Abstract
Diabetic kidney disease (DKD) is a common microvascular complication of diabetes, a global health issue. Hyperglycemia, in concert with cytokines, activates the Janus kinase (JAK)/signal transducer and activator of transcription (STAT) pathway to induce inflammation and oxidative stress contributing to renal damage. There is evidence of microRNA-155 (miR-155) involvement in diabetes complications, but the underlying mechanisms are unclear. In this study, gain- and loss-of-function experiments were conducted to investigate the interplay between miR-155-5p and suppressor of cytokine signaling 1 (SOCS1) in the regulation of the JAK/STAT pathway during renal inflammation and DKD. In experimental models of mesangial injury and diabetes, miR-155-5p expression correlated inversely with SOCS1 and positively with albuminuria and expression levels of cytokines and prooxidant genes. In renal cells, miR-155-5p mimic downregulated SOCS1 and promoted STAT1/3 activation, cytokine expression, and cell proliferation and migration. Conversely, both miR-155-5p antagonism and SOCS1 overexpression protected cells from inflammation and hyperglycemia damage. In vivo, SOCS1 gene delivery decreased miR-155-5p and kidney injury in diabetic mice. Moreover, therapeutic inhibition of miR-155-5p suppressed STAT1/3 activation and alleviated albuminuria, mesangial damage, and renal expression of inflammatory and fibrotic genes. In conclusion, modulation of the miR-155/SOCS1 axis protects kidneys against diabetic damage, thus highlighting its potential as therapeutic target for DKD.
Keywords: MT: Non-coding RNAs: diabetes, renal inflammation, diabetic kidney disease, fibrosis, microRNA, signal transduction, JAK/STAT/SOCS, cytokines, hyperglycemia
Graphical abstract

Gomez-Guerrero and colleagues report that miR-155-5p promotes renal damage through targeting SOCS1, leading to JAK/STAT activation, cytokine expression, and cell proliferation and migration. In diabetic mice, both miR-155-5p inhibition and SOCS1 overexpression attenuate kidney damage, inflammation, and fibrosis, thus supporting the therapeutic potential of the miR-155/SOCS1 axis in human diabetic kidney disease.
Introduction
Diabetes mellitus is a common and serious metabolic disease with significant clinical impact and economic costs and a leading cause of disability, morbidity, and premature mortality worldwide. These outcomes are due largely to macrovascular complications (e.g., coronary heart disease, peripheral arterial disease, stroke) and microvascular complications including diabetic kidney disease (DKD), peripheral neuropathy, and retinopathy.1 DKD affects about one-third of patients with diabetes, is the primary cause of end-stage renal disease, and is a risk factor for macrovascular complications.2 Furthermore, hyperlipidemia contributes to the progression of diabetic chronic kidney disease, indicating an interrelationship between the two processes.3
Because of extensive research in recent years, our understanding of the metabolic and hemodynamic alterations as the main cause of renal injury in diabetes has been transformed into a more complex scenario. In fact, a diabetic milieu promotes the accumulation of immune cells, whose mediators can activate resident renal cells by direct contact or paracrine signaling to secrete additional pro-inflammatory cytokines and activate oxidative stress responses, thereby amplifying the effects of hyperglycemia and potentiating renal damage.4,5
Current management of DKD relies on the control of blood glucose, hypertension, and lipid levels. Because many patients still progress to advanced DKD and end-stage renal disease, there is a need to identify diabetes-induced mechanisms regulating inflammation and oxidative stress genes that may be relevant in the search of alternative therapies.2,5
MicroRNAs (miRNAs) are fundamental post-transcriptional regulators of gene expression involved in many biological processes, including differentiation, development, signaling, and immune response. miRNAs are small non-coding RNAs that bind primarily to the 3′ UTR of target mRNAs and mediate mRNA degradation, translational repression, or mRNA destabilization.6 Dysregulated expression of specific miRNA-mRNA target pairs has relevance not only to tumorigenesis but also to developmental and metabolic diseases.7 Previous studies have explored the diagnostic and therapeutic value of miRNAs in kidney diseases. For example, increased levels of miR-21, miR-27a, miR-146, and miR-216 contribute to renal fibrosis by regulating mesangial cell hypertrophy, extracellular matrix accumulation, epithelial-to-mesenchymal transition and transforming growth factor β (TGF-β) signaling,8,9 while miR-27b, miR-29, and miR-192 are found to be downregulated and revealed inhibitory effects on kidney damage and fibrosis.10,11,12 The present work focusses on the role and regulation of miR-155 during renal inflammation and diabetes.
miR-155 is considered a master mediator of immune and inflammatory responses in different pathologies.13 The pre-miR-155 hairpin is processed into two mature strands (miR-155-3p and miR-155-5p), of which miR-155-5p is the most abundant and functionally relevant form (miRBase: hsa-mir-155, MI0000681 and mmu-mir-155, MI0000177). miR-155 increases the expression of genes involved in inflammation and renal fibrosis, such as interleukin (IL)-1β, IL-6, IL-12, tumor necrosis factor α (TNF-α), TGF-β, inducible nitric oxide synthase, angiotensin II receptors, and metalloproteinases.14 Elevated miR-155 levels have been found in urine samples from patients with acute and chronic kidney diseases.9,11,15 In rodent models of type 1 diabetes (T1D) and type 2 diabetes (T2D), miR-155 expression associates with the progression of kidney disease,16,17 while constitutive miR-155 gene deficiency alleviates renal damage.18,19
miR-155 expression is controlled by multiple signaling pathways and transcription factors.14 Among them is the Janus kinase (JAK)/signal transducer and activator of transcription (STAT) pathway, which mediates the effects of many cytokines and inflammatory molecules.20 In the canonical signaling, STAT transcription factors are activated by JAK-mediated phosphorylation, then dimerize and translocate into the nucleus to regulate target gene expression.21 Suppressor of cytokine signaling 1 (SOCS1) is an important negative regulator of cytokine signaling by inhibiting JAK activation, STAT dimerization, or targeting ubiquitinated signal transduction factors.22 Dysregulated functionality of the JAK/STAT/SOCS pathway contributes to the harmful effects of hyperglycemia by inducing the expression of genes implicated in leukocyte infiltration, cell growth, and fibrosis,23 and hence therapies counteracting JAK/STAT hyperactivation in diabetes are being considered.24,25 Previous studies have revealed that miR-155-associated inflammatory response to lipopolysaccharide (LPS) in the kidney is mediated by SOCS1 repression.26 However, the role of miR-155/SOCS1 as a functional axis in the pathogenesis of DKD is not fully defined. Therefore, the aim of this study is to analyze the interplay between the JAK/STAT/SOCS1 pathway and epigenetic regulation by miR-155-5p in renal cells and to test in vivo the inhibition of miR-155-5p as a potential therapy against DKD.
Results
miR-155-5p is induced by inflammation and hyperglycemia in renal cells and damaged kidney and correlates inversely with SOCS1 expression
We analyzed the expression of miR-155-5p in cultured renal cells exposed to inflammatory and hyperglycemic conditions. In glomerular mesangial cells, miR-155-5p expression was induced by stimulation with cytokines (TNF-α, IL-6, and interferon-γ [IFNγ]), LPS alone, a combination of LPS plus IFNγ, and high glucose concentration, compared with basal conditions (Figure 1A). In proximal tubular epithelial cells, time-response studies revealed a gradual increase of miR-155-5p expression by LPS or high-glucose treatment, whereas IL-1β and TGF-β induced an earlier expression, peaking at 3 and 6 h, respectively (Figure 1B). We further analyzed SOCS1 expression, which was identified as a direct target of miR-155-5p in other cell types.27 SOCS1 mRNA was transiently induced by inflammatory stimuli and hyperglycemia in tubular epithelial cells (Figure 1B), showing an inverse pattern of expression with that of miR-155-5p.
Figure 1.
Inverse relationship between miR-155-5p and SOCS1 in murine renal cells and damaged kidneys
(A) miR-155-5p expression in mesangial cells stimulated with inflammatory stimuli (10 ng/mL TNF-α, 2 ng/mL IL-6, 200 ng/mL IFNγ, and 1 μg/mL LPS) for 2 h or high glucose concentration (HG; 30 mM D-glucose) for 24 h. (B) Evolution of miR-155-5p and SOCS1 expression in tubular epithelial cells after 3–24 h of incubation with 10 ng/mL IL-1β, 100 ng/mL LPS, 10 ng/mL TGF-β, and HG. (C and D) Analysis of samples from the mouse model of mesangial proliferative glomerulonephritis. (C) Renal expression of miR-155-5p and SOCS1 and levels of albuminuria (urine albumin-to-creatinine ratio, UACR) at days 0 (n = 5), 5 (n = 7), and 7 (n = 6) of disease induction. (D) Correlation analysis of miR-155-5p versus SOCS1 at day 5 and 7. (E and F) Analysis of samples from the T1D model in WT and ApoE mice. (E) Levels of miR-155-5p, SOCS1, and UACR in normoglycemic (WT-Co, n = 5; ApoE-Co, n = 6) and diabetic (WT-DM, n = 6, ApoE-DM, n = 7) mouse groups. (F) Correlation analysis of miR-155-5p levels versus SOCS1. The qPCR values were normalized by RNU6B (miR-155-5p) or 18S rRNA (mRNAs) and expressed as fold increases versus basal (A and B) and control mouse group (C – F). Results are presented as individual data points and the mean ± SD of n = 4 (in vitro experiments) and the total number of animals per group. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 vs. basal cells or indicated groups.
