Abstract
The topoisomerase IV parC and parE genes from the wall-less organism Mycoplasma hominis PG21 were cloned and sequenced. The coupled genes are located far from the DNA gyrase genes gyrA and gyrB. They encode proteins of 639 and 866 amino acids, respectively. As expected, the encoded ParE and ParC proteins exhibit higher homologies with the topoisomerase IV subunits of the gram-positive bacteria Staphylococcus aureus and Streptococcus pneumoniae than with their Escherichia coli counterparts. The conserved regions include the Tyr residue of the active site and the region involved in quinolone resistance (quinolone resistance-determining region [QRDR]) in ParC and the ATP-binding site and the QRDR in ParE.
The intracellular targets of fluoroquinolones are considered to be the bacterial type II topoisomerases (23, 30). These enzymes are responsible for the control of the topological state of DNA in the cell. In Escherichia coli, two type II topoisomerases have been identified, DNA gyrase (23) and topoisomerase IV (28). DNA gyrase is composed of two A and two B subunits encoded by the gyrA and gyrB genes, respectively. The tetrameric enzyme catalyzes ATP-dependent negative supercoiling of DNA (50). Topoisomerase IV also is a heterotetramer consisting of two C and two E subunits encoded by the parC and parE genes, respectively. This enzyme is essential for chromosome segregation (2).
Mutations in the quinolone resistance-determining regions (QRDRs) of gyrA (15, 22, 55) and parC (25, 30, 31) have been described as the major mechanism for quinolone resistance. In gram-negative bacteria such as E. coli and Neisseria gonorrhoeae, DNA gyrase is thought to be the primary target of quinolones, since amino acid changes in ParC or ParE could be detected only when GyrA mutations were present (10, 12, 25, 31). Inversely, in ciprofloxacin-resistant strains of the gram-positive bacteria Staphylococcus aureus (17, 18, 51) and Streptococcus pneumoniae (38, 48), GyrA mutations could be detected only when a ParC mutation was present, indicating that, in these gram-positive bacteria, topoisomerase IV was the primary target of ciprofloxacin.
However, the primacy of topoisomerase IV over DNA gyrase as the quinolone target is not a conserved feature of gram-positive bacteria. Studies of fluoroquinolone-resistant mutants of S. pneumoniae (40) showed that the target preference depends on the quinolone structure. Similarly, we recently reported that, in Mycoplasma hominis, DNA gyrase is the primary target of sparfloxacin whereas topoisomerase IV is the primary target of pefloxacin, ofloxacin, and ciprofloxacin (9), indicating that topoisomerase IV as well as DNA gyrase is an important target for fluoroquinolones in this organism. Indeed, characterization of fluoroquinolone-resistant strains of M. hominis selected in vitro showed that they harbored mutations not only in gyrA but also in parC and parE genes (7, 9). It is noteworthy that in clinical isolates also, we found resistant strains that harbored alterations in both DNA gyrase and topoisomerase IV subunits (8). Thus, the widespread use of fluoroquinolones had also led to the emergence of quinolone resistance in vivo in M. hominis, a genital mycoplasma involved in endometritis, salpingitis, postpartum septicemia (6), and some extragenital infections (36).
For mycoplasmas, complete genome sequencing projects led to the identification of the gyrA, gyrB, parC, and parE genes of Mycoplasma genitalium (20) and Mycoplasma pneumoniae (26). In these mycoplasmas, as in most of the gram-positive bacteria including Bacillus subtilis (37) and S. aureus (35), the adjacent gyrA and gyrB genes are located very close to the replication origin downstream of the dnaA-dnaN genes, whereas parC and parE, which also are contiguous, are quite distant from the origin.
Previous studies of M. hominis by Ladefoged and Christiansen (33) showed the gyrB gene not to be in the immediate vicinity of the replication origin. In addition, the gyrA gene was found to map 35 kbp from gyrB on the chromosome. While the gyrB gene has been fully sequenced (33), only the QRDR sequence of gyrA has been determined (7).
Here we report the cloning and characterization of the parC and parE genes of the M. hominis topoisomerase IV that were previously shown to be associated with quinolone resistance in this organism (8, 9). The primary structures of the parC- and parE-encoded polypeptides are compared to their counterparts in other bacteria.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The reference strain PG21 (ATCC 23114) of M. hominis was grown in Hayflick modified broth medium supplemented with arginine (21). The E. coli strain TG1 and the pBS+ vector (Stratagene cloning systems) were used to construct libraries and to subclone DNA inserts. For transformation with plasmid DNA or ligation mixtures, competent E. coli cells were prepared in accordance with the Hanahan procedure (24).
DNA isolation.
Mycoplasmal genomic DNA was isolated as previously described (7). Large-scale and small-scale preparations of plasmid DNA amplified in E. coli were carried out in accordance with standard procedures (44).
Restriction mapping of the parC and parE region.
