Abstract
Mutations within viral epitopes can result in escape from T cells, but the contribution of mutations in flanking regions of epitopes in SARS-CoV-2 has not been investigated. Focusing on two SARS-CoV-2 nucleoprotein CD8+ epitopes, we investigated the contribution of these flanking mutations to proteasomal processing and T cell activation. We found decreased NP9-17-B*27:05 CD8+ T cell responses to the NP-Q7K mutation, likely due to a lack of efficient epitope production by the proteasome, suggesting immune escape caused by this mutation. In contrast, NP-P6L and NP-D103 N/Y mutations flanking the NP9-17-B*27:05 and NP105-113-B*07:02 epitopes, respectively, increased CD8+ T cell responses associated with enhanced epitope production by the proteasome. Our results provide evidence that SARS-CoV-2 mutations outside the epitope could have a significant impact on proteasomal processing, either contributing to T cell escape or enhancement that may be exploited for future vaccine design.
1. Introduction
Defining CD8+ T cell responses to SARS-CoV-2 has been a priority since the virus first emerged in 2019 as these responses are instrumental in clearing viral infections [1]. Multiple immunodominant CD8+ T cell epitopes have been identified [[2], [3], [4], [5], [6], [7]]. Generation of CD8 epitopes initially requires antigen processing through the proteasome, a multi-subunit cytosolic machinery that can degrade intracellular proteins following ubiquitination. The proteasome is composed of a catalytic core particle (20s) and one or two terminal 19s regulatory particles. The 20s catalytic core is capable of cleaving at the C-terminal side of acidic, basic and hydrophobic amino acid residues [[8], [9], [10]]. As part of this proteolytic process, a small fraction of peptide fragments will contain correctly cleaved C-termini of CD8 epitopes, whereas the majority contain extended N-termini that require further trimming. This can occur in the cytoplasm by various peptidases or, more commonly, in the endoplasmic reticulum (ER) following translocation of peptides from the cytosol via the transporter associated with antigen processing complex TAP1/TAP2 [11,12].
An essential peptidase responsible for trimming peptides in the ER is ERAP1; an aminopeptidase optimised to generate peptides of 8–10 amino acids for binding to MHC Class I molecules [13]. ERAP1 activity varies depending on the N-terminal amino acid with little ability to cleave after proline residues and high ability to cleave after hydrophobic amino acids [14]. Another related aminopeptidase, ERAP2, can also help to shape the MHC Class I peptide repertoire, but has a slightly different trimming capacity preferring positively charged amino acids [15].
Once cleaved to optimal length, peptides will bind to MHC Class I molecules and those with sequences that bind strongly into the peptide-binding groove, so-called high affinity peptides, will induce transport of peptide-MHC complexes to the cell surface for activation of CD8+ T cells [16]. Thus, the sequence of the peptides is important for ensuring strong binding to MHC, and mutations within these sequences can alter the affinity of this interaction. Using exogenous peptide assays, the effect of mutations within epitope sequences have been previously shown to lead to escape from viral T cell responses in various settings [[17], [18], [19]] including SARS-CoV-2 [20].
In addition, components of the antigen processing pathway have preferences for amino acid sequences for their optimal processing as discussed above. Indeed, previous studies [21] looking into the effect of viral mutations in the flanking regions of epitopes on antigen processing of HIV-1 found evidence of disruption of proteasomal cleavage [22] and endoplasmic reticulum aminopeptidase 1 (ERAP1) trimming within the ER [23].
Traditionally, single-stranded RNA viruses have a high propensity for mutation due to a lack of proof-reading ability by the RNA-dependent RNA polymerases that replicate their genomes [24]. However, coronaviruses have adapted proofreading capabilities that reduce their mutation rate [25]. Although several variants of concern have still emerged since the start of the pandemic. Currently, the effects of SARS-CoV-2 epitope-flanking mutations on viral epitope antigen processing are unknown. Our aim of this proof-of-concept study was to identify whether mutations are occurring in these regions with possible effects on CD8+ T cell activation.
2. Results
2.1. Identification of mutations in flanking regions of SARS-CoV-2 nucleoprotein CD8 epitopes
Focusing on two immunodominant SARS-CoV-2 nucleoprotein (NP) epitopes previously identified, NP9-17 and NP105-113 [2,26], we searched sequences derived from the GISAID database (www.gisaid.org; July 6, 2020 (EPI_SET_230323 zm) for mutations within six amino acids either side of these epitopes through sequence alignment with the originally identified Wuhan sequence (hCoV-19/Wuhan/WIV04/2019 (WIV04) (EPI_ISL_402,124)), which yielded 29 mutations (Fig. 1A). Mutations found in five or fewer sequences (<10%) were excluded from further investigation to eliminate mutations that were due to sequencing artefacts, resulting in 19 mutations. We subjected these sequences through the IEDB NetChop 20S 3.0 prediction tool for proteasomal cleavage [27] and compared cleavage against wild-type (WT) NP sequence derived from the viral strain originally isolated in Wuhan, China (Fig. 1B–C). The mutations that showed the greatest predicted differences in proteasomal cleavage as compared to WT NP sequence for each epitope were selected for further investigation (n = 7) (Fig. 1D). While the frequency of these mutations was extremely low at the time of analysis, they have been steadily rising since July 2020 (Fig. 1E–F). The frequency of each mutation compared to the vast number of sequences now available shows that these mutations are still relatively rare but 4 out of 7 mutations have increased in frequency across the 2 years studied (Supplementary Table 1). Up until March 2022, they have not appeared in any variants of interest or concern identified by the WHO.
Fig. 1.
