Abstract
PURPOSE
Synthetic data are artificial data generated without including any real patient information by an algorithm trained to learn the characteristics of a real source data set and became widely used to accelerate research in life sciences. We aimed to (1) apply generative artificial intelligence to build synthetic data in different hematologic neoplasms; (2) develop a synthetic validation framework to assess data fidelity and privacy preservability; and (3) test the capability of synthetic data to accelerate clinical/translational research in hematology.
METHODS
A conditional generative adversarial network architecture was implemented to generate synthetic data. Use cases were myelodysplastic syndromes (MDS) and AML: 7,133 patients were included. A fully explainable validation framework was created to assess fidelity and privacy preservability of synthetic data.
RESULTS
We generated MDS/AML synthetic cohorts (including information on clinical features, genomics, treatment, and outcomes) with high fidelity and privacy performances. This technology allowed resolution of lack/incomplete information and data augmentation. We then assessed the potential value of synthetic data on accelerating research in hematology. Starting from 944 patients with MDS available since 2014, we generated a 300% augmented synthetic cohort and anticipated the development of molecular classification and molecular scoring system obtained many years later from 2,043 to 2,957 real patients, respectively. Moreover, starting from 187 MDS treated with luspatercept into a clinical trial, we generated a synthetic cohort that recapitulated all the clinical end points of the study. Finally, we developed a website to enable clinicians generating high-quality synthetic data from an existing biobank of real patients.
CONCLUSION
Synthetic data mimic real clinical-genomic features and outcomes, and anonymize patient information. The implementation of this technology allows to increase the scientific use and value of real data, thus accelerating precision medicine in hematology and the conduction of clinical trials.
Artificial intelligence–based synthetic data with high fidelity and privacy enhance precision hematology and clinical trials.
INTRODUCTION
Personalized medicine combines established clinical-pathologic parameters with advanced genomic profiling to develop innovative diagnostic, prognostic, and therapeutic strategies.1 Hematology has been rapidly transformed by genome characterization and is the forefront to reap the benefits of personalized medicine for patient management.1
CONTEXT
Key Objective
Are synthetic data able to recapitulate real clinical-genomic features and clinical outcomes, and to guarantee privacy preservability? Can this technology accelerate clinical/translational research in hematology?
Knowledge Generated
We developed a new technology on the basis of generative artificial intelligence that allows to generate synthetic patient cohorts with high clinical fidelity and privacy performances. We created a prototype web portal for synthetic data generation to help clinicians to be familiar with this new technology.
Relevance
This technology allows the resolution of lack/incomplete information and data augmentation starting from real patients. Synthetic data generate new knowledge to accelerate both translational research and the conduction of clinical trials.
The clinical implementation of personalized medicine requires the availability of a great amount of real-world data, including clinical features, genomics, treatments, and outcomes.2-4 Collecting such information in large patient populations is challenging, especially when facing rare diseases with heterogeneous clinical/molecular background. Additionally, real data often have imbalances or lack/incomplete information.5,6 Finally, there are many issues concerning patient privacy that may prevent use of data outside specific contexts and that are to be accounted for.7
One approach that can circumvent these issues is the creation of synthetic data. Synthetic data are artificial data generated by a model trained to learn the essential characteristics of a real source data set.8,9 Synthetic data building techniques attempt to ensure that the generated data are neither a copy nor a representation of the real data, setting the grounds to data sharing without violating the current legislation on privacy.8,9 Moreover, synthetic data allow to increase insufficient information obtained from real patients by data augmentation and data integration, thus potentially solving issues related with small sample size and clinical/molecular class imbalance.10
Overall, synthetic data may overcome many of the pitfalls of real data, allowing for faster, less expensive, and more scalable access to information that is representative of the underlying source and privacy-preserving.8-11 Synthetic data is a growing technology8 and it is expected that in the next 2-3 years, >60% of the data used in research and development process across different domains (including life sciences) will be synthetically generated.12
In this project, we addressed the issue of clinical validation and research utility of synthetic data in hematology. To this purpose, we aimed to (1) apply innovative synthetic data generation methods to real-world data sets of different hematologic malignancies including comprehensive clinical and genomic information; (2) develop a synthetic validation framework (SVF) to evaluate data fidelity and privacy preservability; and (3) test the capability of synthetic data to accelerate translational and clinical research.
As a paradigmatic use case, we focused on myeloid malignancies, which are rare neoplasms with high clinical heterogeneity and complex genomic background and that include patients with unmet clinical needs.13
METHODS
Study Populations
The study was conducted by GenoMed4All and Synthema European consortia and supported by EuroBloodNET, the European Reference Network on rare hematologic diseases. Written informed consent was obtained from each participant. The Humanitas Ethics Committee approved the study. This study was registered at ClinicalTrials.gov (identifier: NCT04889729).
All the study procedures were compliant with the 2021 WHO guidance on ethics and governance of artificial intelligence for health.14
Inclusion criteria were age ≥18 years, a diagnosis of myeloid neoplasm (either myelodysplastic syndromes [MDS] or AML) according to WHO 2016 criteria,15 and information available on demographics, clinical features, mutational screening/chromosomal abnormalities, treatment, and survival. Overall, 7,133 patients were included.
Generative Model for Synthetic Data
Artificial intelligence (AI)–based generative models are characterized by multi-layer neural networks that are able to generate samples (patients) by learning the distribution of a set of real data.16 In this context, generative adversarial networks (GANs)17 create simulation scenarios where models and processes interact to create completely new data sets of events. GANs consist of two networks: the generator and the discriminator. These two networks are trained adversarially. The generator creates artificial outputs that are passed to the discriminator along with real data, while the discriminator is tasked to identify which outputs were real and which were fake. The final goal here is to reach equilibrium, in which the generated samples follow the same distribution as the real data. When this happens, the discriminator can do no better than random guessing.16 Conditional GANs are variants of GANs where a label is added as a parameter to the input of the models to create more realistic data by learning specific correlations.18 In this study, we implemented a conditional Wasserstein's tabular GAN18 with gradient penalty19 that ensures high performance in modeling large data sets with complex distribution and interactions among different features. We adopted different preprocessing steps and training strategies to properly prepare the input data and optimize the training steps.
Development of a Synthetic Validation Framework
A SVF was developed to evaluate fidelity and privacy preservability of the newly generated synthetic data.
We assessed the quality of the following data types: demographics, clinical features, genomics (evaluated as categorical variables), and clinical outcomes (probability of overall survival and leukemia-free survival). Distribution, correlation, and principal component analysis evaluation were then assessed on all data types. Descriptive statistics and pairwise association analyses were carried out. We calculated a clinical synthetic fidelity (CSF) and a genomic synthetic fidelity (GSF) as the average of multiple metric tests adopted; optimal threshold was considered ≥85% in both systems.
Real and synthetic patients were stratified by hierarchical Dirichlet clustering20 to identify genomic associations and subgroups. Survival analyses were performed with Kaplan-Meier curves. We implemented Cox proportional hazard and L1-penalized Cox regression models to define features with significant impact on survival probability.20,21 Model discrimination was assessed using Harrell's concordance index.22
To assess the privacy preservability and evaluate the risk associated with synthetic datasets of resampling a patient from a synthetic record, we first measured the exact matches between synthetic and original data (identical match share [IMS]). Moreover, we calculated the distance to closest record that measures the absolute distances between synthetic records to their nearest original records, and we then calculated the nearest neighbor distance ratio (NNDR), that is, the ratio of the distances of each synthetic record to the nearest and to the second nearest neighbors, that allows to compare inliers and outliers in the population on an equal base.23 Optimal range for NNDR was considered from 0.60 to 0.85 (value closer to 0.50 indicating a significant loss of similarity of the synthetic patients compared with the real ones that can affect the fidelity of synthetic data; value closer to 1.00 indicating an excess of similarity of synthetic data with respect to the real ones, thus possibly affecting the privacy preservability).23
Explainability of AI algorithms was assessed by Shapley Additive Explanations (SHAP), a method to explain individual predictions on the basis of the game theoretically optimal Shapley values.24
Experimental Setup
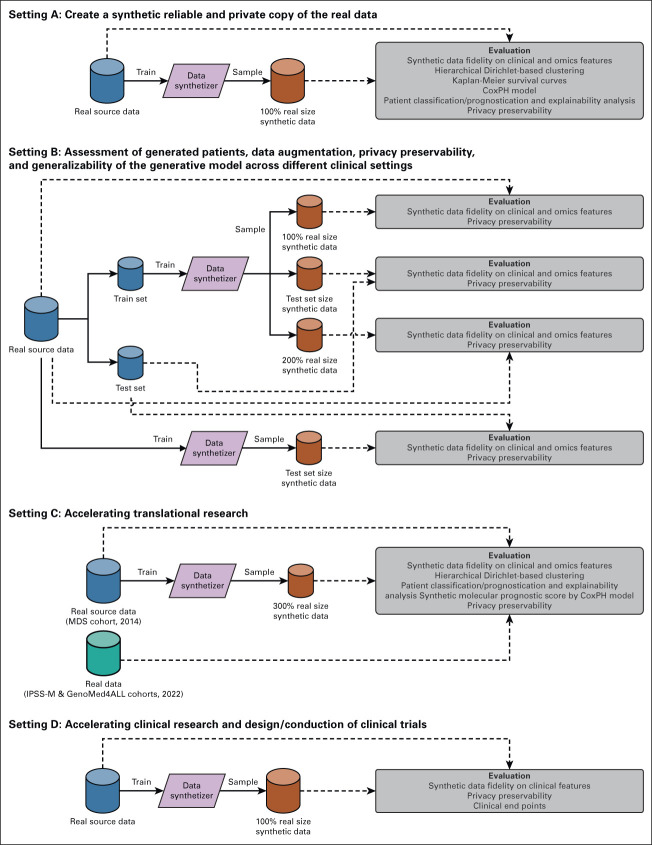
We tested synthetic data generation process in different experimental settings (Fig 1).
FIG 1.
Overview of experimental settings to validate synthetic data. Setting A: Create a synthetic reliable and private copy of the real data. Setting B: Assessment of generated patients, data augmentation, privacy preservability, and generalizability of the generative model across different clinical settings. Setting C: Accelerating translational research. Setting D: Accelerating clinical research and design/conduction of clinical trials. IPSS-M, Molecular International Prognostic Scoring System; MDS, myelodysplastic syndromes.
In setting A, we investigated the capability of the generative model to create a synthetic reproduction of real data with high grade of fidelity on clinical/genomic features, clinical outcomes, and with high privacy preservability. We used 2043 patients with MDS from GenoMed4All cohort20 to train and test the model.
