Abstract
Numerous studies have shown that stress in plant cells and organelles with transport electron chains is related to RNA editing. The ATP synthase complex present in mitochondria plays a crucial role in cellular respiration and consists of several subunits. Among them is the b subunit, which is encoded by the mitochondrial atp4 gene. Computing-based analysis of the effects of RNA editing of the Withania somnifera atp4 gene in mitochondria leading to alterations in the b subunit of ATP synthase. Using the CLC Genomic Workbench 3, RNA editing analysis between the control and salt stress conditions was not significantly different. Depending on RNA editing, the tertiary structure model revealed a change in the states of the b subunit, reflecting differences in the central stalk and F1-catalytic domain. The study found that polar edits in the N-terminus of the b subunit allow for efficient H + ion selectivity and introduce a new coiled-coil alpha-helical structure that may help stabilize the complex. The most noteworthy finding of this study was the strong impact of these editing events on the tertiary structure of the b subunit, which has the potential to affect the ATPase activity and indicate that the editing in this subunit aimed to restore the original active protein and not as a response to salt stress.
Keywords: RNA editing, Mitochondrial atp4 gene, Withania somnifera, 3D protein model
1. Introduction
Due to its promise as a therapeutic and stress-resistant herb, Withania somnifera, also known as Indian ginseng, has attracted attention in RNA editing studies. In order to control gene expression and produce protein variety, RNA editing is essential, and a recent report has shown that RNA editing events can significantly affect protein structure and function (Hao et al. 2021).
The RNA editing process comprises the deletion, insertion, and modification of nucleotides after transcription, but it does not involve splicing, polyadenylation or capping (Nishikura, 2006, Farajollahi and Maas, 2010). This mechanism was investigated initially in Trypanosoma mitochondria (Benne et al. 1986). The common type of this process is converting cytosine (C) to uracil (U), but adenosine (A) can also be modified to inosine (I) in animals (Nishikura 2006). An example of RNA editing, which is carried out by the cytidine deaminase (apobec/ACF complex) editosome, involves the conversion of C-to-U and has been identified in the apolipoprotein-B gene (Maris et al. 2005). In the plant kingdom, RNA editing is frequently observed in transcripts of plastids and mitochondria, where the conversion of C to U nucleotides occurs as a result of pentatricopeptide repeat (PPR) proteins (Castandet and Araya, 2011, Takenaka et al., 2019). Studies have reported that Arabidopsis thaliana exhibits 43 and 619 editing sites in the chloroplast and mitochondria, respectively (Ichinose and Sugita, 2016; Bentolila et al. 2013). This process can occur in both non-coding and coding regions (Licht and Jantsch, 2016, Picardi et al., 2014) and become more effective in biological functions if it is released in the 1st or 2nd position of codons (Edera et al. 2018). Environmental factors, developmental stages, tissue type, and ecotypes can all influence RNA editing (Tseng et al. 2013). In terrestrial environments, research has concentrated on editing of mitochondria transcripts such as CoxIII, Cytb, CoxII and NadI (Takenaka et al. 2008). The atp4 transcript undergoes RNA editing, specifically the cytidine-to-uridine modification (Kong et al. 2019). Complex V, which includes the ATP synthase subunits, is the final enzyme in the transfer electron chain. ATPases are enzymes that hydrolyze ATP (adenosine triphosphate) to release energy for cellular processes. An ATPase is classified into four types: F-ATPase, V-ATPase, and A-ATPase. Each of them has a unique structure and function, and they play important roles in various cellular processes, including metabolism, transport, and signaling. F-ATPases are reversible ATPases found in bacteria, plants (chloroplasts) and eukaryotes (mitochondria). They can hydrolyze ATP by creating a proton gradient or use a proton gradient to synthesis ATP (Sobti et al. 2021).
ATP is generated within the matrix of the mitochondria by an enzyme called ATP synthase. The structure of ATP synthase has been extensively studied, and it consists of two main domains: the F1 domain, which is the catalytic unit responsible for ATP synthesis, and the F0 domain, which is the membrane-embedded proton channel that generates the electrochemical gradient used to drive ATP synthesis. The catalytic site of ATP synthesis or hydrolysis is the F1, while the rotor-like motor part is the F0, which works as a channel to transfer the proton through ATP synthesis. The F1 domain is composed of five subunits, namely alpha, beta, gamma, delta, and epsilon, arranged in a hexagonal pattern and extending up into the F0 part. It mainly works as the axle of the complex. The F0 domain consists of several subunits, including a ring of c-subunits, one a-subunit that is anchored to the membrane and whose rotation powers the synthesis of ATP, and one b subunit that stabilizes the complex as it extends from the inner membrane to the matrix (Pinke et al. 2020). The structure of mitochondrial ATPase is highly conserved across different eukaryotic organisms and is essential for cellular energy production.
The b subunit is a membrane-bound protein that plays a key role in ATP synthesis. It is encoded by the mitochondrial atp4 gene and located in F-ATPase. In addition to the transmembrane domain, this protein also contains an extramembrane domain. B subunit N-terminal is bonded with the complex membrane part through two antiparallel α-helicals (in mitochondria), and the remaining protein extends from the membrane in the form of an electrically charged coil structure. The protein is highly important for F1 to bind the membrane part F0. Subunit B has a long helix extending from the F1 domain to the membrane surface domain F0 and probably no break in it across the membrane (Sobti et al., 2021, Pinke et al., 2020). The b subunit also contains a regulatory domain that is implicated in ATP synthase complex assembly and regulates the enzyme's activity.
