Abstract
Background
Gu-ben-hua-shi (AESS) formula is a clinical experienced prescription from Guangdong Hospital of Traditional Chinese Medicine (TCM), which is used to treat atopic dermatitis (AD). Our previous work has shown that AESS has therapeutic effect on AD by regulating yes-associated protein (YAP). AESS formula has multi-component and multi-target characteristic, and need to be analyzed by systematic chemical profiling and network pharmacology technology, as well as verification of key signaling pathways. Therefore, this study aimed at investigating the efficacy and effect of AESS formula in the treatment of AD and its effect on NLRP3 signaling pathway.
Methods
The components of AESS formula were analyzed and identified by ultra high performance liquid chromatography/tandem mass spectrometry (UHPLC- MS/MS), and the potential mechanism of AESS formula in the treatment of AD was predicted by network pharmacology approach, with detected main components, and the potential components targeted NOD-like receptor thermal protein domain associated protein (NLRP3) signaling pathway [Direct binding with NLRP3, apoptosis-associated speck-like protein (ASC) and Caspase-1] were assessed using molecular docking. AD-like symptoms were constructed by DNCB induced BALB/c mice. The effect of AESS formula on dorsal skin structure in AD-like mice was observed using H&E staining. Furthermore, the western blotting experiment explored the expression of the NLRP3 pathway protein.
Results
By UHPLC-MS/MS analysis, 91 compounds were detected in AESS formula, and 76 of them were identified, while by network pharmacological analysis, 1500 component targets were obtained, and 257 of them were obtained by intersection with eczema targets. Then one of the key pathways, nucleotide-binding oligomerization domain (NOD)-like signaling pathway was obtained by KEGG enrichment analysis. Molecular docking results showed 24 main components could effectively combine with ASC and Caspase-1 (≤−7 kcal/mol). The animal experiment results further showed that AESS formula alleviates symptoms in AD-like mice. ELISA kit results showed that the expression of IL-1β and IL-18 in serum was inhibited after AESS treatment. Additionally, western blotting analysis showed that the expressions of ASC, Caspase-1 and NLRP3 protein expression in the skin tissue of mice were down-regulated after AESS treatment. The experimental results show that AESS formula inhibited the expression of NLRP3 signaling pathway for the treatment of AD.
Conclusions
AESS formula can improve AD symptoms in mice by inhibiting the activation of NLRP3 inflammasome and the expression of the related downstream inflammatory cytokines.
Keywords: Atopic dermatitis, Traditional Chinese medicine, UHPLC-MS/MS, NLRP3 signaling pathway
1. Introduction
Atopic dermatitis (AD), also known as eczema, is a common clinical inflammatory skin disease caused by a variety of factors, clinically manifested by severe pruritus, erythema, papules, lichen skin lesions and so on (Jiang et al., 2018). Due to the rapid growth of population and the aggravation of environmental pollution, the incidence of AD has increased. Each year, up to 17.1% of adults and 22.6% of children were diagnosed with AD, including 9.6% new cases of AD in children (Bylund et al., 2020). So the prevention and treatment of AD is one of the hot topics of medical focus.
It is reported that nucleotide-binding and oligomerization domain-like receptors (NLRs) play essential roles in various autoimmune diseases. NLRs are divided into five subfamilies, namely NLRA, NLRB, NLRC, NLRP and NLRX. NLRP and NLRC are the two major subfamilies that are found to be relevant to allergic diseases, including AD and allergic asthma, while NLRP3 is the most important inflammasome in the NLRP subfamily. NLRP3 usually forms inflammatory bodies with the adaptor protein apoptosis-associated speck-like protein (ASC) and an inactive zymogen pro-Caspase-1 (Tsang et al., 2021). The assembled inflammasome induces the processing of the effector protein Caspase-1 and subsequently promotes the cleavage of inactive precursors pro-IL-1β and pro-IL-18 into pro-inflammatory cytokines IL-1β and IL-18 (Swanson et al., 2019). IL-1β is significantly elevated in skin blisters of patients with AD that have filaggrin gene (FLG) mutation and and in the skin of Flgft/ft mice (Schwartz et al., 2018), and IL-18 level was found to be elevated in the serum of AD patients and NC/Nga mice during the development of AD (Tanaka et al., 2001). It has been demonstrated that activating the NLRP3 inflammatories in keratinocytes and promoting the release of downstream pro-inflammatory cytokines may be a risk factor for AD (Zheng et al., 2021). Therefore, inhibition of NLRP3 inflammasome may be an effective treatment strategy for AD.
At present, the clinical drugs treatment of AD is mainly divided into chemical synthetic drugs and traditional folk medicine. Chemical medicine usually uses topical drugs, such as glucocorticoids for external use, which has anti-inflammatory, anti-allergic, and immunosuppressive effects, may cause serious side effects, such as sleep disorders, skin pain, drug dependence, etc. Therefore, an increasing number of patients are asking for plant-based therapeutic products as complementary dermatologic therapy (Reuter et al., 2010). Traditional Chinese Medicine (TCM) is an alternative therapy that can be used in the treatment for dermatologic disorders, including AD and psoriasis (Koo et al., 2003). The Gu-ben-hua-shi (AESS) formula is the clinical experience prescription of professor Chen Dacan from Guangdong Hospital of TCM, and has been used to treat AD. According to TCM theory, Lonicerae Japonicae Flos was acted as the ‘Monarch drug’, Sophorae Flos and Curcumae Rhizoma were acted as ‘minister drugs’, Atractylodis Macrocephalae Rhizoma, Atractylodis Rhizoma, Rehmanniae Radix and Coicis Semen were acted as ‘assistant drug’, Saposhnikoviae Radix was acted as the ‘guide drug’, but the specific mechanism of action is still unclear. Our team's previous work has shown that AESS formula can regulate Th1/Th2/Th17/Treg immune balance and through the interaction of YAP and NF-κB signaling pathway (Jia et al., 2022). The purpose of this study was to further investigate the therapeutic material basis and intervention mechanism of AESS formula in the treatment of AD, and to verify the mechanism through network pharmacology and pharmacological experiments in vivo.
2. Material and methods
2.1. Preparation of AESS decoction
The prescribed herbs were purchased from different Chinese herbal medicine markets (Table 1). The plant names in AESS are checked with https://www.theplantlist.org. The standard information is shown in Supplemental Table S1. According to traditional decoction preparation process, the herbs were added to 10 times volume of water and boiled for 1 h, then concentrated at 60 °C under reduced pressure, and freeze-dried to obtain lyophilized powder. 5.0 mg of the freeze-dried powder was accurately weighed, 80% MeOH was added, and dissolved by ultrasound for 30 min. Then the final solvent was diluted with equal volume ultra-pure water, and the final solvent was 40% MeOH. After centrifugation at 13000 rpm for 10 min, the supernatant solution was filtered through a 0.22 µm filter membrane, and taken into sample bottle (5 mg/ml). Then placed at 4 °C for use. The standard was accurately weighed and prepared into 5 μg/ml standard solution with 40%MeOH, which was placed at 4 °C for reserve.
Table 1.