In setting B, we tested the capability of the model to overcome lack/incomplete information in real data and to allow data augmentation; moreover, we assessed the generalizability of the model's performances across different clinical settings. We considered three different populations: 2,043 MDS from GenoMed4All cohort20; 2,957 MDS from the International Working Group for Prognosis in MDS (IWG-PM) cohort,21 and 1,002 AML from GenoMed4All cohort.20 In all experiments, we calculated fidelity and privacy metrics.
In setting C, we investigated if the generation of synthetic data can accelerate translational research. Starting from a MDS cohort available in 2014 (N = 944),25 we generated a 300% augmented synthetic data set. We aimed to recapitulate and anticipate in this cohort of synthetic patients the most relevant and recent insights in personalized medicine (ie, the definition of a new molecular MDS classification and of a molecular scoring system, developed on 2,043 and 2,957 real patients in 2022, respectively).20,21
In setting D, we generated synthetic patients to be used as a control arm in clinical trials, thus possibly accelerating clinical development of new drugs/new indications of existing drugs. Starting from 187 MDS treated with luspatercept into a multicenter clinical trial,26 we generated a new synthetic cohort of the same size. Then, we tested the capability of newly generated synthetic patients to recapitulate all the clinical end points of the original study.
RESULTS
Creation of a Synthetic, Reliable, and Private Reproduction of Real Data (Setting A)
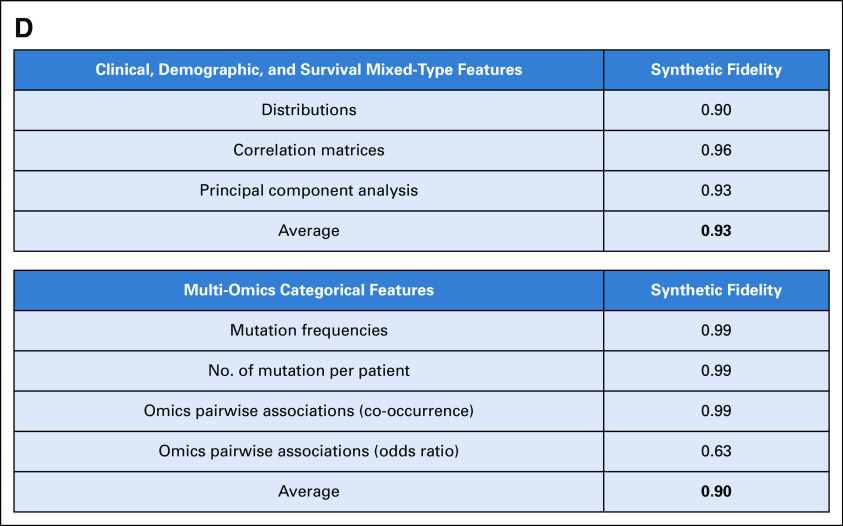
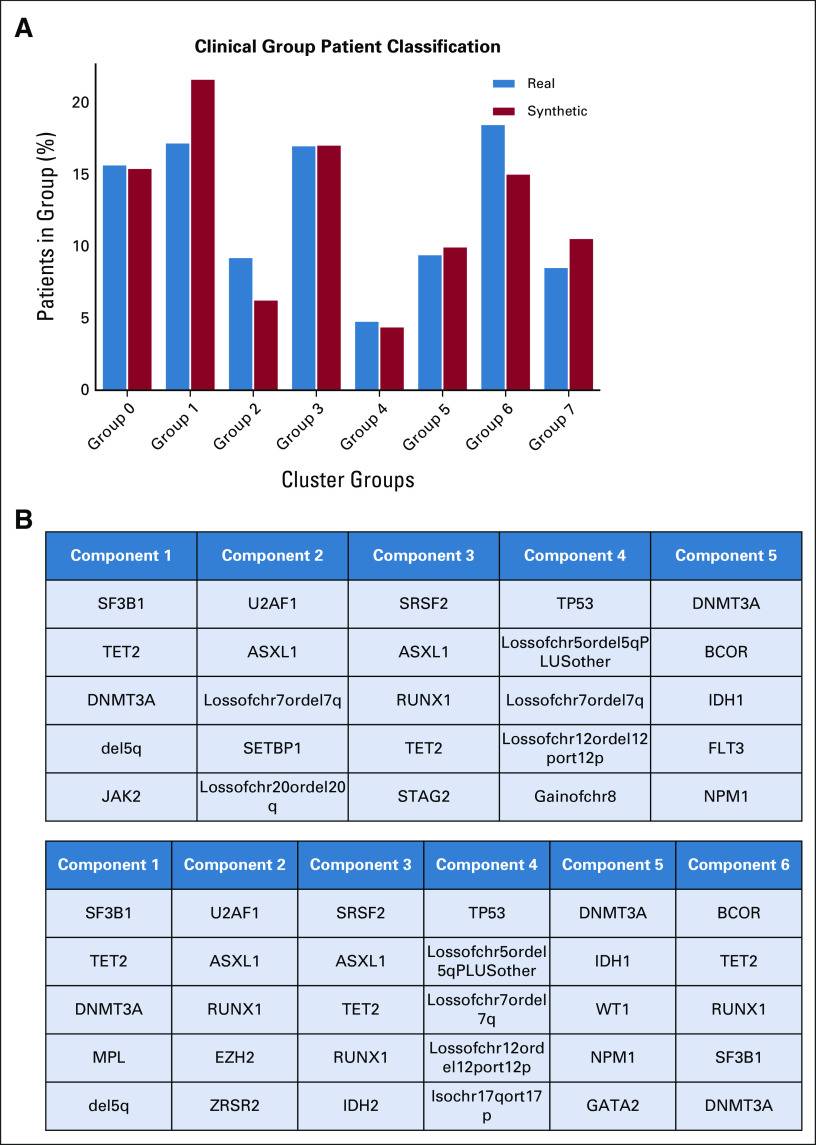
We used 2,043 real MDS from GenoMed4All cohort20 to generate a new cohort of 2,043 synthetic patients. The model showed high-fidelity performances for both clinical and genomic features (CSF = 93%; GSF = 90%; Fig 2 and Appendix Fig A1). We then applied Dirichlet processes to compare complex interactions and broad dependencies among genomic features in real versus synthetic patients and we obtained highly comparable results; explainability analysis (SHAP) showed that similar features drive patients' classification in both data sets (Appendix Fig A2).
FIG 2.
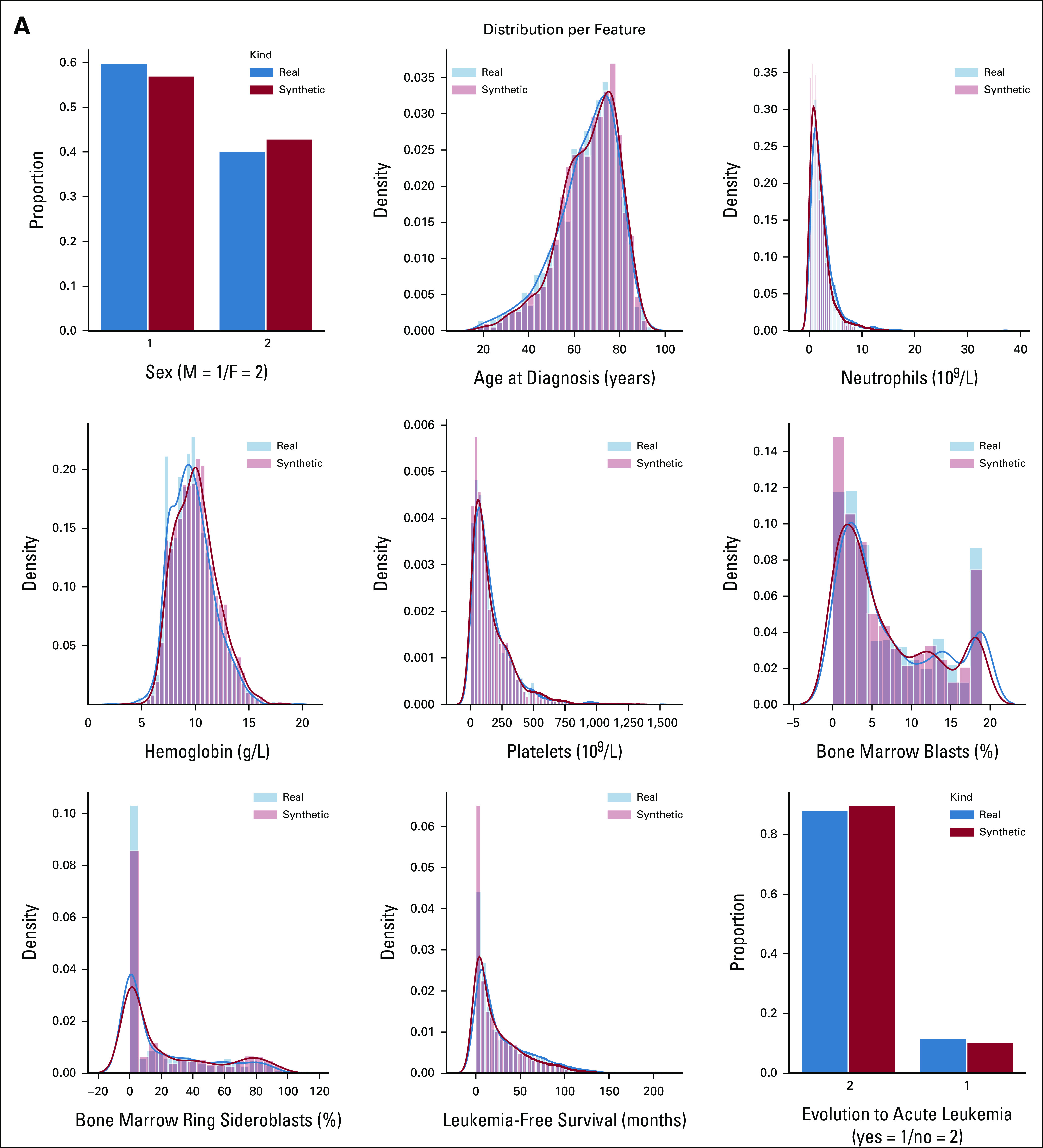
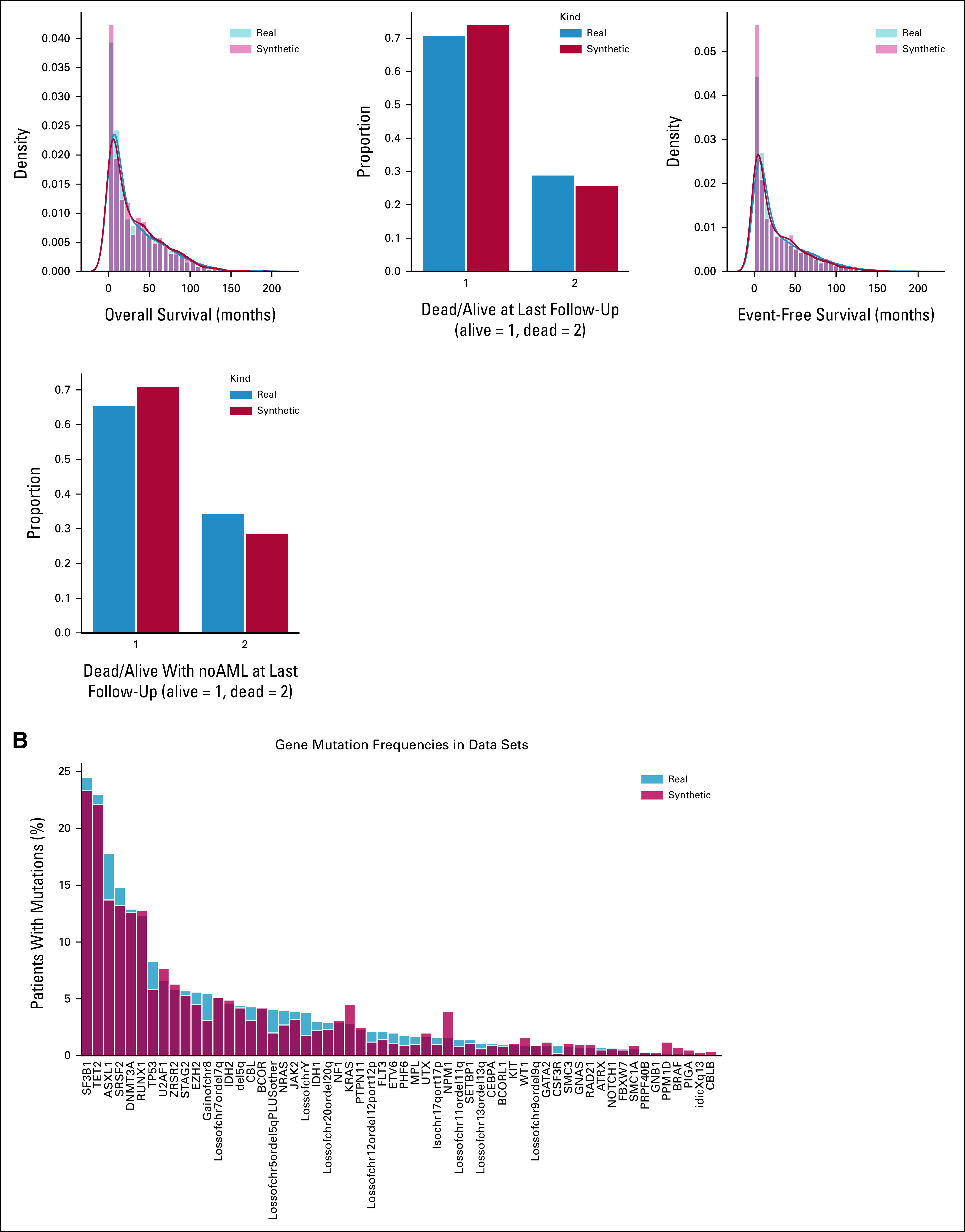
SVF on synthetic MDS cohort (N = 2,043), as performed in setting A. (A) Distributions for clinical, demographic, and survival features. Blue illustrates the real data, while red illustrates the synthetic data. (B) Frequency of recurrently mutated genes and chromosomal abnormalities. (C) Pairwise association among genes and/or cytogenetics abnormalities. In the upper triangle, for each couple of genomic abnormalities, the numbers of patients showing mutation co-occurrences are illustrated using a blue and white color scale. In the lower triangle, the gene-gene co-occurrence and mutual exclusivity is assessed using odds ratio, illustrated using a green and yellow color scale according to odds ratio values. All results in (A), (B), and (C) are referring to one MDS synthetic data set of 2,043 patients generated. Detailed results are reported in the Data Supplement. (D) Synthetic data fidelity calculated by SVF on clinical, demographic, and genomic features and patient survival. Average over three training and sampling replications on MDS cohort of 2,043 patients. MDS, myelodysplastic syndromes; SVF, synthetic validation framework.
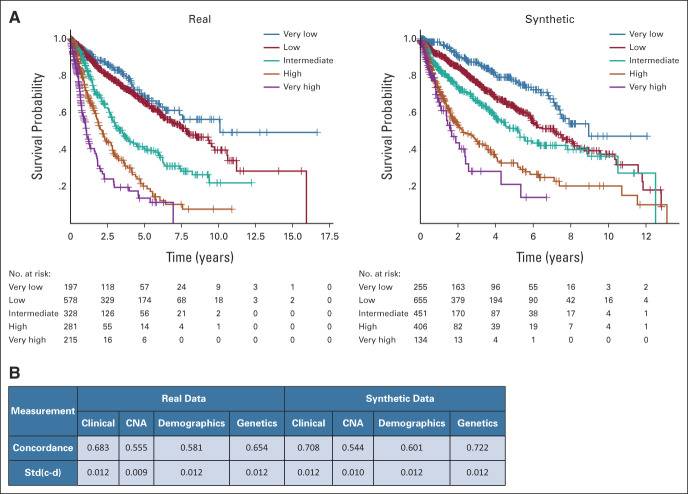
Synthetic patients had comparable survival outcomes with respect to the real ones. When applying the reference scoring system for MDS prognostication (Revised International Prognostic Scoring System), the probability of survival of the five risk categories between synthetic and real patients was comparable (Fig 3).
FIG 3.
