-angle shot (FLASH) MRI sequence on a Siemens 7 T MRI scanner using a custom-built 31-channel head coil (15). The MRI data were processed to correct for geometric distortions due to B0 field inhomogeneities (16, 17), contrast variations due to B1 transmit field inhomogeneities, and intensity bias due to B1 receive coil sensitivity variations (18). Figure 2A illustrates the 150-μm isotropic root mean squared (RMS) FLASH MR image of the sample presented in this paper. The RMS was calculated across four echo times for a flip angle of 20°. Figure 2 (B to D) demonstrates the improvement in vessel and laminar contrast quality attained by performing various artifact correction steps. The artifact-corrected whole-hemisphere MRI data presented here provide a reliable laminar framework, improved quality of vessel landmarks, and a reference space to which downstream modalities can be registered. Figure 2A zooms into Broca’s area where a clear contrast between gray and white matter and the transition from the infra- to supragranular cortical layers are visible in these images. These laminar boundaries are critical for tabulating information about cell types and their distributions within the characteristic architectural infrastructure that defines the cortical sheet. Figure 2B shows a zoomed-in portion of the frontal lobe that is severely affected by B0 and B1 transmit distortions. The B0 inhomogeneity blurred the vessels and the cortex in the RMS image due to misalignment between their locations in the different echo images. B1 transmit field variation reduced the overall vessel contrast. Vessels are important anatomical landmarks used to aid cross-modality registration, and since the distortion corrected images demonstrate improved vessel sharpness and contrast, they aid in providing accurate landmarks essential for registration. Figure 2 (C and D) shows axial and sagittal slices before and after intensity bias correction. The bias-corrected images show improved laminar and white matter contrast.
Fig. 2. MRI for global reference.
The figure shows results for 150-μm isotropic RMS FLASH MR images with RMS calculated across four echo times (TE = 5.65, 11.95, 18.25, and 24.55 ms) for a flip angle of 20°. (A) A sagittal section that is zoomed into Broca’s area showing infra- and supragranular gray matter contrast. (B) A zoomed-in sagittal frontal section of the same brain sample with vessels that are blurred and have reduced contrast. The blurring is due to B0 field inhomogeneity, while the reduced contrast is due to B1 transmit field inhomogeneity. We also show vessels postdistortion correction that are well preserved and have high contrast, demonstrating the effectiveness of the correction methods. (C and D) An axial and sagittal slice before and after B1 receive intensity bias correction. The intensities are visibly more uniform after bias removal. a.u., arbitrary units.
OCT 3D reconstruction for tissue registration
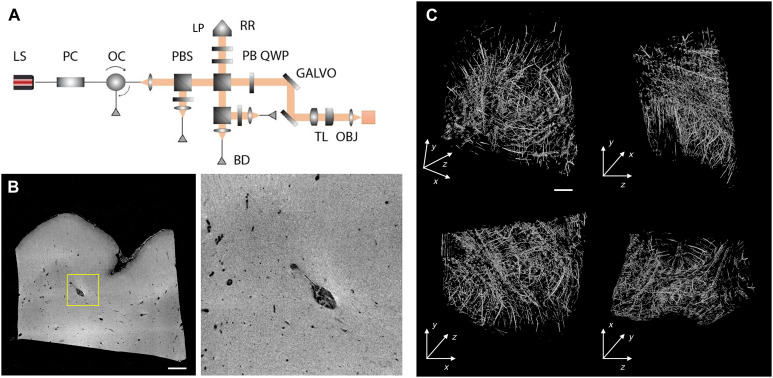
The excised block (approximately 1.5 cm by 1.3 cm by 0.8 cm) was imaged with a home-built serial sectioning OCT system at 5-μm isotropic resolution (11). To enhance the penetration of light deep inside the sample for OCT, we exploited a clearing procedure, based on the organic solvent 2,2′-thiodiethanol (TDE), to reach an imaging depth of up to 500 μm (19). Sectioning the 500-μm-thick slices was performed using a custom-built vibratome that is capable of slicing sections up to 6 cm in width (20). By capturing the intrinsic back-scattering properties of the tissue, OCT elucidated features such as vasculature to be used as registration landmarks (Fig. 3 and movie S1). In addition, as OCT imaging precedes sectioning, tissue deformations from sectioning were almost completely eliminated.
Fig. 3. OCT analysis.
(A) Schematic rendering of the OCT apparatus. LS, light source; PC, polarization controller; OC, optical circulator; PBS, polarizing beam splitter; PBS, polarization beam splitter; RR, retroreflector; LP, linear polarizer; QWP, quarter wave plate; GALVO, galvo mirror; BD, balanced detector; TL, telescope; OBJ, objective. (B) An example XY slice of the OCT volume intensity (left) and zoom-in view of the highlighted window (right, dimension = 1.5 mm by 1.5 mm). Scale bar, 1 mm. (C) 3D rendering and orthogonal views of the blood vessel segmentation of the OCT volume using the Frangi filtering method. Top left: 3D rendering. Top right: YZ view. Bottom left: XY view. Bottom right: XZ view. Scale bar, 1 mm.
Molecular phenotyping reconstruction with LSFM
Fluorescence imaging of the 16 slices was obtained using a custom-built dual-view inverted confocal LSFM with a voxel size of 0.55 μm × 0.55 μm × 3.3 μm that results in a 3.3-μm isotropic resolution after postprocessing (Fig. 4A) (21). Molecular specificity was achieved by combining LSFM with an advanced tissue transformation protocol called SHORT (13). The protocol rendered the sample completely transparent to light by refractive index matching (Fig. 4B) and allowed homogeneous colabeling of large 3D volumes with different markers (Fig. 4C and fig. S1).
Fig. 4. Multicolor imaging with LSFM.
(A) Schematic rendering of the LSFM apparatus. LS, laser source; TL, telescope; M, mirror; DC, dichroic; BS, beam splitter; GALVO, galvo mirror; AOTF, acousto-optical tunable filters; CAM, camera; OBJ, objective. (B) A 500-μm-thick slice in PBS before (left) and after (right) TDE tissue clearing. (C) A representative slice of a middle plane (~200-μm depth) of a 500-μm-thick slice stained with CR in blue (λexc = 488 nm), PI in green (λexc = 561 nm), and NeuN in red (λexc = 638 nm). Vessels are visible in the blue and green channels due to the presence of autofluorescence signals. Scale bar, 1 mm. (D) Single channels of slice in (C). Scale bar, 1 mm. (E) High-resolution insets showing the different cellular markers used in the study: calretinin (CR), propidium iodide (PI), and neuronal nuclear antigen (NeuN). Scale bar, 50 μm.
To perform the cell census in the area 44/45, we used immunofluorescence to label specific neuronal populations. The use of neurochemical markers enables the definition of region-specific staining patterns and generally results in a high definition of cortical areas that complement traditional Nissl and myelin preparations. In this context, calcium-binding proteins have been shown to be reliable cellular markers for cytoarchitectural studies of the primate neocortex, in which they are present principally in distinct populations of inhibitory neurons that exhibit recognizable patterns of regional and laminar specialization (5, 22, 23). We used an anti–neuronal nuclear antigen (NeuN) antibody to stain all neurons and an anti–calretinin (CR) antibody to identify a subpopulation of γ-aminobutyric acidergic (GABAergic) interneurons. To detect the nuclei of all cells, we used an exogenous dye [propidium iodide (PI)] obtaining a three-color costaining in the same tissue (Fig. 4D and movie S2). Vessels were identified from autofluorescence signals generated by retained blood.
Stereological cell counting on 3D LSFM reconstruction
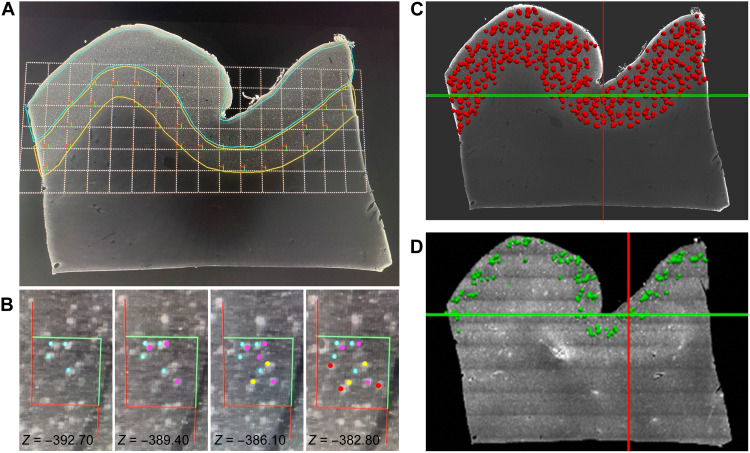
While LSFM enables the investigation of brain structures from the subcellular to the mesoscale, by recording different distributions of neuronal populations in large, cleared, specifically immunolabeled tissues with micrometric resolution and reasonably efficient acquisition times, no quantifications of such population-level imaging data currently exist. Hence, we performed stereological assessments on LSFM 3D reconstructions of identified neuronal populations using an Optical Fractionator probe (24) adapted to cleared materials (Fig. 5 and fig. S2). The tridimensionality of the acquisition permits us to visualize the neurons in their entirety allowing a precise quantification, avoiding over- or underestimation bias present in 2D (13) evaluation. We performed the analysis on the slices acquired with LSFM obtaining the results presented in Table 1.
Fig. 5. Stereological evaluation on LSFM reconstructions.
