Abstract
Clarithromycin resistance in Helicobacter pylori is mainly due to A-to-G mutations within the peptidyltransferase region of the 23S rRNA. In the present study, cross-resistance to macrolide, lincosamide, and streptogramin B (MLS) antibiotics (MLS phenotypes) has been investigated for several clinical isolates of H. pylori. Two major types of MLS resistance were identified and correlated with specific point mutations in the 23S rRNA gene. The A2142G mutation was linked with high-level cross-resistance to all MLS antibiotics (type I), and the A2143G mutation gave rise to an intermediate level of resistance to clarithromycin and clindamycin but no resistance to streptogramin B (type II). In addition, streptogramin A and streptogramin B were demonstrated to have a synergistic effect on both MLS-sensitive and MLS-resistant H. pylori strains. To further understand the mechanism of MLS resistance in H. pylori, we performed in vitro site-directed mutagenesis (substitution of G, C, or T for A at either position 2142 or 2143 of the 23S rRNA gene). The site-directed point mutations were introduced into a clarithromycin-susceptible strain, H. pylori UA802, by natural transformation followed by characterization of their effects on MLS resistance in an isogenic background. Strains with A-to-G and A-to-C mutations at the same position within the 23S rRNA gene had similar levels of clarithromycin resistance, and this level of resistance was higher than that for strains with the A-to-T mutation. Mutations at position 2142 conferred a higher level of clarithromycin resistance than mutations at position 2143. All mutations at position 2142 conferred cross-resistance to all MLS antibiotics, which corresponds to the type I MLS phenotype, whereas mutations at position 2143 were associated with a type II MLS phenotype with no resistance to streptogramin B. To explain that A-to-G transitions were predominantly observed in clarithromycin-resistant clinical isolates, we propose a possible mechanism by which A-to-G mutations are preferentially produced in H. pylori.
Helicobacter pylori is a microaerophilic, gram-negative bacterium that colonizes the human gastric mucosa and that causes gastritis and peptic ulceration (8). It is also associated with the development of gastric cancer (22). Clarithromycin is a potent macrolide that has frequently been used in combination with other antimicrobial agents for the treatment of H. pylori infections (23, 32). However, the development of clarithromycin resistance among H. pylori strains has become a predominant cause of the failure of therapy incorporating clarithromycin (3, 15). Previous studies have examined clarithromycin-resistant H. pylori isolates from various geographic locations and have revealed that mutations responsible for alterations in the 23S rRNA gene are the mechanism of clarithromycin resistance (7, 21, 28, 29, 34, 35). Specifically, adenine-to-guanine transitions at either position 2058 or position 2059 (Escherichia coli coordinates) in the peptidyltransferase region of the 23S rRNA were in most cases associated with clarithromycin resistance. Recently, two identical copies of the 23S rRNA have been sequenced and the transcription start site of the gene from a clarithromycin-susceptible strain, strain UA802, was determined (30). According to the new numbering scheme for H. pylori 23S rRNA, E. coli bases 2058 and 2059 correspond to H. pylori positions 2142 and 2143, respectively (see Fig. 3).
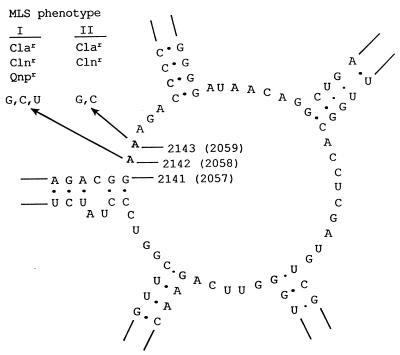
FIG. 3.
Secondary structure of the central part of domain V (peptidyltransferase loop) of the H. pylori 23S rRNA gene based on the model of Egebjerg et al. (9)), with indication of the mutations that confer MLS resistance. The mutation sites are numbered according to the newly proposed numbering system (30), and the equivalent positions in E. coli are indicated in parentheses. The base substitutions made by site-directed mutagenesis in this work are indicated by arrows, and the associated MLS phenotypes are indicated. Abbreviations for antibiotics: Cla, clarithromycin; Cln, clindamycin; Qnp, quinupristin.
In E. coli, as well as in some other bacteria, it is well known that the base equivalent to base A2058 in the 23S rRNA of E. coli is the target of ribosomal methyltransferase (products of erm genes which are frequently plasmid encoded) and the binding site for macrolide antibiotics (5, 39). Methylation or mutation at this position confers complete cross-resistance to the macrolide, lincosamide, and type B streptogramin (MLS) antibiotics (MLS resistance), suggesting that these structurally distinct antibiotics have similar effects in inhibiting ribosomal function. Mutations within the vicinity, at position 2059 or 2057, have also been associated with resistance to the macrolide group of antibiotics (20, 24, 33). To date, the MLS resistance phenotypes associated with mutations in the peptidyltransferase region of the 23S rRNA have not yet been investigated in H. pylori.
This study was initiated to characterize the MLS phenotypes and the associated genotypes of several clinical isolates of H. pylori. Furthermore, to demonstrate the cause-effect relationship between particular types of mutations and MLS resistance phenotypes, we performed in vitro site-directed mutagenesis. The site-directed point mutations in the 23S rRNA gene were introduced into an MLS-susceptible strain of H. pylori by natural transformation, followed by characterization of their effects on MLS resistance in an isogenic background.
