The widespread use of broad-spectrum antimicrobial agents poses a significant challenge to bacteria pathogenic for humans, a challenge that has been answered by the acquisition and spread of a variety of antimicrobial resistance determinants. Bacterial resistance to antibiotics results from the mutation of normal cellular genes, the acquisition of foreign resistance genes, or a combination of these two mechanisms. Resistance to some antimicrobials or classes of antimicrobials, notably, rifampin or the fluoroquinolones, occurs primarily through point mutations and is not transferable. Other forms of resistance, such as multiple antibiotic resistance efflux systems and inducible β-lactamase-mediated resistance, result from regulatory mutations in normal cellular processes and are also not transferable. For many bacteria-antibiotic combinations, however, such intrinsic mechanisms of resistance are not available, so resistance genes must be imported. To accomplish this end, bacteria use a complex array of mechanisms to share and disseminate useful resistance determinants. Mechanisms frequently cited as potentially important for the dissemination of antibacterial resistance determinants include natural transformation (penicillin resistance in Streptococcus pneumoniae and Neisseria spp.) (16, 60), bacteriophage-mediated transduction (β-lactamase-mediated penicillin resistance in staphylococci) (33), and plasmid- or transposon-mediated conjugation (ampicillin resistance in members of the family Enterobacteriaceae, tetracycline resistance in enterococci, and many others) (3, 20).
With the widespread availability of techniques for analyzing the molecular genetics of bacterial pathogens, understanding of the role of transposons in the dissemination of antimicrobial resistance has expanded significantly. Among the more interesting and important classes of transposons are the conjugative transposons. As their name implies, conjugative transposons are mobile elements that possess the genetic machinery to facilitate their own transfer between bacterial cells. Several prototypes of conjugative transposons have been described. Conjugative transposons encoding the tet(Q) tetracycline resistance determinants are widespread in clinical Bacteroides isolates and have been the subject of several recent reviews (54, 55). Tn5276-like transposons are large (70- to 80-kb) elements that encode genes for nisin production (44, 45). These elements have not been shown to encode antimicrobial resistance determinants, appear to be restricted to lactococci, and will not be addressed further in this minireview. The present minireview focuses on the Tn916 and Tn1545 family of conjugative transposons that predominates in clinically important gram-positive bacteria. The basic mechanisms of movement of these transposons have recently been reviewed (57). The present work focuses on recent developments in the basic science of conjugative transposons and more specifically on the actual and potential role of these elements in the dissemination of important resistance determinants in pathogenic bacteria.
STRUCTURE AND MECHANISM OF TRANSPOSITION
The two most extensively studied conjugative transposons are Tn916 from Enterococcus faecalis (21) and Tn1545 from S. pneumoniae (14). These two transposons differ in size (18 versus 25.2 kb, respectively) and in the antimicrobial resistance determinants that they encode {tetracycline-minocycline [tet(M)] by Tn916 and erythromycin [ermAM], kanamycin [aphA-3], and tetracycline-minocycline [tet(M)] by Tn1545}. Despite these differences, the two transposons are similar and are even identical in many respects. Their termini are identical for at least 250 bp on each end (6, 12). Moreover, their integrase and excisase genes that encode transposition functions differ by only one nucleotide over ca. 2,000 bp (19, 42). The tet(M) genes from the two transposons exhibit roughly 94.5% nucleotide identity over 2 kb (40), a difference that probably reflects divergent evolution of the tet(M) determinant in disparate genera. Supporting this hypothesis is the observation that the tet(M) gene of Tn5251 (a pneumococcal conjugative transposon otherwise structurally indistinguishable from Tn916) has a 688-bp segment that is only 90% identical to the corresponding region of tet(M) from Tn916 but 100% identical to the same region from Tn1545 (43). The nucleotide sequence of the entire 18 kb of Tn916 has been published (19) and reveals that 70% of the restriction sites represented only once in the transposon are found within the 2-kb region of the tet(M) gene (40). In addition, the deduced amino acid sequence of a Tn916 open reading frame (orf-18) to the right of tet(M) (Fig. 1A) exhibits significant local homology to restriction and modification inhibitors from self-transmissible plasmids in gram-negative bacteria (19). The relative dearth of restriction sites in the non-tet(M) portion of Tn916, combined with evidence for restriction modification resistance, suggest that conjugative transposons are elements highly evolved for broad-host-range transfer and that the tet(M) genes are only relatively recent arrivals.
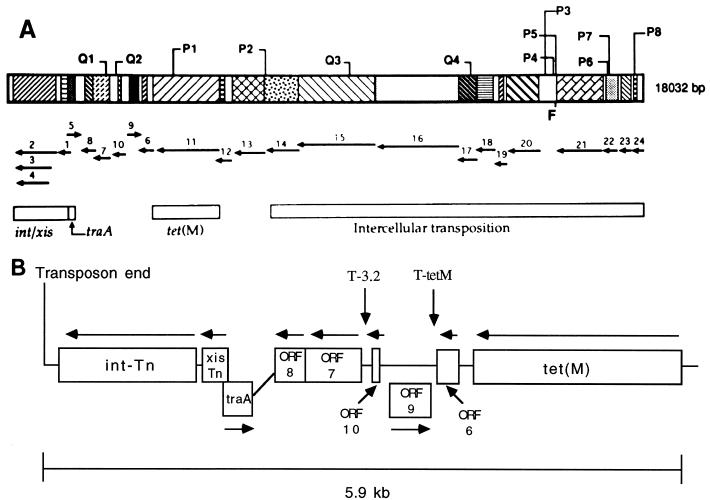
FIG. 1.
(A) Map of Tn916 (18,032 bp). Identified open reading frames and the directions of their transcription are indicated by the arrows below the map. Areas that have been identified as important for specific functions are identified by open boxes below the map. For more detail on the left end of Tn916, refer to panel B. Points designated by the letters F, P, or Q, with or without numbers following them, represent sequences with homology to the nic sites of plasmid IncF, IncP, or IncQ, respectively. The region between open reading frames 20 and 21 allows nonconjugative plasmids to be mobilized by chromosomal copies of Tn916, and therefore represents a functional origin of transfer for Tn916. Adapted with permission from reference 27. (B) Schematic portrayal of the left end of Tn916. The terminus of the transposon is marked. The open reading frames (ORFs) are designated by names [xis-Tn, int-Tn, traA, tet(M)] if such designations have been given. Otherwise they are listed as numbered open reading frames. Arrows above or below the boxes represent the direction of transcription of the indicated gene. tet(M) confers tetracycline and minocycline resistance. Open reading frames 6, 9, and 10 have no identified function but appear to be related to the tet(M) gene since tet(M) transcripts of extended length (corresponding to the length added by these open reading frames) have been identified after tetracycline exposure. T-tetM indicates the terminator of the tet(M) transcript. T-3.2 indicates a potential termination site for the extended 3.2-kb transcript seen after tetracycline exposure (see text). Open reading frames 7 and 8 have no identified function. traA is thought to be a positive activator of conjugation functions, all of which are located on the opposite side of tet(M) within Tn916. int-Tn encodes the integrase that is responsible for transposition; xis-Tn encodes the excisase that increases the frequency of excision, but is not required.