(A) A slab from the LSFM imaging dataset stained with the anti-NeuN antibody, displaying a systematic-random counting grid used during stereologic analyses. The grid is placed over the infragranular layers (yellow outline), and optical disector frames are placed in all segments of the grid that are located within the region of interest. (B) Different z-levels within a disector with color markers indicating NeuN+ neurons that have been sampled, according to stereologic rules (see Materials and Methods), at different focal planes in the disector thickness (24). The neurons are marked, from left to right, in blue, purple, yellow, and red as they appeared in focus moving up in the z axis. Neurons that touch or cross the inclusion (green) sides of the disector are included in the sampling at this site, whereas those touching or crossing the exclusion (red) sides of the disector are not, giving each NeuN+ neuron in this region of interest the same probability to be counted only once. (C and D) The registration of stereologic data onto the LSFM dataset for the entire slab. Each red dot in (C) corresponds to NeuN+ neurons sampled through the slab within an optical disector depth during stereologic analysis and used to generate an estimate of the total population number. The green dots in (D) represent CR+ neurons that were analyzed only in layers 2 and 3 as they are very sparse in the deep layers of the neocortex. Grid size in (A) = 800 μm, and counting frame in (B) = 150 μm.
Table 1. Stereologic estimates of laminar volumes and CR+ and NeuN+ cell numbers and densities in LSFM images for each slice from the Broca’s area evaluated in the study.
| Calretinin | NeuN | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Slice | Layer 3 estimated population | Cavalieri layer 3 estimated volume (mm3) | Layer 3 CR+ cell density (cells/mm3) | Slice | Layer 3 estimated population | Cavalieri layer 3 estimated volume (mm3) | Layer 3 NeuN+ cell density (cell/mm3) | Layer 5 estimated population | Cavalieri layer 5 estimated volume (mm3) | Layer 5 NeuN+ cell density (cells/mm3) |
| A | 29,752.80 | 10.14 | 2,934.55 | A | 83,530.61 | 13.0596 | 6,396.11 | 62,109.34 | 14.2373 | 4,362.44 |
| B | 26,950.00 | 10.58 | 2,546.61 | B | 83,544.02 | 12.3819 | 6,747.27 | 63,195.51 | 14.7354 | 4,288.69 |
| C | 21,344.40 | 8.99 | 2,374.55 | C | 84,187.67 | 11.0245 | 7,636.42 | 62,337.3 | 13.8289 | 4,507.76 |
| D | 24,075.33 | 8.75 | 2,750.57 | D | 82,985.28 | 13.0852 | 6,341.92 | 61,775.89 | 13.4359 | 4,597.82 |
| E | 18,110.40 | 8.56 | 2,115.54 | E | 83,128.32 | 11.1208 | 7,475.03 | 61,707.05 | 11.646 | 5,298.56 |
| F | 21,416.27 | 8.68 | 2,468.03 | F | 80,365.96 | 9.91782 | 8,103.19 | 57,912.16 | 11.8368 | 4,892.55 |
| G | 22,026.50 | 7.03 | 3,134.99 | G | 80,607.33 | 9.3727 | 8,600.23 | 57,617.15 | 10.5244 | 5,474.63 |
| H | 18,182.27 | 8.44 | 2,153.41 | H | 79,977.08 | 8.73192 | 9,159.16 | 58,247.39 | 10.443 | 5,577.65 |
| I | 24,499.20 | 7.86 | 3,118.48 | I | 90,412.38 | 10.7255 | 8,429.67 | 65,681.63 | 8.46014 | 7,763.66 |
| J | 22,667.33 | 6.76 | 3,353.66 | J | 95,556.27 | 9.79419 | 9,756.42 | 7,4060.8 | 8.18111 | 9,052.66 |
| K | 21,697.12 | 8.03 | 2,701.96 | K | 84,646.95 | 9.69507 | 8,730.93 | 62,176.39 | 8.75804 | 7,099.35 |
| L | 22,903.47 | 9.48 | 2,414.91 | L | 93,678.93 | 9.84716 | 9,513.29 | 66,926.93 | 8.44643 | 7,923.69 |
| M | 20,744.53 | 7.87 | 2,635.57 | M | 83,729.06 | 9.45871 | 8,852.06 | 53,785.6 | 7.96777 | 6,750.4 |
| N | 20,744.53 | 8.08 | 2,568.81 | N | 94,242.13 | 11.1953 | 8,418.01 | 70,493.87 | 8.34508 | 8,447.36 |
| O | 21,806.40 | 6.56 | 3,325.91 | O | 79,223.47 | 8.50373 | 9,316.32 | 60,731.73 | 7.51905 | 8,077.05 |
| Total | 336,920.55 | 125.80 | 40,597.55 | Total | 1,279,815.46 | 157.91 | 123,476.03 | 938,758.74 | 158.37 | 94,114.27 |
| Mean | 22,461.37 | 8.39 | 2,706.50 | Mean | 85,321.03 | 10.53 | 8,231.74 | 62,583.92 | 10.56 | 6,274.28 |
| SD | 3,011.73 | 1.14 | 393.39 | SD | 5,434.89 | 1.45 | 1,099.85 | 5,148.73 | 2.56 | 1,670.17 |
Multimodal registration for MRI-referenced cell census
A defining feature of this project is processing the same human brain sample through each of the imaging techniques and quantitative tools described above. This enables us to place the cellular anatomical information within a cortical coordinate system. Cross-modality registration of cytoarchitectural properties is complicated due to distortions introduced by histological imaging techniques. While registration is facilitated by leveraging the fact that OCT is performed on the tissue block before sectioning, slices imaged with LSFM show important distortions when compared to the OCT data. Hence, we developed a nonlinear registration method that uses the segmentation of common features visible across all imaging modalities, such as blood vessels, to overcome this challenge. Vessels that have a diameter larger than 150 μm were manually segmented in MRI and OCT data and in each LSFM slice (fig. S3). The resulting labels were used in a composite objective function that optimizes intensity similarity and label distance to drive registration between MRI and OCT on one hand and LSFM and OCT on the other hand.
All deformations are fully invertible and allow any one modality to be warped to the space of any other modality. In addition to vasculature, the boundary between the cortical infra- and supragranular layers was segmented in MRI data and in a subset of LSFM slices to measure registration accuracy. The resulting transforms were used to warp the MRI segments to each manually segmented LSFM section (fig. S4), where the 95th (527 ± 137 μm) and 75th (306 ± 96 μm) percentiles of the minimum distance from the LSFM boundary to the MRI boundary were computed. It should be noted that these distances were computed in 2D sections and are therefore upper bounds on the 3D distances and that 75% of boundary points have less than two-voxels errors in the 150-μm isotropic MRI space.
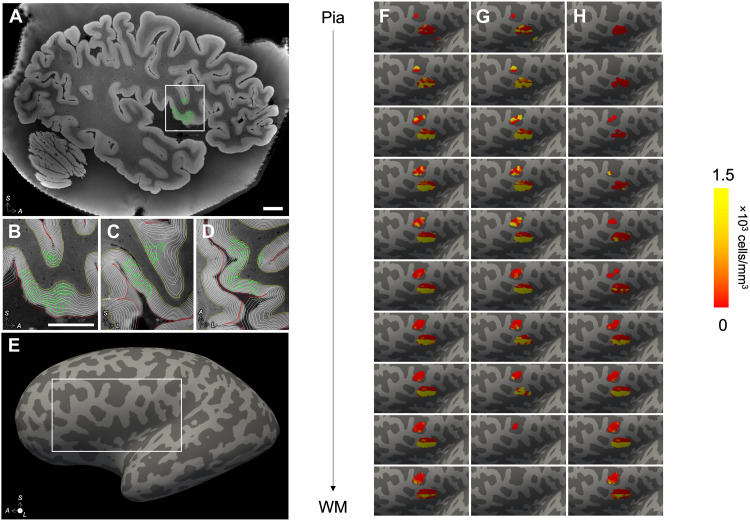
Last, we demonstrate instantiation of microscopic cellular and stereologic information from distorted LSFM histological images within the MRI volume with OCT data serving as a critical intermediary modality with mesoscopic resolution and minimal distortion (movies S3 and S4). The stereological annotations were warped to MRI space and used to compute local cell densities on 10 intermediate surfaces spanning the depth of the cortex (Fig. 6), allowing to map the stereological annotations performed at the microscopic scale back into the space of the intact brain.
Fig. 6. Instantiation of stereological counting sites in MRI space.
(A) The stereological counting sites (green) for the NeuN markers were mapped to the MRI space by composing all spatial transformations from MRI to OCT to LSFM. (B to D) The MRI was segmented using SAMSEG, and the white matter (WM) and pial surfaces were computed using FreeSurfer; 10 equidistant intermediate surfaces were generated between the WM and pial surfaces, and counting sites were projected to their nearest vertex across all 10 surfaces (B to D: sagittal, coronal, and axial views). (E and F) The number of counted cells per vertex and total Jacobian-modulated volume of the projected sites were smoothed on the surface (three consecutive averages over the 1-ring neighbors), and their ratio yielded the density of NeuN+ neurons associated with each vertex. These densities are shown on the inflated surface. Note that only cells from layers 3 (G) and 5 (H) were counted and are included in the surface density map. Scale bars, 1 cm.
