Abstract
KRAS G12C mutation (mKRAS G12C) is the most frequent KRAS point mutation in non-small cell lung cancer (NSCLC) and has been proven to be a predictive biomarker for direct KRAS G12C inhibitors in advanced solid cancers. We sought to determine the prognostic significance of mKRAS G12C in patients with NSCLC using the meta-analytic approach. A protocol is registered at the International Prospective Register for systematic reviews (CRD42022345868). PubMed, EMBASE, The Cochrane Library, and Clinicaltrials.gov.in were searched for prospective or retrospective studies reporting survival data for tumors with mKRAS G12C compared with either other KRAS mutations or wild-type KRAS (KRAS-WT). The hazard ratios (HRs) for overall survival (OS) or Disease-free survival (DFS) of tumors were pooled according to fixed or random-effects models. Sixteen studies enrolling 10,153 participants were included in the final analysis. mKRAS G12C tumors had poor OS [HR, 1.42; 95% CI, 1.10–1.84, p = 0.007] but similar DFS [HR 2.36, 95% CI 0.64–8.16] compared to KRAS-WT tumors. Compared to other KRAS mutations, mKRAS G12C tumors had poor DFS [HR, 1.49; 95% CI, 1.07–2.09, p < 0.0001] but similar OS [HR, 1.03; 95% CI, 0.84–1.26]. Compared to other KRAS mutations, high PD-L1 expression (>50%) [OR 1.37 95% CI 1.11–1.70, p = 0.004] was associated with mKRAS G12C tumors. mKRAS G12C is a promising prognostic factor for patients with NSCLC, negatively impacting survival. Prevailing significant heterogeneity and selection bias might reduce the validity of these findings. Concomitant high PD-L1 expression in these tumors opens doors for exciting therapeutic potential.
Keywords: non-small cell lung cancer, NSCLC, KRAS, KRAS G12C, systematic review, meta-analysis
1. Introduction
Approximately 236,000 new lung cancer cases are expected to be diagnosed in the United States in 2022, contributing to 12.3% of all new cancer cases and 21.4% of all cancer-related deaths [1]. Of these, 85% are non-small cell lung Cancer (NSCLC), and 75% of NSCLC cases present at either the advanced or relapsed stage [2]. Kirsten rat sarcoma viral oncogene homolog (KRAS) is one of the most common oncogenic drivers in NSCLC, seen in over 30% of lung adenocarcinomas (LUAD), depending on ethnicity and tumor stage and associated with smoking and female patients [3,4,5]. Notwithstanding this, KRAS testing is not included in the routine genomic panel for NSCLC, probably due to its less well-established efficacy in daily clinical practice based on current evidence [The European Society for Medical Oncology (ESMO) Scale for Clinical Actionability of Molecular Targets (ESCAT) recommendation category 2B] [6,7]. Despite exhaustive exploration, the prognostic role of KRAS status remains contentious [8,9,10,11].
KRAS status harbors a spectrum of distinctly mutated KRAS substitutions [12]. In NSCLC, codon 12 is the most common site for KRAS point mutations, and glycine-to-cysteine (mKRAS G12C) (40–50% of all KRAS mutations) is present in 10–13% of LUAD patients [13,14]. Other common point mutations are glycine-to-aspartic acid (mKRAS G12D) and glycine-to-valine (mKRAS G12V) substitutions, which are observed in approximately 5% and 4% of LUAD patients, respectively [12]. Epidemiologically, mKRAS G12C and mKRAS G12V are associated with a history of smoking, whereas mKRAS G12D is associated with non-smokers [15,16]. These mutations can uniquely alter downstream effector molecules, leading to mutation-specific signal transduction, which can eventually modify clinical outcomes and treatment responses [17,18]. Molecular dynamics studies have shown that KRAS mutant proteins differ from wild-type proteins and have mutation subtype-specific differences [19].
Recently, leveraging the peculiar structural and biochemical properties of mKRAS G12C, various selective KRAS G12C small-molecule inhibitors have been developed [20]. The clinical efficacy of these newer agents is being explored in multiple clinical trials for advanced solid tumors [21,22,23]. Direct KRAS G12C inhibitors, sotorasib and adagrasib, received accelerated approval from the U.S. Food and Drug Administration (FDA) for previously treated advanced NSCLC patients with mKRAS G12C [24,25]. Similarly, based on the results of the CodeBreak200 trial (n = 345), ESMO guidelines recommend sotorasib as second-line therapy for advanced NSCLC patients with mKRAS G12C [26,27]. Based on its relatively ubiquitous epidemiology and distinctive biological behavior, this study aimed to quantitatively synthesize evidence on the prognostic role of mKRAS G12C in patients with NSCLC. Specifically, the primary aim of this study was to evaluate whether the survival rate of NSCLC patients with mKRAS G12C is similar to that of patients with tumors with either other KRAS mutations or KRAS-wild type (KRAS-WT). The secondary aim was to determine the association between frequent KRAS-comutations and mKRASG12C tumors. In addition, the correlation between PD-L1 expression levels and the KRAS subtype was investigated.
2. Materials and Methods
2.1. Search Strategy and Study Selection
Preferred Reporting Items for Systematic Review and Meta-analysis (PRISMA) guidelines were followed to conduct this study [28] (Table S1). The protocol was registered in the International Prospective Register of Systematic Reviews (PROSPERO identifier: CRD42022345868). A systematic and comprehensive search was conducted by two reviewers (DW and PH) of PubMed, Embase, Cochrane Library, and ClinicalTrials.gov from inception until April 2023. The following combinations of keywords and Medical Subject Headings (MeSH)/EMTREE terms were used: ‘KRAS OR Kirsten rat sarcoma viral homolog’, ‘mutation* OR mutated’, and ‘lung* OR pulmonary’, ‘cancer* OR tumor* or tumour* OR carcinom* OR neoplas* or malignan*’ and ‘prognos* or survival or recurren* or mortality or predict* or outcome* or death’. The search was restricted to articles published in English.
Inclusion criteria for eligible studies were defined as follows:
-
(a)
Studies investigating adult patients (>18 years) with pathologically confirmed NSCLC;
-
(b)
Studies assessing the mutation status of the KRAS gene in primary lung cancer tissue;
-
(c)
Studies reporting time-to-event data, including overall survival (OS), and disease-free survival (DFS) for the individual groups of interest, i.e., KRAS G12C, other point mutations of KRAS gene and KRAS-WT;
-
(d)
Original studies, including randomized-controlled (RCT) and non-randomized-controlled (NRCTs) studies, enrolling more than five individuals in the KRAS G12C group;
-
(e)
Studies published in English in a peer-reviewed journal.
The exclusion criteria for the study selection were as follows:
-
(a)
Studies investigating small cell lung cancers;
-
(b)
KRAS mutation analysis performed in plasma;
-
(c)
Studies reporting mixed data for KRAS G12C and other G12 substitutions;
-
(d)
Conference abstracts and preprint articles;
-
(e)
Narrative and systematic reviews, meta-analyses, expert opinions, editorial letters, case reports, and case series with a sample size of fewer than five individuals.
Studies exported from the databases were deduplicated, and unique studies were screened based on title, keyword, and abstract information. Subsequently, eligible studies were screened for full-text information, and data extracted from the studies met the eligibility criteria.
2.2. Data Extraction and Quality Assessment
Two reviewers independently retrieved the following data: authors, patient source, study design, histology, stage, mutation analysis methods, the total number of patients and patients with KRAS G12C mutation, and median follow-up duration in patients with KRAS WT, mKRAS G12C, other KRAS mutations. In addition, statistics extraction for time-to-event analyses (OS and DFS) were Hazard Ratio (HR) and 95% Confidence Interval (CI), median survival, and p-values. The OS and DFS were defined per the definition in each primary study. In case of a lack of reported survival outcomes, data were reconstructed from Kaplan–Meier curves using Tierney’s or Parmar’s methods [29,30]. We also assessed whether the survival outcomes were adjusted for clinicopathological covariates through univariate or multivariate analysis. We would utilize the latter if the author reported univariate and multivariate survival analysis results. Items were listed as “not available” (NA) when data from any of the categories mentioned above was unavailable”.
