Abstract
Background
Intramuscular fat (IMF) and intramuscular connective tissue (IMC) are often seen in human myopathies and are central to beef quality. The mechanisms regulating their accumulation remain poorly understood. Here, we explored the possibility of using beef cattle as a novel model for mechanistic studies of intramuscular adipogenesis and fibrogenesis.
Methods
Skeletal muscle single‐cell RNAseq was performed on three cattle breeds, including Wagyu (high IMF), Brahman (abundant IMC but scarce IMF), and Wagyu/Brahman cross. Sophisticated bioinformatics analyses, including clustering analysis, gene set enrichment analyses, gene regulatory network construction, RNA velocity, pseudotime analysis, and cell–cell communication analysis, were performed to elucidate heterogeneities and differentiation processes of individual cell types and differences between cattle breeds. Experiments were conducted to validate the function and specificity of identified key regulatory and marker genes. Integrated analysis with multiple published human and non‐human primate datasets was performed to identify common mechanisms.
Results
A total of 32 708 cells and 21 clusters were identified, including fibro/adipogenic progenitor (FAP) and other resident and infiltrating cell types. We identified an endomysial adipogenic FAP subpopulation enriched for COL4A1 and CFD (log2FC = 3.19 and 1.92, respectively; P < 0.0001) and a perimysial fibrogenic FAP subpopulation enriched for COL1A1 and POSTN (log2FC = 1.83 and 0.87, respectively; P < 0.0001), both of which were likely derived from an unspecified subpopulation. Further analysis revealed more progressed adipogenic programming of Wagyu FAPs and more advanced fibrogenic programming of Brahman FAPs. Mechanistically, NAB2 drives CFD expression, which in turn promotes adipogenesis. CFD expression in FAPs of young cattle before the onset of intramuscular adipogenesis was predictive of IMF contents in adulthood (R 2 = 0.885, P < 0.01). Similar adipogenic and fibrogenic FAPs were identified in humans and monkeys. In aged humans with metabolic syndrome and progressed Duchenne muscular dystrophy (DMD) patients, increased CFD expression was observed (P < 0.05 and P < 0.0001, respectively), which was positively correlated with adipogenic marker expression, including ADIPOQ (R 2 = 0.303, P < 0.01; and R 2 = 0.348, P < 0.01, respectively). The specificity of Postn/POSTN as a fibrogenic FAP marker was validated using a lineage‐tracing mouse line. POSTN expression was elevated in Brahman FAPs (P < 0.0001) and DMD patients (P < 0.01) but not in aged humans. Strong interactions between vascular cells and FAPs were also identified.
Conclusions
Our study demonstrates the feasibility of beef cattle as a model for studying IMF and IMC. We illustrate the FAP programming during intramuscular adipogenesis and fibrogenesis and reveal the reliability of CFD as a predictor and biomarker of IMF accumulation in cattle and humans.
Keywords: Adipogenesis, Fibro/adipogenic progenitor, Fibrogenesis, Intramuscular adipose tissue, Single‐cell RNAseq
Introduction
Sarcopenia and muscular dystrophies are frequently associated with the accumulation of intramuscular fat (IMF) and intramuscular connective tissue (IMC), which can significantly interfere with muscle contraction. 1 In both mice and humans, IMF and IMC originate from fibro/adipogenic progenitors (FAPs), a type of bipotent progenitor that can differentiate into both adipocytes and myofibroblasts, the main cell types in adipose and fibrotic tissues, respectively [S1, S2]. However, the understanding of mechanisms regulating FAP differentiation remains limited. While current animal studies on intramuscular adipogenesis and fibrosis mainly rely on mouse models, some limitations have been identified, such as less robust fatty infiltration in rodents. 2 , 3 The accumulation of IMF and IMC is also observed in beef cattle and affects the quality of beef. Although IMF is positively correlated with the juiciness, flavour, and tenderness of beef, excess IMC increases background toughness and negatively impacts beef quality. 4 The more prominent IMF and IMC accumulation in bovine muscle and the existence of beef cattle breeds with differential IMF and IMC contents make cattle a perfect candidate model for studying the general mechanisms underlying intramuscular adipogenesis and fibrogenesis. In addition, the large body size of cattle ensures ample experimental materials that can be collected through biopsies without the need for euthanasia.
Wagyu cattle are renowned for abundant IMF. In contrast, the beef from Brahman is generally of low quality due to its low IMF content and tenderness. 5 Previous mechanistic studies at the whole muscle level revealed limited differences among breeds [S3–S6]. One possible reason is the complicated cell type composition of skeletal muscle (SkM). Here, for the first time, we report a single‐cell atlas of bovine SkM using single‐cell RNAseq (scRNAseq). Significant heterogeneities were identified in all bovine SkM‐resident cell types, especially in FAPs. Our analyses suggested mechanisms contributing to the programming differences among bovine FAP subpopulations and between Brahman and Wagyu FAPs. Comparison studies with published human and non‐human primate datasets suggest conserved pathways regulating intramuscular adipogenesis and fibrogenesis in larger mammals. Our study not only unveils novel targets for IMF and IMC manipulation but also provides a new model for studying IMF and IMC accumulation.
Methods
Study approval
All experiments involving cattle and mice were approved by the IACUC at LSU (approval numbers A2018‐04, A2021‐07, 18‐024, and 21‐034).
Cell isolation for scRNAseq
Muscle tissue was minced and digested in Dulbecco's modified Eagle's medium (DMEM) containing 0.75 U/mL collagenase D and 1 U/mL of Dispase II for 40 min at 37°C. The tissue slurry was filtered through a 100 μm cell strainer and pelleted through centrifugation. Debris and dead cells were removed using a debris removal solution (130‐109‐398) and a dead cell removal kit (130‐090‐101) from Miltenyi Biotec, respectively.
scRNAseq library construction, sequencing, and data analysis
scRNAseq was performed using cells isolated from bovine muscle biopsy samples (two animals of each breed). Libraries were constructed using Chromium Single Cell 3′ Reagent Kits v3 (10× Genomics) at the LSU Gene lab. The cDNA libraries were sequenced on the Illumina NovaSeq platform using 150 bp paired‐end sequencing at a depth of more than 20 000 read pairs per cell. Raw sequencing data were processed using Cell Ranger 6.0.0 (10× Genomics) and ARS‐UCD1.2 bovine genome. Processed data were analysed using Seurat (version 4.0.4) in R. Cells with mitochondrial gene expression >5% of total gene expression were removed from the analysis. The resulting dataset was then normalized with a global‐scaling normalization method (‘LogNormalize’) with default parameters. Highly variable features were identified by directly calculating the mean–variance relationship using the FindVariableFeatures function (Seurat). For other detailed methods, please refer to the supporting information.
Data availability
All necessary data to evaluate the paper's conclusions are available in the paper and supporting information. Raw genomic sequencing datasets have been deposited at GEO (GSE205347). Additionally, we used publicly available human SkM microarray datasets from GEO (GSE6011, GSE38417, and GSE136344), scRNAseq data of human muscle tissue from GEO (GSE143704), scRNAseq data of Macaca fascicularis muscle tissue from CNGB Nucleotide Sequence Archive (CNP0001469), scRNAseq data of WT and DMD mouse muscle tissue from GEO (GSE156497), and scRNAseq data for glycerol‐injected mouse muscle from a published study [S7].
Statistics
All data except for scRNAseq data are expressed as mean ± SEM unless otherwise stated. Data were analysed using GraphPad Prism 9 (GraphPad Software, Inc.). Two‐tailed t test was used to determine the significance of differences between the two groups. One‐way ANOVA with post hoc Tukey's test was used to determine the significance of the difference when more than two groups were compared. Simple linear regression was used to determine the correlations between CFD expression and IMF content and between the expression of two genes. P < 0.05 was considered significant.
Results
A single‐cell atlas of bovine skeletal muscle tissue
To determine the cellular composition of bovine SkM, we performed scRNAseq using cells isolated from the Longissimus dorsi (LD) muscle of 4‐month‐old full‐blood Wagyu, purebred Brahman, and Wagyu/Brahman crossbred female calves. A total of 32,708 cells passed quality control, including 11,765 from Wagyu, 5,903 from Brahman, and 15 040 from crossbred. Unsupervised clustering identified 21 clusters (Figure 1A, Table S1). Clusters 1 and 3 (C1 and C3) expressed PDGFRA, a FAP marker (Figure 1B). C2, C9, and C11 expressed the endothelial cell (EndoC) marker, PECAM1 (Figure 1B). Moreover, C11 specifically expressed MMRN1, a lymphatic EndoC (LEndoC) marker (Figure 1B), whereas C9 specifically expressed the venular EndoC (VEndoC) marker, AQP1 (Figure 1B) [S8–S9]. C6 and C14 expressed ACTA2, the gene encoding smooth muscle alpha‐actin (SMαA), and were likely mural cells (MuCs), including vascular smooth muscle cells (VSMCs) and pericytes. C8 expressed myogenic cell (MC) marker, MYF5 (Figure 1B). Interestingly, a small cluster (C20) expressing the tenocyte marker gene, COMP, was also identified (Figure 1B), likely represented an interstitial tenocyte population identified in a recent mouse study. 6 C17 expressing the type I muscle fibre myosin gene, MYH7, and C18 expressing the type II muscle fibre myosin gene, MYH2, were also identified (Figure 1B). Clusters expressing various haematopoietic cell markers were identified as well (Figure 1B). FAP was the most abundant muscle‐resident cell type (Figure 1C). Interestingly, the proportion of FAPs was greater in Brahman, while vascular cells were more abundant in Wagyu (Figure 1C), which may contribute to the lower adipogenic activity in Brahman muscle as vascularization supports adipogenesis. 7 Consistently, the capillary abundance was greater in Wagyu versus Brahman muscle, and gene set enrichment analysis (GSEA) showed a mild enrichment of gene ontology biological process (GOBP) term ‘blood vessel development’ in Wagyu versus Brahman blood vessel EndoCs (Figure S1A–C).
Figure 1.