DISCUSSION
In this work, we create a micrometer-resolution cell census atlas of an identified region of the human cerebral cortex. By combining different techniques, we were able to overcome the inherent limitations associated with any single modality. To achieve this goal, we implemented an imaging and image processing approaches to correlate the four modalities involved: MRI, OCT, LSFM, and stereology. By first establishing a reference coordinate system for the cellular atlas, through volumetric MRI, we greatly expand the utility of the cellular atlas and provide a basis for in vivo inferences with our analysis. Existing 3D models of the human brain with cellular resolution such as the Big Brain project (4) include sectioning distortions inherent to histology and do not permit through-plane tracing of features of interest across slices such as connecting axons, vasculature and laminar surfaces. However, the ability to build undistorted laminar models of the human cerebral cortex is a critical component of any cellular atlas. In the past decade, the combination of tissue clearing and high-throughput microscopy techniques, such as LSFM, has paved the way for the investigation of brain anatomy in 3D (25) with subcellular resolution. However, clearing methods and physical sectioning of the human brain sample introduce tissue distortions that complicate the coregistration between MRI reference data and fluorescence reconstructions. Attempts to perform MRI on cleared samples showed a contrast loss that prevents visualization of tissue microstructure (26), suggesting that MRI should be performed before tissue clearing. Nevertheless, the modifications undergone by the sample prevents accurate laminar registration. Images that are obtained in a single preferred orientation (typically coronal), which have fundamentally differing in-plane and through-plane resolution due to slice thickness and distortions, are exceedingly difficult if not impossible to use to directly detect 3D boundaries due to the complex geometry of cortical folding patterns (27). To facilitate the alignment between MRI and LSFM reconstruction, we decided to use OCT as an intermediate method to enable registration of the distorted fluorescence images of cleared volumes to the MRI-based atlas coordinate system. We demonstrate accurate alignment of OCT and whole-hemisphere MRI of the brain at the vasculature level. As OCT data are acquired before slicing, it provides a critical, minimally distorted, intermediate reference between the MRI and LSFM.
The choice of using immunofluorescence is an important aspect of our integrative approach, as it allows identification of specific cellular markers. The precise immunoreactivity in neurons for neurochemical markers allows us to define region-specific staining patterns and generally results in a better definition of the cortical areas compared to Nissl or myelin preparations. In this context, markers like calcium-binding proteins are reliable cellular markers for the mammalian neocortex that identify distinct neuronal populations exhibiting recognizable patterns of regional and laminar specialization (5, 23). Calcium-binding proteins are expressed in morphologically non-overlapping subgroups of GABAergic interneurons and display a highly specific regional distribution (28). While these markers define generic classes of neurons, they also identify neuronal groups known to be differentially affected in many neuropsychiatric conditions (22, 29).
We performed analytical neurostereology with layer-specific resolution on the large volume LSFM acquisitions (24). A fundamental aspect of stereologic approaches is the accurate delineation of the region of interest. For the Broca’s area, we established reliable cyto- and chemoarchitectural criteria based on published studies (30). This approach is rigorous, validated, and has been extensively applied to the study of the human brain (30, 31). We analyzed multiple fluorescence channels on the same counting frame location (31). Such morphometric parameters can be used together with a combinatorial expression profiling analysis of cell classes to provide a comprehensive morphomolecular characterization of the cortical cellular typology (32). Using this approach, we successfully created a cell census of NeuN+ and CR+ neurons in Broca’s area that was registered to a within-subject MRI coordinate system. We analyzed 15 slices of 500-μm thickness (for a total volume of 1.5 cm × 1.3 cm × 0.75 cm) and we obtained, on average, 22,461 CR+ neurons in layer 3 across the slabs, 85,321 NeuN+ neurons in layer 3, and 62,584 NeuN+ in layer 5, with corresponding densities of 2707, 8232, and 6274 neurons/mm3, respectively.
A final aspect of the overall approach is the big data management, which was optimized to allow the combination of the different techniques. Special attention was devoted to the choice of the nomenclature, reference data type format, compression, and postprocessing methods to achieve a standardized procedure for data storage and sharing, replicable on multiple samples. We published all the data on the DANDI platform, an open access Brain Research Through Advancing Innovative Neurotechnologies (BRAIN) Initiative supported data archive for publishing and sharing data. The datasets have an associated specific DOI that facilitate discovery and reuse by other researchers; moreover, they are made available using the Datalad management platform, a tool that treats data just like software and can track their version over time including reorganization, without duplicating the data. This is of fundamental importance since we are applying the pipeline to 10 different subjects to evaluate the biological variability of neuronal distributions in Broca’s area, and the data are made available through the same link, allowing an easy access to them. Data were prepared following the Brain Imaging Data Structure (BIDS) standards, a formal way to organize neuroimaging data in a file system that simplifies communication of information about the dataset to other investigators and enables easier software development through consistent naming and location of file data.
Several challenges remain to be addressed further to improve the current approach. The 3D stereological analysis used in this study to obtain a reliable counting of cells is highly time-consuming. To obtain the census of neurons in the entire acquired volumes, the raw images must be automatically analyzed. In particular, we are exploring the possibility of using a convolutional neural network for pixel classification, previously proposed to analyze two-photon fluorescence microscopy images (33), and automatic cell detection obtained on mouse brain reconstruction with LSFM (34, 35). After careful validation, such approaches can be used to automatically count the different neuronal populations stained in the samples, providing a comprehensive characterization of the whole imaged volume. In this context, the stereological count provided in this work will provide a gold standard against which automated approaches, such as machine learning–based methods, can be verified.
To expand the molecular phenotyping of the tissue, multiround staining on a cleared sample (13, 32, 36) could be implemented to characterize human brain cytoarchitecture in greater detail. In this context, a quantitative database of morphofunctional neuronal types in identified cortical regions represents a crucial normative resource for the study of cellular changes in brain disorders. The consideration of differential cortical vulnerability in brain diseases can also be used for targeting key cortical domains to carry out future analyses. Also, to speed up the alignment process between the different techniques, eliminating the need to use manual landmarks as performed in this study, microstructure-informed automatic nonlinear registration tools are needed. In this regard, the development of an accurate automated vascular segmentation approach (37) is required to avoid manual segmentation. The reference labels generated in this study can serve as examples to train segmentation models that will be used on future samples, bypassing manual annotation toward the creation of a scalable human brain cellular atlas.
The cell census produced in this study is registered within a whole-hemisphere coordinate space, which is critical for its future integration with other cross-sectional atlases, as intersubject registration is notably driven by the gross cortical geometry. Similarly, the availability of both microscopic and macroscopic features allows this atlas to be accurately integrated with myeloarchitecture data and cerebral microvasculature distributions obtained on other specimens. MRI atlas data will enable the determination of patterns of molecular cytoarchitecture and connectivity at multiple scales, from single layers to columnar domains of cortex within a cortical region identified based on specific cytoarchitectural characteristics.
Moreover, there are multiple implications of this approach to studying brain disorders in an efficient way. Selective vulnerability of certain brain areas and neuronal subpopulations has long been reported in many brain disorders. However, the pathogenic and compensatory mechanisms driven by heterogeneity within cell types and states, cell-to-cell interactions, and spatial contributors are only beginning to be unveiled with the availability of single-cell and spatial approaches. For example, among age-related neurodegenerative disorders, principal neurons in layer II of the entorhinal cortex are the earliest to exhibit Alzheimer’s disease pathology in the aging brain, while corticocortically projecting pyramidal neurons in layers III and V are progressively affected in a regionally specific manner when Alzheimer’s disease develops (38, 39). Disease-associated gene expression in glial cells, immune cells, microvasculature, and their cross-talk with neighboring neuronal subtypes may unveil clues to regional vulnerability in disorders like autism spectrum disorder, epilepsy, stroke, and dementias (40–42). The extension of a multimodal cell type atlas of select regions in the human brain to multiple brain regions, particularly those housing vulnerable cell populations, will generate openly available datasets crucial to future studies comparing cell types within their spatial context in the normative brain to those in neuropsychiatric disease, with the addition of transcriptional, epigenetic, morphological, and neurophysiological datasets from postmortem brains. As techniques for spatial detection of proteins and metabolites achieve multiplexing capabilities as well as cellular resolution, the addition of such data will help to uncover disease mechanisms that may explain functional changes detected in specific cell types and brain areas in a given disease.
Last, working with axonal markers and myelin markers at this level of resolution will aid in vivo and postmortem MRI/Diffusion-Weighted magnetic resonance Imaging (DWI) studies to refine the knowledge of the human brain connectome. Direct measurement of axonal orientations across the intact human brain at the single-micrometer scale will enable the study of the connectivity between different areas that have not been comprehensively mapped due to highly packed fibers densities. The successful realization of this cell census atlas represents a substantial scientific step forward in terms of rigor and reproducibility for human brain high-resolution 3D characterization. We believe that the widespread application of our multimodal approach will provide a deeper understanding of the entire human brain architecture across resolution levels. Our imaging technology pipeline will ultimately enable the automated reconstruction of undistorted 3D microscopic models of not only a brain area but also of an entire human brain, enabling the assessment of intra- and intersubject variability.
MATERIALS AND METHODS
Human brain sample collection
The samples used in this project were obtained from control subjects who died of natural causes with no clinical diagnoses or neuropathology. The brain hemisphere imaged for this paper was from a 79-year-old male donor. A standard fixation protocol was used in which the brain was fixed in 10% formalin for a minimum of 90 days. The sample was packed in a 2% buffered paraformaldehyde solution for MRI scanning. The human brain tissue sample was procured from the Department of Neuropathology at the Massachusetts General Hospital Autopsy Service (Boston, USA). Written consent was obtained from healthy participants before death, following institutional review board–approved tissue collection protocols from Partners Institutional Biosafety Committee (protocol 2003P001937).
