Abstract
Background: The diagnostic performance of numerous clinical specimens to diagnose COVID-19 through RT-PCR techniques is very important, and the test result outcome is still unclear. This review aimed to analyze the diagnostic performance of clinical samples for COVID-19 detection by RT-PCR through a systematic literature review process. Methodology: A compressive literature search was performed in PubMed/Medline, Scopus, Embase, and Cochrane Library from inception to November 2022. A snowball search on Google, Google Scholar, Research Gate, and MedRxiv, as well as bibliographic research, was performed to identify any other relevant articles. Observational studies that assessed the clinical usefulness of the RT-PCR technique in different human samples for the detection or screening of COVID-19 among patients or patient samples were considered for this review. The primary outcomes considered were sensitivity and specificity, while parameters such as positive predictive value (PPV), negative predictive value (NPV), and kappa coefficient were considered secondary outcomes. Results: A total of 85 studies out of 10,213 non-duplicate records were included for the systematic review, of which 69 articles were considered for the meta-analysis. The meta-analysis indicated better pooled sensitivity with the nasopharyngeal swab (NPS) than saliva (91.06% vs. 76.70%) and was comparable with the combined NPS/oropharyngeal swab (OPS; 92%). Nevertheless, specificity was observed to be better with saliva (98.27%) than the combined NPS/OPS (98.08%) and NPS (95.57%). The other parameters were comparable among different samples. The respiratory samples and throat samples showed a promising result relative to other specimens. The sensitivity and specificity of samples such as nasopharyngeal swabs, saliva, combined nasopharyngeal/oropharyngeal, respiratory, sputum, broncho aspirate, throat swab, gargle, serum, and the mixed sample were found to be 91.06%, 76.70%, 92.00%, 99.44%, 86%, 96%, 94.4%, 95.3%, 73.63%, and above 98; and 95.57%, 98.27%, 98.08%, 100%, 37%, 100%, 100%, 97.6%, and above 97, respectively. Conclusions: NPS was observed to have relatively better sensitivity, but not specificity when compared with other clinical specimens. Head-to-head comparisons between the different samples and the time of sample collection are warranted to strengthen this evidence.
Keywords: COVID-19, diagnosis, sensitivity, specificity
1. Introduction
The recent global pandemic was caused by a respiratory tract infection in the Wuhan province of China in December 2019. The causative organism was recognized as a severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The coronavirus disease 2019 (COVID-19) has spread across the world and contributed to many deaths in a huge proportion of the population. Fast and accurate detection of viruses and/or diseases is essential to controlling the sources of infection and having a better patient outcome through inhibiting disease progression [1,2].
According to the Foundation of Innovative New Diagnostics in collaboration with the WHO, the sensitivity and specificity of several available kits for molecular detection of SARS-CoV-2 by the PCR technique are around 92–100% and 98–100%, respectively [3]. However, the diagnostic accuracy of RT-PCR was studied in several reports, and it is less than the standard optimum value (100%) for an ideal diagnostic biomarker [4]. The RT-PCR showed false negative results in 3% of 167 confirmed cases of COVID-19 by chest CT typical criteria, which turned positive after repeating RT-PCR testing at an interval of about 5.0 ± 2.7 days [5]. Likewise, on a larger cohort of 1014 suspected COVID-19 patients, 88% of all studied cases showed positive chest CT findings of COVID-19, while only 59% had positive RT-PCR testing. Remarkably, 93% of that cohort turned into positive RT-PCR results within 5.1 ± 1.5 days after preliminarily being negatively tested, although they showed suggestive chest CT findings of COVID-19 [6]. In fact, RT-PCR detection is dependent on viral load, so it may show initial negative results during the incubation period, especially when the viral load is low [7].
According to experts, the results of real-time RT-PCR tests must be cautiously interpreted, along with the suggestive clinical presentations. Repeated tests can be considered when the clinical presentations resemble the diagnostic criteria of COVID-19 and the test is negative. A combination of objective evidence such as chest CT, C-reactive protein, and d-dimer, along with RT-PCR, could help in better patient management and outcomes [2].
The literature indicates that saliva is superior to the nasopharyngeal swab (NPS) for the detection of SARS-CoV-2, whereas other research evidence suggests that NPS may be more suitable than the oropharyngeal swab (OPS) for the detection of COVID-19 through RT-PCR. Hence, identifying the most suitable sample for the detection of disease, especially in the case of a pandemic, is crucial [8,9,10].
Although RT-PCR is considered the gold standard for COVID-19 diagnosis and is mandatory in our daily lives, there is variable evidence on the clinical performance when screening among various samples [11,12].
We, therefore, aimed to identify all the currently available literature and assess the clinical usefulness of RT-PCR in different COVID-19 samples through a comprehensive systematic literature review process and meta-analysis.
2. Materials and Methods
We followed pre-defined inclusion and exclusion criteria for selecting the studies in this review and adapted the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) Guidelines to report this systematic review [13]. The protocol for this meta-analysis is submitted to The International Prospective Register of Systematic Reviews (PROSPERO) with a registration ID of CRD42023449573.