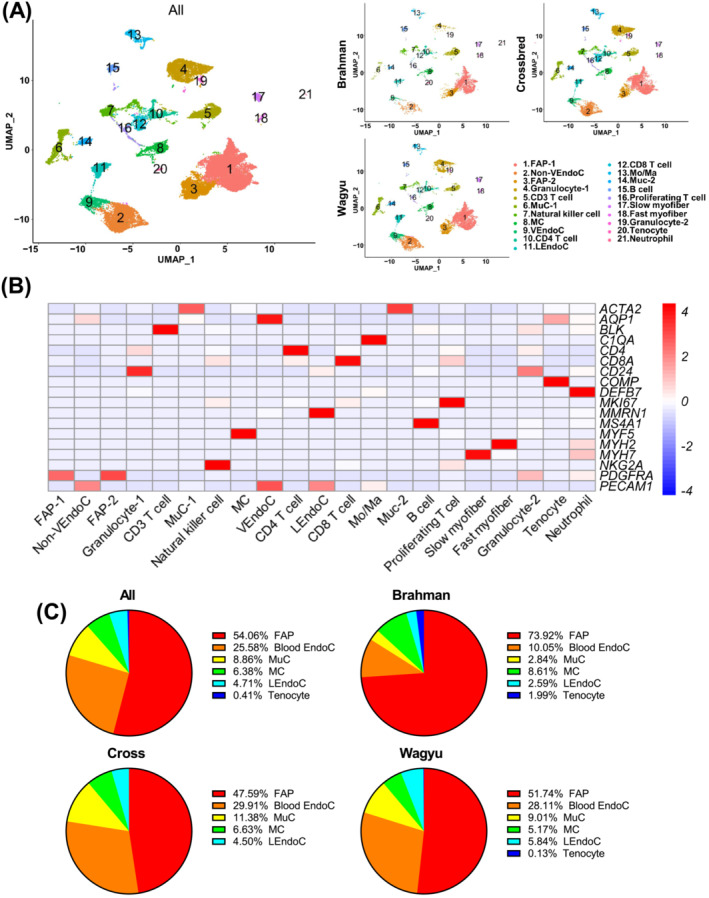
scRNAseq reveals multiple muscle‐resident and circulating cell types in bovine SkM. (A) UMAP graphs show unsupervised clustering of cells from all breeds and individual breeds identified by scRNAseq. (B) A heatmap shows the expression of representative marker genes used in cell‐type annotation. (C) Pie graphs show the non‐immune muscle‐resident cell type composition in all breeds and individual breeds revealed by scRNAseq. FAP, fibro/adipogenic progenitor; VEndoC, venular endothelial cell; MuC, mural cell; MC, myogenic cell; LEndoc, lymphatic endothelial cell; Mo/Ma, monocyte/macrophage.
Crosstalk between different cell types in bovine skeletal muscle
Cell–cell communication analysis identified various interactions between major cell types, many of which involved FAPs (Figures 2A and S2A–C). Strong communications from EndoCs and MuCs to FAPs were identified, which involved pathways known to regulate adipogenesis and fibrogenesis, such as PDGF, FGF, and IGF signallings, suggesting the important regulatory roles of vascular cells in FAP programming (Figure S2B). Of note, an interaction was also identified between FAPs and MCs through laminins and dystroglycan 1 encoded by DAG1 (Figure S2C). DAG1 was recently identified as a satellite cell (SC)‐specific gene playing an important role in the asymmetric division of SCs. 8 A comparative analysis showed that FAP‐1 was a generally stronger sender than FAP‐2 (Figure S2C). Cell–cell communications involving FAP‐1 but not FAP‐2 as the senders included those of the VEGF, CCL, PTN, THBS, and ICAM1 signalling pathways (Figure S2C). These pathways have known functions in angiogenesis, cell activation, and inflammation [S10–S13]. Moreover, FAP‐1 had stronger communications with other cell types through the expression of IGF2, an important cell proliferation and differentiation regulator (Figure S2C). Another comparative analysis identified stronger communication from FAPs to EndoCs in Wagyu versus Brahman through VEGF pathways and stronger communication from FAPs to MCs in Brahman versus Wagyu through DAG1 pathways (Figure 2B; Table S2). The results suggest that Wagyu FAPs are pro‐angiogenic, whereas Brahman FAPs are pro‐myogenic.
Figure 2.
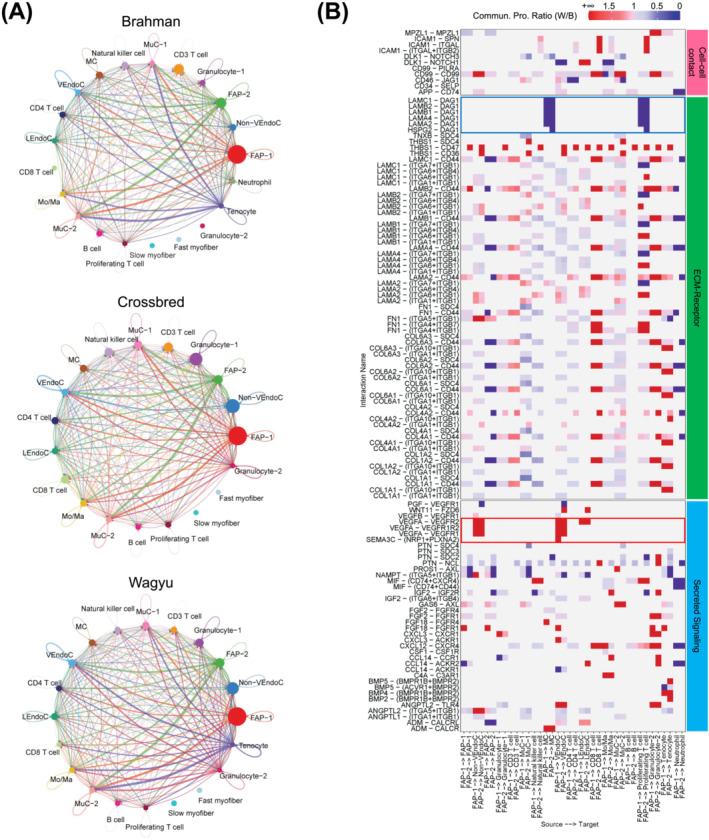
scRNAseq reveals communications between FAPs and other cell types in Wagyu muscle. (A) Cell–cell communication networks include all identified significant communications between different cell types in individual breeds identified by scRNAseq. (B) A heatmap shows the differential cell–cell communications in which FAP‐1 or FAP‐2 functions as the sender between Wagyu and Brahman. The red box highlights VEGF signalling communications from FAPs to EndoCs only detected in Wagyu. The blue box highlights DAG1 signalling communications from FAPs to MCs only detected in Brahman. Values are the ratios of communication probability scores in Wagyu to those in Brahman.
Bovine fibro/adipogenic progenitors are a heterogeneous population
To explore bovine FAP heterogeneity, FAP‐1 and FAP‐2 (C1 and C3) were further separated into 6 clusters (FAP C1–C6) (Figure 3A; Table S3). The analysis showed that COL1A1 and COL3A1, major fibrillar collagen genes, were enriched in FAP C4 (Figure 3B). 9 In contrast, basement collagen genes, COL15A1 and COL4A1, were enriched in FAP C3 (Figure 3B). 9 Enriched genes in FAP C4 also included pro‐fibrogenic genes, FMOD, ADAMTS2, S100A4, TGFBR3, and POSTN [S14–S19] (Figure 3B). Other genes enriched in FAP C3 included MME, CFD, SNAI2, SFRP1, PPARG, and ZC3H12A, all known for their pro‐adipogenic activities [S20–S24] (Figure 3B). These cells also expressed a relatively high level of pro‐angiogenic genes such as BTG1, BMP2, and BMP4 [S25, S26] (Figure 3B). GSEA showed that FAP C3 was enriched for GOBP terms related to mesenchymal cell differentiation, proliferation, and fat cell differentiation, and GOBP terms related to extracellular matrix proteins deposition and remodelling were enriched in FAP C4 (Figure 3C). Wikipathways analysis also revealed differential enrichment of adipogenesis‐related genes between FAP C3 and C4 (Figure 3C). These results indicate that FAP C3 and C4 might represent major adipogenic and fibrogenic FAP subpopulations, respectively.
Figure 3.
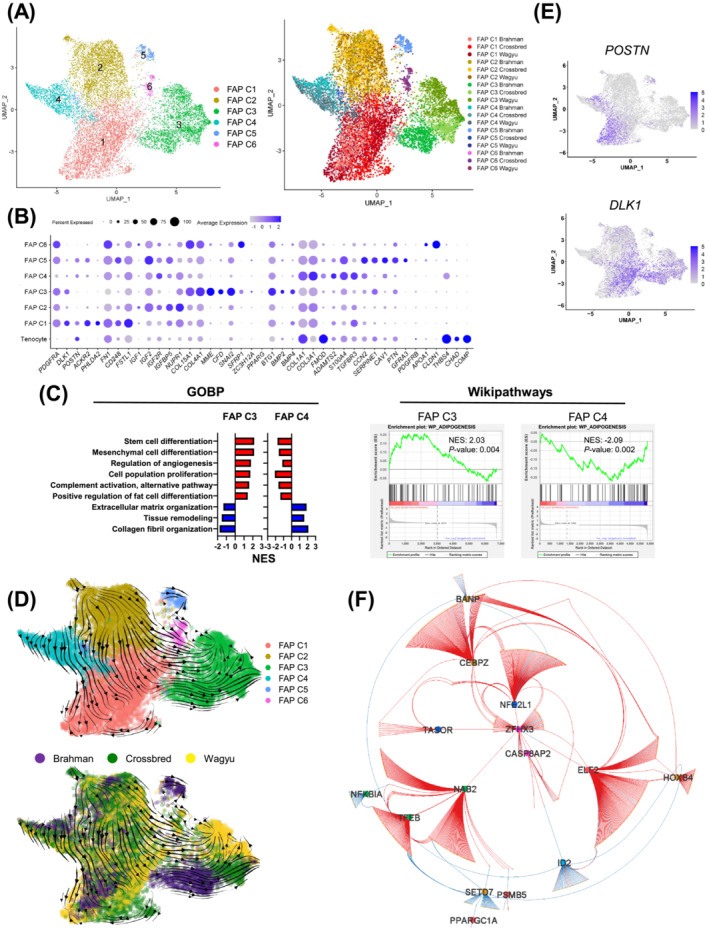
Bovine FAPs are a heterogeneous population. (A) UMAP graphs show unsupervised clustering of bovine FAPs from all breeds identified by scRNAseq. (B) A dot plot shows the expression of select DEGs among FAP subpopulations. (C) Graphs show the enrichment of select GOBP and Wikipathways terms in FAP C3 and C4. (D) RNA velocity graphs in which different FAPs subpopulations or FAPs of different breeds are differentially labelled. (E) Feature plots show select DEGs among FAP subpopulations. (F) A GRN regulates the differential gene expression among FAP clusters.