Magnetic resonance imaging
Ex vivo MRI was performed on the whole human hemisphere using multi-echo fast low-angle shot (ME FLASH or MEF) sequence at 150-μm isotropic resolution with bipolar readout gradients, on a 7 T Siemens MR scanner. Specific scan parameters were as follows: Repetition Time (TR) = 34 ms; Time to Echo (TE) = 5.65, 11.95, 18.25, and 24.55 ms; field of view (FOV) = 192 mm by 81.3 mm; and slice thickness = 150 μm. We acquired three MEF images with flip angles of 10°, 20°, and 30°. Bipolar readout gradients improve the signal-to-noise ratio (SNR) efficiency of the MEF scan. However, in the presence of B0 inhomogeneity, the odd and even echoes of the MEF are distorted in opposite directions because they are acquired with opposite polarity readout gradients, leading to misalignment between them (17, 43). To address the misalignment issue, an optimization framework was developed to enhance SNR and to correct geometric and intensity distortions without needing additional high-resolution scans. We collected a 2D-encoded B0 field map, which estimated the amount of inhomogeneity at every voxel and in turn gave us a measure of displacement at every voxel of the 150-μm isotropic MEF. The field map acquisition was a standard gradient echo scan consisting of two gradient echoes with TR = 5 s, FOV = 192 mm by 144 mm, matrix size = 160 × 120, and slice thickness = 1.2 mm. Geometric distortions were then corrected using a group sparsity-based edge preserving intensity correction algorithm that used the field map and all the FLASH images jointly to perform the correction (16). In addition, B1+ variations were estimated by acquiring multiple single echo FLASH sequences with short TE (2.7 ms), long TR (5 s), flip angles varying from 20° to 340°, FOV = 192 mm by 156 mm, matrix = 96 × 78, and slice thickness = 2 mm. We fit the frequency of the sinusoid at each voxel to estimate the multiplicative bias in our flip angle measurements. The estimated flip angle bias map was then used to correct the flip angle value at each voxel location. We fit the T1 relaxation time at each voxel using a dictionary look-up procedure and the corrected flip angle map. We synthesized and implemented FLASH MRI scans by incorporating the corrected T1 estimate within the FLASH MRI physics forward model to remove variations caused by spatially nonuniform B1+ field (17, 44).
The whole hemisphere was imaged using a custom-built 31-channel phased array coil (15, 18), the intrinsic sensitivity profile of which can cause nonuniform intensity. The lack of a body coil with a uniform receive sensitivity profile on the 7 T scanner makes acquisition-based receive bias estimation non-applicable to our scenario. We use a model-based B1− bias correction method that jointly segments tissue type at each voxel and estimates the intensity bias using a likelihood-based cost function. The method assumes that the voxels classified under the same tissue class will have the same FLASH intensity value (18). These acquisition and artifact correction methods provided us with a brain hemisphere structural image that is used to generate a brain surface and provide a reference coordinate system for registration purposes explained in the later sections.
Serial sectioning OCT imaging
A custom-built polarization-sensitive OCT (PSOCT) system was used to acquire the volumetric data; however, only the intensity signal was analyzed as the polarization part is beyond the scope of this study. Therefore, we use the terminology of OCT instead of PSOCT in the results. The system was built on the basis of a previously reported setup (45) with a schematic as shown in Fig. 3A. Specifically, a swept light source (AxsunTech) was used in the OCT system, which has an A-line rate of 100 kHz, a center wavelength of 1310 nm, and a bandwidth of 135 nm. The axial resolution was estimated to be 5.6 μm in tissue (with a refractive index of 1.4). The sample arm consists of a pair of XY scanning mirrors, a 4× telescope, and a 4× air objective [Olympus, UPLFLN4x, numerical aperture (NA) 0.13] to obtain a lateral resolution of 6 μm. The interference fringe from the sample and the reference arms was collected by a balanced detector. The post-objective power was measured to be 3.7 mW, achieving a 95-dB SNR. For this study, we used a 3 mm by 3 mm FOV with a 3-μm lateral step size and 10% overlap between tiles, with each tile taking about 16 s.
For large human brain blocks, the embedded samples are usually a few centimeters in each dimension, which is over 10 times greater than that of the FOV of a single image tile. Therefore, the whole sample surface was divided into a grid of views, and the images from all views were stitched together to form a full surface. Motorized xyz stages (x and y stages: LTS150, Thorlabs; z stage: MLJ150, Thorlabs) were incorporated to translate the samples under the OCT scanning head to all the imaging locations. The maximum travel distance for x, y, and z stages was 150, 150, and 50 mm with correspondent one-direction moving accuracy of 2, 2, and 10 μm. A customized vibratome slicer was mounted adjacent to the OCT imaging head to cut off a superficial slice of the tissue block of 500 μm of thickness upon completion of the scanning of the sample surface. A 6.35 cm custom sapphire blade (DDK CO.) was vibrated at 50 Hz and 1.2 mm peak-to-peak amplitude while slicing, with 0.1 mm/s stage feed rate. Custom software written in LabVIEW was used to control OCT imaging and vibratome slicing. To extend the depth of focus in the ex vivo human brain blocks, the optical index matching was applied to the samples before the serial sectioning. Specifically, human brain blocks were first washed for 1 month in phosphate-buffered saline (PBS) solution 0.01 M at room temperature (RT) while gently shaking. Then, the human brain blocks were embedded with agarose. The index matching was performed with serial incubations in 100 ml of 20, 40, and 60% (v/v) TDE in 0.01 M PBS (TDE/PBS) each for 24 hours at RT while gently shaking. For large human brain blocks, the index matching process usually takes about 1 week for each TDE concentration. The equilibrium of the index matching was determined when the floating embedded brain blocks sank down in the TDE solution.
The data acquisition computer was a high-performance local computer with a four-core processor, 64 GB memory, a high-speed digitizer (ATS9350, Alazar), a GPU (RTX4000, NVIDIA), and a 10 Gb/s high-speed Ethernet card. Using the k-clock from the light source, the signal was digitized in even-k space. The GPU fetched the spectral domain data and performed real-space reconstruction on the fly, which included dispersion compensation (46), Fourier transform for depth-profile creation, and rough trimming in depth. The reconstructed data were then saved to a local storage server with 28-TB space through the 10 Gb/s Ethernet. Compared with the 0.2 Gb/s data acquisition rate, the Ethernet transfer rate was much faster and helped avoid any data loss. For high-speed postprocessing, data saved in local server were automatically uploaded to Boston University Shared Computing Cluster (SCC), a high-performance computing resource located at the site of Massachusetts Green High Performance Computing Center at Holyoke, Massachusetts. Upon completion of the experiment, a parallelized postprocessing script was executed on SCC, which included distortion correction, volume stitching, and various features extraction. We used both ImageJ plug-ins (47) and customized functions to correct distortion that is introduced during OCT acquisition, such as shading effect and grid distortion. To stitch the OCT volume in 3D, we first stitched tiles in 2D using the average intensity projection. After the coordination for each tile was obtained, we linearly blended them in x and y and then stacked them in z. Once the OCT volume is reconstructed, various features, such as vessels, axonal bundles, and cortical laminar structure, can be extracted using feature enhancement algorithms (48).
After each sectioning, the 500-μm brain slices were manually captured to reserve the sequence. Each slice was kept in between two TDE-rinsed gauze pads in a plastic shell. The gauze pads created a semidry environment to keep the slices moisturized while convenient for transportation. Each plastic shell was then labeled with the slice sequence and stacked and sealed in a plastic bag for transportation of the slices to the other sites.
Tissue clearing and labeling for LSFM imaging
Brodmann’s areas 44/45 brain 500-μm-thick slices were treated with SHORT (13), a modified version of the SWITCH/TDE tissue transformation protocol from Costantini et al. (33) that combines the SWITCH technique (32) with the TDE clearing method (49). Briefly, each slice was incubated in a SWITCH-off solution, consisting of 50% PBS titrated to pH 3 using HCl, 25% 0.1 M HCl, 25% 0.1 M potassium hydrogen phthalate, and 4% glutaraldehyde. After 24 hours, the solution was replaced with PBS (pH 7.4) with 1% glutaraldehyde. After three washes for 2 hours each in PBS at RT, the specimens were inactivated by overnight incubation in a solution consisting of 4% glycine and 4% acetamide at 37°C. Following inactivation, the samples were washed in PBS three times for 2 hours at RT and then incubated in the clearing solution containing 200 mM SDS, 10 mM lithium hydroxide, and 40 mM boric acid for 4 days at 55°C. After the clearing process, the samples were washed three times in PBS + 0.1% Triton X-100 (PBST) at 37°C for 24 hours. The presence of lipofuscin in the cellular soma (50) and the free-aldehyde double-bounds introduce high autofluorescence signals (51). To decrease such spurious and nonspecific signals, the specimens were treated with hydrogen peroxide (30%, v/v) for 1 hour at RT. After three washes in PBS each for 1 hour at RT, antigen retrieval was performed using preheated tris-EDTA buffer [10 mM tris base (v/v), 1 mM EDTA solution (w/v), and 0.05% Tween 20 (v/v) (pH 9)] for 10 min at 95°C. After cooling down to RT, the specimens were washed in deionized (DI) water for 5 min each and then equilibrated with PBS for 1 hour. To perform the multicolor staining, the samples were incubated with primary antibodies against NeuN (Merck ABN91 chicken) and CR (Proteintech 12278-1-AP rabbit) at 37°C for 7 days in PBST. Dilutions for the anti-NeuN and anti-CR antibodies were 1:100 and 1:200, respectively. Following three washes in PBST each for 30 min at 37°C, the samples were incubated for 5 days at 37°C with the secondary antibodies conjugated with different Alexa Fluor (AF) dyes with a dilution of 1:200 [anti-chicken immunoglobulin Y (IgY), AF 647 Abcam ab150171, anti-rabbit IgG AF 568 Abcam ab175470, goat anti-chicken IgY AF 488 Abcam ab150169, donkey anti-rabbit IgG AF 647 Abcam ab150075, and donkey anti-rabbit IgG AF 488 Abcam ab150073] and then washed three times for 1 hour each at 37°C. The slices were rendered transparent by soaking the samples in increasing solutions of 20, 40, and 68% (v/v) of TDE in PBS, each added with PI (dilution of 1:100) or SytoxGreen (dilution of 1:100) for 1 day each step at RT with gentle shaking. Samples were mounted in a sandwich-like configuration between a 250-μm-thin quartz coverslip (for refractive index matching at a refractive index of 1.46) and a standard glass sample holder, with a 500-μm-thick steel spacer in between (13, 52). Glycerol (91%) in distilled water was used outside the sandwich for the LSFM objective immersion. This step allowed us to achieve high penetration depth and to avoid any optical aberration by matching the refractive index of the brain samples.