To corroborate the findings in renal cells under inflammatory and hyperglycemic conditions, we next explored in vivo the renal expression of miR-155-5p and SOCS1 in two complementary experimental models of acute and chronic kidney disease. First, in the mouse model of mesangial proliferative glomerulonephritis characterized by acute glomerulopathy and inflammation, we observed a progressive induction of miR-155-5p over 7 days in parallel with the increase of albuminuria (urine albumin-to creatinine ratio, UACR), while SOCS1 mRNA expression was maximal at day 5 and declined thereafter (Figure 1C). Pearson analysis revealed an inverse correlation between miR-155-5p and SOCS1 levels (Figure 1D) and positive association with albuminuria (r = 0.7983, p = 0.0011) and the gene expression levels of renal damage marker KIM1 (kidney injury molecule 1; r = 0.6939, p = 0.0085) and C-C motif ligand (CCL) 2 chemokine (r = 0.7364, p = 0.0041).
Second, we studied the model of chronic DKD induced by streptozotocin (STZ) injection in aged wild-type (WT) and apolipoprotein E knockout (ApoE) mice. We observed that the miR-155-5p expression in diabetic mice was significantly higher than in respective non-diabetic controls, with the ApoE diabetic group displaying the highest levels of miR-155-5p (5-fold vs. control) and albuminuria (Figure 1E). Moreover, SOCS1 gene expression was found decreased in diabetic mice compared with controls (Figure 1E). Correlation analysis showed an inverse relationship between miR-155-5p and SOCS1 among all mouse groups (Figure 1F). There was also a direct association of miR-155-5p with albuminuria, markers of renal damage (KIM1) and inflammation (CCL2 and CCL5), and prooxidant enzymes (NADPH oxidase subunits NOX2 and NOX4), and a negative correlation with antioxidant genes (superoxide dismutase 1 and catalase) (Table S1). These data suggest a reciprocal relationship between miR-155-5p and SOCS1 expression in cultured renal cells and damaged kidneys.
The JAK/STAT/SOCS axis regulates miR-155-5p expression in vitro and in vivo
To evaluate the direct involvement of the JAK/STAT family in miR-155-5p regulation, gene silencing and overexpression experiments were performed in tubular epithelial cells using small interfering RNA (siRNA) against STAT1 or SOCS1 and plasmid transfection, respectively (Figures 2 and S1). Effective silencing of STAT1 (Figure S1) resulted in a decreased expression of miR-155-5p in response to LPS plus IFNγ stimulation compared with non-transfected, Lipofectamine-treated, or scramble siRNA-transfected controls (Figure 2A). In contrast, SOCS1 silencing enhanced STAT1 activation/phosphorylation (Figures 2B and 2C) and miR-155-5p expression (Figure 2A) compared with controls. On the other hand, SOCS1 enforced expression (by transfection with hemagglutinin-tagged pSocs1 plasmid) attenuated STAT1 and miR-155-5p induction by inflammatory stimuli, compared with control and cells transfected with p513-empty vector (Figures 2A–2C). According to this, the gene expression of chemokines (CCL2 and C-X-C motif chemokine 10 [CXCL10]) and pro-inflammatory cytokines (TNF-α and IL-6), was downregulated by both STAT1 silencing and SOCS1 overexpression and was found upregulated in SOCS1-silenced cells (Figure 2D).
Figure 2.
JAK/STAT/SOCS pathway regulates miR-155-5p expression in vitro
Tubular epithelial cells were transfected with STAT1 siRNA (siStat1), SOCS1 siRNA (siSocs1), or SOCS1 overexpression plasmid (pSocs1) and then incubated for 2 h with 1 μg/mL LPS and 200 ng/mL IFNγ (LPS + IFNγ). Non-transfected cells (Ctrl), Lipofectamine (Lipo), scramble siRNA (siSCr), or empty plasmid (p513) were used as negative controls. (A) Analysis of miR-155-5p expression after silencing and overexpression. (B) Upper panel: Immunodetection of SOCS1, P-STAT1, total STAT1, and α-tubulin in SOCS1-silenced cells. Two bands (≈26 kDa) were detected by SOCS1 antibody, consistent with the unmodified and phosphorylated forms reported previously.35 Lower panel: Western blot analysis in SOCS1-overexpressing cells showing expression of exogenous hemagglutinin-tagged SOCS1 protein (upper bands in pSOCS1), endogenous SOCS1 (lower bands), and STAT1 (phosphorylated and total). (C) Quantitative analysis of the SOCS1/α-tubulin and P-STAT1/STAT1 ratios from SOCS1 silencing and overexpression experiments. (D) Analysis of pro-inflammatory gene expression in transfected cells. The qPCR values were normalized by corresponding RNU6B (A) or 18S rRNA (D). Results expressed as fold increases versus basal conditions are presented as individual data points and the mean ± SD of n = 4–6 independent experiments. &p < 0.05 vs. Ctrl/Lipo, $p < 0.05 vs siScr, and #p < 0.05 vs. p513.
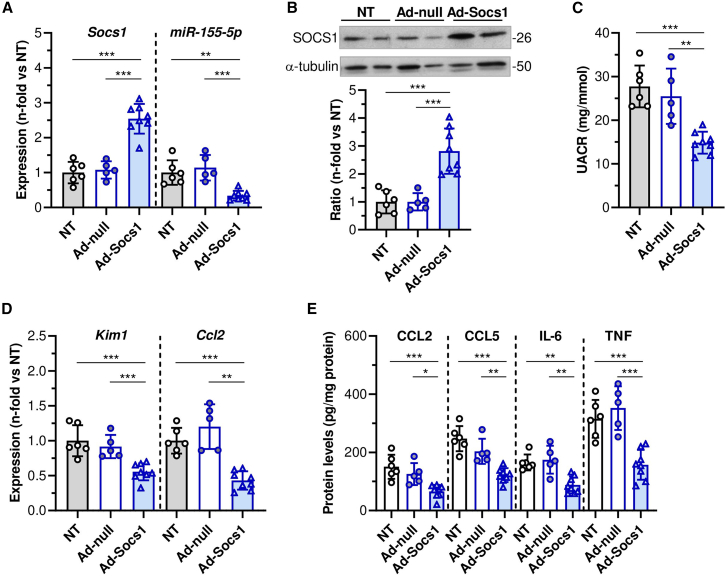
To corroborate the in vitro findings, we explored in vivo the effects of adenovirus-mediated SOCS1 gene transfer in diabetic ApoE mice, as a clinically relevant model of advanced DKD. Compared with non-treated and empty vector groups, SOCS1-overexpressing mice showed lower expression miR-155-5p (Figures 3A and 3B). SOCS1 gene therapy significantly reduced albuminuria (Figure 3C) and markers of kidney injury and inflammation (KIM1 and CCL2; Figure 3D). Pearson analysis confirmed an inverse relationship between miR-155-5p and SOCS1 and direct association with renal parameters (Table S2). Moreover, protein levels of chemokines (CCL2 and CCL5) and pro-inflammatory cytokines (IL-6 and TNF-α) in kidney tissue were found significantly decreased in mice receiving SOCS1 adenovirus (Figure 3E). Together, these results evidence a critical role of JAK/STAT1/SOCS1 axis in miR-155-5p expression in kidney disease.
Figure 3.