Co‐expression analysis identified modules enriched in FAP C1 (modules 3 and 13) and C2 (module 2) (Figure S3A,B). GOBP terms related to positive and negative developmental regulations were enriched in module 2 genes (Figure S3C). Correspondingly, upregulated genes in FAP C2 included pro‐development IGF signalling genes, such as IGF1, IGF2, and IGF2R, 10 and genes inhibiting development, such as IGFBP5 and NUPR1 [S27, S28] (Figure 3B). GOBP terms ‘negative regulation of cell differentiation’ and ‘positive regulation of cell population proliferation’ were also enriched in module 2 genes (Figure S3C). These indicate that FAP C2 was a relatively undetermined population. Module 3 and 13 genes showed GOBP term enrichments related to cell migration and adhesion (Figure S3C). Representative genes in these modules that were also upregulated in FAP C1 included migration‐related genes, such as ACKR2, PHLDA2, FN1, CD248, and FSTL1, suggesting the relatively high motility of these cells [S29–S33] (Figure 3B). FAP C5 expressed pericyte‐expressed genes CCN2, SERPINE1, and CAV1 [S34–S36] but did not express the pericyte marker PDGFRB (Figure 3B; Table S3). Moreover, FAP C5 included some cells expressing GFRA3, a gene encoding a receptor for glial cell line‐derived neurotrophic factor 11 (Figure 3B). The top 2 enriched genes in FAP C6 were APOA1 and CLDN1 (Figure 3B; Table S3). APOA1 has been previously shown to promote EndoC proliferation. 12 CLDN1 is highly expressed in perineurial cells, a specialized type of fibroblasts. 13 It is possible that FAP C5 and C6 comprised some FAPs proximal to EndoCs or neurons.
RNA velocity suggests that FAP C2 could give rise to FAP C1, C3, and C4 through two paths (Figure 3D). FAP C2 directly differentiates into FAP C4, which further differentiates into POSTN + cells in FAP C1 (Figure 3D,E). In the other path, FAP C2 appears to indirectly give rise to FAP C3 through DLK1 + cells in FAP C1 as an intermediate state (Figure 3D,E). DLK1 inhibits multiple mesenchymal cell differentiation processes, including adipogenesis, through Notch‐related and ‐unrelated mechanisms [S37–S41]. The transient expression of DLK1 during this process likely inhibits premature differentiation. Pseudotime analysis largely agreed with RNA velocity (Figure S3D). To explore the transcriptional regulation of FAP heterogeneity, a gene regulatory network (GRN) was constructed through the integration of GSEA GTRD transcription factor (TF) targets analysis and co‐expression correlation (see Methods section for details), which identified some key TFs, including NAB2, ELF2, ZFHX3, and CEBPZ (Figure 3F; Table S4).
Clustering analysis of all cells showed adjacent locations of FAPs and tenocytes (Figure 1A). FAP C4 also expressed tenocyte marker genes THSB4 and CHAD but not COMP (Figures 3B and S3E). Pseudotime and RNA velocity analyses of FAP and tenocytes suggest that tenocytes may contribute to FAP C4 (Figure S3F,G).
scRNAseq reveals myogenesis in cattle at 4‐month of age
MCs are critical for muscle growth and regeneration. To explore their diversity, MCs were further separated into six clusters (MC C1–C6) (Figure 4A; Table S5). MC C1 and C2 expressed high levels of SC marker genes such as MYF5, CHODL, NOTCH3, and CHRDL2 [S42–S43] (Figures 4B and S4A,B). MC C2 expressed slightly lower MYF5 and ACTA2, two activated SC markers (Figures 4B and S4A,B). Moreover, MC C2 expressed high TGFBR3, a gene downregulated by MYF5 14 (Figures 4B and S4A,B). MC C2 might represent a more quiescent SC population. In contrast, MC C1 expressed a higher level of SC proliferation gene LIX1 15 and might represent a slightly more active population (Figures 4B and S4A,B). Interestingly, PAX7, a specific SC marker, was not identified as a significantly enriched gene in MC C1 and C2 because of an average log2FC lower than the 0.25 threshold. This was primarily due to its relatively sparse expression (Figures 4B and S4A). MC C3, which expressed MYOD1 and its predicted target gene MT3, 16 was found between MC C2 and C5 (Figures 4A,B and S4A,B). MC C3 also expressed PFN1, a gene inhibiting myogenesis‐associated cytoskeletal remodelling, 17 and thus was likely composed of activated myoblasts (Figures 4B and S4A,B). MC C5 expressed high levels of myogenic differentiation markers, MYOG, MYMK, and ACTC1, and likely represented a differentiating myoblast/myocyte population (Figures 4B and S4A,B). MC C4 and C6 expressed some genes that were upregulated in EndoCs and FAPs, such as MAL, SNCA, COL6A1, and SFRP4, and likely represent some rare doublets that were not removed by the quality control process. Interestingly, MC C3 was identified as the origin in RNA velocity and pseudotime analyses, which gave rise to MC C1 and C2 and MC C5 through separated paths (Figure 4C,D). These results suggest that in these animals, some committed myoblasts were undergoing myogenic differentiation while the other population was reverting to quiescence.
Figure 4.
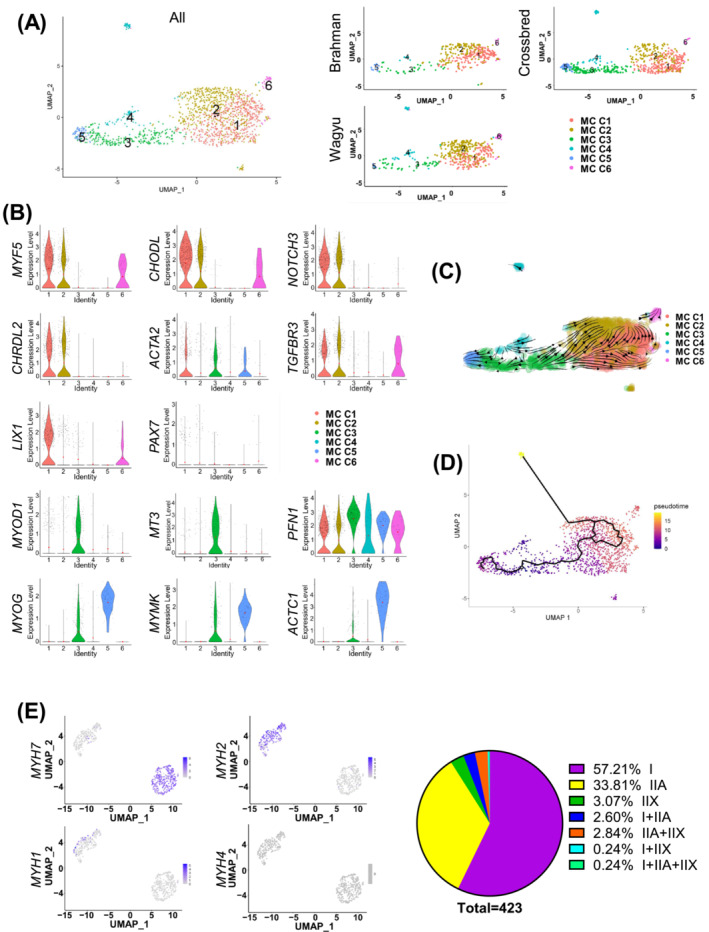
scRNAseq suggests active myogenesis in cattle at 4 months of age. (A) UMAP graphs show unsupervised clustering of bovine MCs from all breeds and individual breeds identified by scRNAseq. (B) Violin plots show the expression of select DEGs among different MC subpopulations. (C) RNA velocity analysis of MCs. (D) Pseudotime analysis of MCs. (E) Feature plots show the expression of indicated myosin heavy chain genes in myonuclei identified by scRNAseq, and a pie graph shows the muscle fibre type composition revealed by scRNAseq.
Clustering analysis of all cells identified clusters expressing MYH7 (C17, type I muscle fibre) and MYH2 (C18, type IIA muscle fibre) (Figure 1B). Re‐clustering analysis also identified type IIX muscle fibre fragments expressing MYH1 and hybrids expressing more than one myosin gene (Figure 4E). Consistent with the previously reported lack of type IIB fibres in most bovine muscles, 18 no expression of MYH4 was identified (Figure 4E). Separate analyses of muscle fibre fragments from different breeds suggest more type IIX fibres in Brahman versus Wagyu (Figure S4C). A similar trend was identified by fibre typing (Figure S4D,E). The larger size of type IIX fibres likely contributed to their lower abundance detected in scRNAseq as larger fragments were preferentially removed during sample processing (Figure S4F).
