Highlights
-
•
Increased SPN content in a mixed protein system enhances the strength of the sono-physico-chemical action.
-
•
A ratio of SPI to SPN of 1:3 gives the greatest sono-physical–chemical effect.
-
•
Excessive sono-physico-chemical effects can significantly reduce coupling rates and cause protein reaggregation.
-
•
The high SPN group's grafting rate rose from 39.13% to 55.26% in comparison to the high SPI content group.
Keywords: Meat, Sarcoplasmic protein (SPN), Ultrasound (UID), Protein structure, Atomic force microscope (AFME)
Abstract
Since it may be employed to guide the production of high-quality plant protein as a partial substitute for animal protein using sono-physico-chemical effects, it is important to investigate the mixing of animal and plant protein in ultrasound (UID)-assisted processing systems. A study group of sono-physico-chemical processing with five distinct soy protein isolate (SPI)/ sarcoplasmic protein (SPN) ratios was developed in this work. The results showed that adding additional SPN to the mixed protein can increase its sono-physico-chemical impact, and this effect is greatest when the ratio of SPI to SPN is 1:3. The high SPN group's grafting rate rose from 39.13% to 55.26% in comparison to the high SPI content group. Quercetin (Que) may more readily modify SPN than SPI in the “dual protein” system used in this work, highlighting the critical function of plant protein in controlling the effects of UID-assisted processing in the “dual protein” system. Changes in apparent viscosity and microstructure are the primary parameters that affect the severity of sono-physico-chemical effects in SPI/SPN mixed protein systems, in addition to structural variables.
1. Introduction
The sarcoplasmic protein (SPN) in muscle is a type of high-quality protein that is less abundant than myofibrillar protein, comprising 20–30 % of the total protein [1]. Compared to myofibrillar protein, SPN can be extracted from muscle tissue more readily due to its high solubility in water or low ionic strength solution [2]. In addition, the high amino acid content of SPN suggested that it could be developed as a protein-containing beverage shortly. However, the utilization of SPN as a by-product is considerably lower than that of myofibrillar protein. This may be because its deficient gelling or emulsifying ability frequently results in a decline in product quality [2], [3], [4]. More so, the release of huge amounts of the solution containing SPN into the environment can lead to an excessive amount of nitrogen and sulfur in the water, which corresponds to eutrophication and environmental contamination. Therefore, it is essential to maximize the utilization of SPN by-products.
It may be more appealing to consumers to partially replace animal protein with plant protein as opposed to using SPN directly. Multiple factors are responsible for this: First, excessive intake of animal protein without ingesting other proteins may be detrimental to health, which has spurred a surge of research on the partial replacement of animal protein with plant-based proteins [5], [6], [7]. Additionally, recent studies have demonstrated that plant-based meat products cannot fully replace animal-based meat products in terms of nutritional value [8], [9]. Consequently, a “dual protein” system comprised of animal and plant proteins may be the preferable option. It should be noted that the “dual protein” system can prevent the unbalanced nutrition of a single protein and imbue the product with more robust sensory qualities [6], [10]. In this investigation, soy protein isolate (SPI) was chosen as a partial replacement for SPN to build a “dual protein” system. Protein isolate from soybeans is a common plant protein with a high nutritional value and a low price. By using ultracentrifugation, SPI can be separated into four different parts, including the 2S, 7S, 11S, and 15S fractions [11]. Moreover, SPI has been recognized as an excellent substitute for animal protein. For example, modified SPI has the potential to improve myofibrillar protein gel texture and water-holding capacity. This may be due to its excellent manufacturability and distinct microstructure [12]. Therefore, it is feasible to produce protein beverages by substituting SPI for a portion of SPN. However, the lack of structural changes for 7S and 11S globulins under normal meat processing conditions can limit the interaction between SPI and meat proteins.
With the assistance of ultrasound (UID) technology, food scientists have endeavored to enhance the flavor and functionality of protein-based beverages recently [13], [14]. Notably, the covalent modification of polyphenols on proteins facilitated by UID is a classic example. As reported, the grafting of polyphenols with the aid of UID was a promising technique for reducing thermally-induced protein beverage denaturation [14], [15]. In addition, the stability and antioxidant properties of protein-based beverages could be significantly enhanced through the use of UID processing [16], [17]. However, the sono-physico-chemical effect and processing efficacy of UID treatment appeared to be more complicated in a “dual protein” system composed of SPN and SPI. In a “dual protein” system, the presence of SPI could impact the grafting efficacy of polyphenol to SPN during UID. It can be observed that the sono-physico-chemical impact exhibited a close relationship with environmental factors [13]. In this context, various composition ratios of SPN and SPI might have a significant impact on the sono-physico-chemical effect, resulting in distinct polyphenol grafting effects. It should be noted that various proteins may show distinct responses to the sono-physico-chemical of UID. Therefore, it is important to evaluate the sono-physico-chemical and processing effects of the SPN/SPI system. In particular, quercetin (Que) was selected for UID-assisted processing in this study. This was due to its high antioxidant activity and abundance of sources. According to previous reports [18], [19], [20], [21], Que is abundant in the stem bark, flowers, leaves, seeds, and fruits of a variety of plants, and it possesses numerous protein-reactive sites.
The purpose of the research was to analyze the sono-physico-chemical impacts in the combination SPN/SPI system and their grafting effect on Que. First, the sono-physico-chemical effects and grafting sites of SPI/SPN samples with five different ratios (3:1, 2:1, 1:1, 1:2, and 1:3) were evaluated. Following that, the SPI/SPN sample's structural characteristics were examined. Additional assessments were the mixed SPI/SPN system's microstructure and apparent viscosity. With UID-assisted processing, this work advanced the development of “dual protein” diets that partly substitute some animal proteins with plant proteins.
2. Materials and methods
2.1. Materials
Soy protein isolate (SPI), Que (molecular weight: 302.24, light yellow powder), and sodium chloride were supplied by Solarbio Technology Co., Ltd. (Beijing, China). After achieving a homogeneity (PD500-TP, PRIMA, England) (7,000 rpm, 5 min), sarcoplasmic protein (SPN) was extracted from the chicken breast via centrifugation (Avanti JC, Beckman Coulter, California, USA) at 10,100 g for 20 min. Additionally, SPN was filtered to remove lipid particles without additional purification. Sodium hydroxide, hydrochloric acid, and other reagents were provided by Nanjing Chemical Reagent Co., Ltd. (Nanjing, China).
2.2. Preparation of protein mixtures
Within the mixture system, the total concentration of protein (including SPI and SPN) was approximately 30 mg/mL, and they were separated into five distinct groups. The ratios of SPI to SPN in these five groups were set to 3:1, 2:1, 1:1, 1:2, and 1:3, with the corresponding labels being SSP1, SSP2, SSP3, SSP4, and SSP5, respectively. Before homogenization (6,000 rpm for 45 s) in the SSP1-SSP5 groups, Que solutions (1 mL, 0.45 mM, ethanol dissolution) were added to mixed proteins containing SPI and SPN (159 mL). To initiate covalent reactions of mixed SPI and SPN, the pH value of the liquid system was adjusted to about 9.0. During the processing time of 6 min, UID (400 W/L) was undertaken in these SSP1-SSP5 groups. The sample's pH was then readjusted to 7.0 after the covalent reaction for 60 min and its temperature was kept at 4 °C.
2.3. Confirmation of various UIDs
2.3.1. Degradation rate of crystal violet
Initially, the change rate of crystal violet was utilized to confirm the UID impacts of the SSP1–SSP5 groups [13], [16]. In brief, 0.8 mg of crystal violet was dissolved uniformly in 50 mL of variously mixed SSP samples with 500 rpm of agitation. The SSP1-SSP5 classes' absorbance was determined at 580 nm. Then, the identical UID operation was separately applied to distinct SSP samples. Again, the absorbance of the SSP1-SSP5 groups at 580 nm was measured to determine crystal violet alterations.
2.3.2. Decomposition of potassium iodide
The rate of potassium iodide change was also used to validate the sono-physico-chemical results of the SSP1–SSP5 groups [14], [22]. Simply, 0.83 g of potassium iodide was dispersed uniformly at 500 rpm in 50 mL of variably mixed SSP samples. The absorbance of the SSP1-SSP5 classes at 355 nm was measured. A similar UID operation is then applied separately to distinct SSP samples. Once again, the absorbance of the SSP1-SSP5 groups was measured at 355 nm to ascertain potassium iodide changes.
2.4. Grafting effect
The grafting effect of different SSP1–SSP5 groups was evaluated using the absorbance method. Mix various protein samples containing SPI and SPN with Folin Ciocalteu reagent in a ratio of 5:1 and facilitate the mixture to react for approximately 6 min. Then, a solution containing 7.5 % sodium carbonate at a volume of 40 % was added to the reaction solution and allowed to stand for 119 min. The sample was centrifuged extensively at 5, 500 rpm for 14 min before absorbance detection at 760 nm (SpectraMax M2, Molecular Devices Ltd., Sunnyvale, CA, USA). Que's standard curve was used to calculate the conjugate effect.
2.5. Liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS)
LC-MS/MS was used to determine the grafting sites for SSP1–SSP5 groups. Samples of different SSPs were mixed with trypsin in a ratio of 50 to 1 and stored at 37 °C for 12 h overnight. The resulting peptide mixture containing SPI and SPN was centrifuged for 15 min at 8,000 rpm. The supernatant was subsequently desalinated through a C18 column. After vacuum centrifugation, the sample was redissolved in a 0.1 % formic acid solution for LC-MS/MS analysis. The peptide sample's loading quantity is 5 μg. In the experiment, buffer B consisted of 80 % acetonitrile and 0.1 % formic acid. When searching for peptide data in the Chicken Uniprot database, the fragment ion error was 20 ppm.