MATERIALS AND METHODS
H. pylori strains, growth medium, and antibiotics.
H. pylori A, B, D, E, and MQ are clarithromycin-resistant clinical isolates which originated in Europe (30). Clarithromycin-susceptible strain UA802 was an isolate from the University of Alberta Hospital and has been used extensively in this laboratory (16). H. pylori strains were grown on BHI-YE agar (3.7% brain heart infusion agar base with 0.3% yeast extract and 5% animal serum) at 37°C under microaerobic conditions (5% CO2, 5% H2, and 90% N2). The antibiotics used in this study were obtained as follows: Clarithromycin was from Bayer, Leverkusen, Germany; clindamycin was from the Upjohn Company of Canada, Don Mills, Ontario, Canada; and quinupristin (streptogramin B) and dalfopristin (streptogramin A) were provided by both Sylvia Pong-Porter (Department of Microbiology, Mount Sinai Hospital, Toronto, Ontario, Canada) and Rhone-Polenc Rorer, Collegeville, Pa.
MIC test.
H. pylori cells were grown for 2 days and were suspended in sterile BHI-YE liquid medium, and the turbidity of the suspensions was adjusted to that of a 2.0 McFarland standard. The suspended cells were inoculated (8 μl/spot) onto BHI-YE agar plates containing different concentrations of antibiotics obtained by serial twofold dilution. The plates were incubated as described above, and the growth was examined after 3 days.
DNA manipulation.
Chromosomal DNAs from the H. pylori strains were isolated by a previously described method (13). DNA sequencing was carried out with the thermocycling sequencing system with Thermo-Sequenase purchased from Amersham Life Sciences, Cleveland, Ohio. Other DNA manipulations including PCR and gel electrophoresis were performed by standard methods (25).
Site-directed mutagenesis.
A series of point mutations at position 2142, 2143, or 2141 of the H. pylori 23S rRNA gene were generated by a sequential PCR method (1). Table 1 lists all of the primers that were used. To create a particular point mutation, two common primers, primers DP1 and ZGE23, and a pair of primers containing site-specific mutation were used (Fig. 1). For example, to make an A2142G mutation, two fragments were amplified by PCR with primer pairs DP1-GW11 and ZGE23-GW1 in the first step. In the second PCR step, these two fragments encompassing the mutation were annealed with each other and were extended by mutually primed synthesis. The final products were 307-bp PCR fragments containing a point mutation in the center. The PCR products were gel purified with Spin-X (purchased from Corning Costar Corporation, Cambridge, Mass.), and the site-directed mutations in the PCR fragments were verified by DNA sequencing.
TABLE 1.
Oligonucleotides used in this study
| Designation | Sequence (5′ to 3′)a | Positionsb | Strandc | Purposed | Reference or source |
|---|---|---|---|---|---|
| DP1 | ACGGCGGCCGTAACTATA | 1985–2002 | + | Commonly used for SDM, sequencing | 30 |
| ZGE23 | ACAGGCCAGTTAGCTA | 2292–2277 | − | Commonly used for SDM, sequencing | 30 |
| GW1 | GACGGGAAGACCCCGTGGA | 2137–2155 | + | SDM (A2142G) | This work |
| GW11 | GGTCTTCCCGTCTTGCCGC | 2148–2130 | − | SDM (A2142G) | This work |
| GW2 | GACGGCAAGACCCCGTGGA | 2137–2155 | + | SDM (A2142C) | This work |
| GW12 | GGTCTTGCCGTCTTGCCGC | 2148–2130 | − | SDM (A2142C) | This work |
| GW3 | GACGGTAAGACCCCGTGGA | 2137–2155 | + | SDM (A2142T) | This work |
| GW13 | GGTCTTACCGTCTTGCCGC | 2148–2130 | − | SDM (A2142T) | This work |
| GW4 | GACGGAGAGACCCCGTGGA | 2137–2155 | + | SDM (A2143G) | This work |
| GW14 | GGTCTCTCCGTCTTGCCGC | 2148–2130 | − | SDM (A2143G) | This work |
| GW5 | GACGGACAGACCCCGTGGA | 2137–2155 | + | SDM (A2143C) | This work |
| GW15 | GGTCTGTCCGTCTTGCCGC | 2148–2130 | − | SDM (A2143C) | This work |
| GW6 | GACGGATAGACCCCGTGGA | 2137–2155 | + | SDM (A2143T) | This work |
| GW16 | GGTCTATCCGTCTTGCCGC | 2148–2130 | − | SDM (A2143T) | This work |
| GW7 | AGACGAAAAGACCCCGTGG | 2136–2154 | + | SDM (G2141A) | This work |
| GW17 | GTCTTTTCGTCTTGCCGCG | 2147–2129 | − | SDM (G2141A) | This work |
The bases different from those in the wild-type 23S rRNA gene sequence are underlined.
Positions refer to the new numbering scheme for the H. pylori 23S rRNA (30).
The plus (+) strand is equivalent to the rRNA sequence, and the minus (−) strand is the complementary one.
SDM, site-directed mutagenesis.
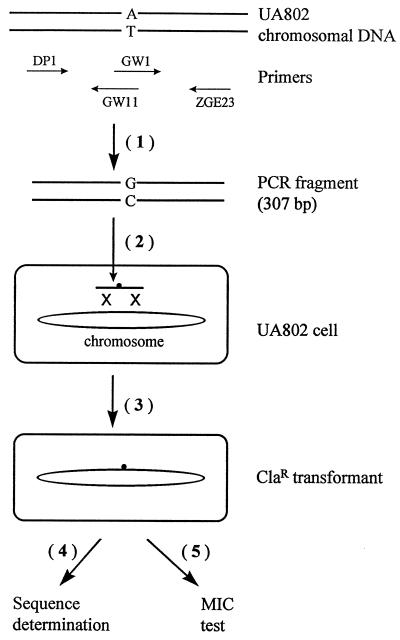
FIG. 1.