2.1. Criteria for Considering the Studies for This Review
The observational studies assessed the clinical usefulness of the RT-PCR technique in various human samples for the detection or screening of COVID-19 among the patients or patient samples that were considered for this review. Only the studies with full-text availability in the English language were considered. Studies comparing the numerous samples were also considered for this review. The primary outcomes considered were sensitivity and specificity, while parameters such as positive predictive value (PPV), negative predictive value (NPV), and kappa coefficient were considered secondary outcomes. We have considered all types of RT-PCR techniques in our review, per the author’s discretion. Any studies that used RT-PCR as a reference to assess the performance of other screening techniques were excluded. Studies such as reviews, descriptive studies, non-clinical studies, non-COVID-19 participants, commentary, guidelines, and qualitative analyses were excluded.
2.2. Search Methods for Identification of Studies
PubMed/Medline, Scopus, Embase, and Cochrane Library) were accessed through a comprehensive search strategy using all the possible keywords and entry terms from inception to November 2022. We also performed a snowball search on Google, Google Scholar Research Gate, and MedRxiv to identify any relevant articles. The reference lists of potential articles were also screened to identify additional potentially relevant citations. A detailed search strategy in various databases is provided in Supplementary File S1.
2.3. Study Selection
All the identified records through a database literature search were retrieved in an Excel sheet and screened against the pre-defined criteria. The studies were screened by reading the title and abstracts in the initial stage, followed by the full text. Only the studies passing these two stages were considered for final inclusion in the review. Two independent reviewers were involved in the study selection to limit bias, and discrepancies were resolved through consensus or discussion with another member of the research team.
2.4. Data Extraction
The data were extracted to a well-defined data extraction form by two independent reviewers. The author’s first name and year of publication were used to identify the studies. The study detailed information such as year, country, study design, and study settings; the participants’ information including the total number of samples/participants, age and gender of cohort, and clinical presentation or characteristics; type of specimen; and the characteristics of RT-PCR techniques were captured from the studies. The outcomes were collected from the studies or calculated from the available data in terms of percentage with a 95% confidence interval. The highest values of primary and secondary outcomes were captured in the case of multiple RT-PCR kits used in the same study. Two independent reviewers were involved in the data extraction, and disagreements were resolved through discussion or consultation with another reviewer.
2.5. Evidence Synthesis and Meta-Analysis
All the evidence extracted through the systematic process was summarized narratively and presented in tabular form. The studies that have sufficient homogenous data or if there is a sufficient number of studies to perform meta-analysis were only considered for meta-analysis. Review Manager 5.4 was used to conduct the meta-analysis [14]. The available data were converted into percentage and standard error and presented as pooled outcomes with a 95% confidence interval. We used the random effect model, as there was substantial heterogeneity (I2 > 50%; p < 0.10) in all analyses.
2.6. Publication Bias and Sensitivity Analysis
The visual inspection of the funnel plot for the sensitivity of RT-PCR in COVID-19 diagnosis was used to check publication bias using RevMan 5.4, which was further assessed for statistical significance with Egger’s and Begg’s test using comprehensive meta-analysis (trial version). A probability of less than 0.05 was considered to be statistically significant [15,16]. The sensitivity analysis was performed to check the robustness of the findings by removing the study with the lowest weight in the analysis [17].
3. Results
3.1. Study Selection Process
A total of 32,006 records were identified from literature sources and 10,213 records were screened by title and abstract following duplicate removal. A total of 8265 records (animal studies and case reports: 403; non-diagnostic and treatments: 303; guidelines and protocols: 42; non-English: 72; not RT-PCR: 6925; pediatric: 315; qualitative research and reviews: 205) were excluded at this stage, and the remaining 1948 full texts were considered for their eligibility. Following the exclusion of 1863 articles with numerous reasons (animal studies and case reports: 78; duplicate: 1; not outcome of interest: 1248; non-English: 43; not RT-PCR: 398; review: 95), 85 studies [18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102] were considered for this systematic review. Hence, a total of 69 articles with homogenous data were used for the meta-analysis. A detailed description of the study selection process is depicted in Figure 1.
Figure 1.
The PRISMA flow diagram for study selection.
3.2. Study Characteristics
The studies were published between the years 2020 and 2022 from different parts of the world with a major contribution from the USA, the UK, and India. The studies were observational making them retrospective, prospective, and cross-sectional in nature. The studies were from hospital settings or sample collection centers. The human samples were analyzed across the included studies. The majority of the studies included adult participants with an average age of 18 to 65 years. The participants were asymptomatic or symptomatic, severe or non-severe, and positive or negative at the time of sample collection. A detailed description of the studies and participant characteristics are provided in Table 1.
Table 1.