Transposition genes and their functions.
The tet(M) gene is located toward the left end of Tn916 (open reading frame extending from bp 12,108 to bp 14,040 of the 18,032-bp transposon) (19). The genetic structure of Tn916 is shown in Fig. 1A. Several virtually identical structures are present within what has been designated the right end of Tn1545. Located within the approximately 4 kb extending from the stop codon of the tet(M) gene to the left terminus of Tn916 are open reading frames designated int-Tn and xis-Tn that encode transposition functions (28). These open reading frames have been so designated on the basis of their homology with the lambda integrase (Int) family of proteins and the xis gene of P22 and Pin (a Hin-related invertase) (42). xis-Tn and int-Tn are located downstream of the tet(M) gene; all three genes are transcribed from the same DNA strand. Int-Tn mediates the specific DNA cleavage required for excision and integration of Tn916 (63). The amino terminus of Int-Tn binds to direct repeats located ca. 120 bp from the ends of the element, while the carboxy terminus binds to the terminal nucleotides of Tn916 and flanking sequences. Expression of the Xis-Tn protein increases the frequency of excision by binding to a region between the Int-Tn-binding sites and may be involved in bending the DNA to facilitate the action of Int-Tn (52). Recent in vitro data indicate that Xis-Tn is required for excision of Tn916-like molecules at high salt concentrations (150 mM NaCl) but that Int-Tn alone can catalyze limited excision at low salt concentrations (37.5 mM NaCl) (51). At low salt concentrations, Int-Tn-mediated excision was stimulated by the presence of Xis-Tn.
Excision of Tn916 and the circular intermediate of transposition.
The first step in Tn916 transposition is excision of the molecule to form a nonreplicative circular intermediate. Several factors have been associated with increased numbers of detectable excised forms within a culture (although not within a specific organism). The sequences flanking the inserted transposon (coupling sequences) appear to exert an important influence on the frequency of excision from a specific site. Differences in conjugative transposition frequency of 10,000-fold were related to different coupling sequences in one study of Tn916 (26). The insertions exhibiting increased conjugation frequency also yielded more excised molecules from exposed strains in PCR amplification studies, suggesting that excision is the rate-limiting step for Tn916-mediated conjugation (26). Other studies have also suggested an association between transfer frequency and the ability to detect circular intermediates either by plasmid analysis or by PCR amplification (35, 49). In a recent study by Celli et al. (8), a linear correlation between rates of excision by a trans-complementation assay and the conjugal transfer frequency of intact Tn916 was not observed. However, since the molecule transferred in those studies (Tn916) and the molecule used for excision-rate reporter studies (a modified Tn916-like molecule unable to manufacture Int-Tn or Xis-Tn) were different, it is difficult to draw firm conclusions. Some studies have suggested that exposure to tetracycline increases the level of excision of conjugative transposons (35, 49), although this has not been a universal finding (8). The mechanism by which increased quantities of circular forms may occur after incubation of Tn916-containing cells with tetracycline is not known. However, since tet(M) transcripts are increased in quantity and length following tetracycline exposure, the positioning of xis-Tn and int-Tn downstream of tet(M) may result in increased levels of expression of these genes by mechanisms similar to those resulting in increased levels of expression of tet(M) (61). Rates of transfer to recipient cells have also been shown to be increased in vitro and in vivo after preincubation of donor cells with tetracycline (15, 49, 64). Although the commonly held view is that the increased levels of transfer of conjugative transposons after tetracycline exposure are merely by-products of increased levels of excision of the molecule, Celli et al. (8) have suggested that a tetracycline-induced transcript extends through the newly formed attachment at the ends of the transposon, activating conjugation genes. At the present time, there are no data to support this hypothesis. This tetracycline-induced or -selected transfer may represent an important mechanism by which the tet(M) gene disseminates among bacteria under the selective pressure of tetracycline in the clinical setting. Moreover, in the setting of the frequent use of tetracyclines for the treatment of human infections and as a growth enhancer in animals, an increased prevalence of conjugative transposons may have the additional effect of promoting the spread of multidrug resistance in the bacterial population (see below).
In 1989, Caparon and Scott (7) reported on studies of the excision of Tn916 in Escherichia coli indicating that the circular intermediate was joined by a 5-bp sequence representing one strand from each coupling sequence flanking the previous insertion site. Since target duplications are not generated by the insertion of Tn916, the nucleotide sequence of the two strands of this “joint” region were mismatched, forming a heteroduplex. Concurrent studies by a PCR amplification strategy suggested that the joint sequences were composed of six nucleotides on each strand (58, 65). In those studies, each strand was represented equally in the experimental results. Rice et al. (49) reported the first direct evidence of a circular form of Tn916 in E. faecalis in 1992. Subsequent studies of the joint region by Rice and Carias (46) in 1994 indicated that the sequences of the two strands of the heteroduplex could be mismatched not only in nucleotide content but also in nucleotide number, a possibility that had been suggested in previous work by Poyart-Salmeron et al. (42). For both Tn916 and the closely related conjugative transposon Tn5381, it was shown that roughly 75 to 80% of the amplified joint sequences represented the 6-bp coupling sequence on the left side of the previous insertion, whereas the remaining amplification products reflected the 5-bp coupling sequence on the right side (46).