Patient classification and survival analysis on the synthetic MDS cohort (N = 2,043), as performed in setting A. (A) Kaplan-Meier survival probability curves obtained from the real (left) and synthetic (right) populations, stratified according to IPSS-R risk categories. The P values of the log-rank test are calculated, confirming the hypothesis of no difference in survival probabilities between real and synthetic patients for every IPSS-R risk group. (B) Partial concordance and standard error for each category of variables obtained from the mixed-effect CoxPH models fitted on the real and synthetic cohorts. CNA, copy number alteration; IPSS-R, Revised International Prognostic Scoring System; MDS, myelodysplastic syndromes.
We build a CoxPH model including all features of prognostic relevance with a unique binary covariate (indicating the belonging of the patient to the real or the synthetic data set) that obtained a P value of .742, suggesting that there is no significant difference in the survival probability between the two cohorts in a multivariable setting (Fig 3). Concordances obtained for the different category included in the model (demographics, clinical, and genomics) were comparable in both cohorts. Considering the global concordance of the model, we obtained similar results with the model fitted on real versus synthetic data (0.736 ± 0.012 v 0.769 ± 0.012; Fig 3).
In terms of privacy metrics, the IMS analysis showed that none of the real patients were copied in the synthetic dataset; moreover, we obtained good results for NNDR (0.64), indicating adequate distance to real data and poor privacy risk.23
Resolution of Lack/Incomplete Information, Data Augmentation, Privacy Preservability, and Generalizability of the Model Across Different Clinical Settings (Setting B)
Starting from the MDS GenoMed4all cohort (N = 2,043),20 we trained the model with a set of a smaller size (including 70% of the patients) and then with a set with 30% of missing information across all features. We obtained the same high-fidelity performances as in setting A, in which synthetic patients were generated form the whole real data set (CSF and GSF were >90% in both experiments).
Then we generated a 200% augmented data set of synthetic MDS patients, resulting into a high fidelity of the model (CSF = 91%; GSF = 89%) that was maintained when comparing the synthetic data sets with the real test set never seen by the model during the training phase (CSF = 90%; GSF = 88%).
When considering a more complex data set (IWG-PM MDS cohort, N = 2,604) including a higher number of genomic features (245 v 65), we obtained comparable fidelity performances to the previous experiments (CSF = 93%; GSF = 93%).
Importantly, a similar trend was noted by replicating all these experiments in a cohort of 1,002 synthetic patients with AML generated form an equal number of real subjects (CSF > 90%; GSF > 88% in all cases), thus providing evidence for a generalizability of the generative model across different clinical settings.
In terms of privacy metrics, in all experiments on the three different synthetic patient populations, the IMS analysis showed that none of the real patients were copied in the synthetic data sets; moreover, we obtained similar good distance results in all experiments for NNDR (values from 0.60 to 0.71).
Accelerating Translational Research by Synthetic Data (Setting C)
Starting from a MDS cohort available in 2014 (N = 944),25 we generated a 300% augmented synthetic data set of 2,832 patients. Fidelity and privacy performances were comparable with previous experiments (CSF = 92%; GSF = 89%; NNDR = 0.62). We aimed to recapitulate and anticipate in this cohort of synthetic patients the most relevant insights in the field of personalized medicine (ie, the definition of new molecular MDS classification provided on a cohort of 2,043 real patients20 and the definition of the Molecular International Prognostic Scoring Systems [IPSS-M], defined on a cohort 2,897 real patients21).
First, Dirichlet processes were applied to synthetic data to define genomic-based clinical entities, resulting in the identification of the same eight disease categories described in a real cohort of 2,043 patients in 2022. Patients' classification into clinical groups followed a similar distribution as the real cohort, and explainability analysis (SHAP) also showed that similar features drive the patients' classification in both data sets (Fig 4 and Appendix Fig A3).
FIG 4.