Outline of procedures for construction and characterization of site-directed mutations. The steps are numbered in series. (Step 1) Site-directed mutagenesis. Wild-type H. pylori UA802 chromosomal DNA was used as the template for sequential PCR. The figure shows an example for constructing an A2142G mutation, and the primers used (Table 1) are indicated. (Step 2) Natural transformation. It includes DNA uptake, as illustrated by a heavy arrow, into the cell and subsequent homologous DNA recombination into the chromosome of a recipient cell, as depicted by a double crossover event (X X symbol). (Step 3) Selection for clarithromycin resistance. This step includes 3 to 4 days of incubation for the formation of single colonies and subsequent subculturing to obtain genetically stable Clar transformants. (Step 4) Genotypic identification of Clar mutants by DNA sequencing. (Step 5) Characterization of MLS phenotypes of Clar mutants by the MIC test.
Natural transformation.
The PCR fragments containing a site-specific mutation were introduced into clarithromycin-susceptible strain H. pylori UA802 by natural transformation as described previously (13), and the transformants were selected for clarithromycin resistance. Briefly, recipient cells were heavily inoculated on cold BHI-YE agar plates and were grown for 5 h, followed by the addition of 0.2 to 0.5 μg of DNA (307-bp PCR fragment) onto the bacterial lawn. After incubation for 16 to 24 h under microaerobic conditions the transformed cells were streaked onto BHI-YE agar plates containing the selective antibiotic (2, 0.5, 0.1, or 0.02 μg of clarithromycin per ml), and the transformants (single colonies) were obtained after incubation for 3 to 4 days.
RESULTS
Characterization of MLS phenotypes of clinical H. pylori isolates and association with specific mutations in the 23S rRNA gene.
To test the MLS phenotypes of several clarithromycin-resistant H. pylori strains, clarithromycin, clindamycin, and quinupristin were used as the representative antibiotics for macrolide, lincosamide, and type B streptogramin, respectively. On the basis of the MICs of the three antibiotics presented in Table 2, two phenotypes of MLS resistance were identified. Strains that are highly resistant to clarithromycin (MICs, ≥8 μg/ml) exhibit high-level resistance to clindamycin (MICs, >512 μg/ml) and quinupristin (MIC, 128 μg/ml) (type I). Other strains, strains A and B, for which the MIC of clarithromycin ranged from 1 to 4 μg/ml (intermediate level of resistance) have intermediate-level resistance to clindamycin (MIC, 256 μg/ml) but minimal resistance to quinupristin (MIC, 4 μg/ml, which is identical to that for susceptible strain UA802) (type II).
TABLE 2.
Mutations in the 23S rRNA gene and associated MLS phenotypes of clinical H. pylori isolates
| H. pylori straina | 23S rRNA gene mutation | MIC (μg/ml)b
|
MLS pheno- typec | ||||
|---|---|---|---|---|---|---|---|
| Cla | Cln | Qnp | Dfp | RP59500 | |||
| UA802 | Wild type | 0.004 | 32 | 4 | 8 | 2 | S |
| A | A2143G | 4 | 256 | 4 | 8 | 2 | R-II |
| B | A2143G | 1 | 256 | 4 | 8 | 0.5 | R-II |
| D | A2142G | 8 | >512 | 64 | 8 | 1 | R-I |
| MQ | A2142G | 16 | >512 | 128 | 8 | 1 | R-I |
| E | A2142G | 16 | >512 | 128 | 8 | 2 | R-I |
| Eryr-1 | A2142G | 16 | >512 | 128 | 8 | 2 | R-I |
| Clar-1 | A2142G | 16 | >512 | 128 | 8 | 2 | R-I |
Eryr-1 and Clar-1 are erythromycin- and clarithromycin-resistant strains of UA802, respectively, obtained by transformation of the erythromycin or clarithromycin resistance determinant from strain E (30).
Abbreviations for antibiotics: Cla, clarithromycin; Cln, clindamycin; Qnp, quinupristin (streptogramin B); Dfp, dalfopristin (streptogramin A); RP59500, mixture of quinupristin and dalfopristin.
S, susceptible; R-II, type II resistance; R-I, type I resistance.
To examine the genetic basis of these MLS phenotypes, we determined the nucleotide sequence of the 23S rRNA gene coding for the peptidyltransferase region of the 23S rRNA from all of these strains. Briefly, a PCR fragment was amplified from the chromosomal DNA with primers DP1 and ZGE23 (Table 1), followed by sequencing of the fragment with primer DP1. By comparing the DNA sequences from different strains, specific mutations at position 2142 or 2143 were associated with the two MLS phenotypes: A2142G for type I MLS resistance and A2143G for type II MLS resistance (Table 2).
In addition to the mutation at position 2142 or 2143, other mutations were observed in certain strains (in H. pylori coordinates, A2085G in strain A and A2223G in strain E). Notably, although strains A and B have the same phenotypic resistance to clindamycin and quinupristin, there are minor but significant differences in the MICs of clarithromycin for the two strains (4 versus 1 μg/ml; Table 2). To find out whether the additional mutation accounts for the observed difference in the MICs of clarithromycin for strains A and B, we introduced the PCR fragments containing the relevant mutations amplified with primer pair DP1-ZGE23 from the chromosomal DNA of strain A or B into H. pylori UA802. The relevant mutations in both transformants were confirmed by DNA sequencing to be the same as those in the donor strain. By examining their effects in an isogenic background, it was found that the MIC of clarithromycin was the same for both transformants (4 μg/ml). Therefore, it is likely that the observed difference in the clarithromycin MICs for strains A and B is not due to the additional A2085G mutation but, rather, reflects other host effects. The additional mutations observed in the present study seem to be unrelated to clarithromycin resistance but, rather, represent microdiversity in the sequences of 23S rRNA genes from different H. pylori strains.
