FGFR3-TACC3 (F3T3) fusions are seen in 3.1–11.7% of glioblastoma (GBM) cases with better survival [1, 4, 5, 7–9]. We endeavored to characterize methylation profiles in detail, using a cohort of 79 F3T3-positive GBMs from 71 patients (seven patients with primary and recurrent tumor resections, Table S1). Compared to gliomas without a documented fusion, dimensionality reduction showed that F3T3-positive GBMs nested predominantly to GBM-mesenchymal and RTK-II subclasses (Fig. 1a). Transcriptomic analysis on 9 F3T3-positive and 31 negative patients (Fig. 1b) confirmed a previous finding of activated mitochondrial activity and altered vascular activity [2, 6].
Fig. 1.
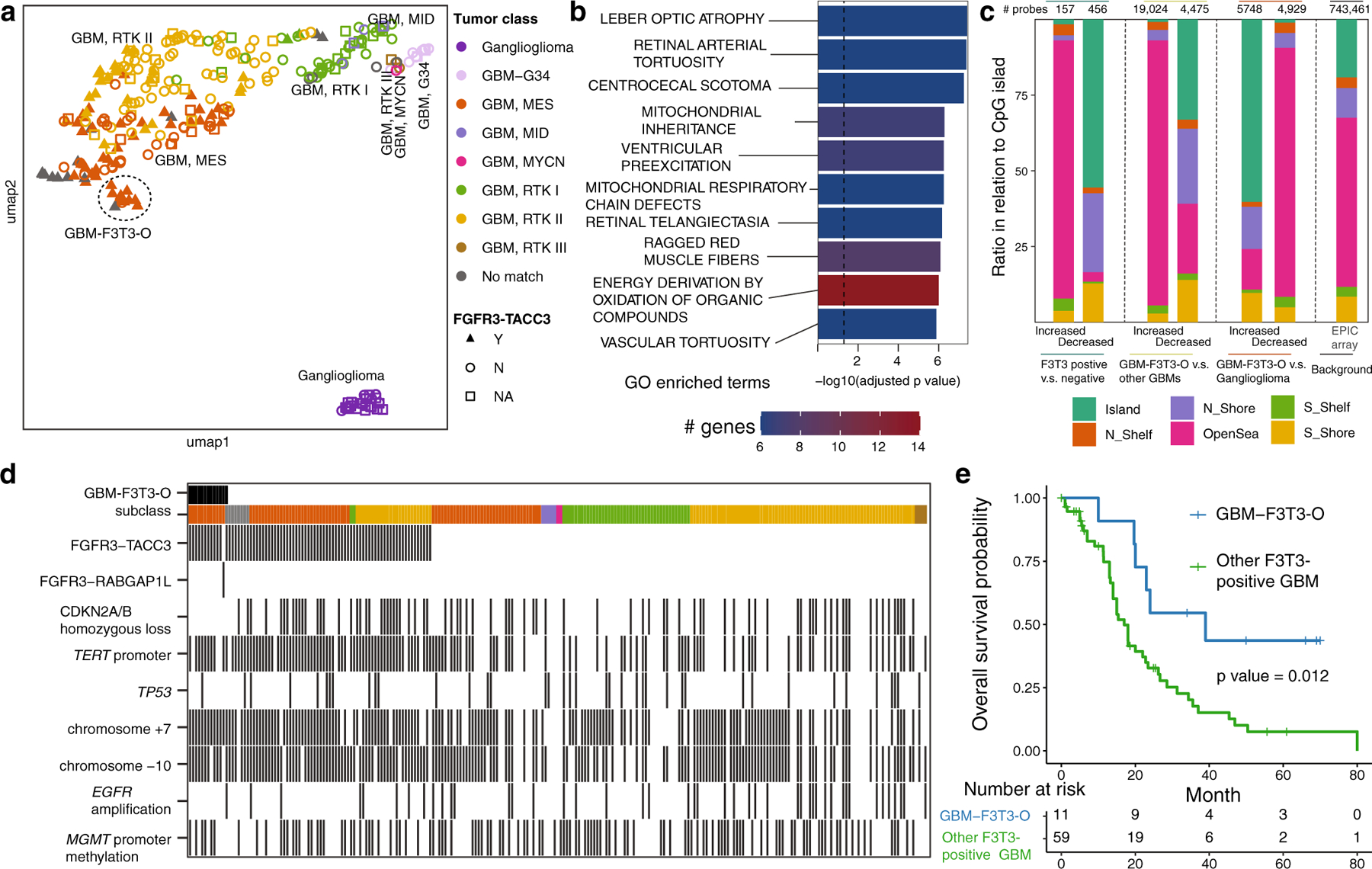
Molecular features of FGFR3-TACC3 fusion (F3T3) glioblastomas (GBMs). a Uniform manifold approximation and projection (UMAP) on a collection of in-house and published F3T3-positive GBMs and F3T3-negative/unknown GBMs and other brain tumors. b Gene ontology enrichment of over-expressed genes in F3T3-positive GBMs. c Differentially methylated sites in relation to CpG island between F3T3 fusion-positive vs. negative GBMs, ‘outlier’ GBMs F3T3-positive group (GBM-F3T3-O) vs. other GBMs, GBM-F3T3-O vs. gangliogliomas and the background. d Molecular features of GBMs in this study, samples were ordered by GBM-F3T3-O, F3T3 status and methylation subclass. e Kaplan–Meier plot of survival for GBM-F3T3-O and other F3T3-positive GBMs
We identified an outlier subgroup of 11 F3T3-positive patients (12 samples) and an additional FGFR3-RABGAP1 fusion-positive case (Fig. 1a, Table S1). These samples were originally classified as GBM-mesenchymal subtype by the Heidelberg/DKFZ classifier version 11.b6 [3]. Interestingly, on recently released version 12.5 of the DKFZ classifier, top scores for seven of these cases were ganglioglioma, whereas five cases remained GBM-mesenchymal as the top score with ganglioglioma as the second hit in four of these five cases. We henceforth referred these ‘outlier’ F3T3-positive GBM tumors as GBM-F3T3-O. GBM-F3T3-O had higher global methylation than other GBMs (0.57 vs. 0.55, p value = 5.10e-3), and had more differentially methylated sites with other GBMs than with gangliogliomas (Fig. 1c).