The publication that provided the most recent or informative data for studies with multiple publications was selected. Discrepancies between reviewers were resolved by consensus or involving a third investigator. Survival outcomes were compared between KRAS G12C vs. other KRAS mutations or KRAS G12C vs. KRAS-WT. In studies where survival data for individual point mutations were provided, we compared KRAS G12C with KRAS G12D mutation. The risk of bias assessment was done using the Newcastle–Ottawa Scale (NOS) for observational studies [31].
2.3. Statistical Methods
The pooled HR evaluated the prognostic role of KRAS G12C for OS and DFS using the generic inverse variance method. The statistical heterogeneity within studies was tested with the Cochrane Q test and measured using I2 indices. If HRs were found to have mild (I2 ≤ 30%) to moderate (I2 = 30–60%) heterogeneity, a fixed-effects model was used. In case of significant heterogeneity (I2 > 60%), a random-effects model was used. By convention, an observed HR > 1 implies worse survival for the group with KRAS G12C mutations. The impact of KRAS status on survival was considered statistically significant if the 95% CI did not overlap with 1. Statistical significance was set at p < 0.05, and all tests were two-sided. Sensitivity analyses were conducted using the leave-one-out method to determine the undue influence of an individual study on the summary estimate or heterogeneity.
We conducted subgroup analyses for outcomes with ≥5 studies and >1 study in each subgroup [32]. The studies were stratified according to ethnicity (Asian/non-Asian), testing methodology (next-generation sequencing (NGS)/polymerase-chain-reaction (PCR)), and adjustment for clinical covariates (HR derived from multivariate (MV)/univariate or recreated from survival curves (UV)). In order to decrease the likelihood of chance differences arising from multiple testing in the subgroup analyses, we used 99% CI for the study estimates and 95% CI for the summary estimate.
Pooled Odds Ratio (OR) with 95% CI for binary variables and standardized mean difference for continuous variables were generated to investigate the relationship of mutant KRAS G12C tumors with PD-L1 expression status (<1%, 1–50%, >50%) and co-occurring mutations (TP53, STK11).
Publication bias was assessed by Begg’s funnel plots and Egger’s test [33]. In case of publication bias, the trim-and-fill method was used to determine an adjusted pooled estimate [34]. Primary analyses were performed using the Review Manager version 5.4 (The Cochrane Collaboration, Copenhagen, Denmark), and publication bias evaluation was performed in the JASP software (JASP 0.16, the JASP team) [35].
3. Results
3.1. Study Selection and Characteristics
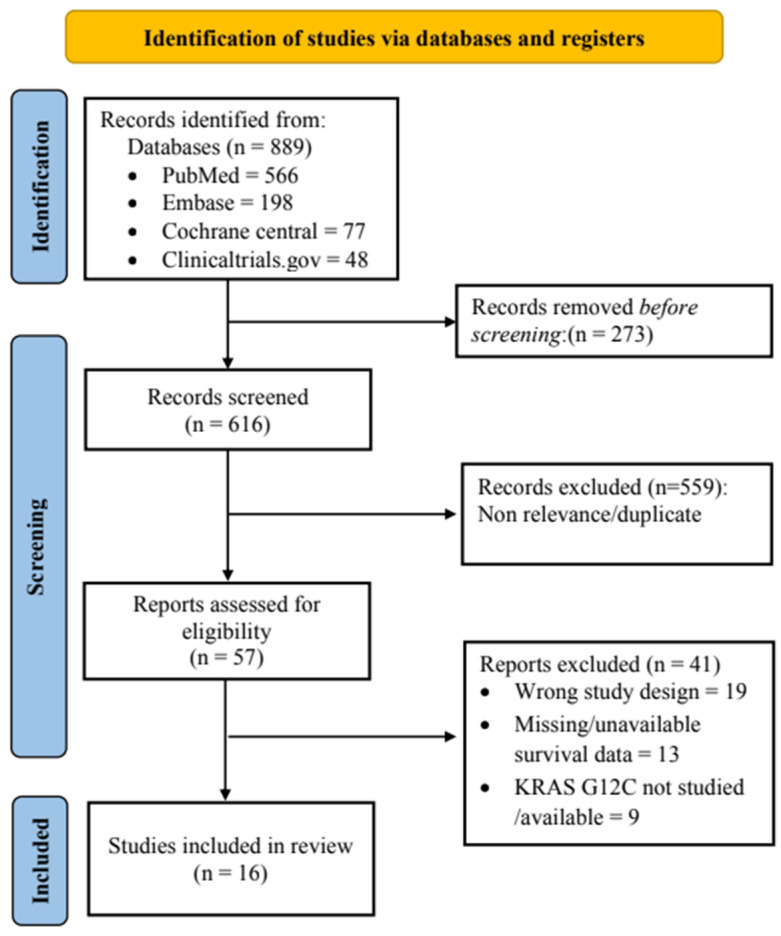
Eight hundred ninety-nine relevant studies were identified from electronic databases, and forty studies were selected for the full-text assessment. A manual search of the references did not yield any relevant study. Finally, sixteen studies were eligible for the systematic review, which were published between 2013 and 2021 [36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51]. The study selection process is presented in Figure 1.
Figure 1.
PRISMA flow diagram for the study selection process.
The study characteristics are reported in Table 1. The total number of patients was 10,153 (84 to 2055), with the median age ranging from 61 to 69. Female patients represented 36.5% of the pooled study population. Three studies were conducted in Asia, representing 8.7% of the pooled study population [38,44,48]. Eight studies reported data specifically on advanced stages (IIIb-IV), representing approximately 49.7% of the study population [36,38,39,41,44,45,50,51]. Seven studies exclusively included non-squamous NSCLC [40,41,42,43,47,49,50]. Next Generation Sequencing (NGS) was the predominant method used for KRAS mutation analysis. The median follow-up duration ranged from 9 to 95 months. In six studies, survival data were retrieved from survival curves to rebuild the HR estimates and their variance [36,38,39,40,42,45]. Ten studies reported HRs derived from multivariate analyses, and most of these studies adjusted for covariates such as age, sex, smoking status, stage, and various treatment modalities.
Table 1.
Study characteristics.