The summarised information of composed herbs for AESS.
| Botanical name | English name | Batch number | Place of origin | Dose (/g)* |
|---|---|---|---|---|
| Saposhnikovia divaricata (Turcz. ex Ledeb.) Schischk. | Saposhnikoviae Radix | 2007001 | Heilongjiang province | 10 |
| Coix lacryma-jobi L. var. mayuen (Roman.) Stapf | Coicis Semen | 181105921 | Guizhou province | 20 |
| Curcuma kwangsiensis S.G.Lee & C.F.Liang | Curcumae Rhizoma | 191203761 | Guangxi province | 10 |
| Atractylodes macrocephala Koidz. | Atractylodis Macrocephalae Rhizoma | 1810002 | Zhejiang province | 15 |
| Lonicera japonica Thunb. | Lonicerae Japonicae Flos | 1911002 | Shandong province | 10 |
| Rehmannia glutinosa Libosch. | Rehmanniae Radix | 191000039 | Henan province | 10 |
| Sophora japonica L | Sophorae Flos | 200901 | Jiangsu province | 10 |
| Atractylodes lancea (Thunb.) D C. | Atractylodis Rhizoma | 191004471 | Inner Mongolia Autonomous Region | 10 |
Drug usage (g) for each herb in AESS formula.
2.2. UHPLC-MS conditions
An Dionex Ultimate HPLC 3000 coupled with QE-Orbitrap mass spectrometer was used in the analytical method development. (Thermo-Fisher, USA). Chromatographic methanol and acetonitrile were produced by Merck (Darmstadt, Germany), and formic acid was produced by Kermeo Chemical Reagent Co., Ltd. (Tianjin,China). The Hypersil GOLD (100 × 2.1 mm, 1.9 μm) column (Thermo Fisher Scientific,USA) was used. Mobile phase: 0.1% formic acid water (A)–acetonitrile (B). Gradient elution: 0–2 min, 5%B; 2–42 min, 5–95%B; 42–47 min, 95%B; 47–47.1 min, 95–5%B; 47.1–50 min, 5%B. Flow rate: 0.3 ml/min. The sample injection volume was 5 μL. The temperature of the column was 40 °C. The mass spectrometer was equipped with an electrospray ionization (ESI) interface operated in both positive and negative ion mode. Scanning range: 100.0–1500.0 m/z. Ion source temperature: 320 °C; The resolution was set at 70,000; The capillary voltage was 3.5 kV in the (+) ESI mode and −3.2 kV in the (−) ESI mode.
2.3. Network pharmacological analysis
2.3.1. Collection and screening of active components and targets of AESS
According to the results of composition analysis of serum after administration (Jia et al., 2022) and the qualitative results of UHPLC-MS/MS analysis, 37 compounds were selected for network pharmacological analysis. Component targets were collected using TCMSP (https://old.tcmspe.com/tcmspsearch.php/), SymMap (https://www.symmap.org/search/), BATMAN-TCM (https://bionet.ncpsb.org.cn/batman-tcm/), Swiss Target Pred-iction (https://www.swissadme.ch/) and TCMIP (https://www.tcmip.cn/TCMIP/index.php/Home/Analysis/personal_center.html). Then input them into Uniport database (https://www.uniprot.org/) to convert the target protein information into standard gene marker na-mes.
2.3.2. Collection and screening of AD targets
The disease target of AD was obtained from GeneCards database (https://www.genecards.org/), DisGeNET database (https://www.disgenet.org/), TTD database (https://data-base.idrb.cqu.edu.cn/), TCMIP database and CTD database (https://ctdbase.org) with the keyword “eczema” as the search term. Then input them into Uniport database (https://www.uniprot.org/) to convert the target protein information into standard gene marker names.
2.3.3. Construction of protein–protein interaction network (PPI)
Input the disease target and component target into the draw Venn Diagram website (https://bioinformatics.psb.ugent.be/webtools/Venn/), establish the Venn Diagram and obtain the intersection targets. The intersection targets of AESS were analyzed in the STRING database (https://string-db.org), selected the Multiple proteins option, and Homo sapiens in Organization, adjusted the confidence to 0.7 and hid the disconnected nodes in the network. Then obtained the PPI and downloaded the target interaction network diagram and data. Input it into Cytoscape 3.8.2 software and used Network Analyzer function to analyze network topology.
2.3.4. GO enrichment analysis and KEGG enrichment analysis
GO and KEGG enrichment analysis of key target proteins were performed using DAVID database (https://david.ncifcrf.gov/), according to P < 0.05 was screened, and the results were analyzed visually.
2.3.5. Network of herbs -active components -targets -main pathways of AESS
The herbs, components, key targets, and key pathways were input into Cytoscape 3.8.2 software to build the “herbs- components-targets-pathways” Network, and Network Analyzer was used to analyze the network topology.
2.3.6. Molecular docking
The 3D structure of target protein, human NLRP3 (Uni Prot ID:Q96P20), human ASC (Uni Prot ID: Q9ULZ3) and Caspase-1 (Uni Prot ID: P29466), were downloaded from the Uni Prot database (https://www.uniprot.org), then the software Pymol was used to remove ligands and solvent molecules, and AutoDock4.2 software was used for hydrogenation and electron addition. The binding energy was defined as ≤−7 kcal/mol as the screening criterion.
2.4. Experimental verification of influence of NLRP3 pathway in AD
2.4.1. Animals
48 SPF healthy male BALB/c mice (20 ± 2 g, age: 6–8 weeks) were raised in Guangdong Provincial Hospital of Chinese Medicine, and the temperature was maintained at 22 ± 3 °C, and the relative humidity was 55 ± 5%. Adaptive feeding was carried out seven days before the experiment, and water and food were freely eaten. Random number method was used to group 6 groups, each group of 8 mice. It included control group, model group (AD), AESS low dose group (AESS0.5), AESS medium dose group (AESS1), AESS high dose group (AESS2) and dexamethasone group (DEX). The experimental animal ethics committee of the GuangdongProvincial Hospital of Chinese Medicine approved all experiments (No. 2021086).
2.4.2. Establishment of the AD-like mouse model
Before modeling, all mice's back skin tissues were depilated. After hair removal, the back skin was sensitized with 200 μL 1% DNCB (Sigma-Aldrich, USA) dissolved in acetone-olive oil (AO, 3:1). Control group were treated with vehicle. Then, the back skin was then sensitized with 200 µL 0.5% DNCB three times a week. The dosage of AESS formula is 14.41/kg/d, which is set as the middle dosage group, the low dosage group is 1/2 of the middle dosage group, and the high dosage group is 2 times of the middle dosage group. The positive group was given dexamethasone solution, 3.0 mg/kg orally daily. Start with the first stimulation once a day for 3 weeks. The changes of body weight and dermatitis score of mice were monitored during modeling.
2.4.3. Experimental model evaluation
The changes of body weight and dermatitis score of mice were monitored during modeling. On the 1st, 7th, 14th and 21st days of the experiment, the back skin lesions of mice in each group were observed, including erythema/haemorrhage, oedema, excoriation/erosion, and scaling/dryness and photographed and recorded. The severity of back lesions was graded on the following scale: 0 = none, 1 = mild, 2 = moderate, 3 = s evere.
2.4.4. Hematoxylin-eosin (H&E) staining
The back skin of mice in each group was fixed in 4% paraformaldehyde (the ratio of skin to fixed liquid was 1:8–1:10), and sent to the company for routine dehydration, fixation, paraffin embedding, sectioning, H&E staining, sealing, observation under the microscope, photo preservation.
2.4.5. Enzyme linked immunosorbent assay (ELISA)
According to the ELISA kit instructions, after the reaction was terminated, the optical density (OD value) of each hole was measured with an enzyme marker at 450 nm, and the data was input into Origin 2018 to establish a standard curve, calculate the sample concentration and statistical results.
2.4.6. Western blotting analysis
A certain amount of skin tissue was weighed and cleaved on ice with RIPA lysate containing PMSF for 30 min and centrifuged at 13000 rpm at 4 °C for 15 min. The protein concentration in skin tissue was determined by BCA protein concentration determination kit. After denatalization, the protein was separated by SDS-PAGE and transferred to PVDF membrane. 5% skim milk (dissolved in BSA reagent) was sealed for 2 h, and the corresponding diluted antibody was added for overnight incubation at 4 °C. The antibody was: ASC/TMS1(1:2000, CST, USA), Caspase-1(1:1000, CST, USA), NLRP3(1:500, CST, USA), and corresponding secondary antibodies were added and incubated at room temperature for 1 h, and imaging was performed by Image Lab 5.2.1 software. GAPDH (1:1000, CST, USA) as the internal standard.