Next-generation sequencing (NGS) has emerged as an effective tool for discovering RNA editing in organelles (Bentolila et al. 2013). This technology is a useful tool for detecting RNA editing at low level, making it a reliable tool for achieving research objectives. Furthermore, NGS and bioinformatics tools have facilitated the identification of numerous novel Arabidopsis editing sites (Ichinose and Sugita, 2016; Lo Giudice et al. 2019).
The present investigation employs RNA sequencing data to investigate the editing sites of the mitochondrial atp4 gene in Withania somnifera and how they affect the tertiary structure of the gene. Furthermore, gene editing patterns are studied in relation to salinity.
2. Materials and methods
2.1. RNA sequencing data
The Withania somnifera RNA-seq data was collected from the NCBI database. The data consists of samples collected in two conditions: a control group and a group subjected to one week of salinity treatment. The control group samples were labeled as SRR10985100, SRR10985101, and SRR10985102, while the salinity-treated group samples were labeled as SRR10985103, SRR10985104, and SRR10985105.
2.2. Site-specific analysis of RNA editing
This process is achieved according to Ramadan et al. (Ramadan et al. 2023). Withania somnifera's mitochondrial atp4 gene's editing sites were identified by CLC Genomic Workbench 3.6.5. With a 0.98 similarity and length fraction, unwanted sequences were removed by adjusting the mapping settings. The reads were matched to the Alkekengi officinarum mitochondrial genome (Accession no. OL467322). The frequency of each conversion site over all salt exposure periods was computed (Chen et al. 2017).
2.3. Exploring atp4 gene amino acids and conserved domain database
The CLC genomic workbench version 3.6.5 software tool was utilized for the purpose of identifying RNA editing sites within the genomic atp4 genes and cDNAs of Withania somnifera. Additionally, the same software tool was employed to predict any potential changes in the secondary structure of proteins resulting from RNA editing events. Accession numbers were measured using the NCBI Conserved Domain database (Lu et al. 2020).
2.4. Validation of RNA editing sites
RNA editing sites were validated by analyzing the leaves of Withania somnifera plants treated with 100 mM NaCl for one week, as well as control samples (in biological triplicates). For RNA extraction, Qiazol (Qiagen, cat. No. 79306) was utilized, and the cDNA synthesis procedure used 1 µg of total RNA and 1 mM poly dT oligonucleotide. Using the Mx3005P qPCR system (Stratagene) and primers designed by PRIMER 3 (Table S1), qRT-PCR experiment was performed according to Wang et al. (Wang et al. 2015). The actin gene was used for normalization purposes (acc. No. OQ291286). RNA editing percentage calculated as following (Rodrigues et al. 2017):
2.5. Statistical analysis
The ANOVA test in the SPSS version 20 was used to conduct the statistical analysis of bioinformatics outputs as well as qRT-PCR result. Tukey's HSD (Tukey, 1949) was used for multiple comparisons.
2.6. Protein structural modeling
Protein modeling of both wild-type and mutated proteins was performed using I-Tasser (Zhang, 2008). To calculate the RMSD between the two models, TM-align was employed (Zhang and Skolnick, 2005). Through repetitions of dynamic programming, the TM-Align approach optimizes residue alignment according to structural similarity. DynaMut2 and I-Mutant + were used to evaluate protein stability (Rodrigues et al., 2021, Capriotti and Fariselli, 2017). Additionally, the coiled-coil structure was predicted using DeepCoil (Ludwiczak et al. 2019), CCHMM (Bartoli et al. 2009) and Scorer 2 (Chari et al. 2017) independently based on the sequences. Using the ConSurf website (Ashkenazy et al. 2016), which shows the arrangement of both structural and functional residues throughout the structure, the conservation study of amino acids was carried out.
3. Results
3.1. Identification of Withania somnifera atp genes and corresponding CDS
It was possible to obtain the genomic DNA and the cDNA of the mitochondrial atp4 gene from Withania somnifera by utilizing the Alkekengi officinarum mitochondrial atp4 gene (accession no. OL467322) as a reference to get atp4 gene (OQ238716), transcript data from control (acc. no. OQ238717) and salinity-stressed (acc. no. OQ238718) samples. To identify transcripts of the atp genes, a total of 128,140,192 pair-end reads were utilized for the control sample, and 117,692,344 reads were used for the one-week salt stress (100 mM NaCl) sample.
3.2. Nucleotide editing and amino acid changes
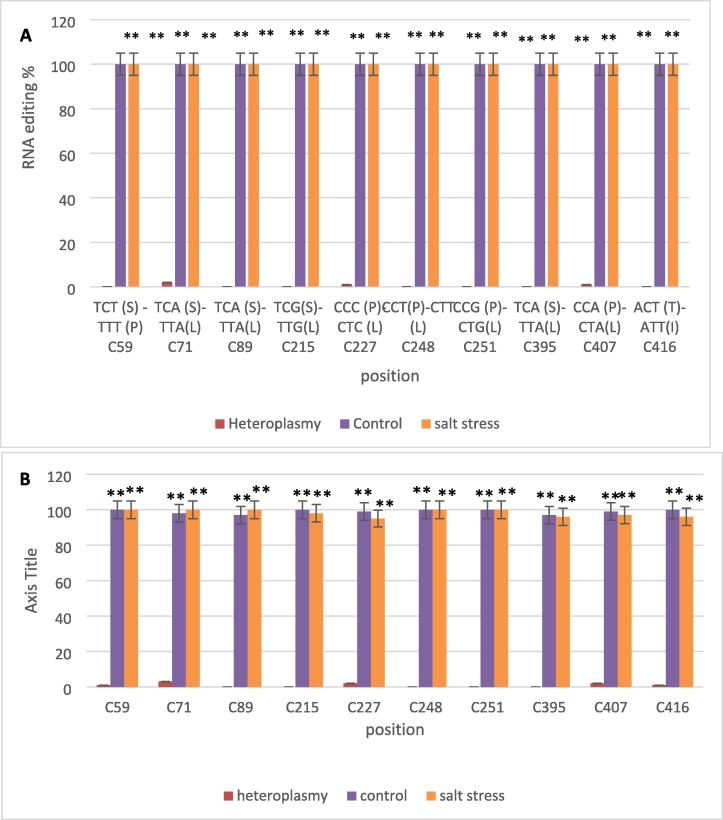
Multiple alignments of atp4 genomic sequence and the two cDNA treatment sequences (Fig. S1) revealed ten C- to- U editing sites (C59, C71, C89, C215, C227, C248, C251, C395, C407, C416) (Fig. 1A, Table S2).