Genomic DNA of M. hominis PG21 was single- or double-digested with various restriction enzymes. Restriction fragments were separated by electrophoresis in 0.8% agarose gels, blotted onto positively charged nylon membranes by the alkali transfer procedure, and hybridized to 32P-labelled probes under standard stringent conditions (44). The parC and parE probes were the 310-bp parC and 297-bp parE DNA fragments generated by PCR amplification of the M. hominis genomic DNA with primer pairs MH11-MH13 and MH28-MH29, respectively (see Fig. 1 and 2). Experimental conditions for PCR amplification were described previously (9). The amplification products were purified by using the Wizard PCR Preps DNA purification system (Promega) and were labelled by the random priming procedure with [α-32P]dATP as the labelled nucleotide. A restriction map was constructed from the hybridization patterns of genomic DNA with the parC and parE probes.
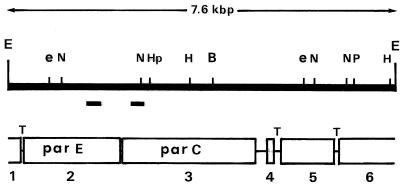
FIG. 1.
Restriction map of DNA insert of plasmid pMHE6. Positions of the parE and parC probes are indicated by the short, solid rectangles. E, EcoRI; N, NsiI; Hp, HpaI; B, BglII; P, PstI; H, HindIII; e, EcoRV. ORFs are numbered 1 to 6. T, putative transcription terminator.
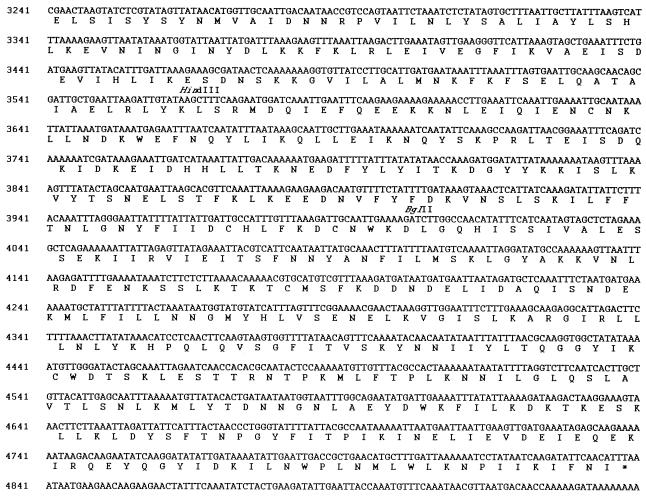
FIG. 2.
DNA sequence of the M. hominis parE-parC locus and deduced amino acid sequences of the ParC and ParE subunits of topoisomerase IV. Nucleotides are numbered according to the 7,603-bp sequence submitted to GenBank (accession no. AF036961). Primers MH11 (11), MH13 (13), MH28 (28), and MH29 (29), RBS, and restriction sites NsiI, HindIII, and BglII are indicated.
Construction and screening of the M. hominis genomic DNA library.
Genomic DNA of M. hominis PG21 was digested to completion with EcoRI, and the fragments were ligated to the EcoRI-linearized pBS+ vector. The recombinant clones containing the parC and parE sequences were selected by in situ hybridization of colonies with a mixture of the 32P-labelled parC and parE probes (see above) in accordance with standard procedures (44). Hybridization-positive clones were selected, and their plasmid content was determined. The recombinant plasmid pMHE6, containing a 7.6-kbp DNA insert hybridizing with both the parC and parE probes, was selected for sequencing studies.
DNA sequence analysis.
Both strands of double-stranded DNA were sequenced by using the AmpliTaq DNA polymerase FS Dye Terminator Cycle Sequencing Ready Reaction kit and an ABI-Prism 377 sequencer (Applied Biosystems Division, Perkin-Elmer) in accordance with the manufacturer’s instructions. Forward and reverse primers flanking the multiple-cloning-site polylinker of the pBS+ vector as well as internal primers were used to obtain the complete sequence of the DNA insert of the recombinant plasmid pMHE6.
Sequence alignments were done with the GAP and PILEUP modules of the Genetics Computer Group software package (16), and searches for similar sequences in the GenBank database were performed by using the BLAST program (3).
PFGE.
Pulsed-field gel electrophoresis (PFGE) and identification of gene positions were performed essentially as described previously for Spiroplasma citri (53). The restriction endonucleases BamHI, SalI, SmaI, and XhoI were used as indicated by the manufacturers. The restriction fragments were separated by using transverse alternating field electrophoresis. The separation conditions were as follows: 5s, 6 h; 10 s, 6 h; 20 s, 6 h; at a voltage of 350 V and a temperature of 8°C. Sizes of separated DNA fragments were determined by comparison with lambda phage DNA concatemers as molecular weight markers.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper will appear in the GenBank nucleotide sequence database under accession no. AF036961.
RESULTS
Cloning the parC and parE genes of M. hominis.