Characteristics of the included studies and patients.
| Study ID, Year, Country | Study Design | Study Settings | Total Number of Samples/ Participants |
Age of Cohort |
Male/ Female |
Clinical Presentation/ Characteristics |
|---|---|---|---|---|---|---|
| Escobar et al., 2021; Chile [18] | Cohort selection of a cross-sectional study | Multi-specialized Guillermo Grant Benavente Hospital (HGGB) and three Family Health Centers (FHCs) in the Chilean city of Concepción | 127 saliva and 127 NPS | <18: 5; 18–34: 69; 35–50: 29; 51–65: 20; >65: 4 | 68/59 | Symptomatic: 111; Asymptomatic:15; No information: 1; RT-PCR positive: 104; RT-PCR negative: 150 |
| Singh et al., 2021; India [19] | Quality audit study | Medical college institution | 92 samples and 60 controls | NR | NR | RT-PCR positive: 92; Healthy individuals: 30; Other respiratory disease: 30 |
| Figueroa et al., 2021; Ecuador [20] | Case–control study | NR | 242 clinical specimens and 11 negative controls | NR | NR | 122 SARS-CoV-2-positive and 120 SARS-CoV-2-negative |
| LeGof et al., 2021; France [21] | Prospective observational study | Two community COVID-19 screening centers | 1718 | 37 (26–52) a | 774/944 | +NPS RT-PCR: 117; Symptomatic: 530 |
| Villota et al., 2021; Ecuador [22] | Cross-sectional study | Two centers from Ecuador and USA | 192 clinical samples (NPS: 132; sputum: 60) | NR | NR | Positive: 142; Negative: 50 |
| De Pace et al., 2021; Italy [23] | Consecutive prospective observational study | Intensive Care Units of San Martino Hospital (Genoa, Italy |
75 patients | 65 (31–81) b | 56/19 | BAS: 43 (57.3%); Negative: 30.2%; Positive: 69.8%; BAL: 32 (42.7%); Negative: 37.5%; Positive: 62.5% |
| Kanwar et al., 2021; USA [24] | Prospective salvage sample study | University of Kansas Health System (TUKHS) | 201 samples | 57 (15–92) a | 103/98 | Positive: 99; Negative: 102 |
| Michel et al., 2021; Germany [25] | NR | Robert Koch Institute | 424 specimens | NR | NR | Positive: 424 |
| Wu et al., 2021; China [26] | Cross-sectional study | Shenzhen Third People’s Hospital and a compulsory quarantine facility | 52 (throat: 30; nasal: 7; NPS: 7; sputum: 8 | NR | NR | Positive: 26; Negative: 26 |
| Lee et al., 2021; UK [27] | Prospective, multi-center, cohort study | Secondary and tertiary care hospitals in Scotland |
1368 patients with 3822 tests | 68 (53–80) b | 731/637 | Confirmed positive: 496 |
| Borkakoty et al., 2021; India [28] | NR | State of Assam | 240 random samples | NR | NR | Positive: 120; Negative: 120 |
| Hata et al., 2021; USA [29] | NR | Mayo Clinic | 135 participants | 20–83 c | NR | Positive: 28; Negative: 106 |
| Wang et al., 2020; China [30] | NR | The Second Xiangya Hospital | 242 samples | NR | NR | Positive: 42 (34 throat swabs and 8 fecal samples); Negative: 200 |
| Mollaei et al., 2020; Iran [31] | NR | Kerman Reference Laboratory | 30 infected patients | NR | NR | Varies based on the gene chosen |
| Pierri et al., 2022; Italy [32] | Post-analysis of a GENCOVID study | GENCOVID people in direct contact with positive patients from the Campania region, Italy | 258 samples | NR | NR | Positive: 164 |
| Torres et al., 2021; Ecuador [33] | Descriptive-correlating, retrospective, cross-sectional study |
Santo Domingo General Hospital (Santo Domingo de los Tsáchilas, Ecuador) | 773 samples | 1–14 years: 38; 15–19 years: 33; 20–49 years: 461; 50–64 years: 127; >65 years: 74 | 344/389 | Symptomatic: 515; Asymptomatic: 218 |
| Pearson et al., 2021; Canada [34] | NR | MSH/UHN clinical diagnostics lab | 59 samples | NR | NR | Positive: 29; Negative: 30 |
| Kriegova et al., 2021; Czech Republic [35] | Large prospective cohort | University Hospital Olomouc and Sumperk Hospital, Czechia | 1038 subjects | NR | NR | Positive: 297; Negative 741 |
| Onyilagha et al., 2021; Canada [36] | Cross-sectional study | NR | 90 samples | NR | NR | Negative: 40; Positive: 50 |
| Desmet et al., 2021; Belgium [37] | Prospective observational study | Ghent University Hospital | 36 patients | 61 (22–90) b | 21/25 | NP or OP/N positive: 35; Combined positive: 31; Mild: 7; Moderate: 10; Severe: 13; Critical care: 5; Pre-symptomatic: 1 |
| Kanji et al., 2021; Canada [38] | Prospective cross-sectional study | Province of Alberta, Canada | 49 patients | 72 (25–97) b | 15/34 | Positive: 49; Negative: 52 |
| Gómez-Romero et al., 2021; Mexico [39] | Prospective database study | Epidemiology department of the Health Ministry of the State of Morelos (Secretaría de Salud Morelos, SSM) |
140 healthcare workers/sample | NR | NR | Positive: 36; Negative: 104 |