Further clarification of the content of the joint in Tn916 circular intermediates was recently published by Manganelli et al. (34). Those investigators used a limiting-dilution PCR amplification strategy to determine the sequence of joint regions of single circular intermediate molecules excised from E. coli or E. faecalis. Consistent with prior studies, they identified heteroduplex molecules in the E. coli excisants, although such heteroduplexes represented only 50% of the molecules studied. In E. faecalis all molecules were homoduplexes, with roughly 80% of the joints reflecting the left coupling sequence and 20% reflecting the right coupling sequence. These data suggest that in E. faecalis the mismatched joint region undergoes repair to a homoduplex and that the repair system used to facilitate this change may favor the preservation of one strand over another. The observation that heteroduplexes were present in 50% of the excised molecules in E. coli suggests that the repair system may not be as efficient in this background. Alternatively, the high frequency of Tn916 excision in E. coli may simply overwhelm the capacity of the system to repair the heteroduplex (34).
Transfer and insertion of Tn916.
The precise excision of Tn916 suggests a conservative mechanism of transposition, or at least one in which replication does not result in the persistence of a transposon copy at the donor site. Despite this presumed conservative mechanism, a consistent result of in vitro mating experiments is the presence of multiple transposons in the chromosome of the recipient strain, suggesting the possibility of replicative transposition (49). Although this observation remains unexplained, one reasonable hypothesis is that intracellular transposition of Tn916 occurs from one of two daughter chromosomes after passage of the replication fork. If subsequent insertion into a chromosomal region that has not yet replicated occurs, the presence of multiple Tn916 copies without the invocation of a replicative transposition mechanism could be explained. Alternative explanations include the cotransfer or the independent transfer of Tn916 copies during a mating event. In this regard, it is noteworthy that the presence of Tn916 within a recipient chromosome does not alter the frequency of transfer of a second copy, suggesting that these transposons do not confer transposition immunity or entry exclusion to the cells that they occupy (39). Despite the frequency of multiple insertions after in vitro mating experiments, multiple conjugative transposons within clinical strains are rarely if ever reported. The potential for recombination between multiple copies within a chromosome and the subsequent deletion of important structural and functional genes may select against multiple copies of Tn916 in clinical strains. Alternatively, failure to observe multiple copies in clinical strains may simply reflect the limited number of strains examined.
Locations for insertion of Tn916 are not entirely random. Consensus sequences for Tn916 and Tn1545 targets have been proposed and in general reflect regions in which homology exists between the target site and sequences within the termini of the transposon (56, 66). One “hot spot” that has been consistently identified is within the hemolysin-bacteriocin gene of pheromone-responsive E. faecalis plasmids (24). Depending on the precise site and orientation of the insertion, either nonhemolytic or hyperhemolytic phenotypes can result. An important consequence of the presence of a hot spot on pheromone-responsive plasmids is that integration into these elements, which transfer at a much higher frequency than Tn916, can promote the dissemination of resistance determinants encoded by the transposon. Moreover, studies performed with animals indicate that the hemolysin determinant is a virulence factor for enterococcal infections (29). Insertions resulting in the hyperhemolytic phenotype have been shown to yield bacteria that are more virulent than nonhemolytic or normally hemolytic isogenic strains.
An open reading frame within the left end of Tn916, designated traA, is required for conjugative transposition. It has been suggested that this gene is a positive regulator of conjugation genes (the precise functions of which remain to be defined) located to the right of the tet(M) gene within Tn916 (28). The start of the traA open reading frame overlaps the start of xis-Tn on the opposite strand (Fig. 1A and B). This intimate association may explain the observed relationship between the excision and transfer frequencies of conjugative transposons.
By analogy with the transfer of plasmid DNA, it is generally presumed that Tn916 transfers to recipient cells as a single strand. The primary experimental evidence in support of this mechanism is the observation that sequences flanking new insertions in transconjugants reflect only one of the prior coupling sequences (56). The recent evidence suggesting that the heteroduplex undergoes preferential repair in the donor (34) diminishes the impact of these data by raising the question of whether enough transconjugants were examined to assess the possibility that both strands are transferred. In addition, it is not clear to what extent and with what preference mismatch repair occurs in Bacillus subtilis, which was used as a transposon donor in these experiments (56). Cell-to-cell contact is required for the transfer of conjugative transposons. In general, mating experiments for the detection of transfer of conjugative transposons are performed on solid media, although transfer in broth has been observed from high-frequency donors. It is not clear exactly how much time is required for Tn916 transfer, but transconjugants are readily observed after 4-h matings (49).
Tn916 family of conjugative transposons that have been described in the literature and their species of origin are listed in Table 1. All of these elements express tetracycline-minocycline resistance encoded by the tet(M) gene. Some encode additional antimicrobial resistance genes as well, most commonly, genes for erythromycin resistance. Several conjugative transposons are incorporated within larger transferable elements that encode additional antimicrobial resistance genes. These larger elements have been analyzed to various degrees and represent several different classes of elements, some of which are discussed below.
TABLE 1.
Tn916 family of transposons
| Transposon designation (resistance) | Reference | Size (kb) | Species of origin | Larger element (additional resistance determinants) |
|---|---|---|---|---|
| Tn916 (Tcr) | 21 | 18 | E. faecalis | None |
| Tn918 (Tcr) | 11 | 16 | E. faecalis | None |
| Tn919 (Tcr) | 17 | 15.4 | Streptococcus sanguis | None |
| Tn920 (Tcr) | 37 | 23 | E. faecalis | None |
| Tn925 (Tcr) | 9 | 18 | E. faecalis | None |
| Tn1545 (Emr, Kmr, Tcr) | 14 | 25.4 | S. pneumoniae | None |
| Tn3702 (Tcr) | 23 | 18 | E. faecalis | None |
| Tn3703 (Emr, Tcr) | 10 | 19.7 | S. pyogenes | Tn3701 |
| Tn3704 (Emr, Tcr) | 10 | 20.3 | Streptococcus anginosus | Tn3705 |
| Tn5251 (Tcr) | 1 | 18 | S. pneumoniae | Tn5252 (Cmr) |
| Tn5381 (Tcr) | 49 | 18 | E. faecalis | Tn5385 (β-lactamase, Emr, Gmr, Merr, Smr) |
| Tn5383 (Tcr) | 49 | 18 | E. faecalis | Tn5385 (β-lactamase, Emr, Gmr, Merr, Smr) |
| Tn5397 (Tcr) | 36 | >10.4 | Clostridium difficile | None |
| Unnamed (Tcr) | 25 | ? | S. agalactiae | Tn3951 (Cmr) |
Cmr, chloramphenicol resistance; Emr, erythromycin resistance; Merr, mercuric chloride resistance; Kmr, kanamycin resistance; Tcr, tetracycline resistance.