Southern blot analysis of genomic DNA from M. hominis PG21 revealed that the parC and parE probes both hybridized with a 6.9-kbp HindIII fragment, a 5-kbp EcoRV fragment, a 7.6-kbp EcoRI fragment, and 3.6-kbp EcoRI-HindIII fragment (data not shown). The 7.6-kbp EcoRI fragment was recovered from a genomic DNA library of M. hominis by in situ colony hybridization, and its nucleotide sequence was determined. A partial restriction map is shown in Fig. 1. DNA sequence analysis (see below) indicated that the recombinant plasmid, named pMHE6, contained the entire parE and parC genes of M. hominis.
Sequence analysis of the pMHE6 DNA insert.
Sequencing the DNA insert of plasmid pMHE6 showed that it comprises 7,603 nucleotides with a G+C content of 28.9% that is almost identical to that (29%) of the full genome of M. hominis PG21. By taking into account the fact that, in mycoplasmas, UGA codes for tryptophan (11, 52), we found this sequence to contain six open reading frames (ORF1 to ORF6), each starting with an ATG codon located downstream of a putative ribosome binding site (RBS). In the sequenced DNA fragment, the 3′ end of ORF1 and the 5′ part of ORF6 are truncated (Fig. 1).
The first ORF (ORF1) starts at nucleotide 1 of the submitted sequence and ends at nucleotide 258 with a TAA stop codon. It encodes the 86-amino-acid C-terminal end of a polypeptide which has significant homology, 26.7% identity and 39.5% similarity in an 86-amino-acid overlap, with the glucose-1-phosphatase of E. coli (GenBank accession no. P19926).
The second and third ORFs, nucleotides 315 to 2231 (ORF2) and nucleotides 2240 to 4837 (ORF3), are separated by only 9 nucleotides. The RBS sequence upstream of the ORF3 initiation codon overlaps the TAG termination codon of ORF2. ORF2 and ORF3 were identified, respectively, as the parE and parC genes of M. hominis (see below).
ORF4 (nucleotides 5077 to 5187) codes for a putative polypeptide for which no appreciable homology with known proteins was found.
ORF5 (nucleotides 5347 to 6429) encodes a putative 361-amino-acid polypeptide with a 27-amino-acid N terminus characteristic of signal peptide sequences. This polypeptide shows significant homology (23.9% identity and 29.2% similarity) with a B. subtilis RNase precursor (GenBank accession no. Q03091). However, it should be noticed that regardless of our careful sequencing of both strands, we found a TAA stop codon within the reading frame at position 6172 of the submitted sequence. However, amino acid sequences upstream and downstream of this stop codon both have homology with the B. subtilis RNase precursor.
The predicted amino acid sequence of ORF6 (starting at nucleotide 6508) has striking homology with the sequences of dipeptidases. In particular, it has 22.8% identical amino acids with a putative peptidase of B. subtilis (GenBank accession no. AF008220). However, the highest homology score (40.4% identity and 52.3% similarity) was found with a hypothetical protein of Mycoplasma capricolum (GenBank accession no. S48588).
Downstream of ORF1, the presence of imperfect inverted repeat sequences (nucleotides 255 to 274 and 281 to 299) followed by a stretch of uridine residues (in the mRNA) strongly suggests a rho-independent terminator. Similar structures were also found between ORF4 and ORF5 (nucleotides 5222 to 5231 and 5236 to 5245) and between ORF5 and ORF6 (nucleotides 6426 to 6438 and 6442 to 6453) (Fig. 1).
No such a structure but, instead, a 230-bp intergenic region was found downstream of ORF3. These observations suggest that ORF1, -4, -5, and -6 would be individually transcribed whereas ORF2 and ORF3 carrying the parE and parC genes of M. hominis would be cotranscribed to one single mRNA.
Sequence analysis of M. hominis ParE and ParC subunits.
As indicated above, ORF2 and ORF3 were identified as the parE and parC genes of M. hominis, respectively. The nucleotide and the derived amino acid sequences of these two ORFs are presented in Fig. 2.
The predicted ParE polypeptide contains 639 amino acids with a calculated molecular mass of 71.7 kDa. As shown in Table 1, the ParE polypeptide exhibits a higher percentage of identity with the GrlB (52%) than with the GyrB (46.1%) subunits of S. aureus (18, 35). The M. hominis ParE exhibits similar percentages of identity with the GyrB (37.5%) and ParE (35.7%) subunits of E. coli (1, 28, 41). The highest level of homology (55.5% identity) was found with the ParE protein of S. pneumoniae (39). In a comparison with other mycoplasmas, the M. hominis ParE was found to have 54.7, 46.5, and 46% amino acid identity with the ParE subunits of M. gallisepticum (46), M. pneumoniae (26), and M. genitalium (20), respectively. From these data, we designated the ORF2-encoded polypeptide the ParE subunit of M. hominis. An overall identity of 44.2% was found between the ParE and GyrB peptidic sequences of M. hominis.
TABLE 1.
