Abstract
Twenty-three norfloxacin-selected first-step mutants of Streptococcus pneumoniae showed low-level fluoroquinolone resistance. Their susceptibility to norfloxacin in the presence or absence of reserpine and known efflux pump substrates was determined by an agar dilution method. Five mutants showed four- to eightfold increases in their susceptibility to norfloxacin in the presence of reserpine and four- to eightfold decreases in their susceptibility to acriflavine and ethidium bromide. This phenotype is suggestive of an efflux mechanism of resistance. A representative of these mutants, 1N27, accumulated significantly less ethidium bromide than the parent strain; reserpine abolished these differences. No changes in the quinolone resistance-determining regions of parC, parE, gyrA, or gyrB were found in this mutant. By our validated agar dilution method, the efflux phenotype was sought in clinical isolates of S. pneumoniae. Of 1,037 clinical isolates examined from the United Kingdom, 273 showed reduced susceptibility to norfloxacin or ciprofloxacin. Of these, 45.4% showed the efflux phenotype. Our findings suggest that an efflux mechanism may be a frequent cause of clinically significant fluoroquinolone resistance in pneumococci.
Streptococcus pneumoniae is an important pathogen and a major etiologic agent of community-acquired pneumonia. Effective treatment of pneumococcal infections has until recently relied upon the use of beta-lactam antibiotics, but with the emergence of antibiotic resistance, their use is now increasingly compromised (2, 4, 9). There is considerable interest in the use of alternative antimicrobials, such as the fluoroquinolones. Although presently available fluoroquinolones, such as ciprofloxacin, are limited in their effectiveness, newer developmental compounds show greater promise (6, 8).
A problem associated with the use of fluoroquinolones is the selection of spontaneous resistant mutants. Several studies with pneumococci have shown that low-level resistance can result from mutations in topoisomerase IV (10, 13, 16, 17). Increased levels of resistance occur following the acquisition of additional mutations in gyrA, which encodes the A subunit of DNA gyrase (10, 13, 16).
Recently, studies in our laboratories and elsewhere have described an efflux mechanism as a further cause of low-level resistance in pneumococci (7, 20). However, these descriptions are restricted to laboratory-generated mutants, and no data are available on the occurrence of this form of fluoroquinolone resistance in clinical isolates. Therefore, we undertook a study to determine the prevalence of the efflux mechanism of resistance in clinical strains of S. pneumoniae. In order to examine a large number of clinical strains a phenotypic method was used. This was based upon the effect of reserpine, a known efflux pump inhibitor (12, 15), on the activity of norfloxacin, and on the susceptibility of the strains to unrelated efflux pump substrates. The effectiveness of this method for the detection of the efflux mechanism was validated with an in vitro mutant which had been examined for antimicrobial susceptibility, altered levels of ethidium bromide accumulation, and changes in topoisomerase IV and DNA gyrase.
MATERIALS AND METHODS
Antibiotics and chemicals.
Norfloxacin was supplied by Merck & Co Inc., Rahway, N.J.; ciprofloxacin and moxifloxacin (BAY 12-8039) were supplied by Bayer AG, Wuppertal, Germany; and sparfloxacin was supplied by Rhône Poulonc Rorer, Vitry-sur-Seine, France. All chemicals were from Sigma-Aldrich Company Ltd., Poole, United Kingdom.
Selection and susceptibility of resistant mutants.
First-step mutants of S. pneumoniae ATCC 49619 were selected by inoculating 108 CFU onto Columbia agar (Oxoid, Basingstoke, United Kingdom) supplemented with 5% horse blood and containing norfloxacin at a concentration of 4× the MIC. Following incubation at 35 to 37°C in an atmosphere of 4 to 6% CO2 for 3 days, mutants were subcultured onto antimicrobial agent-free medium. Their susceptibilities to norfloxacin (in the presence or absence of 10 μg of reserpine/ml), ciprofloxacin, sparfloxacin, moxifloxacin (BAY 12-8039), ethidium bromide, and acriflavine were determined by an agar dilution method with Iso-Sensitest agar (Oxoid) supplemented with 5% horse blood with an inoculum of 104 CFU/spot (18). Reserpine was freshly prepared before use, and media containing it were used immediately. After incubation at 35 to 37°C in 4 to 6% CO2 for 18 to 20 h, the MIC was recorded as the lowest antibiotic concentration inhibiting growth. The norfloxacin susceptibility of several strains, in the presence or absence of reserpine, was also determined by a broth MIC method (18).