Fig. 1.
RNA editing percentage for atp4 gene. A) RNA editing percentage for atp4 gene from RNA-seq data as compared with DNA sequences using CLC program, B) qRT-PCR confirmation. The replicates' worth of data are presented as means with standard deviations (black bars). A statistically significant symbol is ** at P < 0.01.
3.3. Atp4 gene editing validation
In order to validate the identified editing sites of the atp4 gene and to verify the usefulness of RNA-seq as a characterization tool, the editing sites were confirmed using qRT-PCR. The study quantified and measured the levels of atp4 editing positions (C59, C71, C89, C215, C227, C248, C251, C395, C407, C416) in two different treatments. The results of this analysis are presented in Fig. 3, which likely shows the relative expression levels of each editing position in both treatments. This experiment was carried out to confirm the accuracy of the RNA-seq data by comparing it to the RT-qPCR results (Fig. 1B).
Fig. 3.
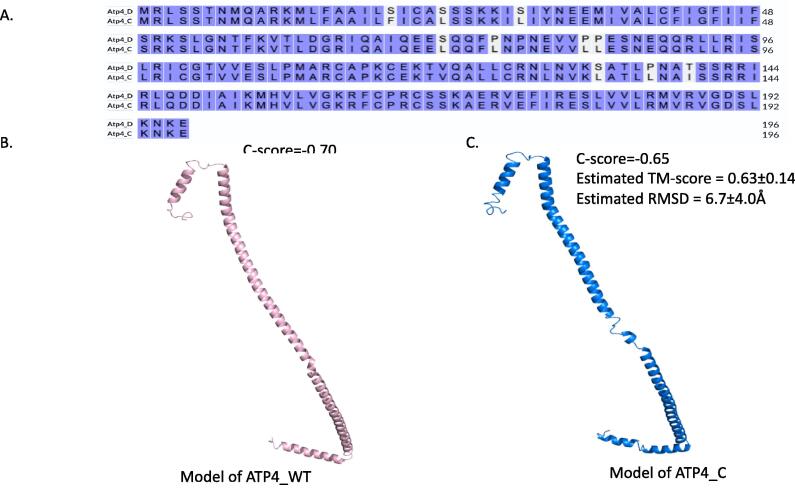
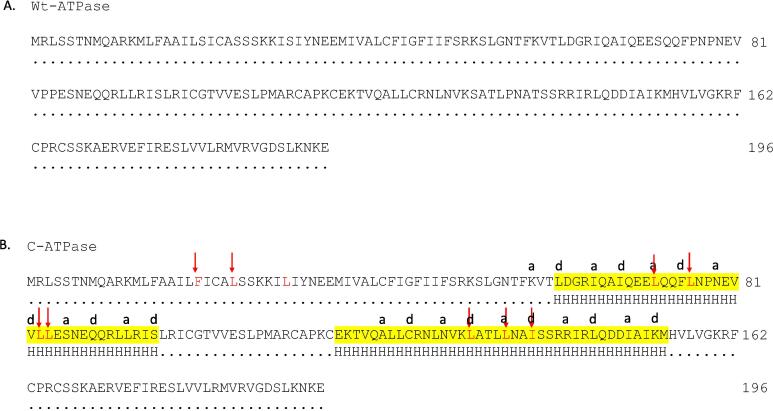
Comparison of the b subunit of ATPase from Withania somnifera. A) Sequences of the b subunit from Withania somnifera before and after editing. The blue purple regions are identical, while the unlabeled ones are the edits. B) The structure model from I-Tasser generated from the wild type of atp4 DNA sequence. The long pink helix represents the b subunit of the ATP complex. C) The structure model from I-Tasser generated from edits of atp4 DNA sequence. The long pink helix represents the b subunit of the ATP complex.
3.4. Conserved domain analysis of atp4 synthase subunits b and secondary structure
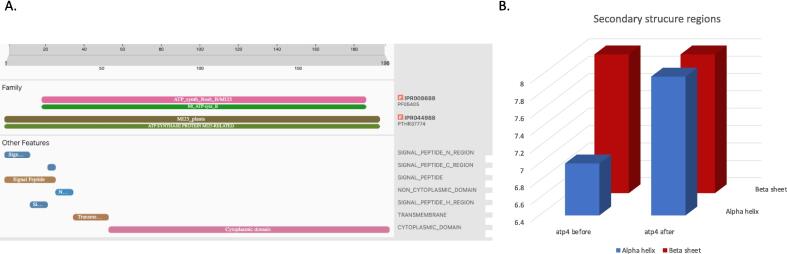
Domain analysis was performed on the discovered ATP synthase subunits 196aa, and their accession numbers were determined using the NCBI Conserved Domain database. Two protein families was detected as following; ATP synthase, F0 complex, subunit B/MI25 was predicted at position 20–148 (IPR008688, PF05405), ATP synthase, F0 complex, subunit MI25, plants at positions 1–191 (IPR044988, PTHR37774). Other features was detected; N-terminal region of a signal peptide (1–13), C-terminal region of a signal peptide (23–26), signal peptide region (1–26),Phobius Region of a membrane-bound protein predicted to be outside the membrane, in the extracellular region (27–35), Hydrophobic region of a signal peptide (14–22), Region of a membrane-bound protein predicted to be embedded in the membrane (36–53) and finally Region of a membrane-bound protein predicted to be outside the membrane, in the cytoplasm (54–196) (Fig. 2A).