In vivo effect of SOCS1 gene therapy in experimental DKD
(A) qPCR analysis of SOCS1 and miR-155-5p in renal samples from diabetic ApoE mice non-treated (NT; n = 6) and treated with empty control vector (Ad-null; n = 5) or SOCS1-encoding adenovirus (Ad-Socs1; n = 8). Normalized expression values are given as fold increases versus NT control group. (B) Western blot analysis of SOCS1 and α-tubulin in kidney samples. Representative immunoblots and quantitative analysis of SOCS1/α-tubulin ratio relative to NT mice are shown. (C) Albuminuria levels. (D) Gene expression of KIM1 and CCL2 in renal samples. (E) ELISA analysis of chemokines and cytokines in kidney lysates. Results are presented as individual data points of the total number of animals per group. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
miR-155-5p activates the JAK/STAT pathway by regulating SOCS1 expression in renal cells
We performed gain- and loss-of-function experiments in renal cells to assess the influence of miR-155-5p on JAK/STAT pathway activation and target gene expression. In mesangial cells, transfection with miR-155-5p mimic reduced the gene and protein expression of SOCS1 (Figures 4A and 4B) and activated STAT1/3 by phosphorylation (Figures 4B and 4C). Consequently, miR-155-5p mimic enhanced the expression of pro-inflammatory genes (CCL2, CXCL10, TNF-α, and IL-6) both in basal conditions (Figure S2A) and upon stimulation with LPS plus IFNγ (Figure 4D). Mimic transfection also promoted the secretion of CCL2 and TNF-α proteins into the culture medium (Figure 4E). In a complementary experiment, renal cells were co-transfected with miR-155-5p mimic and SOCS1 expression plasmid before induction. Compared with miR-155-5p mimic transfection, tubular epithelial cells overexpressing SOCS1 displayed significant decreases in chemokine gene expression under both basal and inflammatory stimulation (Figure 4F). Similarly, co-transfection of mesangial cells with pSOCS1, but not p513 empty vector, effectively reversed the effect of miR-155-5p mimic on pro-inflammatory gene expression in both normal and high-glucose conditions (Figure 4G). These results indicate that SOCS1 antagonizes miR-155-5p in renal cells.
Figure 4.
miR-155-5p overexpression targets SOCS1 and enhances inflammatory response in renal cells
(A) Expression levels of miR-155-5p and SOCS1 in mesangial cells transfected with miR-155-5p mimic or negative control (NC) and stimulated for 2 h with 1 μg/mL LPS and 200 ng/mL IFNγ (LPS + IFNγ). Normalized expression values are given as fold increases versus NC (basal conditions). (B) Representative immunoblots and quantitative analysis of SOCS1, P-STAT1/3, total STAT1/3, and β-actin (as loading control) in non-transfected (Ctrl) and transfected (NC and mimic) mesangial cells under basal and stimulation conditions. The graph represents the SOCS1/β-actin, P-STAT1/STAT1, and P-STAT3/STAT3 ratios in stimulated cells (NC and mimic groups), normalized by respective basal and expressed as relative fold versus Ctrl. (C) Time course of STAT1 activation quantified as P-STAT1/STAT1 ratio by cell-based ELISA. (D) qPCR analysis of pro-inflammatory genes in non-transfected (Ctrl) and transfected (NC and mimic) cells under LPS + IFNγ stimulation. (E) Protein levels of CCL2 and TNF-α in cell supernatants at 24 h were measured by ELISA. (F) qPCR analysis of chemokine genes in tubular epithelial cells co-transfected with miR-155-5p mimic, SOCS1 overexpression plasmid (pSocs1), or empty plasmid (p513) under basal conditions and LPS + IFNγ stimulation. (G) Pro-inflammatory gene expression analysis in mesangial cells without/with high-glucose stimulation (HG; 24 h). Normalized qPCR values (D, F, and G) are expressed as fold increases versus non-transfected and untreated cells (Ctrl basal). Results are presented as individual data points and mean ± SD of n = 4–6 experiments. &p < 0.05 vs. NC basal, #p < 0.05 vs. NC stimulation, ∗p < 0.05 vs. Ctrl, $p < 0.05 vs. mimic, and ¶p < 0.05 vs. p513.
On the other hand, mesangial cell transfected with miR-155-5p inhibitor showed increased gene and protein expression of SOCS1 (Figures 5A and 5B) and impaired STAT1/3 phosphorylation (Figure 5B). In non-stimulated cells, negative control showed a slight but unexpected increase of pro-inflammatory genes, which may be associated with mild off-target effects on cell survival at the dose used and are compatible with the observed trend in cell viability assay (Figures S2B and S2C). Remarkably, transfection with miR-155-5p inhibitor did not affect the viability of mesangial and tubular cells (Figures S2C and S2D) but significantly downregulated the expression of pro-inflammatory genes at both basal levels (Figure S2B) and upon stimulation with LPS plus IFNγ (Figure 5C) and elevated glucose (Figure 5D).
Figure 5.
miR-155-5p regulates inflammation through JAK/STAT pathway
(A) Expression levels of miR-155-5p and SOCS1 in mesangial cells transfected with miR-155-5p inhibitor or negative control (I-NC) and stimulated with LPS plus IFNγ. Normalized qPCR values are expressed as fold increases versus I-NC basal conditions. (B) Representative immunoblots and quantitative analysis of SOCS1, P-STAT1/3, total STAT1/3, and β-actin in non-transfected (Ctrl) and transfected (I-NC and inhibitor) mesangial cells under basal and stimulation conditions. The graph represents the SOCS1/β-actin, P-STAT1/STAT1, and P-STAT3/STAT3 ratios in stimulated cells (I-NC and inhibitor groups), normalized by respective basal and expressed as relative fold versus Ctrl. Pro-inflammatory gene expression analysis in non-transfected (Ctrl) and transfected (I-NC and inhibitor) mesangial cells under LPS + IFNγ stimulation (C) and in tubular epithelial cells under high-glucose (HG) conditions (D). Values normalized by 18S rRNA are expressed as fold increases versus non-transfected and untreated cells (Ctrl basal). (E and F) MTT cell proliferation assay. Mesangial cells were transfected (miR-155-5p inhibitor, miR-155-5p mimic, or negative controls) and co-transfected (miR-155-5p mimic, SOCS1 overexpression plasmid, or p513 empty plasmid) and maintained for the indicated times in medium containing 5% FBS (E) and high glucose concentration (F). Values expressed as fold increases compared with basal conditions. (G) Wound healing assay in mesangial cells. Left: representative images of cells migrating into the wounded area (dotted lines) at 0 and 24 h of incubation. Right: quantitative analysis of covered healing areas expressed as percentage of the initial wound size. Results are presented as individual data points and mean ± SD of n = 4–6 experiments. &p < 0.05 vs. basal or I-NC basal, ∗p < 0.05 vs. Ctrl, #p < 0.05 vs. respective negative control stimulation, $p < 0.05 vs. mimic, and ¶p < 0.05 vs. p513.
We next explored the impact of miR-155-5p on mesangial cell proliferation and migration, which are key findings in DKD. Compared with control conditions, transfection with miR-155-5p inhibitor significantly reduced the proliferative response of mesangial cells to growth medium (Figure 5E) and high-glucose condition (Figure 5F). Suppression of miR-155-5p also decreased the migration ability of mesangial cells, as determined by wound healing assay (Figure 5G). Conversely, miR-155-5p mimic increased cell proliferation under normal and hyperglycemic conditions (Figures 5E and 5F) and promoted cell migration (Figure 5G). These effects were partially reversed by SOCS1 overexpression (Figures 5E–5G), thus indicating a mutual regulatory loop between mir-155-5p and SOCS1 in renal cells.
Therapeutic inhibition of miR-155-5p enhances SOCS1 expression and alleviates renal damage and inflammation in diabetic mice
To complete the study of the regulatory loop between miR-155-5p and the JAK/STAT/SOCS1 axis in DKD, diabetic ApoE mice were administered mmu-miR-155-5p inhibitor or negative control and analyzed for renal function, histology, and gene expression. Compared with vehicle and negative control groups, treatment with miR-155-5p inhibitor resulted in significant knockdown of miR-155-5p (Figure 6A) and upregulated gene and protein expression of SOCS1 (Figures 6A and 6B). Furthermore, the phosphorylation of STAT1/3 in kidney tissue decreased following miR-155-5p inhibition, as determined by western blot (Figure 6B) and immunohistochemistry (Figure 6C).
Figure 6.
miR-155-5p inhibitor reduces JAK/STAT pathway activation in mouse diabetic kidney
(A) qPCR analysis of miR-155-5p and SOCS1 in kidney samples from diabetic ApoE mice treated for 6 weeks with vehicle (Veh; n = 7), miR-155-5p inhibitor (miR-155 Inh; n = 7), or negative control (I-NC; n = 7). Normalized expression values are given as fold increases over vehicle. (B) Representative immunoblots and quantitative analysis of SOCS1/α-tubulin, P-STAT1/STAT1 and P-STAT-3/STAT3 ratios in renal cortex. Values expressed as fold increases versus vehicle. (C) Representative images (scale bars, 25 μm) of P-STAT1/3 immunohistochemistry and quantification of positive stained area in renal sections. Results are presented as individual data points and mean ± SD of the total number of animals per group. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
No significant differences in blood glucose, body weight, or serum cholesterol and triglycerides were observed among diabetic groups at the end of the study (Table 1). Interestingly, albuminuria levels decreased gradually over time by miR-155-5p inhibitor therapy compared with vehicle and negative control groups, reaching significant difference at the end of the study (44% ± 8% and 51% ± 10%, respectively; Figure 7A). Mice receiving miR-155-5p inhibitor also showed a reduction in relative kidney weight and serum creatinine levels (Table 1) as well as lower expression of renal injury markers KIM1 and lipocalin-2 (LCN2; Figure 7B), thus confirming an improved renal function by treatment.
