Abstract
How steroid hormone receptors (SHRs) regulate transcriptional activity remains partly understood. Upon activation, SHRs bind the genome together with a co-regulator repertoire, crucial to induce gene expression. However, it remains unknown which components of the SHR-recruited co-regulator complex are essential to drive transcription following hormonal stimuli. Through a FACS-based genome-wide CRISPR screen, we functionally dissected the Glucocorticoid Receptor (GR) complex. We describe a functional cross-talk between PAXIP1 and the cohesin subunit STAG2, critical for regulation of gene expression by GR. Without altering the GR cistrome, PAXIP1 and STAG2 depletion alter the GR transcriptome, by impairing the recruitment of 3D-genome organization proteins to the GR complex. Importantly, we demonstrate that PAXIP1 is required for stability of cohesin on chromatin, its localization to GR-occupied sites, and maintenance of enhancer-promoter interactions. In lung cancer, where GR acts as tumor suppressor, PAXIP1/STAG2 loss enhances GR-mediated tumor suppressor activity by modifying local chromatin interactions. All together, we introduce PAXIP1 and STAG2 as novel co-regulators of GR, required to maintain 3D-genome architecture and drive the GR transcriptional programme following hormonal stimuli.
INTRODUCTION
Hormones such as glucocorticoids (GCs), androgens, or estrogens – among others- bind with high affinity to different steroid hormone receptors (SHRs), leading to modulation of transcriptional networks related to various cellular functions. Upon activation, SHRs bind the chromatin by engaging with hormone-responsive elements (HREs), predominantly found in enhancers, located distally from promoters (1,2). Once localized to their binding sites, SHRs are able to recruit a large complex of co-regulators required for initiation of SHR-dependent transcription (3,4). SHRs, together with their co-regulator complex, loop toward their target promoters, establishing enhancer-promoter interactions and subsequently leading to gene regulation (3,4). SHRs play pivotal roles in human disease, including cancer. This is best exemplified in breast cancer, prostate cancer, and childhood leukemia where estrogen receptor alpha (ERα), androgen receptor (AR) and glucocorticoid receptor (GR), respectively (5–7) serve as focal points for targeted therapy that successfully reduces disease burden (2,8–9).
SHR-mediated transcription relies on the assembly of a transcriptional complex on HREs. First, pioneer transcription factors such as FOXA1 bind to condensed chromatin, displacing linker histones, demarcating and rendering regulatory elements accessible for SHRs to bind (6,10). Once SHRs occupy their cognate regulatory elements, a coordinated recruitment of co-regulators occurs, including chromatin remodeling factors, mediator complex, histone modifiers including histone lysine (de)-methyl transferases (KMT/KMDs), histone acetyltransferases (HATs) and ultimately RNA polymerase II (RNAPII) to drive gene transcription (11–17). Altogether, SHR activation ultimately results in a large-scale transcriptional complex that loops towards targeting promoters to regulate gene expression, relying on 3D genome architectural proteins, such as the cohesin complex (18–20). As a result, SHRs and their interactors, together with the transcriptional machinery and 3D-genome organization proteins converge to either induce or repress specific target genes (12,17,21,22).
Although different studies addressed the cross-talk, interaction and co-localization of SHRs with their co-regulators (12,17,21–23), little is known about which interactors are required for SHR activity, versus those that act redundantly. By using Rapid Immunoprecipitation of Mass spectrometry of Endogenous proteins (RIME) (24), or quantitative-multiplexed RIME (qPLEX-RIME) (25) most interactors of different SHRs including ERα, AR and GR have been described (21,25–31). However, the latter only provides information of all interactors but not of the functional contribution nor essentiality of every recruited SHR-interactor on the transcriptional output.
In this study, we sought to comprehensively identify which co-regulators are essential to drive SHR-dependent transcription in a gene-specific manner, using GR as a model. We performed a genome-wide CRISPR screen followed by FACS sorting to identify which proteins affect GR-mediated activity. Surprisingly only a small number of hits were identified, suggesting that vast majority of GR interactors act redundantly. We did, however, discover a novel functional cross-talk between PAXIP1 and the cohesin complex, that to our knowledge, has never been described before. PAXIP1, a subunit of the histone modifier complex KMT2D/C (32,33), acts in a non-canonical manner to regulate GR-mediated gene expression by functionally interacting with STAG2; a member of the 3D-genome architecture complex cohesin. Finally, we report PAXIP1 to be essential for stability of cohesin on chromatin and its localization to GR-occupied sites, maintenance of enhancer-promoter interactions following hormonal stimuli, and to ensure a fully-functional GR transcriptional program.
MATERIALS AND METHODS
Cell lines
Lung cancer cell line A549 was obtained from American Type Culture Collection (ATCC). A549 cells were cultured in Dulbecco's modified Eagle's medium (DMEM)/F12 (1:1) (1×) + glutamax (Life technologies) supplemented with 10% fetal calf serum (FCS), 1% penicillin–streptomycin (Pen/Strep) (5000 U/ml, life technologies), unless otherwise stated. All cell lines were cultured at 5% CO2 and 37°C. All cell lines were genotyped and tested negative for mycoplasma.
FACS-based genome-wide CRISPR screen
The human CRISPR Brunello library (34,35) was used in this study, which was a kind gift from Roderick Beijersbergen (The Netherlands Cancer Institute, NKI). The Brunello CRISPR lentiviral library was transduced into A549 cells at a low multiplicity of transduction (MOI) of ∼0.3 to ensure that only one sgRNA was incorporated per cell. Transduced cells were subsequently selected with 2μg/ml of puromycin for 3 days and left untreated for 14 days. After that, cells were incubated in DMEM/F12 (1:1) (1×) + Glutamax, supplemented with dextran-coated charcoal-treated FCS (DCC) for 24 h and then treated with 2.75 μM of hydrocortisone (HC, HY-N0583, MedChemExpress) for additional 24 h. After treatment, cells were fixed in 70% ethanol and left overnight at 4°C. Fixed cells were washed with PBS and permeabilized with PBS containing 0.25% triton X-100 (Sigma-Aldrich) and left on ice for 15 min. Cells were stained with a recombinant anti-FKBP5 antibody (AlexaFluor 647, ab198979, 1:400) in 1% bovine serum albumin (BSA)-PBS. Stained cells were washed with PBS and resuspended in 1% BSA–PBS. Cells were then FACS sorted for 7.5% FKBP5high or 7.5% FKBP5low of the population, with ∼3 500 000 cells/arm to ensure a 500× coverage of the library. Genomic DNA was subsequently isolated using Gentra Puragene Cell kit according to manufacturer's instructions. gRNAs were amplified by two consecutive PCRs as previously described (36). DNA libraries were sequenced on HiSeq 2500 platform (single-read; 65 bp). Read counts were normalized, relative total sizefactors were calculated and that the values within a sample were divided by the respective sample sizefactor. Reads of each replicate were pooled together and subsequent analyses were performed using MaGeCK, using default parameters (v0.5.9.4 (37)). Putative GR regulators were identified comparing the sgRNA abundance amongst the 7.5% FKBP5high versus 7.5% FKBP5low populations and a robust rank aggregation (RRA) score was determined for each gene. Those genes with an FDR <0.15, log2(fold change) >2.5 and –log10(P-value) >2 were considered as putative hits. Results of the screen can be found on Supplementary Table 1.
Gene set over-representation analyses
To perform gene set over-representation analyses, we used the GSEA browser to analyze REACTOME and GO_TERM pathways. The analysis was carried out on the top 100 enriched genes identified in the screen, excluding essential genes and those with fewer than two high-quality sgRNAs.
Genome editing
CRISPR knockout cell lines
For the generation of CRISPR knockout cell lines, guide RNAs were selected from the Brunello or GeCKOv2 human CRISPR KO libraries to target PAXIP1 (CACCGGTGATTCTGTCCGTTCAGTG) STAG2 (AGTCCCACATGCTATCCACA), non-targeting (NT-1) (CACCGAACTACAAGTAAAAGTATCG), non-targeting 2 (NT-2) (GTATTACTGATATTGGTGGG), NR3C1 (GTGAGTTGTGGTAACGTTGC) and KMT2D (TTCGGGGTAGACCTCCATAG). Guide RNA targeting CDKN1B (GGGTTAGCGGAGCAATGCGC) was a kind gift from the Jos Jonker′s lab (NKI, Amsterdam, the Netherlands). The selected gRNAs were cloned into the lentiCRISPRv2 plasmid as previously described (38). All constructs were verified by Sanger sequencing. Lentivirus was produced by transfection of viral packaging vectors and Lentiv2-CRISPR constructs in HEK293T cells. On the first day, 3.5 million HEK293T cells were plated in 10 cm2 dishes and incubated overnight. On day two, viral vectors were produced by mixing three packaging constructs in a 1:1:1 ratio: pRC/CMV-rev 1B, pHDM-G, and pHDM-Hgpm2, after which 10.5 μg of the packaging mix was added to 17.5 μg of every single CRISPR construct together 105 μl of polyethylenimine (PEI, 1 mg/ml), incubated for 15 min and added to HEK293T cells. After an overnight incubation, cells were refreshed with 8 mL of medium. Next day, the supernatant was harvested and added onto A549 cells together with polybrene (200×, 1.6 g/l). After 48hrs, cells were selected with either puromycin (2 μg/ml) or blasticidin (1 mg/ml)
SMC1-EGFP tagged cell lines
In order to generate endogenously expressing EGFP-tagged SMC1A cell lines, we first selected the most suitable 20 bp-long sequence close to the C-terminal end of the SMC1A gene for gRNA binding. The gRNA sequences were designed by using an online algorithm (http://crispor.tefor.net) and the gRNAs closest to the SMC1’s stop-codon were chosen (FW: AAAATACTGCTACTGCTCAT; RV: ATGAGCAGTAGCAGTATTTT). The selected gRNAs were ordered as single-stranded DNA oligos (IDT) and annealed according to an earlier described protocol (39). A PX459 vector (pSpCas9-2A-Puro V2.0; Addgene 62988) (40) was selected for inducing DSBs via CRISPR/Cas9. To do this, the PX459 vector was first digested by BbslI restriction enzyme (NEB: R0539S) and afterwards the oligos were ligated by T4 DNA ligase (NEB: M0202S). Plasmids were sequenced by Sanger sequencing to confirm the cloning of the gRNA sequences.
In parallel, gene fragments were synthesized (IDT) containing 470 bp long homology arms flanked on both sides of the gRNA target site and excluding SMC1’s stop-codon. Subsequently, a donor template with the EGFP-stop-codon cassette including a 6 times glycine-alanine spacer sequence was cloned in between the two homology arms by using MluI (NEB: R0198S) and BglII (Roche: 10567639001) restriction enzymes. Fragments were ligated and afterwards inserted into a pCR-Blunt II TOPO backbone vector (Thermofisher: K280002). Donor template plasmid sequences were confirmed by Sanger sequencing. Next, PX459 and donor template were transfected using lipofectamine 3000 (Thermofisher) according to manufacturer’s instructions. Next, cells were selected with puromycin (2 ug/ml), clones were selected and genotyped.