Fig. 2.
Secondary structure and domain analysis of b subunit of atp4. A. The Domain analysis of b subunit. showing various protein signatures and domains identified within the sequence. Key features include signal peptide regions, transmembrane regions, cytoplasmic domains, and the presence of the ATP synthase protein MI25-related family. B. Number of secondary structure regions in the b- subunit before and after RNA editing. The beta sheet (brown) and the alpha helix (blue).
Protein secondary structure indicated differences in the quantity and size of alpha helix and beta sheets. In atp4 before editing, there were 7 alpha helixes and 8 beta sheets, however, after editing, there were 8 alpha helixes and 8 beta sheets (Fig. 2B, Fig. S2).
3.5. 3D structure of atp4 encoded protein subunit
3.5.1. Structural differences in b subunit
The results indicate that RNA edits occurred at multiple gene locations during the experiment, which resulted in amino acid changes in critical locations of the protein (Fig. 3). Ten different changes in the amino acid sequence were observed, and most of these edits affected the overall structure of the b subunit.
The model exhibited a longer N-terminus α-helical structure in both wild-type ATPase (expected protein from the DNA sequence before editing) and C-ATPase (edited protein from control mRNA sequence as well as salt stress mRNA sequence). This finding is different from the reported ATPases of F-APT structures (Fig. 4).
Fig. 4.
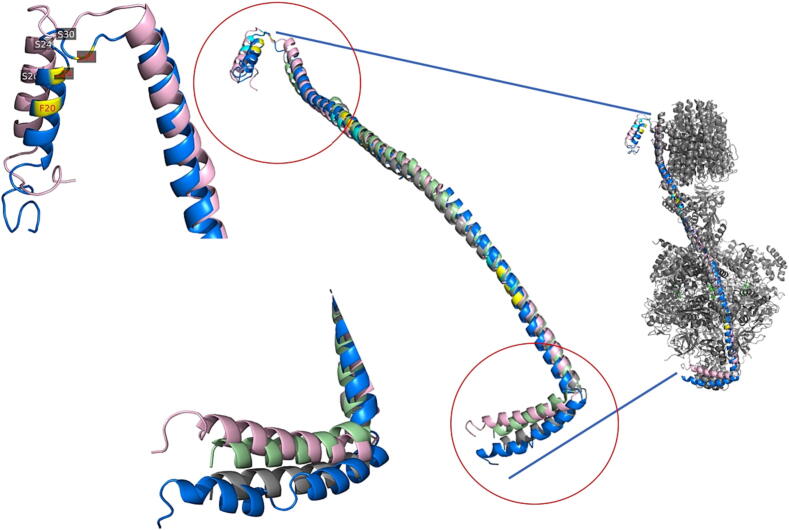
Superpose the structures of subunit b from Withania somnifera_DNA and Withania somnifera Control. The long pink helix represents the b subunit of the Wild type-ATP (Wt-ATPase) complex, and the long blue helix represents the b subunit of the Control-ATP (C-ATPase) complex. the C-ATPase in blue, state 1 in gray and state 3 in mint green. The model is shown along with the F-ATPase complex structure (PDB: 5LQZ) from Pichia angusta in gray color. The labeled yellow and cyan colors represent the sequence differences between Wt-ATPase and C-ATPase.
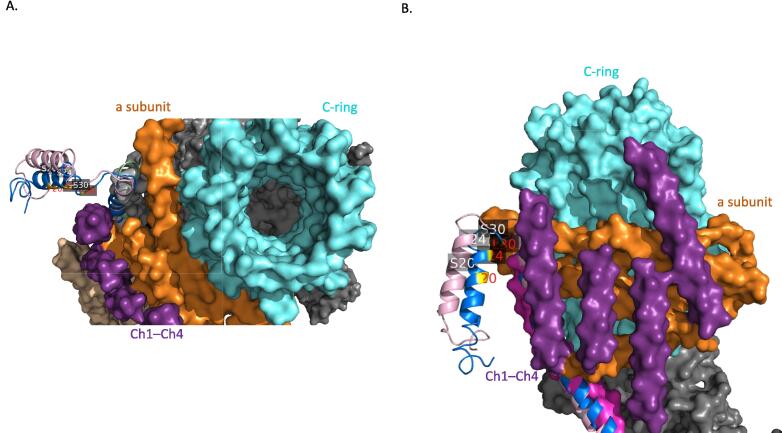
The reported sequence from Pichia angusta (PDB: 5LQZ) are missing the N-terminus domain (the first 33 amino acids are forming external N-terminus loop) that never been noticed in previous structures of b subunit. These external helices are clearly hanging outside the reported b subunit complex structure (Fig. 5).
Fig. 5.
Exposure of N-terminal alpha helical structures in subunit b (Wt-ATPase in pink and C-ATPase in blue helices). The edits are labeled in yellow at both of the helices. A. A Top view of the ATPase complex structure (PDB: 5LQZ) from Pichia angusta. B. A side view of the ATPase complex structure (PDB: 5LQZ) from Pichia angusta. The C-ring in cyan, a subunit in orange and Ch1-Ch4 protein subunits are in purple.
