Abstract
The oral bacterium Streptococcus mutans was transformed by electroporation with a shuttle vector (pCSS945) containing insect luciferase gene from a click beetle (Pyrophorus plagiophthalamus) resulting in a bioluminescent phenotype. This S. mutans strain was used in experiments in which light emission was used as a rapid and, compared to conventional CFU counting, more convenient means of estimating the effects of various antimicrobial treatments. The basic parameters affecting in vivo light production by the strain were studied. It was found that pH 6.0 was optimal for incorporation of the substrate d-luciferin for the luciferase reaction. The optimum concentration of d-luciferin was approximately 150 μM at room temperature. Under optimum conditions the light emission in vivo increased rapidly to a constant level and thereafter had a decay of 0.6%/min when logarithmic-growth-phase cells were used. The light emission closely paralleled the numbers of CFU, giving a detectable signal from 30,000 cells and having a dynamic measurement range over 4 log CFU/relative light unit. The cells were treated with various antimicrobial agents, and the emitted bioluminescence was measured. With the bioluminescent measurements, the results were obtained within hours compared to the days required for agar plates, and also, the kinetics of the antibacterial actions could be followed. Thus, the light emission was found to be a reliable, sensitive, and real-time indicator of the bacteriostatic actions of the antimicrobial agents tested.
The gram-positive oral streptococcus Streptococcus mutans is considered the most important cariogenic species in the human oral microbial flora (for a review, see reference 21). Due to the role of this microorganism in the formation of caries lesions, there is constant interest in the antistreptococcal effects of a wide variety of substances, such as antibiotics and oral hygiene products. In our laboratory the susceptibility of S. mutans to innate salivary antimicrobial proteins (lysozyme, lactoferrin, and the peroxidase system of proteins) and the combined effects of these innate factors with immunoglobulin A (IgA) or other antibacterial agents such as fluoride have been studied (19, 20, 22, 25, 28). All such studies generally require methods like plate dilution, which is laborious and time-consuming, especially with mutans group streptococci, for which growth on agar plates normally takes several days.
Insect luciferase is an enzyme produced by, for instance, the Jamaican click beetle (Pyrophorus plagiophthalamus) (24) or the North American firefly (Photinus pyralis) (23). This enzyme uses d-luciferin, O2, and ATP as substrates and produces AMP, oxyluciferin, inorganic pyrophosphate, water, and light (547 to 617 nm) as end products (1). The gene coding for the green light-emitting luciferase from the click beetle has been expressed in Escherichia coli (32) and in insect Spodoptera frugiperda cells (10). The gene for the better-known homolog firefly luciferase already has an established record of being a standard in reporter gene studies and has therefore been expressed in a wide variety of hosts from procaryotic to eucaryotic cells (for a recent review, see reference 8). In each case the new host for the genes was able to produce light which could be very sensitively measured. Therefore, many applications for luminescent cells have been created during the last few years, for instance, an assay for the extremely sensitive detection of the drug susceptibility of Mycobacterium tuberculosis (9). We have used bacteria cloned either with bacterial luciferase (lux) genes or with insect luciferase (luc) genes as general detectors of toxic substances (17), in studies concerning the killing of E. coli cells with nitric oxide-donating chemicals (29), and for the specific detection of heavy metals such as mercuric ions (30).
Due to its linkage to ATP metabolism, luciferase activity in vivo is a good indicator of the intracellular state of cells. In this report we describe the formation of light-emitting S. mutans obtained by transforming the bacteria with a shuttle vector (pCSS945) containing the luciferase gene from P. plagiophtalamus for the fast and sensitive measurement of antibacterial activity. The conditions affecting the light emission of the transformed bacteria and the suitability of using bioluminescent S. mutans to obtain antimicrobial activity measurements are also examined.
MATERIALS AND METHODS
Bacterial strains, plasmids, and reagents.
The bacterial strains and plasmids used in this study are listed in Table 1. All enzymes used in the study except calf intestinal alkaline phosphatase (CIP) were from New England Biolabs (Beverly, Mass.); CIP was from Pharmacia (Uppsala, Sweden). The growth media brain heart infusion (BHI) broth and mitis salivarius agar were from Oxoid (Basingstoke, United Kingdom), and tryptone and yeast extract were from Difco Laboratories (Detroit, Mich.). The antibiotics that we tested were from Sigma (St. Louis, Mo.).