2.6. Micro-Fourier transform infrared (M-FTIR)
Thermo In 10 (Thermo Fisher, USA) was used for examining the M-FTIR data of distinct SSP1–SSP5 groups. Before being transferred to the sample stage, various SSP samples were freeze-dried at −80 °C. The built-in microscope was then used to ascertain the area of microscopic infrared photography. Lastly, the infrared spectral information (800–3800 cm) of a 50–50 µm square was captured.
2.7. Three-dimensional fluorescence
The data of three-dimensional fluorescence of various SSP1-SSP5 systems were collected using a traditional RF-6000 spectrophotometer. Several protein solutions consisting of SPI and SPN were adjusted to an optimized concentration of about 1 mg/mL. Then, approximately 1.1 mL of these protein samples (SSP1-SSP5) were to transferred a 10-mm-long and 10-mm-wide quartz colorimetric dish. Finally, the SSP's three-dimensional fluorescence information between 280 and 580 nm (excitation wavelength: 220 to 320 nm) was collected.
2.8. Circular dichroism (CD)
The data of the secondary structure for distinct SSP1-SSP5 systems were obtained via a circular dichroism (CD) spectrometer (Jasco, Japan). Some protein-based solutions composed of SPI and SPN were concentrated to around 0.2 mg/mL. Following that, about 1.1 mL of these protein samples (SSP1-SSP5) were transferred into a 10-mm-long and 10-mm-wide quartz colorimetric dish. Ultimately, data from the spectral range between 191 and 259 nm was acquired for the SSP1-SSP5 samples. The final secondary structure was given instantaneously from their internal software.
2.9. Tryptophan fluorescence
The tryptophan fluorescence across the SSP1-SSP5 systems was determined based on their inside fluorescence measurements. All protein-based products comprising SPI and SPN were diluted to a dosage level of 0.5 mg/mL. After that, about 1.1 mL of these protein samples (SSP1-SSP5) were moved to 96-well plates. For the SSP1-SSP5 samples, statistics from the spectral variation throughout 320 and 420 nm (excitation wavelength: 270 nm) were ultimately present.
2.10. X-ray diffraction (XRD)
The X-ray diffraction (XRD) values throughout the SSP1-SSP5 systems were identified through the Bruker D8 instrument. Distinctive examinations of SSP samples were deposited in an experiment plate. Subsequently, the aforementioned substances were precisely set within the XRD device. All SSP1-SSP5 systems' XRD images were captured between 5 and 85 degrees of a diffraction angle (2θ).
2.11. Apparent viscosity
The Antonpa rheometer was used to evaluate the apparent viscosity of the SSP1-SSP5 systems. Putting the protein solution containing SPI and SPN into an Anton rheometer with a 40-millimeter-diameter probe with attention. Under static shear mode operation, the apparent viscosity of the protein solution was determined at various protein ratios. The initial and ultimate static shear rates are 0.001 and 100 1/s, respectively.
2.12. Thermogravimetric analysis (TGA)
Thermogravimetric analysis (TGA) of SSP1–SSP5 samples was evaluated through temperature-mass changes. Before being transmitted to the sample stage of the STA2500 TGA analyzer (NaiChi, Germany), several SSP samples were freeze-dried at −80 °C. The sample's weight was recorded after increasing the temperature of the sample from ambient temperature to 750 °C.
2.13. System density
The tensiometer (KRUSS, Germany) was utilized to determine the SSP1-SSP5 system's density. Initially, various SSP samples were poured into a container. The tensiometer's hook then approached the protein sample to be evaluated. Following the execution of the K100 program, the density results were displayed directly in the Results section.
2.14. Microstructure
The scanning electron microscope (SEME) and atomic force microscope (AFME) were both employed to evaluate the SSP1–SSP5 systems' microstructure. Various SSP samples were freeze-dried at −80 °C before being transferred to the sample stage in SEME (HITACHI, Japan). After the gold spraying application, the morphology of SSP1–SSP5 samples was captured. The image of AFME was obtained by striking a sheet of mica. The torque of the specimen was optimized to about 0.4 N/m.
2.15. Statistical analysis
The significant changes (p < .05) of SSP1–SSP5 samples were assessed using a one-way analysis of variance (SAS software, USA). At this point, the various SPI and SPN ratios were selected as factors of variation, and the obtained data were established as the results to be compared. Furthermore, the significant changes (p < .05) of SSP1–SSP5 samples were also compared through Tukey’s method.
3. Results and discussion
3.1. Verification of sono-physico-chemical effects
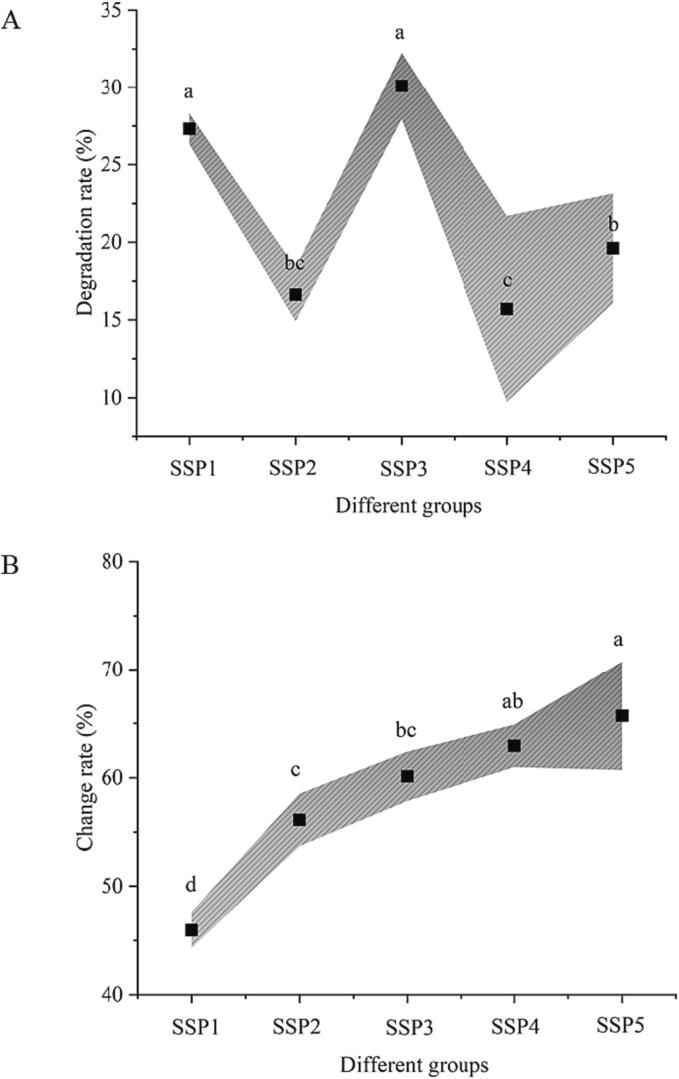
The impact of external elements on the mixed systems containing SPI and SPN can be roughly determined by identifying the strength of the sono-physico-chemical effects. According to previous findings [13], [14], processing effectiveness, particularly in protein systems, can be enhanced by relatively higher sono-physico-chemical impacts. This is understandable given that more cavitation bubbles will act on the protein surface. Meanwhile, more free radicals are produced inside the system as a result of greater sono-physico-chemical effects. Hence, the effects of the protein system alteration can be improved by strong sono-physico-chemical effects. Interestingly, iodometry and crystal violet measurements are not consistent in this paper (Fig. 1). The crystal violet results indicate that the increase in the ratio of SPN to SPI may lessen the crystal violet degradation impact during UID-aided processing. Iodometry's findings, however, indicated that the sono-physico-chemical action was enhanced by the rise in SPN levels.
Fig. 1.
The impacts of various SPI to SPN ratios on the sono-physico-chemical effectiveness of UID-assisted processing systems (A: crystal violets, B: iodometry).
Based on the outcomes of Gao et al. (2023) [23], the natural protein structure may explain why animal protein is processed more efficiently with UID assistance than SPI or whey protein. As a result of the interaction between the two proteins, systems made up of SPI and SPN mixes are more complicated. Thus, it is necessary to validate the impact of the rise in SPI and SPN ratio on the modification effects in the following sections. Previous studies conducted by Zhang et al. (2023) [22] showed that the phthalic acid approach was more accurate in measuring the sono-physico-chemical effects than iodometry. However, in this investigation, the effect of iodometry outperformed the crystal violet degradation approach. This could be related to the “dual protein” system's partial adsorption of Crystal violet. Therefore, it is important to carefully consider the benefits and drawbacks of various measuring techniques while designing distinct studies. It should be underlined that the impact of variations in the ratio of SPI and SPN on the sono-physico-chemical impacts in the “dual protein” mixture system will be verified in the structural sections.