THE BROAD HOST RANGE OF CONJUGATIVE TRANSPOSONS
Transfer of Tn916 in vitro.
There appear to be few if any limits to the types of bacterial hosts into which conjugative transposons will transfer in vitro. Tn916 and Tn1545 transfer readily to a wide variety of gram-positive and gram-negative bacteria in the laboratory. In one report, E. faecalis readily transferred plasmid-borne Tn916 to Alcaligenes eutrophus, Citrobacter freundii, and E. coli, conferring selectable levels of tetracycline resistance (>10 μg/ml) (2). It was unclear from that report whether the donor plasmid was also present in the transconjugants, since no plasmid was detectable but the restriction fragments hybridizing to Tn916 were the same size in donors and transconjugants. In one A. eutrophus transconjugant, two additional hybridizing bands were visible, suggesting transposition of Tn916 into the recipient chromosome. The only species into which these investigators failed to transfer Tn916 was Acetobacterium woodii. In addition, they observed significant rates of Tn916 transfer from the chromosome of E. coli to B. subtilis, Clostridium acetobutylicum, E. faecalis, and Streptococcus lactis subsp. diacetylactis. A more recent study also observed the transfer of Tn916 and Tn1545 from E. faecalis to E. coli and Pseudomonas fluorescens, but those investigators were unable to select directly for Tn916 transfer using tetracycline (41). Rather, they used a mutant of Tn916 in which the tet(M) gene had been replaced by a gene for kanamycin resistance. Similar transfer characteristics were noted for Tn1545, which encodes kanamycin resistance naturally. It is unclear why Tn916 has never been found in clinical E. coli isolates. When cloned into high-copy-number E. coli plasmids, Tn916 excises at a high frequency (58). This increased excision rate, if generally true within E. coli, may result in the instability of these elements in this background.
Not all species are able to serve as Tn916 donors. Lactococcus lactis, for example, can serve as a recipient of Tn916, but it is unable to serve as a donor (5). Failure of secondary transfer after Tn916 integration into a recipient may also be due to deletions of important segments of the element. Tetracycline resistance plasmids in Neisseria gonorrhoeae have been shown to include various portions of Tn916 flanking the tet(M) gene (62). Similar deletion events have been documented experimentally during transformation experiments with Tn916 (62). In such settings, the tet(M) gene can no longer undergo conjugative transposition but may still transfer if the plasmid is capable of independent transfer or conjugative mobilization.
Transfer of Tn916 in vivo.
Evidence for the transfer of conjugative transposons to disparate species in vivo exists as well. Doucet-Populaire et al. (15) have demonstrated the transfer of Tn1545 from E. faecalis to Listeria monocytogenes in the digestive tracts of gnotobiotic mice. In this vein, it is of interest that conjugative transposons are widely prevalent in bacteria known to colonize the gastrointestinal tract in significant numbers. Specifically, conjugative transposons are most frequently described in enterococci and Bacteroides species (54, 57). The availability of broad-host-range transfer systems in a microbial environment that evolves in response to antimicrobial selective pressure confers a significant survival advantage on those bacteria able to take advantage of the systems.
The host range of Tn916-like transposons in nature does not precisely reflect the host range observed in in vitro experiments. For example, although Tn916 is readily transferred to E. coli in the laboratory, similar elements have never been described in clinical isolates of members of the family Enterobacteriaceae. Recently, large transferable chromosomal elements have been described in Salmonella and Vibrio spp., but these have not encoded tet(M) and are therefore unlikely to be closely related to Tn916 (22, 67). A list of genera in which the tet(M) determinant (and, in many cases, Tn916-like elements) have been described appears in Table 2. Much of this dissemination is presumably due to Tn916-mediated intercellular transfer. The genera into which Tn916-like elements have been transferred in the laboratory are also listed in Table 2.
TABLE 2.
Genera in which the tet(M) gene has been found and/or genera which are within the host range of Tn916- and Tn1545-like transposonsa
| Genus | tet(M) found in clinical strains | Tn916 and Tn1545 host range | Genus | tet(M) found in clinical strains | Tn916 and Tn1545 host range | |
|---|---|---|---|---|---|---|
| Acetobacterium | + | Gardnerella | + | |||
| Acholeplasma | + | Gemella | + | |||
| Actinobacillus | + | Haemophilus | + | + | ||
| Actinomyces | + | Kingella | + | |||
| Aerococcus | + | Lactobacillus | + | |||
| Alcaligenes | + | Lactococcus | + | |||
| Bacillus | + | Leuconostoc | + | |||
| Bifidobacterium | + | Listeria | + | + | ||
| Butyrivibrio | + | Mycoplasma | + | + | ||
| Campylobacter | + | Neisseria | + | + | ||
| Citrobacter | + | Peptostreptococcus | + | + | ||
| Clostridium | + | + | Prevotella | + | ||
| Corynebacterium | + | Staphylococcus | + | + | ||
| Eikenella | + | Streptococcus | + | + | ||
| Enterococcus | + | + | Thermus | + | ||
| Escherichia | + | Ureaplasma | + | |||
| Eubacterium | + | + | Veillonella | + | + | |
| Fusobacterium | + | + |
CONJUGATIVE TRANSPOSONS AND THE DISSEMINATION OF ANTIBIOTIC RESISTANCE
Transfer of resistance encoded by genes within the conjugative transposon.
There are several mechanisms by which conjugative transposons can have a significant impact on the dissemination of antimicrobial resistance determinants among pathogenic bacteria. The first and most obvious of these mechanisms is the direct transfer of resistance determinants that are encoded by genes within the transposons themselves. Since most conjugative transposons encode resistance to tetracycline or minocycline alone, the most obvious impact of conjugative transposons has been on the spread of resistance to these antimicrobial agents. With the growing availability of a variety of broadly active and safe oral and intravenous antimicrobial agents, the importance of tetracyclines as therapeutic alternatives has decreased. Nevertheless, tetracyclines remain first-line agents for the treatment of urogenital infections caused by Chlamydia or Mycoplasma species and for diseases caused by Rickettsia, Ehrlichia, and Brucella species and as second-line therapy for Francisella tularensis infections. Tetracycline resistance has not been reported in all of these species, but the continued dissemination of broad-host-range transferable elements encoding tet(M) through the microbial environment is cause for concern.
Mobilization of other replicons.