Flow cytometry
Cells were cultured in DMEM/F12 (1:1) (1×) + glutamax, supplemented with dextran-coated charcoal-treated FCS (DCC) with DMSO or 2.75 μM of HC (HY-N0583, MedChemExpress) for 24 h. Cells were fixed with 70% ethanol at 4°C overnight. Cells were washed with PBS, permeabilised with 0.25% Triton X-100 in PBS-T for 15 min on ice, and stained with primary recombinant Alexa Fluor® Anti-FKBP51 antibody (ab198979, Abcam, 1:100) diluted in 1% BSA–PBS or left unstained. Cells were washed and resuspended in 1% BSA/PBS solution. Cells were sorted using Attune TM NxT Acoustic Focusing Cytometer (Thermo Fisher Scientific). Single-cell flow cytometry analysis was performed using FlowJo™ Software version 10.7.1 (BD Biosciences)
RNA isolation, reverse-transcription and quantitative real-time PCR (RT-qPCR)
Cells were cultured in DMEM/F12 (1:1) (1×) + Glutamax, supplemented with dextran-coated charcoal-treated FCS (DCC) with DMSO or 2.75 μM HC (HY-N0583, MedChemExpress) for 24 h. Total RNA was isolated using Invitrogen™ TRIzol™ Reagent (15596026, Thermo Fisher Scientific) according to manufacturer′s instructions. First-strand cDNA was synthesized from 1 μg of isolated RNA by using SuperScript® III First-Strand Synthesis System for Reverse Transcriptase-PCR (18080-051, Life Technologies). RT-qPCR was performed using SensiMix™ SYBR® No-ROX Kit (QT650-05, Bioline) in a QuantStudio™ 6 Flex System (Thermo Fisher Scientific), and analyzed using the QuantStudio Software. Primers can be found in Supplementary Table 2.
Western blot
Cells were lysed using 2× Laemmli buffer (120 mM Tris, 20% glycerol, 4% SDS) supplemented with protease inhibitor (1:100) and phenylmethylsulfonyl fluoride (PMSF, 1:200) upon overnight treatment with 100nM dexamethasone (HY-14648, MedChemExpress) or 2.75 μM HC (HY-N0583, MedChemExpress) or left untreated. Lysates were sonicated (EpiShear Probe Sonicatore, Active Motif) for 10 cycles with one second intervals and a 20% amplitude. Equal amounts of protein per lysate were run for one hour at 100 V on an 8% acrylamide gel (MilliQ, 40% acrylamide, 1.5 M Tris pH 6.8, 10% SDS, 10% APS, TEMED) in SDS-PAGE 1× Running buffer (25 mM Tris, 0.25 M glycine, 0.1% SDS). Proteins were transferred on ice at 100 V for 90 min or at 0.9 mA overnight at 4°C on a nitrocellulose membrane in cold 1× Transfer buffer (24 mM Tris, 192 mM glycine). Membranes were stained with Ponceau S (Thermo Fisher) and subsequently blocked in 3% BSA (A8022, Sigma/Merck) in 1× PBS–Tween (137 mM NaCl, 10 mM Na2HPO4, 1.5 mM KH2PO4, 2.6 mM KCl, 0.1% Tween-20) for 1 h and incubated with primary antibodies against GR (12041, Cell Signaling Technology, 1:1000), PAXIP1 (ABE1877, Merck, 1:1000), STAG2 (A300-158A, Bethyl, 1:1000), Actin (MAB1501R, Merck, 1:1000), HSP90 (sc-13119, Santa Cruz Biotechnologies, 1:1000), P27 (610242, BD Biosciences, 1:1000), SMC1 (Bethyl:A300-055A; 1:1000) diluted in 3% BSA/PBS-T for 2 h. After three washing steps in PBS-T, membranes were incubated with secondary antibodies donkey-α-mouse 680 RD (926-68073, LI-COR Biosciences, 1:10 000), donkey-α-rabbit 800 CW (926-32213, LI-COR Biosciences, 1:10 000) and donkey-α-goat 680 RD (926-68074, LI-COR Biosciences, 1:10 000), diluted in 3% BSA/PBS-T for 1 h. Membranes were scanned and analysed using an Odyssey® CLx Imaging System (LI-COR Biosciences) and ImageStudio™ Lite v.5.2.5 software (LI-COR Biosciences).
Rapid immunoprecipitation of endogenous proteins (RIME)
RIME was performed as previously described (24). In brief, cells were treated for 2 h with 2.75 μM HC. Subsequently, cells were fixed and crosslinked with 1% formaldehyde (15714, Electron Microscopy Sciences) for exactly 10 min, quenched with glycine (0.125 M) and washed three times with PBS. Cells were collected in 1xPBS supplemented with 1× complete EDTA-free protease inhibitor cocktail (PI tablets, 5056489001, Roche) on ice. Cells were lysed as previously described (24), and cell lysates were sonicated for six cycles (30 s on/30 s off) using the Bioruptor® Pico (B01060001, Diagenode). Per RIME, 50 μl of magnetic Protein A beads (10008D, Thermo Fisher Scientific) beads were conjugated with either 15 μl anti-GR (12041, Cell Signaling Technology) rotating overnight at 4°C.
For mass spectrometry, peptide mixtures were prepared and measured as previously described (27), with the following exceptions. For GR RIMEs in NT versus PAXIP1-KO cells, peptide mixtures (10% of total digest) were loaded directly onto the analytical column and analyzed by nanoLC-MS/MS on an Orbitrap Fusion Tribrid mass spectrometer equipped with a Proxeon nLC1200 system (Thermo Scientific). Solvent A was 0.1% formic acid/water and solvent B was 0.1% formic acid/80% acetonitrile. Peptides were eluted from the analytical column at a constant flow of 250 nl/min in a 120-min gradient, containing a 104-min stepped increase from 6% to 32% solvent B, followed by a 16-min wash at 90% solvent B. For GR RIMEs in NT versus STAG2-KO cells, peptide mixtures (10% of total digest) were loaded directly onto the analytical column and analyzed by nanoLC-MS/MS on an Orbitrap Exploris 480 Mass Spectrometer equipped with a Proxeon nLC1200 system (Thermo Scientific). Solvent A was 0.1% formic acid/water and solvent B was 0.1% formic acid/80% acetonitrile. Peptides were eluted from the analytical column at a constant flow of 250 nl/min in a 90-min gradient, containing a 74-min stepped increase from 6% to 32% solvent B, followed by a 16-min wash at 90% solvent B.
Raw data were analyzed by MaxQuant (GR RIMEs in NT versus PAXIP1-KO cells: version 2.0.1.0; GR RIMEs in NT versus STAG2-KO cells:version 1.6.17.0) (41) using standard settings for label-free quantitation (LFQ). MS/MS data were searched against the Swissprot Human database (GR RIMEs in NT vs PAXIP1-KO cells: 20 395 entries, release 2021_04; GR RIMEs in NT versus STAG2-KO cells: 20,379 entries, release 2021_01) complemented with a list of common contaminants and concatenated with the reversed version of all sequences. The maximum allowed mass tolerance was 4.5 ppm in the main search and 0.5 Da for fragment ion masses. False discovery rates for peptide and protein identification were set to 1%. Trypsin/P was chosen as cleavage specificity allowing two missed cleavages. Carbamidomethylation was set as a fixed modification, while oxidation and deamidation were used as variable modifications. LFQ intensities were Log2-transformed in Perseus (GR RIMEs in NT versus PAXIP1-KO cells: version 1.6.15.0; GR RIMEs in NT versus STAG2-KO cells: version 1.6.14.0), after which proteins were filtered for at least three out of four valid values in at least one sample group. Missing values were replaced by imputation based on a normal distribution (width: 0.3 and downshift: 1.8). Differentially expressed proteins were determined using a Student's t-test.
Chromatin immunoprecipitation followed by sequencing (ChIP-seq)
Chromatin immunoprecipitation followed by sequencing (ChIP)-seq was performed as previously described (42). Cells were treated with 100nM Dexamethasone (HY-14648, MedChemExpress) for 2 h. Cells were fixed in 1% formaldehyde (1039991000, Merck) for 10 min and quenched with 0.125 M glycine. Nuclear lysates were extracted as previously described (42) and sonicated for 13 cycles (30 s on/30 s off) using the Bioruptor® Pico (B01060001, Diagenode). The antibodies that were used are the following: GR (12041, Cell Signaling Technology), RAD21 (05-908, Merck), PAXIP1 (ab70434, abcam), H3K4me1 (ab8895, abcam). Per ChIP, 50 μl of magnetic Protein A beads (10008D, Thermo Fisher Scientific) beads were conjugated to 7.5 μl of GR antibody, or 5 μg of RAD21, PAXIP1, and H3K4me1 antibodies. Immunoprecipitated DNA was processed for library preparation (KAPA library preparation kit, KK8234, Roche). In the case of GR and RAD21 ChIPs, generated libraries were sequenced on the Illumina HiSeq2500 platform (single-end, 65 bp reads). For PAXIP1 and H3K4me1 ChIPs, samples were sequenced on Illumina Novaseq 6000 (paired-end, 51 bp). For those samples processed in Novaseq, adapter trimming was performed before alignment using seqpurge (43). All reads were aligned to the Human Reference Genome (GRCh38.102) using Burrows-Wheeler Aligner (v0.7.17; (44)) and mem algorithm. In samples processed with Novaseq, duplicates were marked umi-aware, using rumidup (https://github.com/NKI-GCF/rumidup). For samples processed in HiSeq, data did not have an UMI, and based in coordinates, biological duplicates were removed. Reads were filtered based on MAPQ quality ≥20 (samtools v1.9), and duplicate reads were removed. Peak calling over input was generated by using MACS2 (v2.1.2; (45)), by using the peakcalling pipeline https://github.com/csijcs/snakepipes. Consensus peaks between biological replicates were generated using mspc tool (46).For visualization, mapped reads of replicate samples were merged using SAMtools (v1.10; (47)). Genome browser snapshots, average density plots and tornado plots were generated using Easeq (v1.101; (48)). Genomic distribution analyses were performed using ChIPseeker (v1.26.2; (49)) under R 4.0.3. Motif enrichment analyses were performed using HOMER Motif analyses tool (50).
Cell proliferation analyses
Cells were plated in a 384-well plate at a density of 250 cells/well. Cells were treated with 100 nM of Dexamethasone (HY-14648, MedChemExpress). Cells were imaged every 4 h by using an IncuCyte ZOOM Live-Cell Analysis System, and cell confluency percentage was calculated using the incucyteZoom software.
Circularized chromosome conformation capture sequencing (4C-seq)
Cells were treated with 100 nM Dexamethasone (HY-14648, MedChemExpress) for 2 h. Experiments of 4C-seq were performed as previously described (51). Briefly, for each restriction enzyme (RE) combination (RE1 NlaIII, RE2 DpnII; New England Biolabs), replicate and treatment about 10 × 106 cells were collected, pelleted and cross-linked by 2% methanol-free formaldehyde for 10 min. Nuclei were isolated and permeabilized to allow digestion of the chromatin by the primary RE (NlaIII, New England Biolabs). Chromatin fragments were then diluted and ligated before crosslinking reversion. Purified DNA was digested by the secondary RE (DpnII, New England Biolabs) and circularized again by ligation. Re-purified circular fragments were amplified by PCR with View-Point specific primers (see Supplementary Table 2) using the Expand Long Template PCR System (Roche). Resulting amplicons were purified with a 0.8 × ratio of AMPure XP beads (Beckman Coulter) and amplified using standard indexed Illumina primers as previously described (51) using the Expand Long Template PCR System (Roche).
Second-round PCR products were purified with PCR purification columns (Qiagen) and quantified by 2100 Bioanalyzer (Agilent, DNA 7500 kit). 4C library was prepared by equimolar mix of the samples. The pooled library was cleaned-up by AMPure XP beads (0.8 × ratio) to remove PCR dimers before sequencing through Illumina MiSeq with a 75 bp Single-End reads setup. Fastq files have been demultiplexed by Cutadapt (http://journal.embnet.org/index.php/embnetjournal/article/view/200) and mapped on Hg38/GRCm38 genome assembly and signal normalized by pipe4C (v1.1) R-package (51) in “cis” mode and with default parameters. Score mean of the replicates and plots of normalized mean 4C-seq and ChIP-seq signal were generated in an R v4.0.3 environment by using get.single.base.score.bw and genomic.track functions from Rseb (v0.3.0) (https://github.com/sebastian-gregoricchio/Rseb)(52) package in combination with ggplot2 (v3.3.5) and ggforce (v0.3.3) packages.