LSFM imaging
In our custom-made inverted light-sheet fluorescence microscope (21), two identical objectives were inclined at 90° relative to each other and were spaced such that their FOVs were orthogonal and overlap in the center. They alternately played excitation and detection roles. The objectives were a pair of LaVision Biotec LVMI-Fluor 12× PLAN with ×12 magnification, Numerical Aperture (NA) 0.53, Working Distance (WD) 8.5 to 11 mm, spherically and chromatically corrected in the visible range, with a correction collar for refractive index matching with the immersion solution. They were inclined at 45° relative to the sample holder plane to allow for the largest possible lateral sample size while not interfering with its extension within the plane. These objectives were carefully chosen to maximize the optical resolution (1.1 μm lateral and 3.7 μm axial) and FOV (1.1 mm by 1.1 mm) while respecting the geometrical constraints and allowing immersion in any refractive index matching medium. The microscope was equipped with four laser sources (Cobolt) Diode Laser Module (MLD) 405 nm/100 mW, MLD 488 nm/60 mW, Diode-Pumped Laser (DPL) 561 nm/100 mW, and MLD 638 nm/180 mW), each emitting a Gaussian beam that had its width adjusted by a dedicated telescope, before combining all of them through a set of three dichroic mirrors. This combined beam was split by a 50 to 50% beam splitter in two equal parts that were conveyed into the two identical excitation pathways of the light-sheet microscope. In each pathway, the beam was modulated in intensity, timing, and transmitted wavelength by an acousto-optical tunable filter (AOTF, AAOptoelectronic AOTFnC-400.650-TN) and then was scanned by a galvo mirror (Cambridge Technology 6220H), to realize the digitally scanned light sheet planar illumination (53). A scanning lens (Edmund Optics #45-353, focal length (fl) = 100 mm, achromat), placed after the galvo mirror, converted the angular deflection into a lateral displacement of the incident light. The beam was then directed by the excitation tube lens (Edmund Optics #45-179, fl = 200 mm, achromat) to the objective’s pupil, through which it sequentially illuminated neighboring lines within a single plane of the sample. Each objective was held on a motorized stage (PI L-509.14 AD00) to adjust its focal plane position. The sample was held in a custom quartz sample holder inserted into a plastic tray filled with refractive index matching medium (glycerol, 91%). The sample was positioned using a 3-axis motorized stage system (two PI M-531.DDG and a PI L-310.2ASD for a motion range within 30 cm × 30 cm × 2.5 cm with submicrometric repeatability) and was imaged by translating it along the horizontal direction. The image velocity through the volume was 47 frames/s, corresponding to a volumetric rate of 0.5 cm3/hour. The fluorescence emitted by the sample was collected by the other objective lens and then was separated from the reflected laser excitation light by a multiband dichroic beam splitter (Semrock Di03-R405/488/561/635-t3-55x75), before being directed by the detection tube lens (Edmund Optics #45-179, fl = 200 mm, achromat) on a scientific Complementary Metal-Oxide-Semiconductor (sCMOS) camera (Hamamatsu OrcaFlash4.0 v3). Each camera operated in confocal detection mode by having the rolling shutter sweep in synchrony with the galvo scan of the digital light sheet (35, 54). Five sets of band-pass filters were mounted in front of each camera on a motorized filter wheel (Thorlabs FW102C) to image selectively the differently labeled cells or structures within the tissue sample.
The operation of the microscope hardware was controlled by a workstation running a custom multithreaded software, developed in C++ using Qt with a flexible and modular architecture and composed now of approximately 9000 lines of code. Our software ensured hardware synchronization and triggering by using a National Instruments PCIe-6363 card and controlled the automatic image acquisition from the two sCMOS cameras in confocal detection mode, with a sustained data rate of 800 MB/s at 47 fps and storage on a 16-TB SSD RAID.
The acquisition procedure for any sample started by determining its edges. Each image stack was acquired by moving the sample along the x axis through the fixed FOVs of the two objectives, and then the sample was shifted by 1 mm along the y axis and the next stack was acquired. This sequence continued until the whole volume had been acquired. Contiguous stacks had an overlap of 100 μm that allowed fusion in postprocessing to form the whole volume. Appropriate metadata was saved jointly with the acquired stacks.
The two identical optical pathways of the LSFM alternately served as excitation and detection arms, with a time delay of a half frame that was introduced between the two roles to avoid exposing the active rolling-shutter rows on the acquiring camera to stray light coming from the illumination beam on the same side. The two AOTFs allowed shuttering each illumination pathway independently to avoid introducing stray light and, furthermore, enabled selection of which laser wavelengths and intensities were impinging on the sample.
LSFM data management
To visualize the reconstruction of an entire slice, a postprocessing pipeline was applied to the data. First, as the objectives acquire images of the moving sample at 45° relative to the slide plane, an affine transform was applied to compensate for the motion and the 45° rotation thus bringing the acquired volume back to the sample’s coordinate system. The affine transform also performs a spatial down-sampling to 3.3-μm isotropic resolution. Then, custom-made stitching software, ZetaStitcher (G. M., “ZetaStitcher: a software tool for high-resolution volumetric stitching” https://github.com/lens-biophotonics/ZetaStitcher), allowed us to fuse the contiguous stacks, using the overlapping regions, to obtain a representation of the whole sample. Only for visualization purposes, an illumination intensity homogenization algorithm was applied to the stitched volume or to single slices to compensate for variations in the laser beam power across the FOV and among stacks. For each fluorescence band, the observed intensity along the light propagation axis was averaged, attaining a smooth intensity profile. By dividing each image for this reference, illumination intensity artefacts occurring across the transversal sample extension were mitigated. To store and share the information, data were compressed using the JPEG2000 lossy approach with a 1:20 compression ratio. The data analysis pipeline was written in Python. The 561-nm wavelength 3D reconstructions were used to visualize and segment the blood vessels to perform the alignment of the three modalities as described in the section below.
Stereology
Stereologic tools provide a set of simple rules and formulas to estimate numerical quantities of various morphologic parameters in tissue, with precision, accuracy, and a design free of observer-induced bias [for review see (24)]. Parameters such a number, density, volume, surface area, or length can be easily estimated based on the systematic sampling of a region of interest (e.g., a layer of a cytoarchitecturally defined region of cortex, defined as a “volume of reference”), with an observer-independent random design, that gives each object under study (e.g., a neuronal population identified by a specific protein marker) the same probability to be sampled once and only once in its volume of reference, based on strict sampling criteria that are kept constant for a given object throughout the analysis (24, 30, 31).
Stereologic analysis was performed on each cleared slice of Broca’s area, 500-μm thick, imaged at 3 μm by 3 μm by 3 μm pixel dimensions, and immunostained for NeuN and CR, using the MBF Bioscience Stereo Investigator Cleared Tissue software (version 2020.1.1) with an Optical Fractionator design (24). The counting frame size was 150 μm by 150 μm, the grid size was 800 μm by 800 μm, and the disector height was 15 μm for all sections of tissue examined generating >600 sampling sites. Layers 3 and 5 were outlined, and their boundaries were used to estimate laminar surface areas and volume. There were 10 virtual 49.5-μm-thick sections for the tissue sample, and layers 3 and 5 were contoured at the widest part of each subslab, with a 400% zoom. Markers were placed at the top of each sampled cell, as it came into focus within the depth of the disector. Sixteen consecutive slices were generated with LSFM, and data were obtained from 15 of them, the last slice being damaged. Data were expressed as means ± SD. The coefficients of error of the estimates were obtained as previously described (24, 30, 31) and were less than 2%.
Image registration
Intermodality deformations were modeled using a combination of affine and nonlinear transformations. The affine transformation was encoded in the Lie algebra of the corresponding affine Lie group. Nonlinear transformations were modeled by stationary velocity fields and exponentiated using a scaling and squaring algorithm, which ensures—under mild smoothness conditions—that the resulting transformations are invertible diffeomorphisms (55). The parameters of the deformations were optimized by minimizing a combination of losses on intensity images [local cross-correlation (LCC) (56)] and manually segmented landmarks [Dice and/or mean Euclidean distance (MED)]. Specifically, for the MED loss, we computed the Euclidean distance transform (57) d ∈ of the segmented vessels in the fixed modality, and this distance was evaluated within the segmented vessels v ∈ [0,1]N of the moving modality and averaged, i.e., ℒ = (dTv)/(1Tv), where 1 is a vector of ones. Stationary velocity fields were regularized with a combination of penalties on their bending and linear elastic energies. When registering a whole volume (in this case, MRI) with a sub-block (e.g., OCT), the larger volume was always deformed to the space of the smaller block, where the objective function was computed. Because the transformation model is diffeomorphic, the resulting transforms could nevertheless be inverted and used to warp the block back to the space of the larger volume. The same transforms were also used to warp stereology coordinates extracted from the stereology software (MBF Bioscience Stereo Investigator Cleared Tissue software version 2020.1.1). The registration model was implemented in PyTorch and efficiently optimized by Gauss-Newton, using analytical first and (approximate) second derivatives of the objective function. For additional robustness, parameters were progressively optimized in a coarse-to-fine fashion (rigid only, then affine only, and then affine and nonlinear). In details, for MRI to OCT registration, the OCT volume was downsampled to 100 μm × 100 μm × 50 μm, and LCC was computed within patches of 4 mm × 4 mm × 2 mm. The objective function combined LCC (weight: 1), Soft Dice between gray matter segments (weight: 1), and MED between vessels segments (weight 0.1). Here, no nonlinear registration was necessary. For OCT to LSFM registration, the OCT volume was downsampled to 96 μm × 96 μm × 12 μm, and the LSFM volumes were downsampled to 40 μm × 40 μm × 10 μm. The objective function combined LCC (weight: 1), Dice between vessels segments (weight: 1), and regularization of the SVF (bending energy: 50, divergence: 10, shears: 10).