A second mechanism by which conjugative transposons can disseminate antimicrobial resistance determinants is by mobilizing plasmids or other transposons that encode antimicrobial resistance determinants. This phenomenon has been best demonstrated in Bacteroides mobile elements, in which conjugative transposons have been shown to mobilize nonreplicative bacteroides units, some of which have encoded determinants of resistance to important antibiotics such as cefoxitin (54). The ability of Tn916-like transposons to mobilize other replicative elements is not as clearly defined as it is for Bacteroides transposons, but it is well established that the excision and transfer of one Tn916-like element enhances the transfer of another conjugative transposon that is residing in the same cell (18). If the two elements are encoding different resistance determinants, multidrug resistance transfer can occur in a single mating event. Tn916 has also been shown to mobilize nontransferable plasmids in experimental settings, although it is not clear how frequently this occurs in vivo (38). Finally, Tn916-like elements could confer transfer capabilities onto nontransferable plasmids into which they insert. The ability of Tn916-like elements to transpose to plasmids in the laboratory is well established. Tn925 was first identified on transferable, pheromone-responsive plasmid pCF10 in E. faecalis (9). In this instance, two independent conjugation systems were present on the same plasmid, so the contribution of Tn925 to plasmid transfer was difficult to assess. A recent report by Jaworski and Clewell (27) identified a region of Tn916 that appears to serve as the origin of transfer (oriT) of these elements. Importantly, those investigators were able to show that a chromosomal copy of Tn916 could mobilize a nonconjugative shuttle plasmid into recipient strains, as long as that plasmid possessed oriT sequences. These data suggest that a conjugative transposon present within a nonconjugative plasmid in a clinical isolate could confer transferability on this plasmid (and its other associated resistance determinants) if a second copy of the element was located in the donor chromosome.
Large, multidrug resistance composite elements.
Tn916-like elements contribute to the dissemination of multidrug resistance by integrating into larger conjugative transposons that encode additional antimicrobial resistance determinants. Perhaps the best-studied example of this phenomenon is the integration of Tn5251 (Tn916-like) into Tn5252 in S. pneumoniae (1). Tn5252 is a large chromosomal element that encodes chloramphenicol resistance and transfers in a site-specific manner to recipient pneumococcal chromosomes (30). Integration of Tn5251 into Tn5252 results in the composite element Tn5253, which transfers between pneumococcal strains at a relatively high frequency (ca. 10−4/recipient CFU). Independent conjugative transfer of Tn5251 from its position within Tn5253 has not been documented; however, it can transpose from Tn5253, excising and integrating elsewhere. Once independent of Tn5252, Tn5251 behaves and transfers like a fully functional conjugative transposon (43). It is not clear whether Tn5251 is truly incapable of conjugative transfer from its position within Tn5253 or whether its transfer frequency is too low to be detected in the presence of the high frequency of transfer of Tn5253. Several similar composite elements have now been described from other strains of pneumococci, as well as from Streptococcus pyogenes and Streptococcus agalactiae (10, 25, 31, 32).
Resistance exchange by chromosomal recombination.
A final mechanism by which Tn916-like elements can influence the transfer of multiple antimicrobial resistance determinants is by allowing homologous recombination between donor and recipient chromosomes during mating events. Torres et al. (64) reported on matings using B. subtilis and E. faecalis strains as donors in which unlinked chromosomal markers were transferred when conjugative transposon Tn925 was present in the donor chromosome. This transfer occurred even in the absence of the documented transfer of the conjugative transposon itself. Those investigators hypothesized that the presence of Tn925 in the donor chromosome resulted in mating events that resembled cell fusion wherein the donor and the recipient chromosomes could recombine across regions of homology. In contrast to these findings, studies comparing Tn925 and Tn916 failed to detect the retrotransfer of plasmids from recipients to donors during conjugative transposition of either transposon, arguing against cell fusion as a mechanism of Tn916 transfer (59).
Chromosomal recombination has recently been observed during studies of enterococcal conjugative elements Tn5381 and Tn5385 (47). Tn5381 is a Tn916-like element integrated into the chromosome of E. faecalis CH19. Tn5381 lies within Tn5385, a transferable composite element that encodes resistance to penicillin (via β-lactamase production), erythromycin, gentamicin, mercuric chloride, streptomycin, and tetracycline-minocycline (48). Tn5385 is a chromosomally integrated vestige of a member of the broad-host-range pAMβ1 family of plasmids. It differs significantly from pAMβ1, however, in that its origin of replication has been deleted and it does not possess conjugation genes related to pAMβ1 (4). The only region within Tn5385 that encodes genes capable of stimulating the transfer of genetic material are the conjugation genes within conjugative transposon Tn5381 (49). In contrast to the observations of Tn5253 in pneumococci, Tn5381 transfers at a much higher frequency than the larger element within which it is incorporated (ca. 10−6 versus <10−8/recipient CFU, respectively). Chromosomal insertion of Tn5381 after transfer to recipient cells is relatively nonselective in its location and is frequently into multiple sites within the same transconjugant, typical of the Tn916 family of conjugative transposons. In contrast, insertion of Tn5385 into the recipient chromosome most commonly occurs by recombination across sequences flanking Tn5385 in the donor and homologous sequences in the recipient.
The mechanism for the initiation of the conjugation event that allows Tn5385 transfer is not known. Recombination between donor and recipient chromosomes, however, is consistent with the occurrence of an intermediate cell fusion event in which close contact between donor and recipient chromosomes occurs. Given the data published by Torres et al. (64) regarding the transfer of disparate chromosomal markers in association with the presence of Tn925 in the donor chromosome, it is reasonable to hypothesize that Tn5385 transfer occurs by chromosomal recombination during a Tn5381-induced cell fusion event. If some or all conjugative transposons are able to create mating events that allow chromosomal recombination, their ultimate impact on the dissemination of antimicrobial resistance determinants will extend far beyond the spread of resistance to tetracycline-minocycline.
CONCLUSION
Conjugative transposons are now widely prevalent in gram-positive bacteria and have made substantial inroads into certain species of gram-negative bacteria as well. Their role in the spread of resistance to tetracycline and minocycline and, to a lesser extent, erythromycin and kanamycin is well established and indisputable. Their ability to increase the level of expression of tetracycline resistance and their own transferability in response to environmental exposure to tetracycline has positioned them to thrive in human and animal gastrointestinal tracts, where significant quantities of tetracycline and its analogs may frequently be found. Moreover, the ability of these novel elements to establish genetic connections between widely disparate species makes them likely to be principal players in the ongoing dissemination of a wide variety of antimicrobial resistance determinants. A thorough understanding of the mechanisms by which these elements facilitate genetic exchange and the environmental signals that stimulate their activity will be an important first step in efforts to forestall the epidemic spread of resistance determinants in human pathogens.