TABLE 1.
Bacterial strains and plasmids used in the study
| Strain or plasmid | Genotype | Reference |
|---|---|---|
| Strains | ||
| E. coli MC 1061 | cI+ Δ(ara leu)7697 ΔlacX74 galU galK hsr hsm+strA araD139 | 2 |
| S. mutans NCTC 10449 | ||
| Plasmids | ||
| p602/22 | Kmr Cmr Ori− Ori+ | 18 |
| pCSS810 | lucGR from P. plagiothalamus Kmr Cmr Ori− Ori+ | 15 |
| pGK13 | Emr Cmr | 27 |
| pCSS945 | lucGR from P. plagiophthalamus Cmr Ori− Ori+ | This study |
Plasmid constructions.
Plasmid pCSS945 containing the luciferase gene from a click beetle (P. plagiophthalamus) was constructed as follows. Plasmid pLucGR (tac) (33) was digested with the restriction enzyme BspHI and filled in with deoxynucleotides and the Klenow fragment of DNA polymerase I. The 1.6-kb DNA fragment was ligated with T4 DNA ligase to the vector p602/22, which was first digested with the restriction enzyme BamHI, and treated with the Klenow enzyme, deoxynucleotides, and CIP. The resulting vector contained the lucGR gene of P. plagiophthalamus under the control of a strong phage T5 promoter and lac operator.
The promoter-operator-luciferase fragment was excised with restriction enzymes XhoI and HindIII, filled in with Klenow enzyme and deoxynucleotides, and ligated to the vector pGK13 which was cut with HindIII, filled in with the Klenow fragment and deoxynucleotides, and treated with CIP. Plasmid pGK13 (27) is a derivative of pGK12. Plasmid pGK13 contains both erythromycin and chloramphenicol resistance markers, like the parent plasmid pGK12, and it has been shown to transform Streptococcus sanguis and Streptococcus pyogenes to erythromycin resistance (27). The ligation mixtures were introduced into electrocompetent E. coli MC1061 cells by electroporation (4) and plated onto L-agar plates (10 g of tryptone, 5 g of yeast extract, 5 g of NaCl, and 15 g of agar in 1 liter) containing chloramphenicol (10 μg/ml). Plasmid minipreparations were made from several colonies and were verified for expected structure by restriction enzyme digestions. One correct structure was named pCSS945.
Transformation.
S. mutans NCTC 10449 cells were transformed with vector pCSS945 by electroporation as follows. The cells were cultivated in BHI broth to an optical density (OD) at 600 nm (OD600) of 0.6, centrifuged at 4,000 × g at 4°C for 15 min, washed twice with ice-cold buffer (10 mM HEPES buffer [pH 7.0] with 15% glycerol), and suspended in 5% sucrose containing 15% glycerol. The electroporation was performed in Bio-Rad (Richmond, Calif.) electroporation cuvettes with a 0.1-cm distance between electrodes with 1 μl of plasmid (10 ng) and 40 μl of ice-cold electrocompetent cells. A single electric pulse of 4.5 ms with settings of 1.25 kV, 25 μF, and 200 Ω was given, and the cells were then rapidly removed from the electroporation apparatus (Bio-Rad) and suspended in 960 μl of BHI broth. After 1 h the cells were plated onto mitis salivarius agar plates containing 10 μg of chloramphenicol and grown for 2 days at 35°C.
Light emission measurements.
Light emission from growing S. mutans cells was measured in vivo by adding 50 μl of substrate solution (1 mM d-luciferin in 0.1 M sodium citrate buffer [pH 5.0]) to 200 μl of cells in a luminometer cuvette. After the addition of substrate solution the cuvette was immediately placed in a manual tube luminometer (1250; LKB-Wallac, Turku, Finland), and light emission was recorded on a chart recorder (LKB-Bromma, Stockholm, Sweden). Some of the light emission measurements were performed with a 96-well microtitration plate luminometer (Luminoscan, Labsystems, Finland) by using a constant measurement mode. If not otherwise stated, all the bioluminescence measurements were performed at room temperature (RT; 20 to 24°C). The light emission as a function of the amount of cells was studied by making dilutions of cell suspensions and measuring both bioluminescence and the numbers of CFU from the samples. To see whether the light emission remains stable for longer periods of time, washed logarithmic-phase cells were stored in buffer (10 mM phosphate buffer containing 74 mM NaCl, 6 mM KCl, 2 mM CaCl2, 0.5 mM MgCl2, 0.5 mM NH4Cl, and 1 g of glucose per liter [pH 6.5]) at different temperatures (0 and 4°C, RT, and 30°C), and the light emission was measured from aliquots withdrawn during 8 h of incubation.