Accumulation of ethidium bromide.
Measurement of the level of ethidium bromide accumulation and efflux in S. pneumoniae ATCC 49619 (parent strain) and strain 1N27 (norfloxacin-selected mutant) was based on a previously described method (5, 14, 15). Briefly, for measurement of the level of accumulation, bacterial suspensions with an optical density at 550 nm of 0.2 were prepared in uptake buffer (NaCl, 110 mM; KCl, 7 mM; NH4Cl, 50 mM; Na2HPO4, 0.4 mM; Tris base, 52 mM; glucose, 0.2% adjusted to pH 7.5 with HCl) and were then exposed to ethidium bromide at a concentration of 2 μg/ml. The increase in fluorescence as ethidium bromide entered the cells was recorded fluorometrically with a Perkin-Elmer model LS50 spectrofluorimeter (excitation λ, 530 nm; emission λ, 600 nm) at 30°C. The effect of reserpine on the level of accumulation was determined in a similar way, except that reserpine was added to the uptake buffer at a concentration of 10 μg/ml.
For determining ethidium bromide loss, bacterial suspensions were first exposed to ethidium bromide (2 μg/ml) in the presence of reserpine (10 μg/ml) for 20 min at 37°C. The cells were then pelleted by centrifugation and were resuspended in fresh uptake buffer. The loss of ethidium bromide from the cells was measured as a decrease in fluorescence.
DNA amplification and sequencing.
Chromosomal DNA was extracted from the parent strain and mutant 1N27 by a standard procedure (3). Using primers and reaction conditions described previously (16), the quinolone resistance-determining regions (QRDRs) of parC, parE, gyrA, and gyrB were amplified by PCR with chromosomal DNA used as the template. PCR products were sequenced by automated fluorescence sequencing.
Detection of efflux phenotype in clinical isolates of S. pneumoniae.
A total of 1,037 clinically significant nonreplicated strains of S. pneumoniae, which had been isolated during 1996 and 1997 from patients in the United Kingdom, were studied. Their susceptibilities to norfloxacin and ciprofloxacin were determined by an agar dilution method. Strains for which norfloxacin MICs were >8 μg/ml and ciprofloxacin MICs were >1 μg/ml were further examined for the effect of reserpine (10 μg/ml) on the activity of norfloxacin and their susceptibilities to ethidium bromide and acriflavine. The activities of several other fluoroquinolones were also determined.
RESULTS
Antimicrobial susceptibilities of fluoroquinolone-resistant mutants.
A total of 23 resistant mutants were selected with norfloxacin. For these mutants, four- to eightfold increases in the MICs of norfloxacin and ciprofloxacin compared with those for the parent strain were found (Table 1). However, the activity of norfloxacin against five of the mutants was increased four- to eightfold in the presence of reserpine. For these same five mutants the MICs of ethidium bromide and acriflavine increased four- to eightfold compared with the MICs for the parent strain. This reduction in susceptibility to ethidium bromide was also reversed by reserpine (data not shown). The activities of sparfloxacin and moxifloxacin against these mutants were unchanged. Reserpine alone did not show any inhibitory activity.
TABLE 1.
Susceptibilities of S. pneumoniae ATCC 49619 (parent strain) and norfloxacin-selected mutants to fluoroquinolones and efflux pump substrates
| Compound | Range of (median) MICs (μg/ml)
|
||
|---|---|---|---|
| Parent strain | Presumptive efflux mutants (n = 5) | Non-efflux mutants (n = 18) | |
| Norfloxacin | 4 | 16–32 (16) | 16–32 (16) |
| Norfloxacin + reserpine | 4 | 4–8 (4) | 16 |
| Ciprofloxacin | 0.5 | 2–4 (2) | 2–4 (2) |
| Sparfloxacin | 0.5 | 0.25–0.5 (0.5) | 0.5 |
| Moxifloxacin | 0.12 | 0.12–0.25 (0.12) | 0.12 |
| Ethidium bromide | 4 | 16–32 (16) | 4 |
| Acriflavine | 4 | 32 | 4 |
Sequencing of QRDRs of parC, parE, gyrA, and gyrB.
No sequence differences were found between the parent strain and the presumptive efflux mutant, 1N27, at any previously described position within the QRDRs of parC, parE, gyrA, and gyrB.
Accumulation and efflux of ethidium bromide.