| Study | Source of Patients | No. of Patients (n) | Histology | Stage | Testing Method | KRAS G12C Mutation Frequency (%) |
KRAS-Comutations (%) | Median Follow-Up Time (Months, Range) | Survival Parameters | NOS Score | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TP53 | STK-11 | KEAP-1 | Outcome | Adjusted Variables | |||||||||
| Arbour, 2021 [36] | USA | 772 | NSCLC | IIIB-IV | NGS-MSK IMPACT assay | 46 | 41.8 | 28.4 | 23.4 | 13.8 | OS | NA | 8 |
| Aredo, 2019 [37] | USA | 186 | NSCLC | I-IV | NGS- STAMP assay |
35 | 38.7 | 11.8 | 8.1 | 15 | OS | Age, sex, smoking, stage, co-mutations, treatment modalities (localized and systemic) | 9 |
| Cai, 2019 [44] | China | 84 | NSCLC | IV | PCR-seq | 28 | 29 | NA | 5 | NA | OS | Age, sex, ECOG PS, smoking pack years, histology, KRAS comutations | 6 |
| Cui, 2020 [45] | Australia | 346 | NSCLC | IV | NGS-PMCC lung panel mutation analysis | 19 | NA | NA | NA | 9 | OS | NA | 5 |
| Finn, 2021 [46] | Multicentric (Europe) | 2055 | NSCLC | I-III | PCR-allele-specific multiplex test | 10.5 | NA | NA | NA | 57.1 (44.3–72.3) | OS, DFS | Sex, ethnicity, smoking, age, Adj CT, Adj R.T., h/o cancer, ECOG PS, stage, primary tumor localization, tumor size, histology, surgery year, technique, and anatomy | 9 |
| Jones, 2021 [47] | USA | 604 | ADC | I-III | NGS- MSK-IMPACT | 16 | 25 | 22 | 7 | 30 (IQR 21–40) | DFS | Tumor SUVmax, DLCO, LVI, VPI, STAS, stage | 8 |
| Liu, 2020 [48] | China | 434 | NSCLC | I-IV | NGS | 9.6 | NA | NA | NA | NA | OS | Age, sex, Smoking, histology, stage, | 6 |
| Nadal, 2014 [49] | USA | 179 | ADC | I-IV | PCR-seq | 19.5 | 85 | 30.5 | NA | 95 | OS, DFS | Age, sex, stage, smoking, adjuvant therapy, tumor differentiation, | 9 |
| Osta, 2019 [50] | USA | 1655 | ADC | IV | NGS | 10.6 | 52 | 18 | NA | 25.8 | OS | Age, sex, race, smoking, ECOG PS, prior surgical/R.T. treatment, systemic therapy, KRAS comutations | 9 |
| Sebastian, 2021 [51] | Germany | 1039 | NSCLC | IIIB-IV | NGS (75%) | 15.4 | NA | NA | NA | NA | OS | Age, sex, BMI, Charlton score, ECOG PS, PD-L1 expression | 6 |
| Sun, 2013 [38] | South Korea |
304 | NSCLC | IIIB-IV | PCR-seq | 3 | NA | NA | NA | 30 | OS | NA | 6 |
| Svaton, 2016 [39] | Czechia | 129 | NSCLC | IIIB-IV | PCR-seq | 11.6 | NA | NA | NA | NA | OS | NA | 5 |
| Tao, 2020 [42] | USA | 254 | ADC | I-IV | Pyrosequencing-based test (PyroMark Q24 System; Qiagen) | 46.1 | NA | NA | NA | 17.2 (0.17–74.9) | OS, DFS | NA | 6 |
| Villacruz, 2013 [40] | USA | 318 | ADC | I-IV | PCR-seq | 43.7 | NA | NA | NA | 24.3 (0–78) | OS, DFS | NA | 6 |
| Wahl, 2021 [43] | Norway | 1117 | ADC/ADSCC | I-IV | NGS | 17 | NA | NA | NA | 52.7 (45.7–59.6) | OS | Age, sex, smoking, ECOG PS, Stage, Surgery, Curative/palliative CT/RT, TKI | 9 |
| Yu, 2015 [41] | USA | 677 | ADC | IV | Standard Sanger sequencing/ | 39 | NA | NA | NA | 17 (1–207) | OS | NA | 6 |
HR: Hazard ratio, CI: Confidence Interval NGS: Next generation sequencing, NOS: Newcastle–Ottawa Scale, PCR-seq: Polymerase chain reaction-direct nucleotide sequencing, OS: Overall survival, DFS: Disease-free survival, P.S.: performance status, Adj: adjuvant, DLCO: diffusing capacity for carbon monoxide, LVI: lymphovascular invasion, VPI: visceral pleural invasion, STAS: spread through air spaces, BMI: Body mass index, ADC: adenocarcinoma, ADSCC: adenosquamous, CT: chemotherapy, R.T.: radiotherapy, TKI: tyrosine kinase inhibitor, NA: not available, STAMP: Solid Tumor Actionable Mutational Panel.
In the overall NSCLC cohort, the rate of KRAS G12C mutation was 11.7%, whereas it was 40.9% in the mutant KRAS (mKRAS) population. Among the studies that reported KRAS-comutations, the frequency of TP53, STK11, and KEAP1 mutation ranged from 25–85%, 8–30%, and 5–23%, respectively. The information on PD-L1 expression status was available for 28–100% of tumor samples from five studies. In the mKRAS cohort, the frequency of tumors with negative (<1%), positive (>1%), and high PD-L1 expression (>50%) was 35% (7.6–49%), 41.2% (39–76%), and 33% (27–38.3%), respectively. Similarly, in the mKRAS G12C cohort, PD-L1 expression was found to be negative in 34% (3–47%) of the study participants, positive in 63% (45–90.5%), and high in 40.6% (18.6–47.6%).
3.2. Quality of Studies
The Newcastle-Ottawa Scale indicated that nine studies had a low risk of bias, and the remainder had a moderate risk of bias (Table 1 and Table S2). The overall quality of studies was moderate to high, with an average score of 7 (range 5–9). A few studies failed to report the duration and adequacy of follow-up. In a few studies, comparability between the two groups could not be ascertained due to the lack of adjustment of confounding variables that were likely to affect the survival outcomes.
3.3. Prognostic Role of KRAS G12C
3.3.1. KRAS G12C versus Other KRAS Mutations
Fourteen studies with 4352 patients were included in the primary meta-analysis for OS [36,37,38,39,40,41,42,43,44,45,46,49,50,51]. Five studies with 3140 patients were included in the meta-analysis for DFS.
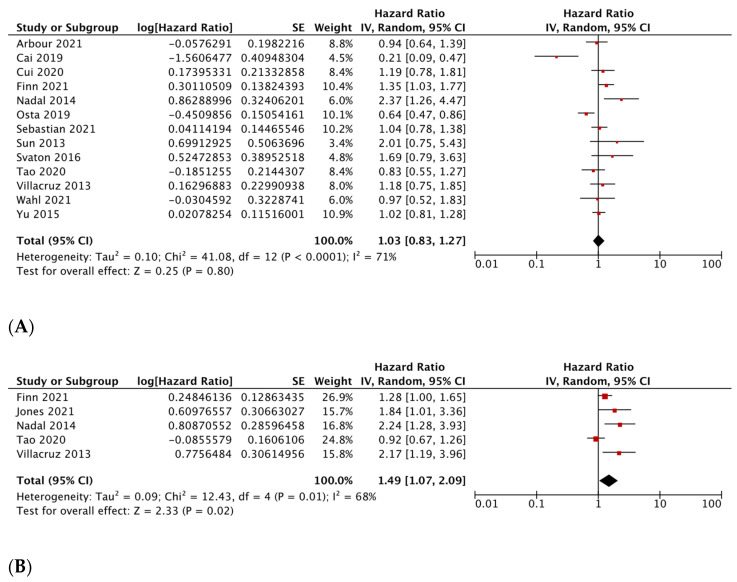
The summary HR for OS showed no statistically significant survival difference between patients with KRAS G12C and non-KRAS G12C mutations [HR 1.03, 95% CI: 0.84–1.26, p = 0.79], though between-study heterogeneity was high (I2 = 68%, p < 0.0001) (Figure 2A)
Figure 2.
Association between KRAS mutation status and survival in non-small cell lung cancer. (A) Overall Survival for KRAS G12C mutant tumors versus other KRAS mutant tumors. Association between KRAS mutation status and survival in non-small cell lung cancer [36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51] (B). Disease-free survival for KRAS G12C mutant tumors versus other KRAS mutant tumors [40,42,46,47,49].
Studies that compared KRAS G12C to KRAS G12D mutated tumors also showed similar outcomes [HR, 0.93, 95% CI, 0.67–12.9; p = 0.66] (Figure S1). Sensitivity analysis did not identify any undue influence of individual studies on effect size or heterogeneity. An asymmetrical right-skewed funnel plot (Figure S2) and significant Egger’s test (p = 0.013) suggested the presence of publication bias. The trim-and-fill method led to the addition of three studies; however, the combined HR after adjustment remained statistically non-significant [adjusted HR (aHR) = 0.92 95% CI 0.73–1.17] (Figure S3).
Studies were stratified according to the patients’ sources, testing methods, and survival data sources. The combined HR for Asians was 0.64 (95% CI 0.07–5.83, p = 0.69), and for non-Asians was 1.07 (95% CI 0.89–1.28, p = 0.47) (Figure S4). Among the studies with NGS-based testing methods, the combined HR was 0.73 (95% CI 0.49–1.07, p = 0.11), whereas, for PCR-based methods, it was 1.20 (95% 0.71–2.01, p = 0.50) (Figure S5). Finally, studies with HRs derived from multivariate analyses [pooled HR = 0.91 95% CI 0.59–1.42, p = 0.68] and HRs derived from univariate analyses/extracted from survival curves [pooled HR = 1.05 95% CI 0.91–1.22, p = 0.29] had non-significant results (Figure S6).
We conducted analyses on studies that included non-squamous NSCLC exclusively and found similar OS in patients with mKRAS G12C and other KRAS mutations [HR = 1.06 95% CI 0.81–1.40, p = 0.66] (Figure S7). Furthermore, an analysis of studies with advanced-stage NSCLC found a comparable outcome between the two arms [HR = 0.92 95% CI 0.69–1.22, p = 0.57] (Figure S8).