Fluorescence recovery after photobleacing (FRAP)
FRAP was performed on a Leica TCS SP8 microscope equipped with a 63×/1.40 NA HC PL APO CS2 oil immersion objective in combination with a 50 mW Argon excitation laser using the 488 nm line. FRAP measurements were conducted on approximately 50% of the nuclear area (344 pixel by 344 pixel, 100 nm pixel size, 2 times line averaging). After 40 scans (500 ms interval), a high intensity laser beam at 488 nm was utilized for three iterations with a 197 ms interval to photobleach all GFP locally inside the selected ROI. Subsequently, the bleached ROI was continuously scanned for 1200 iterations with a 500 ms interval. Quantitative FRAP analysis was performed using a Monte Carlo simulation environment for modeling complex biological molecular interaction networks (53).
Computer modelling used to generate FRAP curves for fitting was based on Monte Carlo simulation of diffusion and binding to immobile elements (representing chromatin binding) in an ellipsoidal volume (representing the nucleus). Bleaching simulation was based on experimentally derived three-dimensional laser intensity profiles, which determined the probability for each molecule to become bleached considering their 3D position relative to the laser beam. Diffusion was simulated at each new time step t + Δt by deriving a new position (xt+Δt, yt+Δt, zt+Δt) for all mobile molecules from their current position (xt, yt, zt) by xt+Δt= xt + G (r1), yt+Δt= yt + G (r2), and zt+Δt= zt + G (r3), where ri is a random number (0 ≤ ri ≤ 1) chosen from a uniform distribution, and G (ri) is an inversed cumulative Gaussian distribution with μ = 0 and σ2 = 2DΔt, where D is the diffusion coefficient. Immobilization was derived from simple binding kinetics: kon/koff = Fimm / (1–Fimm), where Fimm is the fraction of immobile molecules. The probability per unit time to be released from the immobile state was given by Pmobilise = koff = 1 / Timm, where Timm is the characteristic time spent in immobile complexes expressed in unit time steps. The probability per unit time for each mobile particle to become immobilized (representing chromatin-binding) was defined as Pimmobilise = kon = (koff ċ Fimm) / (1 – Fimm), where koff = 1/Timm. Note that kon and koff in this model are effective rate constants with dimension s−1.
In all simulations, the size of the ellipsoid was based on the size of the measured nuclei, and the region used in the measurements determined the size of the simulated bleach region. The laser intensity profile using the simulation of the bleaching step was derived from confocal images stacks of chemically fixed nuclei containing GFP that were exposed to a stationary laser beam at various intensities and varying exposure times. The unit time step Δt corresponded to the experimental sample rate of 100 ms.
For quantitative analysis of the FRAP data, raw FRAP curves were normalized to pre-bleach values and the best fitting curves (by ordinary least squares) were selected from a large set of computer simulated FRAP curves in which three parameters representing mobility properties were varied: diffusion rate, immobile fraction and time spent in immobile state. Because individual curves generated by Monte Carlo modelling, in contrast to analytically derived curves, show the slight variation typical for diffusion of a limited number of molecules in a small volume, we did not use the best-fitting curve only, but took the ten best-fitting curves and calculated the average diffusion coefficients and rate constants corresponding to these curves.
Genetic dependency analyses
The dependency data used in this study was obtained from DepMap 21Q4 public (https://depmap.org/portal/). Dependency data for PAXIP1 and STAG2 genes across 1046 cell lines was downloaded. Pearson correlation coefficient was calculated between CERES scores of PAXIP1 and STAG2. Moreover, top100 co-dependencies for PAXIP1 were downloaded from https://depmap.org/portal/. Data on GR levels in all cell lines was obtained from Expression Public 22Q4 (https://depmap.org/portal/). All data was plotted using ggplot2 (v.3.3.5) and ggpubr (v.0.4.0).
sevenC analyses
Prediction of loops was performed using sevenC (v1.19.0) (54) run with default parameters. We used CTCF sites identified using JASPAR 2022 database in the hg38 human genome (“AH104716”), filtered for those with a P-value < 1e–6. As ChIP input, we used peak called files from the RAD21 ChIP performed in NT and PAXIP1-KO cells in GC-treated conditions (See ChIP section for more details). One sample t-test was used for analyses.
RNA-seq
Cells were treated with Dexamethasone (100nM) for 8 h. Total RNA was extracted using RNeasy Mini kit (Qiagen, Germany) following manufacturer′s instructions. The quality and quantity of the total RNA were assessed by the 2100 Bioanalyzer using a Nanochip (Agilent, USA). NT-1, PAXIP1-KO and GR-KO RNA-seq series was sequenced in the Illumina HiSeq2500 platform. NT-2, STAG2-KO series was sequenced in Illumina Novaseq 6000. Sequencing data were aligned to the human reference genome hg38 using HISAT2 (v2.1.0; (55)), and the number of reads per gene was measured with HTSeq count (v0.5.3; (56)). Read counting, normalization and differential gene expression were performed using R package DESeq2 (v.1.30.1; (57))
Immunofluorescence
Cells were fixed for 10 min using 2% paraformaldehyde (PFA, 103999, Merck), washed twice with PBS, and subsequently permeabilized with 0.5% Triton/PBS (X100, Sigma Aldrich). After two additional washing steps with PBS, cells were blocked in 1% BSA/PBS solution before being incubated with primary antibodies PAXIP1 (ABE1877l, Merck, 1:100) and STAG2 (A300-158A, Bethyl, 1:1000) for 2 h. Laser confocal microscopy (SP5, Leica) was used to detect PAXIP1 and STAG2 using secondary antibodies Alexa Fluor™ 647 goat anti-rabbit IgG (H+L) Cross-Adsorbed and Alexa Fluor™ 647 donkey anti-goat IgG (H+L) Cross-Adsorbed, respectively.
RESULTS
Genome-wide CRISPR screen identifies PAXIP1 and STAG2 as essential regulators of GR-mediated transcription
GR activation results in either induction or repression of gene expression due to either trans-activation or trans-repression mechanisms. The recruitment of either co-activators or co-repressors to the GR complex plays an important role in its function as a trans-activator or trans-repressor. Previously, we comprehensively annotated the GR-interacting protein repertoire in various lung cancer cell lines, including A549 cells (31). However, which of these GR interacting proteins are essential for GR trans-activation function remains unknown. To identify proteins critical for GR-action in a comprehensive manner, we set up a FACS-based genome-wide CRISPR screen, in which we used the expression of a classical GR target gene -FKBP5- as proxy of GR activity (Figure 1A and Supplementary Figure 1A–D). Exposure of the lung cancer cell line A549 to glucocorticoids (GCs) increased GR binding at the FKBP5 locus (Supplementary Figure 1A), and induced transcription of FKBP5 over time (Supplementary Figure 1B). Importantly, FKBP5 mRNA and protein levels were not induced upon GC treatment in GR-knockout cells (Supplementary Figure 1C, D). Jointly, these controls confirm the GR-dependent status of FKBP5, reinforcing its role as marker gene for GR activity. In this screen, A549 cells were transduced with the CRISPR genome-wide Brunello library, including 77,441 sgRNAs, with an average of four targeting sgRNAs per gene, and 1000 non-targeting controls (34,35). Cells were subsequently selected with puromycin, and cultured for 14 days prior to stimulation. In order to induce GR-mediated transcription, A549 cells were treated with GCs for 24 h, subsequently fixed and stained for protein expression of FKBP5. Cells were FACS-sorted for approximately 7.5% of low (FKBP5low) and 7.5% high FKBP5 expression (FKBP5high) populations (Figure 1A). Next, genomic DNA was isolated and sequenced to detect abundance of sgRNAs in both populations. Through MaGECK analyses (37), we compared the sorted FKBP5high and FKBP5low populations, to identify which genes are individually essential to drive GR activity (Figure 1B). These analyses identified two critical positive controls – NR3C1 (encoding for GR) and FKBP5 – as top hits (Figure 1B), demonstrating the robustness of our screen. We observed that few GR-interacting proteins were among the top hits (Supplementary Figure 1E), suggesting a level of functional redundancy between GR co-regulators and only some proteins being essential for GR-mediated transcription. Moreover, over-representation analyses of the top screen hits identified signaling pathways that are involved in SHR-mediated signaling and active transcription, among others (Supplementary Figure 1F). Importantly, we identified PAXIP1 and STAG2 among the top-enriched hits in the FKBP5low population (Figure 1B), implying a critical role of these proteins in regulating GR action. To validate our findings, we generated PAXIP1 and STAG2 CRISPR-mediated gene disruption models (Figure 1C and Supplementary Figure 1G). Loss of either PAXIP1 or STAG2 significantly reduced the expression of FKBP5, both at the protein (Figure 1D) and RNA level (Figure 1E and Supplementary Figure 1H). Importantly, GR protein levels remained unaltered in both PAXIP1-KO and STAG2-KO cells (Supplementary Figure 1I), demonstrating that the observed reduction of FKBP5 expression is not a consequence of altered GR protein stability or transcription.
Figure 1.
PAXIP1 and STAG2 are crucial for GR-mediated transcription (A) Schematic representation of the FACS-based genome-wide CRISPR screen to identify regulators of GR function. (B) Screen results: scatter plot of log2(fold change [FKBP5low versus FKBP5high expression]) versus −log10 (P-value). Dotted lines indicate threshold values (log2(fold change) > 2.5 and −log10 (P-value) > 2). Analyses were performed using MaGECK. Red highlighted dots depict NR3C1 and FKBP5, serving as positive controls. Blue highlighted dots indicate other significant hits of the screen. (C) Western blot analyses showing PAXIP1 (left) and SA2 (protein encoded by STAG2, right) expression, with actin as control, in NT, PAXIP1-KO and STAG2-KO cells (n = 2). Cells have been treated with DMSO (empty dot) or GCs (filled dot) for 24 h. (D) FKBP5 protein levels in A549 cells depleted of PAXIP1 and STAG2 expression assessed by Flow Cytometry analyses. Quantification of Mean-Fluorescence Intensity (MFI) of FKBP5 expression (n = 4 in NT versus STAG2-KO, n = 3 in NT versus PAXIP1-KOs). Cells have been treated with DMSO (empty dot) or GCs (filled dot) for 24 h. Mean values ± SD are depicted. Two-way ANOVA test was performed. (E) Relative FKBP5 mRNA levels (normalized to geometric mean of housekeeping genes GAPDH and ACTIN) of DMSO (empty dot) and GC-treated (filled dot) in cells depleted of PAXIP1 and STAG2 expression. Mean values ± SD are depicted (n = 4 biological replicates in NT versus STAG2-KO, n = 3 technical replicates in NT versus PAXIP1-KOs). Two-way ANOVA test was performed.
PAXIP1 as a novel functional interactor of STAG2
Having identified PAXIP1 and STAG2 as novel regulators of GR function, we next aimed to elucidate the mechanism behind this observation. For this purpose, we first assessed a possible impact of these proteins on GR/DNA interactions, through chromatin immunoprecipitation followed by sequencing (ChIP-seq) analyses for GR in non-target (NT), PAXIP1-KO and STAG2-KO cells, both in vehicle and GC-treated conditions. As expected for NT cells, GR chromatin interactions were induced by GC-treatment (Figure 2A and B). Interestingly, GR binding to the FKBP5 locus was not affected in neither PAXIP1-KO or STAG2-KO cells (Figure 2A), suggesting that PAXIP1 and STAG2 regulate GR functionality downstream of its capacity to associate with the chromatin. The latter was, importantly, also observed on a genome-wide scale (Figure 2B). As expected, genomic distribution analyses showed that GR binding occurs mainly at introns and distal intergenic regions in all cell lines (Supplementary Figure 2A). However, we did observe a slight enrichment of GR localization at promoters and promoter-proximal regions, relative to most-closely located transcription start site (TSS) in PAXIP1-KO cells when compared to NT control (Supplementary Figure 2B). In addition, motif analyses at peaks identified in NT, PAXIP1-KO and STAG2-KO cells showed, as expected, glucocorticoid response elements (GRE), other HREs and forkhead transcription factors (FOX) among the top most enriched motifs, with no major differences between models (Supplementary Figure 2C).