2.4.7. Statistical analysis
Statistical software IBM SPSS Statistics 25 was used for data analysis, and Graph Pad Prism 8.0 (San Diego, CA, USA) was used for data mapping. The quantitative data are expressed as the mean ± SD. One-way analysis of variance and LSD test were used. P < 0.05 is considered statistically significant.
3. Results
3.1. Chemical composition analysis of AESS
A total of 91 components were detected and 76 components were identified in AESS, including 9 flavonoids, 11 organic acids, 6 phenylethanoid glycosides, 24 terpenoids, 12 diarylheptanoids and 14 other components. 47 of them were compared with standards. The other compounds were identified according to literature and database comparison, and the results are shown in Table 2. The total ion chromatography (TIC) in positive and negative ion modes is shown in Fig. 1. The main components include chlorogenic acid, cryptochlo- rogenic acid, isochlorogenic acid A, isochlorogenic acid C, rutin, narcissoside, cmicifugoside, kaempferol-3-rutinoside and 4′-O-beta-D-Glucosyl-5-O-methylvisamminol, etc.
Table 2.
Analysis results of chemical compounds in AESS.
| No. | tR (min) | Experimental mass | Detection mode | Error (ppm) | Main fragment ions | Molecular Formula | Identification | Category | Resources | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| 1# | 1.24 | 191.01872 | – | −5.27 | 173.00784,129.01794,111.00735, | C6H8O7 | Citric Acid | Organic acids | AMR | * |
| 2 | 1.27 | 268.10455 | + | 1.94 | 136.06207,268.10208,119.03558 | C10H13N5O4 | Adenosine | Nucleoside | CS | * |
| 3# | 2.06 | 166.08658 | + | −0.45 | 120.08120,103.05477 | C9H11NO2 | L-Phenylalanine | Amino acids | AR | * |
| 4 | 2.58 | 153.01790 | – | −2.20 | 153.01801,109.02808 | C7H6O4 | Protocatechuic acid | Organic acids | SF | * |
| 5 | 2.62 | 393.13977 | – | −5.61 | 393.13803,127.03858,234.52377 | C15H24O9 | Leonuride | Terpenoids | RR | TCMSP |
| 6 | 3.09 | 127.03885 | + | −2.30 | 127.03935,109.02878 | C6H6O3 | 5-hydroxymethyl furfural | Furaldehyde | Ad | * |
| 7# | 3.14 | 353.08737 | – | −1.23 | 191.05499,135.04381,161.02293,179.03884 | C16H18O9 | Neochlorogenic acid | Organic acids | LJF | * |
| 8 | 3.79 | 205.09714 | + | −1.80 | 146.06038,118.06561,188.07050 | C11H12O2N2 | D-tryptophan | Amino acids | AR | * |
| 9 | 4.12 | 137.02307 | – | −2.41 | 137.02307,109.02784,119.01232 | C7H6O3 | Protamine sulfate | Organic acids | LJF | Li et al. (2019) |
| 10# | 4.24 | 345.11859 | – | −1.49 | 299.11224,119.04854,59.01255 | C14H20O7 | Salidroside | Glycosides | LJF | * |
| 11# | 4.46 | 375.12927 | – | −1.07 | 151.07533,69.03310,95.04869 | C16H24O10 | 8-Epi-Loganic acid | Terpenoids | RR | * |
| 12 | 5.24 | 449.12933 | – | −1.63 | 416.35358,150.26289,139.00277 | C18H26O13 | Unknown | Unknown | LJF | |
| 13# | 5.32 | 375.12903 | – | −1.71 | 375.12943,113.02301,95.04888 | C16H24O10 | Loganic acid | Terpenoids | LJF | * |
| 14 | 5.40 | 471.22098 | + | 1.87 | 453.20734,291.15701,203.05284 | C21H36O10 | Atractyloside A | Glycosides | AR | Zhang et al. (2019b) |
| 15 | 5.42 | 493.22864 | – | −0.83 | 447.22415,285.16980,89.02297 | C22 H38 O12 | (1R,4S,6R)-1,3,3-trimethyl-2-oxabicyclo[2.2.2]oct-6-yl-6-O-β-D-glucopyranosyl-β-D-glucopyranoside | Glycosides | AMR,AR | Yang et al. (2012) |
| 16# | 5.74 | 353.08701 | – | −2.17 | 191.05504,85.02805,173.04411 | C16H18O9 | Chlorogenic acid | Organic acids | LJF | * |
| 17# | 5.85 | 451.14462 | – | −2.42 | 155.03357,243.09,101.02298 | C17H26O11 | Morroniside | Terpenoids | RR | * |
| 18# | 6.09 | 353.08734 | – | −1.32 | 191.05499,135.04384,173.04424,161.02293 | C16 H18 O9 | Cryptochlorogenic acid | Organic acids | LJF | . * |
| 19 | 6.11 | 375.12900 | – | −0.67 | 375.12866,201.01515,121.06453,89.02291,59.01252 | C16 H24 O10 | Unknown | Unknown | LJF | |
| 20 | 6.12 | 197.08124 | + | 2.05 | 179.07062,127.03935,111.08093 | C10 H12 O4 | Xanthoxylin | Phenols | LJF | Li et al. (2022a) |
| 21 | 6.45 | 389.10852 | – | −1.07 | 69.03304,95.04860,121.06445,389.10806,314.66028 | C16 H22 O11 | Unknown | Unknown | LJF | |
| 22# | 6.71 | 373.11322 | – | −2.14 | 59.01252,97.02798,193.04890 | C16H22O10 | Secologanic acid | Terpenoids | LJF | * |
| 23 | 7.06 | 507.17133 | – | −1.18 | 507.17093,191.05510,95.04880 | C21H32O14 | Unknown | Unknown | LJF | |
| 24 | 7.18 | 697.21808 | – | −2.10 | 697.21765,341.10855,179.05511 | C28 H43O20 | Unknown | Unknown | LJF | |
| 25 | 7.22 | 353.08731 | – | −1.40 | 191.05515,179.0339885.02806 | C16 H17O9 | Unknown | Organic acids | LJF | |
| 26 | 7.39 | 337.09192 | – | −0.97 | 191.05511,119.04886,163.03883,93.03311 | C16 H18O8 | 5-p-coumaroylquinic acid | Organic acids | LJF | Li et al. (2022a) |
| 27 | 7.40 | 447.15012 | – | −1.25 | 401.14505,269.10620,101.02297 | C19 H28 O12 | Unknown | Unknown | LJF | |
| 28# | 7.60 | 433.13443 | – | −1.66 | 387.20322,225.07568,101.02296 | C17H24O10 | Vegeloside | Terpenoids | LJF | * |
| 29# | 7.73 | 435.15002 | – | −1.37 | 101.02304,227.09172,127.03857 | C17H26O10 | Loganin | Terpenoids | LJF | * |
| 30# | 7.76 | 403.12381 | – | −1.92 | 125.02296,81.03316,376.25455 | C16H22O9 | Sweroside | Terpenoids | LJF | * |
| 31 | 7.78 | 345.15503 | – | −1.77 | 165.09082,59.01247,71.01229,89.02283,179.05455,345.15460 | C16H26O8 | RehMapicroside | Terpenoids | RR | Zhao et al. (2007) |
| 32 | 8.23 | 367.103 | – | −0.46 | 191.05505,173.04446,134.03596,93.03311 | C17H20O9 | 3-O-feruloylquinic acid | Organic acids | LJF | Li et al. (2022a) |