Analysis of mural cells reveals distinct vascular smooth muscle cell and pericyte populations
Re‐clustering analysis of MuCs identified five clusters (MuC C1–C5) (Figure 5A; Table S6). MuC C1 likely represented a pericyte population, evidenced by the enrichment for pericyte markers, such as KCNJ8, RGS5, and NDUFA4L2 [S44–S46], and the lack of VSMC contractile protein genes, MYH11 and ACTG2 (Figure 5B). In contrast, MuC C4 expressed high MYH11, ACTG2, and DES but lacked PDGFRB expression, indicating its mature VSMC identity (Figure 5B). MuC C2, located between MuC C1 and C4, expressed a high level of DKK1, a pro‐angiogenic gene, 19 and SERPINE2, which limits VSMC proliferation 20 and was weakly positive for MYH11 (Figure 5B). Taken together, MuC C2 might include some MuC progenitors. MuC C3 expressed a low level of MYH11 and ACTG2 and were also positive for RGS16 and ID4, negative regulators of VSMC differentiation [S47, S48] (Figure 5B). More importantly, it was the only major MuC cluster without CSPG4 expression, an arterial‐specific gene, suggesting its venous VSMC identity 21 (Figure 5B). The lower expression of select contractile protein genes in venous versus arterial VSMCs reflects their less robust contraction. Interestingly, similar ACTA2 expression in arterial and venous VSMCs was identified, and the expression of MYH9 was even greater in arterial VSMCs, demonstrating distinct actomyosin machinery in arterial versus venous VSMCs (Figure 5B). Interestingly, RNA velocity and pseudotime analyses suggest that arterial pericytes (MuC C1) and VSMCs (MuC C4) might both be derived from MuC C2 (Figure 5C,D). In addition, a small cluster (MuC C5) related to MuC C2 and C4 was identified. These cells expressed a high level of PECAM1, SOX17, and SOX18, which were also expressed in EndoCs (Figure 5B; Table S1), and possibly represent a rare population with both VSMC and EndoC signatures, which has been reported in some previous studies [S49, S50].
Figure 5.
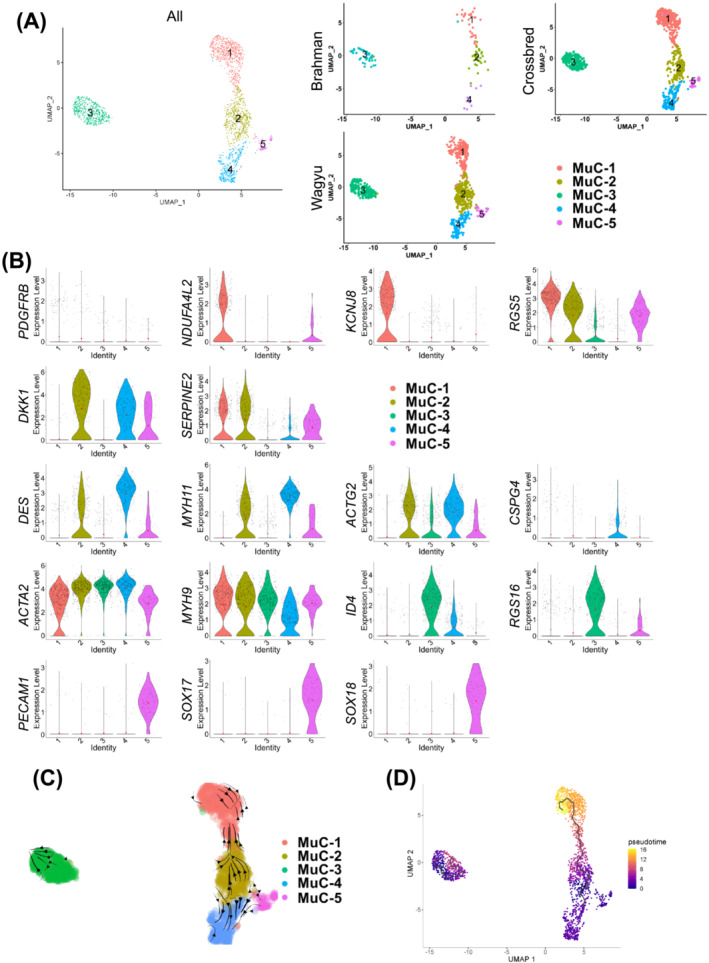
An analysis of MuCs reveals distinct VSMC and pericyte populations. (A) UMAP graphs show unsupervised clustering of bovine MuCs from all breeds and individual breeds identified by scRNAseq. (B) Feature plots show the expression of select DEGs among different MuC subpopulations. (C) RNA velocity of MuCs. (D) Pseudotime analysis of MuCs.
scRNAseq suggests differential activities between Wagyu and Brahman fibro/adipogenic progenitors
A comparison of Wagyu and Brahman FAPs identified 472 DEGs (Table S7). More DEGs between FAPs of the two breeds were identified in FAP C3 and C4, the adipogenic and fibrogenic FAP clusters (637 and 457, respectively), including many specific to FAP C3 or C4 (Figure S5A–C; Tables S8 and S9).
The most upregulated gene in Wagyu versus Brahman FAP C3 was CFD (Figures 6A and S5A,D). CFD was also an upregulated gene in FAP C3 versus other FAP clusters (Figure 3B). Other genes upregulated in Wagyu versus Brahman FAP C3 with reported pro‐adipogenic or anti‐fibrotic activities included PTX3, HSPA1A, CXCL3, TWIST1, GPX3, and CXXC5 22 , 23 , 24 , 25 [S51, S52] (Figures 6A and S5A,D). In contrast, many genes upregulated in Brahman versus Wagyu FAP C3 have anti‐adipogenic or pro‐fibrogenic functions, such as MSMO1, PEG3, DLK1, and HMGCS1 26 , 27 [S37, S53, S54] (Figures 6A and S5A,D). RNA velocity analysis of all FAP clusters suggests that Wagyu FAP C3 and C4 was developmentally more advanced than Brahman FAP C3 and C4 (Figure 3E). Consistently, pseudotime analysis placed Wagyu FAP C3 after Brahman FAP C3 while crossbred FAP C3 was between the two breeds (Figure 6B). RNA velocity analysis of FAP C3 alone showed a similar trend even though the path was less clear (Figure S5F). The opposite expression patterns of DLK1, an anti‐differentiation gene highly enriched in FAP C1, and CFD, a gene not expressed in FAP C1, on the pseudotime trajectory again suggests that Brahman FAP C3 was more closely resembled FAP C1 and was in a less advanced differentiation state compared with Wagyu FAP C3 (Figures 3B and 6B).
Figure 6.
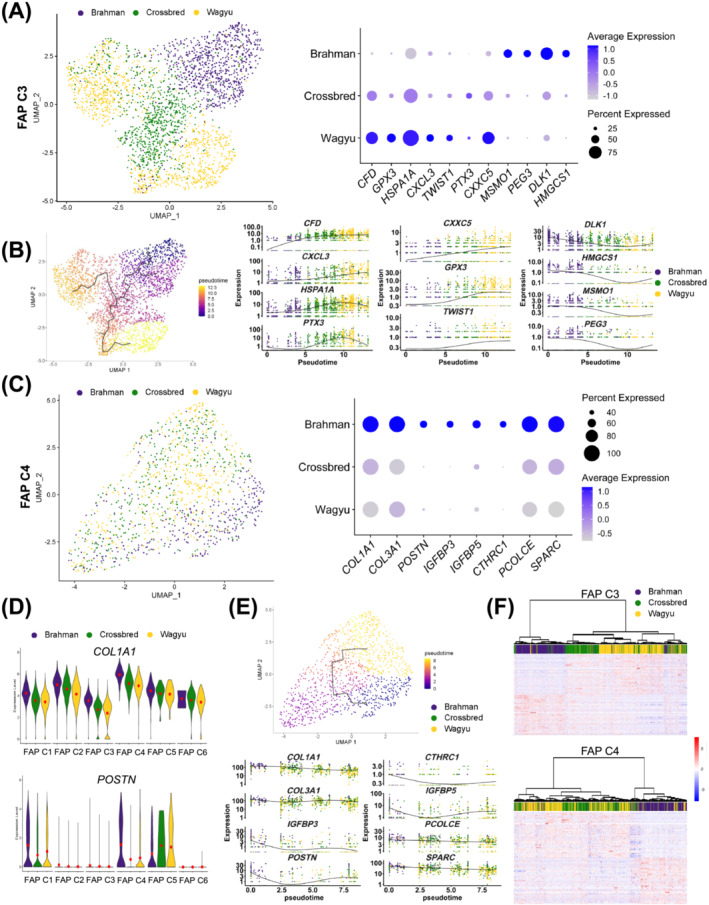
scRNAseq suggests differential activities between Wagyu and Brahman FAPs. (A) A UMAP graph shows bovine FAP C3 from different breeds and a dot plot shows the expression of select DEGs in Wagyu, crossbred, and Brahman FAP C3. (B) Pseudotime analysis of FAP C3 from all breeds and graphs showing the expression of select DEGs along the FAP C3 pseudotime trajectory. (C) A UMAP graph shows bovine FAP C3 from different breeds and a dot plot shows the expression of select DEGs in Wagyu, crossbred, and Brahman FAP C4. (D) Violin plots show the expression of COL1A1 and POSTN in different FAP subpopulations of different breeds. (E) Pseudotime analysis of FAP C4 from all breeds and graphs showing the expression of select DEGs along the FAP C4 pseudotime trajectory. (F) Heatmaps with dendrograms show the expression of the top 50 upregulated genes in Wagyu and Brahman FAP C3 or C4 in Wagyu, Brahman, and crossbred C3 or C4. Individual cells were clustered and connected based on similarities in gene expression.
Genes upregulated in Brahman versus Wagyu FAP C4 included COL1A1 and COL3A1 and the myofibroblast‐specific extracellular matrix gene, POSTN (Figures 6C and S5B,E). Also among these genes were other pro‐fibrogenic genes, such as IGFBP3, IGFBP5, CTHRC1, PCOLCE, and SPARC 28 [S55–S58] (Figures 6C and S5B,E). In addition to the elevated fibrogenic gene expression in Brahman versus Wagyu FAP C4, we also identified greater COL1A1 and POSTN expression in other major FAP clusters of Brahman as compared with Wagyu, suggesting systemically stronger fibrogenic programming of Brahman FAPs than Wagyu FAPs (Figure 6D). Similar to FAP C3, RNA velocity and pseudotime analysis of FAP C4 showed that Wagyu FAP C4 was developmentally more advanced than Brahman FAP C4, owing to lower fibrogenic gene expression, which was consistent with RNA velocity (Figures S5F and 6E).
As expected, the gene expression profile of crossbred FAP C3 and C4 fell between the two purebred breeds (Figure S5D,E). Moreover, a greater similarity was observed between crossbred and Wagyu than between crossbred and Brahman, suggesting that the Wagyu sire had a stronger influence than Brahman dams on FAP C3 and C4 gene expression in crossbred cattle (Figure 6F).