ACKNOWLEDGMENTS
This research was supported by the Medical Research Service of the U.S. Department of Veterans Affairs.
I thank Don B. Clewell for helpful suggestions and informative discussions during the writing of the manuscript.
REFERENCES
- 1.Ayoubi P, Kilic A O, Vijayakumar M N. Tn5253, the pneumococcal Ω (cat tet) BM 6001 element, is a composite structure of two conjugative transposons, Tn5251 and Tn5252. J Bacteriol. 1991;173:1617–1622. doi: 10.1128/jb.173.5.1617-1622.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertram J, Stratz M, Durre P. Natural transfer of conjugative transposon Tn916 between gram-positive and gram-negative bacteria. J Bacteriol. 1991;173:443–448. doi: 10.1128/jb.173.2.443-448.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bingen E H, Desjardins P, Arlet G, Bourgeois F, Mariana-Kurkdjian P, Lambert-Zechovsky N Y, Denamur E, Philippon A, Elion J. Molecular epidemiology of plasmid spread among broad-spectrum β-lactamase-producing Klebsiella pneumoniae isolates in a pediatric hospital. J Clin Microbiol. 1993;31:179–184. doi: 10.1128/jcm.31.2.179-184.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonafede M E, Carias L L, Rice L B. Enterococcal transposon Tn5384: evolution of a composite transposon through cointegration of enterococcal and staphylococcal plasmids. Antimicrob Agents Chemother. 1997;41:1854–1858. doi: 10.1128/aac.41.9.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bringel F, Van Alstine G L, Scott J R. A host factor absent from Lactococcus lactis subspp. lactis MG1363 is required for conjugative transposition. Mol Microbiol. 1991;5:2983–2993. doi: 10.1111/j.1365-2958.1991.tb01858.x. [DOI] [PubMed] [Google Scholar]
- 6.Caillaud F, Courvalin P. Nucleotide sequence of the ends of conjugative shuttle transposon Tn1545. Mol Gen Genet. 1987;209:110–115. doi: 10.1007/BF00329844. [DOI] [PubMed] [Google Scholar]
- 7.Caparon M G, Scott J R. Excision and insertion of the conjugative transposon Tn916 involves a novel recombination mechanism. Cell. 1989;59:1027–1034. doi: 10.1016/0092-8674(89)90759-9. [DOI] [PubMed] [Google Scholar]
- 8.Celli J, Poyart C, Trieu-Cuot P. Use of an excision reporter plasmid to study the intracellular mobility of the conjugative transposon Tn916 in gram-positive bacteria. Microbiology. 1997;143:1253–1261. doi: 10.1099/00221287-143-4-1253. [DOI] [PubMed] [Google Scholar]
- 9.Christie P J, Korman R Z, Zahler S A, Adsit J C, Dunny G M. Two conjugation systems associated with plasmid pCF10: identification of a conjugative transposon that transfers between Streptococcus faecalis and Bacillus subtilis. J Bacteriol. 1987;169:2529–2536. doi: 10.1128/jb.169.6.2529-2536.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clermont D, Horaud T. Genetic and molecular studies of a composite chromosomal element (Tn3705) containing a Tn916-modified structure (Tn3704) in Streptococcus anginosus F22. Plasmid. 1994;31:40–48. doi: 10.1006/plas.1994.1005. [DOI] [PubMed] [Google Scholar]
- 11.Clewell D B. Movable genetic elements and antibiotic resistance in enterococci. Eur J Clin Microbiol Infect Dis. 1990;9:90–102. doi: 10.1007/BF01963632. [DOI] [PubMed] [Google Scholar]
- 12.Clewell D B, Flannagan S E, Ike Y, Jones J M, Gawron-Burke C. Sequence analysis of the termini of conjugative transposon Tn916. J Bacteriol. 1988;170:3046–3052. doi: 10.1128/jb.170.7.3046-3052.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clewell D B, Flannagan S E, Jaworski D D. Unconstrained bacterial promiscuity: the Tn916-Tn1545 family of conjugative transposons. Trends Microbiol. 1995;3:229–236. doi: 10.1016/s0966-842x(00)88930-1. [DOI] [PubMed] [Google Scholar]
- 14.Courvalin P, Carlier C. Tn1545: a conjugative shuttle transposon. Mol Gen Genet. 1987;206:259–264. doi: 10.1007/BF00333582. [DOI] [PubMed] [Google Scholar]
- 15.Doucet-Populaire F, Trieu-Cuot P, Dosbaa I, Andremont A, Courvalin P. Inducible transfer of conjugative transposon Tn1545 from Enterococcus faecalis to Listeria monocytogenes in the digestive tracts of gnotobiotic mice. Antimicrob Agents Chemother. 1991;35:185–187. doi: 10.1128/aac.35.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dowson C G, Coffey T J, Spratt B G. Origin and molecular epidemiology of penicillin-binding-protein-mediated resistance to β-lactam antibiotics. Trends Microbiol. 1994;2:361–366. doi: 10.1016/0966-842x(94)90612-2. [DOI] [PubMed] [Google Scholar]
- 17.Fitzgerald G F, Clewell D B. A conjugative transposon (Tn919) in Streptococcus sanguis. Infect Immun. 1985;47:415–420. doi: 10.1128/iai.47.2.415-420.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flannagan S E, Clewell D B. Conjugative transfer of Tn916 in Enterococcus faecalis: trans activation of homologous transposons. J Bacteriol. 1991;173:7136–7141. doi: 10.1128/jb.173.22.7136-7141.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flannagan S E, Zitzow L A, Su Y A, Clewell D B. Nucleotide sequence of the 18-kb conjugative transposon Tn916 from Enterococcus faecalis. Plasmid. 1994;32:350–354. doi: 10.1006/plas.1994.1077. [DOI] [PubMed] [Google Scholar]
- 20.Franke A, Clewell D B. Evidence for conjugal transfer of a Streptococcus faecalis transposon (Tn916) from a chromosomal site in the absence of plasmid DNA. Cold Spring Harbor Symp Quant Biol. 1980;45:77–80. doi: 10.1101/sqb.1981.045.01.014. [DOI] [PubMed] [Google Scholar]