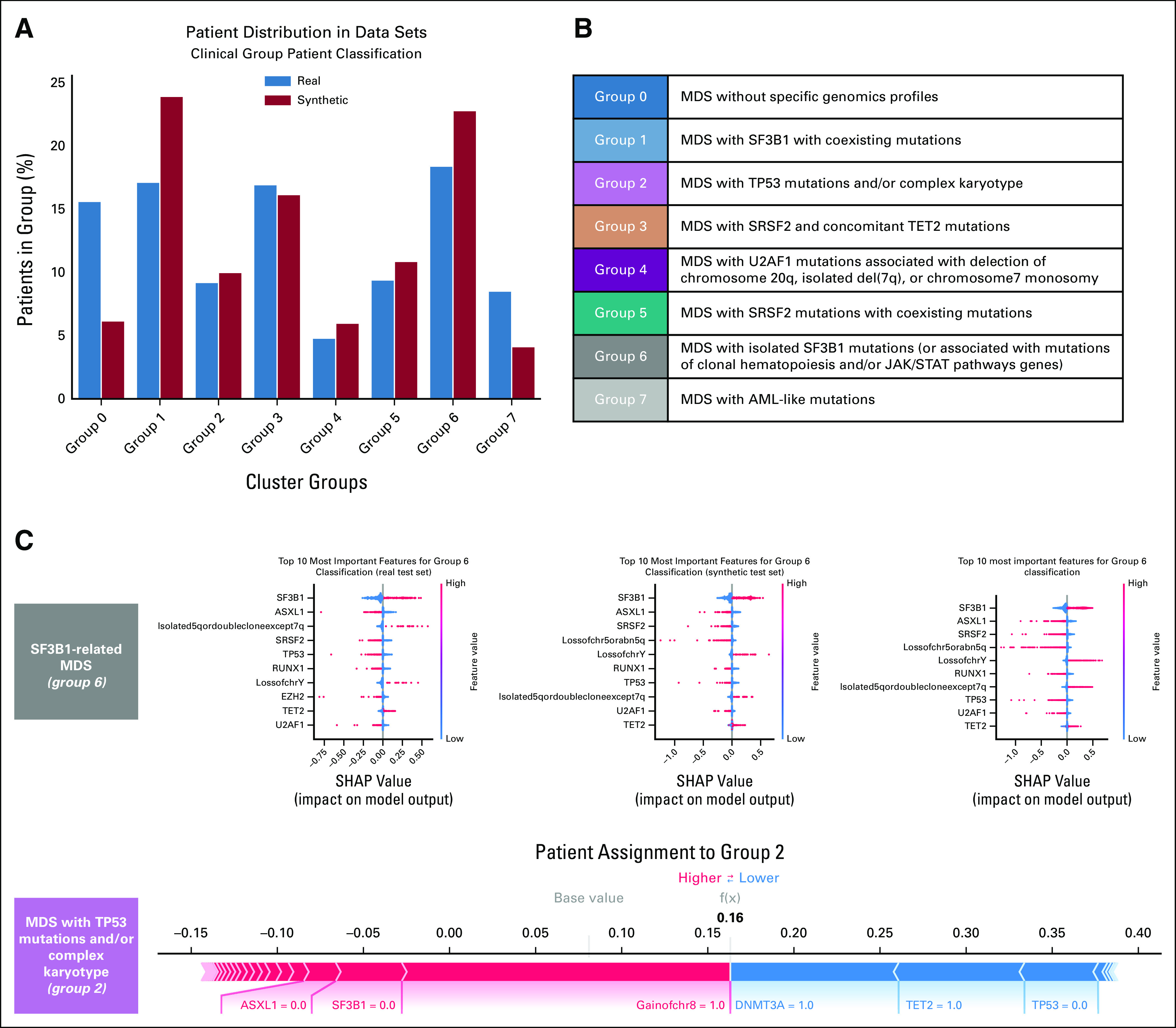
Definition of a molecular classification on augmented synthetic MDS cohort starting from 944 patients available in 2014, as performed in setting C. (A) Evaluation of the real (blue) and synthetic (red) patients' distribution considering genomic groups classification. (B) Genomic group definition according to Bersanelli et al.20 (C) SHAP summary plot analysis on the top 10 most important features for a real test set, a synthetic test set, and a complete augmented synthetic data set for the genomic group 6. Below is the force plot showing the importance of the most relevant features in assigning a synthetic patient to genomic group 2. MDS, myelodysplastic syndromes; SHAP, Shapley Additive Explanations.
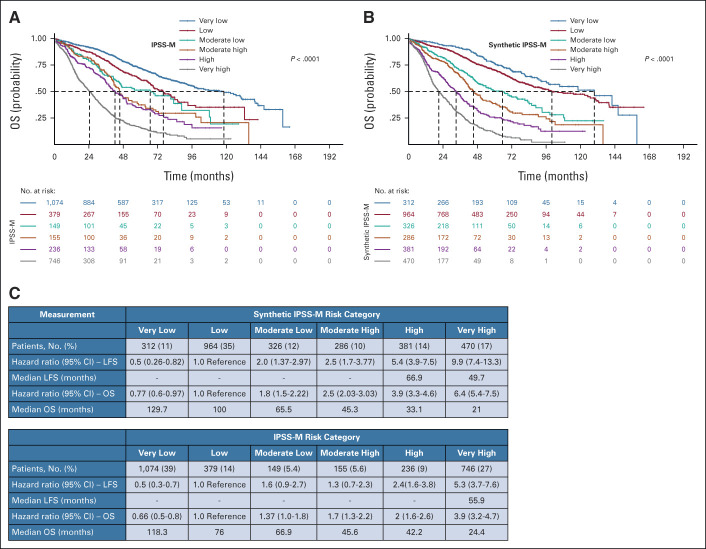
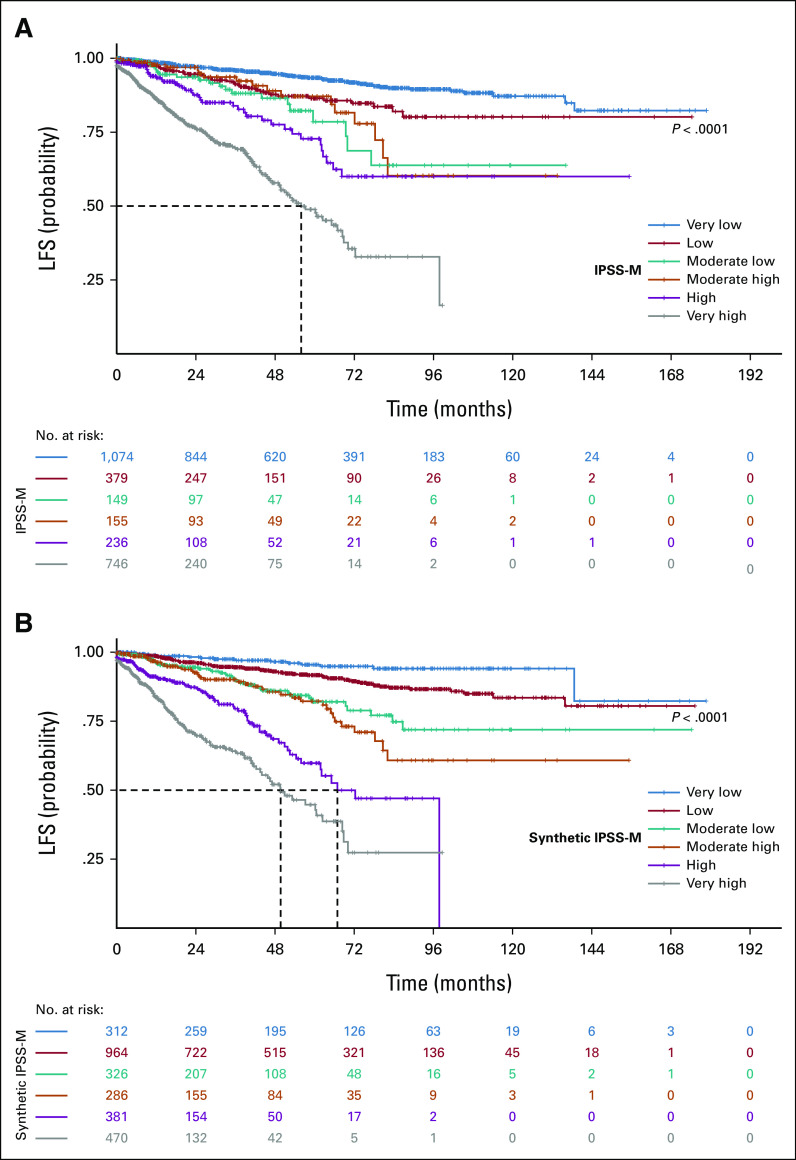
As a second experiment, we applied a L1-penalized Cox regression model to the synthetic data set of 2,832 patients to generate a molecular prognostic score (synthetic IPSS-M). After feature selection, we developed a prognostic tool on the synthetic cohort and compared it with IPSS-M developed on real patients. The comparison of the two scores reveals the same feature extraction and the identification of six risk categories with comparable probability of overall survival and leukemia-free survival (Fig 5 and Appendix Fig A4).
FIG 5.
Survival analysis on synthetic molecular prognostic score generated (synthetic IPSS-M) performed in setting C. (A) Kaplan-Meier probability estimates of OS for synthetic patients with MDS are represented and stratified by IPSS-M risk categories as defined by Bernard et al.21 P value is from log-rank test. (B) Kaplan-Meier probability estimates of OS for synthetic patients with MDS are represented and stratified by synthetic IPSS-M risk categories. P value is from log-rank test. (C) Percentage of patients in each IPSS-M risk category (both synthetic and original) with the HRs for each outcome, and the median survival for each patient class, where values could be calculated. HR, hazard ratio; IPSS-M, Molecular International Prognostic Scoring System; LFS, leukemia-free survival; MDS, myelodysplastic syndromes; OS, overall survival.
Accelerating Clinical Research and Conduction of Clinical Trials by Using Synthetic Data (Setting D)
We investigated the possibility to use a synthetic data set as a comparison group in a clinical trial. We therefore aimed to replicate a real patient cohort from a multicenter study including 187 patients with MDS who were treated with luspatercept.26
Eligible patients were age 18 years or older and had an MDS with ring sideroblasts; were receiving regular red blood cells transfusions; and were refractory to erythropoiesis-stimulating agent therapy. Primary end point was transfusion independence (TI) for ≥8 weeks during weeks 1-24; key secondary end point was TI for ≥12 weeks during both weeks 1-24 and 1-48.
We generated a synthetic cohort (N = 187) from the patients included in the study using all data for training, and we compared the synthetic end points with the original study results. All the characteristics and metrics of the synthetic cohort were comparable with respect to the original data set, as shown in Figure 6 and Appendix Figure A5, with high efficient coefficient of privacy preservability (NNDR = 0.71).
FIG 6.