Sequence identities between M. hominis ParC and ParE and the topoisomerase II subunits of M. genitalium, S. aureus, and E. coli
| Subunit | % Amino acid identity with:
|
|
|---|---|---|
| M. hominis ParC | M. hominis ParE | |
| M. genitalium | ||
| GyrA | 32.6 | |
| GyrB | 44.3 | |
| ParC | 34 | |
| ParE | 46 | |
| S. aureus | ||
| GyrA | 31.3 | |
| GyrB | 46.1 | |
| GrlA | 37.6 | |
| GrlB | 52 | |
| E. coli | ||
| GyrA | 30.2 | |
| GyrB | 37.5 | |
| ParC | 28.2 | |
| ParE | 35.7 | |
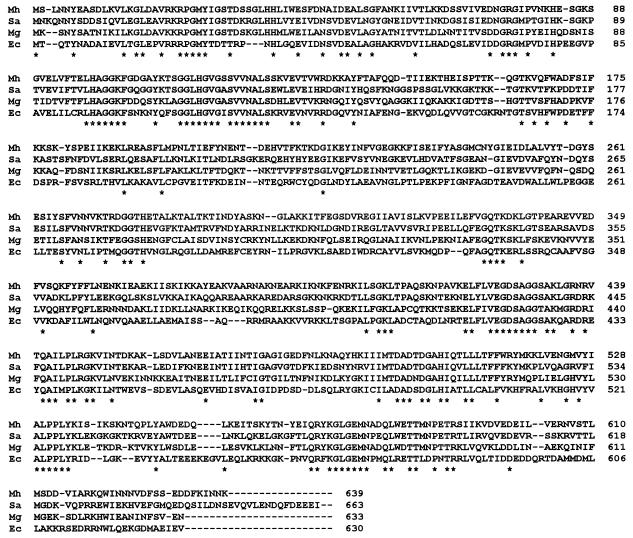
In Fig. 3, the ParE amino acid sequence of M. hominis is compared to those of S. aureus, M. genitalium, and E. coli (18, 20, 28, 41). Like topoisomerase II B subunits, M. hominis ParE contains the highly conserved motifs VEGDSAGG (positions 423 to 430 of the M. hominis ParE sequence) and PL(R/K)GK (positions 445 to 449) in the C-terminal region of the protein (Fig. 3). Interestingly, the Asp426→Asn substitution, which was associated with the resistance to fluoroquinolones of M. hominis mutants (8, 9), is located within the first motif. In the N-terminal part, the ATP-binding region (positions 1 to 150 approximately) containing a glycine-rich segment is also conserved (Fig. 3).
FIG. 3.
Alignment of M. hominis (Mh) ParE sequence with its counterparts from S. aureus (Sa), M. genitalium (Mg), and E. coli (Ec). An asterisk indicates an identical residue for all four protein sequences. The ParE (GrlB) sequences of S. aureus, M. genitalium, and E. coli are from references 18, 20, and 28 and 41, respectively.
The predicted ParC polypeptide of M. hominis comprises 866 amino acids with a calculated molecular mass of 92.2 kDa. It has 37.6% identity with the GrlA (ParC) subunit (18) but only 31.3% identity with the GyrA protein of S. aureus (35) (Table 1). When compared to the ParC subunit of S. pneumoniae (39), another gram-positive bacterium, M. hominis ParC shows 37.3% sequence identity. As in the case of ParE, similar identities were found between M. hominis ParC and the ParC (28.2%) or GyrA protein (30.2%) of E. coli (28, 41, 47) (Table 1). Among mycoplasmas, the M. hominis sequence has 34 and 31.3% identity with the ParC subunit sequences of M. genitalium and M. pneumoniae, respectively (20, 26). From these sequence homologies, we identified ORF3 as the parC gene of the topoisomerase IV of M. hominis. Compared to its counterparts in S. aureus, M. genitalium, and E. coli (Fig. 4), the ParC protein shows greatest homology in its N-terminal breakage-reunion region, which includes the most conserved stretches DGLKPV (positions 47 to 52 of the M. hominis ParC sequence), YHPHGD (positions 85 to 90), and AAMRYTE (positions 126 to 132) (Fig. 4). The tyrosine active site, Tyr130, is located within the third conserved domain, AAMRYTE, of the protein (Fig. 4). The second consensus sequence, YHPHGD, maps within the QRDR. Amino acid residues Ser91 and Glu95 are the equivalents of Ser80 and Glu84 of the S. aureus and E. coli ParC subunits, which have been shown to be hot spots for quinolone resistance (17, 18, 25, 30, 31, 51). Indeed, we have demonstrated substitutions of these two amino acids in fluoroquinolone-resistant mutants of M. hominis selected in vivo and in vitro as well (8, 9). As shown in Fig. 4, the C-terminal region of the protein is much less conserved.
FIG. 4.