3.2. Grafting effects
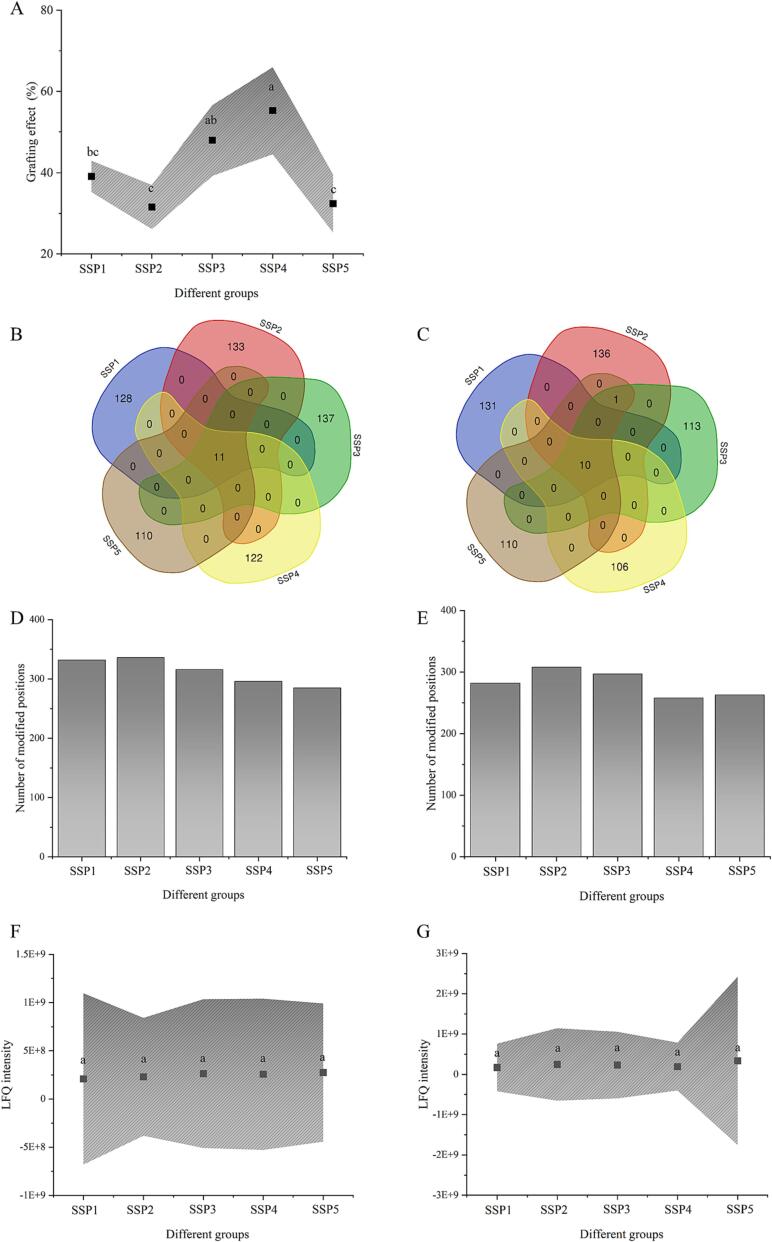
The covalent strength of the bond between Que and proteins (SPI and SPN) can be determined using the grafting rate as an indication [24]. After UID-assisted alteration, the grafting rates of SSP1 to SSP5 are 39.13 %, 31.56 %, 47.92 %, 55.26 %, and 32.42 %, respectively (Fig. 2A). The findings show that SPI and SPN protein molecules can bind covalently to Que. Additionally, when the ratio of SPI and SPN was altered from 2:1 to 1:1 or 1:2, the grafting rate rose by 51.82 % and 75.06 %, respectively. Typically, more free radicals can be employed to oxidize SPI and SPN owing to increased sono-physico-chemical effects, leading to higher grafting rates. This can be attributed to the generation of SPI-Que and SPN-Que. As reported by Pan et al. (2020) [25], free radical oxidation is the primary pathway in UID-aided processing. In this regard, the enhancement of sono-physico-chemical effects is the key factor contributing to the rise in the grafting rate. Additionally, higher sono-physico-chemical impacts can also produce larger turbulent pressures to lessen protein aggregation, which has been widely documented in previous research [16], [26]. The findings are consistent with those of iodometry, indicating that the sono-physico-chemical impact dramatically increased as SPI content dropped (from SSP1 to SSP5).
Fig. 2.
The effects of various SPI to SPN ratios on the grafting effect (A) of UID-assisted processing systems. The quantitative evaluation of sequence variations (B-C), the number of altered locations (D-E), and LFQ intensity (F-G) in the protein system containing SPI (B, D, and F) and SPN (C, E, and G).
Unexpectedly, the coupling rate dropped significantly in the SSP5 group, which may be related to the protein refolding in the mixed system (Fig. 2A). A relatively high protein aggregation at this step may hinder the covalent binding of Que on proteins. According to the findings of Zhang et al. (2022) [27], excessive UID treatment could cause severe, out-of-control responses and internal hotspots that might lead to protein reaggregation. Furthermore, a “dual protein” imbalance may weaken the interaction between SPI and SPN, which can be attributed to the large amount of SPN in SSP5. In the SSP5 group, SPN may also be more likely to combine with SPI to form bigger aggregates, which would prevent Que from binding covalently in considerable amounts. It should be considered that the produced SPN-Que-SPN or SPI-Que-SPI complexes can prevent the increase in the Que grafting rate because of the cascade effect. In other words, the covalent binding of Que on SPI and SPN may be limited by these dense complexes. Han et al. (2022) [28] discovered that covalent crosslinking accelerated the growth of protein molecular weight and that, at this point, increased steric hindrance and prevented further covalent reactions. Additionally, under an alkaline condition, Que could bind and lower the reactivity of protein nucleophilic groups, such as histidine, tryptophan, or tyrosine [29]. This effect was strongly related to the alteration of the protein's active sites.
3.3. Distribution of grafting site
The coupling rate cannot, however, be used to identify sensitive proteins in the “dual protein” system. In this work, the grafting sites of Que on proteins were carefully analyzed. As can be seen in Fig. 2B-C, about 11 shared peptide segments were found in the five groups (from SSP1 to SSP5) after Que modification. When the ratio of SPI to SPN decreases from 3:1 to 1:1, more modified peptides can be detected in SPI samples. More so, the amount of changed peptide segments in SPI appeared to be declining as the SPN concentration increased (Fig. 2B). In this context, there are more available locations for SPN when the ratio of SPI to SPN decreases from 1:1 to 1:3. The findings show that some similar peptide in the SPI of the “dual protein” system exhibits multiple modifications. It should be highlighted that the higher SPN ratio of the “dual protein” system results in a decrease in the number of SPI sites accessible for Que grafting. As a consequence, a significant association between the protein content of SPI and the number of changed locations was observed in Fig. 2D. However, SPI possessed the poorest modifying impact when compared with whey protein and myofibrillar protein in a prior study [23]. This might be a beneficial effect of the “dual protein” system where SPN facilitates the covalent binding of SPI to Que. It is interesting to note that SPN appears to be more oxidation-sensitive than SPI. Fig. S1 shows that the number of modification sites in SPN is significantly higher than that in SPI under oxidation conditions.
Notably, the sensitivity of various proteins to the UID effects depends on their structure and experimental context. According to the outcomes of Wang et al. (2022) [30], compared to animal protein, SPI is more sensitive to environmental factors. Moreover, myofibrillar proteins exhibit a loose structure that is particularly favorable to covalent interactions, which is another reason why they can increase the effectiveness of UID processing [17], [23]. In comparison, SPN's spherical form and denser structure than that of myofibrillar protein, however, restrict the extent of its response. Finally, SPI is more readily changed by Que than SPN in the “dual protein” system used in this investigation.
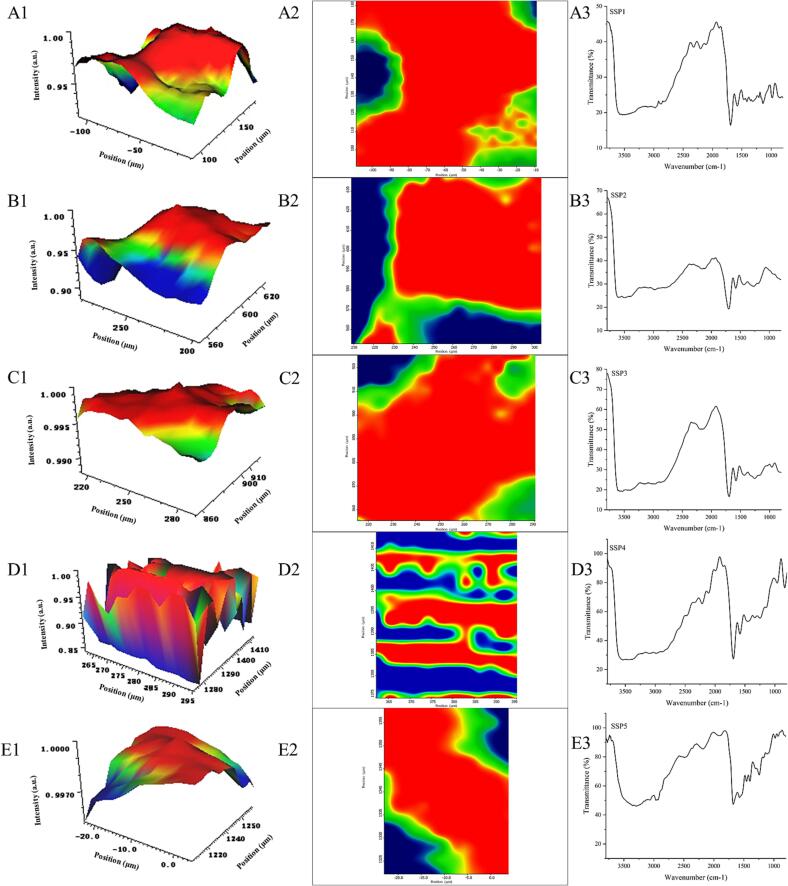
3.4. M-FTIR
Different SSP samples were examined using M-FTIR to determine their chemical composition and micrometer-scale spatial distribution. In the present study, the infrared spectrum data of distinct SSP samples (SSP1 to SSP5) were first scanned. Then, the spectral correlation was scanned in the predetermined region, resulting in the development of matching 3D and 2D imaging maps. The correlation is greater in M-FTIR pictures if their colors are darker. The maximum height values of the 3D picture vary between distinct SPN and SPI mixed groups because of the variability of the spatial distribution of SPI and SPN (Fig. 3 A1-E1). In Fig. 3 A3-E3, two apparent bands were displayed at 1693.3 cm−1, and 1581.4 cm−1, corresponding to amide I and II, respectively. Proteins were recognized to display a strong peak between 1500 and 1600 cm−1 [31], and changes in the ratio of SPI and SPN also significantly altered the strength of this distinctive peak. When compared to other treatment groups, the blue and red sections of the SSP4 group displayed an almost symmetrical, distinctive shape (Fig. 3 A2-E2), demonstrating that SPI and SPN were the most compatible in this group. Following the findings of Li et al. (2022) [31], the M-FTIR's dark blue and red areas independently displayed unique two-phase separation states. At this stage, the blue region kept growing while the red area steadily shrank, suggesting a significant alteration in the internal structure [32]. As summarized by Ling et al. (2013) [33], the conformational transition was often poor when the noncovalent contact between two substances in the system is significantly stronger than the interaction inside the same molecule. In this study, when the ratio of SPI to SPN reaches 1:2, the distinctive conformational shift of the SSP4 group may be connected with the distinctive structural modifications. This was consistent with unexpected transitions in the coupling sites and grafting rate in the SSP4 group mentioned above.