Figure 2.
PAXIP1 functionally interacts with cohesin. (A) Snapshot of GR binding around the FKBP5 locus in non-targeting (NT), PAXIP1-KO, and STAG2-KO cells, comparing vehicle (DMSO) and treated (GC) conditions. Data represent the average of three biological replicates (n = 3). (B) Heatmap (top) and average density plot (bottom) of GR ChIP-seq signal in NT, PAXIP1-KO, STAG2-KO cells in vehicle (DMSO) or treated (GC) conditions. Data are centered at GR-peaks shared in NT, PAXIP1-KO and STAG2-KO in GC-treated arm, depicting a ±10 kb (heatmap) and ±2.5 kb (average plot) window around the peak center. Data represent the average of three biological replicates. (C) RIME analyses showing the composition of the GR protein complex in PAXIP1-KO cells. Volcano plot comparing GR-IP in NT cells over PAXIP1-KO. Cells were treated with GCs for 2 h. Proteins considered to be interacting with GR are highlighted in orange. Significance cut-off is depicted with a dotted line (LFQ > 0.8, −log (P-value) > 1.3) (n = 4). (D) RIME analyses depicting the composition of the GR protein complex in STAG2-KO cells. Volcano plot comparing GR-IP in NT cells over STAG2-KO. Cells were treated with GCs for 2 h. Proteins considered to be interacting with GR are represented in orange. Significance cut-off is represented with a dotted line (LFQ > 0.8, −log(P-value) > 1.3) (n = 4). (E) GSEA enrichment profiles for Sister chromatid cohesion reactome (M27181), comparing NT versus PAXIP1-KO (top) or NT versus STAG2-KO (bottom). Ranking is based on the LFQ values of GR-RIME data. Nominal P-value and NES were determined with GSEA. (F) Correlation plot between those proteins that were commonly significantly (−log (P-value) > 1.3) differentially recruited to the GR complex (Pearson correlation R = 0.89, P-value < 0.001). (G) Correlation between the DepMap CERES values of STAG2 and PAXIP1 across all 1043 cell lines from the DepMap database (Pearson correlation R = 0.61, P < 0.001). (H) DepMap analyses showing top 100 co-dependencies for PAXIP1. Pearson correlation values are depicted.
Due to the critical role of these co-regulators on GR transcriptional activity, we next investigated whether the composition of the GR complex was altered upon PAXIP1 or STAG2 depletion. To do so, we performed RIME analyses (24) for GR in both PAXIP1 (Figure 2C) and STAG2 (Figure 2D) deficient cells. PAXIP1 is a subunit of the multiprotein complex KMT2D/C; a histone methyl transferase complex, responsible for methylation of H3K4, demarcating enhancers (32,33,58). Surprisingly, GR-mediated recruitment of the KMT2D/C complex (DPY30, ASH2L, KMT2D, WDR5, RBBP5) was not affected when PAXIP1 was knocked out (Figure 2C). In line with this, depletion of a main subunit of the complex -KMT2D- did not affect expression of FKBP5, as measured by flow cytometry analyses (Supplementary Figure 2D). Jointly, these findings suggest that PAXIP1 impacts GR action in a KMT2D/C-independent manner in lung cancer cells.
Surprisingly, GR RIME analyses showed diminished recruitment of members of the 3D-genome organization-related cohesin complex in cells lacking PAXIP1, including SMC1A and SMC3 (Figure 2C). These proteins serve to facilitate promoter/enhancer interactions and enable long-range gene regulation by cis-acting transcription factors (58). These results suggest that PAXIP1 may regulate association of cohesin to GR-bound regulatory sites on the genome. Analogous to our PAXIP1 findings, knockout of STAG2 perturbed the capacity of GR to interact with SMC1A, SMC3 and RAD21 (Figure 2D). The strong similarities of PAXIP1-KO and STAG2-KO impacting the GR protein interactome were further highlighted in Gene Set Enrichment Analyses (GSEA), in which the proteins group in the sister chromatid reactome dataset were significantly lost upon either PAXIP1 or STAG2 knockouts (Figure 2E and Supplementary Figure 2E). Reduced recruitment of cohesin subunits to the GR complex upon depletion of either PAXIP1 or STAG2, suggested a possible novel functional cross-talk of PAXIP1 with STAG2. These findings were further confirmed in quantitative analyses on our RIME data, demonstrating that core GR-associated proteins remained recruited even upon PAXIP1 and STAG2 ablation (Supplementary Figure 2F). Furthermore, high correlation was found in the levels of differential protein recruitment to the GR complex in NT when compare to STAG2-KO and PAXIP1-KO cell lines, including cohesin subunits (Figure 2F). To gain further insights into a possible PAXIP1-STAG2 functional interaction, we analyzed the essentiality of STAG2 and PAXIP1 in a publicly available CRISPR-screen dataset containing data for 1,043 cell lines from different tumor types and derived from various organs (DepMap 21Q4 public; https://depmap.org/portal/) (Figure 2G). Interestingly, we found a high level of co-dependency between PAXIP1 and STAG2 across all cell lines (R = 0.61, P-value < 0.001) (Figure 2G), implying that PAXIP1 and STAG2 might be involved in regulating similar cellular processes in different cell types. Importantly, we observed that cells originating from lymphocytes, liver, plasma cell and blood lineages were the most dependent on STAG2 and PAXIP1, with no clear correlation with GR expression levels (Supplementary Figure 2G), implying a broader STAG2/PAXIP1 functional cross-talk not restricted to GR function, which warrants further investigation. Next to STAG2, cohesin loaders MAU2 and NIPBL (59), as well as the subunit of cohesin SMC3, were all found amongst the highest co-dependencies for PAXIP1 in this dataset (Figure 2H), providing further evidence for a functional cross-talk between PAXIP1 and STAG2. All together, we find that loss of both PAXIP1 and STAG2 does not affect GR chromatin binding, but does diminish the recruitment of proteins involved in 3D-genome organization to the GR complex, which is associated with a loss of GR target–gene regulation, as measured by expression of GR-activity marker FKBP5.
PAXIP1 is required for cohesin stability on the genome, its localization to GR-bound enhancers and enhancer-promoter interactions
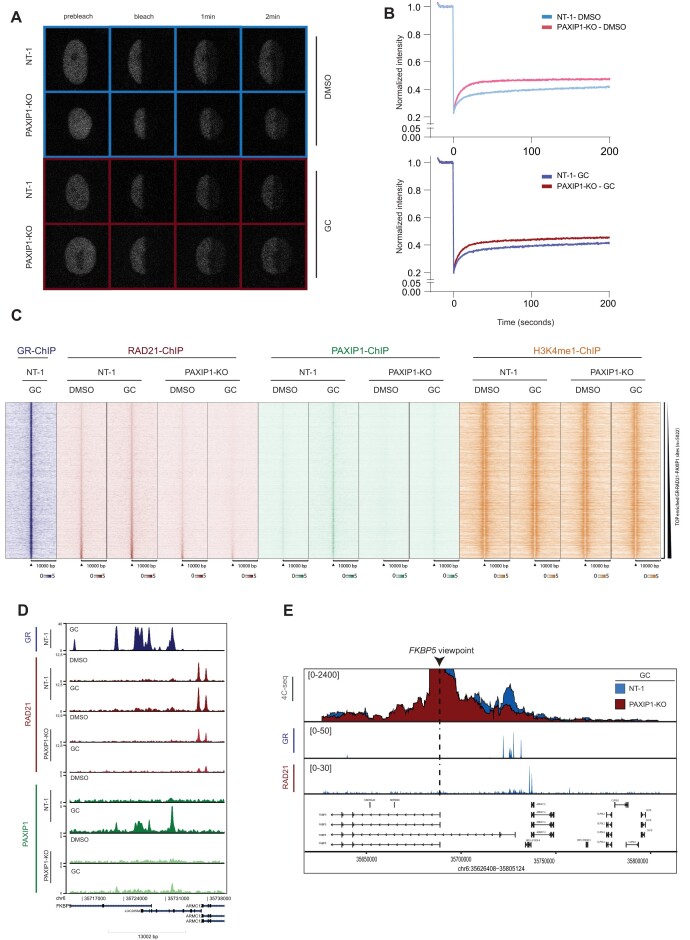
Our findings indicated that PAXIP1 may regulate cohesin recruitment to the chromatin-bound GR complex. To further explore this, we endogenously tagged cohesin subunit SMC1 with an EGFP-tag on the C-terminal end of the protein (SMC1-EGFP) in both wild-type and PAXIP1 deficient cells (Supplementary Figure 3A), and performed recovery after photobleaching (FRAP) experiments to determine SMC1 mobility in live cells (Figure 3A and B; Supplementary Figure 3B). PAXIP1 knockout increased SMC1-EGFP mobility in these cells, both under vehicle and GC treatment (Figure 3A and B), indicating that cohesin chromatin stability was impaired upon PAXIP1 loss (Figure 3A and B). We performed computer simulations of the FRAP curves (53) and calculated the different immobile and mobile fractions of SMC1-EGFP (Supplementary Figure 3B). Upon knocking out PAXIP1, the immobile fraction of cohesin was decreased in both DMSO and GC-treated cells, indicating reduced stability of cohesin on the genome (Supplementary Figure 3B). In addition, when cells were exposed to GCs, the immobile fraction in PAXIP1-KOs was also lower than in NTs, with short and intermediate immobile fractions being more prominent in PAXIP1-KOs when compared to NT (Supplementary Figure 3B). All together, we observed that depletion of PAXIP1 results in a general reduction of cohesin stability on the chromatin, both in DMSO and GC-treated cells.
Figure 3.
PAXIP1 is required for cohesin function. (A) Representative image of FRAP experiments in NT-SMC1-EGFP and PAXIP1-KO-SMC1-EGFP cells with vehicle (top, blue) and GC-treated (bottom, red) cells. (B) Quantification of FRAP experiments over time in NT-SMC1-EGFP and PAXIP1-KO-SMC1-EGFP cells with vehicle (above) and GC-treated cells (below). Average ± SD for 30 cells quantified over three independent biological experiments. Intensity is pre-bleached normalized. (C) Heatmap of RAD21, PAXIP1 and H3K4me1 ChIP-seq signal in NT, PAXIP1-KO cells in vehicle (DMSO) or treated (GC) conditions. Data are centered at commonly bound RAD21 and GR-peaks in NT cells treated with GC, depicting a ±10kb window around the peak center (n = 2). (D) Snapshots of GR, RAD21 and PAXIP1 ChIP-seq tracks around the FKBP5 locus. (E) 4C-seq experiments in NT and PAXIP1-KO cells using the FKBP5 TSS as a viewpoint. Cells were treated with GCs for 2 h. 4C data is shown as the mean of three independent biological experiments. ChIP tracks of RAD21 and GR in GC-treated NT-1 cells are depicted. ChIP-tracks are a representative snapshot of one biological replicate.