| 33 | 8.48 | 785.25000 | – | −1.23 | 161.02321,785.24902,133.02814 | C35 H46 O20 | Echinacoside | Phenylethanoid glycosides | RR | * |
| 34 | 8.57 | 435.14999 | – | −1.86 | 101.02304,157.04935,69.03306 | C17H26O10 | Unknown | Unknown | LJF | |
| 35# | 8.62 | 433.13437 | – | −1.80 | 123.04407,155.03326,424.50870 | C17H24O10 | Epivogeloside | Terpenoids | LJF | * |
| 36# | 8.70 | 403.12378 | – | −2.00 | 121.02808,59.01253,95.04877 | C17H24O11 | Secoxyloganin | Terpenoids | LJF | * |
| 37# | 8.98 | 469.17148 | + | −1.30 | 469.16974,307.11700 | C22 H28O11 | Cimicifugoside | Chromones | SR | * |
| 38 | 9.35 | 799.26544 | – | −1.62 | 179.05460,623.21759,161.02341,461.16934,315.10806,135.04376 | C36 H48O20 | Jionoside A1 | Phenylethanoid glycosides | RR | * |
| 39 | 9.47 | 538.22980 | + | 0.77 | 141.05482,228.10222,376.17453 | C26H35O11 N | Unknown | Unknown | CS | |
| 40# | 9.51 | 609.14484 | – | −2.08 | 609.14496,301.03574,271.02460,151.00282 | C27H30O16 | Rutin | Flavonoids | SF | * |
| 41# | 9.78 | 463.08780 | – | −0.86 | 463.08777,271.02448,300.02707,255.02933,151.00250 | C21H20O12 | Isoquercitrin | Flavonoids | SF | * |
| 42# | 9.99 | 447.09265 | – | −1.42 | 285.03998,447.09256 | C21H20O11 | Luteoloside | Flavonoids | LJF | * |
| 43 | 10.00 | 438.23969 | + | −0.30 | 147.04393,204.10175 | C25H31O4N3 | Unknown | Unknown | CS | |
| 44# | 10.15 | 593.15051 | – | −1.15 | 593.15045,285.03995 | C27H30O15 | Lonicerin | Flavonoids | LJF | * |
| 45 | 10.16 | 435.22433 | – | −0.95 | 435.22433,383.97205,71.01240 | C20H36O10 | Unknown | Unknown | RR | |
| 46# | 10.22 | 623.19714 | – | −1.61 | 161.02318,135.04381,623.19666,461.16678,315.10797 | C29H36O15 | Verbascoside | Phenylethanoid glycosides | RR | * |
| 47# | 10.33 | 307.11810 | + | −0.80 | 259.05975,235.05974,307.11694 | C16 H18 O6 | Cimifugin | Chromones | SR | * |
| 48# | 10.43 | 593.15063 | – | −0.83 | 255.02942,284.03229,227.03423 | C27H30 O15 | Kaempferol-3-Rutinoside | Flavonoids | SF | * |
| 49 | 10.53 | 743.23975 | – | −0.19 | 743.23926,203.03419,101.02301 | C46 H36O8 N2 | Unknown | Unknown | LJF | |
| 50# | 10.58 | 813.28070 | – | −1.93 | 175.03888,160.03537,134.03596,637.23291,497.17712 | C37H50O20 | Jionoside B1 | Phenylethanoid glycosides | RR | * |
| 51# | 10.66 | 623.16003 | – | −2.77 | 315.05060,300.02695,272.03085,151.00206 | C28 H32O16 | Narcissoside | Flavonoids | RR | * |
| 52# | 10.66 | 515.11847 | – | −2.12 | 191.05507,135.04379 | C25H24O12 | Isochlorogenic acid A | Organic acids | LJF | * |
| 53 | 10.77 | 507.20776 | – | −1.09 | 461.20074,255.02832,268.19434 | C21H34O11 | Unknown | LJF | ||
| 54# | 10.77 | 623.19702 | – | −1.15 | 161.02318,133.02811,623.19775 | C29H36O15 | Isoacteoside | Phenylethanoid glycosides | SF | * |
| 55 | 11.08 | 403.16064 | + | −0.6 | 109.02896,165.05496 | C18H26O10 | Icariside F2 | Flavonoids | LJF | |
| 56# | 11.15 | 453.17645 | + | 2.25 | 291.12317,273.11249,231.06560,245.11766,203.07080 | C22H28O10 | 4′-O-beta-D-Glucosyl-5-O-methylvisamminol | Chromones | SR | * |
| 57# | 11.40 | 515.11871 | – | −1.28 | 191.05508,135.04381,173.04436 | C25H24O12 | Isochlorogenic acid C | Organic acids | LJF | * |
| 58 | 11.54 | 803.26044 | – | −3.32 | 525.16046,285.03989 | C34H46O19 | Centauroside or isomer | Terpenoids | LJF | TCMSP |
| 59 | 11.64 | 331.15469 | – | −1.23 | 331.15518,96.95868,122.03590 | C19H24O5 | 4-(3,5-dihydroxy-7-(4-hydroxyphenyl)heptyl)benzene-1,2-diol | Diarylheptanoids | CR | Chang et al. (2020) |
| 60 | 11.81 | 361.16550 | – | −0.45 | 151.03873,346.14185,361.16531 | C20H26O6 | Unknown | RR | ||
| 61 | 11.84 | 331.15479 | – | −0.93 | 331.15491,122.03595 | C19H24O5 | 4-(3,5-dihydroxy-7-(4-hydroxyphenyl)heptyl)benzene-1,2-diol isomer | Diarylheptanoids | CR | |
| 62 | 12.01 | 803.26080 | – | −1.28 | 101.02301,525.16174,595.20209 | C34H46O19 | Centauroside or isomer | Terpenoids | LJF | TCMSP |
| 63 | 12.13 | 523.18170 | – | −0.76 | 134.03600,160.01537,193.04961,175.03883 | C25 H32O12 | 6-O-E-Feruloylajugol | Terpenoids | RR | Lu et al. (2023) |
| 64 | 12.37 | 577.15619 | – | −0.47 | 145.02818,119.04876,163.03880 | C27H30O14 | Unknown | Unknown | SF | |
| 65 | 12.99 | 651.22845 | – | −1.53 | 160.01537,175.03885,651.22821 | C31H40O15 | Martynoside/ Isomartyside | Phenylethanoid glycosides | RR | Wei et al. (2022) |
| 66 | 13.01 | 317.06278 | + | 1.56 | 317.06278,281.15466,187.11211,147.08072,133.06508,107.04976 | C19H24O4 | 1,7-bis(4-hydroxyphenyl)heptane-3,5-diol | Diarylheptanoids | CR | Chang et al. (2020) |
| 67# | 13.26 | 301.03485 | – | −1.75 | 151.00240,301.03491 | C15H10O7 | Quercetin | Flavonoids | SF | * |
| 68# | 14.01 | 439.16101 | + | 2.59 | 259.09689,277.10745,231.10150,205.04994 | C21 H26O10 | Sec-O-glucosylhamaudol | Chromones | SR | * |
| 69 | 14.02 | 389.15994 | – | −1.64 | 329.13895,389.15985 | C21H26O7 | 1,7-bis(3,4-dihydroxyphenyl)-5-hydroxyheptan-3-yl acetate | Diarylheptanoids | CR | Li et al. (2021) |
| 70 | 15.26 | 313.14404 | – | −1.57 | 313.14386,121.02812,165.05424 | C19 H22O4 | (E)-4-(3-hydroxy-7-(4-hydroxyphenyl)hept-6-enyl)benzene-1,2-diol | Diarylheptanoids | CR | Chang et al. (2020) |
| 71# | 15.53 | 375.18085 | + | −0.92 | 297.14828,357.16925 | C21H26O6 | 1-(3,4-dihydroxyphenyl)-5-hydroxy-7-(4-hydroxyphenyl)heptan-3-yl acetate | Diarylheptanoids | CR | * |
| 72 | 15.60 | 315.05063 | – | −1.35 | 300.02707,315.09036,151.00240,272.03253 | C16H12O7 | Isorhamnetin | Flavonoids | CR | * |
| 73 | 16.70 | 431.17020 | – | −2.18 | 311.12845,431.17053,189.09099 | C23H28O8 | 1,7-bis(3,4-dihydroxyphenyl)heptane-3,5-diyl diacetate | Diarylheptanoids | CR | Chang et al. (2020) |