The greater adipogenic potential of Wagyu fibro/adipogenic progenitors is at least partially due to higher CFD expression and less robust fibrogenic programming
Consistent with scRNAseq data, isolated Wagyu FAPs were more efficient in adipogenesis indicated by more abundant Oil Red O positive lipid droplets, whereas Brahman FAPs were more fibrogenic suggested by stronger SMαA expression, a myofibroblast marker (Figure 7A). A gain of function study was then performed to study the effect of CFD, the most significant adipogenic DEG upregulated in Wagyu FAPs. LipidTox, a dye for neutral lipid droplets [S59], staining showed that overexpression of HA‐tagged CFD significantly enhanced Brahman FAP adipogenesis (Figure 7B). CFD encodes adipsin, a key enzyme in the alternative pathway of complement activation that generates C3a. 29 Mice lacking C3ar1, the C3a receptor, are resistant to diet‐induced obesity. 30 As expected, treatment with a C3aR inhibitor attenuated Wagyu FAP adipogenesis (Figure 7C). The expression of CFD and C3AR1 increased after adipogenic induction, supporting their important function in bovine FAP adipogenesis (Figure S6A). Interestingly, at 2 years of age, the IMF content of cattle was positively correlated with their FAP CFD expression at 4 months of age (Figures 7D and S6B). No correlation was identified between IMF content and CFD expression in other cell types (Figures 7D and S6B,C). These results suggest that CFD is an early adipogenic programming indicator in bovine FAPs. To explore possible mechanisms promoting CFD expression in adipogenic FAPs, we screened the target genes of TFs included in the GRN for CFD (Figure 3F). The analysis identified NAB2 as a possible driver of CFD expression (Figure S6D). NAB2 was also upregulated in Wagyu versus Brahman FAP C3 (Table S8). Knockdown of NAB2 reduced CFD expression in Wagyu FAPs (Figure 7E). NAB2 does not bind to DNA directly but functions as a co‐factor for the EGR family TFs. The upregulation of EGR1 expression and enrichment of its target genes were identified in FAP C3 (Figure S6D, Table S3). Motif analysis located multiple EGR1 motifs immediately upstream of the bovine CFD gene (Figure S6E). These results suggest that CFD is regulated by NAB2/EGR1 in bovine FAPs.
Figure 7.
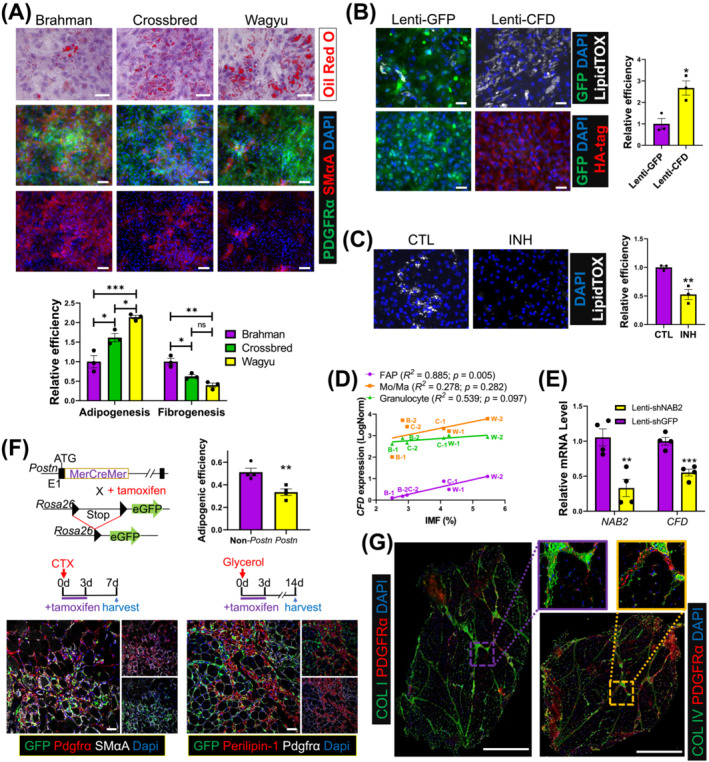
The greater adipogenic potential of Wagyu FAPs is at least partially due to higher CFD expression and less robust fibrogenic programming. (A) Oil Red O staining and ICC show the adipogenic efficiency and the myofibroblast differentiation capacities of FAPs indicated by SMαA signal strength of FAPs from different breeds, respectively. The purity of FAPs was verified by PDGFRα staining. Scale bar: 50 μm (Oil Red O), 100 μm (ICC). n = 3 for each breed. (B) Brahman FAPs were transduced with Lenti‐GFP or Lenti‐CFD and induced for adipogenesis. Adipogenesis was evaluated by LipidTOX staining. The expression of GFP and CFD was verified by ICC using anti‐GFP and anti‐HA tag antibodies. n = 3 for each treatment. Scale bar: 50 μm. (C) Wagyu FAPs were treated with C3aR inhibitor (INH) or vehicle (CTL) and induced for adipogenesis. Adipogenesis was evaluated by LipidTOX staining. n = 3 for each treatment. (D) The correlation between IMF content and CFD expression in FAPs, Mo/Ma, and granulocytes. (E) Wagyu FAPs were transduced with Lenti‐shGFP or Lenti‐shNAB2. The expression levels of NAB2 and CFD were measured 2 days after transduction by realtime PCR. (F) Postn MCM/+ ;R26 eGFP mice were treated with tamoxifen from day 0 to day 3 after CTX or glycerol injections, followed by sample collection on day 7 after the CTX injection or day 14 after the glycerol injection. Muscle samples were subjected to IHC to identify Postn lineage cells (GFP+), FAPs (Pdgfrα+), myofibroblasts (Pdgfrα+;SMαA+, CTX injected), and adipocytes (perilipin‐1+, glycerol injected). The adipogenic efficiencies of Postn‐lineage FAPs [(GFP+;perilipin‐1+ cell number)/(GFP+;Pdgfrα+ cell number)] and non‐Postn‐lineage FAPs [(GFP−;perilipin‐1+ cell number)/ (GFP−;Pdgfrα+ cellnumber)] were calculated. n = 4 for each breed. Scale bar: 50 μm. (G) Representative IHC images show the locations of collagen I, collagen IV, and FAPs (PDGFRα+) in Wagyu and Brahman muscles. *P < 0.05; **P < 0.01; ***P < 0.001. COL I, collagen I; COL IV, collagen IV.
scRNAseq identified higher POSTN expression, a myofibroblast marker gene, in Brahman versus Wagyu FAP C4 (Figure 6C). To confirm the fibrogenic cell identity of Postn‐expressing cells in SkM, we took advantage of the Postn MCM/+ ;R26 GFP mouse line, in which Cre recombinase driven by the Postn locus induces permanent GFP expression upon tamoxifen treatment, and a cardiotoxin (CTX)‐induced muscle injury and transient fibrosis model [S2] (Figure 7F). Seven days after CTX injection, many Postn‐lineage myofibroblasts (GFP+;Pdgfrα+) expressing SMαA were identified (Figure 7F). Fourteen days after glycerol injection, an injury‐induced intramuscular adipogenesis model [S2], staining of perilipin‐1, a protein expressed on lipid droplets [S59], showed that the adipogenic efficiency of Postn‐lineage FAPs was significantly lower than that of non‐Postn‐lineage FAPs (Figure 7F). Moreover, FAPs isolated from CTX‐injected muscle of Postn MCM/+ ;R26 GFP mice showed reduced adipogenic efficiency but increased myofibroblast differentiation efficiency compared with FAPs from the uninjured muscle (Figure S6F). These data further indicate that Postn is a marker for fibrogenic programming.
Our analysis revealed dominant collagen IV and collagen I gene expression in FAP C3 (adipogenic) and C4 (fibrogenic), respectively (Figure 3B). IHC showed higher collagen I content in the perimysium and stronger collagen IV intensity in the endomysium, both of which had abundant FAPs (Pdgfrα+) (Figure 7G). These data suggest the respective perimysial and endomysial locations of fibrogenic and adipogenic FAPs.
Mechanisms identified in bovine fibro/adipogenic progenitors may also regulate fatty infiltration and fibrosis in human myopathies
To explore the common mechanisms in IMF and IMC accumulation across species, we first re‐analysed published scRNAseq datasets, which showed the strong expression of CFD/Cfd in FAPs of aged human SkM and non‐human primate SkM, but not in FAPs in mouse models of intramuscular adipogenesis, including glycerol‐injected mouse muscle and muscle of a mouse model of DMD [S7, S60–S62] (Figure S7A–D). Clustering analyses identified similar adipogenic and fibrogenic FAP clusters in human (C1 and C3, respectively) and monkey (C2 and C3, respectively) SkM (Figures 8A and S7E). While FAP clusters enriched for Col15a1 and Col4a1 (C1 and C2) were also identified in mouse SkM, no upregulation of adipogenic genes was identified in these clusters (Figure 8A). The results indicate greater similarities in FAPs among larger mammals.
Figure 8.