Table 1.
Biochemical and metabolic parameters in diabetic mice treated with miR-155-5p inhibitor
| Veh | I-NC | miR-155 Inh | |
|---|---|---|---|
| Blood glucose (mg/dL) | 533 ± 56 | 554 ± 39 | 530 ± 61 |
| Cholesterol (mg/dL) | 513 ± 110 | 508 ± 90 | 442 ± 62 |
| Triglycerides (mg/dL) | 119 ± 23 | 125 ± 34 | 97 ± 23 |
| Body weight change (g) | 5.1 ± 1.5 | 5.5 ± 2.1 | 4.9 ± 1.5 |
| Kidney-to-body weight ratio (mg/g) | 24.0 ± 2.4 | 24.9 ± 3.4 | 20.4 ± 2.1a,b |
| Creatinine (mg/dL) | 0.86 ± 0.16 | 0.83 ± 0.17 | 0.63 ± 0.07a,b |
Results from diabetic mice treated for 6 weeks with vehicle (Veh), negative control (I-NC), or mmu-miR-155-5p inhibitor (miR-155 Inh). Mean ± SD of n = 7 animals per group.
P<0.05 vs. Veh.
P<0.05 vs. I-NC.
Figure 7.
In vivo inhibition of miR-155-5p attenuates kidney damage, inflammation, and fibrosis in diabetic mice
(A) Evolution of UACR levels in diabetic ApoE mice treated with vehicle (Veh; n = 7), miR-155-5p inhibitor (miR-155 Inh; n = 7), or negative control (I-NC; n = 7). (B) Gene expression of kidney injury markers in renal cortex of diabetic mice. Normalized qPCR values are expressed as fold increases over vehicle. (C) Representative images of PAS (scale bars, 25 μm) and Sirius red (scale bars, 20 μm) staining and quantification of glomerular mesangial expansion and collagen content in renal samples from diabetic groups. (D) Representative images (scale bars, 25 μm) of F4/80 macrophage immunodetection and quantitative assessment of positive staining. (E) Normalized qPCR analysis of inflammatory and profibrotic genes in renal cortex. Results are presented as individual data points and mean ± SD of the total number of animals per group. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Kidney histology with periodic acid-Schiff (PAS) and Sirius red staining revealed a significant decrease of mesangial expansion and collagen content in mice treated with miR-155-5p inhibitor (percentage vs. vehicle 54% ± 4% and 46% ± 5%, respectively; Figure 7C). Moreover, therapeutic inhibition of miR-155-5p reduced the number of F4/80+ infiltrating macrophages (percentage vs. vehicle 43% ± 4%; Figure 7D) and downregulated the gene expression of inflammatory (CCL2, CCL5, and IL-6) and profibrotic (TGF-β) mediators (Figure 7E) in diabetic mice. These results indicate that miR-155-5p inhibition exerts a protective effect in experimental DKD by improving renal function and reducing inflammation and fibrosis.
Discussion
The present study demonstrates that the reciprocal regulation of miR-155 and SOCS1 in renal cells plays a critical role in the pathogenesis of renal inflammation and DKD. We provide evidence that (1) miR-155-5p expression is induced by hyperglycemia and inflammation and correlates inversely with SOCS1 expression in cultured renal cells and damaged kidneys, (2) SOCS1 is a functional target of miR-155-5p and antagonizes its effects on renal cells, and (3) therapeutic miR-155-5p inhibition and SOCS1 induction mitigate JAK/STAT signaling and renal damage in diabetic mice.
The multifunctional molecule miR-155 is induced in response to inflammatory signals in different cell types and tissues and can be suppressed by anti-inflammatory cytokines and pro-resolving factors.13,14,28 Previous studies have explored the functional role of miR-155 in renal pathology. Elevated miR-155 levels have been found in kidney tumors;29 in urine samples from patients with acute kidney injury, immune nephritis, and T1D11,15; and in renal biopsies from patients with DKD.11,16,30 Preclinical studies have linked miR-155 to mesangial cell proliferation in lupus nephritis,31 Th17 immune response in crescentic glomerulonephritis,15 tubular cell apoptosis in renal ischemia-reperfusion,32 and podocyte injury in T2D.17
Our study analyzes the miR-155 regulation in two complementary models. First, cytokine-stimulated mesangial cells and mice with mesangial proliferative glomerulonephritis provided a model of renal inflammation and acute glomerulopathy. Second, high-glucose-stimulated cells and STZ-induced diabetic ApoE mice provided a valuable model of chronic DKD. We demonstrate an upregulated intra-renal expression of miR-155-5p and a tight correlation with the severity of the disease, as assessed by albuminuria, renal injury markers, and inflammatory genes. In vitro, a time-dependent induction of miR-155 was noted in renal cells exposed to elevated glucose and inflammatory stimuli such as LPS and cytokines. Importantly, the induction of miR-155-5p in renal cells and damaged kidneys associates with downregulation of SOCS1. This supports an inverse relationship between miR-155 and SOCS1 in kidney inflammation, as previously reported in the setting of ulcerative colitis,33 inflammatory lung injury,34 and sepsis-associated kidney injury.26
SOCS1 protein negatively regulates JAK/STAT signaling by suppressing JAK-mediated STAT1/3 phosphorylation. Altered expression and activity of SOCS1 affects multiple cellular responses to cytokines and thereby contributes to the pathogenesis of inflammatory diseases.22 Other groups and ours have reported that ectopic expression of SOCS1 promotes renal injury repair by regulating STAT1/335,36,37 and Toll-like receptor38 signaling. We have also demonstrated that administration of SOCS1 peptidomimetic effectively delays the progression of inflammation, oxidative stress, and kidney injury in T1D and T2D mouse models.25,36,39 Previous evidence showed that miR-155 and SOCS1 interactions mediate the inflammatory response in neutrophils,40 T lymphocytes,41 macrophages,42 myofibroblasts,33 and hepatic epithelial cells43 exposed to cytokines. In line with this, our study shows that miR-155-5p mimic transfection in renal cells decreases SOCS1 expression and activates STAT1/3 by phosphorylation, thereby promoting the expression of CCL2, CXCL10, TNF-α and IL-6, which are key mediators involved in cell recruitment, migration and activation in the setting of kidney injury.20 Through gene-silencing experiments, we demonstrate that the upstream transcription factor STAT1 is a key regulator of miR-155-5p expression in response to inflammatory signals. Importantly, SOCS1 gene therapy downregulates miR-155-5p expression in renal cells and diabetic kidneys, which results in anti-inflammatory effects and renal damage improvement. These findings support the existence of active interactions of miR-155/STAT1/SOCS1 in the diabetic environment, where STAT1 positively regulates its function by upregulating miR-155 and downregulating SOCS1 in renal cells. A similar mechanism is proposed in a murine model of renal fibrosis after unilateral ureteral obstruction, where the profibrotic effect of miR-155 is associated with STAT3 activation by directly targeting SOCS1/6.44
One limitation of this study is that we did not localize the expression of miR-155-5p and SOCS1 using other techniques (e.g., combined in situ hybridization and immunostaining in renal samples), which would have confirmed the cellular origin in the damaged kidney. Nevertheless, the in vitro experiments underline the importance of miR-155-5p/SOCS1 interactions in the function of mesangial cells and tubular epithelial cells, which are important for the development of kidney diseases and particularly in the setting of DKD. Besides podocyte damage, mesangial cell injury participates indirectly in the development of albuminuria by mesangial expansion leading to reduced glomerular filtration rate, while tubulointerstitial injury and fibrosis contribute to albuminuria by impairing proximal tubular protein reabsorption.45 Previous studies have indicated that mesangial cells activated by hyperglycemia, cytokines, and oxidative stress exhibit a higher proliferative and migration activity and excess production of matrix proteins, thus contributing to mesangial expansion and glomerulosclerosis.46 miR-155 has been shown to regulate mesangial cell growth and extracellular matrix synthesis.31,47 Moreover, miR-155-5p upregulation in podocytes and glomerular endothelial cells contributes to glomerular filtration barrier impairment in diabetes.16,17 Our functional studies expand these findings by underscoring the role of miR-155/SOCS1 interplay in mesangial expansion. We show that miR-155-5p promotes the proliferation and migration of mesangial cells under inflammatory and hyperglycemic conditions, an effect prevented by SOCS1. Conversely, miR-155-5p inhibition upregulates SOCS1 and declines the proliferation and migration rates, which is in line with previous studies in different cell types, such as renal cancer cells29 and fibroblasts.48
Our preclinical data reveal that pharmacological silencing of miR-155-5p successfully increases SOCS1 and attenuates STAT1/3 activation in diabetic kidneys, resulting in a dramatic improvement of albuminuria, mesangial expansion, tubular damage, fibrosis, and inflammation, thus highlighting the miR-155/SOCS1 axis as a potential therapeutic target for DKD. However, this cannot be extended to other renal diseases, because we did not explore the miR-155-5p/SOCS1 therapy in the mouse model of mesangial proliferative glomerulonephritis. Our findings agree with those of a previous study in diabetic miR-155 knockout mice, in which the improvement of proteinuria and podocyte injury was associated with inhibition of Th17 immune response and SOCS1 induction.18 Gene deletion of miR-155 can also prevent diet-induced obesity and improve insulin sensitivity,49 which are hallmarks of T2D patients. In mouse models, miR-155 inhibition has been shown to reduce cardiac fibrosis, wound inflammation, and nerve injury induced by diabetes.50,51,52 miR-155 antagonism can alleviate podocyte damage in KK-Ay mice and Adriamycin-injected rats,17,19 and prevent renal fibrosis induced by unilateral ureteral obstruction.44 Nanoliposomal delivery of miR-155 inhibitor successfully reduces acute kidney injury through cytokine suppression and SOCS1 induction,53 while intravitreal administration prevents retinal degeneration.54 In the clinical setting, a miR-155 inhibitor is being investigated in mycosis fungoides (NCT02580552 and NCT03713320),55 but no studies are currently available in diabetes.