Synergistic effect of type A and B streptogramins on H. pylori.
It has been shown in E. coli and some other bacteria that type A and B streptogramins can block the peptidyltransferase activity of the 50S ribosomal subunit and can have synergistic effects resulting from conformational changes imposed upon the peptidyltransferase center by streptogramin A and by inhibition of both early and late stages of protein synthesis (4). For the H. pylori strains mentioned above, we also characterized the MICs of streptogramin A (dalfopristin) and a mixture of streptogramin B and A (quinupristin-dalfopristin in a 30:70 ratio; RP59500) (2). All the strains tested in this study were inhibited by 8 μg of dalfopristin per ml (Table 2), regardless of their susceptibility or resistance to MLS antibiotics. By using RP59500, the MICs for all the strains further decreased to 0.5 to 2 μg/ml (Table 2), demonstrating a synergistic effect of streptogramins A and B on H. pylori. These effects are similar to those previously observed for both Staphylococcus aureus (12) and Enterococcus faecium (11).
In vitro site-directed mutagenesis in the 23S rRNA gene.
Of all the clarithromycin-resistant clinical isolates of H. pylori reported so far, the resistance in most strains is associated with A-to-G transition mutations (at position 2142 or 2143, according to the revised numbering system; see Fig. 3) in the 23S rRNA gene (7, 21, 28, 29, 34, 35). To find out if other types of mutations (A to C or A to T) also confer clarithromycin resistance, we performed in vitro site-directed mutagenesis. Since a suitable replicative or integrative vector is generally unavailable for H. pylori, we used the natural transformation process to introduce site-directed mutations into the chromosome. This corresponds to the situation of clarithromycin resistance in clinical isolates in which mutations are chromosomal rather than plasmid borne. The natural transformation process includes DNA uptake and homologous DNA recombination (Fig. 1). After clarithromycin selection, only those mutations that confer clarithromycin resistance and that are incorporated into the chromosome can give rise to transformants. In addition, we took advantage of the low fidelity of Taq DNA polymerase for use in sequential PCR to construct mutants with site-directed mutations. Using a strategy similar to that described by Kok et al. (17), we sought to obtain other types of mutations (random mutations) that confer Clar and that may be screened out by natural transformation. Using the primers listed in Table 1 and the method described in Materials and Methods, we obtained the PCR fragments containing the following specific point mutations: A2142G, A2142C, A2142T, A2143G, A2143C, A2143T, and G2141A. These were verified by DNA sequencing. Note that at this stage a very minor fraction of the PCR products may possibly contain certain random mutations, produced by Taq DNA polymerase errors, that could not be detected by DNA sequencing. These site-specific mutations were introduced into UA802 by natural transformation so that we could characterize their effects on MLS resistance in an isogenic background.
First, different concentrations of clarithromycin (2, 0.5, 0.1, and 0.02 μg/ml in agar medium) were tested for transformant selection. We found that 0.1 μg/ml is the lowest concentration that can be used to give unambiguous transformation results. With this concentration of antibiotic, no transformants were obtained for the control (wild-type) DNA or for DNA with the A2143T or G2141A mutation (Table 3), suggesting that these mutations do not confer any resistance to up to 0.1 μg of clarithromycin per ml. In contrast, more than 1,000 transformants with each of the A2142G, A2142C, A2142T, A2143G, and A2143C mutations were obtained. This corresponds to a transformation frequency of 10−5 per viable cell. Subsequently, some of the transformants were colony purified. Upon subculturing of the colonies, the Clar phenotypes of these clones were shown to be stable, suggesting that the corresponding mutations were incorporated into the chromosome.
TABLE 3.
In vitro site-directed mutagenesis of H. pylori 23S rRNA gene and MLS phenotypes of the mutants obtained
| Mutation in PCR fragment | No. of transformants obtained | No. of transformants sequenced | Mutation in 23S rRNA gene | MIC (μg/ml)a
|
||
|---|---|---|---|---|---|---|
| Cla | Cln | Qnp | ||||
| Wild type | 0 | Wild type | 0.004 | 32 | 4 | |
| G2141A | 0 | NK | NK | NK | ||
| A2142G | >1,000 | 8 | A2142G | 16 | >512 | 128 |
| A2142C | >1,000 | 3 | A2142C | 16 | >512 | 128 |
| A2142Cb | 1 | A2142C/T | 8 | >512 | 128 | |
| A2142T | >1,000 | 4 | A2142T | 4 | 512 | 64 |
| A2143G | >1,000 | 4 | A2143G | 4 | 256 | 4 |
| A2143C | >1,000 | 3 | A2143C | 4 | 256 | 4 |
| A2143Cc | 1 | A2143C + A2142G | 16 | >512 | 128 | |
| A2143T | 0 | NK | NK | NK | ||
Abbreviations: Cla, clarithromycin; Cln, clindamycin; Qnp, quinupristin; NK, not known.
Of the four transformants picked up for sequencing, one contained an A2142C/T mutation (both C and T bands of equal intensities were revealed on the sequencing gel [see Fig. 2]).
Of the four transformants picked up for sequencing, one contained a double mutation (A2143C and A2142G).