3.5.2. Coiled-coil effect on b subunit
The RNA edits were introduced at critical positions of the α-helical structure of the b subunit. Most of them are leucine edits to support the strong coiled-coil structure. Some of the leucine was introduced at a, d junctions (Fig. 6). The “a” and “d” positions within a coiled-coil structure are critical for the stability and packing of the alpha helices that form the coiled coil. These positions are typically occupied by hydrophobic or nonpolar amino acids, which engage in important interactions to maintain the coiled-coil structure's integrity. The arrangement of amino acids at these positions is a key determinant of the coiled coil's stability and function in various proteins.
Fig. 6.
Coiled-coil heptad repeats identification. A. Wt-ATPase sequence, where no coiled-coil structure was predicted. B. C-ATPase, where the d amino acids are labeled in the sequence. The two predicted coiled-coil structures are identified in the region as L61-S96 and E117–M154, respectively. The edited amino acids were labeled in red within the sequence.
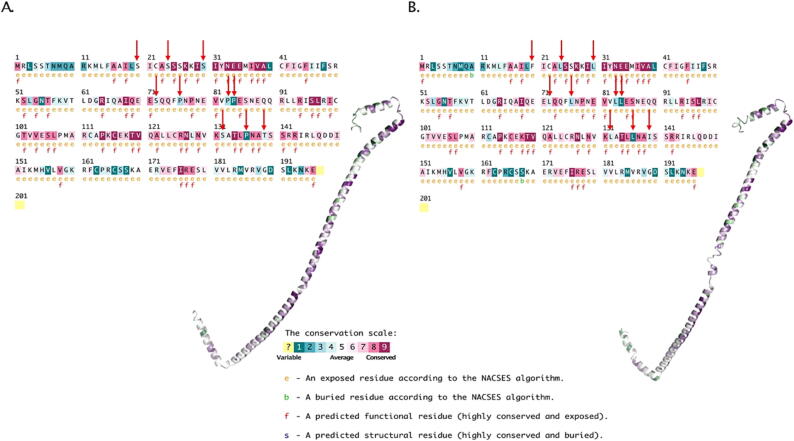
The conservation of amino acids over the structure was investigated and revealed that all the editing positions were typically located in relatively non-conserved positions (Fig. 7). In particular, P84L and P136L are identified as highly variable regions in the protein. To identify functionally important regions in the protein, an automated tool called ConSurf was utilized, which uses surface mapping to determine how each amino acid site position has changed over time. The analysis assumes that residues that are highly conserved across different homologous proteins are likely to be important for the protein's structure, function, or both.
Fig. 7.
Conservation analysis of the protein models. ConSurf results showing the conservation quality of amino acid residues. A. The Wt-ATPase; B. The C-ATPase. Arrows indicate the substitution positions. The conservation scale are ranging from dark teal color (variable) to rosewood color (conserved).
4. Discussion
In most organisms, RNA editing is one of a key of the functional protein’s expression, particularly in organelles and researchers links ecological stress to RNA editing, highlighting its crucial role in maintaining cellular homeostasis (Castandet and Araya, 2011, Yuan and Liu, 2012). When RNA editing is disturbed in Arabidopsis, free oxygen (ROS) production rises, heightened dehydration sensitivity. These studies emphasis how important RNA editing is to plant biology and how it might affect how plants react to environmental stress (Yuan and Liu, 2012).
Depending on the species, different editing sites for the atp4 gene have been reported in various plants. Arabidopsis contained nine editing sites, compared to Onion's 13, Cucumber's 10, wild Carrot's 18, Soybean's 12, and Sunflower's 11 (Edera et al. 2018). Prior to utilize the bioinformatics tools to study RNA editing, it is crucial to eliminate the impact of heteroplasmy on nucleotide variation, as it may interfere with the RNA editing analysis (Fig. 1) (Makki et al. 2019). In this study, we ensured the exclusion of heteroplasmy and identified 10 RNA editing sites (nucleotide no. C59, C71, C89, C215, C227, C248, C251, C395, C407, C416) in Withania somnifera.
Real-time PCR analysis confirmed that these sites were completely altered in control and salt stress (Fig. 1B, Table S2). These results enhance our understanding of the potential involvement of RNA editing systems in shaping plants' responses to abiotic stress. The current study yielded several insights into RNA editing in Withania somnifera. Initially, it was observed that salt stress did not influence any of the editing sites. This raises the possibility that RNA editing in this gene may be intended to restore the conserved protein, raising the possibility that it might have originated to undo previous mutations and highlighting the significance of this editing procedure. Additionally, it was found that there were no synonymous amino acids, which is in contrast to earlier studies (Edera et al. 2018). These findings highlight the unique characteristics of RNA editing in Withania somnifera and provide insights into the functional significance of this process in this species.
RNA editing has a profound impact on the secondary structure of proteins, including changes to the number of alpha helices and beta sheets and their length (Fig. 2B, Fig. S2). At a numeric scale, 7 alpha helix and 8 beta sheet was found before editing, however, 8 alpha helix and 8 beta sheet was found after editing. At length level, the alpha helix in regions (8.0.22, 65.0.71,109.0.111,115.0.125, 130.0.146) at DNA are different in size at control and salinity stress (8.0.23, 64.0.76,109.0.112,115.0.127,142.0.146). Also, beta sheet regions (29…32, 81, 150…158) at DNA are different in size at control (28…32, 81…84, 153…158). In addition substitute part of beta sheet at (150…158) in corresponding DNA to alpha helix in control (150…152). These findings align with our earlier discovery in the Ccmfn gene (Ramadan et al. 2023) and underscore the substantial impact of RNA editing on the integrity and quantity of alpha helices and beta sheets.