- 21.Franke A E, Clewell D B. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of “conjugal” transfer in the absence of a conjugative plasmid. J Bacteriol. 1981;145:494–502. doi: 10.1128/jb.145.1.494-502.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hochhut B, Jahreis K, Lengeler J W, Schmid K. CTnscr94, a conjugative transposon found in Enterobacteria. J Biol Chem. 1997;179:2097–2102. doi: 10.1128/jb.179.7.2097-2102.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horaud T, Delbos F, de Cespédès G. Tn3702, a conjugative transposon in Enterococcus faecalis. FEMS Microbiol Lett. 1990;72:189–194. doi: 10.1016/0378-1097(90)90370-6. [DOI] [PubMed] [Google Scholar]
- 24.Ike Y, Flannagan S E, Clewell D B. Hyperhemolytic phenomena associated with insertions of Tn916 into the hemolysin determinant of Enterococcus faecalis plasmid pAD1. J Bacteriol. 1992;174:1801–1809. doi: 10.1128/jb.174.6.1801-1809.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inamine J M, Burdett V. Structural organization of a 67 kilobase streptococcal conjugative element mediating multiple antibiotic resistance. J Bacteriol. 1985;161:620–626. doi: 10.1128/jb.161.2.620-626.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaworski D D, Clewell D B. Evidence that coupling sequences play a frequency-determining role in conjugative transposition of Tn916 in Enterococcus faecalis. J Bacteriol. 1994;176:3328–3335. doi: 10.1128/jb.176.11.3328-3335.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaworski D D, Clewell D B. A functional origin of transfer (oriT) on the conjugative transposon Tn916. J Bacteriol. 1995;177:6644–6651. doi: 10.1128/jb.177.22.6644-6651.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaworski D D, Flannagan S E, Clewell D B. Analysis of traA, int-Tn, and xis-Tn mutations in the conjugative transposon Tn916 in Enterococcus faecalis. Plasmid. 1996;36:201–208. doi: 10.1006/plas.1996.0047. [DOI] [PubMed] [Google Scholar]
- 29.Jett B D, Huycke M M, Gilmore M S. Virulence of enterococci. Clin Microbiol Rev. 1994;7:462–478. doi: 10.1128/cmr.7.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kilic A O, Vijayakumar M N, Al-Khaldi S F. Identification and nucleotide sequence analysis of a transfer-related region in the streptococcal conjugative transposon Tn5252. J Bacteriol. 1994;176:5145–5150. doi: 10.1128/jb.176.16.5145-5150.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Bouguenec C, De Cespedes G, Horaud T. Molecular analysis of a composite chromosomal conjugative element (Tn3701) of Streptococcus pyogenes. J Bacteriol. 1988;170:3930–3936. doi: 10.1128/jb.170.9.3930-3936.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le Bouguenec C, De Cespedes G, Horaud T. Presence of chromosomal elements resembling the composite structure Tn3701 in streptococci. J Bacteriol. 1990;172:727–734. doi: 10.1128/jb.172.2.727-734.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lyon B R, Skurray R. Antimicrobial resistance in Staphylococcus aureus: genetic basis. Microbiol Rev. 1987;51:88–134. doi: 10.1128/mr.51.1.88-134.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manganelli R, Ricci S, Pozzi G. The joint of Tn916 circular intermediates is a homoduplex in Enterococcus faecalis. Plasmid. 1997;38:71–78. doi: 10.1006/plas.1997.1300. [DOI] [PubMed] [Google Scholar]
- 35.Manganelli R, Romano L, Ricci S, Zazzi M, Pozzi G. Dosage of Tn916 intermediates in Enterococcus faecalis. Plasmid. 1995;34:48–57. doi: 10.1006/plas.1995.1032. [DOI] [PubMed] [Google Scholar]
- 36.Mullany P, Wilks M, Lamb I, Clayton C, Wren B, Tabaqchali S. Genetic analysis of a tetracycline resistance element from Clostridium difficile and its conjugal transfer to and from Bacillus subtilis. J Gen Microbiol. 1990;136:1343–1349. doi: 10.1099/00221287-136-7-1343. [DOI] [PubMed] [Google Scholar]
- 37.Murray B E, An F Y, Clewell D B. Plasmids and pheromone response of the β-lactamase-producer Streptococcus (Enterococcus) faecalis HH22. Antimicrob Agents Chemother. 1986;32:547–551. doi: 10.1128/aac.32.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naglich J G, Andrews R., Jr Tn916-dependent conjugal transfer of pC194 and pUB110 from Bacillus subtilis into Bacillus thuringensis subsp. israelensis. Plasmid. 1988;20:113–126. doi: 10.1016/0147-619x(88)90014-5. [DOI] [PubMed] [Google Scholar]
- 39.Norgren M, Scott J R. The presence of conjugative transposon Tn916 in the recipient strain does not impede transfer of a second copy of the element. J Bacteriol. 1991;173:319–324. doi: 10.1128/jb.173.1.319-324.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oggioni M R, Dowson C G, Smith J M, Provvedi R, Pozzi G. The tetracycline resistance gene tet(M) exhibits mosaic structure. Plasmid. 1996;35:156–163. doi: 10.1006/plas.1996.0018. [DOI] [PubMed] [Google Scholar]
- 41.Poyart C, Celli J, Trieu-Cuot P. Conjugative transposition of Tn916-related elements from Enterococcus faecalis to Escherichia coli and Pseudomonas fluorescens. Antimicrob Agents Chemother. 1995;39:500–506. doi: 10.1128/aac.39.2.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poyart-Salmeron C, Trieu-Cuot P, Carlier C, Courvalin P. The integration-excision system of the conjugative transposon Tn1545 is structurally and functionally related to those of lambdoid phages. Mol Microbiol. 1990;4:1513–1521. doi: 10.1111/j.1365-2958.1990.tb02062.x. [DOI] [PubMed] [Google Scholar]
- 43.Provvedi R, Manganelli R, Pozzi G. Characterization of conjugative transposon Tn5251 of Streptococcus pneumoniae. FEMS Microbiol Lett. 1996;135:231–236. doi: 10.1111/j.1574-6968.1996.tb07994.x. [DOI] [PubMed] [Google Scholar]