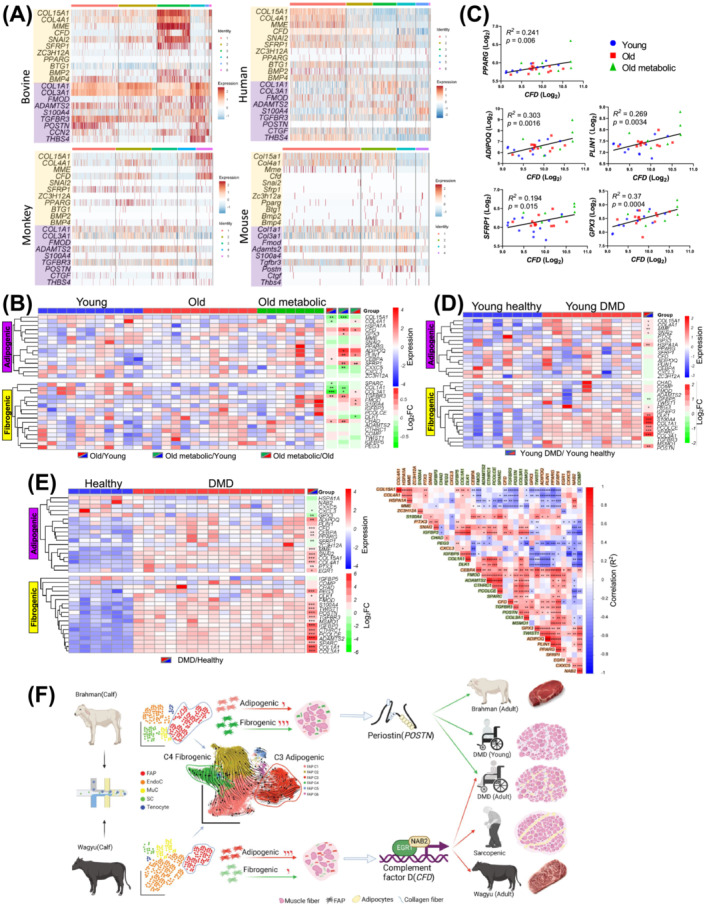
An analysis reveals common mechanisms regulating bovine and human IMF and IMC accumulation. (A) Heatmaps show the expression of select adipogenic and fibrogenic genes in bovine, human, monkey, and uninjured WT mouse FAP clusters. (B, C) The differential expression of select adipogenic and fibrogenic genes in human SkM among young, old, and old with metabolic syndrome individuals (B). The correlation between the expression of select genes and the expression of CFD in human SkM across all groups (C). (D) The differential expression of select adipogenic and fibrogenic genes in human SkM between DMD patients and healthy controls aged 1 year or less. (E) The differential expression of select adipogenic and fibrogenic genes in SkM between DMD patients and healthy controls aged 2 to 8 years (left) and expression correlation between these genes (right). (F) A schematic diagram of the proposed mechanism regulating IMF and IMC accumulation in cattle and humans. *P < 0.05; **P < 0.01; ***P < 0.001.
Re‐analysis of published human SkM microarray data, including young, old, and old with metabolic syndrome individuals, showed that CFD expression increased with aging‐related metabolic disorders (Figure 8B). 31 In these individuals, positive correlations were identified between CFD expressions and pro‐adipogenic gene expressions, such as ADIPOQ, PLIN1, PPARG, SFRP1, and GPX3 (Figure 8C). Moreover, a meta‐analysis including 3000 human SkM samples showed a positive correlation between CFD expression and aging. 32 No significant increase in the expression of fibrogenic genes, such as COL1A1 and COL3A1, was identified in aged groups, suggesting the absence of fibrosis (Figure 8B). We then re‐analysed two human DMD datasets. Interestingly, the upregulation of CFD and many adipogenic markers was identified in older DMD patients but not in DMD patients aged 1 year or less despite the common upregulation of fibrogenic genes (Figure 8D,E). 33 The discrepancy agrees with the earlier onset of intramuscular fibrosis than adipogenesis in DMD. 2 , 34 Moreover, in older DMD patients, the expression of CFD was positively correlated with adipogenic genes (Figure 8E). As such, CFD likely plays a central role in human intramuscular adipogenesis. These results also suggest common mechanisms regulating intramuscular adipogenesis and fibrogenesis in larger mammals.
Discussion
Here, we report the first comprehensive scRNAseq analysis of Wagyu and Brahman SkM, two opposite extremes in IMF accumulation.
One of the most important findings is the great diversity among bovine FAPs, which are mainly composed of a fibrogenic, an adipogenic, an intermediate subpopulation, and an unspecified subpopulation. The differences in adipogenic and fibrogenic programming between Brahman and Wagyu FAPs are consistent with the established IMF content difference between beef of the two breeds. The conclusion is further supported by the validation of the pro‐adipogenic function of CFD and the confirmation of POSTN as a fibrogenic indicator. The endomysial localization of CFD + FAPs likely contributes to the unique accumulation of endomysial IMF in Wagyu cattle. 35 The positive correlation between early CFD expression in FAPs and IMF content in mature animals and the strong influence of Wagyu genetics on CFD expression in FAPs of crossbred cattle also indicate the potential use of this gene in marker‐assisted breeding. In addition, targeting CFD may promote IMF formation in beef cattle, as a CFD‐inducing small molecule was shown to promote adipogenesis [S21].
Also interesting is the widespread expression of CFD in human and non‐human primate FAPs and the positive correlation between CFD expression and aging‐ and DMD‐associated intramuscular adipogenesis in humans, suggesting an important role of CFD as a biomarker in human myopathies‐associated fatty infiltration and the possibility of targeting CFD in muscle fatty infiltration and obesity treatments. Indeed, serum adipsin levels have been reported to be positively correlated with adiposity in certain animal obesity models and obese humans. 36 , 37 Loss of C3aR, the receptor for C3a whose production is dependent on CFD/adipsin, transiently prevented diet‐induced obesity. 30 , 36 , 37 However, CFD seems to play a role in β cell function regulation as mice lacking CFD have reduced insulin secretion after a prolonged high‐fat diet treatment, leading to glucose intolerance. 37 Thus, caution needs to be made when systematically inhibiting CFD or other components of the alternative complement pathway. Rather, specific activation of CFD/adipsin in adipogenic progenitors may promote de novo adipogenesis and ameliorate the pathological expansion of adipose tissue, which requires further study. The absent expression of Cfd in mouse FAPs suggests that they are less predisposed to adipogenesis than human FAPs and likely contributes to the less robust fatty infiltration in dystrophic mice compared with dystrophy patients, which further suggests an advantage of bovine models over mouse models in IMF studies. 2 , 38 Interestingly, despite the lack of Cfd expression in mouse FAPs, Cfd was recently reported to promote bone marrow adipogenesis, suggesting the conserved pro‐adipogenic function of Cfd/CFD across species. 39
In conclusion, our study identifies distinct tissue‐resident cell types in bovine SkM and unveils the mechanisms contributing to differential IMF and IMC accumulation between Wagyu and Brahman cattle, which may also contribute to the pathological changes in human aging and myopathies (Figure 8F). Further studies are required to uncover the precise pathway through which CFD promotes FAP adipogenesis. The investigation of the genetic and epigenetic mechanisms responsible for the enhanced CFD expression in adipogenic FAPs is also an area for future research. Finally, our study provides a novel tool for studying intramuscular adipogenesis and fibrogenesis.
Conflict of interest
The authors declare no conflict of interests.
Supporting information
Figure S1. More robust angiogenic activity in Wagyu muscle (related to Figure 1). (A) The capillary density was quantified by isolectin B4 staining. n = 4 for each breed. Scale bar: 50 μm. (B‐C) GSEA was performed using genes differentially expressed between Wagyu and Brahman blood vessel EndoCs. A graph shows the enrichment of GOBP term ‘blood vessel development’ in Wagyu blood vessel EndoCs compared to Brahman blood vessel EndoCs (B). A dot plot shows the expression of core genes included in (B) in Brahman, Wagyu, and crossbred blood vessel EndoCs (C). *, p < 0.05; ***, p < 0.001.
Figure S2. ScRNAseq reveals communications between FAPs and other cell types in Wagyu muscle (related to Figure 2). (A) A cell–cell communication network includes all identified significant communications between different cell types in all breeds. (B) A dot plot shows all identified significant communications in which FAP‐1 or FAP‐2 functions as the signal receiver. Cell–cell communications are clustered based on the types of communication. Each column indicates a sender‐receiver pair. Each row indicates a ligand‐receptor pair. (C) A dot plot shows all identified significant communications in which FAP‐1 or FAP‐2 functions as the signal sender. Cell–cell communications are clustered based on the types of communication. Each column indicates a sender‐receiver pair. Each row indicates a ligand‐receptor pair. Red boxes highlight interactions that are specific to FAP‐1 or stronger between FAP‐1 and other cell types than between FAP‐2 and other cell types.
Figure S3. Bovine FAPs are a heterogeneous population (related to Figure 3). (A) Module analysis identified 42 modules. (B) UMAP graphs show the differential enrichment of select gene modules among FAPs subpopulations. (C) Graphs show GOBP terms enriched in module 2 and in modules 3 and 13. (D) Pseudotime analysis of FAPs. (E) A UMAP graph shows FAP C1–6 and tenocytes from all breeds, and feature plots show the expression of THBS4, CHAD, and COMP in FAPs and tenocytes. (F‐G) RNA velocity (F) and pseudotime analyses (G) of FAP C1–6 and tenocytes from all breeds.
Figure S4. More abundant type IIX muscle fibres are present in Brahman muscle (related to Figure 4). (A) Feature plots show the expression of select DEGs among different MC subpopulations. (B) A Heatmap shows the expression of top‐15 upregulated genes in each MC subpopulation. (C) The proportions of each type of myonuclei identified in Brahman, Wagyu, and crossbred by scRNAseq. (D‐F) IHC identifies different types of muscle fibres (D), the quantification of muscle fibre type composition (E), and average minimum diameters of different types of muscle fibres (F) in Brahman, Wagyu, and crossbred SkM. n = 4 for each breed. Scale bar: 100 μm.
Figure S5. ScRNAseq suggests differential activities between Wagyu and Brahman FAPs (related to Figure 6). (A) A volcano plot shows DEGs between Wagyu and Brahman FAP C3. (B) A volcano plot shows DEGs between Wagyu and Brahman FAP C4. Red dots indicate DEGs with Log2FC > 0.5 and p < 0.001. (C) A Venn diagram shows the overlap among DEGs in all FAPs and FAP C1–4 between Wagyu and Brahman. (D) A heatmap shows the expression of the top 50 upregulated genes in Wagyu FAP C3 and the top 50 upregulated genes in Brahman FAP C3 in FAP C3 of Wagyu, Brahman, and crossbred cattle. (E) A heatmap shows the expression of the top 50 upregulated genes in Wagyu FAP C4 and the top 50 upregulated genes in Brahman FAP C4 in FAP C4 of Wagyu, Brahman, and crossbred cattle. (F) RNA velocity of FAP C3 and C4.