Electron cryo-microscopy was used to determine the three states of mitochondrial ATP synthase in Pichia angusta (Vinothkumar et al. 2016). Our models also reveal a clear change in the states of the b subunit (Fig. 4). The two different states of the structure reflecting differences in the central stalk and F1-catalytic domain where F1 is bound into a different catalytic interface in each state. The inhibitor protein's position relative to the peripheral stalk (Dickson et al., 2006, Nirody et al., 2020). A bound inhibitor at state 3 can be oriented either towards the peripheral stalk or away from it, while a bound inhibitor at state 1 will be closest to it, whereas an inhibitor at state 1 will be further away from it (Murphy et al. 2019). This study provides valuable insights into the structural changes that occur in the mitochondrial ATP synthase during different states, which can be important for understanding the molecular mechanism underlying the function of this enzyme. It also highlights the role of F1 in regulating ATP synthesis by inhibiting the ATP synthase when the cellular energy charge is high, and thereby, preventing the wasteful hydrolysis of ATP. These findings have implications for understanding the bioenergetics of cells.
In our knowledge, a b subunit C-terminal domain binds to an OSCP C-terminal domain (Moralesrios et al. 2015), however in our study, we observed a clear difference in the c-terminus between the wild-type ATPase (Wt-ATPase) and control ATPase (C-ATPase) models, which we believe may be related to the regulation of overall rotation and binding. It is possible that the edits that occurred could have a role in how the ATP pump is regulated. Our model is perfectly fitting with Walker study (Walker and Dickson, 2006), as we identified α-helical structure from residues 12–26 while they mentioned α-helical structure 19–26 that is exposed in the mitochondrial matrix. Furthermore, we identified a second transmembrane α-helix 33–47 followed by small turn in the long a-helix at residues 48–54.
The polar edits (S20F, S24L and S30L) in b subunit N-terminus indicates the “switching” between these two states (Fig. 6). This localized amino acid changes in composition from serine to phenylalanine and leucine is believed to affect the proton flow mechanism, instead of significant structural changes (Leone et al. 2015). These minor changes could be responsible for the strong H + ion selectivity to reduce the excessive salt in the environment (Leone et al. 2015). The N-terminus of the b subunit is responsible for proton translocation, which is a crucial step in ATP synthesis. The switching mechanism between two states allows the ATP synthase to adapt to changes in the proton motive force, which in turn can be affected by changes in the environment. This mechanism is suspected to be modulated by polar edits, specifically the substitution of serine with phenylalanine and leucine. These changes likely affect the interactions between the b subunit and other components of the ATP synthase, such as the OSCP and the peripheral stalk, which are involved in regulating the overall rotation and binding of the ATP synthase. Ultimately, these changes in the proton flow mechanism allow for efficient H + ion selectivity, which is important for the ATP synthase power functioning in high-sodium environments.
Our analysis revealed an additional alpha helix structure in the N-terminus region of the mitochondrial ATPase that is not embedded within the membrane (Fig. 5). This feature is quite different than the other previously reported structures of mitochondrial ATPases. ATPases is exposed to the mitochondria matrix (Walker and Dickson, 2006). The presence of this α-helical structure is believed to stabilize the long helical axis of b subunit.
The polar edits at positions S72L, P76L, P83L, P84L, S131L, P136L and T139I were observed to introduce a new coiled-coil alpha-helical structure that may play a crucial role in stabilizing the entire complex. This coiled-coil alpha-helix tends to dimerism with the b subunit for better stabilization and therefore, prevent the dissociation of ATPase complex for a longer effect during the stress (Fig. 4).
When the cells are exposed to environmental stress such as drought or salt, the b subunit plays a critical role in providing a flexible elastic link for a smoothly rotating torque. Therefore, the stability of the b subunit is crucial under these conditions. We assume that the edits we figured out in this manuscript are giving the b subunit the stability required during ATP synthesis and hydrolysis to provide the adequate energy required either for immediate usage or for storage as backup in the cell. As a consequence, the proton channel interface remains intact. It was always discussed before the support for the long α-helical axis of b subunit in the middle. C-terminal is supported by interactions with OSCP C-terminal and stabilized, while N-terminal is integrated into the membrane sub-complex, stabilizing it, along with other interactions with the d-subunit and intact peripheral stalks. Only the lower part of the helix appears to be lacking in support from other subunits. Coild-coil domains before and after this area provide the required support for the overall complex (Fig. 6) in addition to the other single α-subunit (residues 1–30).
The study compared the RNA editing frequency in the mitochondrial genes of control and salt-stressed plants in Withania somnifera. The findings suggest that there were no significant differences in RNA editing between the two conditions. This result contrasts with most studies that have reported differences in the editing sites frequency in mitochondrial genes between normal and abiotic stress conditions. We suggest that our findings may be due to the specific conditions of the salt stress applied in this study. Therefore, we recommend that future studies should consider investigating salt stress in a narrower time frame, as was done in a previous study by Ramadan et al. (Ramadan et al. 2021), to confirm the effect of salt stress on RNA editing in Withania somnifera. Overall, these findings provide insight into the complex regulatory mechanisms involved in plant responses to environmental stress and suggest that the impact of stress on RNA editing may be context-dependent and influenced by various factors.