Identification of mutations in SARS-CoV-2 CD8+ T cell nucleoprotein epitope flanking regions. (A) 53,996 SARS-CoV-2 sequences were downloaded from the GISAID database on July 6, 2020. Mutations in the flanking regions (±6 amino acids) were assessed either side of identified CD8 nucleoprotein epitopes (NP9-17/NP105-113) and analysed the potential effect these mutations may have on antigen processing by the proteasome. (B) Heatmap results from NetChop 20s prediction tool for the effect of mutations on proteasomal cleavage of NP9-17 region. Red = increase in cleavage, blue = decrease. (C) Heatmap results from NetChop 20s prediction tool for the effect of mutations on proteasomal cleavage of NP105-113 region. Red = increase in cleavage, blue = decrease. (D) Mutations taken forward for further investigation. (E) Frequency of mutations for NP4-22 region expressed as cumulative number of sequences with each mutation since January 2020. (F) Frequency of mutations for NP100-119 region expressed as cumulative number of sequences with each mutation since January 2020. Data downloaded from COVID-19 Viral Genome Analysis Pipeline.
2.2. Unaltered HLA expression in SARS-CoV-2 NP-transduced B cells
Having identified mutations of interest, we next created stable B cell lines expressing SARS-CoV-2 WT or mutated NP through lentiviral transduction. B cells from patient C–COV19-005 isolated and immortalised as previously described in Ref. [2] were chosen for initial experiments as they expressed both HLA-B*27:05 and HLA-B*07:02, the correct MHC class I restriction for NP9-17 and NP105-113, respectively (full HLA typing for donor C–COV19-005 is available in the methods section). Each NP construct also contained a GFP gene separated by a viral self-cleavage T2A site, resulting in one long transcript but two translated proteins, GFP and NP. NP-transduced cells showed no significant differences in GFP, SARS-CoV-2 NP or HLA expression (Supplementary Fig. 1).
2.3. Flanking mutations alter CD8+ T cell responses
Our NP-transduced B cell system allowed us to evaluate the effect of one amino acid mutation on T cell responses to NP. We recognise that this is not a complete surrogate for viral infection as NP protein expression in transduced cells may be greater than that of a SARS-CoV-2 infection. However, NP is the most abundant viral protein at all stages of the infection cycle, especially in the initial stages [28,29], and a recent study showed that comparable levels of NP could be detected following virus infection as compared to transfection of HEK293T cells [30].
Using NP-transduced cell lines as target cells, we co-cultured with bulk CD8+ T cells-specific for NP9-17 or NP105-113 (Supplementary Fig. 2) for 4 h prior to analysis of CD107a, IFNγ and TNFα expression (Fig. 2A). To show the specificity of our bulk T cell lines, we show a peptide-pulsed control for each T cell line. We included the mutation NP-P13L as a control in our experiments as previous work had shown this to be a CD8+ T cell escape mutant within the epitope NP9-17 [20]. This mutation has subsequently been found within the Omicron variant of SARS-CoV-2. As expected, the NP-P13L mutation was shown to lead to complete escape from all NP9-17 CD8+ T cell responses measured when compared to WT NP (Fig. 2B).
Fig. 2.
Mutations in flanking regions of epitopes alter CD8+ T Cell responses. (A) Intracellular cytokine staining of NP9-17 or NP105-113 epitope-specific bulk CD8+ T cells following incubation with GFP- or NP-transduced B cells. NP9-17 or NP105-113 peptide-pulsed B cells are also shown. (B) CD8+ T Cell responses to mutations flanking NP9-17 were analysed by flow cytometry at an E:T ratio of 2:1. (C) CD8+ T Cell responses to mutations flanking NP105-113 were analysed by flow cytometry at an E:T ratio of 2:1. (D–E) Selected mutations were investigated in more detail by altering the E:T ratio from 2:1 to 0.1:1. Data is presented as mean ± SEM and was analysed by one-way ANOVA and Dunnett's multiple comparisons test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Interestingly, we also saw significant differences in T cell responses to several flanking mutations (Fig. 2B–C). NP-Q7K significantly decreased IFNγ+ NP9-17-specific CD8+ T cells (p = 0.0448). In contrast, NP-P6L was shown to significantly increase the proportion of IFNγ+ NP9-17-specific T cells (p = 0.0107), while NP-D103Y significantly increased TNFα+ NP105-113-specific T cells (p = 0.0424).
Addressing the mutations that showed significant differences, in addition to NP-D103 N as it affects the same amino acid position as NP-D103Y, we investigated how these mutations modulate CD8+ T cell responses in more detail. The previous experiment was performed at an effector (CD8+ T Cell) to target (NP-transduced B cell) ratio (E:T) of 2:1. We next used a range of E:T from 2:1 to 0.1:1 to expose differences in CD8+ T cell responses (Fig. 2D–E). For the mutations flanking NP9-17, there was clear separation between the results for wild type (WT) vs mutant, with NP-P6L always showing higher and NP-Q7K showing lower T cell responses. For NP105-113, the differences were more subtle, but there is separation between WT and mutant proteins. Taken together, these results clearly demonstrate that flanking mutations, particularly N-terminal mutants, can significantly alter CD8+ T cell responses, presumably by altering the antigen load of the epitopes on the cell surface.
2.4. Flanking mutations alter epitope generation by the proteasome
CD8+ T cell epitopes are generated through degradation of proteins by the proteasome, machinery that selectively cleaves after certain amino acid residues. Therefore, we investigated whether our flanking mutations alter proteasomal digestion of NP epitopes. To do this, we digested long peptide precursors (NP1-22 and NP99-126), containing the epitopes NP9-17 and NP105-113 and either WT or mutated flanking regions with constitutive (20S) or immunoproteasome (i20S) for up to 3 h in vitro, followed by analysis of the digestion products via quantitative mass spectrometry (MS).
The immunoproteasome is constitutively expressed in haematopoietic cells or induced in response to IFNγ or TNFα and can produce antigenic peptides at a faster rate than the constitutive proteasome [31,32]. As a haematopoietic cell line, we expected that our B cell lines would express a combination of 20s and i20s proteasomes, which was confirmed through detection of constitutive proteasome (PSMB6) and immunoproteasome (LMP2, LMP7) subunits (Supplementary Fig. 3).