To confirm the FRAP results, we employed an orthogonal approach assessing PAXIP1 and the cohesin subunit RAD21 genome interactions in both wild-type and PAXIP1-depleted cells, by ChIP-seq analyses (Figure 3C and Supplementary Figure 3C). Both PAXIP1 and RAD21 were found to occupy GR-binding sites, which was dependent on GC-treatment, suggesting both PAXIP1 and RAD21 are recruited to these sites following GR chromatin binding (Figure 3C and Supplementary Fig 3C). Importantly, RAD21 recruitment to GR-bound sites was lost when PAXIP1 was knocked out (Figure 3C, D and Supplementary Figure 3C), confirming a critical role of PAXIP1 for RAD21 binding to the genome. Genomic distribution analyses showed that GR-PAXIP1-RAD21 co-occupancy on the genome occurs mainly at introns and distal intergenic regions (Supplementary Figure 3D). Motif analyses at GR-PAXIP1-RAD21 sites showed, as expected, GREs, HREs and FOX among the top most enriched motifs (Supplementary Figure 3E). Interestingly, when interrogating all RAD21 binding sites across the genome in GC-treated cells, we observed reduced promoter signal for RAD21 upon PAXIP1 knockout (Supplementary Fig 3F), implicating that PAXIP1 enables recruitment of RAD21 to the TSS of genes. In addition, we performed motif analyses on these sites, which were enriched for CTCF, but also for classical SHR motifs (Supplementary Figure 3G). In parallel, we also performed ChIP-seq experiments for H3K4me1, which is a histone modification marking for enhancers, generally deposited by KMT2D/C complex (58,60) (Figure 3C and Supplementary Figure 3H). Interestingly, enrichment for H3K4me1 at these GR-bound sites was not affected by PAXIP1 knockout at the FKBP5 locus (Supplementary Figure 3H) and on a genome-wide scale (Figure 3C). This is in line with the unaltered KMT2D complex recruitment to the GR complex in PAXIP1-KO cells (Figure 2C). Jointly, these data suggests that cohesin recruitment to GR-binding sites is dependent on non-canonical and novel function of PAXIP1.
As we demonstrated that PAXIP1 is required to localize and stabilize cohesin on the genome, and due to the importance of cohesin for 3D genome organization, we next sought to investigate whether PAXIP1 is necessary to maintain local enhancer-promoter interactions. For this purpose, we performed 4C-seq experiments in cells expressing or lacking PAXIP1 expression, selecting the TSS of FKBP5 gene as view-point (Figure 3E). In wild-type cells, we found a significant increase of interactions between the FKBP5 promoter and GR-bound enhancer regions upon stimulation with GCs (Figure 3E and Supplementary Fig 3I), which is in line with our previous observations reporting GR-stimulated promoter-enhancer interactions (31). The interacting regions spanned the genomic coordinates in which we observed co-binding of PAXIP1, RAD21 and GR (Figure 3E and Supplementary Figure 3I). However, in cells depleted for PAXIP1, promoter/enhancer interactions were not increased, remaining at basal levels despite GC treatment (Figure 3E and Supplementary Figure 3I). Cumulatively, these data illustrate a novel role of PAXIP1 in facilitating stable cohesin-chromatin dynamics, cohesin localization to GR-binding sites and GC-induced enhancer-promoter interactions.
PAXIP1 deficiency mimics the transcriptional program of STAG2 depleted cells
Since PAXIP1 and STAG2 appear to have a functional cross-talk that is reflected at the level of 3D genome organization, we next examined whether PAXIP1 and STAG2 also control similar transcriptional programs upon GR activation. For this, we generated RNA sequencing (RNA-seq) data in PAXIP1-KO and STAG2-KO cells, cultured in vehicle or GC-treated conditions (Figure 4A). Both PAXIP1 and STAG2 knockouts affected baseline expression of various genes, suggesting a broader significance of both these factors for gene expression regulation (Figure 4A). Importantly, we observed that GC-mediated gene expression was altered in PAXIP1-KO and STAG2-KO cells (Figure 4A), implying the importance of PAXIP1 and STAG2 in regulating only a subset of GR-target genes. Importantly, we observed a high correlation (R = 0.86, P-value < 0.001) on the level of directionality and magnitude of affected genes following knockout of PAXIP1 and STAG2 (Figure 4B and Supplementary Figure 4A), with a highly significant (P-value < 2.2e–16) overlap in the transcriptional changes in GC-treatment conditions (Supplementary Fig 4B). As STAG2 is crucial for enhancer-promoter interactions (61), we interrogated whether the similarities on PAXIP1- and STAG2-dependent transcriptional programs were due to regulation of chromatin looping by PAXIP1 on a genome-wide scale. To test this hypothesis in silico, we used sevenC analyses, which allows for the prediction of enhancer-promoter interactions based on CTCF sites and proximal RAD21 binding (54). Using RAD21 ChIP-seq data in NT and PAXIP1-KO cells, we observed the number of predicted loops in GC-treated cells were significantly decreased in PAXIP1-KO cells (Figure 4C and D; Supplementary Figure 4C). Next, we aimed to validate these predictions experimentally, and confirm that the changes in gene expression were due to perturbed enhancer-promoter interactions. For this purpose, we selected a GR-target gene -P4HA3- whose expression was significantly reduced upon PAXIP1-KO in GC-treated cells (Supplementary Figure 4D). 4C-seq analyses were performed, using the P4HA3 promoter as a viewpoint and confirmed a loss of enhancer-promoter interactions in the area spanning RAD21 binding, which also coincides with the genomic locus of PPME1 gene (Supplementary Figure 4E). Interestingly, GR-mediated expression of genes in close proximity of the P4HA3 locus -such as PPME1- was also significantly reduced (Supplementary Figure 4F), which could suggest regional transcriptomic effects when enhancer-promoter interactions are lost. All together, we demonstrate that PAXIP1 and STAG2 control a similar transcriptional landscape by maintaining enhancer-promoter interactions.
Figure 4.
PAXIP1 and STAG2 control similar transcriptional programs. (A) MA plots depicting differential gene expression between DMSO- and GC-treatment in NT, PAXIP1-KO and STAG2-KO cells, NT versus PAXIP1-KO in GC-treated cells, and NT versus STAG2-KO in GC-treated cells. Red and blue dots indicate significantly differentially expressed genes with log2(fold change) >1 and adjusted P-value <0.05. Data represent the average of three independent biological replicates. (B) Correlation of differentially expressed genes in STAG2-KO cells with PAXIP1-KO cells, both in GC-treated conditions. Genes differentially expressed in PAXIP1-KO cells (Padj < 0.05) were ranked based on log2(fold change) and differential expression of those genes was plotted for STAG2-KOs. (C) sevenC analyses of WT and PAXIP1-KO based on RAD21 ChIP-seq and CTCF motifs. Number of predicted genomic interactions per chromosome in GC-treated conditions are depicted. One sample t-test was used. P-value < 0.0001. (D) Snapshot of sevenC predicted chromosomal interactions in chr 11, both in NT and PAXIP1-KO cells.
Loss of PAXIP1 enhances the tumor suppressor action of GR by modifying local chromatin interactions
In this study, we used the lung cancer model A549 to study GR co-regulators. Importantly, we (31) and others (8,62–64) have previously shown that GC treatment of these lung cancer cells reduces tumor cell proliferation, for which we recently reported the mechanisms underlying GR-mediated cancer cell dormancy (31). As we observed major changes in the transcriptional landscape of PAXIP1- and STAG2-deficient cells, we next interrogated whether this would result in alterations of tumor cell growth following GR activation. For these purposes, we performed cell proliferation analyses in NT, PAXIP1-KO and STAG2-KO cells, treated with GCs. In line with our previous findings, GC-treatment diminished tumor cell proliferation capacity (Figure 5A and Supplementary Figure 5A). Interestingly, knockout of PAXIP1 and STAG2 further strengthened the anti-proliferative effects of GC treatment, while not affecting the cell growth capacity in absence of GCs (Figure 5A and Supplementary Figure 5A). These data suggest an enhanced GR-mediated tumor suppressive capacity upon loss of PAXIP1 or STAG2. Based on these results, we hypothesized that loss of PAXIP1 or STAG2 leads to the activation of newly acquired GR-responsive tumor suppressor genes, dampening cell cycle progression upon PAXIP1 and STAG2 depletion. To test this hypothesis, we selectively analyzed genes with significantly gained expression in GC-treated PAXIP1-KO and STAG2-KO cells, and tested these for overlap with a Cell Cycle geneset (hsa04110), resulting in the identification of two candidate driver genes: CDKN1B and CDKN2C (Figure 5B). Interestingly, both CDKN1B and CDKN2C are known to mediate cell cycle arrest (65,66). As PAXIP1 depletion resulted in increased GR chromatin binding around the CDKN1B locus, in contrast to CDKN2C (Supplementary Figure 5B), we selectively focused follow-up experiments on CDKN1B (Supplementary Fig 5B). Importantly, PAXIP1 knockout resulted in increased CDKN1B expression, both on mRNA (Figure 5C), and protein level with increased P27 -encoded by CDKN1B- expression (Figure 5D). To confirm that GR-mediated expression of CDKN1B was responsible for the enhanced sensitivity to GCs in PAXIP1-KO cells, we generated PAXIP1/CDKN1B double knockout (DKO) cells (Supplementary Figure 5C). We observed that PAXIP1/CDKN1B-DKO cells restored the tumor cell proliferative phenotype that was lost upon PAXIP1 knockout, rendering the cells again less responsive to GCs (Figure 5E). Importantly, our observations indicate that cells only depleted for PAXIP1 (Supplementary Figure 5D) were unable to rescue the sensitivity to GCs. Similarly, PAXIP1/CDKN1B-DKO cells did not have any proliferation advantage in DMSO conditions (Figure 5E), suggesting that the increased levels of CDKN1B in PAXIP1-KO cells are solely responsible for enhancing the response to GCs.
Figure 5.
Loss of STAG2 and PAXIP1 enhance the tumor suppressor action of GR by modifying local chromatin interactions. (A) Cell proliferation of NT and PAXIP1-KO cells following DMSO or GC treatment over time (n = 3 up to 168 h, n = 2 from 168 h to 216 h, error bars represent 95% confidence interval). (B) Overlap of significantly differentially expressed genes in PAXIP1-KO and STAG2-KO cells following GC stimulation (log2(fold change) > 0.75; Padj < 0.05), with a cell cycle gene set (hsa04110). (C) Boxplot showing normalized CDKN1B expression in NT, PAXIP1-KO and GR-KO cells upon vehicle or GC treatment for 8 h (n = 3). P-value was calculated using U Mann–Whitney t-test. (D) P27 protein expression levels in NT, PAXIP1-KO and GR-KO cells upon vehicle or GC treatment for 24 h. Actin was used as a loading control (n = 2). (E) Cell proliferation of PAXIP1-KO cells and PAXIP1/CDKN1B-DKO following DMSO or GC treatment over time (n = 3 up to 168 h, n = 2 from 168 h to 216 h, error bars represent 95% confidence interval). (F) 4C-seq experiments in NT and PAXIP1-KO cells using the CDKN1B TSS as a viewpoint showing NT versus PAXIP1-KO in GC-treated conditions. Cells were treated for 2 h with GCs. 4C data is shown as the mean of three independent biological experiments. ChIP tracks of RAD21 and GR in GC-treated NT-1 cells are depicted. ChIP-tracks are a representative snapshot of one biological replicate.
As PAXIP1 is required for enhancer-promoter interactions around the FKBP5 locus (Figure 3E), we next investigated impact of PAXIP1 knockout on 3D genome regulation around the CDKN1B locus. For that, we performed 4C-seq experiments using the CDKN1B promoter as viewpoint and observed again alterations upon PAXIP1 loss, but now this was associated with increased target gene expression (Figure 5F). A loss of long-range chromatin interactions was observed stemming from the CDKN1B promoter in PAXIP1-KO cells, spanning a region containing both GR and RAD21 binding sites. These data suggest that this loop may act as an insulator between the promoter and the GR binding site, allowing GR-mediated transcription of CDKN1B upon PAXIP1 loss due to loop disruption. Follow-up studies are needed to corroborate this model.