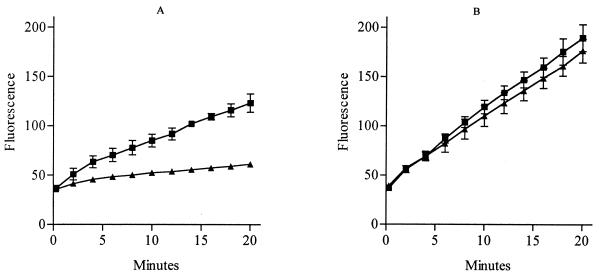
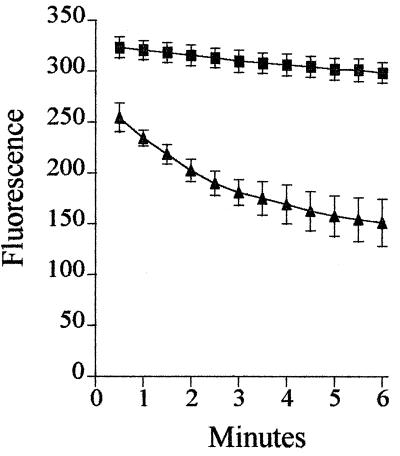
Figure 1 compares the levels of accumulation of ethidium bromide in the parent strain and mutant 1N27. The rate of accumulation in the mutant was significantly slower, and after 20 min the level of accumulation in the mutant was 69.5% less than that in the parent strain. In the presence of reserpine there was no significant difference between the levels of accumulation in the parent strain and the mutant, although the overall level of ethidium bromide accumulation increased in both strains. The rate of ethidium bromide loss from 1N27 was significantly increased compared with that from the parent strain (Fig. 2).
FIG. 1.
Accumulation of ethidium bromide by the parent strain (■) and a mutant strain, 1N27 (▴), of S. pneumoniae in the absence of reserpine (A) and in the presence of reserpine (B). Each point is the mean of at least three experiments. Bars represent standard deviations.
FIG. 2.
Efflux of ethidium bromide from the parent strain (■) and a mutant strain, 1N27 (▴), of S. pneumoniae. Each point is the mean of at least three experiments. Bars represent standard deviations.
Antimicrobial susceptibilities of the clinical isolates.
Of the 1,037 clinical isolates, 273 strains showed reduced susceptibility to norfloxacin or ciprofloxacin (MIC of norfloxacin, >8 μg/ml; MIC of ciprofloxacin, >1 μg/ml). These strains could be divided into two groups on the basis of the effect of reserpine on their susceptibility to norfloxacin. Group A contained strains for which the MIC of norfloxacin was reduced only twofold or less by reserpine (Table 2). Group B contained the 45.4% of strains for which the MIC of norfloxacin was reduced fourfold or greater by reserpine. Ninety-eight of the group B strains had susceptibility patterns identical to those of the norfloxacin-selected efflux mutants, and for the 98 group B strains the MICs of ethidium bromide and acriflavine were ≥16 μg/ml. However, the remaining 25 strains showed variations in their susceptibilities to ethidium bromide and acriflavine. For 8 strains the MICs of ethidium bromide and acriflavine were ≤8 μg/ml, for 11 strains the MICs of acriflavine were ≥16 μg/ml but the MICs of ethidium bromide were ≤8 μg/ml, and for 6 strains the MICs of ethidium bromide were ≥16 μg/ml and the MICs of acriflavine were ≤8 μg/ml.
TABLE 2.
Susceptibilities of clinical isolates of S. pneumoniae to fluoroquinolones and efflux pump substrates
| Group | Compound | MIC (μg/ml)a
|
|||
|---|---|---|---|---|---|
| 50% | 90% | Range | Geometric mean | ||
| A (n = 149) | Norfloxacin | 16 | 32 | 4–256 | 14.0 |
| Norfloxacin + reserpine | 8 | 16 | 4–128 | 7.7 | |
| Ciprofloxacin | 2 | 4 | 1–128 | 2.4 | |
| Sparfloxacin | 0.5 | 1 | 0.25–32 | 0.6 | |
| Moxifloxacin | 0.25 | 0.5 | 0.12–8 | 0.3 | |
| Ethidium bromide | 8 | 16 | 2–32 | 7.0 | |
| Acriflavine | 8 | 16 | 4–32 | 7.4 | |
| B (n = 124) | Norfloxacin | 16 | 32 | 8–128 | 18.7 |
| Norfloxacin + reserpine | 4 | 8 | 2–32 | 4.3 | |
| Ciprofloxacin | 2 | 4 | 2–8 | 2.5 | |
| Sparfloxacin | 0.5 | 0.5 | 0.25–8 | 0.4 | |
| Moxifloxacin | 0.25 | 0.25 | 0.12–0.5 | 0.2 | |
| Ethidium bromide | 16 | 32 | 4–32 | 16.6 | |
| Acriflavine | 16 | 32 | 4–64 | 17.9 | |
50% and 90%, MICs at which 50 and 90% of isolates are inhibited, respectively.