The products of NP1-22 with a 3-h immunoproteasome digestion showed a distinct pattern of products for NP-WT, NP-P6L and NP-Q7K (Fig. 3A). Strikingly, for both NP9-17 and NP105-113 epitopes, proteasomal cleavage alone was sufficient to produce an abundance of correctly C-terminally cleaved peptides (Fig. 3). Additionally, while N-terminally extended versions were also present, the complete nine amino acid (9mer) epitopes were identified from proteasomal cleavage alone. This is unusual [33,34] and warrants further investigation to confirm whether these 9mer peptides can be transported into the ER through TAP for direct loading on HLA Class I molecules, or whether these peptides are not immunogenic at all. In either case, the effect of mutations on proteasomal cleavage is clearly demonstrated.
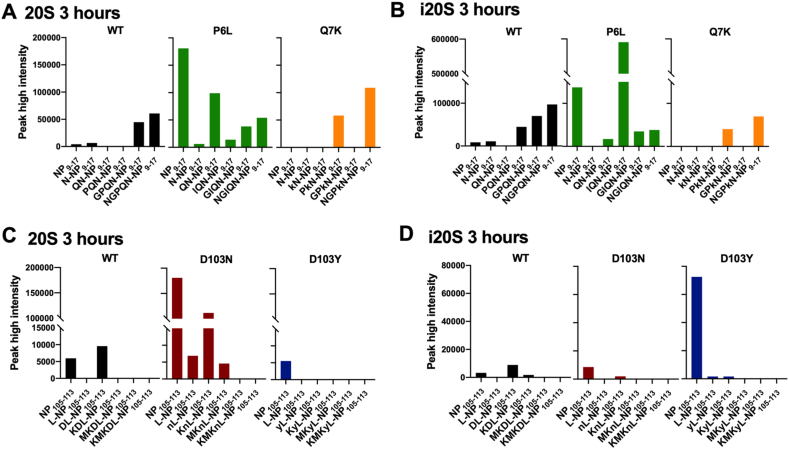
Fig. 3.
Mutations in flanking regions affect proteasomal digestion and epitope generation of SARS-CoV-2 CD8+ T cell epitopes. (A) PEAKS data showing the products of NP1-22 peptide digested with immunoproteasome for 3 h. Correctly C-terminally cleaved peptides are labelled for 9-14mer peptides. (B) Chromatograms showing generation of NP9-17 epitope after 3-h incubation of NP1-22 peptide with immunoproteasome provide peak high intensity values. (C) Chromatograms showing generation of NP9-17 epitope after 3-h incubation of NP1-22 peptide with constitutive proteasome provide peak high intensity values. (D) PEAKS data showing the products of NP99-126 peptide digested with immunoproteasome for 3 h. Correctly C-terminally cleaved peptides are labelled for 9-14mer peptides. (E) Chromatograms showing generation of NP105-113 epitope after 3-h incubation of NP99-126 peptide with immunoproteasome provide peak high intensity values. The additional peaks correspond to NP103-110 and NP112-124. (F) Chromatograms showing generation of NP105-113 epitope after 3-h incubation of NP99-126 peptide with constitutive proteasome provide peak high intensity values. The additional peaks correspond to NP103-110 and NP112-124. (G) Time-course of NP9-17 epitope generation from NP1-22 long peptide with variants digested with immuno- or constitutive proteasome. (H) Time-course of NP105-113 epitope generation from NP99-126 long peptide with variants digested with immuno- or constitutive proteasome.
Using the peak-high intensity values from extracted ion chromatograms as shown in Fig. 3B–C, we were able to get a relative quantitative value for each peptide over a time-course of 3 h. Across this 3-h time-course, we observed higher NP9-17 epitope generation with NP-P6L as compared to WT sequences, which was consistent with both immunoproteasome and constitutive proteasome digestions (Fig. 3G). We were unable to detect NP9-17 epitope from NP-Q7K mutated protein sequences with either proteasome (Fig. 3A–C, 3G). We identified proteasomal differences in the response to mutated proteins in the NP99-126 peptide, with NP-D103 N increasing NP105-113 epitope generation by the constitutive proteasome but having an insignificant effect with the immunoproteasome. In contrast, NP-D103Y enhanced NP105-113 epitope generation by immunoproteasome digestion (Fig. 3D–F, 3H).
2.5. Flanking mutations alter N-terminally extended proteasomal products
In addition to differences in 9mer epitope generation by the proteasome, we found that mutations also affected generation of N-terminally extended epitope precursors (Fig. 4). This appears to be particularly prevalent for the mutations NP-P6L (Fig. 4A–B) and NP-D103 N (Fig. 4C). We also observed differences dependent on proteasome species (Fig. 4C–D). N-terminally extended precursors are most commonly trimmed by endoplasmic reticulum aminopeptidase 1 (ERAP1) in the ER to the optimal length for HLA Class I binding [35], confirmed to be nine amino acids for NP9-17 and NP105-113 [2]. The requirement for further processing of epitopes can alter the efficiency of antigen processing and presentation potentially affecting antigen load at the cell surface.
Fig. 4.
Flanking mutations alter N-terminally extended proteasomal products. (A) The generation of 9–14mers (9mer = NP9-17) from NP1-22 peptide following digestion with constitutive proteasome for 3 h. (B) The generation of 9–14mers (9mer = NP9-17) from NP1-22 peptide following digestion with immunoproteasome for 3 h. (C) The generation of 9–14mers (9mer = NP105-113) from NP99-126 peptide following digestion with constitutive proteasome for 3 h. (D) The generation of 9–14mers (9mer = NP105-113) from NP99-126 peptide following digestion with immunoproteasome for 3 h.