Fig. 3.
The effects of various SPI to SPN ratios on the three-dimensional (A1-E1), two-dimensional M-FTIR (A2-E2), and FTIR spectra (A3-E3) of UID-assisted processing systems.
3.5. Three-dimensional fluorescence
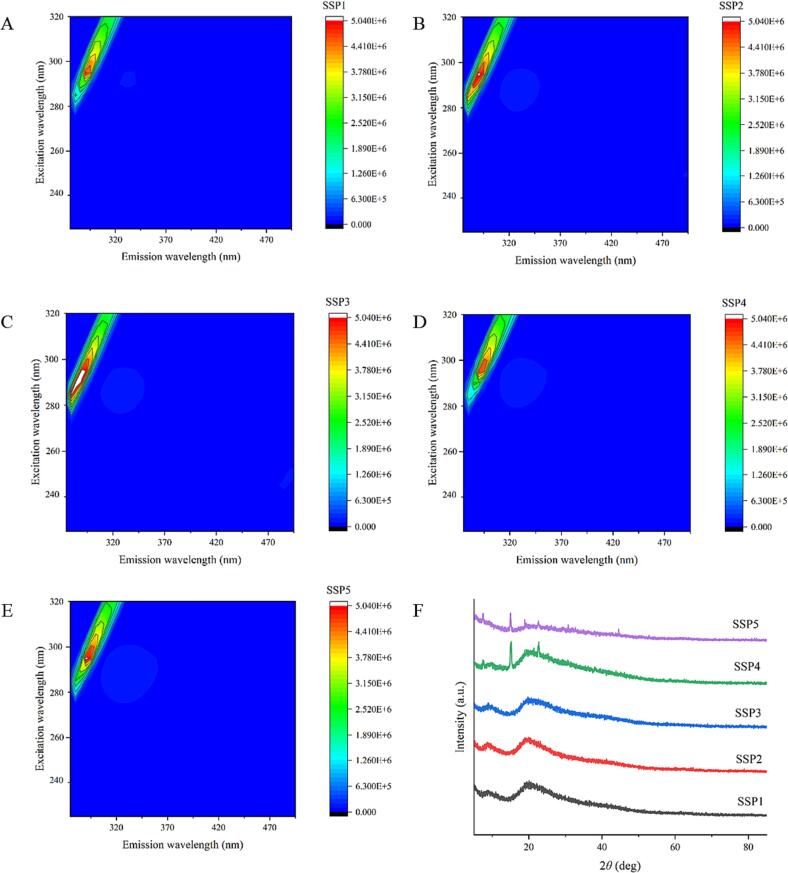
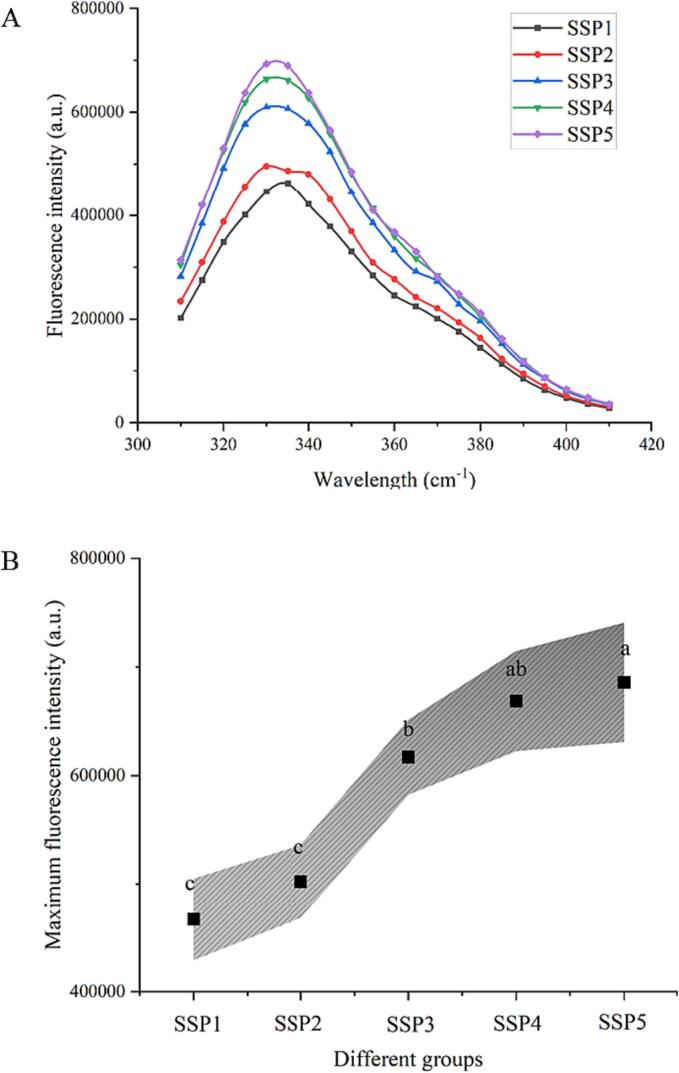
Three-dimensional fluorescence was used to analyze various SSP samples to ascertain their structural characteristics. Fig. 4 depicts the three-dimensional fluorescence diagram of the protein mixture containing SPI and SPN. The fluorescence peak at 290 nm emission wavelength mainly illustrates the fluorescence behavior of aromatic amino acid residues in the SSP samples [34]. As can be seen in Fig. 4 A-E, the highest fluorescence peak significantly enhances with an increase in the amount of SPN in the mixed “dual protein” system. This is explained by the considerably compact spherical shape and amino acid distribution of SPN. The fluorescence intensity of the distinctive peaks in the SSP4 group, however, dramatically decreased in contrast to other groups (Fig. 4D). Following the addition of polyphenols, proteins' fluorescence intensity often decreases noticeably, which is consistent with lighter-colored characteristic peaks in the three-dimensional fluorescence spectrum.
Fig. 4.
Three-dimensional fluorescence spectra (A-E) and XRD patterns (F) of protein mixtures with various SPI to SPN ratios (SSP1-SSP5).
The study conducted by Sui et al. (2018) [35] implied that the strong interaction between SPI and polyphenols may be responsible for the decrease in fluorescence intensity in SPI-polyphenol complexes. Furthermore, covalent connections of proteins with polyphenols are stronger than those of noncovalent contacts. This may be because quinone, which covalently interacts with tryptophan and tyrosine residues on SPI or SPN, is produced when Que undergoes oxidation in an alkaline environment. In summary, the SSP4 group's relatively high grafting rate causes a significant drop in fluorescence value, which is consistent with earlier findings in section 3.2. The results further confirm the effectiveness of iodometry in identifying the sono-physico-chemical impact in this experiment. According to the outcomings of Jia et al. (2016) [36], compact protein molecules promoted the embedding of fluorescent groups, hence limiting the exposure of tryptophan residues in whey protein isolate if they interact with polyphenols. Thus, the conformational changes of various proteins vary in their sources, and these factors should be carefully examined in different tests.