Surface-based cell census
RMS image across the four echo times for the artifact corrected whole-hemisphere MEF MRI with flip angle of 20° was calculated, resampled to 600 μm isotropic, and segmented with Sequence Adaptive Multimodal SEGmentation (SAMSEG). White matter and pial surfaces were obtained with FreeSurfer, intermediate cortical surfaces were generated by growing the white matter toward the pial surface while minimizing variational energy, as described in (27). The center-of-mass coordinate, number of cells, and volume of each stereological sampling site were extracted from the stereology software. The site coordinates were warped to MRI space using the precomputed transformation. To compensate for local volume changes induced by cutting and histological processing, the site volumes were modulated by the Jacobian determinant of transformation. Each site was projected to its closest vertex across all 10 intermediate surfaces, and the local cell density was computed on each surface by smoothing the projected cell numbers and site volumes (three consecutive averages over the 1-ring neighbors) and taking their ratio.
Acknowledgments
We thank J. Glaser and J. McMullen (MBF Bioscience) for all the help during implementation of the Stereo Investigator Cleared Tissue software in our application and M. D. Tisdall who participated in building the MRI data handling infrastructure. Bioretics srl, a company specialized in Machine Learning solutions for Computer Vision, is a subcontractor of LENS in the framework of the BRAIN Initiative Cell Census Network (grant 1U01 MH117023-01). We express our gratitude to the donor involved in the body donation program of the Massachusetts General Hospital who made this study possible by donating his body to science. This publication was supported by and coordinated through NIH's Brain Research through Advancing Innovative Neurotechnologies (BRAIN) Initiative Cell Census Network (BICCN). This publication is part of the Human Cell Atlas, www.humancellatlas.org/publications/.
Funding: Support for this research was provided in part by the BRAIN Initiative Cell Census Network grant U01 MH117023; The National Institute for Biomedical Imaging and Bioengineering P41 EB015896, R01 EB023281, R01 EB006758, R21E B018907, R01 EB019956, P41 EB030006, and R00 EB023993; The National Institute on Aging R56 AG064027, R01 AG064027, R01 AG008122, and R01 AG016495; The National Institute of Mental Health R01 MH123195, R01 MH121885, and RF1 MH123195; The National Institute for Neurological Disorders and Stroke R01 NS0525851, R21 NS072652, R01 NS070963, R01 NS083534, U01 NS086625, U24 NS10059103, R01 NS105820, and R01 NS128843; Eunice Kennedy Shriver National Institute of Child Health and Human Development R21 HD106038; Chan-Zuckerberg Initiative DAF, an advised fund of Silicon Valley Community Foundation grant number 2019-198101; Shared Instrumentation Grants S10 RR023401, S10 RR019307, and S10 RR023043; NIH Blueprint for Neuroscience Research U01 MH093765, part of the multi-institutional Human Connectome Project; European Union’s Horizon 2020 research and innovation Framework Programme under grant agreement no. 654148 (Laserlab-Europe); European Union’s Horizon 2020 Framework Programme for Research and Innovation under the specific grant agreement no. 785907 (Human Brain Project SGA2) and no. 945539 (Human Brain Project SGA3); Marie Skłodowska-Curie grant agreement no. 793849 (MesoBrainMicr); Italian Ministry for Education in the framework of Euro-Bioimaging Italian Node (ESFRI research infrastructure); European Research Council (starting grant 677697, project BUNGEE-TOOLS); Alzheimer’s Research UK (Interdisciplinary grant ARUK-IRG2019A-003); The National Institute of Health R01 AG070988-01 and RF1 MH123195-01; and “Fondazione CR Firenze” (private foundation) Human Brain Optical Mapping. The content of this work is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health and other funding agencies.
Author contributions: D.V. developed the B0 and B1 transmit distortion correction methods and the associated imaging protocols to improve laminar and vessel contrast, optimized the scan protocol to improve overall CNR and SNR of MRI, and integrated intensity bias correction method into the processing pipeline. A.V.D.K. developed the custom sequences used to acquire high-resolution structural MRI data, built the custom data handling infrastructure, and oversaw the development of the MRI scan protocol with B.F. D.V., R.F., and L.M. contributed toward the development of the MRI scan protocol, image acquisition, and analysis. Y.B. contributed toward the development of the multimodal registration pipeline, building on original work done by J.E.I. and A.V.D. M.P.F. provided the human brain samples for this study. J.C.A. consulted on sample storage and treatment for preservation throughout the imaging pipeline, including consulting on histology protocols. L.M., D.C., M.Ve., J.N., K.N., J.M., E.G.P., and K.E. performed the ex vivo imaging and analysis team and are involved in image acquisition, data processing, and developing standards for segmenting vasculature and the infra/supragranular boundary of the cortex in MRI, OCT, and LSFM data. D.C., J.N., K.N., J.M., E.G.P., and K.E. performed manual labeling of these data. Labeling standards built on C.M.’s original groundwork for segmenting features of interest for the purpose of registering MRI to OCT data. C.M. also consulted on OCT imaging and analysis for the BU team. L.M. oversaw the integration of the image and analysis pipeline and data publication for this project. This builds on M.F.’s original project oversight and management. J.Y., S.C., H.W., and I.A.C. developed and performed image acquisition and data processing of OCT measurement. J.Y., S.C., and S.-.C.C built the serial sectioning vibratome. J.Y. coordinated the work on OCT sample preparation and transportation. I.C., L.P., and M.S. developed and performed the clearing and staining protocol; V.G., L.S., and G.M. built the LSFM hardware and software apparatus, and L.P. and V.G. performed light-sheet fluorescence imaging. G.M., F.M.C., V.G., and M.R. contributed to software, image processing, data analysis, and curation. I.C. coordinated the work on sample preparation, image acquisition, and data processing of LSFM measurement. B.W., M.V., S.R., and J.L. processed tissue for stereology and performed the stereologic analyses. B.F. helped design many of the algorithms used to analyze the data. F.S.P., D.A.B., B.F., and P.R.H. conceived and supervised the study. I.C., L.M., J.Y., Y.B., D.V., and P.R.H. wrote the paper with inputs from all authors.
Competing interests: B.F. has a financial interest in CorticoMetrics, a company whose medical pursuits focus on brain imaging and measurement technologies. B.F.’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies. M.R. is the CEO of Bioretics Srl, and F.M.C. is an employee of the company. Bioretics is specialized in Machine Learning solutions for Computer Vision. The authors declare that they have no other competing interests.
Data and materials availability: All the datasets acquired for this study are made available on the DANDI platform with the identification code DANDI:000026 at the link: https://gui.dandiarchive.org/#/dandiset/000026 (14). The protocols used in this study are made available at: https://protocols.io/view/high-resolution-ex-vivo-structural-mri-imaging-amp-q26g7823qlwz/v1, https://protocols.io/view/serial-sectioning-optical-histology-cell-census-ne-j8nlk4rb6g5r/v1, https://protocols.io/view/short-a-human-brain-tissue-clearing-and-labeling-p-kxygxp6nkl8j/v1, and https://protocols.io/view/stereologic-cell-population-estimates-in-cleared-h-4r3l273dqg1y/v1. The whole pipeline has the following RRID:SCR_023000, BICCN Imaging, and analysis Techniques to Construct Cell Census Atlas of Human Brain (DOI:10.1101/2021.10.20.464979). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All the codes used in this work are available on Zenodo, DOI: 10.5281/zenodo.7832049 (https://zenodo.org/record/7832049#.ZDz1cnZBzEY).