3. Discussion
For SARS-CoV-2 infection, it has been demonstrated that mutations within epitope sequences can lead to escape from T cell recognition [20], presumably by disruption of HLA-TCR engagement. The effect of mutations within the flanking regions of SARS-CoV-2 epitopes was recently shown by Motozono et al., where they identified a flanking mutation, G446S, in the Omicron BA.1 variant spike protein which increased CD8+ T Cell responses [36]. Here, we show that mutations within the nucleoprotein also influence CD8+ T cell responses due to differences in the proteasomal processing of viral antigens. Most importantly, we show that mutations such as NP-Q7K are capable of decreasing T cell responses and potentially leading to escape through poor proteasomal processing of the epitope while other mutations are capable of increasing T cell responses. This work highlights the need to study T cell responses in a model system that allows for natural processing and presentation of epitopes.
We established a system which enabled us to evaluate T cell responses to naturally processed epitope peptides and to natural mutations that impact the delivery of epitope antigen to the cell surface even when the epitope sequence is conserved. We, and others [37,38], believe that continual monitoring of potential escape from immunodominant T cell responses in circulating variants before they become dominant strains is critically important for COVID-19 surveillance and future vaccine strategies. Our antigen processing assay was applicable to SARS-CoV-2 encoded NP, but we faced challenges when examining processing of epitopes derived from the Spike protein with very low transduction efficiency despite several optimisation steps. Spike is membrane associated and may be processed via the endocytic-lysosomal pathway, therefore requiring cross-presentation for CD8+ antigen recognition [39,40]. A different experimental approach is necessary to assess the effect of Spike mutations on T cell responses.
While it was encouraging to note that these mutations directly affected generation of the 9mer epitope peptides by the proteasome, in agreement with our T cell data, we accept that we have not established whether the 9mer peptide generated by the proteasome is the most antigenic version. Further studies will be required to determine the transport efficiency of the 9mer or extended peptide antigen variants into the endoplasmic reticulum (ER) via TAP, followed by trimming by ERAP1/2 and loading onto HLA Class I molecules. However, the correlation between 9mer generation by the proteasome and T cell responses are remarkable. For NP-P6L, the proportion of 12mer peptide generated is increased considerably. The mutation of the Proline residue to Leucine may make this 12mer peptide a better substrate for ERAP1/2 trimming, thus increasing the epitope peptide pool, especially as ERAP1/2 do not like to cleave after Proline residues. Potentially, the change in proteasomal cleavage due to the mutation is increasing the pool of peptides suitable for ERAP trimming and thus further increasing the generation of epitope peptides.
We showed that both NP-P6L and NP-D103Y could increase CD8+ T cell responses due to increased efficiency of proteasomal digestion suggesting that some variants identified in the NP sequence could be beneficial for viral fitness, representing an advantage over increasing CD8+ T cell responses. We believe continued identification of such “beneficial” natural mutations within immunodominant T cell epitope flanking regions could provide a new avenue of research for vaccine design, especially as the inclusion of NP into future vaccine designs against SARS-CoV-2 shows promise [[41], [42], [43]]. The fact that our experimental results, particularly for P6L and Q7K, reflected the predicted in silico results suggests that analysis similar to that conducted here could be used on a much larger scale with the most up-to-date sequence information to identify and determine other flanking residue mutations that may also increase T cell responses. Accumulation of these mutations into the next vaccine design could increase T cell immunity and might provide greater protection against the virus.
The number of mutations investigated in this study reflects a small subset due to the early timepoint of sequence sampling, but three out of the twenty-nine mutations (∼10%) originally identified showed significant differences in CD8+ T cell responses. Moreover, four mutations (∼14%) investigated for their effect on proteasomal digestion showed differences in the amount of 9mer epitope peptide generated. This suggests that there are many mutations that may be naturally occurring in the flanking regions of epitopes that have the potential to alter T cell responses. Future studies into the effect of flanking mutations will hopefully create a bank of mutations for multiple HLA Class I allotypes that could be used to investigate whether these mutations have a collective effect on an individual's or population's response to the virus in terms of disease severity or long COVID.
Our results emphasise that the antiviral efficacy and cross-reactive potential of CD8+ T cells should be assessed in response to virus-infected cells, or minimally within systems that allow natural processing of epitopes. Tracking and characterisation of epitope-flanking regions in a system where natural processing of viral antigens occurs will allow us to make more precise judgements of the risk of future viral strains. We also provide a framework for future, larger-scale investigations into the potential effect of flanking residue mutations on T cell responses that could help to create a more robust vaccine in the future.
4. Star methods
4.1. Resource availability
Lead Contact: Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contacts, Tao Dong (tao.dong@ndm.ox.ac.uk) or Dannielle Wellington (dannielle.wellington@rdm.ox.ac.uk).
Data and code availability: The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [44] partner repository with the dataset identifier PXD032054. Any additional information required to re-analyse the data reported in this paper is available from the lead contact upon request.
4.2. Method details
4.2.1. Identification of mutations of interest proximal to SARS-CoV-2 CD8 nucleoprotein epitopes
On July 6, 2020, all available sequences for SARS-CoV-2 that differed from the original Wuhan strain were downloaded from the GISAID database (www.gisaid.org). The regions NP4-22 and NP100-119 were analysed for mutations. Those falling within the epitope sequence region or found in less than 5 sequences were discounted for further analysis.
4.2.2. Prediction tool analysis of proteasomal digestion
Using the NetChop 20S 3.0 prediction tool (www.iedb.org) [27], we evaluated the predicted proteasomal cleavage sites for each peptide region in both WT and mutated sequences (NP4-22: Supplementary Table 2, NP100-119: Supplementary Table 3). Data was presented as fold-change compared to the predicted proteasomal cleavage in WT sequences (Fig. 1B–C).
4.3. Tracking of mutation frequencies
The frequency of mutations over time was assessed from data downloaded from the COVID-19 Viral Genome Analysis Pipeline (https://cov.lanl.gov/content/index) tracking mutations tool [45]. Data was accessed on August 1, 2022.
