To the Editor: Mumbach et al. recently described HiChIP, a novel protein-mediated chromatin-conformation assay that lowers cellular input requirements while simultaneously increasing the yield of informative reads compared to previous methods1. We introduce hichipper (http://aryee.mgh.harvard.edu/hichipper), an open-source HiChIP data preprocessing tool, with features that include bias-corrected peak calling, library quality control (QC), DNA loop calling, and output of processed data for downstream analysis and visualization (Fig. 1a).
Figure 1 |.
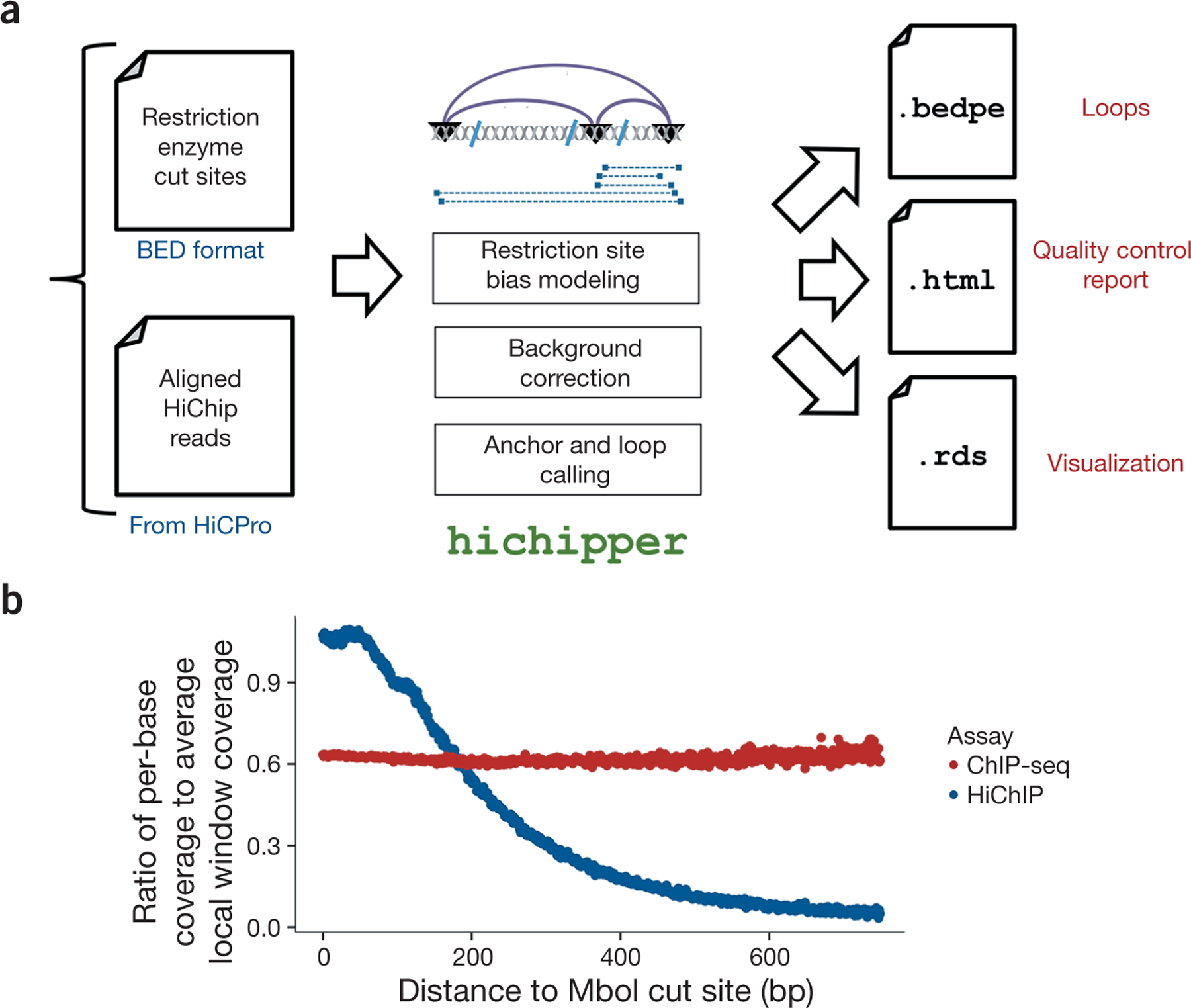
Overview of the hichipper analysis pipeline. (a) hichipper requires aligned and annotated interaction files from preprocessing tools such as Hi-C Pro2 as well as a .bed file of restriction sites. The output of hichipper can be used for quality control, visualization, and downstream topology analysis of HiChIP data. (b) Ratio of per-base coverage to local MACS-estimated local window background signal as a function of distance to nearest MboI cut site for a HiChIP sample (blue) and a ChIP-seq sample (red). Both samples represent published mouse embryonic stem cells (ESCs) with SMC1 (cohesin) ChIP1.
The HiChIP protocol, unlike assays such as ChIA-PET and ChIP-seq, involves a restriction enzyme. This results in a read-density landscape that is biased by proximity to restriction sites (Fig. 1b), hampering identification of peaks representing DNA loop anchors and hence loops. hichipper accounts for this read-density bias by modeling the background read density as a function of proximity to restriction sites, resulting in substantial improvements in identification of loop anchors and overall loop calling. Further, we have found that taking the location of restriction enzyme cut sites into account also boosts sensitivity by increasing the fraction of reads that can be assigned to loops. hichipper outperforms existing non-HiChIP-specific tools in terms of identifying long-range (>100-kb) loops with typical anchors resolved at a 2.5-kb resolution (see Supplementary Methods).
Additionally, hichipper produces a QC report that allows evaluation of library preparation, including factors such as the efficiency of proximity ligation and chromatin immunoprecipitation. We provide example QC reports from both successful and unsuccessful HiChIP experiments to guide new users in assessing their own library quality.
The hichipper package, documentation, and QC report examples are available online at http://aryee.mgh.harvard.edu/hichipper. Details of the background model and implementation are available in the Supplementary Methods.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NSF Graduate Research Fellowship #DGE1144152 (C.A.L.) and an MGH Pathology Startup Fund (M.J.A.).
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Life Sciences Reporting Summary.
Further information on experimental design is available in the Life Sciences Reporting Summary.
References
- 1.Mumbach MR et al. Nat. Methods 13, 919–922 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Servant N et al. Genome Biol. 16, 259 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