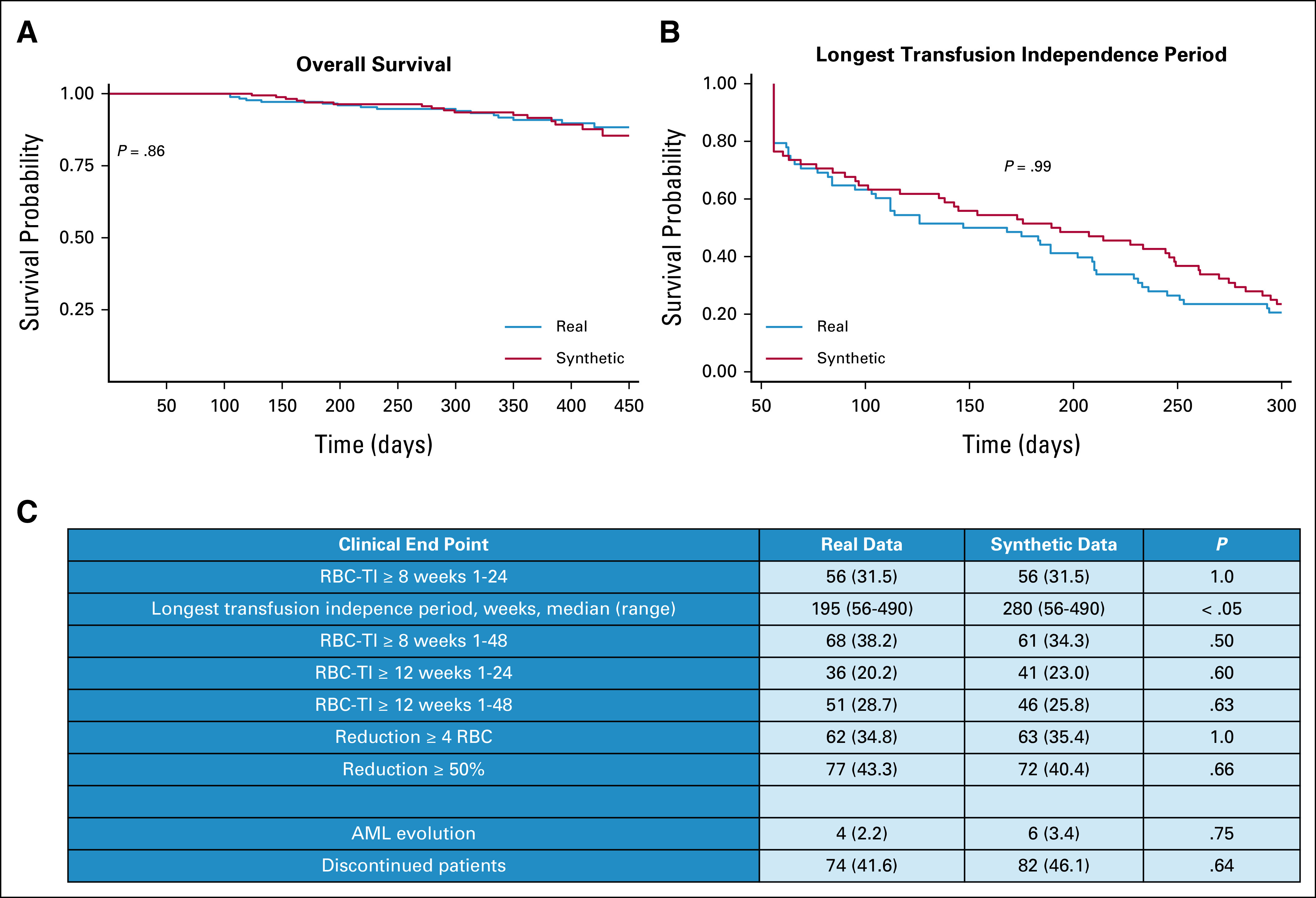
Comparison of clinical trial end points between real and synthetic patients, as performed in setting D. (A) Kaplan-Meier survival probability curves compared for real and synthetic patients' overall survival. (B) Kaplan-Meier curves of longest transfusion independence period for real and synthetic patients. The P values of the log-rank test are calculated, confirming the hypothesis of no difference in survival probabilities between real and synthetic cohorts. (C) Study end point comparison between real and synthetic cohorts. RBC-TI, rate of red blood cell transfusion independence.
Generator of Synthetic Data
To help clinicians to be familiar with generative AI to build synthetic data, we have created a prototype web portal27 that allows to generate synthetic patients starting from 2,957 real MDS of IWG-PM cohort.21 This portal allows to generate synthetic cohorts with different sizes, to verify the performance of the newly generated data (fidelity and privacy preservability), and to download the synthetic data set for research use.
DISCUSSION
In this study, we showed that synthetic data may (1) efficiently recapitulate statistical properties and complex interactions between clinical and genomic features in hematologic malignancies; (2) replicate reliable estimates of survival and effectiveness of specific treatments; (3) overcome lack/imbalance of information of real data; and (4) allow effective data augmentation.
The implementation of this technology may allow to increase the scientific use and value of real data, and it is expected to accelerate precision medicine in hematology and the conduction of clinical trials.
To help clinicians to be familiar with this new technology, we created a prototype web portal that allows to generate synthetic data from a real data set of patients with clinical and genomic information, and that provides a report of the quality of the newly generated synthetic patients.
The implementability of synthetic data in translational and clinical research depends on two main properties: (1) fidelity, ie, the newly generated data should be plausible and preserve structural properties of the real data; (2) privacy, that is, it should be possible to precisely quantify how much information about the original data is revealed through the releasing of the synthetic sample.28,29
The use of generative AI rapidly increased the implementation of synthetic data in life sciences in past years.8-10 As an example, SyntheticMass hosts over one million synthetic patient records from the state of Massachusetts.30 In Europe, synthetic data sets that mimic a part of the Netherlands Cancer Registry and Public Health England's Cancer Registration are now available for research purposes.31,32 The creation of a synthetic data bank makes the information accessible while also streamlining the data sets that medical research teams have to work with. But, there are limitations: the more complex the data query, the more approximate the results; in particular, the generation of high-fidelity synthetic patients with comprehensive clinical and genomic information reproducing complex interactions among different data layers is still a challenge.8-10
In this study, we used an optimized method (conditional GAN)17-19 to recapitulate clinical and genomic properties of real patients with myeloid neoplasms, which are rare diseases characterized by large clinical and biological heterogeneity.13,15 The methodologic advantage of conditional GAN allowed us to face specific challenges in research on rare diseases (such as lack/imbalance of data) and we provided evidence for a high generalizability of the performances of the model across different clinical settings.
Synthetic data require an extensive validation of their reliability in recapitulating properties of real patients.8-10,28-30 We therefore created a SVF to perform a clear fidelity analysis of clinical, survival, and genomic information and that may represent a solid basis to define the quality of a newly generated synthetic data set. Moreover, we implemented a comprehensive approach for data explainability,24 thus facilitating the clinical interpretation of the results of deep learning analysis on synthetic data.
Sharing data has the potential to improve decision making and accelerate research and innovation.2-4,11 At the same time, many data are highly sensitive and sharing them may violate fundamental rights guarded by modern privacy regulations.7,11 Anonymization (where potentially identifiable variables are removed) is one way to make data available; however, intensive anonymization can degrade the data to the extent that they are no longer fit for purpose. Moreover, several reidentification attempts on anonymized data have been successful and have harmed public and regulators' trust in such methods.33,34 We showed that generative AI can guarantee a high privacy preservability of newly generated synthetic data. We focused on analyzing the distance between the real and synthetic patients and we showed that there was enough distance between the real and synthetic patients to avoid the risk of revealing sensitive information from the training data and not too far away to maintain correlations of the source real population.23
We provided evidence that synthetic data can accelerate translational research in hematology. Since the first publication on clinical relevance of gene mutations in MDS, it took several years to collect real large patient populations for defining a molecular classification and molecular prognostic score.20,21 By generating synthetic data from a relative small cohort of patients available in 2014,25 we were able to recapitulate the definition of genomic-based subgroups and of a molecular prognostic score as described in real cohorts many years later.20,21
Finally, synthetic patients could be used in the future to improve the conduction of clinical trials. The use of synthetic control arms may reduce clinical trial costs and duration. Moreover, using a synthetic control arm may ensure that all participants receive the active treatment, thus eliminating patient concerns about treatment assignment.35
Secondary analyses of data from clinical trials can provide new insights compared with the original publications.36 In this context, our findings suggest that generative AI can create synthetic patients that efficiently reproduce clinical characteristics and efficacy end points of the original study and that can be promptly available for secondary analyses.
As a possible improvement of our approach, recently, GAN technology was optimized to generate synthetic patients with time-series records and longitudinal evaluation of treatment response (multilabel time-series GAN [MTGAN]).37 MTGAN can preserve temporal information by developing a temporally correlated generation process, thus finally increasing the generation quality of uncommon diseases and the performance of predictive models.
To maximize the impact of this technology in accelerating precision medicine in hematology, it will be relevant to develop regulatory frameworks involving synthetic data and to define standards for synthetic data quality and privacy preservability.8,12
APPENDIX
FIG A1.