3.6. CD
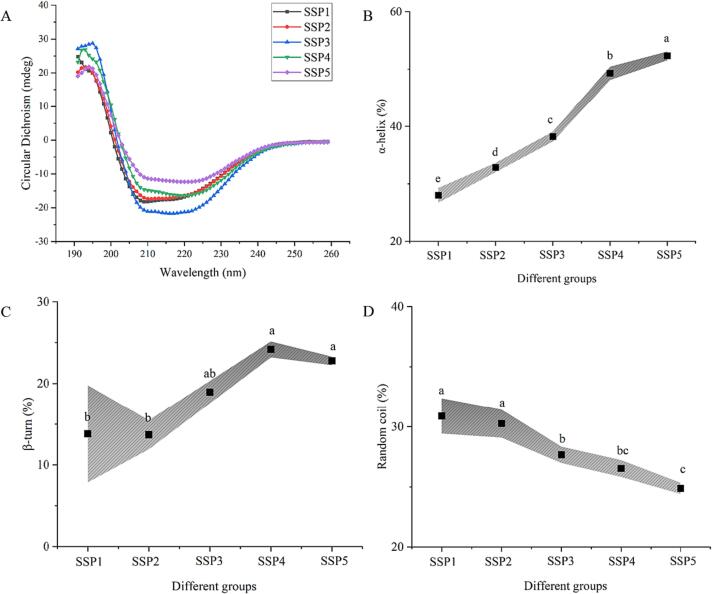
The secondary structural properties of several SSP samples (SSP1-SSP5) were examined using the CD technique. All SSP samples display typical bimodal patterns between 200 and 230 nm, which are illustrated in Fig. 5A. Notably, an increased SPN ratio in the “dual protein” system accelerates a rise in the proportion of α-helix structures. This contradicts the prior findings of sono-physico-chemical effects. In general, strong sono-physico-chemical impacts contribute to the reduction of proteins' dense spiral structure [13], [16]. In this regard, the hydrogen link between the carbonyl and amino groups of the peptide chain was broken by the local shear force and cavitation effect of UID [37]. The increased levels of SPN content in the SSP4 and SSP5 groups in this study demonstrated the higher α-helix when compared with other SSP samples (SSP1-SSP3). A spiral structure of around 14.20 % in SPI was reported by Yan et al. (2021) [38]. However, in the research on the physicochemical and functional characteristics of mussel SPNs, SPN is reported to be at 36.90 % [39]. It should be noted that the peculiar spherical shape of SPN makes it harder to be impacted by UID. In addition, the over-processing of UID causes the folding protein structures to reorganize, which also explains the reduced grafting rate of the SSP5 group in section 3.2. Therefore, the SSP4 and SSP5 groups exhibit higher α-helix content due to the stronger sono-physico-chemical effects.
Fig. 5.
The impacts of various SPI to SPN ratios on the CD spectra (A) and secondary structures (B-D) of UID-assisted processing systems.
Disordered structures, such as β-turn and random coil, display entirely distinct tendencies in different SSP samples (Fig. 5C-D). Damodaran et al. (2007) [40] concluded that the random coil reflected the protein's flexibility, demonstrating that stronger sono-physico-chemical impacts led to a more rigid secondary conformation in the “dual protein” system. The SPI or SPN during the Que reaction may also strengthen their interaction during UID, changing the hydrogen bonding pattern of the mixed proteins [38]. As a consequence, in the “dual protein” system, SPI is more sensitive to the sono-physico-chemical action of UID. Also, a larger SPN content in the mixed protein system contributes to the reburied of the protein's secondary structure.
3.7. Tryptophan fluorescence
The tertiary structural characteristics of numerous SSP samples (SSP1-SSP5) were investigated utilizing tryptophan fluorescence. The decline of tryptophan residue fluorescence is typically regarded as a significant signal of structural alterations in the protein system since tryptophan residues are extremely sensitive to the polarity of the protein environment [41]. Fig. 6 A-B displays the fluorescence spectra of distinct SSP complexes. Each SSP group had intense fluorescence, with the highest intensity remaining between 400,000 and 800,000. As illustrated in Fig. 6 B, varying SSP groups' fluorescence intensities steadily decrease when the ratio of SPI in the “dual protein” mixed system increases, suggesting that Que can interact with SPI or/and SPN. Besides, the SSP1 group presented the least amount of tryptophan fluorescence. The following two explanations can account for this result: The higher proportion of SPI in the SSP1 group, which indicates that SPI is more sensitive to UID-assisted Que processing than SPN, demonstrated the largest change in tryptophan fluorescence under the weakest sono-physico-chemical effect. This resulted in varying degrees of changes in the fluorescence of the final tryptophan residue. This is in line with the earlier findings of the section on the grafting rate. Dai et al. (2022)'s research also demonstrated that polyphenols induced the structure of SPI to unfold [42], leaving a significant amount of the SPI's initial hydrophobic groups exposed to the hydrophilic polar environment. Second, compared to SPN, SPI showed reduced intrinsic tryptophan fluorescence. Although the addition of polyphenols can make the protein's tryptophan residues more exposed and boost its unfolding impact, the actual change in fluorescence intensity is dependent on the protein's structural properties [23]. Afterward, it can be claimed that the proportion of SPN or SPI in the “dual protein” system may affect the sono-physico-chemical impacts on the tertiary structure.
Fig. 6.
The impacts of various SPI to SPN ratios on the endogenous fluorescence (A) and corresponding maximum fluorescence intensity (B) of UID-assisted processing systems.
3.8. XRD
The crystal structural properties of a variety of SSP samples (SSP1-SSP5) were examined utilizing the XRD technique. At angles of around 9° and 21°, two wide peaks can be seen in several SSP groups of proteins' XRD patterns (Fig. 4F). The spiral and folding structure were independently attributed to these broad peaks in a previous study [43]. The positions of the SSP2 and SSP3 groups did not exhibit significant differences when compared with those of SSP1. In contrast to the findings of SSP4 and SSP5, the XRD patterns of other groups are not impacted by the ratio of SPI to SPN. Based on the identical XRD peak patterns in Fig. 4F, the protein–protein interactions in SSP1, SSP2, and SSP3 do not alter noticeably during UID-assisted Que processing. Furthermore, it is plausible to conclude that intense sono-physico-chemical impacts caused changes in the structure and protein–protein interaction of mixed proteins, which were reflected in dramatic XRD spectrum shifts. It should be noted that XRD information modifications in SSP5 are weaker than those in SSP4. The findings show that the SSP5 group's strongest sono-physico-chemical action boosts the refolding of SPI or/and SPN. In addition, a large amount of SPN in the SSP5 system disrupts the balance of the “dual protein” system, which may weaken the interaction between SPI and SPN. These findings are consistent with Zhang et al.'s hypothesis that excessive intensity of sono-physico-chemical effects can change the distribution of sound fields [44]. At this point, SPI or SPN molecules might undergo severe denaturation or re-aggregation into new aggregates, and they also observed a significant increase in sample turbidity. These outcomes agree with those of the grafting rate test.
3.9. Apparent viscosity
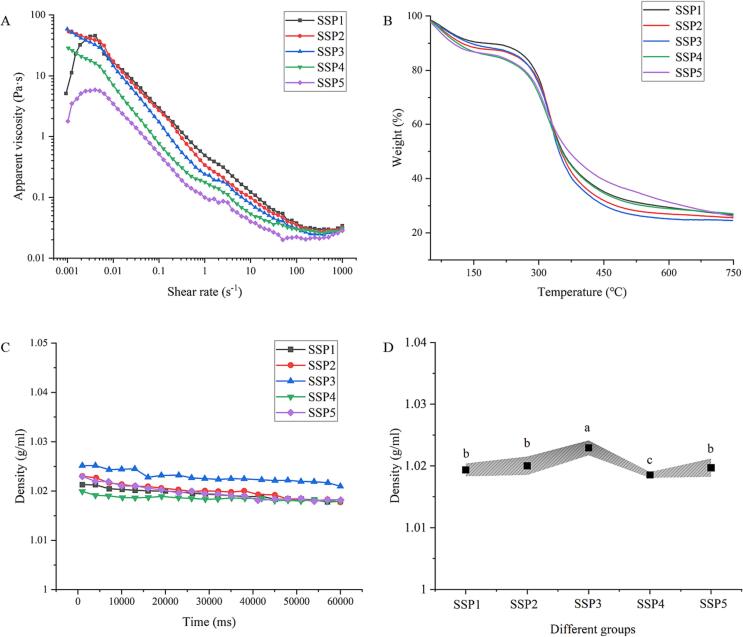
The rheological approach was used to investigate the apparent viscosity properties of numerous SSP samples (SSP1-SSP5). Fig. 7A describes the apparent viscosity-shear rate curve of the mixed protein system after the covalent modification of Que and UID. The consistent drop in apparent viscosity under conditions of increasing shear rate shows that all SSP samples behave in a manner characteristic of pseudo-plastic fluids [45]. The apparent viscosities of the SSP2 and SSP3 groups are significantly higher than those of the SSP4 and SSP5 groups when the shear rate is about 0.01 s−1. Yang et al. (2023) [46] reported that increased sono-physico-chemical effects might reduce the sample's particle size by causing more hydrophobic contacts and homogenous aggregation structures to raise the system's apparent viscosity. The apparent viscosity in Fig. 7A, on the other hand, showed the opposite pattern, probably as a result of the influence of Que covalent modification on protein viscosity. Similar findings were also found in the research of Chen et al. (2023) [26]. A prior study by Chen et al. (2022) [13] concluded that a relatively low apparent viscosity could enhance the sono-physico-chemical effects. This is understandable since the mixed “dual protein” system's strong oscillation impact motivates cavitation bubble production as sample viscosity decreases. The SSP4 and SSP5 groups' lower viscosities suggest that the mixed protein's increased SPN content facilitates cavitation bubble diffusion and amplifies its sono-physico-chemical action. The accuracy of iodometry in identifying sono-physico-chemical impacts in this investigation was once again confirmed by these rheological characteristics.