Supplementary Materials
This PDF file includes:
Figs. S1 to S4
Legends for movies S1 to S4
Other Supplementary Material for this manuscript includes the following:
Movies S1 to S4
REFERENCES AND NOTES
- 1.A. L. Tyson, T. W. Margrie, Mesoscale microscopy and image analysis tools for understanding the brain. Prog. Biophys. Mol. Biol. 168, 81–93 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.BRAIN Initiative Cell Census Network (BICCN) , A multimodal cell census and atlas of the mammalian primary motor cortex. Nature 598, 86–102 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.A. Franceschini, I. Costantini, F. S. Pavone, L. Silvestri, Dissecting Neuronal Activation on a Brain-Wide Scale With Immediate Early Genes. Front. Neurosci. 14, 569517 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.K. Amunts, C. Lepage, L. Borgeat, H. Mohlberg, T. Dickscheid, M. E. Rousseau, S. Bludau, P. L. Bazin, L. B. Lewis, A. M. Oros-Peusquens, N. J. Shah, T. Lippert, K. Zilles, A. C. Evans, BigBrain: An ultrahigh-resolution 3D human brain model. Science 340, 1472–1475 (2013). [DOI] [PubMed] [Google Scholar]
- 5.S.-L. Ding, J. J. Royall, S. M. Sunkin, L. Ng, B. A. C. Facer, P. Lesnar, A. Guillozet-Bongaarts, B. McMurray, A. Szafer, T. A. Dolbeare, A. Stevens, L. Tirrell, T. Benner, S. Caldejon, R. A. Dalley, N. Dee, C. Lau, J. Nyhus, M. Reding, Z. L. Riley, D. Sandman, E. Shen, A. van der Kouwe, A. Varjabedian, M. Wright, L. Zollei, C. Dang, J. A. Knowles, C. Koch, J. W. Phillips, N. Sestan, P. Wohnoutka, H. R. Zielke, J. G. Hohmann, A. R. Jones, A. Bernard, M. J. Hawrylycz, P. R. Hof, B. Fischl, E. S. Lein, Comprehensive cellular-resolution atlas of the adult human brain. J. Comp. Neurol. 524, 3127–3481 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.B. Fischl, A. A. Stevens, N. Rajendran, B. T. T. Yeo, D. N. Greve, K. Van Leemput, J. R. Polimeni, S. Kakunoori, R. L. Buckner, J. Pacheco, D. H. Salat, J. Melcher, M. P. Frosch, B. T. Hyman, P. E. Grant, B. R. Rosen, A. J. W. van der Kouwe, G. C. Wiggins, L. L. Wald, J. C. Augustinack, Predicting the location of entorhinal cortex from MRI. Neuroimage 47, 8–17 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.K. L. Miller, C. J. Stagg, G. Douaud, S. Jbabdi, S. M. Smith, T. E. J. Behrens, M. Jenkinson, S. A. Chance, M. M. Esiri, N. L. Voets, N. Jenkinson, T. Z. Aziz, M. R. Turner, H. Johansen-Berg, J. A. McNab, Diffusion imaging of whole, post-mortem human brains on a clinical MRI scanner. Neuroimage 57, 167–181 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.J. E. Iglesias, R. Insausti, G. Lerma-Usabiaga, M. Bocchetta, K. Van Leemput, D. N. Greve, A. van der Kouwe, B. Fischl, C. Caballero-Gaudes, P. M. Paz-Alonso, A probabilistic atlas of the human thalamic nuclei combining ex vivo MRI and histology. Neuroimage 183, 314–326 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.L. Larsen, L. D. Griffin, D. Grassel, O. W. Witte, H. Axer, Polarized light imaging of white matter architecture. Microsc. Res. Tech. 70, 851–863 (2007). [DOI] [PubMed] [Google Scholar]
- 10.P. Majka, S. Bai, S. Bakola, S. Bednarek, J. M. Chan, N. Jermakow, L. Passarelli, D. H. Reser, P. Theodoni, K. H. Worthy, X.-J. Wang, D. K. Wójcik, P. P. Mitra, M. G. P. Rosa, Open access resource for cellular-resolution analyses of corticocortical connectivity in the marmoset monkey. Nat. Commun. 11, 1133 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.J. F. De Boer, C. K. Hitzenberger, Y. Yasuno, Polarization sensitive optical coherence tomography – a review [Invited]. Biomed. Opt. Express 8, 1838–1873 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.H. R. Ueda, H. U. Dodt, P. Osten, M. N. Economo, J. Chandrashekar, P. J. Keller, Whole-brain profiling of cells and circuits in mammals by tissue clearing and light-sheet microscopy. Neuron 106, 369–387 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.L. Pesce, M. Scardigli, V. Gavryusev, A. Laurino, G. Mazzamuto, N. Brady, G. Sancataldo, L. Silvestri, C. Destrieux, P. R. Hof, I. Costantini, F. S. Pavone, 3D molecular phenotyping of cleared human brain tissues with light-sheet fluorescence microscopy. Commun. Biol. 5, 447 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.G. Mazzamuto, I. Costantini, V. Gavryusev, F. M. Castelli, L. Pesce, M. Scardigli, F. S. Pavone, M. Roffilli, L. Silvestri, P. R. Hof, D. A. Boas, B. Fischl, L. Morgan, J. Yang, S. Chang, J. Laffey, C. Magnain, D. Varadarajan, H. Wang, R. Frost, A. v. d. Kouwe, A. S. Player, A. Atzeni, J. E. I. Gonzalez, Y. Balbastre, M. Vera, D. Cordero, K. Nestor, W. Ammon, J. Nolan, J. Mora, E. G. Pallares, J. Augustinack, B. Diamond, M. Fogarty, E. Boyd, M. Varghese, A. V. Dalca, B. Edlow, M. Frosche, I.-C. A. Chen, B. Wicinski, "https://dandiarchive.org/dandiset/000026/draft" U01MH117023 (Version draft) [Data set] DANDI archive, (2021).
- 15.B. L. Edlow, A. Mareyam, A. Horn, J. R. Polimeni, T. Witzel, M. D. Tisdall, J. C. Augustinack, J. P. Stockmann, B. R. Diamond, A. Stevens, L. S. Tirrell, R. D. Folkerth, L. L. Wald, B. Fischl, A. van der Kouwe, 7 Tesla MRI of the ex vivo human brain at 100 micron resolution. Sci. Data 6, 244 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D. Varadarajan, R. Frost, A. van der Kouwe, L. Morgan, B. Diamond, E. Boyd, M. Fogarty, A. Stevens, B. Fischl, J. R. Polimeni, Edge-preserving B0 inhomogeneity distortion correction for high-resolution multi-echo ex vivo MRI at 7T. Abstract of International Society for Magnetic Resonance in Medicine, 664, (2020).
- 17.B. Fischl, D. H. Salat, A. J. W. van der Kouwe, N. Makris, F. Ségonne, B. T. Quinn, A. M. Dale, Sequence-independent segmentation of magnetic resonance images. Neuroimage 23, S69–S84 (2004). [DOI] [PubMed] [Google Scholar]
- 18.K. Van Leemput, F. Maes, D. Vandermeulen, P. Suetens, Automated model-based bias field correction of MR images of the brain. IEEE Trans. Med. Imaging 18, 885–896 (1999). [DOI] [PubMed] [Google Scholar]
- 19.J. Yang, I. A. Chen, S. Chang, J. Tang, B. Lee, K. Kiliç, S. Sunil, H. Wang, D. Varadarajan, C. Magnain, S.-C. Chen, I. Costantini, F. Pavone, B. Fischl, D. A. Boas, Improving the characterization of ex vivo human brain optical properties using high numerical aperture optical coherence tomography by spatially constraining the confocal parameters. Neurophotonics 7, 045005 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.T. Ragan, L. R. Kadiri, K. U. Venkataraju, K. Bahlmann, J. Sutin, J. Taranda, I. Arganda-Carreras, Y. Kim, H. S. Seung, P. Osten, Serial two-photon tomography for automated ex vivo mouse brain imaging. Nat. Methods 9, 255–258 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.L. Pesce, L. Annunziatina, G. Vladislav, M. Giacomo, S. Giuseppe, S. Marina, R. Matteo, S. Ludovico, C. Irene, P. Francesco Saverio, Fast volumetric mapping of human brain slices. Proc. SPIE 11360, (2020).
- 22.P. R. Hof, J. H. Morrison, Neocortical neuronal subpopulations labeled by a monoclonal antibody to calbindin exhibit differential vulnerability in Alzheimer's disease. Exp. Neurol. 111, 293–301 (1991). [DOI] [PubMed] [Google Scholar]
- 23.M. R. del Río, J. DeFelipe, Colocalization of calbindin D-28k, calretinin, and GABA immunoreactivities in neurons of the human temporal cortex. J. Comp. Neurol. 369, 472–482 (1996). [DOI] [PubMed] [Google Scholar]
- 24.L. Slomianka, Basic quantitative morphological methods applied to the central nervous system. J. Comp. Neurol. 529, 694–756 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.I. Costantini, R. Cicchi, L. Silvestri, F. Vanzi, F. S. Pavone, In-vivo and ex-vivo optical clearing methods for biological tissues: Review. Biomed. Opt. Express 10, 5251–5267 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.M. Morawski, E. Kirilina, N. Scherf, C. Jager, K. Reimann, R. Trampel, F. Gavriilidis, S. Geyer, B. Biedermann, T. Arendt, N. Weiskopf, Developing 3D microscopy with CLARITY on human brain tissue: Towards a tool for informing and validating MRI-based histology. Neuroimage 182, 417–428 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.B. Fischl, M. I. Sereno, Microstructural parcellation of the human brain. Neuroimage 182, 219–231 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.J. DeFelipe, P. L. López-Cruz, R. Benavides-Piccione, C. Bielza, P. Larrañaga, S. Anderson, A. Burkhalter, B. Cauli, A. Fairén, D. Feldmeyer, G. Fishell, D. Fitzpatrick, T. F. Freund, G. González-Burgos, S. Hestrin, S. Hill, P. R. Hof, J. Huang, E. G. Jones, Y. Kawaguchi, Z. Kisvárday, Y. Kubota, D. A. Lewis, O. Marín, H. Markram, C. J. McBain, H. S. Meyer, H. Monyer, S. B. Nelson, K. Rockland, J. Rossier, J. L. R. Rubenstein, B. Rudy, M. Scanziani, G. M. Shepherd, C. C. Sherwood, J. F. Staiger, G. Tamás, A. Thomson, Y. Wang, R. Yuste, G. A. Ascoli, New insights into the classification and nomenclature of cortical GABAergic interneurons. Nat. Rev. Neurosci. 14, 202–216 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.D. A. Lewis, A. A. Curley, J. R. Glausier, D. W. Volk, Cortical parvalbumin interneurons and cognitive dysfunction in schizophrenia. Trends Neurosci. 35, 57–67 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.S. Jacot-Descombes, N. Uppal, B. Wicinski, M. Santos, J. Schmeidler, P. Giannakopoulos, H. Heinsen, C. Schmitz, P. R. Hof, Decreased pyramidal neuron size in Brodmann areas 44 and 45 in patients with autism. Acta Neuropathol. 124, 67–79 (2012). [DOI] [PubMed] [Google Scholar]