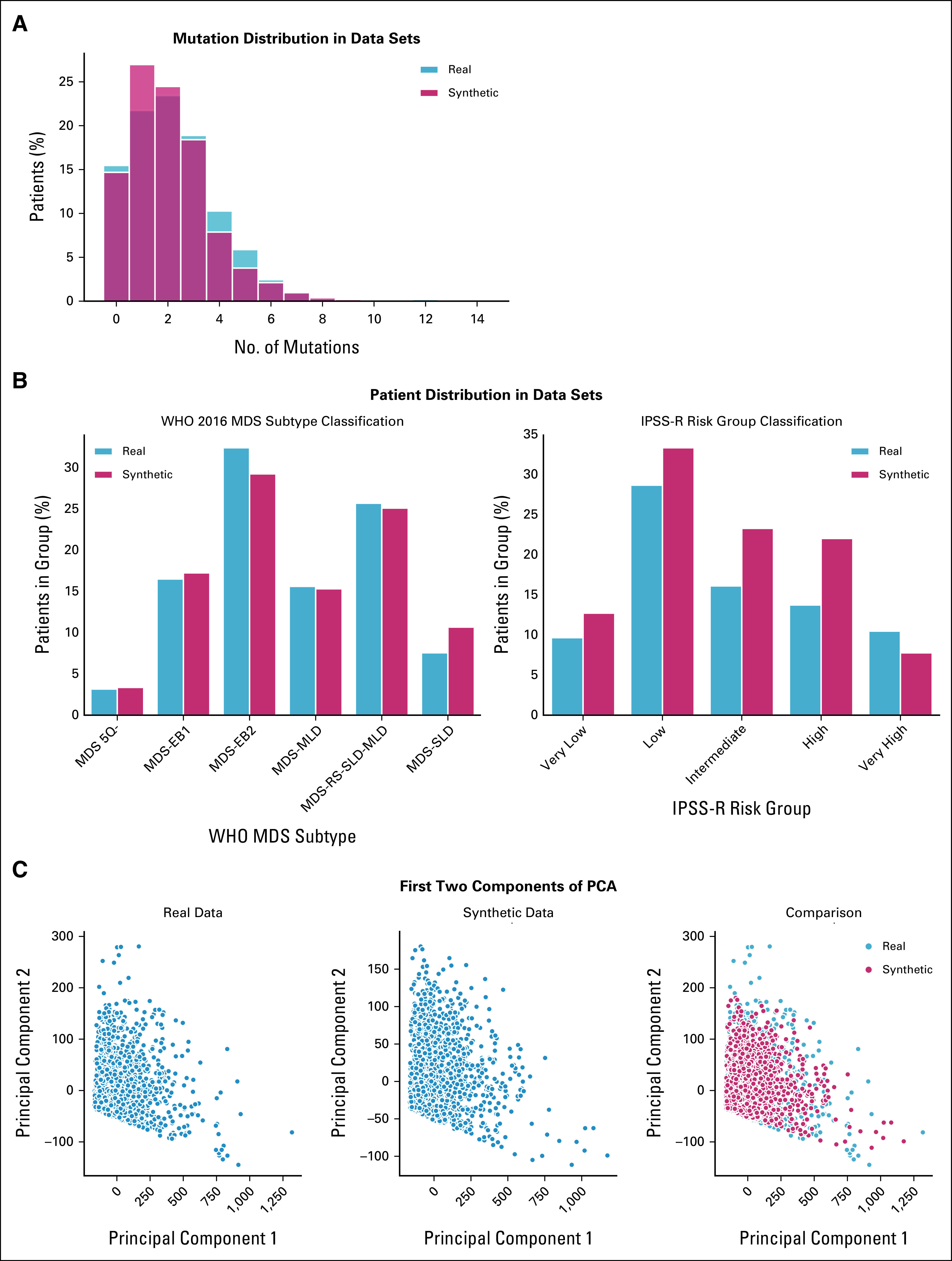
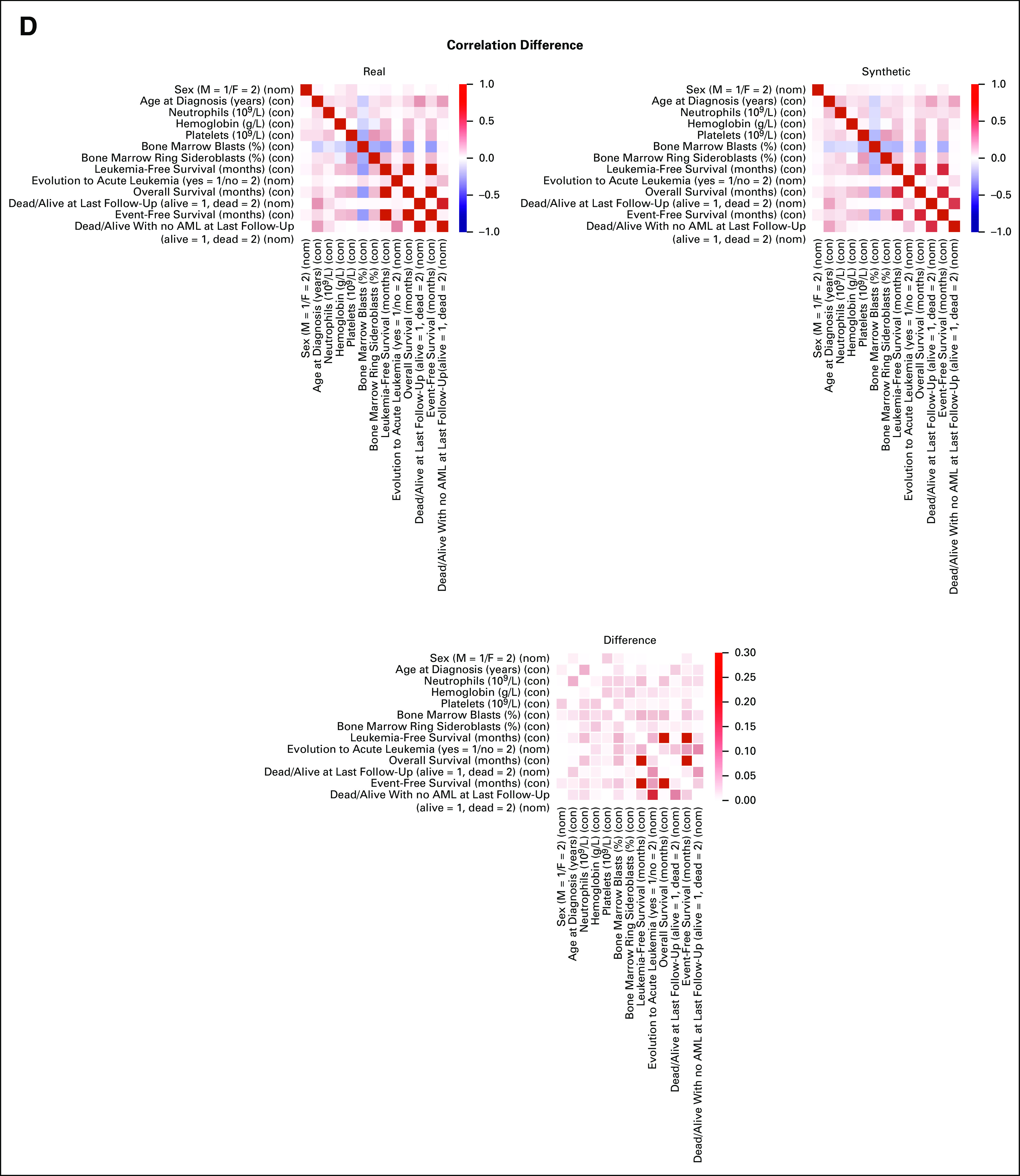
SVF on synthetic MDS cohort (N = 2,043), as performed in setting A. (A) Distributions of the patients according to the number of recurrently mutated genes and chromosomal abnormalities. (B) Evaluation of the real (blue) and synthetic (red) patients' distribution considering WHO 2016 classification and IPSS-R risk value. (C) PCA for clinical, demographic, and survival features. (D) Correlation matrices for clinical, demographic, and survival features, indicating the interdependencies per column on real and synthetic data sets. All results are referring to one MDS synthetic data set of 2,043 patients generated. Detailed results are reported in the Data Supplement. IPSS-R, Revised International Prognostic Scoring System; MDS, myelodysplastic syndromes; PCA, principal component analysis; SVF, synthetic validation framework.
FIG A2.
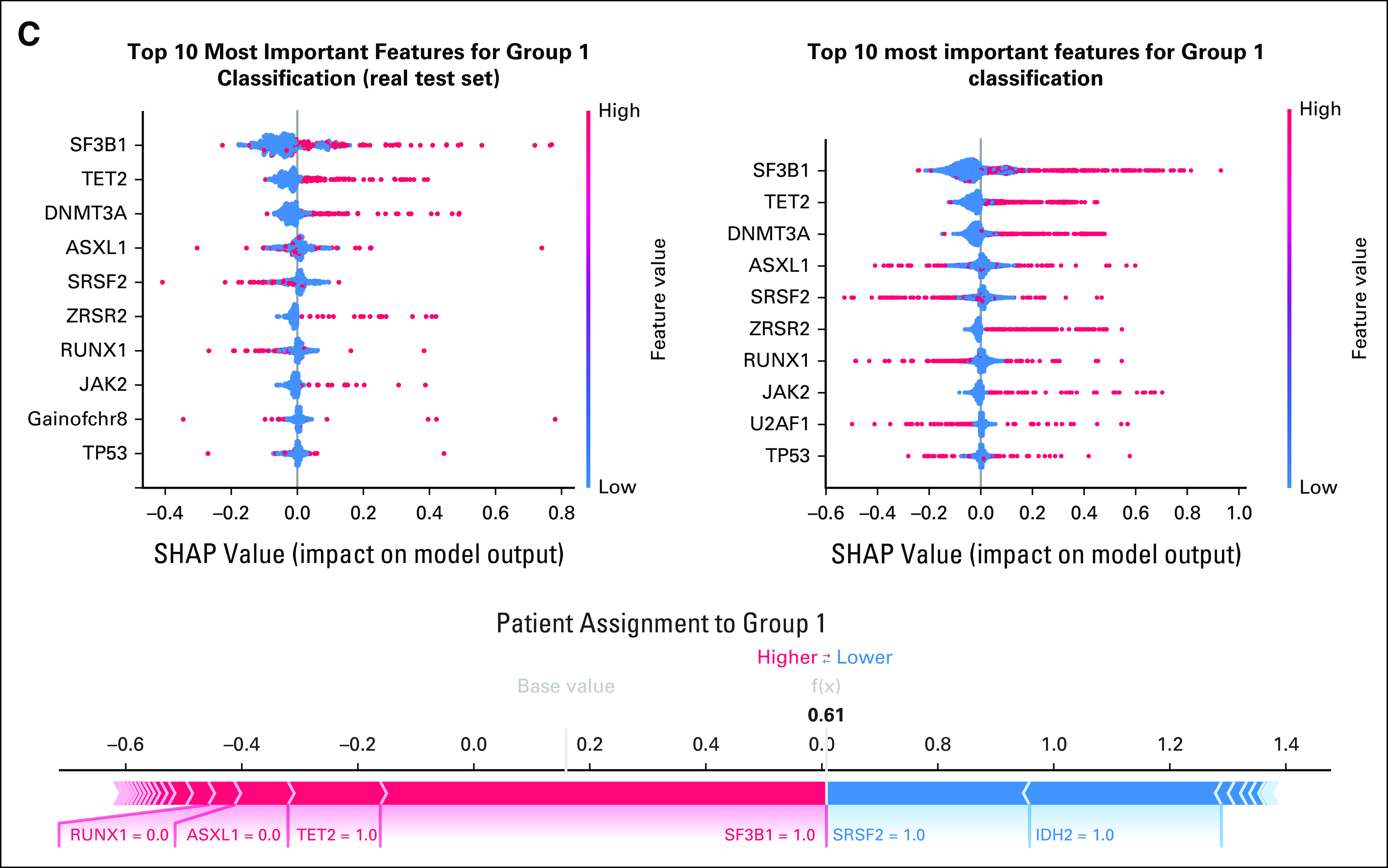
Patient classification and survival analysis on synthetic MDS cohort (N = 2,043), as performed in setting A. (A) Evaluation of the real (blue) and synthetic (red) patients' distribution considering clinical groups. Patient assignment was made by using a multiclass classifier (MLP) trained on the clinical groups identified in the EuroMDS cohort. (B) Components from the Dirichlet process on real data (above) and for the synthetic ones (below). Only the top five anomalies have been reported per cluster, decreasingly sorted by importance. (C) SHAP summary plot analysis on the top 10 most important features for real (left) and synthetic (right) for the genomic group defined as MDS with SF3B1 mutation. Below is the force plot showing the features importance in assigning a synthetic patient to this genomic group. MDS, myelodysplastic syndromes; MLP, multilayer perceptron; SHAP, Shapley Additive Explanations.
FIG A3.