- 44.Rauch P J G, de Vos W M. Identification and characterization of genes involved in excision of the Lactococcus lactis conjugative transposon Tn5276. J Bacteriol. 1994;176:2165–2171. doi: 10.1128/jb.176.8.2165-2171.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rauch P J G, Beerthuyzen M M, de Vos W M. Distribution and evolution of nisin-sucrose elements in Lactococcus lactis. Appl Environ Microbiol. 1994;60:1798–1804. doi: 10.1128/aem.60.6.1798-1804.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rice L B, Carias L L. Studies on excision of conjugative transposons in enterococci: evidence for joint sequences composed of strands with unequal numbers of nucleotides. Plasmid. 1994;31:312–316. doi: 10.1006/plas.1994.1034. [DOI] [PubMed] [Google Scholar]
- 47.Rice L B, Carias L L. Transfer of Tn5385, a composite, multiresistance element from Enterococcus faecalis. J Bacteriol. 1998;180:714–721. doi: 10.1128/jb.180.3.714-721.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rice L B, Eliopoulos G M, Wennersten C, Goldmann D, Jacoby G A, Moellering R C., Jr Chromosomally mediated β-lactamase production and gentamicin resistance in Enterococcus faecalis. Antimicrob Agents Chemother. 1991;35:272–276. doi: 10.1128/aac.35.2.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rice L B, Marshall S H, Carias L L. Tn5381, a conjugative transposon identifiable as a circular form in Enterococcus faecalis. J Bacteriol. 1992;174:7308–7315. doi: 10.1128/jb.174.22.7308-7315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roberts M. Epidemiology of tetracycline resistance determinants. Trends Microbiol. 1994;2:353–357. doi: 10.1016/0966-842x(94)90610-6. [DOI] [PubMed] [Google Scholar]
- 51.Rudy C, Taylor K L, Hinerfeld D, Scott J R, Churchward G. Excision of a conjugative transposon in vitro by the Int and Xis proteins of Tn916. Nucleic Acids Res. 1997;25:4061–4066. doi: 10.1093/nar/25.20.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rudy C K, Scott J R, Churchward G. DNA binding by the Xis protein of the conjugative transposon Tn916. J Bacteriol. 1997;179:2567–2572. doi: 10.1128/jb.179.8.2567-2572.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salyers A A, Amabile-Cuevas C F. Why are antibiotic resistance genes so resistant to elimination? Antimicrob Agents Chemother. 1997;41:2321–2325. doi: 10.1128/aac.41.11.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salyers, A. A., and N. B. Shoemaker. 1996. Resistance gene transfer in anaerobes: new insights, new problems. Clin. Infect. Dis. 23(Suppl 1):S36–S43. [DOI] [PubMed]
- 55.Salyers A A, Shoemaker N B, Stevens A M, Li L-H. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol Rev. 1995;59:579–590. doi: 10.1128/mr.59.4.579-590.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scott J R, Bringel F, Marra D, Van Alstine G, Rudy C K. Conjugative transposition of Tn916: preferred targets and evidence for conjugative transfer of a single strand and for a double stranded circular intermediate. Mol Microbiol. 1994;11:1099–1108. doi: 10.1111/j.1365-2958.1994.tb00386.x. [DOI] [PubMed] [Google Scholar]
- 57.Scott J R, Churchward G G. Conjugative transposition. Annu Rev Microbiol. 1995;49:367–397. doi: 10.1146/annurev.mi.49.100195.002055. [DOI] [PubMed] [Google Scholar]
- 58.Scott J R, Kirchman P A, Caparon M G. An intermediate in the transposition of the conjugative transposon Tn916. Proc Natl Acad Sci USA. 1988;85:4809–4813. doi: 10.1073/pnas.85.13.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Showsh S A, Andrews R E., Jr Functional comparison of conjugative transposons Tn916 and Tn925. Plasmid. 1996;35:164–173. doi: 10.1006/plas.1996.0019. [DOI] [PubMed] [Google Scholar]
- 60.Spratt B G, Zhang Q-Y, Jones D M, Hutchison A, Brannigan J A, Dowson C G. Recruitment of a penicillin-binding protein gene from Neisseria flavescens during the emergence of penicillin resistance in Neisseria meningitidis. Proc Natl Acad Sci USA. 1989;86:8988–8992. doi: 10.1073/pnas.86.22.8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Su Y A, He P, Clewell D B. Characterization of the tet(M) determinant of Tn916: evidence for regulation by transcriptional attenuation. Antimicrob Agents Chemother. 1992;36:769–778. doi: 10.1128/aac.36.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Swartley J S, McAllister C F, Hajjeh R A, Heinrich D W, Stephens D S. Deletions of Tn916-like transposons are implicated in tetM-mediated resistance in pathogenic Neisseria. Mol Microbiol. 1993;10:299–310. doi: 10.1111/j.1365-2958.1993.tb01956.x. [DOI] [PubMed] [Google Scholar]
- 63.Taylor K L, Churchward G. Specific DNA cleavage mediated by the integrase of conjugative transposon Tn916. J Bacteriol. 1997;179:1117–1125. doi: 10.1128/jb.179.4.1117-1125.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Torres O R, Korman R Z, Zahler S A, Dunny G M. The conjugative transposon Tn925: enhancement of conjugal transfer by tetracycline in Enterococcus faecalis and mobilization of chromosomal genes in both Bacillus subtilis and E. faecalis. Mol Gen Genet. 1991;225:395–400. doi: 10.1007/BF00261679. [DOI] [PubMed] [Google Scholar]
- 65.Trieu-Cuot P, Poyart-Salmeron C, Carlier C, Courvalin P. Molecular dissection of the transposition mechanism of conjugative transposons from Gram-positive cocci. In: Dunny G M, Patrick P, Cleary L L, editors. Genetics and molecular biology of streptococci, lactococci, and enterococci. Washington, D.C: American Society for Microbiology; 1991. pp. 21–27. [Google Scholar]
- 66.Trieu-Cuot P, Poyart-Salmeron C, Carlier C, Courvalin P. Sequence requirements for target activity in site-specific recombination mediated by the Int protein of Tn1545. Mol Microbiol. 1993;8:179–185. doi: 10.1111/j.1365-2958.1993.tb01214.x. [DOI] [PubMed] [Google Scholar]
- 67.Waldor M K, Tschape H T, Mekalanos J J. A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J Bacteriol. 1996;178:4157–4165. doi: 10.1128/jb.178.14.4157-4165.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]