DISCUSSION
Several efflux proteins are well described in gram-positive bacteria, including Bmr in Bacillus subtilis (1) and NorA in Staphylococcus aureus (15). These can mediate low-level resistance to hydrophilic fluoroquinolones and a variety of unrelated compounds; both are inhibited by reserpine (14, 15). More recently, reserpine was shown to inhibit the level of accumulation of ethidium bromide in an ethidium bromide-selected mutant of S. pneumoniae which showed cross-resistance to fluoroquinolones (5). For this reason we used reserpine to detect mutants resistant to fluoroquinolones by an efflux mechanism.
We showed that reserpine significantly increased the activity of norfloxacin against several norfloxacin-selected mutants. These presumptive efflux mutants also showed increased levels of resistance to ethidium bromide and acriflavine, which agrees with the description by Zeller et al. (20) of the presence of an efflux pump in pneumococci. Ethidium bromide accumulation in strain 1N27, a presumptive efflux mutant, was significantly reduced compared with that in the parent strain; these differences were abolished by reserpine. Increased loss of ethidium bromide was shown from strain 1N27, suggesting the presence of an efflux mechanism of resistance. The reduced activity of norfloxacin and ciprofloxacin against strain 1N27 occurred in the absence of changes in the QRDR of topoisomerase IV or DNA gyrase. Interestingly, the level of accumulation in the parent strain in the presence of reserpine was significantly higher than that in uptake medium alone. This increase in the level of accumulation may be the result of reserpine inhibiting low levels of a constitutively expressed efflux protein in the parent strain.
We used an agar dilution method to detect the effect of reserpine on the activity of norfloxacin. Studies with Bmr have suggested that reserpine may be inactivated by agar (14). We found no difference in the effect of reserpine on the MIC of norfloxacin when MICs were determined by a broth or an agar dilution method. However, we did find that reserpine is labile and should be used immediately once it is in solution.
Having demonstrated that our agar dilution method was able to detect laboratory mutants with an efflux phenotype, we determined the prevalence of an efflux mechanism of fluoroquinolone resistance among clinical isolates of S. pneumoniae. Reserpine significantly increased the activity of norfloxacin against 45.4% of the clinical strains with reduced susceptibility to norfloxacin or ciprofloxacin. Although the majority of these strains had susceptibility patterns identical to those of the efflux mutants, 20% of the strains showed variations in their susceptibilities to ethidium bromide and acriflavine. It is possible that these strains may be resistant by further efflux mechanisms which have different substrate profiles or that mutations in the efflux pump have changed the spectrum of cross-resistance. Both the mutants and the clinical strains with the efflux phenotype remained susceptible to the more hydrophobic compounds sparfloxacin and moxifloxacin. This suggests that hydrophobic fluoroquinolones are poor substrates for the pneumococcal efflux pump, as they are for NorA (11, 19).
Most studies investigating the mechanism of fluoroquinolone resistance in clinical strains of S. pneumoniae have shown that alterations in the target sites are responsible for resistance. We have presented data that suggest that an efflux mechanism may be a common cause of clinically significant fluoroquinolone resistance. Our study is the first to screen large numbers of clinical isolates for an efflux mechanism of antibiotic resistance. Further work is required to establish the genetic basis of efflux pump resistance in pneumococci.
ACKNOWLEDGMENT
This work was supported by a grant from Bayer AG.