| 74# | 16.96 | 297.14914 | – | −1.61 | 191.10663,132.05673 | C19H22O3 | (3S)-/(3R)-1,7-Bis(4-hydroxyphenyl)-(6E)-6-hepten-3-ol | Diarylheptanoids | CR | * |
| 75 | 17.11 | 357.17010 | – | −1.81 | 315.15979,149.05957,191.10678 | C21H26O5 | (3S)-and (3R)-3-acetoxy-1-(3,4-dihydroxyphenyl)-7-(4-hydroxy-phenyl)heptanes | Diarylheptanoids | CR | Chang et al. (2020) |
| 76 | 17.94 | 295.10040 | – | 5.01 | 187.00595,295.10049 | C19H20O3 | (E)-1,7-bis(4-hydroxyphenyl)hept-6-en-3-one | Diarylheptanoids | CR | Chang et al. (2020) |
| 77# | 18.02 | 293.11807 | – | −0.95 | 119.04884,187.07542 | C19H18O3 | 1,7-bis(4-hydroxyphenyl)hepta-4E-6E-dien-3-one | Diarylheptanoids | CR | * |
| 78# | 18.38 | 415.17551 | – | −1.65 | 415.17514,295.13333 | C23H28O7 | 1-(3,4-dihydroxyphenyl)-7-(4-hydroxyphenyl)heptane-3,5-diyl diacetate | Diarylheptanoids | CR | * |
| 79 | 20.15 | 235.16975 | + | −0.41 | 161.09636,177.12775 | C15H22O2 | Curcumenol | Terpenoids | CR | Li et al. (2022b) |
| 80 | 20.68 | 183.08089 | + | 2.45 | 153.07014,155.41937,115.05470,129.07040 | C13H11O | Atractylodin | Polyacetylene | AR | Zhang et al. (2019c) |
| 81 | 20.69 | 249.14905 | + | −0.65 | 231.13834,203.14326,189.12729,175.07576,163.07558,119.08596,105.07042 | C15 H20O3 | Atractylenolide III | Terpenoids | AMR | Dong et al. (2020) |
| 82# | 20.87 | 247.13348 | + | −0.40 | 139.03929,121.02888,229.12254 | C15H18O3 | Zederone | Terpenoids | CR | * |
| 83 | 22.30 | 779.45746 | – | −1.61 | 779.45685,439.35782 | C42H68O13 | Azukisaponin I | Terpenoids | SF | TCMSP |
| 84 | 23.61 | 233.15315 | + | −0.20 | 233.15343,187.14783 | C15H20O2 | Atractylenolide II | Terpenoids | AR,AMR | * |
| 85 | 23.70 | 217.15919 | + | −2.76 | 217.15926,161.09633,199.14853 | C15H20O | Atractylone | Terpenoids | AMR | Zhou et al. (2020) |
| 86 | 23.83 | 231.13850 | + | −2.39 | 83.08630,231.13853,213.12781 | C15H18O2 | Curzerenone/Furanodienone | Terpenoids | CR | Li et al. (2022b) |
| 87 | 24.27 | 379.15237 | + | 1.63 | 153.07016,128.06241,115.05471,197.09637, | C23H23O5 | Unknown | Unknown | AMR | |
| 88 | 25.20 | 199.07587 | + | −3.18 | 146.96158,125.00292,181.06540 | C13H10O2 | Atractylodinol | Terpenoids | AR | Dong et al. (2020) |
| 89 | 18.39 | 231.13744 | + | 0.19 | 231.13745,213.12738,203.14333,185.13300,143.08588, | C15H18O2 | Atractylenolide I | Terpenoids | AMR,AR | * |
| 90# | 27.30 | 219.17482 | + | −0.05 | 88.03899,146.08095,201.16383 | C15H22O | Germacrone | Terpenoids | CR | Li et al. (2022b) |
| 91 | 32.34 | 203.17993 | + | 0.53 | 203.17993,175.14859,161.13289,147.11716,133.10150 | C15H22 | Dehydrocitrene | Terpenoids | AMR | Zhong et al. (2015) |
Notes: Atractylodis Macrocephalae Rhizoma: AMR. Atractylodis Rhizoma: AR. Curcumae Rhizoma: CR. Lonicerae Japonicae Flos: LJF. Sophorae Flos: SF. Rehmanniae Radix: RR. Coicis Semen: CS. Saposhnikoviae Radix: SR. Ad: Artificial derivative; The column of Ref. “*” represent identified by standards, others referred to references or databases. “#” represent component selected for network pharmacology.
Fig. 1.
Total ion chromatography (TIC) of AESS in negative and positive ion modes with amplified part.
3.2. Network pharmacological analysis
3.2.1. Active components and eczema targets of AESS
Combined with the qualitative analysis results of AESS formula and absorbed components analysis, 37 compounds were selected as potential active components to obtain 1500 component targets, 1396 CE targets, and 257 intersection targets, as shown in Fig. 2A of Venn Diagram. The components related to the intersection target were screened out by using the intersection target, and one component (Isoacteoside) without common target was eliminated.
Fig. 2.
Venn diagram of the relationship between the target of components of AESS and the target of AD (A) and PPI network of key targets (B). A: I1396 disease targets related to AD, II: 1500 related targets of AESS, III: 257 common targets of components and disease; B: Node color;  colored nodes: query proteins and first shell of interactors,
colored nodes: query proteins and first shell of interactors,  white nodes: second shell of interactors, Node content;
white nodes: second shell of interactors, Node content;  empty nodes: proteins of unknown 3D structure,
empty nodes: proteins of unknown 3D structure,  filled nodes: some 3D structure is known or predicted, Known Interactions;
filled nodes: some 3D structure is known or predicted, Known Interactions;  from curated databases,
from curated databases,  experimentally determined, Predicted Interactions;
experimentally determined, Predicted Interactions;  gene neighborhood,
gene neighborhood,  gene fusions,
gene fusions,  gene co-occurrence and Others;
gene co-occurrence and Others;  textmining,
textmining,  coexpression,
coexpression,  protein homology.
protein homology.
3.2.2. The PPI network of AESS
The interaction between targets of AESS in the treatment of AD was analyzed based on String database. Intersection targets were input and data was imported into Cytoscape 3.8.1 for visualization analysis (Fig. 2B). The results showed that there were 227 nodes and 1554 edges. Sorted according to the Degree value, the larger the degree value, the larger the node, the darker the color (green to red), and the wider the line width. It can be seen from the figure that the degree value of AKT1, TP53, TNF, JUN, IL-6, RELA, MAPK3, SRC, EGFR, VEGFA, IL-1β and other targets were larger. They may be the key targets of AESS in the treatment of AD.