The summary HR for DFS showed that patients with KRAS G12C mutations had a higher risk of relapse compared to patients with other KRAS mutations [HR 1.49 95% CI 1.07–2.09, p < 0.0001], and significant heterogeneity was observed [I2 = 68%, p = 0.02] (Figure 2B). However, studies that compared KRAS G12C to KRAS G12D mutated tumors found a non-significant outcome [HR, 1.36, 95% CI, 0.59–3.15; p = 0.48] (Figure S9). Leave-one-out analysis revealed that none of the studies contributed to heterogeneity; however, the exclusion of any of the following studies led to a non-significant outcome: Finn et al., Jones et al., Nadal et al., and Villaruz et al. [40,46,47,49].
3.3.2. KRAS G12C versus Wild-Type KRAS
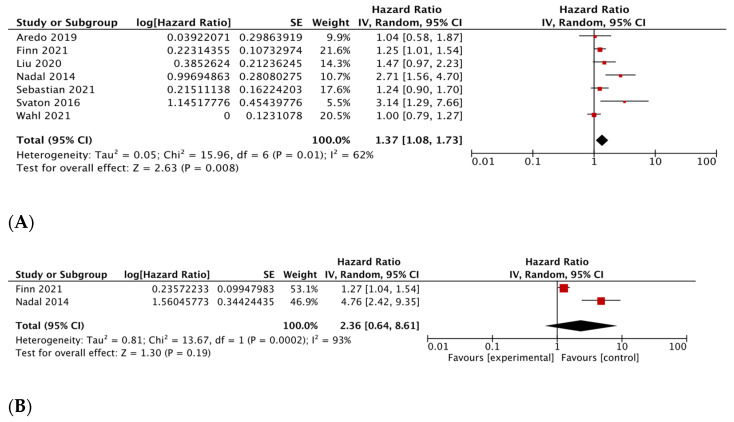
Six studies with 4953 patients were included in the meta-analysis for OS [39,43,46,48,49,51]. Two studies with 2234 patients were included in the meta-analysis for DFS [46,49]. The summary HR for OS comparing KRAS G12C to KRAS-WT showed results favoring KRAS-WT [HR 1.42, 95% CI 1.10–1.84, p = 0.007], though significant heterogeneity was present [I2 = 68%, p = 0.008] (Figure 3A). Sensitivity analysis revealed consistent results. The meta-analysis for DFS comparing KRAS G12C with KRAS-WT showed similar survival outcomes between the two arms [HR 2.36, 95% CI 0.64–8.16, p = 0.19]; however, between-study heterogeneity was high [I2 = 93%, p = 0.0002] (Figure 3B).
Figure 3.
Association between KRAS mutation status and survival in non-small cell lung cancer, (A). Overall Survival for KRAS G12C mutant tumors versus wild-type KRAS tumors [37,39,43,46,48,49,51]; (B). Disease-free survival for KRAS G12C mutant tumors versus wild-type KRAS tumors [46,49].
3.4. Secondary Outcomes
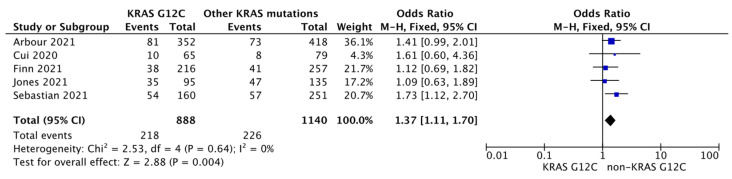
We found that the proportion of patients with tumors expressing PD-L1 > 50% [OR 1.37 95% CI 1.11–1.70, p = 0.004] (Figure 4) were higher in the mKRAS G12C tumors compared to other KRAS mutations; however, tumors with neither negative (<1%) [OR 0.85 95% CI 0.70–1.04, p = 0.12] (Figure S10) nor with moderate PD-L1 expression (1–49%) [OR 0.94 95% CI 0.75–1.18, p = 0.59] (Figure S11) were associated with mKRAS G12C. Frequently encountered comutations did not show associations with either mKRAS G12C or other KRAS mutations [TP53: OR 0.96 95% CI 0.59–1.56, p = 0.88 (Figure S12); STK11: OR 1.01 95% CI 0.77–1.32, p = 0.93 (Figure S13)].
Figure 4.
Association between KRAS status and PD-L1 in non-small cell lung cancer [36,45,46,47,51].
4. Discussion
Small molecule inhibitors of KRAS G12C mutant proteins finally broke the curse of the “undruggable” status of KRAS mutation based on the promising results of CodeBreak 100 and KRYSTAL-1 trials [25,52,53]. There is renewed interest in evaluating long-term oncological outcomes in patients with mKRAS G12C tumors [54]. Our systematic review and meta-analysis of sixteen retrospective studies comprising 10,153 patients showed that the mKRAS-G12C predicts poor survival in patients with NSCLC. The key findings of our study were as follows: First, compared to patients with other KRAS mutations, mKRAS-G12C predicted poor DFS. Second, mKRAS-G12C tumors were at a higher risk of all-cause mortality than KRAS-WT tumors despite similar DFS. Third, mKRAS G12C tumors were associated with high PD-L1 expression (>50%) compared with other KRAS mutations. Finally, TP53 and STK11 were not associated with either mKRAS G12C or other KRAS mutant tumors. Overall, KRAS G12C mutated tumors harbor poor prognosis; however, considerable between-study heterogeneity existed in most of these analyses.
In prior meta-analyses, the overall link between mKRAS and survival in NSCLC patients is relatively weak and primarily restricted to advanced disease [10,11]. Different mutation testing methodologies, variable patient selection criteria, and lack of stratification across stages further muddled the conclusions [55]. Furthermore, the downstream signaling of KRAS mutation subtypes uniquely alters tumor biology and thus may influence distinct clinical behavior that may not be apparent in examining KRAS mutations in toto [17,56]. Our analyses built on these lacunae by reporting the survival outcomes for the most common KRAS mutation subtype in NSCLC and showed consistent results across ethnicities and testing methodologies.
Surgically resected NSCLC are at a higher risk of disease relapse if associated with the mKRAS G12C than other KRAS mutations [47,49]. In contrast, tumors with mKRAS G12C and other KRAS mutations showed comparable OS rates in our study. In a study on early-stage NSCLC (n = 179), mKRAS G12C was associated with poor DFS (p = 0.006) compared with KRAS-WT; however, the statistical strength of this analysis dropped significantly for stage I patients, suggesting the influence of pathological stage on DFS [49]. Notwithstanding this, KRAS G12C inhibitors showed clinical efficacy in eradicating micrometastases and enhanced anti-tumor activity when combined with targeted agents [23]. Based on these results and our findings, adjuvant or neoadjuvant KRAS G12C inhibitor monotherapy or in combination with targeted agents in patients with early-stage mKRAS G12C tumors may open the door for exciting therapeutic paradigms.
KRAS-comutations and PD-L1 expression may also modify the behavior of mKRAS tumors [57,58]. Approximately 50% of mKRAS tumors exhibit co-mutations, of which TP53 (35–50%), STK11 (12–20%), and KEAP1 (7–10%) constitute the majority [59,60]. mKRAS G12C tumors show similar concurrent genomic alterations, although they have also been associated with ERBB2 amplification and ERBB4 mutations [59]. PTEN and PDGFRA mutations are less commonly observed in mKRAS G12C tumors [59,61,62]. KRAS-comutations have been considered an adverse prognostic factor capable of predicting tumor progression and chemo-resistance [63,64,65,66]. Due to a lack of available data, we could not comment on the prognostic role of KRAS-comutations. However, data from clinical trials indicated that patients harboring STK11 and KRAS co-mutations had a higher objective response rate (ORR) (63% vs. 33%), whereas KEAP1 and KRAS co-mutation were associated with a lower ORR (20% vs. 44%) following KRAS G12C inhibitor therapy [52,53]. These genes need further investigation to evaluate their impact on prognosis and therapeutic response in patients with mKRAS. In this context, KRAS G12C inhibitors are being evaluated as a first-line therapeutic option in patients with NSCLC harboring STK11 and KRAS G12C co-mutations (KRYSTAL-1 and CodeBreaK 201).