| Milosevic et al., 2021; United States [40] | Prospective cohort study | Penn State Health Milton S. Hershey Medical Center | 60 samples | NR | NR | Positive: 30; Negative: 30 |
| Pekosz et al., 2021; United States [41] | Prospective cohort study | FDA EUA study samples which occurred across 21 geographically diverse study sites | 251 sample | Symptomatic: 251 | ||
| Ferreira et al., 2021; Brazil [42] | Prospective cohort study | State of Rio de Janeiro and the state of Ceará | 65 patients | NR | NR | NPS: 42; Serum: 12; Saliva: 11; Positive: 51; Negative: 14 |
| Dumaresq et al., 2021; Canada [43] | Prospective cohort study (SPRING study) | Département de microbiologie et d’infectiologie du centre hospitalier universitaire | 2010 sample from 987 patients | 40 (6–91) a | NR | 1005 ONPS and 1005 gargles; Symptomatic: 987; Asymptomatic: 987 |
| Morecchiato et al., 2021; Italy [44] | Prospective cohort study (SPRING study) | Microbiology and Virology Unit of Florence Careggi University Hospital (Florence, Italy) | 139 samples | NR | NR | Positive: 96; Negative: 43 |
| Olearo et al., 2021; Germany [45] | Cross-sectional retrospective study | University Hospital Hamburg. | 7513 HCWs (55,122 samples); 11,192 sample pools | NR | NR | Negative: 11,041; Invalid: 82; Positive: 69 |
| Ghoshal et al., 2021; India [46] | Retrospective observational study | Triage of a dedicated COVID-19 tertiary care center with 180 beds including 30 ICU ventilator beds |
1807 patients | NR | NR | RT-PCR positive; 174; TrueNat: 174 |
| Balaska et al., 2021; Greece [47] | Prospective observational study | AHEPA University Hospital, Thessaloniki | 420 pairs of samples | 44.7 (13) a | 161/259 | Positive diagnostic sample: 27.7%; Screening sample: 5% |
| Watanabe et al., 2021; Japan [48] | NR | Kawasaki Rinko General Hospital and the Matsudo City General Hospital | 96 patients | 49.3 (27.8) a | 45/51 | Positive: 20; Negative: 76 |
| Domnich et al., 2021; Italy [49] | Prospective observational study | San Martino Policlinico Hospital (Genoa, Liguria, Northwest Italy) | 98 samples | NR | NR | Positive: 98 |
| Kim et al., 2021; South Korea [50] | NR | Kyungpook National University | 300 samples | NR | NR | Positive: 260; Negative: 40 |
| Carvalho et al., 2021; Brazil [51] | NR | Municipal medical service | 346 samples | NR | NR | Detectable: 194; Undetectable: 152 |
| Kritikos et al., 2021; Switzerland [52] | Prospective observational study | Tertiary university hospital in Lausanne, Switzerland | 58 patients | 70 (61–77) b | 45/13 | Symptomatic: 49 |
| Brotons et al., 2021; Spain [53] | Three-phase cross-sectional study | Molecular Microbiology Department of Sant Joan de Déu Hospital | 183 samples | NR | NR | Positive: 10; Negative: 173 |
| Laverack et al., 2021; USA [54] | NR | Cornell COVID-19 Testing Laboratory by three other COVID-19 testing laboratories in the United States | 225 samples | NR | NR | NPS: 201; AN: 24; NPS positive: 100; Negative: 101; AN positive: 12; AN negative: 12 |
| Avetyan et al., 2021; Armenia [55] | Cross-sectional study | Institute of Molecular Biology, National Academy of Sciences | NPS: 74; RNA sample: 196 | NR | NR | NPS: Positive: 44; Negative: 30; RNA sample positive: 196 |
| Hernandez et al., USA; 2021 [56] | NR | Clinical Microbiology Laboratory at the Mount Sinai Health System | 60 patients | NR | NR | NR |
| Hernández et al., 2021; Colombia [57] | NR | Not reported | 94 samples | Positive: 49; Negative: 45 | ||
| Leber et al., 2021; UK [58] | Prospective cohort study | GP participating in the National Influenza Surveillance Network in the ski resort of Schladming-Dachstein | 66 patients | NR | NR | Positive: 22; Negative: 44 |
| Gadkar et al., 2021; Canada [59] | NR | Microbiology and virology laboratories of BC Children’s Hospital | 372 samples | NR | NR | Positive: 142 |
| Bruno et al., 2021; Ecuador [60] | NR | INSPI and UDLA | 1036 samples | NR | NR | Positive: 543; Negative: 493 |
| Sun et al., 2021; France [61] | Single center, retrospective, observational study | Radiation therapy department, Gustave Roussy, Paris-Saclay University | 480 patients | 62 (50–70) b | 228/252 | Positive: 26; Negative: 446 |
| Rigo et al., 2021; Pordenone [62] | NR | Microbiology and Virology Department Laboratory | 180 samples | NR | NR | Positive: 93; Negative: 88 |
| Banko et al., 2021; Serbia [63] | NR | Laboratory of Molecular Microbiology, Institute for Biocides and Medical Ecology, Belgrade |
354 samples | NR | NR | Sansure Biotech: Positive: 190; Negative: 164 GeneFinderTM: Positive: 176; Negative: 178 TaqPathTM: Positive: 178; Negative: 176 |
| Tastanova et al., 2021; Switzerland [64] | NR | University Hospital Zurich and at ADMed Laboratory in La Chaux-de-Fonds, Switzerland |