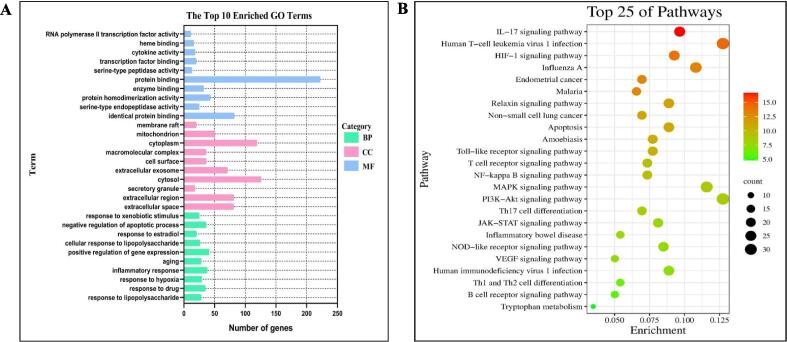
3.2.3. GO analysis and KEGG pathway enrichment analysis
The intersection targets were imported into DAVID online database for GO and KEGG enrichment analysis. 681 GO enrichment analysis results were obtained (P Value < 0.01), including 495 biological processes (BP), 79 cellular components (CC) and 107 molecular functions (MF). The top 10 items of each item are selected for visual analysis, and the results are shown in Fig. 3A. The results suggest that AESS prescription may interact with these outcomes, influencing these biological processes and thereby exerting corresponding molecular functions to achieve the therapeutic effect of eczema. KEGG enrichment analysis showed that 144 pathways (P Value < 0.01), and the first 25 pathways were selected for visual analysis according to the size of P value and the degree of correlation with disease, and the results were shown in Fig. 3B. It mainly involves TNF signaling pathway, IL-17 signaling pathway, HIF-1 signaling pathway, Toll-like receptor signaling pathway, NF-κB signaling pathway, NOD-like receptor signaling pathway, etc.
Fig. 3.
GO enrichment (A) and KEGG pathway enrichment (B) analysis of core target of AD by AESS (A: The different coloured nodes in the graph indicate the different biological types of GO; B: The nodes in the graph represent the type of KEGG signal pathway. The redder the nodes, the stronger the enrichment significance, and the larger the area, the more the number of enriched genes).
3.2.4. Herb-component-target-pathways interactive network
Herbs, components, key targets, and 25 pathways were imported into Cytoscape software for visualization analysis, and the results were shown in Fig. 4. There are 325 nodes and 1108 edges in the figure. The orange hexagon represents the analyzed ingredients in Table 2, the pink square represents the pathway, the yellow triangle represents the herbal medicine, and the green circle represents the target. The larger the node, the greater the corresponding Degree value. The thicker the connection between nodes, the closer the connection between them.
Fig. 4.
Network of the herb-component-target-main pathways of AESS (AMR: Atractylodis Macrocephalae Rhizoma; AR: Atractylodis Rhizoma; CR: Curcumae Rhizoma; LJF: Lonicerae Japonicae Flos; SF: Sophorae Flos; RR: Rehmanniae Radix; SR: Saposhnikoviae Radix.
3.2.5. Molecular docking
The molecular docking results showed that 24 main components could effectively combine with NLRP3, ASC and Caspase-1. Among them, chromones flavonoids, Cimicifugoside (37) had the strongest affinity for NLRP3 and ASC protein (−10.90 and −8.69 kcal/mol, respectively), while its aglycone, Cimifugin (47) had the strongest affinity for Caspase-1 protein (−7.62 kcal/mol) (Fig. 5). Other flavonoids, such as Quercetin(67), Luteoloside(42), Isoquercitrin (41), Narcissoside (51), Kaempferol-3-Rutinoside (48), Lonicerin (44), 4′-O-beta-D-Glucosyl-5-O-methylvisamminol (56), Sec-O-glucosyl- hamaudol (68), iridoid glycosides, including Sweroside (30), Epivogeloside (35), Secologanic acid (22), Morroniside (17), 8-Epi- Loganic acid (11), Loganin (29) and Vegeloside (28), cafeoyl quinic acid derivatives, including Isochlorogenic acid A (52), Isochlorogenic acid C (57), Chlorogenic acid (16), Neochlorogenic acid(7) and Cryptochlorogenic acid (18), and sesquiterpene, Zederone (82) and Germacrone (90), all had good affinity (≤−7 kcal/mol), indicating these components may exert anti-inflammatory activity through targeting NLRP3 signaling pathway.
Fig. 5.
Molecular docking between the representative small molecule ligands (Cimicifugoside and Cimifugin) and protein (human NLRP3, ASC and Caspase-1), on the top shows the 3D structure of ligands and receptors, at the bottom shows the surface of the receptor and 3D structure of the ligands.
3.3. Animal experiment
3.3.1. Body weight change
The body weight change of mice were recorded weekly during modeling, as shown in Fig. 6A. The results showed that the body weight of the administration group increased after the beginning of membrane building, thus the drug had no obvious toxicity to mice.
Fig. 6.
Treatment with AESS formula inhibits DNCB induced AD-like skin inflammation in BALB/c mice. (A) Changes of body weight in mice. (B) Photograph of the Skin on the Back of Mice. (C) Dermatitis scores in mice. (D) Dermatitis scores in mice. *P < 0.05, **P < 0.01, ***P < 0.001, compared with control group; #P < 0.05, ##P < 0.01, ###P < 0.001, compared with AD group (n = 3–6 per group).
3.3.2. Dermatitis score
In order to evaluate the therapeutic effect of AESS formula on eczema-like mice, we took photos and compared the skin lesions of mice in each group, and statistically analyzed the measured results (Fig. 6B, C). No significant changes were observed in the control group before and after membrane construction. In the model group, the back skin of mice gradually showed obvious eczema symptoms such as erythema, oedema, bleeding, dryness, erosion and scab due to DNCB application, and the dermatitis score gradually increased with the test time. In the AESS or DEX group, the symptoms of mice were significantly improved, and on day 21, the dermatitis score of mice was significantly lower than that of the model group, indicating that AESS formula and DEX had a good therapeutic effect on DNCB induced mice.
3.3.3. Histopathology
To investigate whether AESS formula could ameliorate the DNCB-induced animal model of AD, a histological examination (H&E) showed that epidermal thickness was reduced upon treatment with AESS formula. In addition, AESS formula treatment also significantly reduced the infiltration of eosinophils, lymphocytes, mononuclear cells and inflammatory cells into dorsal skin lesions (Fig. 7A). Moreover, in histopathological assessment of skin sections of mice intact structure and clear subcutaneous were observed in control group. Compared with the model group, DNCB application produced marked epidermal thickening. As we expected, AESS formula group (especially the medium dose AESS1 group) and the DEX group were significantly decreased inflammatory cell infiltration. The results indicated that AESS formula suppressed the symptoms of DNCB-induced AD model.
Fig. 7.
(A) HE staining for histopathological features (bar length = 100 μm). (B) The serum levels of total IgE in mice. (C) Serum IL-1β expression level. (D) Serum IL-18 expression level. *P < 0.05, **P < 0.01, ***P < 0.001, compared with control group; #P < 0.05, ##P < 0.01, ###P < 0.001, compared with AD group (n = 3–6 per group).
3.3.4. AESS inhibited the expression levels of IgE, IL-1β and IL-18 in serum
The increase of serum IgE expression level is one of the key features of AD. After treatment with AESS formula, the expression levels of IL-1β, IgE and IL-18 in serum of mice were determined. The results were shown in Fig. 7B–D. The results showed that compared with the control group, the serum expression levels of IgE, IL-1β and IL-18 in the model group were significantly increased, while compared with model group, the expression levels of IL-1β, IgE and IL-18 in serum of AESS formula group and the DEX group were significantly decreased.