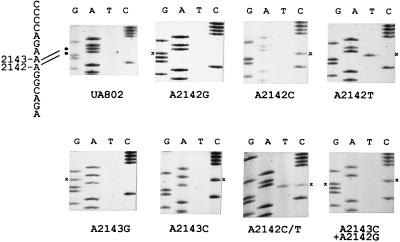
Next, the chromosomal DNA sequences of the region of interest were examined for some of these Clar clones (Fig. 2; Table 3). In the majority of the clones examined, targeted mutations were detected, i.e., substitution of G, C, or T for A2142 and substitution of G or C for A2143. Two untargeted mutations were observed. One was a double mutation at A2143C and A2142G which was obtained from transformation of the A2143C mutation. Another clone obtained from the transformation of A2142C was shown to carry an A2142C/T mutation because both C and T bands at position 2142 were revealed on the sequencing gel at approximately the same intensity (Fig. 2). Since there are two copies of the 23S rRNA gene in H. pylori (16, 30, 31), this mutant may represent a heterozygote with C in one copy of the gene and T in the other copy of the gene. The occurrence of the mutants with untargeted mutations may be due to (i) a mutation randomly introduced in the PCR fragment or (ii) a spontaneous or drug-induced mutation which occurred in recipient cells.
FIG. 2.
Nucleotide sequences of the short region in the 23S rRNA gene from Clas H. pylori UA802 (wild type) and the constructed Clar mutants showing their relevant genotypes. The corresponding sequence for the wild-type strain (see Fig. 3) is indicated on the left, with both adenines at positions 2142 and 2143 highlighted with black dots. The position of a specific base substitution(s) in each particular mutant is marked with an asterisk.
Characterization of MLS phenotypes of the site-directed mutations.
The MICs of clarithromycin, clindamycin, and quinupristin for all the constructed mutants with site-directed mutations are listed in Table 3, from which the following points can be made. (i) Mutations at position 2142 always confer a higher level of clarithromycin resistance than mutations at position 2143. At the same position, an A-to-G or an A-to-C mutation gives rise to similar clarithromycin MICs which are higher than that conferred by the A-to-T mutation. (ii) There is no further increase in the MIC of clarithromycin for the mutant with the double mutation (A2142G plus A2143C) compared to that for the mutant with the single mutation (A2142G) (MIC, 16 μg/ml). The clarithromycin MIC for heterozygous mutant A2142C/T is 8 μg/ml, which is intermediate between those for both homozygous mutants, A2142C (MIC, 16 μg/ml) and A2142T (MIC, 4 μg/ml). (iii) Two kinds of MLS phenotypes were observed for these mutants with site-directed mutations (summarized in Fig. 3). Any mutation (A to G, C, or T) which occurred at position 2142 conferred cross-resistance to all three kinds of antibiotics tested (type I MLS resistance). Substitution of A at position 2143 with G or C gave rise to intermediate levels of resistance to clarithromycin and clindamycin but no resistance to quinupristin (type II MLS resistance). These two phenotypes of MLS resistance are similar to those observed for clinical isolates (Table 2), but distribution of the 23S rRNA genotypes associated with these phenotypes in the constructed mutants are more extensive than those found in clinical isolates, in which A-to-G mutations have predominantly been observed, A-to-C mutations have occasionally been observed, and A-to-T mutations have never been observed.
DISCUSSION
In the first part of this study, we observed the following two major phenotypes of MLS resistance for H. pylori: type I, high-level cross-resistance to all MLS antibiotics, and type II, intermediate-level resistance to clarithromycin and clindamycin but no resistance to streptogramin B. By examining the DNA sequences of these strains, specific mutations in the 23S rRNA gene, A2142G and A2143G, were associated with these two MLS phenotypes, respectively. The observation that the A2142G mutation is associated with cross-resistance to all MLS antibiotics is in agreement with the finding for E. coli (equivalent to the A2058G mutation) and other organisms (27, 36, 39). The A-to-G mutation at the base equivalent to base 2059 in E. coli has been shown to be associated with resistance to macrolides in many organisms including mycoplasmas, mycobacteria (18, 19, 37), and H. pylori, but its association with cross-resistance to all MLS antibiotics was not reported. A recent study with propionibacteria demonstrated that the A2059G mutation gives rise to a high level of resistance to macrolides, a moderate level of resistance to licosamides, and no resistance to type B streptogramins (24). Similar to the findings in that study, we observed that the A2143G mutation in the H. pylori 23S rRNA gene is linked to an intermediate level of resistance to clarithromycin and clindamycin and no resistance to streptogramin B.
In addition, we also determined the MICs of dalfopristin and a combination of quinupristin and dalfopristin (RP59500) and demonstrated a synergistic effect of type A and B streptogramins on H. pylori. Both MLS-sensitive and MLS-resistant H. pylori strains were found to be moderately susceptible to dalfopristin and susceptible to RP59500. RP59500 is a new semisynthetic injectable streptogramin which has been shown to have excellent activity against most gram-positive bacteria including staphylococci, E. faecium, and pneumococci (2). It offers some advantages over the commercially available antimicrobial agents against drug-resistant gram-positive bacteria. In vitro studies have shown that it also has good activity (MIC, <2 μg/ml) against some selected gram-negative pathogens such as Moraxella catarrhalis, Mycoplasma pneumoniae, and Neisseria gonorrhoeae and a moderate level of activity against Haemophilus influenzae (MICs, 2 to 8 μg/ml) (for a review, see reference 2). Our results showed that RP59500 has good activity (MICs, 0.5 to 2 μg/ml) against H. pylori and even against MLS-resistant strains. Thus, our data may prompt consideration of the use of quinupristin-dalfopristin as a possible alternative antibiotic in the case of failure of therapy with a clarithromycin-based treatment regimen.