4.3.1. Lentivirus construction & production
SARS-CoV-2 nucleoprotein (NP) was cloned from pHRSIN (kind gift from Prof. Alain Townsend) into the Addgene 17,448 lentivirus vector by InFusion PCR (TaKaRa) and verified by Sanger sequencing. NP expression was coupled to eGFP expression using a T2A linker. Single nucleotide mutations were introduced using site-directed mutagenesis (SDM) (New England BioLabs, NEB Base Changer kit). Once confirmed, mutated gene cassettes were extracted by restriction digest and cloned into a clean vector to prevent any off-target effects of SDM-PCR.
Lentivirus was produced using HEK293T cells, expression constructs were co-transfected with psPAX2 and pMD2. G using PEIPro. Lentiviral supernatants were filtered using a 0.45 μm CA filter and overlayed on 20% sucrose before ultracentrifugation at 90,000g for 120 min at 4 °C. Pelleted virus was resuspended in PBS, aliquoted and stored at −80 °C.
4.3.2. Lentivirus transduction
Cells from donor C–COV19-005 were used for investigation as they contained the HLA for both NP9-17-HLA-B*27:05 and NP105-113-HLA-B*07:02. Full HLA Class I type for this donor is: A*01:01, A*02:01, B*07:02, B*27:05, C*01:02 and C*07:02. PBMCs were pulsed with neat Epstein-Barr virus (EBV) for 3 h. Cyclosporine A (CsA) was then added to reach a final concentration of 1 μg/ml. Cells were kept in 1 μg/ml CsA (diluted in RPMI 1640 medium with 10% foetal calf serum (FCS), 2 mM glutamine and 100 mg/ml pen/strep) for 14–28 days until B cell colonies appear. After that, CsA was removed and B cells were cultured in RPMI 1640 medium with 10% foetal calf serum (FCS), 2 mM glutamine and 100 mg/ml pen/strep.
EBV-transformed B Cells were then spin inoculated with lentivirus to generate stable cell lines. In brief, B cells were plated at 5 × 105 cells per well of a 96-well U-bottom plate and supplemented with 5 × 106 TU/ml lentivirus and 8 mg/ml polybrene in RPMI +1% FBS. The plate was spun at 500g, 37 °C for 1 h prior to overnight incubation at 37 °C. After 24 h, B cells were washed and cultured in RPMI +10% FBS until enough cells were obtained for cell sorting. Cells were then bulk sorted for GFP expression on a BD FACSAria III Cell sorter.
4.4. Generation of short-term SARS-CoV-2 specific T cell lines
Short-term SARS-CoV-2-specific T cell lines were established as previously described [2,26]. Briefly, 3 × 106 to 5 × 106 PBMCs were pulsed as a pellet for 1 h at 37 °C with 10 μM of peptides containing T cell epitope regions and cultured in R10 (RPMI 1640 medium with 10% foetal calf serum (FCS), 2 mM glutamine and 100 mg/ml pen/strep) at 2 × 106 cells per well in a 24-well Costar plate. IL-2 was added to a final concentration of 100U/mL on day 3 and cultured for a further 10–14 days. NP9-17-specific Bulk T cells and NP105-113 specific Bulk T cells were generated by sorting HLA-B*27:05 NP9-17 and HLA-B*07:02 NP105-113 Pentamer + CD8+ T cells (Proimmune, UK) from short-term cell lines.
The purity of the sorted population was confirmed at 95% purity. The isolated cells were then clonally expanded and cultured in vitro. The sorted cells were cultured with 2 million irradiated cone blood cells (Oxford NHS Blood and Transplant Service) for 14 days in H10 medium (RPMI-1640 supplemented with 10% v/v heat-inactivated human AB serum (National Blood Service), 2 mmol/L l-glutamine, 1% v/v (500 U/mL) penicillin/streptomycin (Sigma-Aldrich)), supplemented with recombinant human IL2 (200 U/mL; PeproTech). The T cells were confirmed for antigen specificity using HLA-B*27:05 NP9-17 and HLA-B*07:02 NP105-113 Pentamer staining, ran on Attune Nxt flow cytometry (Thermo Fisher) and analysed using FlowJo v.10 (TreeStar Inc.). The validated T cells were then stored in several batches for future assays and used for all in vitro functional assays. T-cell cultures were tested for Mycoplasma monthly, maintained for 2 months for every assay, passaged once after every thaw, and reauthenticated again using the tetramers method after every thawing and passage [46].
4.4.1. Cell culture of transduced B cell lines & T cells
B cell lines were cultured in RPMI (Lonza) + 10% foetal calf serum (Sigma) + 2 mM l-glutamine (Sigma) at 37 °C, 5% CO2. Bulk T cells were cultured in RPMI (Gibco) + 10% human serum (Sigma) + 2 mM l-glutamine (Sigma) + Penicillin-Streptomycin (Sigma, 100 units penicillin and 100 μg streptomycin per mL), supplemented with 100 U/ml IL-2 (Sigma).
4.5. Flow cytometry analysis of transduced B cells
Transduced and sorted B cell lines were stained with Zombie-Violet live dead stain (Thermofisher) and PE-anti-HLA-ABC (BioLegend) and run on an Attune NXT Flow cytometer (Thermofisher). Data was analysed using FlowJo v10 (BD Biosciences).
4.6. Western blot analysis of transduced B cells
Transduced B cell lines were homogenized using RIPA lysis buffer (Thermo Fisher Scientific). Anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibody clone 6C5 (Merck Millipore, MAB374) was used as control antibody; anti-SARS-CoV-2 nucleocapsid antibody (2 μg/μl, Sino Biological, 40,143-R001) was used to probe NP expression. Anti-PSBM6 (AbCam), anti-LMP2 (AbCam) and anti-LMP7 (AbCam) were used individually at 1 in 1000 dilution to assess proteasomal subunit expression in the transduced B cells.