In conclusion, we have demonstrated that modulation of the miR-155/SOCS1 axis reduces functional and structural damage in diabetic mice, and future perspectives are directed toward a potential application in the treatment of human DKD.
Materials and methods
Reagents
Recombinant mouse cytokines TNF-α (315-01A) IL-1β (211-11B), IL-6 (216-16), TGF-β (100-21), and IFNγ (315-05) were provided by PeproTech (Rocky Hill, NJ). D-glucose (G7528) and LPS (L2630) were purchased from Sigma-Aldrich (St. Louis, MO). The cloning and production of hemagglutinin-tagged SOCS1 plasmid (pSocs1) and empty plasmid (p513), as well as the adenoviral construct encoding SOCS1 (Ad-Socs1) and empty control vector (Ad-null), were previously described.35,56 Recombinant adenoviruses were propagated on human embryonic kidney 293 cells (CRL-1573; American Type Culture Collection [ATCC], Manassas, VA), then purified and titrated by using Adeno-X Virus Kits (BD Biosciences, Heidelberg, Germany). The siRNA directed against SOCS1 (siSocs1, 4390771) and STAT1 (siStat1, 4390771) and the negative control scramble siRNA (siScr, 4390843) were purchased from Ambion (Austin, TX). The mmu-miR-155-5p mimic (miRCURY LNA miRNA Mimic, YM00470919), the mmu-miR-155-5p inhibitor (miRCURY LNA Power Inhibitor, YCI0201878), and their respective negative controls (YM00479902-ABG and YI00199006-011-ADB) were provided by Qiagen (Hilden, Germany).
Cell cultures and transfections
Mouse mesangial cells (SV40 MES13 cell line, CRL-1927; ATCC) were cultured in a 3:1 mixture of DMEM:F12 medium (D6546 and N4888) supplemented with 5% fetal bovine serum (FBS; F7524), 14 mM HEPES (H3375), 1% L-glutamine (G7513), and 1% penicillin/streptomycin (P0781), all from Sigma-Aldrich. Mouse kidney proximal tubular epithelial cells (SV40 MCT line) were grown in RPMI 1640 medium (R0883; Sigma-Aldrich) containing 10% FBS, 1% L-glutamine, and 1% penicillin/streptomycin.57 Cells were serum deprived overnight in medium with 0%–0.5% FBS and then left untreated or stimulated for different times in medium containing 30 mM D-glucose, 0.1 or 1 μg/mL LPS, 10 ng/mL TNF-α, 10 ng/mL IL-1β, 2 ng/mL IL-6, 5 ng/mL TGF-β, 200 ng/mL IFNγ, or a combination of 1 μg/mL LPS plus 200 ng/mL IFNγ. For gene-silencing experiments, cells were transfected for 16–18 h in OptiMEM medium (31985-070; Thermo Fisher Scientific, Waltham, MA) with specific siRNA (siSTAT1 and siSOCS1, 20 nM) or negative control (siScr) using Lipofectamine RNAiMAX reagent (13778; Thermo Fisher Scientific). For SOCS1 overexpression, 2 μg of plasmids (pSocs1 or empty p513) were transfected to the cells using Lipofectamine 3000 reagent (L3000; Thermo Fisher Scientific). For induction and inhibition of miR-155-5p, cells were transfected with mmu-miR-155-5p mimic (10 nM), mmu-miR-155-5p inhibitor (50 nM) or respective negative controls using Lipofectamine RNAiMAX. Cells without transfection were used as respective control groups. After transfection, medium was changed and then cells were serum deprived for further stimulation.
Cell viability, proliferation, and migration assays
Cell viability and proliferation were measured using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide tetrazolium (MTT) assay. Cells were transfected on 96-well plates (1 × 104 cells/well) and incubated in medium containing 0.5% FBS, 5% FBS, or 30 mM D-glucose. MTT stock solution (0.5 mg/mL; M-5655; Sigma-Aldrich) was added to each well for 60 min, then dimethyl sulfoxide was added to dissolve the reduced MTT crystals, and the absorbance was measured at 570 nm.
Mesangial cell migration was measured by the wound healing assay as previously described.25 Briefly, mesangial cells on 12-well multiplates were transfected with miR-155-5p mimic or miR-155-5p inhibitor alone or co-transfected with miR-155-5p mimic and pSocs1 expression plasmid. After 24 h, cell layers were gently wounded using a sterile 200 μL pipette tip, then washed and allowed to repopulate the scratched area for 24 h. Four randomly selected microscopic fields per well were used for quantification, and remaining wound areas were normalized to time 0 values.
Experimental models and treatments
All the animal procedures in this study were performed under the 3R (replacement, refinement, or reduction) principle in accordance with the European Directive 2010/63/EU and were approved by the Animal Care and Use Committee of IIS-FJD and by the Madrid regional government (Proex 217/19). ApoE knockout mice and WT littermates (Mus musculus, C57BL/6J background, males) were bred and maintained at the Animal Facility of IIS-FJD in ventilated racks (2 or 3 mice/cage) in a conventional temperature-controlled room (20°C–22°C, 12 h light/dark cycle) with environmental enrichment and bedding and free access to water and standard food.
Experimental model of mesangial proliferative glomerulonephritis was induced in 12-week-old WT mice by intravenous injection of sheep anti-murine mesangial cell antiserum (10 μL/g body weight), as described previously.58 Animals were studied on days 0 (n = 5), 5 (n = 7), and 7 (n = 6).
Experimental model of diabetes was induced in 12-month-old WT mice (WT-DM group, n = 6) and ApoE mice (ApoE-DM group, n = 7) with 2 daily consecutive intraperitoneal injections of STZ (125 μg/g body weight in 10mM citrate buffer; S0130; Sigma-Aldrich), as previously described.25,57 Mice were monitored weekly for non-fasting blood glucose (NovaPro glucometer; Nova Biomedical Iberia, Barcelona, Spain) and body weight for 8 weeks. Only animals showing blood glucose levels above 350 mg/dL glucose levels were considered diabetic. Age-matched non-diabetic mice receiving citrate were used as controls (WT-Co, n = 5; ApoE-Co, n = 6).
For in vivo SOCS1 gene transfer, 12-week-old ApoE mice were made diabetic by STZ injection and after 2 weeks were randomly distributed into 3 groups: non-treated controls (NT; n = 6), treated with SOCS1-encoding adenovirus (Ad-Socs1; n = 8) and empty control vector (Ad-null; n = 5). Adenoviruses were administered by tail-vein injection (1 × 109 viral particles/g body weight) and mice were analyzed after 6 weeks of intervention. For in vivo miR-155-5p inhibition, diabetic mice were distributed into 3 groups: vehicle (Veh, n = 7), mmu-miR-155-5p inhibitor (miR-155 Inh, n = 7), and negative control (I-NC, n = 7). Both inhibitor and negative control were diluted following manufacturer instructions and injected (2 μg/g body weight) intraperitoneally twice a week for 6 weeks.
At the end of the experiments, 16 h fasted mice were anesthetized (100 μg/g ketamine plus 15 μg/g xylazine), perfused at physiological pressure with saline solution via left ventricle and euthanized. Blood and urine samples were collected. Kidneys were dissected and processed for histology and RNA expression analysis.