Alignment of M. hominis (Mh) ParC sequence with its counterparts from S. aureus (Sa), M. genitalium (Mg), and E. coli (Ec). An asterisk indicates an identical residue for all four protein sequences. The ParC (GrlA) sequences of S. aureus, M. genitalium, and E. coli are from references 18, 20, and 28 and 41, respectively.
Location of the topoisomerase IV genes on the genomic map of M. hominis PG21.
The parC and parE genes were located on the map of M. hominis PG21 (32) by using PFGE and Southern blot hybridization with the M. hominis parC and parE probes (data not shown). As expected, both probes hybridized with the same fragments, SmaI fragment H, BamHI fragment E, XhoI fragment D, and SalI fragment C, of the previously published genetic map of M. hominis PG21 (32). Hence, the adjacent parC and parE genes are located within the 110-kbp region where these restriction fragments overlap. It is noteworthy that this region is quite distant from the gyrB and gyrA genes (33).
DISCUSSION
Two M. hominis coupled genes encoding proteins homologous to the subunits of DNA gyrase and DNA topoisomerase IV were cloned and sequenced. By use of PFGE and hybridization with appropriate probes, these genes were found to map far away from gyrA and gyrB genes on the chromosome.
In many bacteria, gyrB lies close to the origin of replication, where genes are organized in the following order: dnaA, dnaN, recF, gyrB, and gyrA. A similar gene organization was described for M. pneumoniae (14), M. genitalium (4), and also for the plant mollicute Spiroplasma citri (54). However, gyrA and gyrB were shown not to be coupled in M. hominis. Instead, gyrA was mapped 35 kbp downstream of gyrB. This might have arisen by chromosomal rearrangement, regardless of the phylogenetic evolution (33). In addition, the gyrB gene of M. hominis was not located in the vicinity of the replication origin (33). Such a situation was also described for M. capricolum, an animal mycoplasma, where the gyrB gene is located opposite the chromosomal replication origin (45). In M. hominis, the gyrB gene was identified since it encodes a protein exhibiting higher homology with the E. coli GyrB subunit (50% identity) than with ParE (40% identity) (33).
From these data and from the finding that the encoded proteins were most closely related to the topoisomerase IV subunits of S. aureus, the M. hominis genes that we have cloned were designated the parC and parE genes. It is known that the ParC protein of a given organism has generally lower homology with the ParC proteins of other organisms than does GyrA with other GyrA proteins (27). This is also true for the ParC subunit of M. hominis. In the case of ParE, the similarity to GyrB subunits is significantly lower than that to other ParE proteins.
The M. hominis ParC subunit with 866 residues seems to be the largest ParC subunit sequenced so far. In comparison to ParC subunits of S. pneumoniae (823 amino acids) and to GrlA of S. aureus (800 amino acids), the M. hominis ParC contains additional amino acids both at the N- and the C-terminal ends. However the N-terminal moiety of the protein, containing the active site, is much more conserved than the C-terminal part. In contrast to ParC, the ParE subunit of M. hominis, with 639 amino acids, has a size similar to those of the other ParE proteins. Also, the overall similarities to other ParE proteins, which range from 49% (with E. coli) to 66% (with S. pneumoniae), are higher than those to the ParC subunit, which range from 41% (with E. coli) to 51% (with S. aureus).
In M. hominis, the parC and parE genes were found to be contiguous, as described previously for the topoisomerase IV genes of the gram-positive bacteria S. aureus (18), B. subtilis (43), S. pneumoniae (39), and the mycoplasmas M. genitalium (5, 20), M. pneumoniae (26), and M. gallisepticum (46). In the gram-negative bacteria E. coli and S. typhimurium, in which the gyrase genes are not linked, the topoisomerase IV genes parC and parE are separated by only 6 kbp (28, 34). In S. aureus and M. genitalium, the grlA or parC and grlB or parE genes overlap by 1 nucleotide, suggesting that these genes are transcriptionally coupled (5, 18). A similar situation seems to occur in M. hominis since the RBS sequence of parC overlaps the stop codon of parE and since no terminator-like structure was detected between parE and parC. Hence, it is likely that the topoisomerase IV genes of M. hominis are transcribed as a single message. Although no transcription terminator structure was detected immediately downstream of ParC, such a structure was found 400 bp downstream, at the end of ORF4.
Topoisomerase IV and DNA gyrase are both essential to the cell. Gyrase, through the ability to introduce negative supercoils into DNA, is involved mostly during the initiation and elongation stages of replication. The primary function of topoisomerase IV is the decatenation of chromosomes during the terminal stages of DNA replication. Interestingly, complete sequencing of the Helicobacter pylori genome (49) revealed that this organism does not possess the topoisomerase IV genes parC and parE. In contrast, in the hyperthermophilic archaeon Methanococcus jannaschii, only genes encoding topoisomerase IV and reverse gyrase, no DNA gyrase genes, were found (13). Furthermore, DNA gyrase activity has not yet been detected in vitro in any of the archaea (19). Several studies showed that topoisomerase IV and gyrase could have redundant activity since very high level expression of gyrase subunits can suppress defects in topoisomerase IV, although the reverse is not true (29, 56). From these data, the question of a common ancestor for both gyrase and topoisomerase IV can be raised. Further studies on the distribution of various topoisomerase activities among prokaryotes would help to test this hypothesis.