The major goal of this work was the construction of a series of site-directed mutations in the H. pylori 23S rRNA gene that are associated with clarithromycin resistance and the characterization of their effects on MLS resistance. At first, by introducing a 307-bp PCR fragment containing a specific point mutation into UA802 by natural transformation, a reasonably high efficiency of transformation was obtained, suggesting that a DNA fragment as small as 300 bp is sufficient for the occurrence of a double crossover in homologous recombination in H. pylori (Fig. 1). Using this method, we have constructed the expected mutants except mutant A2143T with base substitutions at position 2142 or 2143. In addition, a double point mutation and a heterozygous mutant (A2142C/T) were obtained. By examining the MLS phenotypes of these constructed mutants, two types of MLS resistance similar to those seen for the clinical isolates were observed. Type I is associated with mutations at position 2142 and type II is associated with mutations at position 2143 (Fig. 3). These data imply that position 2142 is the binding site for all MLS antibiotics and that position 2143 has a binding affinity for macrolides and lincosamides but not for streptogramin B. According to these results, we can consider the different MLS phenotypes as a signature for the specific type of mutation in the 23S rRNA gene in identifying clarithromycin-resistant H. pylori isolates.
G2057A in E. coli conferred low-level resistance to erythromycin (10). A similar result was found recently with propionibacteria (24). It was hypothesized that disruption of the G2057-C2611 base pairing leads to a weaker rearrangement and affects the binding site of macrolide antibiotics (33). To investigate whether there is a similar effect in H. pylori, we also created an equivalent mutation, G2141A. However, after selection with 0.1 μg of clarithromycin per ml, no transformant was obtained. Under identical transformation conditions, the other mutations gave rise to more than 1,000 transformants. The unavailability of G2141A and A2143T mutants gave us at least indirect evidence that such mutations do not confer a significant level of clarithromycin resistance. We also attempted to use even lower concentrations of clarithromycin (0.02 μg/ml) for the selection of resistance with the goal of obtaining these two mutants, but they were not forthcoming.
Another interesting feature that we noted is that most mutants constructed appear to carry a homozygous mutation in both copies of the 23S rRNA gene, because only a single band representing the mutated base was revealed at the relevant position on a sequencing gel (Fig. 2). Certainly, the evidence from sequencing alone is not convincing for the resolution of heterozygosity. Recently, Sander et al. (26) demonstrated that clarithromycin resistance is dominant over sensitivity in Mycobacterium smegmatis, another eubacterium carrying two rRNA operons. Evidence of heterozygosity has also been reported for a few clinical isolates of H. pylori (28, 34). However, most of the Clar H. pylori isolates so far reported are homozygous mutants. Thus, it is possible that the minor fraction of heterozygous mutants escaped our examination since only small numbers of transformants (four clones for each type of mutation) were sequenced. The prevalence of homozygosity over heterozygosity in H. pylori may reflect a high efficiency of DNA recombination in this organism. The mutation in one copy of the 23S rRNA gene may be easily copied to the other 23S rRNA gene by efficient homologous DNA recombination under the selection pressure to produce a diploid mutation that may confer a higher level of resistance.
To date, A-to-G mutations have been predominantly associated with clarithromycin resistance in clinical H. pylori isolates; few mutations from A to C and no mutation from A to T in the 23S rRNA gene were identified (7, 21, 28, 29, 30, 34, 35). Concerning the possible mechanism for this phenomenon in H. pylori, Debets-Ossenkopp et al. (6) proposed that it is due to the relatively higher growth rates and the MIC for the strains with A-to-G mutations. Indeed, our preliminary data suggest that the growth of the A-to-C or A-to-T mutants is significantly slower (a lag of about 1 day) than that of the wild type or the A-to-G mutants (38). The differences in the MIC of clarithromycin that we observed for these mutants (G = C > T >> A) are essentially in agreement with their data (6). However, we found that the A-to-G and the A-to-C mutations at the same position mediate identical MICs; and particularly, the MIC for the strains with the A2142C mutation is higher than that for the strains with the A2143G mutation. We observed that two additional mutations in the 23S rRNA gene from Clar clinical isolates are also A-to-G transitions but are unrelated to clarithromycin resistance. By inferrence from other gene sequences in H. pylori, such as those for α1,3-fucosyltransferase genes (14, 31), we also found that transitions (from the A/T base pair to the G/C base pair or vice versa) account for the majority of intraspecies microdiversity. In general, a particular mutation occurs in two steps: mutation formation and selective accumulation. As mentioned above (6), the relatively higher growth rate and MIC could offer the A-to-G mutation an advantage over other types of mutations in selective accumulation (step 2). As an additional possible mechanism, we propose that the A-to-G transitions may be preferentially formed or produced (in step 1) in H. pylori.
ACKNOWLEDGMENTS
We thank S. Pong-Porter and D. Low, as well as Rhone Poulenc Rorer, for providing the antibiotics quinupristin and dalfopristin. We also thank Z. Ge and Q. Jiang for technical guidance.