Fig. 7.
Apparent viscosity (A), TGA patterns (B), and density (C-D) of protein mixtures with various SPI to SPN ratios (SSP1-SSP5).
3.10. TGA
The TGA was used for investigation into the thermodynamic characteristics of some SSP samples (SSP1-SSP5). The thermos-gravimetric curves of all SSP samples changed similarly from 25 °C to 750 °C during the heating procedure (Fig. 7B). The first stage of weight loss in the mixed protein sample occurs between 50 and 150 °C and is significantly associated with changes in moisture content [14], [47]. At a heating temperature of around 700 °C, the weight of each SSP sample remained unaltered, proving that the majority of the components in SPI or SPN have undergone full degradation. Given that most of the proteins are broken down at a high temperature of 700 °C, the structural change in the SSP sample is positively related to weight loss. At a temperature of approximately 700 °C, the residual rates for the SSP1, SSP2, SSP3, SSP4, and SSP5 samples were 27.44 %, 26.06 %, 24.77 %, 27.72 %, and 27.70 %, respectively. The findings confirmed SSP4 and SSP5 systems' higher structural stability by demonstrating that their thermal deterioration was less pronounced than that of SSP2 and SSP3 groups. This conclusion is understandable, as it takes more energy to break more complex covalent bonds. Also, SPI and SPN underwent substantial denaturation and increased thermal stability in the SSP4 and SSP5 groups as a result of intense sonic cavitation. The cavitation and hyper-mixing effects of UID treatment promoted the creation of mixed complexes, thus improving their thermal stability [48]. The internal structure's compactness of mixed protein can also increase with a rise in SPN concentration. In this sense, excessive sono-physico-chemical impacts may harm the thermal stability of SSP samples, leading to a reduction in the residual rate of SSP5 samples. The results of this experiment further supported the adverse consequences of high SPN levels on the “dual protein” system's structural integrity when subjected to severe sono-physico-chemical effects.
3.11. System density
Fig. 7C-D displayed the system density for all SSP samples (SSP1-SSP5). The groups with increased action intensity (SSP4 and SSP5) exhibit a lower density when compared to the group with reduced sono-physico-chemical effects (SSP2 and SSP3). This could be connected with the physical action of UID on proteins. Wang et al. (2022) [49] reported that UID may be able to decrease the particle size of protein particles and their irregular aggregation. In this regard, the homogeneity of the protein system is finally improved. Furthermore, strong sono-physico-chemical impacts also promote protein unfolding, corresponding to protein expansion in SSP systems. Under the same mass conditions, an increase in volume causes a drop in system density. Therefore, the changes in system density and protein structure in this study are entirely correlated.
3.12. Microstructure
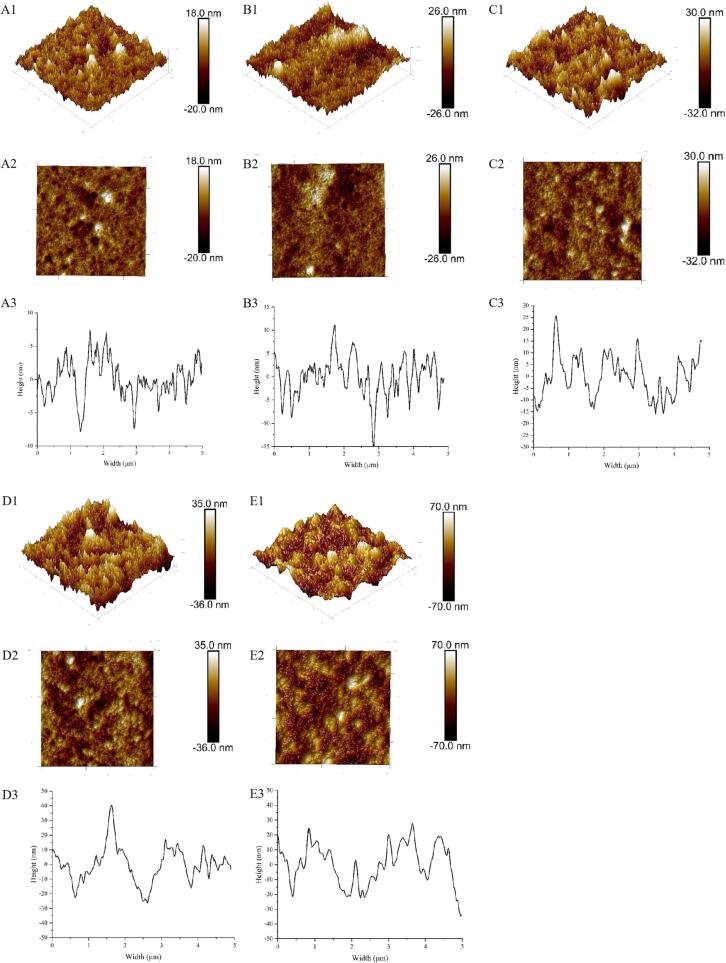
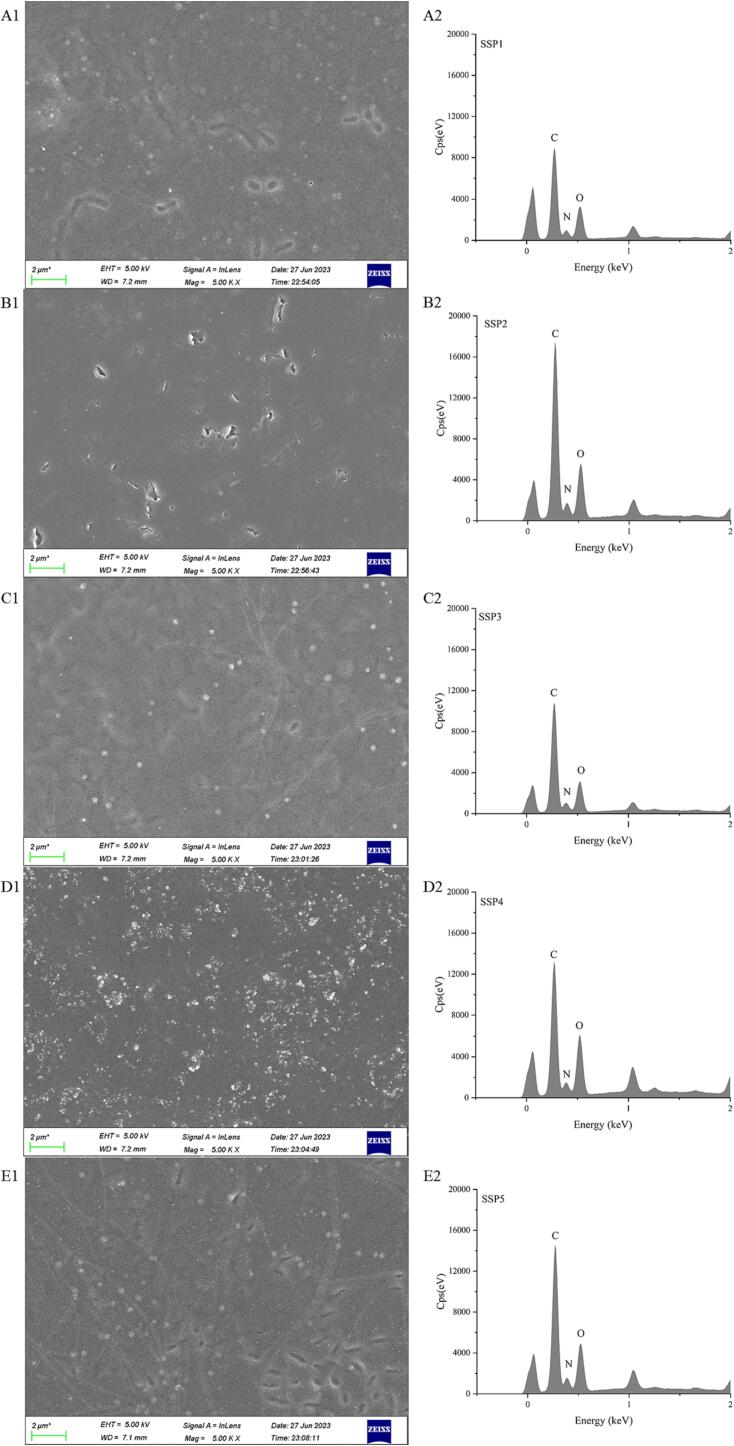
SEME and AFME were used for examining the microstructure across multiple SSP samples (SSP1-SSP5). Fig. 8 illustrates that several SSP samples have a small number of spherical particle structures. The SSP3 and SSP4 group samples were rougher than those of the SSP1 and SSP2 groups, showing an irregular particle structure. More so, SPI and SPN molecules are both spherical particles, as reported by other investigations [50], [51], however, SSP samples show considerable amounts of uneven aggregate structures. The results showed that the mixed proteins were dispersed and broken up into aggregates under UID, changing the protein structure and its properties. These particles reaggregate into numerous bigger aggregates with a height higher than 10 nm when the ratio of SPI and SPN rises from 3:1 to 1:2. This finding suggests that, following UID treatment, the microstructure of SPI or SPN is degraded to varied degrees, displaying varying degrees of looseness [52]. The phenomena of protein reassembly following ultrasonography have also been supported by other investigations [53], [54]. The outcomes of SEME pictures are also exhibited in Fig. 9. It's interesting to note that the SSP4 group contains more proteins in various forms than the other groups. Accordingly, the strong ultrasonic impact facilitates protein structure breakdown and increases the probability that proteins are to come into contact with other molecules. Additionally, the SSP4 group's strong sono-physico-chemical action caused Que molecules to connect to its surface. This explains the growth in protein particle size. The oxygen element in the SSP4 group displays the largest peak value in the element distribution map (Fig. 9A2-E2). This demonstrates that the coupling of SPI and SPN with Que is the process that causes the microstructural alterations in the “dual protein” system. Increasing the SPN ratio shows a considerable impact on protein aggregation in the “dual protein” system, as further evidenced by the examination of the microstructure.