Topoisomerase IV, as well as DNA gyrase, is considered a target of quinolones. In previous studies, we found that the substitutions Ser91→Ile, Ser92→Pro, and Glu95→Lys or Gly in ParC and Asp426→Asn in ParE were associated with fluoroquinolone resistance in M. hominis mutants selected in vivo and in vitro (8, 9). These positions have been found to be frequently altered in quinolone-resistant mutants of E. coli, S. aureus, and S. pneumoniae (17, 18, 25, 30, 31, 38, 39, 42, 51).
On the basis of sequence comparisons and physical mapping, we conclude that we have identified the parC and parE genes of the wall-less organism M. hominis. However, the ATP-dependent DNA decatenase activity of the protein reconstituted from the parE- and parC-encoded subunits is still to be demonstrated.
ACKNOWLEDGMENTS
We thank Patricia Carle for PFGE experiments and Sybille Duret for excellent technical assistance.
REFERENCES
- 1.Adachi T M, Mizuuchi M, Robinson E A, Apella E, O’Dea M H, Gellert M, Mizuuchi K. DNA sequence of the Escherichia coli gyrB gene: application of a new sequencing strategy. Nucleic Acids Res. 1987;15:771–784. doi: 10.1093/nar/15.2.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams D E, Shekhtman E M, Zechiedrich E L, Schmid M B, Cozzarelli N R. The role of topoisomerase IV in partitioning bacterial replicons and the structure of catenated intermediates in DNA. Cell. 1992;71:277–288. doi: 10.1016/0092-8674(92)90356-h. [DOI] [PubMed] [Google Scholar]
- 3.Altshul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Bailey C C, Bott K F. An unusual gene containing a dnaJ N-terminal box flanks the putative origin of replication of Mycoplasma genitalium. J Bacteriol. 1994;176:5814–5819. doi: 10.1128/jb.176.18.5814-5819.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailey C C, Younkins R, Huan W M, Bott K F. Characterization of genes encoding topoisomerase IV of Mycoplasma genitalium. Gene. 1996;168:77–80. doi: 10.1016/0378-1119(95)00718-0. [DOI] [PubMed] [Google Scholar]
- 6.Bébéar C. Mycoplasmas. In: Emmerson A M, Hawkey P M, Gillespie S H, editors. Principles and practice of clinical bacteriology. Chichester, United Kingdom: John Wiley and Sons Ltd.; 1997. pp. 709–722. [Google Scholar]
- 7.Bébéar C M, Bové J M, Bébéar C, Renaudin J. Characterization of Mycoplasma hominis mutations involved in resistance to fluoroquinolones. Antimicrob Agents Chemother. 1997;41:269–273. doi: 10.1128/aac.41.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bébéar, C. M., and J. Renaudin. Unpublished data.
- 9.Bébéar, C. M., H. Renaudin, A. Charron, J. M. Bové, C. Bébéar, and J. Renaudin. Alterations in topoisomerase IV and DNA gyrase in quinolone-resistant mutants of Mycoplasma hominis obtained in vitro. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 10.Belland R J, Morrison S G, Ison C, Huang W H. Neisseria gonorrhoeae acquires mutations in analogous regions of gyrA and parC in fluoroquinolone-resistant isolates. Mol Microbiol. 1994;14:371–380. doi: 10.1111/j.1365-2958.1994.tb01297.x. [DOI] [PubMed] [Google Scholar]
- 11.Bové, J. M. 1993. Molecular features of mollicutes. Clin. Infect. Dis. 17(Suppl. 1):S10–S31. [DOI] [PubMed]
- 12.Breines D M, Ouabdesselam S, Ng E Y, Tankovic J, Shah S, Soussy C J, Hooper D C. Quinolone resistance locus nfxD of Escherichia coli is a mutant allele of the parE gene encoding a subunit of topoisomerase IV. Antimicrob Agents Chemother. 1997;41:175–179. doi: 10.1128/aac.41.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, Fitzgerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J-F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Georghagen N S M, Weidman J F, Fuhrmann J L, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Roberts K M, Hurst M A, Kaine B P, Borodovsky M, Klenk H-P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 14.Colman S D, Hu P C, Bott K F. Mycoplasma pneumoniae DNA gyrase genes. Mol Microbiol. 1990;4:1129–1134. doi: 10.1111/j.1365-2958.1990.tb00687.x. [DOI] [PubMed] [Google Scholar]
- 15.Cullen M E, Wyke A W, Kuroda R, Fisher L M. Cloning and characterization of a DNA gyrase A gene from Escherichia coli that confers clinical resistance to 4-quinolones. Antimicrob Agents Chemother. 1989;33:886–894. doi: 10.1128/aac.33.6.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferrero L, Cameron B, Crouzet J. Analysis of gyrA and grlA mutations in stepwise-selected ciprofloxacin-resistant mutants of Staphylococcus aureus. Antimicrob Agents Chemother. 1995;39:1554–1558. doi: 10.1128/aac.39.7.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrero L, Cameron B, Manse B, Lagneaux D, Crouzet J, Framechon A, Blanche F. Cloning and primary structure of Staphylococcus aureus DNA topoisomerase IV: a primary target of fluoroquinolones. Mol Microbiol. 1994;13:641–653. doi: 10.1111/j.1365-2958.1994.tb00458.x. [DOI] [PubMed] [Google Scholar]