Cumulatively, we observed that loss of PAXIP1 and STAG2 enhances the response of lung cancer cells to GCs, mediated by alterations in long-range 3D-genome contacts and resulting gained expression of a novel target gene.
DISCUSSION
SHRs are ligand-sensing transcription factors that play critical roles in a large variety of cellular processes, including maintenance of cellular homeostasis, metabolism, immune system activity, sex dimorphism and cell proliferation (8). SHRs are expressed throughout the body, often in a highly tissue-selective manner, and their deregulation can result in different human pathologies such as metabolic disorders and cancer development (67). SHRs regulate gene expression through specific DNA regions, generally enhancers, along with a large complex of co-regulators (3). Although it is known that SHRs can interact with a large number of proteins, little is known about which ones are essential for their activity.
In this study, we use GR function in lung cancer as a model system, and interrogate on a genome-wide level which co-regulator proteins are required for GR activity. Due to the design of our screen, we exclusively identified hits that are not replaceable in their ability to shape GR-mediated gene expression, suggesting that many of the classical GR interacting proteins may be functionally redundant, and could be compensated by other components in the complex. Interestingly, we identified both PAXIP1 and STAG2 as two key essential proteins in facilitating GR-mediated gene expression, that converge to control 3D genome architecture. We demonstrate that PAXIP1 is required for enhancer-promoter interactions mediated by STAG2, necessary to drive GR-mediated gene expression.
We propose a mode of the functional cross-talk between PAXIP1 and STAG2 that is as follows: upon activation, GR and PAXIP1 bind together to regulatory elements, inducing enhancer-promoter interactions, for which cohesin is necessary (Figure 6A and B). We identified PAXIP1 as essential for recruitment of cohesin to GR-binding sites and consequently, to maintain enhancer-promoter interactions. Upon loss of PAXIP1, there is a reduction of localization of cohesin to GR binding sites and this results in impaired GR-mediated transcription (Figure 6A). On the other hand, we propose that GR can also bind together with PAXIP1 and cohesin to GR-binding sites resulting in an insulator loop, avoiding enhancer/promoter contacts to prevent GR-mediated transcription, or prevent basal levels of gene expression (Figure 6B). However, in the absence of PAXIP1, cohesin localization to those sites is lost, resulting in a loss of insulator loop formation and consequently gained gene expression (Figure 6B).
Figure 6.
Proposed model. PAXIP1 and STAG2 converge to facilitate enhancer-promoter interactions and fine-tune GR-target gene expression either by enhancing expression of GR-target genes (A) or by blocking expression through insulator loops (B).
STAG2 belongs to the ring-shaped structured cohesin complex, which plays an essential role in sister chromatid cohesion, DNA damage repair, as well as the formation and stability of cis-chromatin loops for transcriptional regulation (68–70). Here, we report a new role of PAXIP1 regulating cohesin function. We demonstrate that PAXIP1 is required for cohesin stability on chromatin and its localization to GR-bound sites. While the functional role of PAXIP1 in 3D genome organization is novel, prior studies have reported an interaction between 3D-genome organization proteins and different SHRs, including GR and ERα, being able to bind specific genomic regions together with cohesin (20,71–74). Moreover, GR activation increases both the number and intensity of enhancer-promoter interactions of their target genes upon hormonal stimulation (20). GR was recently described to interact with NIPBL, which is responsible for cohesin loading onto DNA, localization of cohesin to GR-bound sites, and promotion of long-range genomic interactions (18).
Here we show, that cohesin binding to GR-bound sites is critically dependent on PAXIP1, with its loss altering GR-mediated transcriptome. Importantly, PAXIP1 and STAG2 depletion resulted in similar changes in gene expression, providing further evidence for their functional interplay. PAXIP1 is a component of the KMT2D/C complex (32,33), a histone methyltransferase necessary for H3K4 methylation at enhancers of actively transcribed genes (60,75). Surprisingly, we observed that in PAXIP1-depleted cells, GR-mediated recruitment of the KMT2D complex and its subunits was not altered. Moreover, H3K4me1 levels remained unaffected upon PAXIP1 knockout, and depletion of KMT2D did not alter expression of the GR-target protein FKBP5. Altogether, this suggests that PAXIP1 can function in a KMT2D-independent manner to regulate cohesin binding and stability on the genome and subsequently regulate enhancer-promoter interactions following hormonal-response. In line with this, prior reports describe PAXIP1 to act in a KMT2D/C-independent manner in conjunction with its counterpart PAGR1 (76). PAXIP1 has been involved in immunoglobin class switching and V(D)J recombination (76,77); a biological process in which cohesin-mediated loop extrusion plays a key role (78,79). Moreover, PAXIP1 is critically involved in DNA damage response, localizing together with 53BP1 in DNA damage sites, to enable ATM-mediated phosphorylation of cohesin subunit SMC1 and ensuring a correct DNA damage response (80,81). Finally, in parallel to our study, Van Schie et al, described the interplay of PAXIP1 and PAGR1 to maintain chromatin-bound cohesin during cell cycle progression (82). These observations not only support our findings of PAXIP1 regulating cohesin activity in a KMT2D/C-independent manner, but also highlight a more-general contribution of PAXIP1 function on cohesin action, beyond its role on facilitating promoter/enhancer interactions, and re-positions PAXIP1 as a critical component of cohesin function.
Our study, together with the work from Van Schie et al. (82), provides evidence for interactions between PAXIP1 and members of the cohesin complex. However, it should be noted that these macro-molecular interactions do not necessarily indicate direct protein-protein interactions between the specific components of interest, namely PAXIP1 and STAG2. Future studies can investigate the direct interaction between PAXIP1 and STAG2 using biochemical techniques such as protein pull-down assays, surface plasmon resonance, co-crystallization, or NMR spectroscopy. While outside the scope of our current study, these methods can provide direct evidence of their interaction, as well as insights into the binding affinity, kinetics, and 3D structure of the complex formed between the two proteins.
While our screen was effective in identifying modulators of GR activity, there were certain limitations that should be acknowledged. For instance, we found that depleting PAXIP1 and STAG2 only partially impairs GR-mediated transcription, which suggests that additional co-regulators are necessary for full GR function. Unfortunately, our experimental setup did not allow us to identify these co-regulators. Due to the design of our screen and the dynamic window used to sort FKBP5low cells, we could only identify GR co-regulators whose individual knockout strongly reduces the expression of FKBP5. Consequently, subtle effects following perturbations of individual genes might have been missed. Furthermore, our screen relied on a single gene as a readout of GR activity, and the composition of the GR complex could vary depending on the gene being investigated. Identifying co-regulators that bind in a locus-specific manner together with GR or other SHRs is a persistent challenge. To address this, conducting a genome-wide CRISPR screen with different GR targets as readouts would yield more information about the composition of the GR complex in a context-dependent manner. Other teams have made considerable efforts to identify complexes on a locus-specific manner, and albeit challenging, it presents a valuable opportunity to discover distinct GR-complexes (83–85).
Although our study is focused on the functional cross-talk between PAXIP1 and STAG2 following GR activation in lung cancer, we hypothesize that our results may be extrapolated to other tissues and cis-acting transcription factors, which is supported by the more-general action of cohesin in facilitating transcription factor action in 3D genomic space, beyond GR alone. We observed a high level of co-dependency between PAXIP1 and the cohesin subunits STAG2, SMC1A and cohesin loaders MAU2 and NIBPL across >1000 cell lines, suggesting that the PAXIP1-cohesin cross-talk is underexplored, yet serves a universal synergistic role across many tissues and tumor types. Therefore, interrogating whether the activity of other cis-acting transcription factors—including NF-κB or other SHRs—are dependent on PAXIP1-cohesin functional cross-talk would be of high interest for future studies.
GR is known to function as a tumor suppressor in lung cancer (31). Here we show that depletion of PAXIP1 or STAG2 results in enhanced tumor suppressor activity of GR, due to increased CDKN1B expression. The latter encodes for P27; a well described tumor suppressor, which is often downregulated in cancer and serves as a prognostic marker in lung cancer (66). We describe a possible mechanism, by which changes in enhancer-promoter interactions proximal to the CDKN1B locus, result in acquired regulation of CDKN1B. We hypothesize, that this altered CDKN1B expression a consequence of disruption of an insulator loop following PAXIP1 and STAG2 depletion, which warrants further investigation.
GR is known to function as a tumor suppressor in lung cancer (31). With PAXIP1 serving as genuine GR interactor and functionally relevant for GR activity, we anticipated loss of PAXIP1 to alleviate the tumor-suppressive features of GR. Interestingly, depletion of PAXIP1 had the opposite effects on GR-induced cell cycle arrest. Here we show that depletion of PAXIP1 results in enhanced tumor suppressor activity of GR, due to increased CDKN1B expression. The latter encodes for P27; a well described tumor suppressor, which is often downregulated in cancer and serves as a prognostic marker in lung cancer (66). Our observations on the role of PAXIP1 in maintaining enhancer-promoter contacts prompted us to investigate changes in the 3D-genome organization around the CDKN1B locus. In line with our hypothesis, we found that depletion of PAXIP1 disrupted a pre-established loop between the promoter of CDKN1B and an enhancer. This suggests that this loop may serve as an insulator between the enhancer and the CDKN1B promoter, avoiding CDKN1B expression, and only after loss of PAXIP1, CDKN1B expression is gained. We propose the aforementioned model as an explanation of PAXIP1 role in GR-tumor suppressor activities, but further experiments are required to confirm our observations.
Regulation of gene expression in 3D genomic space is a research field in active development, in which the cohesin complex—originally reported to drive mitotic entry and orchestrate chromosomal segregation—plays a pivotal role. Now, we reveal that PAXIP1—originally reported as part of the KMT2D/C complex – functionally and critically contributes to cohesin action, driving expression of genes under control of a cis-acting transcription factor, the GR.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Zwart and Bergman labs for valuable feedback, suggestions and input. This work was supported by KWF/Alpe d’Huzes and Oncode Institute. We would like to thank the NKI genomics core facility for next-generation sequencing and bioinformatics support. We thank the NKI Flow Cytometry facility for technical support. We thank the NKI Proteomics/Mass Spectrometry facility (M.A. and L.H. are supported by the Dutch NWO X-omics Initiative). We thank Reuven Agami, Julien Champagne and Remco Nagel for fruitful discussions and technical support.
Author contributions: Conceptualization: I.M.P., S.P. and W.Z.; S.P. and W.Z. were responsible for project funding. I.M.P., S.G. and A.K. performed 4C-seq experiments and S.G. analyzed 4C-seq experiments; I.M.P., K.S., S.P., N.A. and S.B. performed RIME, ChIP-seq and RNA-seq experiments; I.M.P. and N.A. performed RT-qPCR and western blot analyses; I.M.P., K.S., A.Z., M.v.B. performed the CRISPR screen; J.S. and T.M.S. provided bioinformatics support for peakcalling; L.H. and M.A. performed and analyzed the proteomic experiments.; C.L. and R.L.B. provided bioinformatics and technical support for the CRISPR screen; S.Y., B.G. and A.B.H. performed FRAP experiments. I.M.P., S.P. and W.Z. wrote the manuscript, with input from all co-authors.
Contributor Information
Isabel Mayayo-Peralta, Division of Oncogenomics, Oncode Institute, The Netherlands Cancer Institute, Amsterdam, The Netherlands.
Sebastian Gregoricchio, Division of Oncogenomics, Oncode Institute, The Netherlands Cancer Institute, Amsterdam, The Netherlands.
Karianne Schuurman, Division of Oncogenomics, Oncode Institute, The Netherlands Cancer Institute, Amsterdam, The Netherlands.
Selçuk Yavuz, Erasmus Optical Imaging Center, Erasmus University Medical Center Rotterdam, Rotterdam, The Netherland.