This work was supported in part by funding from the Canadian Bacterial Diseases Network (Centers of Excellence Program) to D.E.T., who is a medical scientist with the Alberta Heritage Foundation for Medical Research (AHFMR), and by a postdoctoral fellowship from the Canadian Association of Gastroenterology and Astra Canada in association with an MRC-PMAC award to G.W., who also held a fellowship from AHFMR.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1991. pp. 8.5.7–8.5.9. [Google Scholar]
- 2.Bryson H M, Spencer C M. Quinupristin-dalfopristin. Drugs. 1996;52:406–415. doi: 10.2165/00003495-199652030-00006. [DOI] [PubMed] [Google Scholar]
- 3.Cayla, R., F. Zerbib, P. Talbi, F. Megraud, and H. Lamouliatte. 1995. Pre- and posttreatment clarithromycin resistance of Helicobacter pylori strains: a key factor of treatment failure. Gut 37(Suppl. 1):A55.
- 4.Cocito, C., M. D. Giambattista, E. Nyssen, and P. Vannuffel. 1997. Inhibition of protein synthesis by streptogramins and related antibiotics. J. Antimicrob. Chemother. 39(Suppl. A):7–13. [DOI] [PubMed]
- 5.Cundliffe E. Recognition sites for antibiotics within rRNA. In: Hill W E, Dalberg A, Garrett R A, Moore P B, Schlessinger D, Warner J R, editors. The ribosome: structure, function, and evolution. Washington, D.C: American Society for Microbiology; 1990. pp. 479–490. [Google Scholar]
- 6.Debets-Ossenkopp, Y. J., A. B. Brinkman, E. J. Kuipers, J. G. Kusters, and C. M. J. E. Vandenbroucke-Grauls. 1997. Preferential A to G of clarithromycin resistance mutation in 23S rRNA in Helicobacter pylori is due to relative higher growth rates and MIC of these A to G mutations. Gut 41(Suppl. 1):A7.
- 7.Debets-Ossenkopp Y J, Sparrius M, Kusters J G, Kolkman J J, Vandenbroucke-Grauls C M J E. Mechanism of clarithromycin resistance in clinical isolates of Helicobacter pylori. FEMS Microbiol Lett. 1996;142:37–42. doi: 10.1111/j.1574-6968.1996.tb08404.x. [DOI] [PubMed] [Google Scholar]
- 8.Dick J D. Helicobacter (Campylobacter) pylori: a twist on an old disease. Annu Rev Microbiol. 1990;108:70–90. doi: 10.1146/annurev.mi.44.100190.001341. [DOI] [PubMed] [Google Scholar]
- 9.Egebjerg J, Larsen N, Garrett R A. Structural map of 23S rRNA. In: Hill W E, Dalberg A, Garrett R A, Moore P B, Schlessinger D, Warner J R, editors. The ribosome: structure, function, and evolution. Washington, D.C: American Society for Microbiology; 1990. pp. 168–179. [Google Scholar]
- 10.Ettayebi M, Prasad S M, Morgan E A. Chloramphenicol-erythromycin resistance mutations in a 23S rRNA gene of Escherichia coli. J Bacteriol. 1985;162:551–557. doi: 10.1128/jb.162.2.551-557.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fantin B, Leclercq R, Garry L, Carbon C. Influence of inducible cross-resistance to macrolides, lincosamides, and streptogramin B-type antibiotics in Enterococcus faecium on activity of quinupristin-dalfopristin in vitro and in rabbits with experimental endocarditis. Antimicrob Agents Chemother. 1997;41:931–935. doi: 10.1128/aac.41.5.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fantin B, Leclercq R, Merle Y, Saint-Julien L, Veyrat C, Duval J, Carbon C. Critical influence of resistance to streptogramin B-type antibiotics on activity of RP59500 (quinupristin-dalfopristin) in experimental endocarditis due to Staphylococcus aureus. Antimicrob Agents Chemother. 1995;39:400–405. doi: 10.1128/aac.39.2.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ge Z, Taylor D E. H. pylori DNA transformation by natural competence and electroporation. In: Clayton C L, Mobley H L T, editors. Methods in molecular medicine. Helicobacter pylori protocols. Totowa, N.J: Humana Press Inc.; 1997. pp. 145–152. [DOI] [PubMed] [Google Scholar]
- 14.Ge Z, Chan N W C, Palcic M M, Taylor D E. Cloning and heterologous expression of an α1,3 fucosyltransferase gene from the gastric pathogen Helicobacter pylori. J Biol Chem. 1997;272:21357–21363. doi: 10.1074/jbc.272.34.21357. [DOI] [PubMed] [Google Scholar]
- 15.Goddard A F, Logan R P H. Antimicrobial resistance and Helicobacter pylori. J Antimicrob Chemother. 1996;37:639–643. doi: 10.1093/jac/37.4.639. [DOI] [PubMed] [Google Scholar]
- 16.Jiang Q, Hiratsuka K, Taylor D E. Variability of gene order in different Helicobacter pylori strains contributes to genome diversity. Mol Microbiol. 1996;20:833–842. doi: 10.1111/j.1365-2958.1996.tb02521.x. [DOI] [PubMed] [Google Scholar]
- 17.Kok R G, D’Argenio D A, Ornston L N. Combining localized PCR mutagenesis and natural transformation in direct genetic analysis of a transcriptional regulator gene, pobR. J Bacteriol. 1997;179:4270–4276. doi: 10.1128/jb.179.13.4270-4276.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lucier T S, Heitzman K, Liu S-K, Hu P-C. Transition mutations in the 23S rRNA of erythromycin-resistant isolates of Mycoplasma pneumoniae. Antimicrob Agents Chemother. 1995;39:2770–2773. doi: 10.1128/aac.39.12.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meier A, Kirschner P, Springer B, Steingrube V A, Brown B A, Wallace R J, Jr, Böttger E C. Identification of mutations in 23S rRNA gene of clarithromycin-resistant Mycobacterium intracellulare. Antimicrob Agents Chemother. 1994;38:381–384. doi: 10.1128/aac.38.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moazed D, Noller H F. Chloramphenicol, erythromycin, carbomycin, and vernamycin B protect overlapping sites in the peptidyl transferase region of 23S ribosomal RNA. Biochemie. 1987;69:879–884. doi: 10.1016/0300-9084(87)90215-x. [DOI] [PubMed] [Google Scholar]