REFERENCES
- 1.Ahmed M, Lyass L, Markham P N, Taylor S S, Vazquez-Laslop N, Neyfakh A A. Two highly similar multidrug transporters of Bacillus subtilis whose expression is differentially regulated. J Bacteriol. 1995;177:3904–3910. doi: 10.1128/jb.177.14.3904-3910.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appelbaum P C. Antimicrobial resistance in Streptococcus pneumoniae: an overview. Clin Infect Dis. 1997;15:77–83. doi: 10.1093/clinids/15.1.77. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. [Google Scholar]
- 4.Baquero F. Pneumococcal resistance to beta-lactam antibiotics: a global geographic overview. Microb Drug Resist. 1995;1:115–121. doi: 10.1089/mdr.1995.1.115. [DOI] [PubMed] [Google Scholar]
- 5.Baranova N, Neyfakh A A. Apparent involvement of a multidrug transporter in the fluoroquinolone resistance of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:1396–1398. doi: 10.1128/aac.41.6.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barry A L, Fuchs P C, Brown S D. In vitro activities of five fluoroquinolone compounds against strains of Streptococcus pneumoniae with resistance to other antimicrobial agents. Antimicrob Agents Chemother. 1996;40:2431–2433. doi: 10.1128/aac.40.10.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brenwald N P, Gill M J, Wise R. The effect of reserpine, an inhibitor of multi-drug efflux pumps, on the in-vitro susceptibilities of fluoroquinolone-resistant strains of Streptococcus pneumoniae to norfloxacin. J Antimicrob Chemother. 1997;40:458–460. doi: 10.1093/jac/40.3.458. [DOI] [PubMed] [Google Scholar]
- 8.Brueggemann A B, Kugler K C, Doern G V. In vitro activity of BAY 12-8039, a novel 8-methoxyquinolone, compared to activities of six fluoroquinolones against Streptococcus pneumoniae, Haemophilus influenzae and Moraxella catarrhalis. Antimicrob Agents Chemother. 1997;41:1594–1597. doi: 10.1128/aac.41.7.1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doern G V, Brueggemann A, Preston Holley H, Rauch A M. Antimicrobial resistance of Streptococcus pneumoniae recovered from outpatients in the United States during the winter months of 1994 to 1995: results of a 30-center national surveillance study. Antimicrob Agents Chemother. 1996;40:1208–1213. doi: 10.1128/aac.40.5.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janoir C, Zeller V, Kitzis M, Moreau N J, Gutmann L. High-level fluoroquinolone resistance in Streptococcus pneumoniae requires mutations in parC and gyrA. Antimicrob Agents Chemother. 1996;40:2760–2764. doi: 10.1128/aac.40.12.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaatz G W, Seo S M, Ruble C A. Efflux-mediated fluoroquinolone resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1993;37:1086–1094. doi: 10.1128/aac.37.5.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klyachko K A, Schuldiner S, Neyfakh A A. Mutations affecting substrate specificity of the Bacillus subtilis multidrug transporter Bmr. J Bacteriol. 1997;179:2189–2193. doi: 10.1128/jb.179.7.2189-2193.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munoz R, De La Campa A G. ParC subunit of DNA topoisomerase IV of Streptococcus pneumoniae is a primary target of fluoroquinolones and cooperates with DNA gyrase A subunit in forming resistance phenotype. Antimicrob Agents Chemother. 1996;40:2252–2257. doi: 10.1128/aac.40.10.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neyfakh A A, Bidnenko V E, Chen L B. Efflux-mediated multidrug resistance in Bacillus subtilis: similarities and dissimilarities with the mammalian system. Proc Natl Acad Sci USA. 1991;88:4781–4785. doi: 10.1073/pnas.88.11.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neyfakh A A, Borsch C M, Kaatz G W. Fluoroquinolone resistance protein NorA of Staphylococcus aureus is a multidrug efflux transporter. Antimicrob Agents Chemother. 1993;37:128–129. doi: 10.1128/aac.37.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan X, Ambler J, Mehtar S, Fisher L M. Involvement of topoisomerase IV and DNA gyrase as ciprofloxacin targets in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1996;40:2321–2326. doi: 10.1128/aac.40.10.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perichon B, Tankovic J, Courvalin P. Characterization of a mutant in the parE gene that confers fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:1166–1167. doi: 10.1128/aac.41.5.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Working Party of the British Society for Antimicrobial Chemotherapy. 1991. A guide to sensitivity testing. J. Antimicrob. Chemother 27(Suppl. D):22–30. [PubMed]
- 19.Yoshida H, Bogaki M, Nakamura S, Ubukata K, Konno M. Nucleotide sequence and characterization of the Staphylococcus aureus norA gene, which confers resistance to quinolones. J Bacteriol. 1990;172:6942–6949. doi: 10.1128/jb.172.12.6942-6949.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeller V, Janoir C, Kitzis M, Gutmann L, Moreau N J. Active efflux as a mechanism of resistance to ciprofloxacin in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:1973–1978. doi: 10.1128/aac.41.9.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]