3.3.5. AESS reduced the expression of ASC, Caspase-1 and NLRP3 proteins in skin tissue
In order to investigate whether the mechanism of AESS formula in the treatment of eczema is related to NLRP3 inflammasome, western blot analysis was used to detect the expression levels of NLRP3, ASC and Caspase-1 proteins in skin tissues. Compared with the control group, the expression levels of NLRP3, ASC and Caspase-1 in the model group were significantly increased, while compared with model group, the expression levels of ASC protein and NLRP3 protein in AESS formula group and DEX group were significantly decreased, Caspase-1 protein was significantly decreased in medium and AESS2 group and DEX group, and Caspase-1 protein expression level was slightly decreased in AESS 0.5 group (p > 0.05). The results indicated that AESS formula could treat DNCB induced AD like mice by inhibiting the expression of NLRP3 inflammasome (Fig. 8A–D).
Fig. 8.
Western blotting analysis of mouse skin tissue. *P < 0.05, **P < 0.01, ***P < 0.001, compared with control group; #P < 0.05, ##P < 0.01, ###P < 0.001, compared with AD group (n = 3–6 per group).
4. Discussion
AD is a clinical high incidence of dermatological disease, by a variety of factors act together, stimulate the body to produce an immune response, cytokine expression caused by a delayed abnormal reaction. The disorder of Th1/Th2 balance is the main immunological factor, which is mainly Th1 immune response in the chronic stage and Th2 immune response in the acute stage. IFN-γ secreted by Th1 cells can inhibit Th2 cells, and cytokines secreted by Th2 cells, such as IL-4, IL-5, and IL-13, can induce eosinophilia and increased IgE levels, which restrict each other and maintain the balance. When the body is exposed to allergens, it is easy to react with Th2, stimulate the body's immune cells to secrete IgE and inflammatory cytokines, and trigger allergic reactions by identifying and combining allergens (Zhang et al., 2019a). Multiple factors including race, environment, skin barrier dysfunction, immune regulatory abnormalities, and microbiome have been reported to affect the pathophysiology of AD in a summary of the latest research progress on AD. Progress in the understanding of the immunological mechanisms of AD has found that not only Th1, Th2, but Th17, Th22, and type 2 innate lymphoid cells also contribute to AD (Ahn et al., 2020).
Inherent immunity is an important line of defense for the body against pathogens. Pattern recognition receptors (PRRs) recognize invading microorganisms, dead cell debris or environmental stimuli to activate innate immunity. NLRs are a pattern recognition receptor widely found in the cytosol and are known to be associated with the development of autoimmune diseases such as psoriasis, rheumatoid arthritis and inflammatory bowel disease, and the most studied one is NLRP3 inflammasome (Chen et al., 2021). Recent findings showed that the NLRP3 inflammasome not only is an innate responder to pathogenic and danger signals, but also can affect the adaptive immune response (Chen et al., 2011). IL-18 is an important mediator in the Th1 response, primarily by induction of IFN-γsecretion from T cells and natural killer (NK) cells (Nakanishi et al., 2001). The possible involvement of the NLRP3 inflammasome in Th2 responses was suggested by the fact that IL-33, an IL-1 family cytokine, was shown to be processed by the NLRP3 inflammasome (Ogura et al., 2006). NLRP3 might be involved in Th2 responses, but the role of the NLRP3 inflammasome in type II immune response and AD remains unclear. IL-1β, as a key proinflammatory cytokine in inflammation, was also involved in Th17 cell differentiation (Sutton et al., 2006). IL-1β could synergize with IL-6 to promote Th17 cell development via up-regulation of key IL-17 transcription factors, IRF4 and ROR γt (Chung et al., 2009). DNCB -induced mice model is a classic animal model of AD, and it has been shown to activate the NLRP3 signaling pathway (Liu et al., 2022). When the body is stimulated by DNCB, it sensitizes keratinocytes, and then initiates NLRP3 inflammasome assembly, prompting caspase-1 activation, and then caspase-1 cleaves pro-IL-1β and pro-IL-18 as the active forms IL-1β and IL-18, causing inflammatory response and Th2, Th17-related immune imbalance. Since the NOD-like receptor signaling pathway was deduced to be one of the key pathways by network pharmacological analysis of AESS. Our study shows that AESS formula alleviates symptoms in AD-like mice by regulating the NLRP3 signaling pathway through inhibiting the level expression of IL-1β and IL-18. Reduced IL-1β and IL-18 could restore the Th2 and Th17 immune balance, and thereby alleviate delayed hypersensitivity. Our experiments found that AESS formula regulates the NLRP3 signaling pathway in AD for the first time.
5. Conclusions
In this study, based on systematic chemical profiling and the network pharmacology prediction results, TNF, IL-17, HIF-1, Toll-like receptor, NF-κB and NOD-like receptor signaling pathway were mainly involved, and their related cytokines, such as TNF-ɑ, IL-6, IL-1β, IL-17, etc, influence Th1-Th2, Th17-Treg related immune balance. Most of the main potential bioactive ingredients in this formula originated from Lonicerae Japonicae Flos, which further verify its ‘Monarch drug’ rule. Since NOD-like receptor signaling pathway may be an important mechanism for AESS prescription to treat AD, as testified by animal experiments, the protein expression levels of ASC, Caspase-1, and NLRP3 were significantly increased, demonstrating that NLRP 3 activation promoted eczema symptoms in this mice model. By oral administration of AESS prescription, it improved AD-like symptoms, reduced eosinophil infiltration, reduced scratching behavior, and reduced expression levels of IgE and proinflammatory cytokines in serum, as well as protein expression levels of ASC, Caspase-1, and NLRP3 in skin tissue. These results suggest that AESS prescription can reduce the inflammatory response in DNCB induced AD-like mice by inhibiting the activation of NLRP 3 pathway, which may could provide new therapeutic strategies for the medicine treatment of AD.
Financial support
This work was funded by the National Natural Science Foundation of China Youth Fund (82004364); Second Affiliated Hospital of Guangzhou University of Chinese Medicine (SZ2021ZZ17, SZ2021ZZ39); Guangdong-Hong Kong-Macau Joint Laboratory on Chinese Medicine and Immune Disease Research, Guangzhou University of Chinese Medicine (2020B1212030006); Guangdong Provincial Key Laboratory of Chinese Medicine for Prevention and Treatment of Refractory Chronic Diseases (2018B030322012 and MB2020KF04); Guangzhou Science and Technology Plan Project – Municipal School (College) joint funding project (202201020333). Municipal Schools (Colleges) jointly funded projects (SL2024A03J00887). This work was supported by the Key-Area Research and Development Program of Guangdong Province (No. 2020B1111100010).
CRediT authorship contribution statement
Xiumei Mo: Writing – review & editing. Dong Wang: Methodology. Xing Zeng: Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Peer review under responsibility of King Saud University.
Supplementary material to this article can be found online at https://doi.org/10.1016/j.jsps.2023.101792.
Contributor Information
Dacan Chen, Email: cdc@gzucm.edu.cn.
Fenggen Yan, Email: fenggen_yan@gzucm.edu.cn.