Various parameters were studied for optimized light emission in vivo for S. mutans. The effect of pH was studied by adding a constant amount (500 μM) of d-luciferin to 0.1 M phosphate-citrate buffer adjusted to different pHs ranging from 4.5 to 7.0. The bioluminescence reaction was started by adding 100 μl of substrate buffer to a 100-μl sample of living S. mutans cells. The effects of different amounts of d-luciferin at different pHs were studied similarly by using final d-luciferin concentrations of from 5 to 250 μM.
Testing of antimicrobial agents.
Twofold dilution series were made from all tested antimicrobial agents: chlorhexidine, tetracycline, and penicillin G starting from 10, 0.8, and 0.4 μg/ml, respectively. The bacteria were cultivated to the logarithmic phase and were washed twice with 0.9% saline. To prevent the decrease in bioluminescence due to incubation time and temperature and to create an environment similar to that in the agar plate experiments (see below), the bacteria were finally suspended in BHI broth to approximately 107 cells/ml. A 25-μl sample of this cell suspension, 100 μl of the appropriate antimicrobial dilution, and 25 μl of 1 mM d-luciferin were added to the wells of microtiter plates. The plates were incubated at 35°C, and the emitted light was measured after 0, 1, 2, 3, and 4 h with a 1450 Microbeta plus liquid scintillation counter (Wallac, Turku, Finland). The same dilutions of antimicrobial agents were also used to test for growth inhibition on agar plates. An aliquot of 20 μl of a solution of logarithmic-phase bacteria was spread evenly onto a BHI agar plate, and pieces of glass fiber filters containing the antimicrobial solution or 0.9% NaCl for the controls were placed on the bacteria. After 48 h at 35°C the growth inhibition was visually examined.
RESULTS
Light emission measurements.
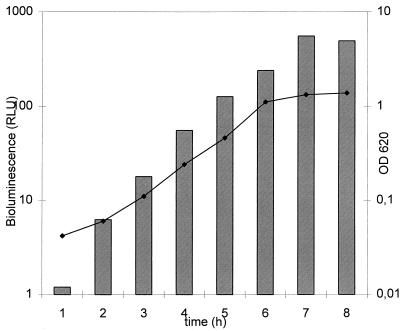
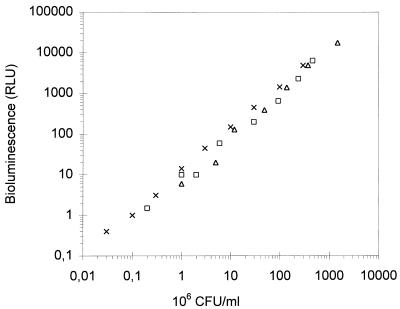
Expression of the luc gene is controlled by a constitutive phage T5 promoter which was found to work in S. mutans, and the light emission measured as a function of cell growth is shown in Fig. 1. The bioluminescence closely correlates with the OD of the culture and also with the cell number over a 4-log range (Fig. 2). The bacterial cells could be stored in buffer for at least 8 h at 4°C without notable changes in light emission, whereas at RT and 30°C the bioluminescence decreased 29 and 62%, respectively, after 1 h of incubation (Table 2).
FIG. 1.
Growth curve for recombinant S. mutans as a function of time and bioluminescence. The growth of S. mutans cells in BHI broth containing 10 μg of chloramphenicol per ml was followed with a spectrophotometer (OD600; curve) and a manual luminometer (relative light units [RLU]; bars) after the addition of d-luciferin at 200 μM and pH 5.0. The values are means of two independent experiments. The variation between the measurements was less than 10%.
FIG. 2.
Linear relationship between amount of cells and the bioluminescence that was produced. Samples from logarithmic-phase bacteria were serially diluted, and light emission (relative light units [RLU]) was determined after the addition of 200 μM d-luciferin. From the same samples the numbers of CFU were also calculated on agar plates. Each symbol represents the results of one independent experiment.