- 19.Forterre P, Bergerat A, Lopez-Garcia P. The unique DNA topology and DNA topoisomerases of hyperthermophilic archea. FEMS Microbiol Rev. 1996;18:137–248. doi: 10.1111/j.1574-6976.1996.tb00240.x. [DOI] [PubMed] [Google Scholar]
- 20.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, Fleischmann R D, Bult C J, Kerlavage A R, Sutton G, Kelley J M, Fritchman J L, Weidman J C, Small K V, Sandusky M, Fuhrmann J, Nguyen D, Utterback T R, Saudek D M, Phillips C A, Merrick J M, Tomb J F, Dougherty B A, Bott K F, Hu P-C, Lucier T S, Peterson S N, Smith H O, Hutchison III C A, Venter J C. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 21.Freundt E A. Culture media for classic mycoplasmas. In: Razin S, Tully J G, editors. Methods in mycoplasmology. Vol. 1. New York, N.Y: Academic Press, Inc.; 1983. pp. 127–135. [Google Scholar]
- 22.Gellert M, Mizuuchi K, O’Dea M H, Itoh T, Tomisawa J. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc Natl Acad Sci USA. 1977;74:4772–4776. doi: 10.1073/pnas.74.11.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gellert M, Mizuuchi K, O’Dea M H, Nash H A. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci USA. 1976;73:3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanahan D. Studies on transformation on Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 25.Heisig P. Genetic evidence for a role of parC mutations in development of high-level fluoroquinolone resistance in Escherichia coli. Antimicrob Agents Chemother. 1996;40:879–885. doi: 10.1128/aac.40.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li B C, Herrmann R. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 1996;24:4420–4429. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang W M. Bacterial diversity based on type II DNA topoisomerase genes. Annu Rev Genet. 1996;30:79–107. doi: 10.1146/annurev.genet.30.1.79. [DOI] [PubMed] [Google Scholar]
- 28.Kato J, Nishimura Y, Imamura R, Niki H, Hiraga S, Suzuki H. New topoisomerase essential for chromosome segregation in Escherichia coli. Cell. 1990;63:393–404. doi: 10.1016/0092-8674(90)90172-b. [DOI] [PubMed] [Google Scholar]
- 29.Kato J, Suzuki H, Ikeda H. Purification and characterization of DNA topoisomerase IV in Escherichia coli. J Biol Chem. 1992;267:25676–25684. [PubMed] [Google Scholar]
- 30.Khodursky A B, Zechiedrich E L, Cozzarelli N R. Topoisomerase IV is a target of quinolones in Escherichia coli. Proc Natl Acad Sci USA. 1995;92:11801–11805. doi: 10.1073/pnas.92.25.11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumagai Y, Kato J-I, Hoshino K, Akasaka T, Sato K, Ikeda H. Quinolone-resistant mutants of Escherichia coli DNA topoisomerase IV parC gene. Antimicrob Agents Chemother. 1996;40:710–714. doi: 10.1128/aac.40.3.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ladefoged S A, Christiansen G. Physical and genetic mapping of the genome of five Mycoplasma hominis strains by pulsed-field gel electrophoresis. J Bacteriol. 1992;174:2199–2207. doi: 10.1128/jb.174.7.2199-2207.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ladefoged S A, Christiansen G. Sequencing analysis reveals a unique gene organization in the gyrB region of Mycoplasma hominis. J Bacteriol. 1994;176:5835–5842. doi: 10.1128/jb.176.18.5835-5842.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luttinger A L, Springer A L, Schmid M B. A cluster of genes that affects nucleoid segregation in Salmonella typhimurium. New Biol. 1991;3:687–697. [PubMed] [Google Scholar]
- 35.Margerrison E E C, Hopewell R, Fisher L M. Nucleotide sequence of the Staphylococcus aureus gyrB-gyrA locus encoding the DNA gyrase A and B proteins. J Bacteriol. 1992;174:1596–1603. doi: 10.1128/jb.174.5.1596-1603.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meyer, R. D., and W. Clough. 1993. Extragenital Mycoplasma hominis infections in adults: emphasis on immunosuppression. Clin. Infect. Dis. 17(Suppl. 1):S243–S249. [DOI] [PubMed]