The degree of PD-L1 expression in tumor cells corresponds to the prognosis in patients with mKRAS tumors, and tumors with high PD-L1 expression were associated with a dismal prognosis [42,67,68]. Our study reported that high PD-L1 expression in tumor cells (>50%) was associated with mKRAS G12C tumors. In preclinical data, KRAS G12C direct inhibitors have been shown to upregulate a pro-inflammatory tumor microenvironment and increase anti-tumor T-cell activity [23]. Furthermore, KRAS G12C inhibitors have been shown to increase intratumoral chemokine concentrations, potentially increasing T-cell infiltration and enhancing immunosurveillance [23]. In CT-26 cell line models, KRAS G12C and anti-PD-1 combination therapy led to a synergistic response compared with either monotherapy [23]. In the CodeBreak 100 trial, 89.8% (53/59) of previously treated NSCLC patients with mKRAS G12C received anti-PD-1/PD-L1 therapy along with sotorasib and demonstrated an ORR of 32.2%, and a median PFS of 6.3 months [52]. Likewise, KRYSTAL-01 (NCT03785249), a phase1/2 multicohort study evaluating adagrasib (monotherapy or platinum-based chemotherapy/anti-PD-1/L1 combination), showed promising efficacy (42.9% ORR and median PFS of 6.5 months) in previously treated NSCLC harboring mKRAS G12C [53]. KRYSTAL-07 (NCT04613596) and CodeBreak 101 (NCT04185883) clinical trials are currently exploring anti-PD-1/KRAS G12C inhibitor combinations for advanced solid tumors.
We found that the frequency of mKRAS-G12C in the NSCLC and mKRAS cohorts reflected the real-world data that were previously reported, suggesting a representative population and thus more likely to reflect prognosis in clinical practice [69,70]. Acknowledging these findings cautiously is warranted. Most analyses involved high heterogeneity, which could not be attributed to an individual study and was presumably due to vast differences in patient characteristics. Publication bias was observed despite following expanded search criteria to include studies with either non-significant or negative results. The trim-and-fill method performs poorly in the presence of substantial between-study heterogeneity [71]. Approximately 49.7% of total patients had an advanced stage. Effect estimates from seven studies were unadjusted, which may influence the summary estimates, particularly by stage and systemic therapy. Lastly, this meta-analysis relied on the summary estimates from observational studies rather than individual patient data, which is considered a statistically superior method.
Trials like ADAURA and PACIFIC have provided sufficient evidence for the role of targeted agents in early-stage NSCLC. In addition, ANVIL (NCT02595944), PEARLS/Keynote-091 (NCT02504372), and IMpower010 (NCT02486718) trials are currently exploring the role of immune checkpoint inhibitors in the adjuvant setting [72,73]. However, the current literature lacks data to determine the prognostic and predictive role of KRAS G12C mutation in early-stage NSCLC; thus, further investigations are needed.
Our findings contribute to the evolving landscape of KRAS mutations in patients with NSCLC. Extrapolating these findings, the following recommendations may be made for clinical practice: 1) Inclusion of KRAS status, especially KRAS G12C mutation, as a routine test using a comprehensive molecular gene panel. Similarly, future studies should focus on 1) Evaluation of KRAS G12C inhibitors-anti-PD-1/PD-L1 inhibitor combination therapy in advanced NSCLC; 2) evaluation of adjuvant KRAS G12C inhibitor in surgically resected NSCLC with mKRAS G12C; and 3) assessment of the impact of KRAS G12C-comutations including TP53, STK11, and KEAP1, on prognosis.
5. Conclusions
Our meta-analysis on NSCLC found that tumors with mKRAS G12C were associated with worse DFS than tumors with other KRAS mutations and worse OS than tumors with KRAS-WT. The presence of significant heterogeneity and publication bias collectively undermines the validity of these findings. However, these outcomes reflect real-world prognoses and may be utilized in clinical practice to stratify high-risk patients and provide more effective therapeutic strategies for patients with NSCLC.
Acknowledgments
The authors sincerely appreciate the statistical advice of Nigar Sofiyeva.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/diagnostics13193043/s1, Figure S1. Additional analysis: Overall Survival, KRAS G12C vs KRAS G12D mutated tumors, Figure S2. Funnel plot: Overall Survival KRAS G12C vs other KRAS mutations, Figure S3 Trim-and-fill analysis Overall Survival KRAS G12C vs other KRAS mutations, Figure S4. Subgroup analysis: ethnicity, Overall Survival KRAS G12C vs other KRAS mutations, Figure S5. Subgroup analysis: Testing methods, Overall Survival KRAS G12C vs other KRAS mutations, Figure S6. Subgroup analysis: data source: Overall Survival KRAS G12C vs other KRAS mutations, Figure S7. Additional analysis: Histology: Overall Survival KRAS G12C vs other KRAS mutations, Figure S8. Additional analysis: Advanced Stage, Overall Survival KRAS G12C vs other KRAS mutations, Figure S9 Additional analysis: Disease-free Survival, KRAS G12C vs KRAS G12D mutated tumors, Figure S10 Association of PD-L1 expression <1% with KRAS G12C, Figure S11. Association of PD-L1 expression 1-49% with KRAS G12C, Figure S12. Association of TP53 mutation with KRAS G12C, Figure S13. Association of STK11 mutation with KRAS G12C; Table S1: PRISMA checklist, Table S2: Newcastle-Ottawa Scale for stdy quality assessment.
Author Contributions
Conceptualization, D.W.; methodology, D.W. and P.H.; formal analysis, D.W., C.B., S.G.; data curation, D.W. and P.H.; writing—original draft preparation, D.W.; writing—review and editing, C.B., S.G. and P.H.; supervision, P.H.; funding acquisition, P.H. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available in the Open Science Framework repository at 10.17605/OSF.IO/AUF2T without third-party permission.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Funding Statement
The authors would like to thank the FHU OncoAge, “Ligue Départementale 06 de Lutte contre le Cancer”, and “Conseii Départemental des Alpes Maritimes” for supporting the fee related to the publication of this manuscript.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.National Cancer Institute SEER. Cancer Stat Facts: Lung and Bronchus Cancer. [(accessed on 11 April 2023)]; Available online: https://seer.cancer.gov/statfacts/html/lungb.html.