Anniek Zaalberg, Division of Oncogenomics, Oncode Institute, The Netherlands Cancer Institute, Amsterdam, The Netherlands.
Aleksandar Kojic, Division of Oncogenomics, Oncode Institute, The Netherlands Cancer Institute, Amsterdam, The Netherlands.
Nina Abbott, Division of Oncogenomics, Oncode Institute, The Netherlands Cancer Institute, Amsterdam, The Netherlands.
Bart Geverts, Erasmus Optical Imaging Center, Erasmus University Medical Center Rotterdam, Rotterdam, The Netherland; Department of Pathology, Erasmus University Medical Center Rotterdam, Rotterdam, The Netherlands.
Suzanne Beerthuijzen, Division of Oncogenomics, Oncode Institute, The Netherlands Cancer Institute, Amsterdam, The Netherlands.
Joseph Siefert, Division of Oncogenomics, Oncode Institute, The Netherlands Cancer Institute, Amsterdam, The Netherlands.
Tesa M Severson, Division of Oncogenomics, Oncode Institute, The Netherlands Cancer Institute, Amsterdam, The Netherlands.
Martijn van Baalen, Flow Cytometry Facility, The Netherlands Cancer Institute, Amsterdam, The Netherlands.
Liesbeth Hoekman, Proteomics Facility, The Netherlands Cancer Institute, Amsterdam, The Netherlands.
Cor Lieftink, Division of Molecular Carcinogenesis, The NKI Robotics and Screening Centre, The Netherlands Cancer Institute, Amsterdam, The Netherlands.
Maarten Altelaar, Proteomics Facility, The Netherlands Cancer Institute, Amsterdam, The Netherlands; Biomolecular Mass Spectrometry and Proteomics, Bijvoet Center for Biomolecular Research, Utrecht Institute for Pharmaceutical Sciences, Utrecht University and Netherlands Proteomics Centre, Utrecht, The Netherlands.
Roderick L Beijersbergen, Division of Molecular Carcinogenesis, The NKI Robotics and Screening Centre, The Netherlands Cancer Institute, Amsterdam, The Netherlands.
Adriaan B Houtsmuller, Erasmus Optical Imaging Center, Erasmus University Medical Center Rotterdam, Rotterdam, The Netherland.
Stefan Prekovic, Division of Oncogenomics, Oncode Institute, The Netherlands Cancer Institute, Amsterdam, The Netherlands; Center for Molecular Medicine, University Medical Center Utrecht, Utrecht, The Netherlands.
Wilbert Zwart, Division of Oncogenomics, Oncode Institute, The Netherlands Cancer Institute, Amsterdam, The Netherlands; Laboratory of Chemical Biology and Institute for Complex Molecular Systems, Department of Biomedical Engineering, Eindhoven University of Technology, Eindhoven, The Netherlands.
DATA AVAILABILITY
RNA-seq, ChIP-seq, 4C-seq has been deposited to the GEO database (GSE221917). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (86) partner repository with the dataset identifier PXD038902.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
KWF Kankerbestrijding/Alpe d’Huzes; Oncode Institute; Dutch NWO X-omics Initiative; NWO Building Blocks of Life program project Genometrack (737.016.014). Funding for open access charge: Financial resources are available to cover publication charges.
Conflict of interest statement. None declared.
This paper is linked to: doi:10.1093/nar/gkad756.
REFERENCES
- 1. Lazar M.A. Maturing of the nuclear receptor family. J. Clin. Investig. 2017; 127:1123–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mayayo-Peralta I., Prekovic S., Zwart W.. Estrogen receptor on the move: cistromic plasticity and its implications in breast cancer. Mol. Aspects Med. 2021; 78:100939. [DOI] [PubMed] [Google Scholar]
- 3. Panigrahi A., O’Malley B.W. Mechanisms of enhancer action: the known and the unknown. Genome Biol. 2021; 22:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aranda A., Pascual A.. Nuclear hormone receptors and gene expression. Physiol. Rev. 2001; 81:1269–1304. [DOI] [PubMed] [Google Scholar]
- 5. Brinkmann A.O., Blok L.J., de Ruiter P.E., Doesburg P., Steketee K., Berrevoets C.A., Trapman J.. Mechanisms of androgen receptor activation and function. J. Steroid Biochem. Mol. Biol. 1999; 69:307–313. [DOI] [PubMed] [Google Scholar]
- 6. Carroll J.S., Meyer C.A., Song J., Li W., Geistlinger T.R., Eeckhoute J., Brodsky A.S., Keeton E.K., Fertuck K.C., Hall G.F.et al.. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 2006; 38:1289–1297. [DOI] [PubMed] [Google Scholar]
- 7. Pottier N., Yang W., Assem M., Panetta J.C., Pei D., Paugh S.W., Cheng C., den Boer M.L., Relling M. v., Pieters R.et al.. The SWI/SNF chromatin-remodeling complex and glucocorticoid resistance in acute lymphoblastic leukemia. J. Natl. Cancer Inst. 2008; 100:1792–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mayayo-Peralta I., Zwart W., Prekovic S.. Duality of glucocorticoid action in cancer: tumor-suppressor or oncogene?. Endocr. Relat. Cancer. 2021; 28:R157–R171. [DOI] [PubMed] [Google Scholar]
- 9. Linder S., van der Poel H.G., Bergman A.M., Zwart W., Prekovic S.. Enzalutamide therapy for advanced prostate cancer: efficacy, resistance and beyond. Endocr. Relat. Cancer. 2019; 26:R31–R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Arruabarrena-Aristorena A., Maag J.L.V., Kittane S., Cai Y., Karthaus W.R., Ladewig E., Park J., Kannan S., Ferrando L., Cocco E.et al.. FOXA1 mutations reveal distinct chromatin profiles and influence therapeutic response in breast cancer. Cancer Cell. 2020; 38:534–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cosma M.P. Ordered recruitment: gene-specific mechanism of transcription activation. Mol. Cell. 2002; 10:227–236. [DOI] [PubMed] [Google Scholar]
- 12. Liu Z., Merkurjev D., Yang F., Li W., Oh S., Friedman M.J., Song X., Zhang F., Ma Q., Ohgi K.A.et al.. Enhancer activation requires trans-recruitment of a mega transcription factor complex. Cell. 2014; 159:358–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yan J., Enge M., Whitington T., Dave K., Liu J., Sur I., Schmierer B., Jolma A., Kivioja T., Taipale M.et al.. Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell. 2013; 154:801–813. [DOI] [PubMed] [Google Scholar]
- 14. Murakami S., Nagari A., Kraus W.L.. Dynamic assembly and activation of estrogen receptor α enhancers through coregulator switching. Genes Dev. 2017; 31:1535–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Voss T.C., Schiltz R.L., Sung M.-H., Yen P.M., Stamatoyannopoulos J.A., Biddie S.C., Johnson T.A., Miranda T.B., John S., Hager G.L.. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell. 2011; 146:544–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Iwafuchi-Doi M., Zaret K.S.. Pioneer transcription factors in cell reprogramming. Genes Dev. 2014; 28:2679–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang F., Ma Q., Liu Z., Li W., Tan Y., Jin C., Ma W., Hu Y., Shen J., Ohgi K.A.et al.. Glucocorticoid receptor: megatrans switching mediates the repression of an ERα-regulated transcriptional program. Mol. Cell. 2017; 66:321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rinaldi L., Fettweis G., Kim S., Garcia D.A., Fujiwara S., Johnson T.A., Tettey T.T., Ozbun L., Pegoraro G., Puglia M.et al.. The glucocorticoid receptor associates with the cohesin loader NIPBL to promote long-range gene regulation. Sci. Adv. 2022; 8:eabj8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. le Dily F., Vidal E., Cuartero Y., Quilez J., Nacht A.S., Vicent G.P., Carbonell-Caballero J., Sharma P., Villanueva-Cañas J.L., Ferrari R.et al.. Hormone-control regions mediate steroid receptor–dependent genome organization. Genome Res. 2019; 29:29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. D’Ippolito A.M., McDowell I.C., Barrera A., Hong L.K., Leichter S.M., Bartelt L.C., Vockley C.M., Majoros W.H., Safi A., Song L.et al.. Pre-established chromatin interactions mediate the genomic response to glucocorticoids. Cell Syst. 2018; 7:146–160. [DOI] [PubMed] [Google Scholar]
- 21. Greulich F., Wierer M., Mechtidou A., Gonzalez-Garcia O., Uhlenhaut N.H.. The glucocorticoid receptor recruits the COMPASS complex to regulate inflammatory transcription at macrophage enhancers. Cell Rep. 2021; 34:108742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Foulds C.E., Feng Q., Ding C., Bailey S., Hunsaker T.L., Malovannaya A., Hamilton R.A., Gates L.A., Zhang Z., Li C.et al.. Proteomic analysis of coregulators bound to ERα on DNA and nucleosomes reveals coregulator dynamics. Mol. Cell. 2013; 51:185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McKenna N.J., Lanz R.B., O’Malley B.W. Nuclear receptor coregulators: cellular and molecular biology. Endocr. Rev. 1999; 20:321–344. [DOI] [PubMed] [Google Scholar]
- 24. Mohammed H., Taylor C., Brown G.D., Papachristou E.K., Carroll J.S., D’Santos C.S. Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) for analysis of chromatin complexes. Nat. Protoc. 2016; 11:316–326. [DOI] [PubMed] [Google Scholar]
- 25. Papachristou E.K., Kishore K., Holding A.N., Harvey K., Roumeliotis T.I., Chilamakuri C.S.R., Omarjee S., Chia K.M., Swarbrick A., Lim E.et al.. A quantitative mass spectrometry-based approach to monitor the dynamics of endogenous chromatin-associated protein complexes. Nat. Commun. 2018; 9:2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mohammed H., D’Santos C., Serandour A.A., Ali H.R., Brown G.D., Atkins A., Rueda O.M., Holmes K.A., Theodorou V., Robinson J.L.L.et al.. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep. 2013; 3:342–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stelloo S., Nevedomskaya E., Kim Y., Hoekman L., Bleijerveld O.B., Mirza T., Wessels L.F.A., van Weerden W.M., Altelaar A.F.M., Bergman A.M.et al.. Endogenous androgen receptor proteomic profiling reveals genomic subcomplex involved in prostate tumorigenesis. Oncogene. 2018; 37:313–322. [DOI] [PubMed] [Google Scholar]
- 28. Hwang J.H., Arafeh R., Seo J.-H., Baca S.C., Ludwig M., Arnoff T.E., Sawyer L., Richter C., Tape S., Bergom H.E.et al.. CREB5 reprograms FOXA1 nuclear interactions to promote resistance to androgen receptor-targeting therapies. Elife. 2022; 11:e73223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Siersbæk R., Scabia V., Nagarajan S., Chernukhin I., Papachristou E.K., Broome R., Johnston S.J., Joosten S.E.P., Green A.R., Kumar S.et al.. IL6/STAT3 signaling hijacks estrogen receptor α enhancers to drive breast cancer metastasis. Cancer Cell. 2020; 38:412–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mohammed H., Russell I.A., Stark R., Rueda O.M., Hickey T.E., Tarulli G.A., Serandour A.A.A., Birrell S.N., Bruna A., Saadi A.et al.. Progesterone receptor modulates ERα action in breast cancer. Nature. 2015; 523:313–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Prekovic S., Schuurman K., Mayayo-Peralta I., Manjón A.G., Buijs M., Yavuz S., Wellenstein M.D., Barrera A., Monkhorst K., Huber A.et al.. Glucocorticoid receptor triggers a reversible drug-tolerant dormancy state with acquired therapeutic vulnerabilities in lung cancer. Nat. Commun. 2021; 12:4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Patel S.R., Kim D., Levitan I., Dressler G.R.. The BRCT-domain containing protein PTIP links PAX2 to a histone H3, lysine 4 methyltransferase complex. Dev. Cell. 2007; 13:580–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cho Y.-W., Hong T., Hong S., Guo H., Yu H., Kim D., Guszczynski T., Dressler G.R., Copeland T.D., Kalkum M.et al.. PTIP associates with MLL3- and MLL4-containing Histone H3 lysine 4 methyltransferase complex. J. Biol. Chem. 2007; 282:20395–20406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F., Smith I., Tothova Z., Wilen C., Orchard R.et al.. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016; 34:184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sanson K.R., Hanna R.E., Hegde M., Donovan K.F., Strand C., Sullender M.E., Vaimberg E.W., Goodale A., Root D.E., Piccioni F.et al.. Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat. Commun. 2018; 9:5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Korkmaz G., Lopes R., Ugalde A.P., Nevedomskaya E., Han R., Myacheva K., Zwart W., Elkon R., Agami R.. Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nat. Biotechnol. 2016; 34:192–198. [DOI] [PubMed] [Google Scholar]