TABLE 2.
Emission of light by S. mutans cells as a function of storage time at different temperaturesa
| Temp (°C) | RLU at the following times (h)b:
|
|||||
|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 5 | 8 | |
| 0 | 38.2 | 40.7 | 40.2 | 36.4 | 29.0 | 24.2 |
| 4 | 37.8 | 39.8 | 41.6 | 44.2 | 36.6 | 29.9 |
| RT | 39.1 | 27.9 | 22.4 | 16.5 | 7.4 | 1.0 |
| 30 | 38.3 | 14.7 | 7.8 | 1.5 | 3.3 | 0.3 |
The bacteria were stored in buffer and were incubated at different temperatures. Aliquots were withdrawn at the indicated time intervals, and the bioluminescence of the bacteria was measured at RT.
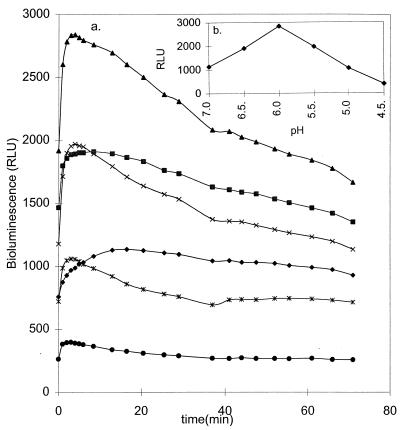
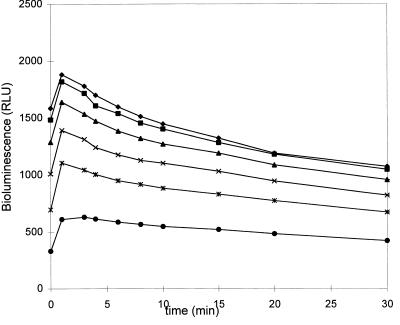
As seen in Fig. 3, pH has a strong effect on the light emission of logarithmic-phase bacteria and pH 6.0 gives the highest emission, with a decay of light of 0.6%/min. Saturating levels of substrate were obtained at about 125 μM for all pHs tested (5.5, 6.0, and 6.5), and again, the cells gave the highest light emission at pH 6.0 (Table 3). The kinetics of light emission of logarithmic-growth-phase cells as a function of the d-luciferin concentration at pH 6.0 are presented in Fig. 4. The kinetics were affected by the growth phase. The decay times became faster as the cells became older (data not shown).
FIG. 3.
Light emission as a function of time and pH. The bioluminescence (in relative light units [RLU]) of logarithmic-phase S. mutans cells was measured at pH 4.5 (•), 5.0 (∗), 5.5 (×), 6.0 (▴), 6.5 (■), and 7.0 (⧫) after the addition of 200 μM d-luciferin (a). (b) Peak relative light units as a function of pH. For details, see Materials and Methods. The values are the means of two independent measurements. The variation between the measurements was less than 10%.
TABLE 3.
Effects of d-luciferin concentrations on bioluminescence of S. mutans cells at pH 5.5, 6.0, and 6.5a
| pH | RLU in the presence of the following d-luciferin concn (μM)b:
|
|||||
|---|---|---|---|---|---|---|
| 5.0 | 12.5 | 25 | 50 | 125 | 250 | |
| 5.5 | 8.8 | 11.2 | 11.8 | 12.8 | 13.3 | 11.9 |
| 6.0 | 6.3 | 10.4 | 13.1 | 15.4 | 17.1 | 17.8 |
| 6.5 | 3.2 | 6.3 | 9.2 | 12.3 | 16.6 | 16.3 |
Different amounts of d-luciferin were added to the S. mutans cells, and the bioluminescence was measured.
The results are the means of two independent measurements. Variation between the measurements was less than 10%.
FIG. 4.
Decay of light emission of logarithmic-phase S. mutans cells at pH 6.0 with different substrate concentrations: 5 (•), 12,5 (∗), 25 (×), 50 (▴), 125 (■), and 250 (⧫) μM. The bioluminescent reaction in cells was triggered by the addition of different concentrations of d-luciferin, and the light emission of the cells was immediately measured with a microtitration plate luminometer. The values are the means of two independent measurements. The variation between the measurements was less than 10%. RLU, relative light units.
Effects of antimicrobial agents.