- 2.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 3.Jordan E.J., Kim H.R., Arcila M.E., Barron D., Chakravarty D., Gao J., Chang M.T., Ni A., Kundra R., Jonsson P. Prospective Comprehensive Molecular Characterization of Lung Adenocarcinomas for Efficient Patient Matching to Approved and Emerging TherapiesMolecular Profiling to Guide Therapy in Lung Adenocarcinoma. Cancer Discov. 2017;7:596–609. doi: 10.1158/2159-8290.CD-16-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prior I.A., Lewis P.D., Mattos C. A Comprehensive Survey of Ras Mutations in Cancer. Cancer Res. 2012;72:2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shepherd F.A., Domerg C., Hainaut P., Jänne P.A., Pignon J.-P., Graziano S., Douillard J.-Y., Brambilla E., Le Chevalier T., Seymour L. Pooled Analysis of the Prognostic and Predictive Effects of KRAS Mutation Status and KRAS Mutation Subtype in Early-Stage Resected Non–Small-Cell Lung Cancer in Four Trials of Adjuvant Chemotherapy. J. Clin. Oncol. 2013;31:2173. doi: 10.1200/JCO.2012.48.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uprety D., Adjei A.A. KRAS: From Undruggable to a Druggable Cancer Target. Cancer Treat. Rev. 2020;89:102070. doi: 10.1016/j.ctrv.2020.102070. [DOI] [PubMed] [Google Scholar]
- 7.Mosele F., Remon J., Mateo J., Westphalen C.B., Barlesi F., Lolkema M.P., Normanno N., Scarpa A., Robson M., Meric-Bernstam F., et al. Recommendations for the Use of Next-Generation Sequencing (NGS) for Patients with Metastatic Cancers: A Report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2020;31:1491–1505. doi: 10.1016/j.annonc.2020.07.014. [DOI] [PubMed] [Google Scholar]
- 8.Mascaux C., Iannino N., Martin B., Paesmans M., Berghmans T., Dusart M., Haller A., Lothaire P., Meert A.-P., Noel S. The Role of RAS Oncogene in Survival of Patients with Lung Cancer: A Systematic Review of the Literature with Meta-Analysis. Br. J. Cancer. 2005;92:131–139. doi: 10.1038/sj.bjc.6602258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Z., Wang T., Zhang J., Cai X., Pan C., Long Y., Chen J., Zhou C., Yin X. Prognostic Value of Epidermal Growth Factor Receptor Mutations in Resected Non-Small Cell Lung Cancer: A Systematic Review with Meta-Analysis. PLoS ONE. 2014;9:e106053. doi: 10.1371/journal.pone.0106053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meng D., Yuan M., Li X., Chen L., Yang J., Zhao X., Ma W., Xin J. Prognostic Value of K-RAS Mutations in Patients with Non-Small Cell Lung Cancer: A Systematic Review with Meta-Analysis. Lung Cancer. 2013;81:1–10. doi: 10.1016/j.lungcan.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 11.Goulding R.E., Chenoweth M., Carter G.C., Boye M.E., Sheffield K.M., John W.J., Leusch M.S., Muehlenbein C.E., Li L., Jen M.-H. KRAS Mutation as a Prognostic Factor and Predictive Factor in Advanced/Metastatic Non-Small Cell Lung Cancer: A Systematic Literature Review and Meta-Analysis. Cancer Treat. Res. Commun. 2020;24:100200. doi: 10.1016/j.ctarc.2020.100200. [DOI] [PubMed] [Google Scholar]
- 12.Consortium A.P.G., Consortium A.P.G., André F., Arnedos M., Baras A.S., Baselga J., Bedard P.L., Berger M.F., Bierkens M., Calvo F. AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 2017;7:818–831. doi: 10.1158/2159-8290.CD-17-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiesweg M., Kasper S., Worm K., Herold T., Reis H., Sara L., Metzenmacher M., Abendroth A., Darwiche K., Aigner C., et al. Impact of RAS Mutation Subtype on Clinical Outcome—A Cross-Entity Comparison of Patients with Advanced Non-Small Cell Lung Cancer and Colorectal Cancer. Oncogene. 2019;38:2953–2966. doi: 10.1038/s41388-018-0634-0. [DOI] [PubMed] [Google Scholar]
- 14.Arbour K.C., Jordan E., Kim H.R., Dienstag J., Yu H.A., Sanchez-Vega F., Lito P., Berger M., Solit D.B., Hellmann M., et al. Effects of Co-Occurring Genomic Alterations on Outcomes in Patients with KRAS-Mutant Non–Small Cell Lung Cancer. Clin. Cancer Res. 2018;24:334–340. doi: 10.1158/1078-0432.CCR-17-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herdeis L., Gerlach D., McConnell D.B., Kessler D. Stopping the Beating Heart of Cancer: KRAS Reviewed. Curr. Opin. Struct. Biol. 2021;71:136–147. doi: 10.1016/j.sbi.2021.06.013. [DOI] [PubMed] [Google Scholar]
- 16.Tomasini P., Walia P., Labbe C., Jao K., Leighl N.B. Targeting the KRAS Pathway in Non-Small Cell Lung Cancer. Oncologist. 2016;21:1450–1460. doi: 10.1634/theoncologist.2015-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ihle N.T., Byers L.A., Kim E.S., Saintigny P., Lee J.J., Blumenschein G.R., Tsao A., Liu S., Larsen J.E., Wang J., et al. Effect of KRAS Oncogene Substitutions on Protein Behavior: Implications for Signaling and Clinical Outcome. JNCI J. Natl. Cancer Inst. 2012;104:228–239. doi: 10.1093/jnci/djr523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li S., Liu S., Deng J., Akbay E.A., Hai J., Ambrogio C., Zhang L., Zhou F., Jenkins R.W., Adeegbe D.O., et al. Assessing Therapeutic Efficacy of MEK Inhibition in a KRASG12C-Driven Mouse Model of Lung Cancer. Clin. Cancer Res. 2018;24:4854–4864. doi: 10.1158/1078-0432.CCR-17-3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pantsar T., Rissanen S., Dauch D., Laitinen T., Vattulainen I., Poso A. Assessment of Mutation Probabilities of KRAS G12 Missense Mutants and Their Long-Timescale Dynamics by Atomistic Molecular Simulations and Markov State Modeling. PLoS Comput. Biol. 2018;14:e1006458. doi: 10.1371/journal.pcbi.1006458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Molina-Arcas M., Moore C., Rana S., Van Maldegem F., Mugarza E., Romero-Clavijo P., Herbert E., Horswell S., Li L.-S., Janes M.R. Development of Combination Therapies to Maximize the Impact of KRAS-G12C Inhibitors in Lung Cancer. Sci. Transl. Med. 2019;11:eaaw7999. doi: 10.1126/scitranslmed.aaw7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ostrem J.M.L., Shokat K.M. Direct Small-Molecule Inhibitors of KRAS: From Structural Insights to Mechanism-Based Design. Nat. Rev. Drug Discov. 2016;15:771–785. doi: 10.1038/nrd.2016.139. [DOI] [PubMed] [Google Scholar]
- 22.Ostrem J.M., Peters U., Sos M.L., Wells J.A., Shokat K.M. K-Ras(G12C) Inhibitors Allosterically Control GTP Affinity and Effector Interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Canon J., Rex K., Saiki A.Y., Mohr C., Cooke K., Bagal D., Gaida K., Holt T., Knutson C.G., Koppada N., et al. The Clinical KRAS(G12C) Inhibitor AMG 510 Drives Anti-Tumour Immunity. Nature. 2019;575:217–223. doi: 10.1038/s41586-019-1694-1. [DOI] [PubMed] [Google Scholar]
- 24.Nakajima E.C., Drezner N., Li X., Mishra-Kalyani P.S., Liu Y., Zhao H., Bi Y., Liu J., Rahman A., Wearne E., et al. FDA Approval Summary: Sotorasib for KRAS G12C-Mutated Metastatic NSCLC. Clin. Cancer Res. 2022;28:1482–1486. doi: 10.1158/1078-0432.CCR-21-3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jänne P.A., Riely G.J., Gadgeel S.M., Heist R.S., Ou S.-H.I., Pacheco J.M., Johnson M.L., Sabari J.K., Leventakos K., Yau E., et al. Adagrasib in Non–Small-Cell Lung Cancer Harboring a KRASG12C Mutation. N. Engl. J. Med. 2022;387:120–131. doi: 10.1056/NEJMoa2204619. [DOI] [PubMed] [Google Scholar]
- 26.de Langen A.J., Johnson M.L., Mazieres J., Dingemans A.-M.C., Mountzios G., Pless M., Wolf J., Schuler M., Lena H., Skoulidis F., et al. Sotorasib versus Docetaxel for Previously Treated Non-Small-Cell Lung Cancer with <em>KRAS</Em>G12C Mutation: A Randomised, Open-Label, Phase 3 Trial. Lancet. 2023;401:733–746. doi: 10.1016/S0140-6736(23)00221-0. [DOI] [PubMed] [Google Scholar]
- 27.Hendriks L.E., Kerr K.M., Menis J., Mok T.S., Nestle U., Passaro A., Peters S., Planchard D., Smit E.F., Solomon B.J., et al. Oncogene-Addicted Metastatic Non-Small-Cell Lung Cancer: ESMO Clinical Practice Guideline for Diagnosis, Treatment and Follow-up☆. Ann. Oncol. 2023;34:339–357. doi: 10.1016/j.annonc.2022.12.009. [DOI] [PubMed] [Google Scholar]
- 28.Page M.J., McKenzie J.E., Bossuyt P.M., Boutron I., Hoffmann T.C., Mulrow C.D., Shamseer L., Tetzlaff J.M., Akl E.A., Brennan S.E., et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ. 2021;372:105906. doi: 10.1136/bmj.n71. [DOI] [PubMed] [Google Scholar]
- 29.Tierney J.F., Stewart L.A., Ghersi D., Burdett S., Sydes M.R. Practical Methods for Incorporating Summary Time-to-Event Data into Meta-Analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parmar M.K.B., Torri V., Stewart L. Extracting Summary Statistics to Perform Meta-Analyses of the Published Literature for Survival Endpoints. Stat. Med. 1998;17:2815–2834. doi: 10.1002/(SICI)1097-0258(19981230)17:24<2815::AID-SIM110>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 31.Wells G., Shea B., O’Connell D., Peterson J., Welch V.L.M. The Newcastle-Ottawa Scale (NOS) for Assessing the Quality of Nonrandomised Studies in Meta-Analyses. [(accessed on 2 May 2023)]. Available online: http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp.