Primary antibodies were probed by IRDye 680LT goat anti-mouse (Li-Cor, 926–68020) and IRDye 800LT goat anti-rabbit (Li-Cor, 925–32210), and visualized using the iBright FL1000 (Invitrogen). Images from Western blot experiments were analysed by Fiji software for band density and expressed in GraphPad Prism as a percentage of the GAPDH expression. A one-way ANOVA was performed along with Tukey's or Sidak's multiple comparisons tests to measure statistical differences between cell lines.
4.7. Intracellular cytokine staining of T cells
Bulk CD8+ T cells specific for NP9-17 or NP105-113 epitope were co-cultured with transduced B cell lines for 4 h with BD GolgiPlug, BD GolgiStop and PE-anti-CD107a. Dead cells were labelled using Fixable Zombie Violet dye from Thermofisher. Cells were then washed, fixed with Cytofix/Cytoperm (BD Biosciences), and stained with PE-Cy7-anti-IFNγ (eBioscience), APC-anti-TNFα (eBioscience) and BV711-anti-CD8 (Biolegend). Cells were fixed with BD CellFix and run on an Attune NXT Flow Cytometer (Thermofisher).
For analysis, live CD8+ T cells were gated on and analysed for their expression of CD107a, IFNγ or TNFα using FlowJo V10 (BD Biosciences). Using each bulk T cell line as a control for the other we calculated the percentage response of NP9-17 over NP105-113, and vice versa. This data was then normalised to a percentage of the response to WT NP. Data was plotted and analysed in GraphPad Prism v9.2.0. Statistical analysis of responses was done by one-way ANOVA and Dunnett's multiple comparisons test.
To assess T cell specificity, WT B cells were pulsed with epitope peptide at 0, 0.0032, 0.016, 0.08, 0.4, 2 or 10 μg/ml for 1-h prior to co-culture with bulk CD8+ T cells specific for NP9-17 or NP105-113 epitope.
4.7.1. Proteasomal digestion of peptides
Peptides were purchased from GenScript (information available in Supplementary Table 4) and reconstituted to 20 mg/ml in DMSO. Recombinant human proteasomes (Human 20S Proteasome Protein, E360-050, bio-techne; Human 20S Immunoproteasome Protein, E370-025, bio-techne) and peptides were combined at a ratio of 50:1 for a period of 15, 60 or 180 min prior to the addition of 0.1 μl formic acid to terminate the function of the proteasome. Incubations were carried out at 37 °C with rotation to increase proteasome activity. Digested samples were frozen and stored at −80 °C until analysis by mass spectrometry.
4.8. Analysis of in vitro proteasome antigen processing products by mass spectrometry
Products generated by in vitro proteasome digestion of antigen precursor peptides were analysed by nano-flow liquid chromatography tandem mass spectrometry (nLC-MS/MS) using a Dionex Ultimate 3000 (Thermo) coupled to a timsTOF Pro trapped ion mobility spectrometry (TIMS) time-of-flight mass spectrometer (Bruker) using a 75 μm × 150 mm C18 column with 1.6 μm particles (IonOpticks) at a flow rate of 400 nL/min at 40 °C. The gradient used was a 17-min increase from 2% buffer B to 30% buffer B, followed by an increase to 95% B in 30 s and a 2.5-min wash and 5 min re-equilibration at 2% B (A: 0.1% formic acid in water. B: 0.1% formic acid in acetonitrile).
The timsTOF Pro mass spectrometer was operated in data dependent acquisition - parallel accumulation, serial fragmentation (ddaPASEF) mode. The TIMS accumulation and ramp times were set to 100 ms, and mass spectra were recorded from 100 to 1700 m/z, with a 0.85–1.30 Vs/cm2 ion mobility range. Precursors were selected for fragmentation from an area of the full TIMS-MS scan that excludes the majority of ions with a charge state of 1+. Those selected precursors were isolated with an ion mobility dependent collision energy, increasing linearly from 27 to 45 eV over the ion mobility range. Four PASEF MS/MS scans were collected per full TIMS-MS scan using a 3-cycle overlap, giving a duty cycle of 0.53 s. Ions were included in the PASEF MS/MS scan if they met the target intensity threshold of 2000 and were sampled multiple times until a summed target intensity of 10,000 was reached. A dynamic exclusion window of 0.015 m/z by 0.015 Vs/cm2 was used, and sampled ions were excluded from reanalysis for 24 s.
Raw MS data files were searched against a database containing the full synthetic antigen peptide precursor sequences using Peaks Studio Version 10 with mass tolerances of 20.0 ppm for precursors and 0.04 Da for fragment ions. Nonspecific cleavage was used and oxidation and deamidation were allowed as variable modifications. Peptide abundance was measured as accumulated ion counts after extraction of the chromatographic peak profiles of the precursor masses using Skyline version 21 and Bruker Compass DataAnalysis Version 5.3. Chromatograms were generated using R software package v4.0.1 and ggplot2 v3.3.2 and ggridges v0.5.2. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [44] partner repository with the dataset identifier PXD032054.
5. quantification and statistical analysis
5.1. Statistical analysis of CD8+ T cell responses
Experiments were repeated to an n of 3. All data with statistical analysis was analysed in GraphPad Prism by one-way ANOVA using Dunnett's multiple comparisons test. ANOVA summaries: NP9-17 CD107a – F = 33.95, P value < 0.0001; NP9-17 IFNγ - F = 17.72, P value < 0.0001; NP9-17 TNFα - F = 17.87, P value < 0.0001; NP105-113 CD107a – F = 4.095, P value = 0.0492; NP105-113 IFNγ - F = 1.323, P value = 0.3328; NP105-113 TNFα - F = 4.492, P value = 0.0397. Brown-Forsythe tests: NP9-17 CD107a – F (DFn, DFd) = 0.6481 (5,12), P value = 0.6685; NP9-17 IFNγ - F (DFn, DFd) = 0.8037 (5,12), P value = 0.5683; NP9-17 TNFα - F (DFn, DFd) = 1.382 (5,12), P value = 0.2981; NP105-113 CD107a – F (DFn, DFd) = 0.5568 (3,8), P value 0.6581; NP105-113 IFNγ - F (DFn, DFd) = 0.5392 (3,8), P value = 0.6686; NP105-113 TNFα - F (DFn, DFd) = 0.9408 (3,8), P value = 0.4650.