184 samples | NR | NR | Positive: 92; Negative: 92 |
| Noor et al., 2021; Bangladesh [65] | Case–control sample study | Department of Biochemistry and Molecular Biology | 240 samples | NR | NR | Positive: 120; Negative: 120 |
| Fitoussi et al., 2021; France [66] | Prospective observational study | entre Cardiologique du Nord-CCN, Saint-Denis, France | 239 patients | NR | NR | Positive: 140; Negative: 99 |
| Freire-Paspuel et al., 2020; Ecuador [67] | Prospective observational study | Laboratory of “Universidad de Las Américas” in Quito (Ecuador) | 89 samples | NR | NR | Positive: 57; Negative: 32 |
| Dierks et al., 2021; Germany [68] | NR | University Medical Center Göttingen | 322 samples | NR | NR | Positive: 21; Negative: 301 |
| Nakura et al., 2021; Japan [69] | NR | Osaka Women’s and Children’s Hospital, Osaka Habikino Medical Center, and Osaka General Medical Center of the Osaka Prefectural Hospital | 213 samples | NR | NR | Sputum: 35; NPS: 124; Saliva: 7 |
| Stockdale et al., 2021; UK [70] | NR | Liverpool University Hospitals NHS Foundation Trust | 429 patients | 67 (55–78) b | 257/172 | Positive: 293; Negative: 136 |
| Kortela et al., 2021; Finland [71] | Population-based retrospective study | Helsinki Capital Region, Finland | 3008 patients | 52.5 (19.7) a; 51 (36–69) b | 1215/1794 | Not suspected: 514; Not excluded: 1318; High suspicion: 516; Laboratory confirmed: 574; Not known: 86; Positive: 585; Negative: 2246 |
| Altamimi et al., 2021; Saudi Arabia [72] | NR | Saudi Center for Disease Prevention and Control (SCDC) Laboratories | 94 samples | NR | NR | Positive: 63; Negative: 31 |
| Visseaux et al., 2021; France [73] | NR | Virology Laboratory of Bichat-Claude Bernard University Hospital, Paris, France | 94 samples | NR | NR | Positive: 69; Negative: 25 |
| Cassinari et al., 2021; France [74] | Prospective observational study | Rouen University Hospital | 130 patients | NR | NR | Positive: 13; Negative: 117 |
| Carrillo et al., 2021; Manila [75] | Prospective cross-sectional diagnostic accuracy study | Philippine General Hospital | 197 patients | 32 (22–64) | 74/123 | Positive: 18; Negative: 179 |
| Girish et al., 2021; India [76] | Cross-sectional, analytical study | BJ Medical College and Civil Hospital | 309 patients | NR | NR | Positive: 55; Negative: 254 |
| Freire-Paspuel et al., 2021; Ecuador [77] | NR | NR | 97 samples | NR | NR | Positive: 43; Negative: 54 |
| Dong et al., 2021; China [78] | NR | Hospitalized patients or close contacts of hospitalized patients tested by Beijing CDC (BJCDC), Wuhan CDC (WHCDC), and a government-designated clinical test laboratory | 196 samples | NR | NR | Febrile suspected patients: 103; Close contacts: 77; Convalescents: 16; Positive: 132; Negative: 64 |
| Gupta-Wright et al., 2021; UK [79] | Retrospective cohort study | Two hospitals within an acute NHS Trust in London, UK | 4008 patients | 69 (56–81) b* | 1142/651 * | Non-COVID-19: 2215; COVID-19 diagnosis: 1793; Positive: 1391; Negative: 283 |
| Dimke et al., 2021; Denmark [80] | NR | Department of Clinical Microbiology, Odense University Hospital | 87 samples | NR | NR | Positive: 57; Negative: 30 |
| Alaifan et al., 2021; Saudi Arabia [81] | NR | Diagnostic laboratories at the Saudi Center for Diseases Control and Prevention | 185 samples | NR | NR | Positive: 121; Negative: 64 |
| Onwuamah et al., 2021; Nigeria [82] | Retrospective study | The Nigerian Institute of Medical Research from people living in Lagos, Nigeria | 63 samples | NR | NR | Positive: 48; Negative: 15 |
| Price et al., 2021; USA [83] | Prospective observational study | University of California, Los Angeles Health System | 10,165 samples from 8948 patients | NR | NR | NPS: 10,215; Bronchoalveolar lavage: 121; Expectorated sputum: 22; Miscellaneous sample types: 35; Positive: 630; Negative: 9535 |
| Trobajo-Sanmartín et al., 2021; Spain [84] | Prospective study | Clinical microbiology department of the Navarra Hospital Complex | 674 pairs of samples (NP and saliva) | 36 (19) b | 300/374 | Positive: 337; Negative: 337; Symptomatic: 333; Non-symptomatic: 341 |
| Omar et al., 2021; South Africa [85] | Retrospective descriptive cross-sectional study | Data from the mobile COVID-19 PCR testing laboratory database and the non-COVID-19 ICU database |
315 samples from 1032 patients | 40 (20.4) a | 551/481 | NPS: 281 Nasal swab: 17; OPS: 1; Tracheal respirate: 7; Not specified: 13; Positive: 51; Negative: 264 |
| Bergevin et al., 2021; Canada [86] | Prospective evaluation | Laval region of Quebec, Canada | 773 pairs | Positive: 44 (31–58) b | Positive: 80/85 | Positive: 165 (symptomatic: 148; asymptomatic: 17) |