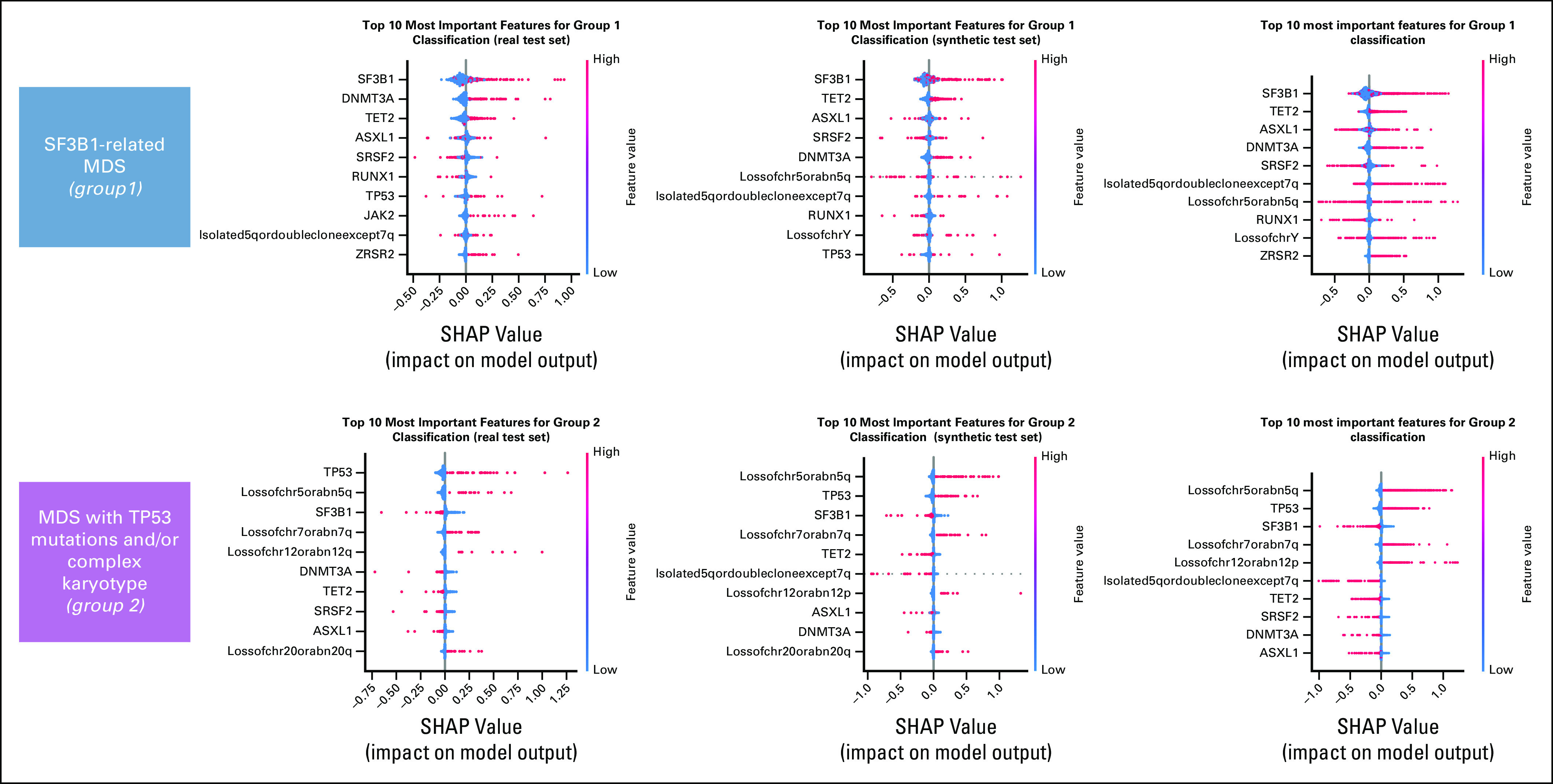
Definition of a molecular classification on augmented synthetic MDS cohort starting from 944 patients available in 2014, as performed in setting C. SHAP summary plot analysis on the top 10 most important features for a real test set, a synthetic test set, and a complete augmented synthetic data set for the genomic groups 1 and 2. MDS, myelodysplastic syndromes; SHAP, Shapley Additive Explanations.
FIG A4.
Survival analysis on synthetic molecular prognostic score generated (synthetic IPSS-M) performed in setting C. (A) Kaplan-Meier probability estimates of LFS for synthetic MDS patients are represented and stratified by IPSS-M risk categories as defined by Bernard et al.21 P value is from log-rank test. (B) Kaplan-Meier probability estimates of LFS for synthetic MDS patients are represented and stratified by synthetic IPSS-M risk categories. P value is from log-rank test. IPSS-M, Molecular International Prognostic Scoring System; LFS, leukemia-free survival; MDS, myelodysplastic syndromes.
FIG A5.
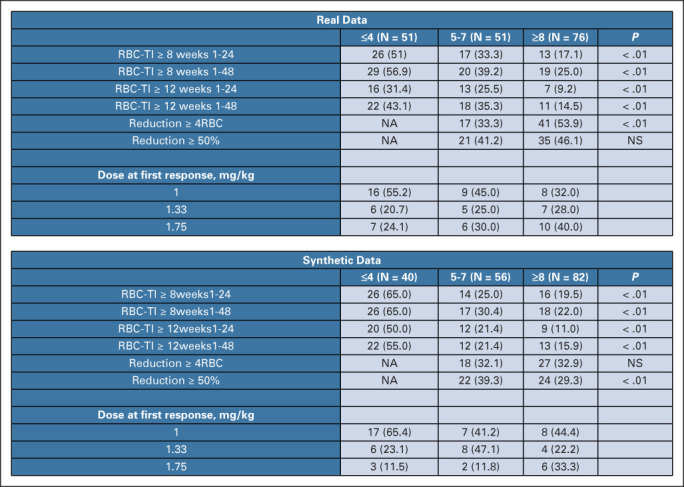
Comparison of clinical trial end points between real and synthetic patients, as performed in setting D. Response rate and dose at first response, stratified by baseline transfusion burden, in both real and synthetic cohorts. RBC-TI, rate of red blood cell transfusion independence.
Antonio Russo
Travel, Accommodations, Expenses: Pfizer, Novartis
Armando Santoro
Consulting or Advisory Role: Bristol Myers Squibb, Servier, Gilead Sciences, Pfizer, Eisai, Bayer, MSD, Sanofi, Incyte
Speakers' Bureau: Takeda, Roche, AbbVie, Amgen, Celgene, AstraZeneca, Lilly, Sandoz, Novartis, BMS, Servier, Gilead Sciences, Pfizer, Eisai, Bayer, MSD
Iñigo Prada-Luengo
Stock and Other Ownership Interests: Novo Nordisk, BioNano Genomics, Lundbeck
Anders Krogh
Employment: AJ Vaccines
Valeria Santini
Honoraria: Celgene/Bristol Myers Squibb, Novartis
Consulting or Advisory Role: Celgene/Bristol Myers Squibb, Novartis, Menarini, Gilead Sciences, AbbVie, Syros Pharmaceuticals, Servier, Geron
Research Funding: Celgene (Inst)
Travel, Accommodations, Expenses: Janssen-Cilag, Celgene
Shahram Kordasti
Honoraria: Beckman Coulter, GWT-TUD, Alexion Pharmaceuticals
Consulting or Advisory Role: Syneos Health, Novartis, Pfizer
Speakers' Bureau: Pfizer
Research Funding: Celgene, Novartis, MorphoSys
Uwe Platzbecker
Honoraria: Celgene/Jazz, AbbVie, Curis, Geron, Janssen
Consulting or Advisory Role: Celgene/Jazz, Novartis, BMS GmbH & Co. KG
Research Funding: Amgen (Inst), Janssen (Inst), Novartis (Inst), BerGenBio (Inst), Celgene (Inst), Curis (Inst)
Patents, Royalties, Other Intellectual Property: Part of a patent for a TFR-2 antibody (Rauner et al Nature Metabolics 2019)
Travel, Accommodations, Expenses: Celgene
Maria Diez-Campelo
Honoraria: Celgene, Novartis
Consulting or Advisory Role: Celgene, Novartis, GlaxoSmithKline, Blueprint Medicines
Travel, Accommodations, Expenses: Gilead Sciences
Pierre Fenaux
Honoraria: Bristol Myers Squibb
Consulting or Advisory Role: Bristol Myers Squibb
Research Funding: Bristol Myers Squibb
Torsten Haferlach
Employment: MLL Munich Leukemia Laboratory
Leadership: MLL Munich Leukemia Laboratory
Consulting or Advisory Role: Illumina
No other potential conflicts of interest were reported.
See accompanying editorial doi: 10.1200/CCI.23.00071
PRIOR PRESENTATION
Presented in part at the 2022 ASH Annual Meeting, New Orleans, LA, December 10–13, 2022.
SUPPORT
Supported by European Union—Horizon 2020/2023 program (GenoMed4All project no. 101017549 to M.G.D.P., A.K., U.P., M.D.-C., P.F., T.H., G.C., Synthema project, no. 1101095530 to M.G.D.P., G.C.); Transcan 7 Horizon 2020—EuroMDS project no. 20180424 to M.G.D.P., U.P., P.F.); AIRC Foundation (Associazione Italiana per la Ricerca contro il Cancro, Milan, Italy—Project no. 22053 to M.G.D.P.; and no. 26216 to G.C.); 5 × 1000 MYNERVA (Myeloid Neoplasms Research Venture AIRC) project to M.G.D.P; PRIN 2017 (Ministry of University & Research, Italy—Project 2017WXR7ZT to M.G.D.P.); Ricerca Finalizzata 2016 and 2018 (Italian Ministry of Health, Italy—Project RF2016-02364918 to M.G.D.P. and Project NET-2018-12365935 to M.G.D.P.); Beat Leukemia Foundation, Milan, Italy (to M.G.D.P.).
G.C. and M.G.D.P. are last senior authors.
DATA SHARING STATEMENT
To help clinicians to be familiar with generative AI to build synthetic data, we have created a prototype web portal (https://sdg-webserver-cloudrun-xkb3corsxq-ew.a.run.app/) that allows to generate synthetic patients starting from 2,957 real MDS of IWG-PM cohort. This portal allows to generate synthetic cohorts with different sizes, to verify the performance of the newly generated data (fidelity and privacy preservability), and to download the synthetic dataset for research use.