Biochemical analysis
Urine samples collected during the experimental models were analyzed for albumin and creatinine (ab108792 and ab65340; Abcam, Cambridge, United Kingdom) to calculate UACR. Serum levels of creatinine (ab65340; Abcam), triglycerides (11528; BioSystems, Barcelona, Spain) and total cholesterol (STA-384; Cell Biolabs, San Diego, CA) were measured by using colorimetric kits.
Histology and immunohistochemistry
Kidney tissues were fixed in 10% buffered formalin and embedded in paraffin. Sample sections (3 μm) were stained with PAS to evaluate renal damage. At least 20 glomeruli randomly selected from each mouse were quantified using Image Pro-Plus software (Media Cybernetics; Bethesda, MD) to assess the percentage of PAS-positive mesangial area.
For immunohistochemistry, samples were deparaffinized and rehydrated through graded xylene and ethanol and subjected to antigen retrieval (0.01 M citrate buffer [pH 6] for 20 min), and blockade of endogenous peroxidase (3% H2O2 in methanol for 30 min) and nonspecific binding (8% host serum for 1 h). Slides were then incubated overnight at 4°C with primary antibodies against F4/80 (MCA497R; Bio-Rad, Hercules, CA), phosphorylated STAT1 (P-STAT1; [Y701], 44-376; Thermo Fisher Scientific), and phosphorylated STAT3 (P-STAT3; [S727], 9134; Cell Signaling Technology, Danvers, MA). After rinsing in PBS, samples were incubated 1 h with biotinylated secondary antibodies (712-065-153 and 111-065-003; Jackson Immunoresearch, West Grove, PA), followed by avidin-biotin complex reagent (PK-4000; Vector Laboratories, Burlingame, CA) for 45 min. Positive signal was visualized by the addition of 3,3′-diaminobenzidine substrate (DAB; ab64238; Abcam) and samples were counterstained with hematoxylin. Positive staining (>10 fields at 20× magnification; 2 slices/mice) was quantified and expressed as percentage of total area.
Quantitative real-time PCR (qPCR) analysis
Total RNA from kidney and cultured cells was extracted using TRI Reagent (Thermo Fisher Scientific). For miRNA expression, miRCURY LNA RT kit (339340) was used for cDNA synthesis and qPCR was made with miRCURY LNA SYBR Green PCR kit (339345) and miR-155-5p assay (YP02119303) using RNU6B (YP00203907) for normalization (all from Qiagen). For mRNA expression assay, cDNA was synthesized with High-Capacity cDNA reverse transcription kit (4368813, Applied Biosystems, Foster City, CA, USA), and then analyzed by qPCR using Premix Ex Taq (RR390; Takara, Shiga, Japan). The TaqMan mouse gene expression assays (Thermo Fisher Scientific) used were: Catalase (Cat, Mm00437992_m1), Ccl2 (Mm00441242_m1), Ccl5 (Mm01302428_m1), Cxcl10 (Mm00445235_m1), Il6 (Mm00446190_m1), Kim1 (Mm00506686_m1), Lcn2 (Mm01324470_m1), Socs1 (Mm00782550_s1), superoxide dismutase 1 (Sod1, Mm01344233_g1), Tgfb (Mm01178819), Tnfa (Mm00443258_m1), and 18S rRNA for normalization (4310893E). PCR primers used for NADPH oxidase (NOX) subunits were as follows: Nox2 (forward, 5′-AACTCAGAATCCGGCCCGCGT-3′; reverse, 5′-AGGGGGCCTGTGTCATTGTGATT-3′), and Nox4 (forward, 5′-CCCTCCTGGCTGCATTAGTC-3′; reverse, 5′-AACCCTCGAGGCAAAGATCC-3′).
Protein immunodetection
Renal tissues and cell cultures were homogenized in cold lysis buffer (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% Triton X-100, 0.5% NP-40, 1 mM EDTA, 1 mM EGTA, 0.2 mM Na3VO4, 10 mM NaF, 0.2 mM PMSF, and protease inhibitor cocktail). A total of 35 μg protein lysates were electrophoresed and immunoblotted for SOCS1 (38-5200; Thermo Fisher Scientific), P-STAT1 (33340; Thermo Fisher Scientific), STAT1 (9172; Cell Signaling), P-STAT3 (9134; Cell Signaling), STAT3 (9139; Cell Signaling), β-actin (A1978; Sigma-Aldrich), and α-tubulin (T9026; Sigma-Aldrich). Immunoblots were quantified (Quantity One; Bio-Rad) and the SOCS1/α-tubulin, SOCS1/β-actin, P-STAT1/STAT1, and P-STAT3/STAT3 ratios were determined in terms of arbitrary units and expressed relative to the ratio for basal conditions or control mice.
A cell-based ELISA was performed to determine STAT1 activation. Briefly, cells (1 × 105/well) on 96-well plates were stimulated with LPS plus IFNγ for different times. Formaldehyde-fixed cells were quenched for endogenous peroxidase activity, then blocked (2% albumin and 5% FBS) and incubated with primary antibodies (anti-P-STAT1 or anti-STAT1) overnight at 4°C, followed by peroxidase-conjugated secondary antibodies and DAB reagent. The absorbance at 450 nm was measured in a microplate reader, and data expressed as the ratio of phosphorylated to total protein. Concentrations of CCL2, CCL5, IL-6, and TNF-α proteins in cell supernatants and kidney lysates were measured by mouse ELISA kits (DY479, DY478, DY406, and DY410; R&D Systems, Minneapolis, MN).
Statistics
Results are presented as individual data points and mean ± SD from separate experiments and mice. Each experimental condition was analyzed in duplicate/triplicate. Statistical analysis was performed using GraphPad Prism version 8 (GraphPad Software, La Jolla, CA). Data passed the D’Agostino and Pearson omnibus normality test and were tested for homogeneity of variance with the Bartlett test. Pearson’s correlation analysis was performed for normally distributed parameters. Statistical significance was established at p < 0.05 using unpaired two-tailed Student’s t test and one- or two-way ANOVA with Tukey’s multiple-comparisons test.
Data and code availability
The authors confirm that the data supporting the findings of this study are available within the article and its supplemental information.
Acknowledgments
This research was funded by grants from Spanish Ministry of Science and Innovation (RTI2018-098788-B-I00 and PID2021-127741OB-I00) to C.G.-G., Instituto de Salud Carlos III (PI20/00487) to J.E, CIBERDEM (postdoctoral contract) to I.P., and Conacyt-Mexico (CB-2015-01 #256639 and FOP02-2022-02 #321869) to O.L.-F. The authors thank Ana Melgar and Patricia Saperas (IIS-FJD, Madrid) for technical support in mouse sample processing and histology, and Carmen Liliana Peña for technical assistant with in vitro experiments.
Author contributions
I.P., L.J.-C., I.L., and O.L.-F. performed the in vivo experiments and ex vivo analysis. I.P., M.K., M.P., I.H.R., M.F.-M., O.L.F., and C.G.-G. performed the in vitro experiments. M.K. and M.P. assisted in ex vivo analysis. J.E. and O.L.-F. critically discussed the data. I.P. and C.G.-G. conceptually designed and interpreted the experimental work and wrote the manuscript.
Declaration of interests
All the authors declared no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2023.102041.
Contributor Information
Oscar Lopez-Franco, Email: oscarlopez01@uv.mx.
Carmen Gomez-Guerrero, Email: cgomez@fjd.es.