6. Limitations of the study
This work investigated the effect of a small number of natural mutations in SARS-CoV-2, none of which have yet been present in variants of interest or concern. However, as discussed above we believe this pilot study provides evidence that further investigation of mutations in flanking regions is valid.
Funding
Chinese Academy of Medical Sciences (CAMS) Innovation Fund for Medical Sciences (CIFMS), China (grant number: 2018-I2M-2-002).
Medical Research CouncilUK programme grant for MRC WIMM Human Immunology Unit.
Additional resources
The following websites were used to access and analyse SARS-CoV-2 sequence data:
https://cov.lanl.gov/content/index
https://covariants.org/variants/21K.Omicron.
The following website was used to predict proteasomal cleavage of sequences:
Author contribution statement
Dannielle Wellington: Conceived and designed the experiments; Performed the experiments; Analysed and interpreted the data; Wrote the paper. Zixi Yin, Zhanru Yu, Raphael Heilig, Simon DavisORCID, Roman FischerORCID, Philip HublitzORCID, Ryan BeveridgeORCID, Danning Dong, Guihai Liu, Yanchun Peng, Xuan Yao: Performed the experiments. Suet Ling Felce and Elie Antoun: Analysed and interpreted the data. Benedikt M Kessler: Conceived and designed the experiments; Performed the experiments; Wrote the paper. Tao Dong: Conceived and designed the experiments; Wrote the paper.
Data availability statement
Data associated with this study has been deposited at The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD032054.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank members of the Dong and Kessler groups for helpful discussions. We thank Prof. Alain Townsend and his group for providing valuable reagents for this work.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e20076.
Contributor Information
Dannielle Wellington, Email: dannielle.wellington@rdm.ox.ac.uk.
Tao Dong, Email: tao.dong@ndm.ox.ac.uk.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Rosendahl Huber S., et al. T cell responses to viral infections - opportunities for Peptide vaccination. Front. Immunol. 2014;5:171. doi: 10.3389/fimmu.2014.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peng Y., et al. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020;21(11):1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grifoni, A., et al., SARS-CoV-2 Human T Cell Epitopes: Adaptive Immune Response against COVID-19.Cell Host & Microbe.. [DOI] [PMC free article] [PubMed]
- 4.Habel J.R., et al. Suboptimal SARS-CoV-2−specific CD8+</sup> T cell response associated with the prominent HLA-A*02:01 phenotype. Proc. Natl. Acad. Sci. USA. 2020;117(39):24384–24391. doi: 10.1073/pnas.2015486117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schulien I., et al. Characterization of pre-existing and induced SARS-CoV-2-specific CD8 T cells. Nat. Med. 2021;27:1–8. doi: 10.1038/s41591-020-01143-2. [DOI] [PubMed] [Google Scholar]
- 6.Ferretti A.P., et al. Unbiased screens show CD8(+) T cells of COVID-19 patients recognize shared epitopes in SARS-CoV-2 that largely reside outside the spike protein. Immunity. 2020;53(5):1095–1107.e3. doi: 10.1016/j.immuni.2020.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grifoni A., et al. Targets of T Cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka K. The proteasome: overview of structure and functions. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009;85(1):12–36. doi: 10.2183/pjab.85.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blum J.S., Wearsch P.A., Cresswell P. Pathways of antigen processing. Annu. Rev. Immunol. 2013;31(1):443–473. doi: 10.1146/annurev-immunol-032712-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sijts E.J., Kloetzel P.M. The role of the proteasome in the generation of MHC class I ligands and immune responses. Cell. Mol. Life Sci. 2011;68(9):1491–1502. doi: 10.1007/s00018-011-0657-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van den Eynde B.J., Morel S. Differential processing of class-I-restricted epitopes by the standard proteasome and the immunoproteasome. Curr. Opin. Immunol. 2001;13(2):147–153. doi: 10.1016/s0952-7915(00)00197-7. [DOI] [PubMed] [Google Scholar]
- 12.Pishesha N., Harmand T.J., Ploegh H.L. A guide to antigen processing and presentation. Nat. Rev. Immunol. 2022;22(12):751–764. doi: 10.1038/s41577-022-00707-2. [DOI] [PubMed] [Google Scholar]
- 13.York I.A., et al. The cytosolic endopeptidase, thimet oligopeptidase, destroys antigenic peptides and limits the extent of MHC class I antigen presentation. Immunity. 2003;18(3):429–440. doi: 10.1016/s1074-7613(03)00058-x. [DOI] [PubMed] [Google Scholar]
- 14.Chang S.-C., et al. The ER aminopeptidase, ERAP1, trims precursors to lengths of MHC class I peptides by a “molecular ruler” mechanism. Proc. Natl. Acad. Sci. USA. 2005;102(47):17107–17112. doi: 10.1073/pnas.0500721102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paladini F., et al. The multifaceted nature of aminopeptidases ERAP1, ERAP2, and LNPEP: from evolution to disease. Front. Immunol. 2020;11:1576. doi: 10.3389/fimmu.2020.01576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neefjes J., et al. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 2011;11(12):823–836. doi: 10.1038/nri3084. [DOI] [PubMed] [Google Scholar]
- 17.Yu H., et al. Conserved sequence preferences contribute to substrate recognition by the proteasome. J. Biol. Chem. 2016;291(28):14526–14539. doi: 10.1074/jbc.M116.727578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abele R., Tampé R. Function of the transport complex TAP in cellular immune recognition. Biochim. Biophys. Acta Biomembr. 1999;1461(2):405–419. doi: 10.1016/s0005-2736(99)00171-6. [DOI] [PubMed] [Google Scholar]