- 37. Li W., Xu H., Xiao T., Cong L., Love M.I., Zhang F., Irizarry R.A., Liu J.S., Brown M., Liu X.S.. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014; 15:554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelsen T.S., Heckl D., Ebert B.L., Root D.E., Doench J.G.et al.. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014; 343:84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cost G.J. Enzymatic ligation assisted by nucleases: simultaneous ligation and digestion promote the ordered assembly of DNA. Nat. Protoc. 2007; 2:2198–2202. [DOI] [PubMed] [Google Scholar]
- 40. Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F.. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013; 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J.. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016; 13:731–740. [DOI] [PubMed] [Google Scholar]
- 42. Schmidt D., Wilson M.D., Spyrou C., Brown G.D., Hadfield J., Odom D.T.. ChIP-seq: using high-throughput sequencing to discover protein–DNA interactions. Methods. 2009; 48:240–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sturm M., Schroeder C., Bauer P.. SeqPurge: highly-sensitive adapter trimming for paired-end NGS data. BMC Bioinformatics. 2016; 17:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W.et al.. Model-based Analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jalili V., Matteucci M., Masseroli M., Morelli M.J.. Using combined evidence from replicates to evaluate ChIP-seq peaks. Bioinformatics. 2015; 31:2761–2769. [DOI] [PubMed] [Google Scholar]
- 47. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lerdrup M., Johansen J.V., Agrawal-Singh S., Hansen K. An interactive environment for agile analysis and visualization of ChIP-sequencing data. Nat. Struct. Mol. Biol. 2016; 23:349–357. [DOI] [PubMed] [Google Scholar]
- 49. Yu G., Wang L.-G., He Q.-Y.. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015; 31:2382–2383. [DOI] [PubMed] [Google Scholar]
- 50. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Krijger P.H.L., Geeven G., Bianchi V., Hilvering C.R.E., de Laat W.. 4C-seq from beginning to end: a detailed protocol for sample preparation and data analysis. Methods. 2020; 170:17–32. [DOI] [PubMed] [Google Scholar]
- 52. Gregoricchio S., Polit L., Esposito M., Berthelet J., Delestré L., Evanno E., Diop M., Gallais I., Aleth H., Poplineau M.et al.. HDAC1 and PRC2 mediate combinatorial control in SPI1/PU.1-dependent gene repression in murine erythroleukaemia. Nucleic Acids Res. 2022; 50:7938–7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Geverts B., van Royen M.E., Houtsmuller A.B.. Analysis of biomolecular dynamics by FRAP and computer simulation. Methods Mol Biol. 2015; 1251:109–133. [DOI] [PubMed] [Google Scholar]
- 54. Ibn-Salem J., Andrade-Navarro M.A.. 7C: Computational chromosome conformation capture by correlation of ChIP-seq at CTCF motifs. BMC Genomics. 2019; 20:777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L.. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019; 37:907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Anders S., Pyl P.T., Huber W.. HTSeq – a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015; 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cenik B.K., Shilatifard A.. 2021) COMPASS and SWI/SNF complexes in development and disease. Nat. Rev. Genet. 22:38–58. [DOI] [PubMed] [Google Scholar]
- 59. Ciosk R., Shirayama M., Shevchenko A., Tanaka T., Toth A., Shevchenko A., Nasmyth K.. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell. 2000; 5:243–254. [DOI] [PubMed] [Google Scholar]
- 60. Lee J.-E., Wang C., Xu S., Cho Y.-W., Wang L., Feng X., Baldridge A., Sartorelli V., Zhuang L., Peng W.et al.. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. Elife. 2013; 2:e01503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kojic A., Cuadrado A., de Koninck M., Giménez-Llorente D., Rodríguez-Corsino M., Gómez-López G., le Dily F., Marti-Renom M.A., Losada A.. Distinct roles of cohesin-SA1 and cohesin-SA2 in 3D chromosome organization. Nat. Struct. Mol. Biol. 2018; 25:496–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Taylor K.M., Ray D.W., Sommer P.. Glucocorticoid receptors in lung cancer: new perspectives. J. Endocrinol. 2016; 229:R17–R28. [DOI] [PubMed] [Google Scholar]
- 63. Balansky R., Ganchev G., Iltcheva M., Steele V.E., de Flora S.. Prevention of cigarette smoke-induced lung tumors in mice by budesonide, phenethyl isothiocyanate, and N -acetylcysteine. Int. J. Cancer. 2009; 126:1047–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lu Y.-S., Lien H.-C., Yeh P.-Y., Kuo S.-H., Chang W.-C., Kuo M.-L., Cheng A.-L.. Glucocorticoid receptor expression in advanced non-small cell lung cancer: clinicopathological correlation and in vitro effect of glucocorticoid on cell growth and chemosensitivity. Lung Cancer. 2006; 53:303–310. [DOI] [PubMed] [Google Scholar]
- 65. Li G.-S., Chen G., Liu J., Tang D., Zheng J.-H., Luo J., Jin M.-H., Lu H.-S., Bao C.-X., Tian J.et al.. Clinical significance of cyclin-dependent kinase inhibitor 2C expression in cancers: from small cell lung carcinoma to pan-cancers. BMC Pulm Med. 2022; 22:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chu I.M., Hengst L., Slingerland J.M.. The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy. Nat. Rev. Cancer. 2008; 8:253–267. [DOI] [PubMed] [Google Scholar]
- 67. Guyton AC &, Hall JE. Textbook of Medical Physiology. 2006; Amsterdam, Netherlands: Elsevier. [Google Scholar]
- 68. Davidson I.F., Peters J.-M.. Genome folding through loop extrusion by SMC complexes. Nat. Rev. Mol. Cell Biol. 2021; 22:445–464. [DOI] [PubMed] [Google Scholar]
- 69. Haarhuis J.H.I., Elbatsh A.M.O., Rowland B.D.. Cohesin and its regulation: on the logic of X-shaped chromosomes. Dev. Cell. 2014; 31:7–18. [DOI] [PubMed] [Google Scholar]
- 70. Li Y., Haarhuis J.H.I., Sedeño Cacciatore Á., Oldenkamp R., van Ruiten M.S., Willems L., Teunissen H., Muir K.W., de Wit E., Rowland B.D.et al.. The structural basis for cohesin–CTCF-anchored loops. Nature. 2020; 578:472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hakim O., Sung M.-H., Voss T.C., Splinter E., John S., Sabo P.J., Thurman R.E., Stamatoyannopoulos J.A., de Laat W., Hager G.L.. Diverse gene reprogramming events occur in the same spatial clusters of distal regulatory elements. Genome Res. 2011; 21:697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Stavreva D.A., Coulon A., Baek S., Sung M.-H., John S., Stixova L., Tesikova M., Hakim O., Miranda T., Hawkins M.et al.. Dynamics of chromatin accessibility and long-range interactions in response to glucocorticoid pulsing. Genome Res. 2015; 25:845–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. le Dily F., Vidal E., Cuartero Y., Quilez J., Nacht A.S., Vicent G.P., Carbonell-Caballero J., Sharma P., Villanueva-Cañas J.L., Ferrari R.et al.. Hormone-control regions mediate steroid receptor–dependent genome organization. Genome Res. 2019; 29:29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Schmidt D., Schwalie P.C., Ross-Innes C.S., Hurtado A., Brown G.D., Carroll J.S., Flicek P., Odom D.T.. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res. 2010; 20:578–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Herz H.-M., Hu D., Shilatifard A.. Enhancer malfunction in cancer. Mol. Cell. 2014; 53:859–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Starnes L.M., Su D., Pikkupeura L.M., Weinert B.T., Santos M.A., Mund A., Soria R., Cho Y.-W., Pozdnyakova I., Kubec Højfeldt M.et al.. A PTIP–PA1 subcomplex promotes transcription for IgH class switching independently from the associated MLL3/MLL4 methyltransferase complex. Genes Dev. 2016; 30:149–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Daniel J.A., Santos M.A., Wang Z., Zang C., Schwab K.R., Jankovic M., Filsuf D., Chen H.-T., Gazumyan A., Yamane A.et al.. PTIP promotes chromatin changes critical for immunoglobulin class switch recombination. Science (1979). 2010; 329:917–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zhang X., Zhang Y., Ba Z., Kyritsis N., Casellas R., Alt F.W.. Fundamental roles of chromatin loop extrusion in antibody class switching. Nature. 2019; 575:385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhang Y., Zhang X., Ba Z., Liang Z., Dring E.W., Hu H., Lou J., Kyritsis N., Zurita J., Shamim M.S.et al.. The fundamental role of chromatin loop extrusion in physiological V (D)J recombination. Nature. 2019; 573:600–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Yi F., Wang Z., Liu J., Zhang Y., Wang Z., Xu H., Li X., Bai N., Cao L., Song X.. Structural maintenance of chromosomes protein 1: role in genome stability and tumorigenesis. Int. J. Biol. Sci. 2017; 13:1092–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wu J., Prindle M.J., Dressler G.R., Yu X.. PTIP regulates 53BP1 and SMC1 at the DNA damage sites. J. Biol. Chem. 2009; 284:18078–18084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. van Schie J.J.M., de Lint K., Molenaar T.M., Gines M.M., Balk J.A., Rooimans M.A., Roohollahi K., Pai G.M., Borghuis L., Ramadhin A.R.et al.. CRISPR screens in sister chromatid cohesion defective cells reveal PAXIP1-PAGR1 as regulator of chromatin association of cohesin. 2022; bioRxiv doi:23 December 2022, preprint: not peer reviewed 10.1101/2022.12.23.521474. [DOI] [PMC free article] [PubMed]
- 83. Knaupp A.S., Mohenska M., Larcombe M.R., Ford E., Lim S.M., Wong K., Chen J., Firas J., Huang C., Liu X.et al.. TINC— a method to dissect regulatory complexes at single-locus resolution— reveals an extensive protein complex at the nanog promoter. Stem Cell Rep. 2020; 15:1246–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Liu X., Zhang Y., Chen Y., Li M., Zhou F., Li K., Cao H., Ni M., Liu Y., Gu Z.et al.. In situ capture of chromatin interactions by biotinylated dCas9. Cell. 2017; 170:1028–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Vermeulen M., Déjardin J.. Locus-specific chromatin isolation. Nat. Rev. Mol. Cell Biol. 2020; 21:249–250. [DOI] [PubMed] [Google Scholar]
- 86. Perez-Riverol Y., Bai J., Bandla C., García-Seisdedos D., Hewapathirana S., Kamatchinathan S., Kundu D.J., Prakash A., Frericks-Zipper A., Eisenacher M.et al.. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022; 50:D543–D552. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq, ChIP-seq, 4C-seq has been deposited to the GEO database (GSE221917). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (86) partner repository with the dataset identifier PXD038902.