Supplemental information
References
- 1.Fowler M.J. Microvascular and Macrovascular Complications of Diabetes. Clin. Diabetes. 2008;26:77–82. doi: 10.2337/diaclin.26.2.77. [DOI] [Google Scholar]
- 2.Sugahara M., Pak W.L.W., Tanaka T., Tang S.C.W., Nangaku M. Update on diagnosis, pathophysiology, and management of diabetic kidney disease. Nephrology. 2021;26:491–500. doi: 10.1111/nep.13860. [DOI] [PubMed] [Google Scholar]
- 3.Russo G., Piscitelli P., Giandalia A., Viazzi F., Pontremoli R., Fioretto P., De Cosmo S. Atherogenic dyslipidemia and diabetic nephropathy. J. Nephrol. 2020;33:1001–1008. doi: 10.1007/s40620-020-00739-8. [DOI] [PubMed] [Google Scholar]
- 4.Pichler R., Afkarian M., Dieter B.P., Tuttle K.R. Immunity and inflammation in diabetic kidney disease: translating mechanisms to biomarkers and treatment targets. Am. J. Physiol. Ren. Physiol. 2017;312 doi: 10.1152/ajprenal.00314.2016. F716-f731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernandez-Fernandez B., Ortiz A., Gomez-Guerrero C., Egido J. Therapeutic approaches to diabetic nephropathy--beyond the RAS. Nat. Rev. Nephrol. 2014;10:325–346. doi: 10.1038/nrneph.2014.74. [DOI] [PubMed] [Google Scholar]
- 6.O'Brien J., Hayder H., Zayed Y., Peng C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018;9:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 8.Barutta F., Bellini S., Mastrocola R., Bruno G., Gruden G. MicroRNA and Microvascular Complications of Diabetes. Internet J. Endocrinol. 2018;2018:6890501. doi: 10.1155/2018/6890501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahtal N., Lenoir O., Tinel C., Anglicheau D., Tharaux P.L. MicroRNAs in kidney injury and disease. Nat. Rev. Nephrol. 2022;18:643–662. doi: 10.1038/s41581-022-00608-6. [DOI] [PubMed] [Google Scholar]
- 10.Conserva F., Barozzino M., Pesce F., Divella C., Oranger A., Papale M., Sallustio F., Simone S., Laviola L., Giorgino F., et al. Urinary miRNA-27b-3p and miRNA-1228-3p correlate with the progression of Kidney Fibrosis in Diabetic Nephropathy. Sci. Rep. 2019;9:11357. doi: 10.1038/s41598-019-47778-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beltrami C., Simpson K., Jesky M., Wonnacott A., Carrington C., Holmans P., Newbury L., Jenkins R., Ashdown T., Dayan C., et al. Association of Elevated Urinary miR-126, miR-155, and miR-29b with Diabetic Kidney Disease. Am. J. Pathol. 2018;188:1982–1992. doi: 10.1016/j.ajpath.2018.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Chen H.Y., Zhong X., Huang X.R., Meng X.M., You Y., Chung A.C., Lan H.Y. MicroRNA-29b inhibits diabetic nephropathy in db/db mice. Mol. Ther. 2014;22:842–853. doi: 10.1038/mt.2013.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahesh G., Biswas R. MicroRNA-155: A Master Regulator of Inflammation. J. Interferon Cytokine Res. 2019;39:321–330. doi: 10.1089/jir.2018.0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jankauskas S.S., Gambardella J., Sardu C., Lombardi A., Santulli G. Functional Role of miR-155 in the Pathogenesis of Diabetes Mellitus and Its Complications. Noncoding. RNA. 2021;7:39. doi: 10.3390/ncrna7030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krebs C.F., Kapffer S., Paust H.J., Schmidt T., Bennstein S.B., Peters A., Stege G., Brix S.R., Meyer-Schwesinger C., Müller R.U., et al. MicroRNA-155 drives TH17 immune response and tissue injury in experimental crescentic GN. J. Am. Soc. Nephrol. 2013;24:1955–1965. doi: 10.1681/asn.2013020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang Y., Liu Y., Li L., Su B., Yang L., Fan W., Yin Q., Chen L., Cui T., Zhang J., et al. Involvement of inflammation-related miR-155 and miR-146a in diabetic nephropathy: implications for glomerular endothelial injury. BMC Nephrol. 2014;15:142. doi: 10.1186/1471-2369-15-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X., Gao Y., Yi W., Qiao Y., Hu H., Wang Y., Hu Y., Wu S., Sun H., Zhang T. Inhibition of miRNA-155 Alleviates High Glucose-Induced Podocyte Inflammation by Targeting SIRT1 in Diabetic Mice. J. Diabetes Res. 2021;2021:5597394. doi: 10.1155/2021/5597394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin X., You Y., Wang J., Qin Y., Huang P., Yang F. MicroRNA-155 deficiency promotes nephrin acetylation and attenuates renal damage in hyperglycemia-induced nephropathy. Inflammation. 2015;38:546–554. doi: 10.1007/s10753-014-9961-7. [DOI] [PubMed] [Google Scholar]
- 19.Lin X., Zhen X., Huang H., Wu H., You Y., Guo P., Gu X., Yang F. Role of MiR-155 Signal Pathway in Regulating Podocyte Injury Induced by TGF-β1. Cell. Physiol. Biochem. 2017;42:1469–1480. doi: 10.1159/000479211. [DOI] [PubMed] [Google Scholar]
- 20.Moreno J.A., Gomez-Guerrero C., Mas S., Sanz A.B., Lorenzo O., Ruiz-Ortega M., Opazo L., Mezzano S., Egido J. Targeting inflammation in diabetic nephropathy: a tale of hope. Expert Opin. Investig. Drugs. 2018;27:917–930. doi: 10.1080/13543784.2018.1538352. [DOI] [PubMed] [Google Scholar]
- 21.Banerjee S., Biehl A., Gadina M., Hasni S., Schwartz D.M. JAK-STAT Signaling as a Target for Inflammatory and Autoimmune Diseases: Current and Future Prospects. Drugs. 2017;77:521–546. doi: 10.1007/s40265-017-0701-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma J., Larkin J., 3rd Therapeutic Implication of SOCS1 Modulation in the Treatment of Autoimmunity and Cancer. Front. Pharmacol. 2019;10:324. doi: 10.3389/fphar.2019.00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y., Jin D., Kang X., Zhou R., Sun Y., Lian F., Tong X. Signaling Pathways Involved in Diabetic Renal Fibrosis. Front. Cell Dev. Biol. 2021;9:696542. doi: 10.3389/fcell.2021.696542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tuttle K.R., Brosius F.C., 3rd, Adler S.G., Kretzler M., Mehta R.L., Tumlin J.A., Tanaka Y., Haneda M., Liu J., Silk M.E., et al. JAK1/JAK2 inhibition by baricitinib in diabetic kidney disease: results from a Phase 2 randomized controlled clinical trial. Nephrol. Dial. Transplant. 2018;33:1950–1959. doi: 10.1093/ndt/gfx377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Recio C., Lazaro I., Oguiza A., Lopez-Sanz L., Bernal S., Blanco J., Egido J., Gomez-Guerrero C. Suppressor of Cytokine Signaling-1 Peptidomimetic Limits Progression of Diabetic Nephropathy. J. Am. Soc. Nephrol. 2017;28:575–585. doi: 10.1681/ASN.2016020237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren Y., Cui Y., Xiong X., Wang C., Zhang Y. Inhibition of microRNA-155 alleviates lipopolysaccharide-induced kidney injury in mice. Int. J. Clin. Exp. Pathol. 2017;10:9362–9371. [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang S., Zhang H.W., Lu M.H., He X.H., Li Y., Gu H., Liu M.F., Wang E.D. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 2010;70:3119–3127. doi: 10.1158/0008-5472.Can-09-4250. [DOI] [PubMed] [Google Scholar]
- 28.Jablonski K.A., Gaudet A.D., Amici S.A., Popovich P.G., Guerau-de-Arellano M. Control of the Inflammatory Macrophage Transcriptional Signature by miR-155. PLoS One. 2016;11:e0159724. doi: 10.1371/journal.pone.0159724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li S., Chen T., Zhong Z., Wang Y., Li Y., Zhao X. microRNA-155 silencing inhibits proliferation and migration and induces apoptosis by upregulating BACH1 in renal cancer cells. Mol. Med. Rep. 2012;5:949–954. doi: 10.3892/mmr.2012.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baker M.A., Davis S.J., Liu P., Pan X., Williams A.M., Iczkowski K.A., Gallagher S.T., Bishop K., Regner K.R., Liu Y., Liang M. Tissue-Specific MicroRNA Expression Patterns in Four Types of Kidney Disease. J. Am. Soc. Nephrol. 2017;28:2985–2992. doi: 10.1681/asn.2016121280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kong J., Li L., Lu Z., Song J., Yan J., Yang J., Gu Z., Da Z. MicroRNA-155 Suppresses Mesangial Cell Proliferation and TGF-β1 Production via Inhibiting CXCR5-ERK Signaling Pathway in. Lupus Nephritis. Inflammation. 2019;42:255–263. doi: 10.1007/s10753-018-0889-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu H., Huang T., Ying L., Han C., Li D., Xu Y., Zhang M., Mou S., Dong Z. MiR-155 is Involved in Renal Ischemia-Reperfusion Injury via Direct Targeting of FoxO3a and Regulating Renal Tubular Cell Pyroptosis. Cell. Physiol. Biochem. 2016;40:1692–1705. doi: 10.1159/000453218. [DOI] [PubMed] [Google Scholar]