Fig. 8.
AFME images, including three-dimensional (A1-E1), two-dimensional pictures (A2-E2), and cross-sections (A3-E3) of protein mixtures with various SPI to SPN ratios (SSP1-SSP5).
Fig. 9.
SEME images (A1-E1) of protein mixtures with various SPI to SPN ratios (SSP1: A1 and A2, SSP2: B1 and B2, SSP3: C1 and C2, SSP4: D1 and D2, SSP5: E1 and E2) and their elemental distribution (A2-E2).
4. Conclusions
The findings of this study demonstrate that the increased level of SPN in the mixed protein system, which can be preliminary assessed by iodometry, increases the strength of the sono-physico-chemical action. This work further supported the finding that SPI has more Que modification sites than SPN, making it more susceptible to the effects of sono-physico-chemical action in the “dual protein” system. However, the number of modification sites in SPN is substantially larger than that in SPI under oxidative conditions, demonstrating the oxidative sensitivity of SPN. A considerable drop in coupling rate and protein reaggregation may result from severe reactions and internal hotspots caused by excessive sono-physico-chemical effects. It should be stressed that the increased SPN concentration in the mixed protein system makes the protein's secondary structure more likely to be reburied. Results from XRD and TGA show that strong sono-physico-chemical impacts can also modify the structure of mixed proteins and protein–protein interactions. According to the results of viscosity and microstructure, a rise in the SPN content of mixed proteins facilitates the impact of sonic cavitation by lowering viscosity and generating aggregates. To process the dual protein system using UID, a ratio of 1:1 or 1:2 for SPI and SPN is preferable.
CRediT authorship contribution statement
Miao Zhang: Conceptualization, Methodology, Data curation, Writing – original draft. Dejiang Xue: Investigation, Formal analysis. Ya Chen: Formal analysis, Data curation. Yanan Li: Visualization. Chunbao Li: Conceptualization, Writing – review & editing, Supervision, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the Youth Project of National Natural Science Foundation of Jiangsu Province (BK20231001), Jiangsu Funding Program for Excellent Postdoctoral Talent (2022ZB335) and the Jiangsu Department of Education (Innovation Group of Meat Nutrition, Health and Biotechnology).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ultsonch.2023.106639.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
The data that has been used is confidential.
References
- 1.Chen Q., Xue C., He Z., Wang Z., Qin F., Chen J., Zeng M. Generation of sarcoplasmic and myofibrillar protein-bound heterocyclic amines in chemical model systems under different heating temperatures and durations. J. Agric. Food Chem. 2021;69:3232–3246. doi: 10.1021/acs.jafc.0c06885. [DOI] [PubMed] [Google Scholar]
- 2.Matak K.E., Tahergorabi R., Jaczynski J. A review: Protein isolates recovered by isoelectric solubilization/precipitation processing from muscle food by-products as a component of nutraceutical foods. Food Res. Int. 2015;77:697–703. [Google Scholar]
- 3.Xia M., Chen Y., Guo J., Huang H., Wang L., Wu W., Xiong G., Sun W. Water distribution and textual properties of heat-induced pork myofibrillar protein gel as affected by sarcoplasmic protein, LWT-Food. Sci. Technol. 2019;103:308–315. [Google Scholar]
- 4.Zhao X., Xing T., Wang Y., Xu X., Zhou G. Isoelectric solubilization/precipitation processing modified sarcoplasmic protein from pale, soft, exudative-like chicken meat. Food Chem. 2019;287:1–10. doi: 10.1016/j.foodchem.2019.02.085. [DOI] [PubMed] [Google Scholar]
- 5.Huang J., Liao L.M., Weinstein S.J., Sinha R., Graubard B.I., Albanes D. Association between plant and animal protein intake and overall and cause-specific mortality. JAMA Intern. Med. 2020;180:1173–1184. doi: 10.1001/jamainternmed.2020.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aiking H., de Boer J. The next protein transition. Trends Food Sci. Tech. 2020;105:515–522. doi: 10.1016/j.tifs.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou T., Wu J., Zhao M., Ke W., Shan K., Zhao D., Li C. Effect of Natural plant extracts on the quality of meat products: A meta-analysis. Food Materials Research. 2023;3 [Google Scholar]
- 8.Sha L., Xiong Y.L. Plant protein-based alternatives of reconstructed meat: Science, technology, and challenges. Trends Food Sci. Tech. 2020;102:51–61. [Google Scholar]
- 9.Day L., Cakebread J.A., Loveday S.M. Food proteins from animals and plants: Differences in the nutritional and functional properties. Trends Food Sci. Tech. 2022;119:428–442. [Google Scholar]
- 10.Gao T., Liu J., Gao X., Zhang G., Tang X. Stability and Digestive Properties of a Dual-Protein Emulsion System Based on Soy Protein Isolate and Whey Protein Isolate. Foods. 2023;12:2247. doi: 10.3390/foods12112247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sui X., Zhang T., Jiang L. Soy protein: Molecular structure revisited and recent advances in processing technologies. Annu. Rev. Food Sci. t. 2021;12:119–147. doi: 10.1146/annurev-food-062220-104405. [DOI] [PubMed] [Google Scholar]
- 12.Zhang T., Dou W., Zhang X., Zhao Y., Zhang Y., Jiang L., Sui X. The development history and recent updates on soy protein-based meat alternatives. Trends Food Sci. Tech. 2021;109:702–710. [Google Scholar]
- 13.Chen J., Chen X., Zhou G., Xu X. New insights into the ultrasound impact on covalent reactions of myofibrillar protein. Ultrason. Sonochem. 2022;84 doi: 10.1016/j.ultsonch.2022.105973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J., Zhang X., Chen X., Bassey A.P., Zhou G., Xu X. Phenolic modification of myofibrillar protein enhanced by ultrasound: The structure of phenol matters. Food Chem. 2022;386 doi: 10.1016/j.foodchem.2022.132662. [DOI] [PubMed] [Google Scholar]
- 15.Chen K., Chen X., Liang L., Xu X. Gallic acid-aided cross-linking of myofibrillar protein fabricated soluble aggregates for enhanced thermal stability and a tunable colloidal state. J. Agric. Food Chem. 2020;68:11535–11544. doi: 10.1021/acs.jafc.0c02059. [DOI] [PubMed] [Google Scholar]
- 16.Chen J., Zeng X., Sun X., Zhou G., Xu X. A comparison of the impacts of different polysaccharides on the sono-physico-chemical consequences of ultrasonic-assisted modifications. Ultrason. Sonochem. 2023;96 doi: 10.1016/j.ultsonch.2023.106427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen J., Chen X., Zhou G., Xu X. Ultrasound: A reliable method for regulating food component interactions in protein-based food matrices. Trends Food Sci. Tech. 2022 [Google Scholar]
- 18.Wang W., Sun C., Mao L., Ma P., Liu F., Yang J., Gao Y. The biological activities, chemical stability, metabolism and delivery systems of quercetin: A review. Trends Food Sci. Tech. 2016;56:21–38. [Google Scholar]
- 19.Singh P., Arif Y., Bajguz A., Hayat S. The role of quercetin in plants. Plant Physiol. Bioch. 2021;166:10–19. doi: 10.1016/j.plaphy.2021.05.023. [DOI] [PubMed] [Google Scholar]
- 20.Cuevas-Bernardino J.C., Leyva-Gutierrez F.M., Vernon-Carter E.J., Lobato-Calleros C., Román-Guerrero A., Davidov-Pardo G. Formation of biopolymer complexes composed of pea protein and mesquite gum–Impact of quercetin addition on their physical and chemical stability. Food Hydrocolloid. 2018;77:736–745. [Google Scholar]
- 21.Sun X., Sarteshnizi R.A., Udenigwe C.C. Recent advances in protein–polyphenol interactions focusing on structural properties related to antioxidant activities. Current Opin. Food Sci. 2022;45 [Google Scholar]
- 22.Zhang M., Gao T., Han Y., Xue D., Jiang S., Li Q., Li C. Improvement of Structural, Rheological, and physicochemical properties of type I collagen by calcium lactate combined with ultrasound. Ultrason. Sonochem. 2023;95 doi: 10.1016/j.ultsonch.2023.106373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao Q., Chen J., Zhou G., Xu X. Different protein-anthocyanin complexes engineered by ultrasound and alkali treatment: Structural characterization and color stability. Food Chem. 2023;136693 doi: 10.1016/j.foodchem.2023.136693. [DOI] [PubMed] [Google Scholar]
- 24.Chen J., Chai J., Chen X., Huang M., Zeng X., Xu X. Development of edible films by incorporating nanocrystalline cellulose and anthocyanins into modified myofibrillar proteins. Food Chem. 2023;417 doi: 10.1016/j.foodchem.2023.135820. [DOI] [PubMed] [Google Scholar]