AUTHOR CONTRIBUTIONS
Conception and design: Saverio D'Amico, Antonio Russo, Maria Elena Bicchieri, Victor Savevski, Anders Krogh, Shahram Kordasti, Pierre Fenaux, Gastone Castellani, Matteo Giovanni Della Porta
Financial support: Gastone Castellani
Administrative support: Shahram Kordasti, Torsten Haferlach
Provision of study materials or patients: Saverio D'Amico, Antonio Russo, Maria Elena Bicchieri, Valeria Santini, Uwe Platzbecker, Maria Diez-Campelo, Pierre Fenaux
Collection and assembly of data: Saverio D'Amico, Elisabetta Sauta, Luca Lanino, Giulia Maggioni, Alessia Campagna, Marta Ubezio, Antonio Russo, Elena Riva, Cristina A. Tentori, Erica Travaglino, Valeria Santini, Uwe Platzbecker, Pierre Fenaux, Torsten Haferlach, Matteo Giovanni Della Porta
Data analysis and interpretation: Saverio D'Amico, Claudia Sala, Elisabetta Sauta, Matteo Zampini, Gianluca Asti, Luca Lanino, Antonio Russo, Pierandrea Morandini, Armando Santoro, Iñigo Prada-Luengo, Valeria Santini, Maria Diez-Campelo, Pierre Fenaux, Torsten Haferlach, Matteo Giovanni Della Porta
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Antonio Russo
Travel, Accommodations, Expenses: Pfizer, Novartis
Armando Santoro
Consulting or Advisory Role: Bristol Myers Squibb, Servier, Gilead Sciences, Pfizer, Eisai, Bayer, MSD, Sanofi, Incyte
Speakers' Bureau: Takeda, Roche, AbbVie, Amgen, Celgene, AstraZeneca, Lilly, Sandoz, Novartis, BMS, Servier, Gilead Sciences, Pfizer, Eisai, Bayer, MSD
Iñigo Prada-Luengo
Stock and Other Ownership Interests: Novo Nordisk, BioNano Genomics, Lundbeck
Anders Krogh
Employment: AJ Vaccines
Valeria Santini
Honoraria: Celgene/Bristol Myers Squibb, Novartis
Consulting or Advisory Role: Celgene/Bristol Myers Squibb, Novartis, Menarini, Gilead Sciences, AbbVie, Syros Pharmaceuticals, Servier, Geron
Research Funding: Celgene (Inst)
Travel, Accommodations, Expenses: Janssen-Cilag, Celgene
Shahram Kordasti
Honoraria: Beckman Coulter, GWT-TUD, Alexion Pharmaceuticals
Consulting or Advisory Role: Syneos Health, Novartis, Pfizer
Speakers' Bureau: Pfizer
Research Funding: Celgene, Novartis, MorphoSys
Uwe Platzbecker
Honoraria: Celgene/Jazz, AbbVie, Curis, Geron, Janssen
Consulting or Advisory Role: Celgene/Jazz, Novartis, BMS GmbH & Co. KG
Research Funding: Amgen (Inst), Janssen (Inst), Novartis (Inst), BerGenBio (Inst), Celgene (Inst), Curis (Inst)
Patents, Royalties, Other Intellectual Property: Part of a patent for a TFR-2 antibody (Rauner et al Nature Metabolics 2019)
Travel, Accommodations, Expenses: Celgene
Maria Diez-Campelo
Honoraria: Celgene, Novartis
Consulting or Advisory Role: Celgene, Novartis, GlaxoSmithKline, Blueprint Medicines
Travel, Accommodations, Expenses: Gilead Sciences
Pierre Fenaux
Honoraria: Bristol Myers Squibb
Consulting or Advisory Role: Bristol Myers Squibb
Research Funding: Bristol Myers Squibb
Torsten Haferlach
Employment: MLL Munich Leukemia Laboratory
Leadership: MLL Munich Leukemia Laboratory
Consulting or Advisory Role: Illumina
No other potential conflicts of interest were reported.
REFERENCES
- 1.Collins FS, Varmus H: A new initiative on precision medicine. N Engl J Med 372:793-795, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Obermeyer Z, Emanuel EJ: Predicting the future—Big data, machine learning, and clinical medicine. N Engl J Med 375:1216-1219, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pencina MJ, Goldstein BA, D'Agostino RB: Prediction models—Development, evaluation, and clinical application. N Engl J Med 382:1583-1586, 2020 [DOI] [PubMed] [Google Scholar]
- 4.Bhinder B, Gilvary C, Madhukar NS, et al. : Artificial intelligence in cancer research and precision medicine. Cancer Discov 11:900-915, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finlayson SG, Subbaswamy A, Singh K, et al. : The clinician and dataset shift in artificial intelligence. N Engl J Med 385:283-286, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trister AD: The tipping point for deep learning in oncology. JAMA Oncol 5:1429-1430, 2019 [DOI] [PubMed] [Google Scholar]
- 7.Hoffman S: Privacy and security—Protecting patients' health information. N Engl J Med 387:1913-1916, 2022 [DOI] [PubMed] [Google Scholar]
- 8.Chen RJ, Lu MY, Chen TY, et al. : Synthetic data in machine learning for medicine and healthcare. Nat Biomed Eng 5:493-497, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rajotte JF, Bergen R, Buckeridge DL, et al. : Synthetic data as an enabler for machine learning applications in medicine. iScience 25:105331, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nikolenko SI: Synthetic Data for Deep Learning. Cham, Switzerland, Springer, 2019 [Google Scholar]
- 11.Bentzen HB, Castro R, Fears R, et al. : Remove obstacles to sharing health data with researchers outside of the European Union. Nat Med 27:1329-1333, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castellanos S: Fake it to make it: Companies beef up AI models with synthetic data. Wall Street Journal, July 23, 2021. https://www.wsj.com/articles/fake-it-to-make-it-companies-beef-up-ai-models-with-synthetic-data-11627032601 [Google Scholar]
- 13.Arber DA, Orazi A, Hasserjian RP, et al. : International consensus classification of myeloid neoplasms and acute leukemias: Integrating morphologic, clinical, and genomic data. Blood 140:1200-1228, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.World Health Organization : WHO Guidance on Ethics and Governance of Artificial Intelligence for Health. Geneva, Switzerland, World Health Organization, 2021. https://www.who.int/publications/i/item/9789240029200 [Google Scholar]
- 15.Arber DA, Orazi A, Hasserjian R, et al. : The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 127:2391-2405, 2016 [DOI] [PubMed] [Google Scholar]
- 16.LeCun Y, Bengio Y, Hinton G: Deep learning. Nature 521:436-444, 2015 [DOI] [PubMed] [Google Scholar]
- 17.Goodfellow IJ, Pouget-Abadie J, Mirza M, et al. : Generative adversarial nets. arXiv:1406.2661 [Google Scholar]
- 18.Xu L, Skoularidou M, Cuesta-Infante A, et al. : Modeling tabular data using conditional GAN. arXiv:1907.00503 [Google Scholar]
- 19.Gulrajani I, Ahmed F, Arjovsky M, et al. : Improved training of Wasserstein GANs. arXiv:1704.00028 [Google Scholar]
- 20.Bersanelli M, Travaglino E, Meggendorfer M, et al. : Classification and personalized prognostic assessment on the basis of clinical and genomic features in myelodysplastic syndromes. J Clin Oncol 39:1223-1233, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernard E, Tuechler H, Greenberg PL, et al. : Molecular International Prognostic Scoring System for myelodysplastic syndromes. NEJM Evid 7, 2022 [DOI] [PubMed] [Google Scholar]
- 22.Harrell FE Jr, Califf RM, Pryor DB, et al. : Evaluating the yield of medical tests. JAMA 247:2543-2546, 1982 [PubMed] [Google Scholar]
- 23.Zhao Z, Kunar A, Birke R, et al. : CTAB-GAN: Effective table data synthesizing. PMLR 157:97-112, 2021 [Google Scholar]
- 24.Lundberg SM, Erion G, Chen H, et al. : From local explanations to global understanding with explainable AI for trees. Nat Mach Intell 2:56-67, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haferlach T, Nagata Y, Grossmann V, et al. : Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 28:241-247, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lanino L, Salutari P, Perego A, et al. : Efficacy and safety of luspatercept in adult patients with transfusion-dependent anemia due to very low, low and intermediate risk myelodysplastic syndromes (MDS) with ring sideroblasts, who had an unsatisfactory response to or are ineligible for erythropoietin-based therapy: A retrospective multicenter study by Fondazione Italiana Sindromi Mielodisplastiche (FiSiM ETS). Blood 140:6945-6948, 2022 [Google Scholar]
- 27. CALR: Synthetic data generation. https://sdg-webserver-cloudrun-xkb3corsxq-ew.a.run.app/
- 28.Azizi Z, Zheng C, Mosquera L, et al. : Can synthetic data be a proxy for real clinical trial data? A validation study. BMJ Open 11:e043497, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.El Emam K, Mosquera L, Fang X, et al. : Utility metrics for evaluating synthetic health data generation methods: Validation study. JMIR Med Inform 10:e35734, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen J, Chun D, Patel M, et al. : The validity of synthetic clinical data: A validation study of a leading synthetic data generator (Synthea) using clinical quality measures. BMC Med Inform Decis Mak 19:44, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Netherlands Comprehensive Cancer Organisation (IKNL) : Synthetic Dataset Netherlands Cancer Registry (NCR). https://iknl.nl/en/ncr [Google Scholar]
- 32.Horvat P, Gray CM, Lambova A, et al. : Comparing findings from a friends of cancer research exploratory analysis of real-world end points with the cancer analysis system in England. JCO Clin Cancer Inform 5:1155-1168, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abay NC, Zhou Y, Kantarcioglu M: Privacy Preserving Synthetic Data Release Using Deep Learning. Cham, Switzerland, Springer, 2018, 510-526 [Google Scholar]
- 34.Ghafur S, Van Dael J, Leis M, et al. : Public perceptions on data sharing: Key insights from the UK and the USA. Lancet Digital Health 2:444-446, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finniss DG, Kaptchuk TJ, Miller F, et al. : Biological, clinical, and ethical advances of placebo effects. Lancet 375:686-695, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilkinson T, Sinha S, Peek N, et al. : Clinical trial data reuse—Overcoming complexities in trial design and data sharing. Trials 20:513, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang L, Chandan R, Ping W, et al. : Multi-label clinical time-series generation via conditional GAN. arXiv:2204.04797 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
To help clinicians to be familiar with generative AI to build synthetic data, we have created a prototype web portal (https://sdg-webserver-cloudrun-xkb3corsxq-ew.a.run.app/) that allows to generate synthetic patients starting from 2,957 real MDS of IWG-PM cohort. This portal allows to generate synthetic cohorts with different sizes, to verify the performance of the newly generated data (fidelity and privacy preservability), and to download the synthetic dataset for research use.