Figure S6. The greater adipogenic potential of Wagyu FAPs is at least partially due to higher CFD expression and less robust fibrogenic programming (related to Figure 7). (A) The expression of CFD and C3AR1 in bovine FAPs with and without adipogenic induction was measured by realtime PCR. n = 3 for each treatment. (B) The expression of CFD in FAPs, Mo/Ma, and granulocytes, and the IMF content (%) of each animal. B, Brahman; C, crossbred; W, Wagyu. (C) A violin plot shows the expression of CFD in different bovine cell types. (D) The enrichment of NAB2 and EGR1 target genes in FAP C3. (E) Motif analysis show predicted EGR1 motifs immediately upstream of CFD. (F) Postn MCM/+ ;R26 eGFP mice were treated with tamoxifen from day 0 to day 3 after CTX injections. Muscle samples were collected without CTX injection and 7 days after the CTX injection. FAPs isolated from collected samples were induced for adipogenic differentiation and subjected to ICC to identify Postn lineage cells (GFP+), myofibroblasts (SMαA+), and adipocytes (perilipin‐1+). The adipogenic and fibrogenic efficiencies of FAPs isolated from uninjured muscles and muscles 7 days after CTX injection were calculated. n = 3 for each breed. Scale bar: 50 μm. **, p < 0.01; ****, p < 0.0001.
Figure S7. CFD is expressed in human FAPs but not mouse FAPs (related to Figure 8). (A‐D) ScRNAseq data from published studies using human muscle (A) [S61], monkey muscle (B) [S62], muscle of DMD mice (C) [S60], and glycerol‐injected mouse muscle (D) [S7] were reanalyzed to identify the expression of PDGFRA/Pdgfra and CFD/Cfd. (E) UMAP graphs show unsupervised clustering of human, Monkey, and uninjured WT mouse FAPs.
Table S1. DEGs among all identified cell types.
Table S2. Comparison of cell–cell communications involving FAPs between Wagyu and Brahman.
Table S3. DEGs among all FAP clusters.
Table S4. GRN regulating differential FAP gene expression. 1 indicates positive regulation. ‐1 indicates negative regulation. 0 indicates no regulation.
Table S5. DEGs among all MC clusters.
Table S6. DEGs among all MuC clusters.
Table S7. DEGs between Wagyu and Brahman FAPs.
Table S8. DEGs between Wagyu and Brahman FAP C3.
Table S9. DEGs between Wagyu and Brahman FAP C4.
Acknowledgements
This project was supported by the National Institute of Food and Agriculture at United States Department of Agriculture through 2020‐67015‐30823 (X.F.) and the National Institutes of Health through R15DK122383 (X.F.), P20GM130555 (X.F.), R01HL157519 (X.F.), R01AR078695 (S.K.), and F31AR077424 (S.O.). Postn‐MerCreMer and R26‐eGFP mice were gifted by Jeffery Molkentin at Cincinnati Children's hospital and David Goldhamer at the University of Connecticut, respectively. pLKO.1‐Cre was a gift from Elaine Fuchs (Addgene plasmid #25997). pLKO.1‐TRC was a gift from David Root (Addgene plasmid # 10878). pLKO.1‐shGFP was a gift from David Sabatini (Addgene plasmid #30323). We thank LSU High Performance Computing for assistance with sequencing data analysis. We also thank Mr. Boo Persica for his help with the ultrasound analysis. The authors of this manuscript certify that they comply with the ethical guidelines for authorship and publishing in the Journal of Cachexia, Sarcopenia and Muscle. 40
Wang L., Gao P., Li C., Liu Q., Yao Z., Li Y., et al (2023) A single‐cell atlas of bovine skeletal muscle reveals mechanisms regulating intramuscular adipogenesis and fibrogenesis, Journal of Cachexia, Sarcopenia and Muscle, 14, 2152–2167, 10.1002/jcsm.13292
Leshan Wang, Peidong Gao, and Chaoyang Li contributed equally to this study.
References
- 1. Rahemi H, Nigam N, Wakeling JM. The effect of intramuscular fat on skeletal muscle mechanics: implications for the elderly and obese. J R Soc Interface 2015;12:20150365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li W, Zheng Y, Zhang W, Wang Z, Xiao J, Yuan Y. Progression and variation of fatty infiltration of the thigh muscles in Duchenne muscular dystrophy, a muscle magnetic resonance imaging study. Neuromuscul Disord 2015;25:375–380. [DOI] [PubMed] [Google Scholar]
- 3. van Putten M, Lloyd EM, de Greef JC, Raz V, Willmann R, Grounds MD. Mouse models for muscular dystrophies: an overview. Dis Model Mech 2020;13:dmm043562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dubost A, Micol D, Picard B, Lethias C, Andueza D, Bauchart D, et al. Structural and biochemical characteristics of bovine intramuscular connective tissue and beef quality. Meat Sci 2013;95:555–561. [DOI] [PubMed] [Google Scholar]
- 5. Phelps K, Johnson D, Elzo M, Paulk C, Gonzalez J. Effect of Brahman genetics on myofibrillar protein degradation, collagen crosslinking, and tenderness of the longissimus lumborum. J Anim Sci 2017;95:5397–5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Giordani L, He GJ, Negroni E, Sakai H, Law JYC, Siu MM, et al. High‐dimensional single‐cell cartography reveals novel skeletal muscle‐resident cell populations. Mol Cell 2019;74:609–21.e6. [DOI] [PubMed] [Google Scholar]
- 7. Cao Y. Angiogenesis modulates adipogenesis and obesity. J Clin Investig 2007;117:2362–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dumont NA, Wang YX, von Maltzahn J, Pasut A, Bentzinger CF, Brun CE, et al. Dystrophin expression in muscle stem cells regulates their polarity and asymmetric division. Nat Med 2015;21:1455–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ricard‐Blum S. The collagen family. Cold Spring Harb Perspect Biol 2011;3:a004978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chao W, D'Amore PA. IGF2: epigenetic regulation and role in development and disease. Cytokine Growth Factor Rev 2008;19:111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Orozco OE, Walus L, Sah DW, Pepinsky RB, Sanicola M. GFRα3 is expressed predominantly in nociceptive sensory neurons. Eur J Neurosci 2001;13:2177–2182. [DOI] [PubMed] [Google Scholar]
- 12. González‐Pecchi V, Valdés S, Pons V, Honorato P, Martinez LO, Lamperti L, et al. Apolipoprotein A‐I enhances proliferation of human endothelial progenitor cells and promotes angiogenesis through the cell surface ATP synthase. Microvasc Res 2015;98:9–15. [DOI] [PubMed] [Google Scholar]
- 13. Folpe AL, Billings SD, McKenney JK, Walsh SV, Nusrat A, Weiss SW. Expression of claudin‐1, a recently described tight junction‐associated protein, distinguishes soft tissue perineurioma from potential mimics. Am J Surg Pathol 2002;26:1620–1626. [DOI] [PubMed] [Google Scholar]
- 14. Ishibashi J, Perry RL, Asakura A, Rudnicki MA. MyoD induces myogenic differentiation through cooperation of its NH2‐ and COOH‐terminal regions. J Cell Biol 2005;171:471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Choi MC, Ryu S, Hao R, Wang B, Kapur M, Fan CM, et al. HDAC4 promotes Pax7‐dependent satellite cell activation and muscle regeneration. EMBO Rep 2014;15:1175–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chang JH, Lin KH, Shih CH, Chang YJ, Chi HC, Chen SL. Myogenic basic helix‐loop‐helix proteins regulate the expression of peroxisomal proliferator activated receptor‐gamma coactivator‐1alpha. Endocrinology 2006;147:3093–3106. [DOI] [PubMed] [Google Scholar]
- 17. Chen M, Zhang L, Guo Y, Liu X, Song Y, Li X, et al. A novel lncRNA promotes myogenesis of bovine skeletal muscle satellite cells via PFN1‐RhoA/Rac1. J Cell Mol Med 2021;25:5988–6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Toniolo L, Maccatrozzo L, Patruno M, Caliaro F, Mascarello F, Reggiani C. Expression of eight distinct MHC isoforms in bovine striated muscles: evidence for MHC‐2B presence only in extraocular muscles. J Exp Biol 2005;208:4243–4253. [DOI] [PubMed] [Google Scholar]
- 19. Glaw JT, Skalak TC, Peirce SM. Inhibition of canonical Wnt signaling increases microvascular hemorrhaging and venular remodeling in adult rats. Microcirculation 2010;17:348–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bouton MC, Richard B, Rossignol P, Philippe M, Guillin MC, Michel JB, et al. The serpin protease‐nexin 1 is present in rat aortic smooth muscle cells and is upregulated in L‐NAME hypertensive rats. Arterioscler Thromb Vasc Biol 2003;23:142–147. [DOI] [PubMed] [Google Scholar]
- 21. Murfee WL, Skalak TC, Peirce SM. Differential arterial/venous expression of NG2 proteoglycan in perivascular cells along microvessels: identifying a venule‐specific phenotype. Microcirculation 2005;12:151–160. [DOI] [PubMed] [Google Scholar]
- 22. Shin M‐K, Choi B, Kim E‐Y, Park J‐E, Hwang ES, Lee HJ, et al. Elevated pentraxin 3 in obese adipose tissue promotes adipogenic differentiation by activating neuropeptide Y signaling. Front Immunol 2018;9:1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kusuyama J, Komorizono A, Bandow K, Ohnishi T, Matsuguchi T. CXCL3 positively regulates adipogenic differentiation. J Lipid Res 2016;57:1806–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pan D, Fujimoto M, Lopes A, Wang Y‐X. Twist‐1 is a PPARδ‐inducible, negative‐feedback regulator of PGC‐1α in brown fat metabolism. Cell 2009;137:73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cheng W, Wang F, Feng A, Li X, Yu W. CXXC5 attenuates pulmonary fibrosis in a bleomycin‐induced mouse model and MLFs by suppression of the CD40/CD40L pathway. Biomed Res Int 2020;2020:7840652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xin Y, Li C, Guo Y, Xiao R, Zhang H, Zhou G. RNA‐Seq analysis reveals a negative role of MSMO1 with a synergized NSDHL expression during adipogenesis of 3T3‐L1. Biosci Biotechnol Biochem 2019;83:641–652. [DOI] [PubMed] [Google Scholar]
- 27. Balaz M, Becker AS, Balazova L, Straub L, Müller J, Gashi G, et al. Inhibition of mevalonate pathway prevents adipocyte browning in mice and men by affecting protein prenylation. Cell Metab 2019;29:901–16.e8. [DOI] [PubMed] [Google Scholar]