- 32.Sedgwick P. Meta-Analyses: Heterogeneity and Subgroup Analysis. BMJ Br. Med. J. 2013;346:f4040. doi: 10.1136/bmj.f4040. [DOI] [Google Scholar]
- 33.Egger M., Smith G.D., Schneider M., Minder C. Bias in Meta-Analysis Detected by a Simple, Graphical Test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duval S., Tweedie R. Trim and Fill: A Simple Funnel-Plot-Based Method of Testing and Adjusting for Publication Bias in Meta-Analysis. Biometrics. 2000;56:455–463. doi: 10.1111/j.0006-341X.2000.00455.x. [DOI] [PubMed] [Google Scholar]
- 35.JASP Team JASP 2022. Published online. 2023. [(accessed on 31 July 2023)]. Available online: https://www.jasp-stats.org.
- 36.Arbour K.C., Rizvi H., Plodkowski A.J., Hellmann M.D., Knezevic A., Heller G., Yu H.A., Ladanyi M., Kris M.G., Arcila M.E. Treatment Outcomes and Clinical Characteristics of Patients with KRAS-G12C–Mutant Non–Small Cell Lung CancerOutcomes of Patients with KRAS-G12C–Mutant Lung Cancer. Clin. Cancer Res. 2021;27:2209–2215. doi: 10.1158/1078-0432.CCR-20-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aredo J.V., Padda S.K., Kunder C.A., Han S.S., Neal J.W., Shrager J.B., Wakelee H.A. Impact of KRAS Mutation Subtype and Concurrent Pathogenic Mutations on Non-Small Cell Lung Cancer Outcomes. Lung Cancer. 2019;133:144–150. doi: 10.1016/j.lungcan.2019.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun J.-M., Hwang D.W., Ahn J.S., Ahn M.-J., Park K. Prognostic and Predictive Value of KRAS Mutations in Advanced Non-Small Cell Lung Cancer. PLoS ONE. 2013;8:e64816. doi: 10.1371/journal.pone.0064816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Svaton M., Fiala O., Pesek M., Bortlicek Z., Minarik M., Benesova L., Topolcan O. The Prognostic Role of KRAS Mutation in Patients with Advanced NSCLC Treated with Second-or Third-Line Chemotherapy. Anticancer Res. 2016;36:1077–1082. [PubMed] [Google Scholar]
- 40.Villaruz L.C., Socinski M.A., Cunningham D.E., Chiosea S.I., Burns T.F., Siegfried J.M., Dacic S. The Prognostic and Predictive Value of KRAS Oncogene Substitutions in Lung Adenocarcinoma. Cancer. 2013;119:2268–2274. doi: 10.1002/cncr.28039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu H.A., Sima C.S., Shen R., Kass S., Gainor J., Shaw A., Hames M., Iams W., Aston J., Lovly C.M., et al. Prognostic Impact of KRAS Mutation Subtypes in 677 Patients with Metastatic Lung Adenocarcinomas. J. Thorac. Oncol. 2015;10:431–437. doi: 10.1097/JTO.0000000000000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tao L., Miao R., Mekhail T., Sun J., Meng L., Fang C., Guan J., Jain A., Du Y., Allen A. Prognostic Value of KRAS Mutation Subtypes and PD-L1 Expression in Patients with Lung Adenocarcinoma. Clin. Lung Cancer. 2021;22:e506–e511. doi: 10.1016/j.cllc.2020.07.004. [DOI] [PubMed] [Google Scholar]
- 43.Wahl S.G.F., Dai H.Y., Emdal E.F., Berg T., Halvorsen T.O., Ottestad A.L., Lund-Iversen M., Brustugun O.T., Førde D., Paulsen E.-E. The Prognostic Effect of KRAS Mutations in Non-Small Cell Lung Carcinoma Revisited: A Norwegian Multicentre Study. Cancers. 2021;13:4294. doi: 10.3390/cancers13174294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cai D., Hu C., Li L., Deng S., Yang J., Han-Zhang H., Li M. The Prevalence and Prognostic Value of KRAS Co-mutation Subtypes in Chinese Advanced Non-small Cell Lung Cancer Patients. Cancer Med. 2020;9:84–93. doi: 10.1002/cam4.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cui W., Franchini F., Alexander M., Officer A., Wong H.-L., IJzerman M., Desai J., Solomon B.J. Real World Outcomes in KRAS G12C Mutation Positive Non-Small Cell Lung Cancer. Lung Cancer. 2020;146:310–317. doi: 10.1016/j.lungcan.2020.06.030. [DOI] [PubMed] [Google Scholar]
- 46.Finn S.P., Addeo A., Dafni U., Thunnissen E., Bubendorf L., Madsen L.B., Biernat W., Verbeken E., Hernandez-Losa J., Marchetti A. Prognostic Impact of KRAS G12C Mutation in Patients with NSCLC: Results from the European Thoracic Oncology Platform Lungscape Project. J. Thorac. Oncol. 2021;16:990–1002. doi: 10.1016/j.jtho.2021.02.016. [DOI] [PubMed] [Google Scholar]
- 47.Jones G.D., Caso R., Tan K.S., Mastrogiacomo B., Sanchez-Vega F., Liu Y., Connolly J.G., Murciano-Goroff Y.R., Bott M.J., Adusumilli P.S. KRASG12C Mutation Is Associated with Increased Risk of Recurrence in Surgically Resected Lung AdenocarcinomaKRASG12C and Genomic Correlates in Lung Adenocarcinoma. Clin. Cancer Res. 2021;27:2604–2612. doi: 10.1158/1078-0432.CCR-20-4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu S.-Y., Sun H., Zhou J.-Y., Jie G.-L., Xie Z., Shao Y., Zhang X., Ye J.-Y., Chen C.-X., Zhang X.-C. Clinical Characteristics and Prognostic Value of the KRAS G12C Mutation in Chinese Non-Small Cell Lung Cancer Patients. Biomark. Res. 2020;8:22. doi: 10.1186/s40364-020-00199-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nadal E., Chen G., Prensner J.R., Shiratsuchi H., Sam C., Zhao L., Kalemkerian G.P., Brenner D., Lin J., Reddy R.M. KRAS-G12C Mutation Is Associated with Poor Outcome in Surgically Resected Lung Adenocarcinoma. J. Thorac. Oncol. 2014;9:1513–1522. doi: 10.1097/JTO.0000000000000305. [DOI] [PubMed] [Google Scholar]
- 50.El Osta B., Behera M., Kim S., Berry L.D., Sica G., Pillai R.N., Owonikoko T.K., Kris M.G., Johnson B.E., Kwiatkowski D.J. Characteristics and Outcomes of Patients with Metastatic KRAS-Mutant Lung Adenocarcinomas: The Lung Cancer Mutation Consortium Experience. J. Thorac. Oncol. 2019;14:876–889. doi: 10.1016/j.jtho.2019.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sebastian M., Eberhardt W.E.E., Hoffknecht P., Metzenmacher M., Wehler T., Kokowski K., Alt J., Schütte W., Büttner R., Heukamp L.C. KRAS G12C-Mutated Advanced Non-Small Cell Lung Cancer: A Real-World Cohort from the German Prospective, Observational, Nation-Wide CRISP Registry (AIO-TRK-0315) Lung Cancer. 2021;154:51–61. doi: 10.1016/j.lungcan.2021.02.005. [DOI] [PubMed] [Google Scholar]
- 52.Hong D.S., Fakih M.G., Strickler J.H., Desai J., Durm G.A., Shapiro G.I., Falchook G.S., Price T.J., Sacher A., Denlinger C.S., et al. KRASG12C Inhibition with Sotorasib in Advanced Solid Tumors. N. Engl. J. Med. 2020;383:1207–1217. doi: 10.1056/NEJMoa1917239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spira A.I., Riely G.J., Gadgeel S.M., Heist R.S., Ou S.-H.I., Pacheco J.M., Johnson M.L., Sabari J.K., Leventakos K., Yau E., et al. KRYSTAL-1: Activity and Safety of Adagrasib (MRTX849) in Patients with Advanced/Metastatic Non–Small Cell Lung Cancer (NSCLC) Harboring a KRASG12C Mutation. J. Clin. Oncol. 2022;40:9002. doi: 10.1200/JCO.2022.40.16_suppl.9002. [DOI] [Google Scholar]