- 19.Zervoudi E., et al. Probing the S1 specificity pocket of the aminopeptidases that generate antigenic peptides. Biochem. J. 2011;435(2):411–420. doi: 10.1042/BJ20102049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Silva T.I., et al. The impact of viral mutations on recognition by SARS-CoV-2 specific T cells. iScience. 2021;24(11) doi: 10.1016/j.isci.2021.103353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Del Val M., et al. Efficient processing of an antigenic sequence for presentation by MHC class I molecules depends on its neighboring residues in the protein. Cell. 1991;66(6):1145–1153. doi: 10.1016/0092-8674(91)90037-y. [DOI] [PubMed] [Google Scholar]
- 22.Ranasinghe S.R.F., et al. The antiviral efficacy of HIV-specific CD8+ T-cells to a conserved epitope is heavily dependent on the infecting HIV-1 isolate. PLoS Pathog. 2011;7(5) doi: 10.1371/journal.ppat.1001341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Draenert R., et al. Immune selection for altered antigen processing leads to cytotoxic T lymphocyte escape in chronic HIV-1 infection. J. Exp. Med. 2004;199(7):905–915. doi: 10.1084/jem.20031982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peck K.M., Lauring A.S. Complexities of viral mutation rates. J. Virol. 2018;92(14) doi: 10.1128/JVI.01031-17. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robson F., et al. Coronavirus RNA proofreading: molecular basis and therapeutic targeting. Mol. Cell. 2020;79(5):710–727. doi: 10.1016/j.molcel.2020.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng Y., et al. An immunodominant NP105–113-B*07:02 cytotoxic T cell response controls viral replication and is associated with less severe COVID-19 disease. Nat. Immunol. 2021 doi: 10.1038/s41590-021-01084-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vita R., et al. The immune epitope database (IEDB): 2018 update. Nucleic Acids Res. 2019;47(D1):D339–d343. doi: 10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grenga L., et al. Shotgun proteomics analysis of SARS-CoV-2-infected cells and how it can optimize whole viral particle antigen production for vaccines. Emerg. Microb. Infect. 2020;9(1):1712–1721. doi: 10.1080/22221751.2020.1791737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finkel Y., et al. The coding capacity of SARS-CoV-2. Nature. 2021;589(7840):125–130. doi: 10.1038/s41586-020-2739-1. [DOI] [PubMed] [Google Scholar]
- 30.Zhao Y., et al. A dual-role of SARS-CoV-2 nucleocapsid protein in regulating innate immune response. Signal Transduct. Targeted Ther. 2021;6(1):331. doi: 10.1038/s41392-021-00742-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murata S., et al. The immunoproteasome and thymoproteasome: functions, evolution and human disease. Nat. Immunol. 2018;19(9):923–931. doi: 10.1038/s41590-018-0186-z. [DOI] [PubMed] [Google Scholar]
- 32.McCarthy M.K., Weinberg J.B. The immunoproteasome and viral infection: a complex regulator of inflammation. Front. Microbiol. 2015;6(21) doi: 10.3389/fmicb.2015.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cascio P., et al. 26S proteasomes and immunoproteasomes produce mainly N-extended versions of an antigenic peptide. EMBO J. 2001;20(10):2357–2366. doi: 10.1093/emboj/20.10.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kloetzel P.-M. The proteasome and MHC class I antigen processing. Biochim. Biophys. Acta Mol. Cell Res. 2004;1695(1):225–233. doi: 10.1016/j.bbamcr.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 35.López de Castro J.A. How ERAP1 and ERAP2 shape the peptidomes of disease-associated MHC-I proteins. Front. Immunol. 2018;9(2463) doi: 10.3389/fimmu.2018.02463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Motozono C., et al. The SARS-CoV-2 Omicron BA.1 spike G446S mutation potentiates antiviral T-cell recognition. Nat. Commun. 2022;13(1):5440. doi: 10.1038/s41467-022-33068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vardhana, S., et al., Understanding T-cell responses to COVID-19 is essential for informing public health strategies. Science Immunology.0(0): p. eabo1303.. [DOI] [PMC free article] [PubMed]
- 38.Wellington D., et al. Immunodominance complexity: lessons yet to be learned from dominant T cell responses to SARS-COV-2. Curr Opin Virol. 2021;50:183–191. doi: 10.1016/j.coviro.2021.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parker R., et al. Mapping the SARS-CoV-2 spike glycoprotein-derived peptidome presented by HLA class II on dendritic cells. Cell Rep. 2021;35(8) doi: 10.1016/j.celrep.2021.109179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qiu C., et al. CD8+ T-cell epitope variations suggest a potential antigen HLA-A2 binding deficiency for spike protein of SARS-CoV-2. Front. Immunol. 2022;12 doi: 10.3389/fimmu.2021.764949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dangi T., et al. Combining spike- and nucleocapsid-based vaccines improves distal control of SARS-CoV-2. Cell Rep. 2021;36(10) doi: 10.1016/j.celrep.2021.109664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matchett W.E., et al. Cutting edge: nucleocapsid vaccine elicits spike-independent SARS-CoV-2 protective immunity. J. Immunol. 2021;207(2):376–379. doi: 10.4049/jimmunol.2100421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silva E.K.V.B., et al. Immunization with SARS-CoV-2 Nucleocapsid protein triggers a pulmonary immune response in rats. PLoS One. 2022;17(5) doi: 10.1371/journal.pone.0268434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perez-Riverol Y., et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022;50(D1):D543–d552. doi: 10.1093/nar/gkab1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Korber B., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abd Hamid M., et al. Self-maintaining CD103+ cancer-specific T cells are highly energetic with rapid cytotoxic and effector responses. Cancer Immunol. Res. 2020;8(2):203–216. doi: 10.1158/2326-6066.CIR-19-0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data associated with this study has been deposited at The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD032054.