- 28. Pilewski JM, Liu L, Henry AC, Knauer AV, Feghali‐Bostwick CA. Insulin‐like growth factor binding proteins 3 and 5 are overexpressed in idiopathic pulmonary fibrosis and contribute to extracellular matrix deposition. Am J Pathol 2005;166:399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Noris M, Remuzzi G. Overview of complement activation and regulation. Semin Nephrol 2013;33:479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mamane Y, Chung Chan C, Lavallee G, Morin N, Xu LJ, Huang J, et al. The C3a anaphylatoxin receptor is a key mediator of insulin resistance and functions by modulating adipose tissue macrophage infiltration and activation. Diabetes 2009;58:2006–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gueugneau M, Coudy‐Gandilhon C, Chambon C, Verney J, Taillandier D, Combaret L, et al. Muscle proteomic and transcriptomic profiling of healthy aging and metabolic syndrome in men. Int J Mol Sci 2021;22:4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Su J, Ekman C, Oskolkov N, Lahti L, Ström K, Brazma A, et al. A novel atlas of gene expression in human skeletal muscle reveals molecular changes associated with aging. Skelet Muscle 2015;5:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pescatori M, Broccolini A, Minetti C, Bertini E, Bruno C, D'Amico A, et al. Gene expression profiling in the early phases of DMD: a constant molecular signature characterizes DMD muscle from early postnatal life throughout disease progression. FASEB J 2007;21:1210–1226. [DOI] [PubMed] [Google Scholar]
- 34. Klingler W, Jurkat‐Rott K, Lehmann‐Horn F, Schleip R. The role of fibrosis in Duchenne muscular dystrophy. Acta Myol 2012;31:184–195. [PMC free article] [PubMed] [Google Scholar]
- 35. Ueda S, Hosoda M, Yoshino KI, Yamanoue M, Shirai Y. Gene expression analysis provides new insights into the mechanism of intramuscular fat formation in Japanese black cattle. Genes (Basel) 2021;12:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Flier JS, Cook KS, Usher P, Spiegelman BM. Severely impaired adipsin expression in genetic and acquired obesity. Science 1987;237:405–408. [DOI] [PubMed] [Google Scholar]
- 37. Lo JC, Ljubicic S, Leibiger B, Kern M, Leibiger IB, Moede T, et al. Adipsin is an adipokine that improves β cell function in diabetes. Cell 2014;158:41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Martins‐Bach AB, Malheiros J, Matot B, Martins PC, Almeida CF, Caldeira W, et al. Quantitative T2 combined with texture analysis of nuclear magnetic resonance images identify different degrees of muscle involvement in three mouse models of muscle dystrophy: mdx, Largemyd and mdx/Largemyd. PLoS One 2015;10:e0117835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Aaron N, Kraakman MJ, Zhou Q, Liu Q, Costa S, Yang J, et al. Adipsin promotes bone marrow adiposity by priming mesenchymal stem cells. Elife 2021;10:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. von Haehling S, Morley JE, Coats AJS, Anker SD. Ethical guidelines for publishing in the Journal of Cachexia, Sarcopenia and Muscle: update 2021. J Cachexia Sarcopenia Muscle 2021;12:2259–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. More robust angiogenic activity in Wagyu muscle (related to Figure 1). (A) The capillary density was quantified by isolectin B4 staining. n = 4 for each breed. Scale bar: 50 μm. (B‐C) GSEA was performed using genes differentially expressed between Wagyu and Brahman blood vessel EndoCs. A graph shows the enrichment of GOBP term ‘blood vessel development’ in Wagyu blood vessel EndoCs compared to Brahman blood vessel EndoCs (B). A dot plot shows the expression of core genes included in (B) in Brahman, Wagyu, and crossbred blood vessel EndoCs (C). *, p < 0.05; ***, p < 0.001.
Figure S2. ScRNAseq reveals communications between FAPs and other cell types in Wagyu muscle (related to Figure 2). (A) A cell–cell communication network includes all identified significant communications between different cell types in all breeds. (B) A dot plot shows all identified significant communications in which FAP‐1 or FAP‐2 functions as the signal receiver. Cell–cell communications are clustered based on the types of communication. Each column indicates a sender‐receiver pair. Each row indicates a ligand‐receptor pair. (C) A dot plot shows all identified significant communications in which FAP‐1 or FAP‐2 functions as the signal sender. Cell–cell communications are clustered based on the types of communication. Each column indicates a sender‐receiver pair. Each row indicates a ligand‐receptor pair. Red boxes highlight interactions that are specific to FAP‐1 or stronger between FAP‐1 and other cell types than between FAP‐2 and other cell types.
Figure S3. Bovine FAPs are a heterogeneous population (related to Figure 3). (A) Module analysis identified 42 modules. (B) UMAP graphs show the differential enrichment of select gene modules among FAPs subpopulations. (C) Graphs show GOBP terms enriched in module 2 and in modules 3 and 13. (D) Pseudotime analysis of FAPs. (E) A UMAP graph shows FAP C1–6 and tenocytes from all breeds, and feature plots show the expression of THBS4, CHAD, and COMP in FAPs and tenocytes. (F‐G) RNA velocity (F) and pseudotime analyses (G) of FAP C1–6 and tenocytes from all breeds.
Figure S4. More abundant type IIX muscle fibres are present in Brahman muscle (related to Figure 4). (A) Feature plots show the expression of select DEGs among different MC subpopulations. (B) A Heatmap shows the expression of top‐15 upregulated genes in each MC subpopulation. (C) The proportions of each type of myonuclei identified in Brahman, Wagyu, and crossbred by scRNAseq. (D‐F) IHC identifies different types of muscle fibres (D), the quantification of muscle fibre type composition (E), and average minimum diameters of different types of muscle fibres (F) in Brahman, Wagyu, and crossbred SkM. n = 4 for each breed. Scale bar: 100 μm.
Figure S5. ScRNAseq suggests differential activities between Wagyu and Brahman FAPs (related to Figure 6). (A) A volcano plot shows DEGs between Wagyu and Brahman FAP C3. (B) A volcano plot shows DEGs between Wagyu and Brahman FAP C4. Red dots indicate DEGs with Log2FC > 0.5 and p < 0.001. (C) A Venn diagram shows the overlap among DEGs in all FAPs and FAP C1–4 between Wagyu and Brahman. (D) A heatmap shows the expression of the top 50 upregulated genes in Wagyu FAP C3 and the top 50 upregulated genes in Brahman FAP C3 in FAP C3 of Wagyu, Brahman, and crossbred cattle. (E) A heatmap shows the expression of the top 50 upregulated genes in Wagyu FAP C4 and the top 50 upregulated genes in Brahman FAP C4 in FAP C4 of Wagyu, Brahman, and crossbred cattle. (F) RNA velocity of FAP C3 and C4.
Figure S6. The greater adipogenic potential of Wagyu FAPs is at least partially due to higher CFD expression and less robust fibrogenic programming (related to Figure 7). (A) The expression of CFD and C3AR1 in bovine FAPs with and without adipogenic induction was measured by realtime PCR. n = 3 for each treatment. (B) The expression of CFD in FAPs, Mo/Ma, and granulocytes, and the IMF content (%) of each animal. B, Brahman; C, crossbred; W, Wagyu. (C) A violin plot shows the expression of CFD in different bovine cell types. (D) The enrichment of NAB2 and EGR1 target genes in FAP C3. (E) Motif analysis show predicted EGR1 motifs immediately upstream of CFD. (F) Postn MCM/+ ;R26 eGFP mice were treated with tamoxifen from day 0 to day 3 after CTX injections. Muscle samples were collected without CTX injection and 7 days after the CTX injection. FAPs isolated from collected samples were induced for adipogenic differentiation and subjected to ICC to identify Postn lineage cells (GFP+), myofibroblasts (SMαA+), and adipocytes (perilipin‐1+). The adipogenic and fibrogenic efficiencies of FAPs isolated from uninjured muscles and muscles 7 days after CTX injection were calculated. n = 3 for each breed. Scale bar: 50 μm. **, p < 0.01; ****, p < 0.0001.
Figure S7. CFD is expressed in human FAPs but not mouse FAPs (related to Figure 8). (A‐D) ScRNAseq data from published studies using human muscle (A) [S61], monkey muscle (B) [S62], muscle of DMD mice (C) [S60], and glycerol‐injected mouse muscle (D) [S7] were reanalyzed to identify the expression of PDGFRA/Pdgfra and CFD/Cfd. (E) UMAP graphs show unsupervised clustering of human, Monkey, and uninjured WT mouse FAPs.
Table S1. DEGs among all identified cell types.
Table S2. Comparison of cell–cell communications involving FAPs between Wagyu and Brahman.
Table S3. DEGs among all FAP clusters.
Table S4. GRN regulating differential FAP gene expression. 1 indicates positive regulation. ‐1 indicates negative regulation. 0 indicates no regulation.
Table S5. DEGs among all MC clusters.
Table S6. DEGs among all MuC clusters.
Table S7. DEGs between Wagyu and Brahman FAPs.
Table S8. DEGs between Wagyu and Brahman FAP C3.
Table S9. DEGs between Wagyu and Brahman FAP C4.
Data Availability Statement
All necessary data to evaluate the paper's conclusions are available in the paper and supporting information. Raw genomic sequencing datasets have been deposited at GEO (GSE205347). Additionally, we used publicly available human SkM microarray datasets from GEO (GSE6011, GSE38417, and GSE136344), scRNAseq data of human muscle tissue from GEO (GSE143704), scRNAseq data of Macaca fascicularis muscle tissue from CNGB Nucleotide Sequence Archive (CNP0001469), scRNAseq data of WT and DMD mouse muscle tissue from GEO (GSE156497), and scRNAseq data for glycerol‐injected mouse muscle from a published study [S7].