Clinical and histologic information was available for 11 and 12 GBM-F3T3-O patients, respectively (Table S2). All showed supratentorial tumors with variable morphology, including five cases with typical GBM histomorphology, and the remainder showing oligodendroglioma-like morphology (p value = 3.35e-4), calcifications and a delicate vascular network, and absence of necrosis (p value = 6.47e-4). None in the GBM-F3T3-O cohort had CDKN2A homozygous loss (p value = 5.07e-3), 10 cases had chromosome + 7/−10, 10 harbored a TERT promoter mutation, and one showed EGFR amplification (Fig. 1d).
GBM-F3T3-O cases showed significantly better patient survival when compared to the remaining F3T3-positive GBMs (Fig. 1e). All cases received the standard glioblastoma treatment consisting of temozolomide and radiation therapy. Three cases received additional agents with either erdafitinib, lomustine or the FGFR inhibitor AZD4547 (Table S2). Overall, our results suggest that a subset of F3T3-positive high-grade tumors with histological low-grade look but molecular features of IDH-wildtype GBM, may exhibit a distinct methylation profile and better patient outcome compared to conventional GBMs.
Supplementary Material
Acknowledgements
This work utilized the computational resources of the NIH HPC Biowulf cluster.
Footnotes
Supplementary Information The online version contains supplementary material available at https://doi.org/10.1007/s00401-022-02430-7.
Data availability
Processed methylation results and raw data are available at the Gene Expression Omnibus (GEO) repository under the accession number GSE200647.
References
- 1.Asif S, Fatima R, Krc R, Bennett J, Raza S (2019) Comparative proteogenomic characterization of glioblastoma. CNS Oncol 10.2217/cns-2019-0003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bielle F, Di Stefano AL, Meyronet D, Picca A, Villa C, Bernier M et al. (2018) Diffuse gliomas with FGFR3-TACC3 fusion have characteristic histopathological and molecular features. Brain Pathol 28:674–683. 10.1111/bpa.12563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D et al. (2018) DNA methylation-based classification of central nervous system tumours. Nature 555:469–474. 10.1038/nature26000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Stefano AL, Picca A, Saragoussi E, Bielle F, Ducray F, Villa C et al. (2020) Clinical, molecular, and radiomic profile of gliomas with FGFR3-TACC3 fusions. Neuro Oncol 22:1614–1624. 10.1093/neuonc/noaa121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferguson SD, Zhou S, Huse JT, de Groot JF, Xiu J, Subramaniam DS et al. (2018) Targetable gene fusions associate with the IDH wild-type astrocytic lineage in adult gliomas. J Neuropathol Exp Neurol 77:437–442. 10.1093/jnen/nly022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frattini V, Pagnotta SM, Tala FJJ, Russo MV, Lee SB, Garofano L et al. (2018) A metabolic function of FGFR3-TACC3 gene fusions in cancer. Nature 553:222–227. 10.1038/nature25171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mata DA, Benhamida JK, Lin AL, Vanderbilt CM, Yang SR, Villafania LB et al. (2020) Genetic and epigenetic landscape of IDH-wildtype glioblastomas with FGFR3-TACC3 fusions. Acta Neuropathol Commun 8:186. 10.1186/s40478-020-01058-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parker BC, Annala MJ, Cogdell DE, Granberg KJ, Sun Y, Ji P et al. (2013) The tumorigenic FGFR3-TACC3 gene fusion escapes miR-99a regulation in glioblastoma. J Clin Invest 123:855–865. 10.1172/JCI67144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh D, Chan JM, Zoppoli P, Niola F, Sullivan R, Castano A et al. (2012) Transforming fusions of FGFR and TACC genes in human glioblastoma. Science 337:1231–1235. 10.1126/science.1220834 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Processed methylation results and raw data are available at the Gene Expression Omnibus (GEO) repository under the accession number GSE200647.