- 33.Pathak S., Grillo A.R., Scarpa M., Brun P., D'Incà R., Nai L., Banerjee A., Cavallo D., Barzon L., Palù G., et al. MiR-155 modulates the inflammatory phenotype of intestinal myofibroblasts by targeting SOCS1 in ulcerative colitis. Exp. Mol. Med. 2015;47:e164. doi: 10.1038/emm.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rao R., Rieder S.A., Nagarkatti P., Nagarkatti M. Staphylococcal enterotoxin B-induced microRNA-155 targets SOCS1 to promote acute inflammatory lung injury. Infect. Immun. 2014;82:2971–2979. doi: 10.1128/iai.01666-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ortiz-Muñoz G., Lopez-Parra V., Lopez-Franco O., Fernandez-Vizarra P., Mallavia B., Flores C., Sanz A., Blanco J., Mezzano S., Ortiz A., et al. Suppressors of cytokine signaling abrogate diabetic nephropathy. J. Am. Soc. Nephrol. 2010;21:763–772. doi: 10.1681/ASN.2009060625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopez-Sanz L., Bernal S., Recio C., Lazaro I., Oguiza A., Melgar A., Jimenez-Castilla L., Egido J., Gomez-Guerrero C. SOCS1-targeted therapy ameliorates renal and vascular oxidative stress in diabetes via STAT1 and PI3K inhibition. Lab. Invest. 2018;98:1276–1290. doi: 10.1038/s41374-018-0043-6. [DOI] [PubMed] [Google Scholar]
- 37.Shi Y., Du C., Zhang Y., Ren Y., Hao J., Zhao S., Yao F., Duan H. Suppressor of cytokine signaling-1 ameliorates expression of MCP-1 in diabetic nephropathy. Am. J. Nephrol. 2010;31:380–388. doi: 10.1159/000286559. [DOI] [PubMed] [Google Scholar]
- 38.Wu X.Y., Yu J., Tian H.M. Effect of SOCS1 on diabetic renal injury through regulating TLR signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2019;23:8068–8074. doi: 10.26355/eurrev_201909_19023. [DOI] [PubMed] [Google Scholar]
- 39.Opazo-Ríos L., Sanchez Matus Y., Rodrigues-Díez R.R., Carpio D., Droguett A., Egido J., Gomez-Guerrero C., Mezzano S. Anti-inflammatory, antioxidant and renoprotective effects of SOCS1 mimetic peptide in the BTBR ob/ob mouse model of type 2 diabetes. BMJ Open Diabetes Res. Care. 2020;8:e001242. doi: 10.1136/bmjdrc-2020-001242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y., Xie Y., Zhang L., Zhao H. MicroRNA-155 Participates in Smoke-Inhalation-Induced Acute Lung Injury through Inhibition of SOCS-1. Molecules. 2020;25 doi: 10.3390/molecules25051022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang D., Tang M., Zong P., Liu H., Zhang T., Liu Y., Zhao Y. MiRNA-155 Regulates the Th17/Treg Ratio by Targeting SOCS1 in Severe Acute Pancreatitis. Front. Physiol. 2018;9:686. doi: 10.3389/fphys.2018.00686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye J., Guo R., Shi Y., Qi F., Guo C., Yang L. miR-155 Regulated Inflammation Response by the SOCS1-STAT3-PDCD4 Axis in Atherogenesis. Mediators Inflamm. 2016;2016:8060182. doi: 10.1155/2016/8060182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao R., Dong R., Yang Y., Wang Y., Ma J., Wang J., Li H., Zheng S. MicroRNA-155 modulates bile duct inflammation by targeting the suppressor of cytokine signaling 1 in biliary atresia. Pediatr. Res. 2017;82:1007–1016. doi: 10.1038/pr.2017.87. [DOI] [PubMed] [Google Scholar]
- 44.Zhang W., Li X., Tang Y., Chen C., Jing R., Liu T. miR-155-5p Implicates in the Pathogenesis of Renal Fibrosis via Targeting SOCS1 and SOCS6. Oxid. Med. Cell. Longev. 2020;2020:6263921. doi: 10.1155/2020/6263921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jefferson J.A., Shankland S.J., Pichler R.H. Proteinuria in diabetic kidney disease: a mechanistic viewpoint. Kidney Int. 2008;74:22–36. doi: 10.1038/ki.2008.128. [DOI] [PubMed] [Google Scholar]
- 46.Thomas H.Y., Ford Versypt A.N. Pathophysiology of mesangial expansion in diabetic nephropathy: mesangial structure, glomerular biomechanics, and biochemical signaling and regulation. J. Biol. Eng. 2022;16:19. doi: 10.1186/s13036-022-00299-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang G., Wu B., Zhang B., Wang K., Wang H. LncRNA CTBP1-AS2 alleviates high glucose-induced oxidative stress, ECM accumulation, and inflammation in diabetic nephropathy via miR-155-5p/FOXO1 axis. Biochem. Biophys. Res. Commun. 2020;532:308–314. doi: 10.1016/j.bbrc.2020.08.073. [DOI] [PubMed] [Google Scholar]
- 48.Zhou X., Yan T., Huang C., Xu Z., Wang L., Jiang E., Wang H., Chen Y., Liu K., Shao Z., Shang Z. Melanoma cell-secreted exosomal miR-155-5p induce proangiogenic switch of cancer-associated fibroblasts via SOCS1/JAK2/STAT3 signaling pathway. J. Exp. Clin. Cancer Res. 2018;37:242. doi: 10.1186/s13046-018-0911-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gaudet A.D., Fonken L.K., Gushchina L.V., Aubrecht T.G., Maurya S.K., Periasamy M., Nelson R.J., Popovich P.G. miR-155 Deletion in Female Mice Prevents Diet-Induced Obesity. Sci. Rep. 2016;6:22862. doi: 10.1038/srep22862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang D., Cui Y., Li B., Luo X., Li B., Tang Y. miR-155 regulates high glucose-induced cardiac fibrosis via the TGF-β signaling pathway. Mol. Biosyst. 2016;13:215–224. doi: 10.1039/c6mb00649c. [DOI] [PubMed] [Google Scholar]
- 51.Moura J., Sørensen A., Leal E.C., Svendsen R., Carvalho L., Willemoes R.J., Jørgensen P.T., Jenssen H., Wengel J., Dalgaard L.T., Carvalho E. microRNA-155 inhibition restores Fibroblast Growth Factor 7 expression in diabetic skin and decreases wound inflammation. Sci. Rep. 2019;9:5836. doi: 10.1038/s41598-019-42309-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen X., Zhang X.B., Li D.J., Qi G.N., Dai Y.Q., Gu J., Chen M.Q., Hu S., Liu Z.Y., Yang Z.M. miR-155 facilitates calcium oxalate crystal-induced HK-2 cell injury via targeting PI3K associated autophagy. Exp. Mol. Pathol. 2020;115:104450. doi: 10.1016/j.yexmp.2020.104450. [DOI] [PubMed] [Google Scholar]
- 53.Chen S., Shan J., Niu W., Lin F., Liu S., Wu P., Sun L., Lu W., Jiang G. Micro RNA-155 inhibitor as a potential therapeutic strategy for the treatment of acute kidney injury (AKI): a nanomedicine perspective. RSC Adv. 2018;8:15890–15896. doi: 10.1039/c7ra13440a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aggio-Bruce R., Chu-Tan J.A., Wooff Y., Cioanca A.V., Schumann U., Natoli R. Inhibition of microRNA-155 Protects Retinal Function Through Attenuation of Inflammation in Retinal Degeneration. Mol. Neurobiol. 2021;58:835–854. doi: 10.1007/s12035-020-02158-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Valipour A., Jäger M., Wu P., Schmitt J., Bunch C., Weberschock T. Interventions for mycosis fungoides. Cochrane Database Syst. Rev. 2020;7:Cd008946. doi: 10.1002/14651858.CD008946.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Recio C., Oguiza A., Mallavia B., Lazaro I., Ortiz-Muñoz G., Lopez-Franco O., Egido J., Gomez-Guerrero C. Gene delivery of suppressors of cytokine signaling (SOCS) inhibits inflammation and atherosclerosis development in mice. Basic Res. Cardiol. 2015;110:8. doi: 10.1007/s00395-014-0458-1. [DOI] [PubMed] [Google Scholar]
- 57.Lazaro I., Oguiza A., Recio C., Mallavia B., Madrigal-Matute J., Blanco J., Egido J., Martin-Ventura J.L., Gomez-Guerrero C. Targeting HSP90 Ameliorates Nephropathy and Atherosclerosis Through Suppression of NF-kappaB and STAT Signaling Pathways in Diabetic Mice. Diabetes. 2015;64:3600–3613. doi: 10.2337/db14-1926. [DOI] [PubMed] [Google Scholar]
- 58.López-Franco O., Suzuki Y., Sanjuán G., Blanco J., Hernández-Vargas P., Yo Y., Kopp J., Egido J., Gómez-Guerrero C. Nuclear factor-kappa B inhibitors as potential novel anti-inflammatory agents for the treatment of immune glomerulonephritis. Am. J. Pathol. 2002;161:1497–1505. doi: 10.1016/s0002-9440(10)64425-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplemental information.