- 37.Moriya S, Ogasawara N, Yoshikawa H. Structure and function of the region of the replication origin of the Bacillus subtilis chromosome. III. Nucleotide sequence of some 10,000 base pairs in the origin region. Nucleic Acids Res. 1985;13:2251–2265. doi: 10.1093/nar/13.7.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan X-S, Ambler J, Mehtar S, Fisher L M. Involvement of topoisomerase IV and DNA gyrase as ciprofloxacin targets in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1996;40:2321–2326. doi: 10.1128/aac.40.10.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan X-S, Fisher L M. Cloning and characterization of the parC and parE genes of Streptococcus pneumoniae encoding topoisomerase IV: role in fluoroquinolone resistance. J Bacteriol. 1996;178:4060–4069. doi: 10.1128/jb.178.14.4060-4069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pan X-S, Fisher L M. Targeting of DNA gyrase in Streptococcus pneumoniae by sparfloxacin: selective targeting of gyrase or topoisomerase IV by quinolones. Antimicrob Agents Chemother. 1997;41:471–474. doi: 10.1128/aac.41.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng H, Marians K. Escherichia coli topoisomerase IV. J Biol Chem. 1993;268:24481–24490. [PubMed] [Google Scholar]
- 42.Périchon B, Tankovic J, Courvalin P. Characterization of a mutation in the parE gene that confers fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:1166–1167. doi: 10.1128/aac.41.5.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rose M, Entian K-D. New genes in the 170° region of the Bacillus subtilis genome encode DNA gyrase subunits, a thioredoxin, a xylanase and an amino acid transporter. Microbiology. 1996;142:3097–3101. doi: 10.1099/13500872-142-11-3097. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 45.Sano K, Miyata M. The gyrB gene lies opposite from the replication origin on the circular chromosome of Mycoplasma capricolum. Gene. 1994;151:181–183. doi: 10.1016/0378-1119(94)90653-x. [DOI] [PubMed] [Google Scholar]
- 46.Skamrov A V, Feoktistova E S, Bibilashvili R S. Cloning and analysis of the nucleotide sequence of the segment in the Mycoplasma gallisepticum genome containing the gene for the ATP-binding subunit of DNA topoisomerase type II (topIIB) Mol Biol. 1995;29:308–316. [PubMed] [Google Scholar]
- 47.Swanberg S L, Wang J C. Cloning and sequencing of the Escherichia coli gyrA gene coding for the A subunit of DNA gyrase. J Mol Biol. 1987;197:729–736. doi: 10.1016/0022-2836(87)90479-7. [DOI] [PubMed] [Google Scholar]
- 48.Tankovic J, Perichon B, Duval J, Courvalin P. Contribution of mutations in gyrA and parC genes to fluoroquinolone resistance of mutants of Streptococcus pneumoniae obtained in vivo and in vitro. Antimicrob Agents Chemother. 1996;40:2505–2510. doi: 10.1128/aac.40.11.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomb J-F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzgerald L M, Lee N, Adams M D, Hickey E K, Berg D E, Gocayne J D, Utterback T R, Peterson J D, Kelley J M, Cotton M D, Weidman J M, Fujii C, Bowman C, Watthey L, Wallin E, Hayes W S, Borodovsky M, Karp P D, Smith H O, Fraser C M, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 50.Wang J C. DNA topoisomerases. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 51.Yamagashi J-I, Kojima T, Oyamada Y, Fujimoto K, Hattori H, Nakamura S, Inoue M. Alterations in the DNA topoisomerase IV grlA gene responsible for quinolone resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1996;40:1157–1163. doi: 10.1128/aac.40.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamao F, Muto A, Kawauchi Y, Iwami M, Iwagami S, Azumi Y, Osawa S. UGA is read as tryptophan in Mycoplasma capricolum. Proc Natl Acad Sci USA. 1985;82:2306–2309. doi: 10.1073/pnas.82.8.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ye F, Laigret F, Whitley J C, Citti C, Finch L R, Carle P, Renaudin J, Bové J M. A physical and genetic map of the Spiroplasma citri genome. Nucleic Acids Res. 1992;20:1559–1565. doi: 10.1093/nar/20.7.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ye F, Renaudin J, Bové J M, Laigret F. Cloning and sequencing of the replication origin (oriC) of the Spiroplasma citri chromosome and construction of autonomously replicating artificial plasmids. Curr Microbiol. 1994;29:23–29. doi: 10.1007/BF01570187. [DOI] [PubMed] [Google Scholar]
- 55.Yoshida H, Bogaki M, Nakamura M, Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob Agents Chemother. 1990;34:1271–1272. doi: 10.1128/aac.34.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zechiedrich E L, Cozzarelli N R. Roles of topoisomerase IV and DNA gyrase in DNA unlinking during replication in Escherichia coli. Genes Dev. 1995;9:2859–2869. doi: 10.1101/gad.9.22.2859. [DOI] [PubMed] [Google Scholar]