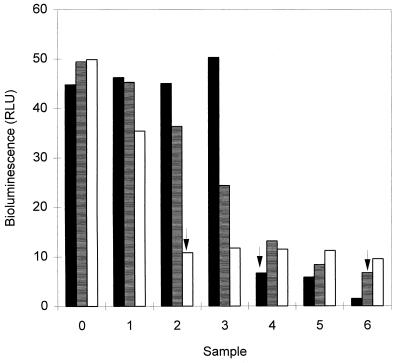
All antimicrobial agents tested clearly inhibited both bacterial light emission and growth on agar plates at the concentrations tested. The inhibition of light emission was time (Table 4) and dose (Fig. 5) dependent and was already seen after 1 h of incubation but was more pronounced after 3 and 4 h. The MICs of chlorhexidine, tetracycline, and penicillin on agar plates were 2.5, 0.8, and 0.025 μg/ml, respectively. The same concentrations of chlorhexidine and penicillin affected both the light emission and growth on agar plates, but lower concentrations of tetracycline were needed to inhibit the light emission than to inhibit growth.
TABLE 4.
Effects of various antimicrobial compounds on the bioluminescence of recombinant S. mutans sensor bacteriaa
| Test agent (concn [μg/ml]) | % of bioluminescence of nontreated bacteria at the following times (h)b:
|
||||
|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | |
| Chlorhexidine (10) | 91 | 39.0 | 15.6 | 5.6 | 3.3 |
| Tetracycline (0.8) | 86 | 55.4 | 31.0 | 17.9 | 13.4 |
| Penicillin (0.4) | 91 | 87.4 | 56.6 | 27.9 | 19.7 |
The bacteria were incubated with the antimicrobial compounds at 35°C in BHI broth, and the bioluminescence was measured at the indicated time intervals.
The results are the means of two independent measurements. Variation between the measurements was less than 10%.
FIG. 5.
Effects of different amounts of chlorhexidine (■), tetracycline (▤), and penicillin (□) on the light emission (relative light units [RLU]) of the bacteria. Twofold dilutions of each antimicrobial agent were made, starting from 10.0 μg/ml for chlorhexidine, 0.08 μg/ml for tetracycline, and 0.40 μg/ml for penicillin (sample 6), and the antimicrobial agents were incubated with the bacteria for 4 h at 35°C in BHI broth. The sample labelled 0 is the nontreated control. The values are the means of two independent measurements. The variation between the measurements was less than 10%. Arrows indicate the concentrations that inhibited growth on agar plates.
DISCUSSION
To our knowledge S. mutans strains have not previously been transformed by electroporation, which in this study was found to be a suitable method for the incorporation of foreign DNA into strain NCTC 10449. The conditions similar to those used with other streptococci (5) also functioned well with S. mutans.
ATP is one of the substrates of the luciferase reaction, and each cell contains a constant intracellular ATP pool which is effectively regulated. The light emission in vivo is therefore a very sensitive indicator of the intracellular state of the cells (26). We have earlier shown that the substrate for the luciferase reaction does not readily pass procaryotic or eucaryotic cell membranes at physiological pH but is efficiently incorporated under slightly acidic conditions in sodium citrate buffer (31), consistent with the results of Wood and DeLuca (32). The optimum pH for E. coli and Bacillus subtilis was shown to be pH 5.0 (16). We tested here the penetration of d-luciferin into S. mutans cells and found the optimum to be at pH 6.0, in contrast to that for another gram-positive bacterium, B. subtilis. At physiological pH (pH 7.0) the substrate also penetrated the membranes of S. mutans cells.