| Yip et al., 2021; China [87] | NR | The University of Hong Kong-Shenzhen Hospital | 296 samples | NR | NR | Positive: 105; Negative: 191 |
| Renzoni et al., 2021; Switzerland [88] | Retrospective analysis | Geneva University Hospitals | 61 samples | NR | NR | Positive: 61; Control: 16 |
| Tsujimoto et al., 2021; Japan [89] | Single-center, prospective study | National Centre for Global Health and Medicine (Tokyo, Japan) | 10 patients (57 sets of NPS, NS, and SS samples) | 47 (30–70) b | 2/8 | Positive: 48; Negative: 9 |
| Mio et al., 2021; Italy [90] | NR | Department of Laboratory Medicine, University Hospital of Udine, Italy | 30 patient samples | NR | NR | Positive: 19; Negative: 11 |
| Lau et al., 2021; Malaysia [91] | NR | Hospital Sungai Buloh, Malaysia | 113 samples | NR | NR | Positive: 78; Negative: 35 |
| Shen et al., 2021; China [92] | NR | Beijing Center for Disease Prevention and Control (BJCDC) | 142 samples | NR | NR | Kit I: Positive: 130; Negative: 12; Kit II: Positive: 116; Negative: 26; Kit III: Positive: 114; Negative: 28; Kit IV: Positive: 129; Negative: 13 |
| Freire-Paspuel et al., (B) 2020; Ecuador [93] | NR | Laboratory of “Universidad de Las Américas” in Quito (Ecuador) | 48 samples | NR | NR | Positive: 30; Negative: 18 |
| Guo et al., 2020; China [94] | NR | Three centers in China | 500 subjects | 0.75–93c | 258/242 | Positive: 242; Negative: 258; OPS: 395; Sputum: 167 |
| Lu et al., 2020; China [95] | NR | Liuzhou People’s Hospital | 118 patients | Cases: 35.94 (16.32); Control: 36.50 (19.93) a | 72/46 | COVD-19: 18; Control: 100 |
| Martín Ramírez et al., 2022; Spain [96] | Retrospective cohort study | Princesa University Hospital | 303 patients | Pre-pandemic control: 73.5 (62.5–85.5); Pandemic control: 69 (62–83); Positive: 64 (56–72) b | Pre-pandemic control: 25/25; Pandemic control: 32/18; positive: 139/64 | Positive: 203; Pre-pandemic control: 50; Pandemic control: 50; |
| Yang et al., 2022; China [97] | NR | Department of Laboratory Medicine, Shengjing Hospital of China Medical University, | 63 samples | NR | NR | Positive: 28; Negative: 35 |
| Sahoo et al., 2021; India [98] | Cross-sectional observational study | Department of Microbiology, ABVIMS, and Dr. RML Hospital | 500 | NR | NR | Positive: 49; Negative: 451 |
| Hofman et al., 2021; France [99] | Prospective cohort study | Downtown free screening centers available to the population of the Nice metropolitan area and the outpatient clinic of the Department of Pulmonary Medicine of the University Hospital of Nice | 112 samples/subjects | 40 (15) b | 69/43 | Positive: 45; Negative: 67 |
| Jamal et al., 2020; Canada [100] | Population-based surveillance of consecutive patients | Six Toronto Invasive Bacterial Disease Network | 91 patients | 66 (23–106) b | 52/39 | Positive: 72; Negative: 19 |
| Ngaba et al., 2021; Cameroon [101] | Cross-sectional and comparative study | Douala Gynaeco-Obstetrics and Pediatric Hospital molecular biology laboratory | 1810 patients | 0–71 + c | 1226/559 | NPS: 1736; Saliva: 2; Throat swab: 1; Positive: 35; Negative: 1775 |
| Procop et al., 2020; USA [102] | NR | Cleveland Clinic | 239 samples | 49.28 (16.86) a | NR | Positive: 168; Negative: 71 |
AN: anterior nares; HCW: healthcare worker; ICMR: Indian Council for Medical Research; INSPI: Instituto Nacional de Salud Pública e Investigación Leopoldo Izquieta Pérez; NIV: National Institute of Virology; NPS: nasopharyngeal swab; NR: not reported; UDLA: Universidad de Las Américas. a indicates mean; b indicates median; c indicates range. * indicates COVID-19-diagnosed patients.
3.2.1. Characteristics of RT-PCR Techniques
Many human samples such as the nasopharyngeal swab, oropharyngeal swab, respiratory tract specimens (bronchoalveolar lavage and broncho aspirates), throat, nasal, saliva, sputum, fecal, gargle, or mixed were used for the detection of COVID-19 using various RT-PCR techniques. The samples were stored at a cool temperature ranging from 40 °C to −800 °C. Many in-house and modified RT-PCR techniques were used by the studies by numerous companies. Detailed information on the RT-PCR techniques is depicted in Supplementary File S2.
3.2.2. The Diagnostic Parameters of RT-PCR in Various Samples
Nasopharyngeal Swabs
Sensitivity and specificity
A meta-analysis of 43 studies indicated a pooled sensitivity of 91.06% (95%CI: 88.91 to 93.21; I2: 100%) for nasopharyngeal swabs using RT-PCR techniques. (Figure 2A). A meta-analysis of 37 studies indicated a pooled specificity of 95.57% (95%CI: 95.19 to 95.96; I2: 100%) for nasopharyngeal swabs using different RT-PCR techniques. (Figure 2B).