- 25.Pan J., Lian H., Jia H., Li S., Hao R., Wang Y., Zhang X., Dong X. Ultrasound treatment modified the functional mode of gallic acid on properties of fish myofibrillar protein. Food Chem. 2020;320 doi: 10.1016/j.foodchem.2020.126637. [DOI] [PubMed] [Google Scholar]
- 26.Chen J., Zeng X., Chai J., Zhou G., Xu X. Improvement of the emulsifying properties of mixed emulsifiers by optimizing ultrasonic-assisted processing. Ultrason. Sonochem. 2023;95 doi: 10.1016/j.ultsonch.2023.106397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J., Liu Q., Chen Q., Sun F., Liu H., Kong B. Synergistic modification of pea protein structure using high-intensity ultrasound and pH-shifting technology to improve solubility and emulsification. Ultrason. Sonochem. 2022;88 doi: 10.1016/j.ultsonch.2022.106099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han G., Xu J., Chen Q., Xia X., Liu H., Kong B. Improving the solubility of myofibrillar proteins in water by destroying and suppressing myosin molecular assembly via glycation. Food Chem. 2022;395 doi: 10.1016/j.foodchem.2022.133590. [DOI] [PubMed] [Google Scholar]
- 29.Xu Y., Han M., Huang M., Xu X. Enhanced heat stability and antioxidant activity of myofibrillar protein-dextran conjugate by the covalent adduction of polyphenols. Food Chem. 2021;352 doi: 10.1016/j.foodchem.2021.129376. [DOI] [PubMed] [Google Scholar]
- 30.Wang C., Zhao F., Bai Y., Li C., Xu X., Kristiansen K., Zhou G. Effect of gastrointestinal alterations mimicking elderly conditions on in vitro digestion of meat and soy proteins. Food Chem. 2022;383 doi: 10.1016/j.foodchem.2022.132465. [DOI] [PubMed] [Google Scholar]
- 31.Li X., Fan M., Huang Q., Zhao S., Xiong S., Yin T., Zhang B. Effect of micro-and nano-starch on the gel properties, microstructure and water mobility of myofibrillar protein from grass carp. Food Chem. 2022;366 doi: 10.1016/j.foodchem.2021.130579. [DOI] [PubMed] [Google Scholar]
- 32.Lu Y., Jia B., Yoon S.-C., Zhuang H., Ni X., Guo B., Gold S.E., Fountain J.C., Glenn A.E., Lawrence K.C. Spatio-temporal patterns of Aspergillus flavus infection and aflatoxin B1 biosynthesis on maize kernels probed by SWIR hyperspectral imaging and synchrotron FTIR microspectroscopy. Food Chem. 2022;382 doi: 10.1016/j.foodchem.2022.132340. [DOI] [PubMed] [Google Scholar]
- 33.Ling S., Qi Z., Knight D.P., Shao Z., Chen X. FTIR imaging, a useful method for studying the compatibility of silk fibroin-based polymer blends. Polym. Chem. 2013;4:5401–5406. [Google Scholar]
- 34.Jiang L., Liu Y., Li L., Qi B., Ju M., Xu Y., Zhang Y., Sui X. Covalent conjugates of anthocyanins to soy protein: Unravelling their structure features and in vitro gastrointestinal digestion fate. Food Res. Int. 2019;120:603–609. doi: 10.1016/j.foodres.2018.11.011. [DOI] [PubMed] [Google Scholar]
- 35.Sui X., Sun H., Qi B., Zhang M., Li Y., Jiang L. Functional and conformational changes to soy proteins accompanying anthocyanins: Focus on covalent and non-covalent interactions. Food Chem. 2018;245:871–878. doi: 10.1016/j.foodchem.2017.11.090. [DOI] [PubMed] [Google Scholar]
- 36.Jia Z., Zheng M., Tao F., Chen W., Huang G., Jiang J. Effect of covalent modification by (−)-epigallocatechin-3-gallate on physicochemical and functional properties of whey protein isolate, LWT-Food. Sci. Technol. 2016;66:305–310. [Google Scholar]
- 37.Li Z., Zheng Y., Sun Q., Wang J., Zheng B., Guo Z. Structural characteristics and emulsifying properties of myofibrillar protein-dextran conjugates induced by ultrasound Maillard reaction. Ultrason. Sonochem. 2021;72 doi: 10.1016/j.ultsonch.2020.105458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan S., Xie F., Zhang S., Jiang L., Qi B., Li Y. Effects of soybean protein isolate− polyphenol conjugate formation on the protein structure and emulsifying properties: Protein-polyphenol emulsification performance in the presence of chitosan. Colloid Surface a. 2021;609 [Google Scholar]
- 39.Zou H., Zhao N., Sun S., Dong X., Yu C. High-intensity ultrasonication treatment improved physicochemical and functional properties of mussel sarcoplasmic proteins and enhanced the stability of oil-in-water emulsion. Colloid Surface a. 2020;589 [Google Scholar]
- 40.Damodaran S., Parkin K.L., Fennema O.R. CRC Press; 2007. Fennema's food chemistry. [Google Scholar]
- 41.Li S., Tang S., Mo R., Li J., Chen L. Effects of NaCl curing and subsequent fermentation with Lactobacillus sakei or Lactobacillus plantarum on protein hydrolysis and oxidation in yak jerky. LWT-Food Sci. Technol. 2023;173 [Google Scholar]
- 42.Dai S., Lian Z., Qi W., Chen Y., Tong X., Tian T., Lyu B., Wang M., Wang H., Jiang L. Non-covalent interaction of soy protein isolate and catechin: Mechanism and effects on protein conformation. Food Chem. 2022;384 doi: 10.1016/j.foodchem.2022.132507. [DOI] [PubMed] [Google Scholar]
- 43.Chen J., Chen X., Zhu Q., Chen F., Zhao X., Ao Q. Determination of the domain structure of the 7S and 11S globulins from soy proteins by XRD and FTIR. J. Sci. Food Agr. 2013;93:1687–1691. doi: 10.1002/jsfa.5950. [DOI] [PubMed] [Google Scholar]
- 44.Zhang L., Wang X., Hu Y., Fakayode O.A., Ma H., Zhou C., Hu Z., Xia A., Li Q. Dual-frequency multi-angle ultrasonic processing technology and its real-time monitoring on physicochemical properties of raw soymilk and soybean protein. Ultrason. Sonochem. 2021;80 doi: 10.1016/j.ultsonch.2021.105803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang S., Lian Z., Wang M., Liao P., Wu H., Cao J., Tong X., Tian T., Wang H., Jiang L. Molecular structural modification of β-conglycinin using pH-shifting with ultrasound to improve emulsifying properties and stability. Ultrason. Sonochem. 2022;90 doi: 10.1016/j.ultsonch.2022.106186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang J., Dou J., Zhu B., Ning Y., Wang H., Huang Y., Li Y., Qi B., Jiang L. Multi-dimensional analysis of heat-induced soybean protein hydrolysate gels subjected to ultrasound-assisted pH pretreatment. Ultrason. Sonochem. 2023;95 doi: 10.1016/j.ultsonch.2023.106403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He L., Gao Y., Wang X., Han L., Yu Q., Shi H., Song R. Ultrasonication promotes extraction of antioxidant peptides from oxhide gelatin by modifying collagen molecule structure. Ultrason. Sonochem. 2021;78 doi: 10.1016/j.ultsonch.2021.105738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu Y., Wang T., Gong Y., Wang W., Wang X., Yu D., Wu F., Wang L. Effect of ultrasound on the structural characteristics and oxidative stability of walnut oil oleogel coated with soy protein isolate-phosphatidylserine. Ultrason. Sonochem. 2022;83 doi: 10.1016/j.ultsonch.2022.105945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y., Li B., Guo Y., Liu C., Liu J., Tan B., Guo Z., Wang Z., Jiang L. Effects of ultrasound on the structural and emulsifying properties and interfacial properties of oxidized soybean protein aggregates. Ultrason. Sonochem. 2022;87 doi: 10.1016/j.ultsonch.2022.106046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi C., He Y., Ding M., Wang Y., Zhong J. Nanoimaging of food proteins by atomic force microscopy. Part II: Application for food proteins from different sources. Trends Food Sci. Tech. 2019;87:14–25. [Google Scholar]
- 51.Zhao X., Xing T., Xu X., Zhou G. Influence of extreme alkaline pH induced unfolding and aggregation on PSE-like chicken protein edible film formation. Food Chem. 2020;319 doi: 10.1016/j.foodchem.2020.126574. [DOI] [PubMed] [Google Scholar]
- 52.Xu B., Yuan J., Wang L., Lu F., Wei B., Azam R.S., Ren X., Zhou C., Ma H., Bhandari B. Effect of multi-frequency power ultrasound (MFPU) treatment on enzyme hydrolysis of casein. Ultrason. Sonochem. 2020;63 doi: 10.1016/j.ultsonch.2019.104930. [DOI] [PubMed] [Google Scholar]
- 53.Tian R., Feng J., Huang G., Tian B., Zhang Y., Jiang L., Sui X. Ultrasound driven conformational and physicochemical changes of soy protein hydrolysates. Ultrason. Sonochem. 2020;68 doi: 10.1016/j.ultsonch.2020.105202. [DOI] [PubMed] [Google Scholar]
- 54.Yan G., Cui Y., Lia D., Ding Y., Han J., Wang S., Yang Q., Zheng H. The characteristics of soybean protein isolate obtained by synergistic modification of high hydrostatic pressure and phospholipids as a promising replacement of milk in ice cream. LWT-Food Sci. Technol. 2022;160 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that has been used is confidential.