The light emission of growing S. mutans cells correlated well with the OD660 as well as the numbers of CFU. However, the kinetics of in vivo bioluminescence were affected by the growth phase. The older the cells became the faster the decay times were, giving an indication of decreased intracellular ATP pools for the luciferase reaction. We have shown earlier that E. coli cloned with Vibrio harveyi luciferase genes luxA and luxB emits bioluminescence at a level that is directly proportional to the numbers of CFU, and hence, a linear relationship can be obtained (11, 14). In the latter study the minimum number of cells detected by their bioluminescent phenotype was 1,000. In this study the linear relationship of bioluminescence versus CFU for the S. mutans cells was shown to give a dynamic measurement range of over 4 logs, and the minimum amount of cells detected over the background amount was roughly 30,000 cells. The same shuttle plasmid construct, pCSS945, in E. coli allows the detection of fewer than 100 cells, indicating that the phage T5 promoter controlling insect luciferase expression is not as strong in S. mutans as it is in E. coli. The extremely high level of expression in E. coli is also reflected in the very slow growth rate and the small colony size on agar plates (unpublished data), but the moderate level of expression in S. mutans keeps these cells in good physiological condition, and hence, the effects of the different antimicrobial treatments shown in this study are probably equal to those for the non-plasmid-bearing cells. Moreover, since the physiological state of the cells remains quite normal, it can be speculated that the cells would also be useful in other studies of agents with activity against streptococci, such as antiadherence studies. The ability of the bacteria to adhere on the hard surfaces is thought to be one of the virulence factors of mutans group streptococci, and the effects of different treatments on the adherence properties of the bacteria are generally studied by the method of Clark et al. (3) with 35S- or 3H-labelled bacteria. The bioluminescent S. mutans phenotype could replace the labelled bacteria, and thus, the use of the radiolabelled compounds in obtaining the adherence measurements could be avoided.
The moderate level of expression can have an influence on the stability of light emission. Cells kept in buffer at 0 or 4°C gave a rather constant level of light emission rate during a prolonged incubation period. Analytically, this situation is good since reproducible and comparable results can be obtained from experiments performed during 1 working day. The higher storage temperatures (RT and 30°C) induced a rapid decrease in the light emission of the bacteria which was probably due to a decrease in the cellular ATP content (7) rather than the proteolytic degradation of the luciferase enzyme or cell death.
An important factor in creating bioluminescent sensor cells is the light emission kinetics as a function of substrate concentration and pH. The decay of the bioluminescence of 0.6%/min at pH 6.0, as measured for S. mutans in this study, is very reasonable and simplifies the analytical performance of the assays. For instance, it is possible for investigators to use a variety of different light-measuring instruments, from fully automated luminometers with substrate dispensers to liquid scintillation counters with a slow rate of transfer of the measuring cuvette in front of the photomultiplier tube.
Luminescent E. coli has previously been used successfully for the measurement of antibacterial activity (29). The light-emitting S. mutans cells also functioned well as biosensors of the antibacterial actions of the antimicrobial agents tested. All antimicrobial agents tested, i.e., penicillin, chlorhexidine, and tetracycline, have different mechanisms of action (12, 13), and especially with tetracycline, which does not cause immediate cell death but which, instead, inhibits the protein synthesis in a cell, lower concentrations clearly affected the light emission but not the growth on the agar plates, indicating disturbances in the intracellular state long before the actual growth is inhibited. With chlorhexidine and penicillin, the bioluminescence and agar plate results correlated well. Thus, the measurement of bioluminescence was found to be a simple, fast, and reliable method for the detection of the antibacterial effects of substances with different mechanisms of action. Moreover, with bioluminescence measurements, the kinetics of the antibacterial effect could also be followed.
However, the ATP content of the S. mutans cells, and thus the emitted light, can decrease without affecting the viability of the cells (6, 7). Thus, by measuring the light emission only, the actual killing of bacteria cannot be determined; rather, the “well-being” of a cell is determined. Greger et al. (6) found that incubation of S. mutans cells in buffer (pH 5.0) with 1 mM sodium fluoride causes a rapid decrease in the cellular ATP content but does not affect the viability for 4 h. We have previously shown that in saliva at pH 5.0 such NaF concentrations reduce the viability of S. mutans during 20 h of incubation (20). We have also measured the light emission of NaF-saliva-bacteria suspensions and found that the bioluminescence has already decreased after 10 min of incubation, while the viability is not affected during the first hours (unpublished data). Thus, a decrease in ATP content does not inevitably cause immediate cell death, but during longer incubation times, without a possibility of recovery, cell death may occur.
To summarize, S. mutans NCTC 10449 cells were found to be transformable by electroporation with a shuttle vector (pCSS945) containing insect luciferase gene resulting in a bioluminescent phenotype. The optimal conditions for light emission by S. mutans were determined, and the bioluminescent bacteria were found to be a versatile tool for the simple, rapid, and sensitive screening of the antimicrobial activities of substances with different mechanisms of action, for example, in human saliva.