Appendix A. Supplementary material
The following are the Supplementary material to this article:
References
- Ahn K., et al. Recent advances in atopic dermatitis. Curr. Opin. Immunol. 2020;66:14–21. doi: 10.1016/j.coi.2020.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylund S., et al. Prevalence and Incidence of atopic dermatitis: a systematic review. Acta Derm. Venereol. 2020;100:adv00160. doi: 10.2340/00015555-3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.L., et al. Anti-tumor activity and linear-diarylheptanoids of herbal couple Curcumae Rhizoma-Sparganii Rhizoma and the single herbs. J. Ethnopharmacol. 2020;250 doi: 10.1016/j.jep.2019.112465. [DOI] [PubMed] [Google Scholar]
- Chen M., et al. Regulation of adaptive immunity by the NLRP3 inflammasome. Int. Immunopharmacol. 2011;11(5):549–554. doi: 10.1016/j.intimp.2010.11.025. [DOI] [PubMed] [Google Scholar]
- Chen L., et al. NOD-like receptors in autoimmune diseases. Acta Pharmacol. Sin. 2021;42(11):1742–1756. doi: 10.1038/s41401-020-00603-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y., et al. Critical regulation of early Th17 cell differentiation by interleukin-1 signaling. Immunity. 2009;30:576–587. doi: 10.1016/j.immuni.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., et al. Study on the change of fingerprint and main components of Rhizoma Atractylodis before and after processing. China Pharmacist. 2020;12:2398–2402. [Google Scholar]
- Jia J., et al. Therapeutic effect of chinese herbal medicine gu-ben-hua-shi (AESS) formula on atopic dermatitis through regulation of yes-associated protein. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.929580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z., et al. Research progress of external treatment of chronic eczema. Med. Rev. 2018;20:4103–4107. [Google Scholar]
- Koo J., et al. Traditional Chinese medicine in dermatology. Dermatol. Therapy. 2003;16:98–105. doi: 10.1046/j.1529-8019.2003.01617.x. [DOI] [PubMed] [Google Scholar]
- Li Y., et al. Rapid identification of chemical constituents of Bushen Huoxue recipe based on UPLC-Q-Exactive quadrupole-electrostatic field orbital trap high resolution mass spectrometry. Chinese J. Pharm. Anal. 2019;01:111–121. doi: 10.16155/j.0254-1793.2019.01.14. [DOI] [Google Scholar]
- Li Z.Y., et al. Research progress of chemical constituents and pharmacological effects of Rhizoma Curcumae in Guangxi and prediction and analysis of its quality marker (Q-Marker) Chinese Herbal Med. 2021;15:4687–4699. [Google Scholar]
- Li H.L., et al. Analysis of chemical constituents in different radix curcumae based on UPLC-Q-TOF/MS. Chinese Herbal Med. 2022;11:2648–2655. doi: 10.13863/j.issn1001-4454.2022.11.021. [DOI] [Google Scholar]
- Li S.F., et al. Analysis of different processing methods of Flos Lonicerae based on ultra-performance liquid chromatography coupled with high-resolution mass spectrometry. Appl. Chem. 2022;05:819–827. doi: 10.19894/j.issn.1000-0518.210160. [DOI] [Google Scholar]
- Liu W., et al. Angelica Yinzi alleviates 1-chloro-2,4-dinitrobenzene-induced atopic dermatitis by inhibiting activation of NLRP3 inflammasome and down-regulating the MAPKs/NF-kB signaling pathway. Saudi Pharm. J.: SPJ: Off. Publ. Saudi Pharm. Soc. 2022;30(10):1426–1434. doi: 10.1016/j.jsps.2022.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X.M., et al. Based on UHPLC-LTQ-Orbitrap MS, the effects of different processing techniques on the chemical constituents of Rehmannia glutinosa were analyzed. China J. Tradition. Chinese Med. 2023;02:399–414. doi: 10.19540/j.cnki.cjcmm.20220816.301. [DOI] [PubMed] [Google Scholar]
- Nakanishi K., et al. Interleukin-18 is a unique cytokine that stimulates both Th1 and Th2 responses depending on its cytokine milieu. Cytokine Growth Fact. Rev. 2001;12(1):53–72. doi: 10.1016/s1359-6101(00)00015-0. [DOI] [PubMed] [Google Scholar]
- Ogura Y., et al. The inflammasome: first line of the immune response to cell stress. Cell. 2006;126(4):659–662. doi: 10.1016/j.cell.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Reuter J., et al. Botanicals in dermatology. Am. J. Clin. Dermatol. 2010;11(4):247–267. doi: 10.2165/11533220-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Schwartz C., et al. Spontaneous atopic dermatitis in mice with a defective skin barrier is independent of ILC2 and mediated by IL-1β. Allergy. 2018:1–14. doi: 10.1111/all.13801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton C., et al. A crucial role for interleukin (IL)-1 in the induction of IL-17–producing T cells that mediate autoimmune encephalomyelitis. JEM. 2006;203(7):1685–1691. doi: 10.1084/jem.20060285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson K.V., et al. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nature Rev. Immunol. 2019;19(8):477–489. doi: 10.1038/s41577-019-0165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., et al. Interleukin-18 is elevated in the sera from patients with atopic dermatitis and from atopic dermatitis model mice, NC/Nga. Int. Arch. Allergy Immunol. 2001;125:236–240. doi: 10.1159/000053821. [DOI] [PubMed] [Google Scholar]
- Tsang M.S., et al. Immunological roles of NLR in allergic diseases and its underlying mechanisms. Int. J. Mol. Sci. 2021;22(4):1507. doi: 10.3390/ijms22041507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M.J., et al. Preparation technology of raw rehmannia juice in the classic prescription Lily Rehmannia Decoction based on high resolution mass spectrometry. Chinese J. Exp. Tradition. Med. Formulae. 2022;09:133–140. doi: 10.13422/j.cnki.syfjx.20220616. [DOI] [Google Scholar]
- Yang E., et al. Research progress on chemical constituents and pharmacological effects of Atractylodes macrocephala. J. Guangdong Pharm. Univ. 2012;02:218–221. [Google Scholar]
- Zhang Q., et al. Research progress on the pathogenesis of atopic dermatitis. World Latest Med. Inf. Abstr. 2019;30:121–123. doi: 10.19613/j.cnki.1671-3141.2019.30.056. [DOI] [Google Scholar]
- Zhang Y., et al. Qualitative and quantitative determination of Atractylodes rhizome using ultra-performance liquid chromatography coupled with linear ion trap-Orbitrap mass spectrometry with data-dependent processing. Biomed. Chromatogr.: BMC. 2019;33(3):e4443. doi: 10.1002/bmc.4443. [DOI] [PubMed] [Google Scholar]
- Zhang Y.H., et al. Shanghai University of Traditional Chinese Medicine; 2019. Study on chemical basis and metabonomics of Atractylodes lancea.https://kns.cnki.net/kcms2/article/abstract?v=QOR9nnE_NmK2DhimfhVwTpLUMVdbLVslx1rbgkA0I-oQ18SFuzBoWAnS_2hY6lLWTEhqd3IXZ57xO5sua8_PlL8b6nV781YUCR4_MfYQrexDaxHLLabQeHz7SNHYWcKUIT1WUQFIFZ8=&uniplatform=NZKPT&language=CHS doctoral thesis. [Google Scholar]
- Zhao X.F., et al. HPLC/ESI-MS analysis of Radix Rehmanniae Preparata. Chinese J. Pharm. Anal. 2007;06:874–876. [Google Scholar]
- Zheng J., et al. A novel function of NLRP3 independent of inflammasome as a key transcription factor of IL-33 in epithelial cells of atopic dermatitis. Cell Death Dis. 2021;12(10):871. doi: 10.1038/s41419-021-04159-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y.M., et al. Study on rapid identification of chemical constituents of Atractylodes macrocephala based on UPLC/Q-TOFMS technology. Acta Mass Spectromet. 2015;01:72–77. [Google Scholar]
- Zhou J., et al. Analysis of chemical constituents of Atractylodes lancea and Atractylodes macrocephala based on UPLC-QTof-MS/MS. Pharm. Clin. Res. 2020;05:321–328. doi: 10.13664/j.cnki.pcr.2020.05.001. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.