- 21.Occhialini A, Urdaci M, Doucet-Populaire F, Bebear C M, Lamouliatte H, Megraud F. Macrolide resistance in Helicobacter pylori: rapid detection of point mutations and assays of macrolide binding to ribosomes. Antimicrob Agents Chemother. 1997;41:2724–2728. doi: 10.1128/aac.41.12.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parsonnet J, Friedman G D, Vandersteen D P, Chang Y, Vogelman J H, Orentreich N, Silbey R K. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991;325:1127–1131. doi: 10.1056/NEJM199110173251603. [DOI] [PubMed] [Google Scholar]
- 23.Rene W M, Hulst V D, Keller J J, Rauws E A J, Tytgat G N J. Treatment of Helicobacter pylori infection: a review of the world literature. Helicobacter. 1996;1:6–19. doi: 10.1111/j.1523-5378.1996.tb00003.x. [DOI] [PubMed] [Google Scholar]
- 24.Ross J I, Eady E A, Cove J H, Jones C E, Ratyal A H, Miller Y W, Vyakrnam S, Cunliffe W J. Clinical resistance to erythromycin and clindamycin in cutaneous propionibacteria isolated from acne patients is associated with mutations in 23S rRNA. Antimicrob Agents Chemother. 1997;41:1162–1165. doi: 10.1128/aac.41.5.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Sander P, Prammananan T, Meier A, Frischorn K, Bottger E C. The role of ribosomal RNAs in macrolide resistance. Mol Microbiol. 1997;26:469–480. doi: 10.1046/j.1365-2958.1997.5811946.x. [DOI] [PubMed] [Google Scholar]
- 27.Sigmund C D, Ettayebi M, Borden A, Morgen E A. Antibiotic resistance mutations in ribosomal RNA genes of Escherichia coli. Methods Enzymol. 1988;164:673–690. doi: 10.1016/s0076-6879(88)64077-8. [DOI] [PubMed] [Google Scholar]
- 28.Stone G G, Shorttridge D, Versalovic J, Beyer J, Flamm R K, Ghoneim A T, Tanaka K. A PCR-oligonucleotide ligation assay to determine the prevalence of 23S rRNA gene mutations in clarithromycin-resistant Helicobacter pylori. Antimicrob Agents Chemother. 1997;41:712–714. doi: 10.1128/aac.41.3.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stone G G, Shorttridge D, Flamm R K, Versalovic J, Beyer J, Idler K, Zulawinski L, Tanaka K. Identification of a 23S rRNA gene mutation in clarithromycin-resistant Helicobacter pylori. Helicobacter. 1996;1:227–228. doi: 10.1111/j.1523-5378.1996.tb00043.x. [DOI] [PubMed] [Google Scholar]
- 30.Taylor D E, Ge Z, Purych D, Lo T, Hiratsuka K. Cloning and sequence analysis of the two copies of 23S rRNA genes from Helicobacter pylori and association of clarithromycin resistance with 23S rRNA mutations. Antimicrob Agents Chemother. 1997;41:2621–2628. doi: 10.1128/aac.41.12.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomb J-F, White O, Kerlavage A R, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 32.Vaira D, Holton J, Miglioli M, Menegatti M, Mule P, Barbara L. Peptic ulcer disease and Helicobacter pylori infection. Curr Opin Gastroenterol. 1994;10:98–104. [Google Scholar]
- 33.Vannuffel P, Di Giambattista M, Morgan E A, Cocito C. Identification of a single base change in ribosomal RNA leading to erythromycin resistance. J Biol Chem. 1992;267:8377–8382. [PubMed] [Google Scholar]
- 34.Versalovic J, Shortridge D, Kibler K, Griffy M V, Bryer J, Flamm R K, Tanaka S K, Graham D Y, Go M F. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob Agents Chemother. 1996;40:477–480. doi: 10.1128/aac.40.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Versalovic J, Osato M S, Spakovsky K, Dore M P, Reddy R, Stone G G, Shortridge D, Flamm R K, Tanaka S K, Graham D Y. Point mutations in the 23S rRNA gene of Helicobacter pylori associated with different levels of clarithromycin resistance. J Antimicrob Chemother. 1997;40:283–286. doi: 10.1093/jac/40.2.283. [DOI] [PubMed] [Google Scholar]
- 36.Vester B, Garrett R A. A plasmid-coded and site-directed mutation in Escherichia coli 23S RNA that confers resistance to erythromycin: implications for the mechanism of action of erythromycin. Biochimie. 1987;69:891–900. doi: 10.1016/0300-9084(87)90217-3. [DOI] [PubMed] [Google Scholar]
- 37.Wallace R J, Jr, Meier A, Brown B A, Zhang Y, Sander P, Onyi G O, Bottger E C. Genetic basis for clarithromycin resistance among isolates of Mycobacterium chelonae and Mycobacterium abscessus. Antimicrob Agents Chemother. 1996;40:1676–1681. doi: 10.1128/aac.40.7.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, G., and D. E. Taylor. Unpublished data.
- 39.Weisblum B. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother. 1995;39:577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]