ACKNOWLEDGMENTS
M. Virta is acknowledged for suggestions concerning using luciferase genes for this study.
This study was supported by the Academy of Finland.
REFERENCES
- 1.Biggley W H, Lloyd J E, Seliger H H. The spectral distribution of firefly light II. J Gen Physiol. 1967;50:1681–1692. doi: 10.1085/jgp.50.6.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casabadan M J, Cohen S N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1978;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 3.Clark W B, Bamman L L, Greenall C, Kemeny D M. Comparative estimates of bacterial affinities and adsorption sites on hydroxyapatite surfaces. Infect Immun. 1978;19:1040–1047. doi: 10.1128/iai.19.3.846-853.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dower W J, Miller J F, Ragsdale C W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6126–6144. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunny G M, Lee L N, LeBlanc D J. Improved electroporation and cloning vector system for gram-positive bacteria. Appl Environ Microbiol. 1991;57:1194–1201. doi: 10.1128/aem.57.4.1194-1201.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greger J E G, Izaguirre-Fernandez E J, Eisenberg A D. Adenosine 5′-triphosphate content of Streptococcus mutans GS-5 during fluoride mediated death at low pH. Caries Res. 1985;19:307–313. doi: 10.1159/000260860. [DOI] [PubMed] [Google Scholar]
- 7.Greger J E G, Eisenberg A D. Adenosine 5′-trisphosphate content of Streptococcus mutans GS-5 during starvation in a buffered salt medium. Caries Res. 1985;19:314–319. doi: 10.1159/000260861. [DOI] [PubMed] [Google Scholar]
- 8.Hill P J, Stewart G S A B, Stanley P E. Bioluminescence and chemiluminescence literature—luciferase reporter genes—lux and luc. J Biolumin Chemilumin. 1993;8:267–291. doi: 10.1002/bio.1170080507. [DOI] [PubMed] [Google Scholar]
- 9.Jacobs W R, Jr, Barletta R G, Udani R, Chan J, Kalkut G, Sosne G, Kieser T, Sarkis G J, Hatfull G F, Bloom B R. Rapid assessment of drug susceptibilities of Mycobacterium tuberculosis by means of luciferase reporter phages. Science. 1993;260:819–822. doi: 10.1126/science.8484123. [DOI] [PubMed] [Google Scholar]
- 10.Karp M, Åkerman K, Lindqvist C, Kuusisto A, Saviranta P, Oker-Blom C. A sensitive model system for in vivo monitoring of baculovirus gene expression in single infected insect cells. Bio/Technology. 1992;10:565–569. doi: 10.1038/nbt0592-565. [DOI] [PubMed] [Google Scholar]
- 11.Korpela M, Karp M. Stable-light producing Escherichia coli. Biotechnol Lett. 1988;10:383–388. [Google Scholar]
- 12.Kral T A, Callaway M D. Penicillin-induced lysis of Streptococcus mutans. Infect Immun. 1984;46:442–447. doi: 10.1128/iai.46.2.442-447.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuyyakanond T, Quesnel L B. The mechanism of action of chlorhexidine. FEMS Microbiol Lett. 1992;79:211–215. doi: 10.1111/j.1574-6968.1992.tb14042.x. [DOI] [PubMed] [Google Scholar]
- 14.Lampinen J, Korpela M, Saviranta P, Kroneld R, Karp M. Use of Escherichia coli cloned with genes encoding bacterial luciferase for evaluation of chemical toxicity. Toxic Assess. 1990;5:337–350. [Google Scholar]
- 15.Lampinen J, Koivisto L, Wahlsten M, Mäntsälä P, Karp M. Expression of luciferase genes from different origins in Bacillus subtilis. Mol Gen Genet. 1992;232:498–504. doi: 10.1007/BF00266255. [DOI] [PubMed] [Google Scholar]
- 16.Lampinen J. Development of recombinant bacterial strains for cytotoxicity testing. Ph.D. thesis. Ser. A1, Tom. 205. Turku, Finland: University of Turku, Annales Universitatis Turkuensis; 1995. [Google Scholar]
- 17.Lampinen J, Virta M, Karp M. Comparison of gram positive and gram negative bacterial strains cloned with different types of luciferase genes in bioluminescence cytotoxicity tests. Environ Toxicol Water Qual. 1995;10:157–166. [Google Scholar]