- 31.T. Bussière, G. Gold, E. Kövari, P. Giannakopoulos, C. Bouras, D. P. Perl, J. H. Morrison, P. R. Hof, Stereologic analysis of neurofibrillary tangle formation in prefrontal cortex area 9 in aging and Alzheimer's disease. Neuroscience 117, 577–592 (2003). [DOI] [PubMed] [Google Scholar]
- 32.E. Murray, J. H. Cho, D. Goodwin, T. Ku, J. Swaney, S. Y. Kim, H. Choi, Y. G. Park, J. Y. Park, A. Hubbert, M. McCue, S. Vassallo, N. Bakh, M. P. Frosch, V. J. Wedeen, H. S. Seung, K. Chung, Simple, scalable proteomic imaging for high-dimensional profiling of intact systems. Cell 163, 1500–1514 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.I. Costantini, G. Mazzamuto, M. Roffilli, A. Laurino, F. Maria Castelli, M. Neri, G. Lughi, A. Simonetto, E. Lazzeri, L. Pesce, C. Destrieux, L. Silvestri, V. Conti, R. Guerrini, F. Saverio Pavone, Large-scale, cell-resolution volumetric mapping allows layer-specific investigation of human brain cytoarchitecture. Biomed. Opt. Express 12, 3684–3699 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.P. Frasconi, L. Silvestri, P. Soda, R. Cortini, F. S. Pavone, G. Iannello, Large-scale automated identification of mouse brain cells in confocal light sheet microscopy images. Bioinformatics 30, i587–593 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.L. Silvestri, M. C. Müllenbroich, I. Costantini, A. P. Di Giovanna, G. Mazzamuto, A. Franceschini, D. Kutra, A. Kreshuk, C. Checcucci, L. O. Toresano, P. Frasconi, L. Sacconi, F. S. Pavone, Universal autofocus for quantitative volumetric microscopy of whole mouse brains. Nat. Methods 18, 953–958 (2021). [DOI] [PubMed] [Google Scholar]
- 36.K. Chung, J. Wallace, S.-Y. Kim, S. Kalyanasundaram, A. S. Andalman, T. J. Davidson, J. J. Mirzabekov, K. A. Zalocusky, J. Mattis, A. K. Denisin, S. Pak, H. Bernstein, C. Ramakrishnan, L. Grosenick, V. Gradinaru, K. Deisseroth, Structural and molecular interrogation of intact biological systems. Nature 497, 332–337 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.A. P. Di Giovanna, A. Tibo, L. Silvestri, M. C. Mullenbroich, I. Costantini, A. L. A. Mascaro, L. Sacconi, P. Frasconi, F. S. Pavone, Whole-brain vasculature reconstruction at the single capillary level. Sci. Rep. 8, 12573 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.T. Bussière, P. Giannakopoulos, C. Bouras, D. P. Perl, J. H. Morrison, P. R. Hof, Progressive degeneration of nonphosphorylated neurofilament protein enriched pyramidal neurons predicts cognitive impairment in Alzheimer's disease: Stereologic analysis of prefrontal cortex area 9. J. Comp. Neurol. 463, 281–302 (2003). [DOI] [PubMed] [Google Scholar]
- 39.J. P. Roussarie, V. Yao, P. Rodriguez-Rodriguez, R. Oughtred, J. Rust, Z. Plautz, S. Kasturia, C. Albornoz, W. Wang, E. F. Schmidt, R. Dannenfelser, A. Tadych, L. Brichta, A. Barnea-Cramer, N. Heintz, P. R. Hof, M. Heiman, K. Dolinski, M. Flajolet, O. G. Troyanskaya, P. Greengard, Selective neuronal vulnerability in Alzheimer's disease: A network-based analysis. Neuron 107, 821–835.e12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.M. Wang, P. Roussos, A. McKenzie, X. Zhou, Y. Kajiwara, K. J. Brennand, G. C. De Luca, J. F. Crary, P. Casaccia, J. D. Buxbaum, M. Ehrlich, S. Gandy, A. Goate, P. Katsel, E. Schadt, V. Haroutunian, B. Zhang, Integrative network analysis of nineteen brain regions identifies molecular signatures and networks underlying selective regional vulnerability to Alzheimer's disease. Genome Med. 8, 104 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D. Velmeshev, L. Schirmer, D. Jung, M. Haeussler, Y. Perez, S. Mayer, A. Bhaduri, N. Goyal, D. H. Rowitch, A. R. Kriegstein, Single-cell genomics identifies cell type-specific molecular changes in autism. Science 364, 685–689 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.C. Munoz-Castro, M. Mejias-Ortega, E. Sanchez-Mejias, V. Navarro, L. Trujillo-Estrada, S. Jimenez, J. A. Garcia-Leon, J. J. Fernandez-Valenzuela, M. V. Sanchez-Mico, C. Romero-Molina, I. Moreno-Gonzalez, D. Baglietto-Vargas, M. Vizuete, A. Gutierrez, J. Vitorica, Monocyte-derived cells invade brain parenchyma and amyloid plaques in human Alzheimer's disease hippocampus. Acta Neuropathol. Commun. 11, 31 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.A. J. W. van der Kouwe, T. Benner, D. H. Salat, B. Fischl, Brain morphometry with multiecho MPRAGE. Neuroimage 40, 559–569 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.S. C. L. Deoni, T. M. Peters, B. K. Rutt, High-resolution T1 and T2 mapping of the brain in a clinically acceptable time with DESPOT1 and DESPOT2. Magn. Reson. Med. 53, 237–241 (2005). [DOI] [PubMed] [Google Scholar]
- 45.J. Golde, F. Tetschke, J. Walther, T. Rosenauer, F. Hempel, C. Hannig, E. Koch, L. Kirsten, Detection of carious lesions utilizing depolarization imaging by polarization sensitive optical coherence tomography. J. Biomed. Opt. 23, 1–8 (2018). [DOI] [PubMed] [Google Scholar]
- 46.M. Wojtkowski, V. J. Srinivasan, T. H. Ko, J. G. Fujimoto, A. Kowalczyk, J. S. Duker, Ultrahigh-resolution, high-speed, Fourier domain optical coherence tomography and methods for dispersion compensation. Opt. Express 12, 2404–2422 (2004). [DOI] [PubMed] [Google Scholar]
- 47.I. Arganda-Carreras, C. O. S. Sorzano, R. Marabini, J. M. Carazo, C. Ortiz-de-Solorzano, J. Kybic, Consistent and elastic registration of histological sections using vector-spline regularization. Conference paper on Computer Vision Approaches to Medical Image Analysis. 4241 (2006). [Google Scholar]
- 48.J. Yang, S. Chang, I. A. Chen, S. Kura, G. A. Rosen, N. A. Saltiel, B. R. Huber, D. Varadarajan, Y. Balbastre, C. Magnain, S.-C. Chen, B. Fischl, A. C. McKee, D. A. Boas, H. Wang, Volumetric characterization of microvasculature in ex vivo human brain samples by serial sectioning optical coherence tomography. I.E.E.E. Trans. Biomed. Eng. 69, 3645–3656 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.I. Costantini, J.-P. Ghobril, A. P. Di Giovanna, A. L. Allegra Mascaro, L. Silvestri, M. C. Mullenbroich, L. Onofri, V. Conti, F. Vanzi, L. Sacconi, R. Guerrini, H. Markram, G. Iannello, F. S. Pavone, A versatile clearing agent for multi-modal brain imaging. Sci. Rep. 5, 9808 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.L. Pesce, A. Laurino, M. Scardigli, J. Yang, D. A. Boas, P. R. Hof, C. Destrieux, I. Costantini, F. S. Pavone, Exploring the human cerebral cortex using confocal microscopy. Prog. Biophys. Mol. Biol. 168, 3–9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.I. Costantini, E. Baria, M. Sorelli, F. Matuschke, F. Giardini, M. Menzel, G. Mazzamuto, L. Silvestri, R. Cicchi, K. Amunts, M. Axer, F. S. Pavone, Autofluorescence enhancement for label-free imaging of myelinated fibers in mammalian brains. Sci. Rep. 11, 8038 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.M. Scardigli, L. Pesce, N. Brady, G. Mazzamuto, V. Gavryusev, L. Silvestri, P. R. Hof, C. Destrieux, I. Costantini, F. S. Pavone, Comparison of different tissue clearing methods for three-dimensional reconstruction of human brain cellular anatomy using advanced imaging techniques. Front. Neuroanat. 15, 752234 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O. E. Olarte, J. Andilla, E. J. Gualda, P. Loza-Alvarez, Light-sheet microscopy: A tutorial. Adv. Opt. Photonics 10, 111–179 (2018). [Google Scholar]
- 54.M. C. Mullenbroich, L. Silvestri, L. Onofri, I. Costantini, M. V. Hoff, L. Sacconi, G. Iannello, F. S. Pavone, Comprehensive optical and data management infrastructure for high-throughput light-sheet microscopy of whole mouse brains. Neurophotonics 2, 041404 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.V. Arsigny, O. Commowick, X. Pennec, N. Ayache, A log-Euclidean framework for statistics on diffeomorphisms. Med. Image Comput. Comput. Assist. Interv. 9, 924–931 (2006). [DOI] [PubMed] [Google Scholar]
- 56.P. Cachier, X. Pennec, 3D non-rigid registration by gradient descent on a Gaussian-windowed similarity measure using convolutions. Proceedings IEEE Workshop on Mathematical Methods in Biomedical Image Analysis. MMBIA-2000 (Cat. No.PR00737), 182–189 (2000).
- 57.P. F. Felzenszwalb, D. P. Huttenlocher, Distance transforms of sampled functions. Theory Comput. 8, 415–428 (2012). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S4
Legends for movies S1 to S4
Movies S1 to S4