- 54.Reck M., Spira A., Besse B., Wolf J., Skoulidis F., Borghaei H., Goto K., Park K., Griesinger F., Font E.F. MO01. 32 CodeBreaK 200: A Phase 3 Multicenter Study of Sotorasib, a KRAS (G12C) Inhibitor, versus Docetaxel in Patients with Previously Treated Advanced Non-Small Cell Lung Cancer (NSCLC) Harboring KRAS p. G12C Mutation. J. Thorac. Oncol. 2021;16:S29. doi: 10.1016/j.jtho.2020.10.137. [DOI] [Google Scholar]
- 55.Bontoux C., Hofman V., Brest P., Ilié M., Mograbi B., Hofman P. Daily Practice Assessment of KRAS Status in NSCLC Patients: A New Challenge for the Thoracic Pathologist Is Right around the Corner. Cancers. 2022;14:1628. doi: 10.3390/cancers14071628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Veluswamy R., Mack P.C., Houldsworth J., Elkhouly E., Hirsch F.R. KRAS G12C–Mutant Non–Small Cell Lung Cancer: Biology, Developmental Therapeutics, and Molecular Testing. J. Mol. Diagn. 2021;23:507–520. doi: 10.1016/j.jmoldx.2021.02.002. [DOI] [PubMed] [Google Scholar]
- 57.Judd J., Karim N.A., Khan H., Naqash A.R., Baca Y., Xiu J., VanderWalde A.M., Mamdani H., Raez L.E., Nagasaka M. Characterization of KRAS Mutation Subtypes in Non–Small Cell Lung Cancer. Mol. Cancer Ther. 2021;20:2577–2584. doi: 10.1158/1535-7163.MCT-21-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schoenfeld A.J., Rizvi H., Bandlamudi C., Sauter J.L., Travis W.D., Rekhtman N., Plodkowski A.J., Perez-Johnston R., Sawan P., Beras A., et al. Clinical and Molecular Correlates of PD-L1 Expression in Patients with Lung Adenocarcinomas. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2020;31:599–608. doi: 10.1016/j.annonc.2020.01.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scheffler M., Ihle M.A., Hein R., Merkelbach-Bruse S., Scheel A.H., Siemanowski J., Brägelmann J., Kron A., Abedpour N., Ueckeroth F. K-Ras Mutation Subtypes in NSCLC and Associated Co-Occuring Mutations in Other Oncogenic Pathways. J. Thorac. Oncol. 2019;14:606–616. doi: 10.1016/j.jtho.2018.12.013. [DOI] [PubMed] [Google Scholar]
- 60.Skoulidis F., Heymach J. V Co-Occurring Genomic Alterations in Non-Small-Cell Lung Cancer Biology and Therapy. Nat. Rev. Cancer. 2019;19:495–509. doi: 10.1038/s41568-019-0179-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saleh M.M., Scheffler M., Merkelbach-Bruse S., Scheel A.H., Ulmer B., Wolf J., Buettner R. Comprehensive Analysis of TP53 and KEAP1 Mutations and Their Impact on Survival in Localized-and Advanced-Stage NSCLC. J. Thorac. Oncol. 2022;17:76–88. doi: 10.1016/j.jtho.2021.08.764. [DOI] [PubMed] [Google Scholar]
- 62.Spira A.I., Tu H., Aggarwal S., Hsu H., Carrigan G., Wang X., Ngarmchamnanrith G., Chia V., Gray J.E. A Retrospective Observational Study of the Natural History of Advanced Non–Small-Cell Lung Cancer in Patients with KRAS p. G12C Mutated or Wild-Type Disease. Lung Cancer. 2021;159:1–9. doi: 10.1016/j.lungcan.2021.05.026. [DOI] [PubMed] [Google Scholar]
- 63.Liu Y., Xu F., Wang Y., Wu Q., Wang B., Yao Y., Zhang Y., Han-Zhang H., Ye J., Zhang L. Mutations in Exon 8 of TP53 Are Associated with Shorter Survival in Patients with Advanced Lung Cancer. Oncol. Lett. 2019;18:3159–3169. doi: 10.3892/ol.2019.10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Izar B., Zhou H., Heist R.S., Azzoli C.G., Muzikansky A., Scribner E.E.F., Bernardo L.A., Dias-Santagata D., Iafrate A.J., Lanuti M. The Prognostic Impact of KRAS, Its Codon and Amino Acid Specific Mutations, on Survival in Resected Stage I Lung Adenocarcinoma. J. Thorac. Oncol. 2014;9:1363–1369. doi: 10.1097/JTO.0000000000000266. [DOI] [PubMed] [Google Scholar]
- 65.Di Federico A., De Giglio A., Parisi C., Gelsomino F. STK11/LKB1 and KEAP1 Mutations in Non-Small Cell Lung Cancer: Prognostic Rather than Predictive? Eur. J. Cancer. 2021;157:108–113. doi: 10.1016/j.ejca.2021.08.011. [DOI] [PubMed] [Google Scholar]
- 66.Krishnamurthy N., Goodman A.M., Barkauskas D.A., Kurzrock R. STK11 Alterations in the Pan-Cancer Setting: Prognostic and Therapeutic Implications. Eur. J. Cancer. 2021;148:215–229. doi: 10.1016/j.ejca.2021.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Falk A.T., Yazbeck N., Guibert N., Chamorey E., Paquet A., Ribeyre L., Bence C., Zahaf K., Leroy S., Marquette C.-H. Effect of Mutant Variants of the KRAS Gene on PD-L1 Expression and on the Immune Microenvironment and Association with Clinical Outcome in Lung Adenocarcinoma Patients. Lung Cancer. 2018;121:70–75. doi: 10.1016/j.lungcan.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 68.Renaud S., Falcoz P.-E., Schaeffer M., Guenot D., Romain B., Olland A., Reeb J., Santelmo N., Chenard M.-P., Legrain M. Prognostic Value of the KRAS G12V Mutation in 841 Surgically Resected Caucasian Lung Adenocarcinoma Cases. Br. J. Cancer. 2015;113:1206–1215. doi: 10.1038/bjc.2015.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guibert N., Ilie M., Long E., Hofman V., Bouhlel L., Brest P., Mograbi B., Marquette C.H., Didier A., Mazieres J. KRAS Mutations in Lung Adenocarcinoma: Molecular and Epidemiological Characteristics, Methods for Detection, and Therapeutic Strategy Perspectives. Curr. Mol. Med. 2015;15:418–432. doi: 10.2174/1566524015666150505161412. [DOI] [PubMed] [Google Scholar]
- 70.Jia Y., Jiang T., Li X., Zhao C., Zhang L., Zhao S., Liu X., Qiao M., Luo J., Shi J. Characterization of Distinct Types of KRAS Mutation and Its Impact on First-line Platinum-based Chemotherapy in Chinese Patients with Advanced Non-small Cell Lung Cancer. Oncol. Lett. 2017;14:6525–6532. doi: 10.3892/ol.2017.7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peters J.L., Sutton A.J., Jones D.R., Abrams K.R., Rushton L. Performance of the Trim and Fill Method in the Presence of Publication Bias and Between-study Heterogeneity. Stat. Med. 2007;26:4544–4562. doi: 10.1002/sim.2889. [DOI] [PubMed] [Google Scholar]
- 72.Wu Y.-L., Tsuboi M., He J., John T., Grohe C., Majem M., Goldman J.W., Laktionov K., Kim S.-W., Kato T., et al. Osimertinib in Resected EGFR -Mutated Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2020;383:1711–1723. doi: 10.1056/NEJMoa2027071. [DOI] [PubMed] [Google Scholar]
- 73.Antonia S.J., Villegas A., Daniel D., Vicente D., Murakami S., Hui R., Kurata T., Chiappori A., Lee K.H., de Wit M., et al. Overall Survival with Durvalumab after Chemoradiotherapy in Stage III NSCLC. N. Engl. J. Med. 2018;379:2342–2350. doi: 10.1056/NEJMoa1809697. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available in the Open Science Framework repository at 10.17605/OSF.IO/AUF2T without third-party permission.