- 18.LeGrice S F J, Beuck V, Mous J. Expression of biologically active human T-cell lymphotrophic virus type III reverse transcriptase in Bacillus subtilis. Gene. 1987;55:95–103. doi: 10.1016/0378-1119(87)90252-6. [DOI] [PubMed] [Google Scholar]
- 19.Lenander-Lumikari M, Månsson-Rahemtulla B, Rahemtulla F. Lysozyme enhances the inhibitory effects of the peroxidase system on glucose metabolism of Streptococcus mutans. J Dent Res. 1992;71:484–490. doi: 10.1177/00220345920710031201. [DOI] [PubMed] [Google Scholar]
- 20.Lenander-Lumikari M, Loimaranta V, Hannuksela S, Tenovuo J, Ekstrand J. Combined inhibitory effect of fluoride and hypothiocyanite on the viability and glucose metabolism of Streptococcus mutans, serotype c. Oral Microbiol Immunol. 1997;12:231–235. doi: 10.1111/j.1399-302x.1997.tb00384.x. [DOI] [PubMed] [Google Scholar]
- 21.Loesche W J. Role of Streptococcus mutans in human dental decay. Microbiol Rev. 1986;50:353–380. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lumikari M, Soukka T, Nurmio S, Tenovuo J. Inhibition of growth of Streptococcus mutans, S. sobrinus and Lactobacillus casei by oral peroxidase systems in human saliva. Arch Oral Biol. 1991;36:155–160. doi: 10.1016/0003-9969(91)90078-9. [DOI] [PubMed] [Google Scholar]
- 23.McElroy W D, Seliger H H. Spectral emission and quantum yield of firefly bioluminescence. Arch Biochem Biophys. 1960;88:136. doi: 10.1016/0003-9861(60)90208-3. [DOI] [PubMed] [Google Scholar]
- 24.Seliger H H, Buck J B, Fastie W G, McElroy W D. The spectral distribution of firefly light. J Gen Physiol. 1964;48:95–104. doi: 10.1085/jgp.48.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soukka T, Lumikari M, Tenovuo J. Combined inhibitory effect of lactoferrin and lactoperoxidase system on the viability of Streptococcus mutans, serotype c. Scand J Dent Res. 1991;99:390–396. doi: 10.1111/j.1600-0722.1991.tb01046.x. [DOI] [PubMed] [Google Scholar]
- 26.Stanley P E. A review of bioluminescent ATP techniques in rapid microbiology. J Biolumin Chemilumin. 1989;4:375–380. doi: 10.1002/bio.1170040151. [DOI] [PubMed] [Google Scholar]
- 27.Suvorov A, Kok J, Venema G. Transformation of group A streptococci by electroporation. FEMS Microbiol Lett. 1988;56:95–100. [Google Scholar]
- 28.Tenovuo J, Moldoveanu Z, Mestecky J, Pruitt K M, Mansson-Rahemtulla B. Interaction of specific and innate factors of immunity: IgA enhances the antimicrobial effect of the lactoperoxidase system against Streptococcus mutans. J Immunol. 1982;128:726–731. [PubMed] [Google Scholar]
- 29.Virta M, Karp M, Vuorinen P. Nitric oxide donor-mediated killing of bioluminescent Escherichia coli. Antimicrob Agents Chemother. 1994;38:2775–2779. doi: 10.1128/aac.38.12.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Virta M, Lampinen J, Karp M. A luminescence-based mercury biosensor. Anal Chem. 1995;34:667–669. [Google Scholar]
- 31.Virta M, Åkerman K E O, Saviranta P, Oker-Blom C, Karp M T. Real-time measurement of cell permeabilization with low-molecular weight membranolytic agents. J Antimicrob Chemother. 1995;36:303–315. doi: 10.1093/jac/36.2.303. [DOI] [PubMed] [Google Scholar]
- 32.Wood K V, DeLuca M. Photographic detection of luminescence in Escherichia coli containing the gene for firefly luciferase. Anal Biochem. 1987;161:501–507. doi: 10.1016/0003-2697(87)90480-5. [DOI] [PubMed] [Google Scholar]
- 33.Wood K V, Lam Y A, Seliger H H, McElroy W D. Complementary DNA coding beetle luciferases can elicit bioluminescence of different colors. Science. 1989;244:700–702. doi: 10.1126/science.2655091. [DOI] [PubMed] [Google Scholar]