Figure 2.
The diagnostic parameters of RT-PCR in the nasopharyngeal samples. (A): Pooled Sensitivity; (B): Pooled Specificity; (C): Pooled PPV and NPV; (D): Pooled kappa coefficient.
PPV and NPV
A meta-analysis of 15 studies indicated a pooled PPV of 95.88% (95%CI: 91.59 to 100.16; I2: 100%) for nasopharyngeal swabs using numerous RT-PCR techniques. (Figure 2C). A meta-analysis of 15 studies indicated a pooled NPV of 91.58% (95%CI: 88.03 to 95.13; I2: 100%) for nasopharyngeal swabs using RT-PCR techniques. (Figure 2C).
Kappa coefficient
A meta-analysis of 16 studies indicated a pooled kappa coefficient of 0.79 (95%CI: 0.71 to 0.87; I2: 98%) for nasopharyngeal swabs using RT-PCR techniques. (Figure 2D). The diagnostic parameters of RT-PCR in the nasopharyngeal samples are provided in Figure 2.
3.3. Saliva Samples
3.3.1. Sensitivity and Specificity
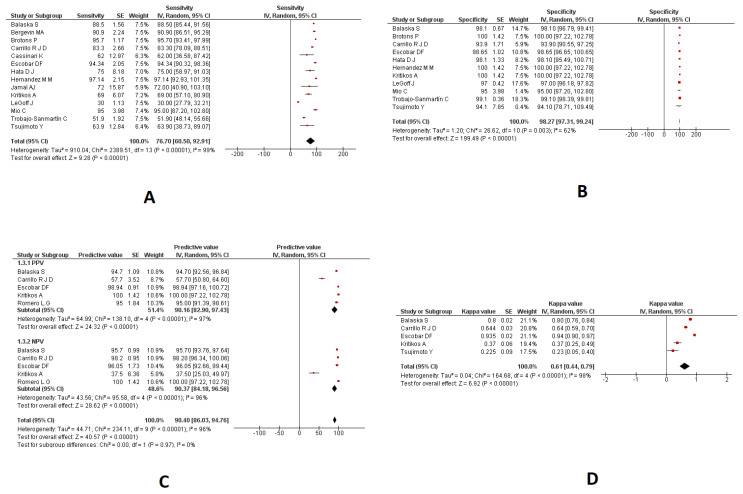
A meta-analysis of 14 studies indicated a pooled sensitivity of 76.70% (95%CI: 60.50 to 92.91; I2: 99%) in the saliva samples using RT-PCR techniques (Figure 3A). A meta-analysis of 11 studies indicated a pooled specificity of 98.27% (95%CI: 97.31 to 99.24; I2: 62%) in the saliva samples using various RT-PCR techniques (Figure 3B).
Figure 3.
The diagnostic parameters of RT-PCR in saliva samples. (A): Pooled Sensitivity; (B): Pooled Specificity; (C): Pooled PPV and NPV; (D): Pooled kappa coefficient.
3.3.2. PPV and NPV
A meta-analysis of five studies indicated a pooled PPV of 90.16% (95%CI: 82.90 to 97.43; I2: 97%) in the saliva samples using RT-PCR techniques (Figure 3C). A meta-analysis of five studies indicated a pooled NPV of 90.37% (95%CI: 84.18 to 96.56; I2: 96%) in the saliva samples using RT-PCR techniques (Figure 3C).
3.3.3. Kappa Coefficient
A meta-analysis of 5 studies indicated a pooled kappa coefficient of 0.61 (95%CI: 0.44 to 0.79; I2: 98%) in the saliva samples using different RT-PCR techniques. (Figure 3D). The diagnostic parameters of RT-PCR in the saliva samples are provided in Figure 3.
3.4. Combined Nasopharyngeal/Oropharyngeal Samples
3.4.1. Sensitivity and Specificity
A meta-analysis of 16 studies indicated a pooled sensitivity of 92.00% (95%CI: 87.57 to 96.43; I2: 100%) in the combined nasopharyngeal/oropharyngeal samples using different RT-PCR techniques (Figure 4A). A meta-analysis of 12 studies indicated a pooled specificity of 98.08% (95%CI: 96.64 to 99.52; I2: 100%) in the combined nasopharyngeal/oropharyngeal samples using RT-PCR techniques (Figure 4B).
Figure 4.
The diagnostic parameters of RT-PCR in the combined nasopharyngeal/oropharyngeal samples. (A): Pooled Sensitivity; (B): Pooled Specificity; (C): Pooled PPV and NPV; (D): Pooled kappa coefficient.
3.4.2. PPV and NPV
A meta-analysis of five studies indicated a pooled PPV of 84.63% (95%CI: 70.14 to 99.12; I2: 100%) in the combined nasopharyngeal/oropharyngeal samples using RT-PCR techniques (Figure 4C). A meta-analysis of five studies indicated a pooled NPV of 96.12% (95%CI: 92.83 to 99.42; I2: 100%) in the combined nasopharyngeal/oropharyngeal samples using RT-PCR techniques (Figure 4C).