5. Conclusion
According to our data, there was no noticeable difference in RNA editing between control and salt-stressed plants of Withania somnifera for the mitochondrial atp4 gene. These findings imply that these locations are critical for b subunit restoration and stability during ATP production and hydrolysis. However, additional experimental validation is required to support our predictions. Also, it demonstrate that RNA editing can have a considerable impact on the structure and function of proteins, including ATPases, by introducing alterations in the amino acid sequence. Our findings pave the way for further research into the role of RNA editing in restoring active enzymes in wild plants that may lack particular gene codons due to mutations, Investigating the development of cellular respiratory genes in wild plants can provide useful insights into their adaption processes to environmental stress.
7. Ethics approval and consent to participate
This article does not contain any studies with human participants or animals performed by the authors.
Funding
Ministry of Education and King Abdulaziz University, Jeddah, Saudi Arabia. Institutional fund projects under grant no. (IFPRC-155–130-2020).
9. Availability of data and material
All data generated or analyzed during this study is included in this published article [and its supplementary information files].
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The research work was funded by institutional fund projects under grant no. (IFPRC-155-130-2020). Therefore, authors gratefully acknowledgment technical and financial supports from the Ministry of Education and King Abdulaziz University, Jeddah, Saudi Arabia.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.sjbs.2023.103817.
Contributor Information
Ahmed M. Ramadan, Email: aamara@kau.edu.sa.
Khalid M. Al-Ghamdi, Email: kalghamdy@kau.edu.sa.
Abdullah J. Alghamdi, Email: agalghamdi1@kau.edu.sa.
Marwa Amer, Email: Marwa.amer@must.edu.eg.
Mona I.M. Ibrahim, Email: mona.ibrahim@must.edu.eg.
Ahmed Atef, Email: abayoumi@kau.edu.sa.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- Ashkenazy H., Abadi S., Martz E., Chay O., Mayrose I., Pupko T., Ben-tal N. Consurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:w344–w350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoli L., Fariselli P., Krogh A., Casadio R. Cchmm_prof: a hmm-based coiled-coil predictor with evolutionary information. Bioinformatics. 2009;25:2757–2763. doi: 10.1093/bioinformatics/btp539. [DOI] [PubMed] [Google Scholar]
- Benne R., Van den Burg J., Brakenhoff J.P., Sloof P., Van Boom J.H., Tromp M.C. Major transcript of the frame shifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986;46(6):819–826. doi: 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
- Bentolila S., Oh J., Hanson M.R., Bukowski R. Comprehensive high-resolution analysis of the role of an Arabidopsis gene family in RNA editing. PLoS Genet. 2013;9:e1003584. doi: 10.1371/journal.pgen.1003584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriotti E., Fariselli P. Phd-snpg: a webserver and lightweight tool for scoring single nucleotide variants. Nucleic Acids Res. 2017;45:w247–w252. doi: 10.1093/nar/gkx369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castandet B., Araya A. RNA editing in plant organelles. Why make it easy? Biochemistry (Mosc.) 2011;76(8):924–931. doi: 10.1134/S0006297911080086. [DOI] [PubMed] [Google Scholar]
- Chari R., Yeo N.C., Chavez A., Church G.M. Sgrna scorer 2.0: a species-independent model to predict crispr/cas9 activity. ACS Synth. Biol. 2017;6:902–904. doi: 10.1021/acssynbio.6b00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T.C., Liu Y.C., Wang X., Wu C.H., Huang C.H., Chang C.C. Whole plastid transcriptomes reveal abundant RNA editing sites and differential editing status in Phalaenopsis aphrodite subsp. formosana. Bot. Stud. 2017;58(1):38 doi: 10.1186/s40529-017-0193-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson V.K., Silvester J.A., Fearnley I.M., Leslie A.G., Walker J.E. On the structure of the stator of the mitochondrial atp synthase. EMBO J. 2006;25:2911–2918. doi: 10.1038/sj.emboj.7601177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edera A.A., Gandini C.L., Sanchez-Puerta M.V. Towards a comprehensive picture of C-to-U RNA editing sites in angiosperm mitochondria. Plant Mol. Biol. 2018;97(3):215–231. doi: 10.1007/s11103-018-0734-9. [DOI] [PubMed] [Google Scholar]
- Farajollahi S., Maas S. Molecular diversity through RNA editing: a balancing act. Trends Genet. 2010;26(5):221–230. doi: 10.1016/j.tig.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao W., Liu G., Wang W., Shen W., Zhao Y., Sun J., Yang Q., Zhang Y., Fan W., Pei S., Chen Z., Xu D., Qin T. RNA editing and its roles in plant organelles. Front Genet. 2021;12 doi: 10.3389/fgene.2021.757109. (PMID: 34659369; PMCID: PMC8511385) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X., Liu D., Zheng J., et al. RNA editing analysis of ATP synthase genes in the cotton cytoplasmic male sterile line H276A. Biol. Res. 2019;52:6. doi: 10.1186/s40659-019-0212-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone V., Pogoryelov D., Meier T., Faraldo-gomez J.D. On the principle of ion selectivity in NA+/H+-coupled membrane proteins: experimental and theoretical studies of an ATP synthase rotor. Proc. Natl. Acad. Sci. U.S.A. 2015;112:e1057–e1066. doi: 10.1073/pnas.1421202112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licht K., Jantsch M.F. Rapid and dynamic transcriptome regulation by RNA editing and RNA modifications. J. Cell Biol. 2016;213(1):15–22. doi: 10.1083/jcb.201511041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Giudice C., Hernández I., Ceci L.R., Pesole G., Picardi E. RNA editing in plants: a comprehensive survey of bioinformatics tools and databases. Plant Physiol. Biochem. 2019;137:53–61. doi: 10.1016/j.plaphy.2019.02.001. [DOI] [PubMed] [Google Scholar]
- Lu S., Wang J., Chitsaz F., Derbyshire M.K., Geer R.C., Gonzales N.R., Gwadz M., Hurwitz D.I., Marchler G.H., Song J.S., Thanki N., Yamashita R.A., Yang M., Zhang D., Zheng C., Lanczycki C.J., Marchler-Bauer A. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 2020;48(D1):D265–D268. doi: 10.1093/nar/gkz991. (PMID: 31777944; PMCID: PMC6943070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwiczak J., Winski A., Szczepaniak K., Alva V., Dunin-horkawicz S. Deepcoil-a fast and accurate prediction of coiled-coil domains in protein sequences. Bioinformatics. 2019;35:2790–2795. doi: 10.1093/bioinformatics/bty1062. [DOI] [PubMed] [Google Scholar]
- Maris C., Masse J., Chester A., Navaratnam N., Allain F.H. Structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor. RNA. 2005;11(2):173–186. doi: 10.1261/rna.7190705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moralesrios E., Montgomery M.G., Leslie A.G., Walker J.E. Structure of atp synthase from paracoccus denitrificans determined by x-ray crystallography at 4.0 a resolution. PNAS. 2015;112:13231–13236. doi: 10.1073/pnas.1517542112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy B.J., Klusch N., Langer J., Mills D.J., Yildiz O., Kuhlbrandt W. Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F(1)-F(o) coupling. Science. 2019;364 doi: 10.1126/science.aaw9128. [DOI] [PubMed] [Google Scholar]
- Nirody J.A., Budin I., Rangamani P. Atp synthase: evolution, energetics, and membrane interactions. J. Gen. Physiol. 2020;152 doi: 10.1085/jgp.201912475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K. Editor meets silencer: crosstalk between RNA editing and RNA interference. Nat. Rev. Mol. Cell Biol. 2006;7(12):919–931. doi: 10.1038/nrm2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picardi E., D'Erchia A.M., Gallo A., Montalvo A., Pesole G. Uncovering RNA editing sites in long non-coding RNAs. Front. Bioeng. Biotechnol. 2014;2:64. doi: 10.3389/fbioe.2014.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinke G., Zhou L., Sazanov L.A. Cryo-EM structure of the entire mammalian F-type ATP synthase. Nat. Struct. Mol. Biol. 2020;27:1077–1085. doi: 10.1038/s41594-020-0503-8. [DOI] [PubMed] [Google Scholar]
- Ramadan A.M., Alnufaei A.A., Khan T.K., et al. The first report of RNA U to C or G editing in the mitochondrial NADH dehydrogenase subunit 5 (Nad5) transcript of wild barley. Mol. Biol. Rep. 2021 doi: 10.1007/s11033-021-06609-1. [DOI] [PubMed] [Google Scholar]
- Ramadan A., Alnufaei A.A., Fiaz S., Khan T.K., Hassan S.M. Effect of salinity on ccmfn gene RNA editing of mitochondria in wild barley and uncommon types of RNA editing. Funct. Integr. Genomics. 2023;23(1):50. doi: 10.1007/s10142-023-00978-5. (PMID: 36707470) [DOI] [PubMed] [Google Scholar]
- Rodrigues C.H.M., Pires D.E.V., Ascher D.B. Dynamut2: assessing changes in stability and flexibility upon single and multiple point missense mutations. Protein Sci. 2021;30:60–69. doi: 10.1002/pro.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobti M., Ueno H., Noji H., Stewart A.G. The six steps of the complete f (1)-atpase rotary catalytic cycle. Nat. Commun. 2021;12:4690. doi: 10.1038/s41467-021-25029-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka M., Verbitskiy D., van der Merwe J.A., Zehrmann A., Brennicke A. The process of RNA editing in plant mitochondria. Mitochondrion. 2008;8(1):35–46. doi: 10.1016/j.mito.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Takenaka M., Jörg A., Burger M., Haag S. RNA editing mutants as surrogates for mitochondrial SNP mutants. Plant Physiol. Biochem. 2019;135:310–321. doi: 10.1016/j.plaphy.2018.12.014. [DOI] [PubMed] [Google Scholar]
- Tseng C.C., Lee C.J., Chung Y.T., Sung T.Y., Hsieh M.H. Differential regulation of Arabidopsis plastid gene expression and RNA editing in non-photosynthetic tissues. Plant Mol. Biol. 2013;82(4–5):375–392. doi: 10.1007/s11103-013-0069-5. [DOI] [PubMed] [Google Scholar]
- Tukey J. Comparing individual means in the analysis of variance. Biometrics. 1949;5(2):99–114. [PubMed] [Google Scholar]
- Vinothkumar K.R., Montgomery M.G., Liu S., Walker J.E. Structure of the mitochondrial ATP synthase from Pichia angusta determined by electron cryo-microscopy. PNAS. 2016;113:12709–12714. doi: 10.1073/pnas.1615902113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J.E., Dickson V.K. The peripheral stalk of the mitochondrial ATP synthase. Biochim. Biophys. Acta. 2006;1757:286–296. doi: 10.1016/j.bbabio.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Wang M., Cui L., Feng K., Deng P., Du X., Wan F., Weining S., Nie X. Comparative analysis of Asteraceae chloroplast genomes: structural organization, RNA editing and evolution. Plant Mol Biol Rep. 2015;33(5):1526–1538. [Google Scholar]
- Yuan H., Liu D. Functional disruption of the pentatricopeptide protein SLG1affects mitochondrial RNA editing, plant development, and responses to abiotic stresses in Arabidopsis. Plant J. 2012;70:432–444. doi: 10.1111/j.1365-313X.2011.04883.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinf. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study is included in this published article [and its supplementary information files].