3.4.3. Kappa Coefficient
A meta-analysis of five studies indicated a pooled kappa coefficient of 0.82 (95%CI: 0.67 to 0.98; I2: 98%) in the combined nasopharyngeal/oropharyngeal samples using various RT-PCR techniques (Figure 4D). The diagnostic parameters of RT-PCR in the nasopharyngeal and oropharyngeal samples are provided in Figure 4. The meta-analysis findings on various samples are provided in Table 2.
Table 2.
The meta-analysis findings on the diagnostic parameters of RT-PCR in various samples.
| Parameter | Number of Studies | Pooled Effect Measure (95%CI) | Heterogeneity |
|---|---|---|---|
| Nasopharyngeal swabs | |||
| Sensitivity | 43 | 91.06% (95%CI: 88.91 to 93.21) | 100% |
| Specificity | 37 | 95.57% (95%CI: 95.19 to 95.96) | 100% |
| PPV | 15 | 95.88% (95%CI: 91.59 to 100.16) | 100% |
| NPV | 15 | 91.58% (95%CI: 88.03 to 95.13) | 100% |
| Kappa coefficient | 16 | 0.79 (95%CI: 0.71 to 0.87) | 98% |
| Saliva samples | |||
| Sensitivity | 14 | 76.70% (95%CI: 60.50 to 92.91) | 99% |
| Specificity | 11 | 98.27% (95%CI: 97.31 to 99.24) | 62% |
| PPV | 5 | 90.16% (95%CI: 82.90 to 97.43) | 97% |
| NPV | 5 | 90.37% (95%CI: 84.18 to 96.56) | 96% |
| Kappa coefficient | 5 | 0.61 (95%CI: 0.44 to 0.79) | 98% |
| Combined nasopharyngeal/oropharyngeal samples | |||
| Sensitivity | 16 | 92.00% (95%CI: 87.57 to 96.43) | 100% |
| Specificity | 12 | 98.08% (95%CI: 96.64 to 99.52) | 100% |
| PPV | 5 | 84.63% (95%CI: 70.14 to 99.12) | 100% |
| NPV | 5 | 96.12% (95%CI: 92.83 to 99.42) | 100% |
| Kappa coefficient | 5 | 0.82 (95%CI: 0.67 to 0.98) | 98% |
3.5. Respiratory Samples
Only two studies [26,69] reported the sensitivity and specificity of RT-PCR in respiratory samples. Wu et al., [26] indicated a sensitivity and specificity of 100% and Nakura Y et al. [69] reported a sensitivity and specificity of 99.44% and 100%, respectively. The studies by Wu S et al. [26] and Pekoz A et al. [41] recorded a PPV of 100% and 73.7; and an NPV of 100% and 100%, respectively. Additionally, Price T K et al. [83] recorded an NPV of 98% among their samples. Wu et al. [26] recorded a kappa coefficient of 1 among 52 samples analyzed. The details are provided in Table 3.
Table 3.
The diagnostic parameters of RT-PCR in different samples.
| Parameter | Study | Total Participants | Effect Measure |
|---|---|---|---|
| Respiratory samples | |||
| Sensitivity | Wu S et al. [26] | 52 | 100 |
| Nakura Y et al. [69] | 213 | 99.44 | |
| Specificity | Wu S et al. [26] | 52 | 100 |
| Nakura Y et al. [69] | 213 | 100 | |
| PPV | Wu S et al. [26] | 52 | 100 |
| Pekoz A et al. [41] | 251 | 73.7 | |
| NPV | Wu S et al. [26] | 52 | 100 |
| Pekoz A et al., [41] | 251 | 100 | |
| Price T K et al. [83] | 10,165 | 98 | |
| Kappa Coefficient | Wu S et al. [26] | 52 | 1 |
| Sputum samples | |||
| Sensitivity | Villota S D et al. [22] | 50 | 90 |
| Torres A et al. [33] | 229 | 86 | |
| Specificity | Villota S D et al. [22] | 50 | 100 |
| Torres A et al. [33] | 229 | 37 | |
| PPV | Torres A et al. [33] | 229 | 38 |
| NPV | Torres A et al. [33] | 229 | 85 |
| Kappa Coefficient | Torres A et al. [33] | 229 | 0.73 |
| Broncho aspirate samples | |||
| Sensitivity | Pace V D et al. [23] | 75 | 96 |
| Specificity | Pace V D et al. [23] | 75 | 100 |
| Kappa Coefficient | Pace V D et al. [23] | 75 | 0.94 |
| Throat swab samples | |||
| Sensitivity | Wang B et al. [30] | 42 | 97.62 |
| Lu Y et al. [95] | 18 | 94.4 | |
| Specificity | Wang B et al. [30] | 158 | 100 |
| Lu Y et al. [95] | 18 | 100 | |
| PPV | Wang B et al. [30] | 200 | 100 |
| Lu Y et al. [95] | 18 | 100 | |
| NPV | Wang B et al. [30] | 200 | 98.52 |
| Lu Y et al. [95] | 18 | 99 | |
| Kappa Coefficient | Wang B et al. [30] | 200 | 0.985 |
| Lu Y et al. [95] | 18 | 0.996 | |
| Gargle samples | |||
| Sensitivity | Dumaresq J et al. [43] | 1005 | 95.3 |
| Kappa Coefficient | Dumaresq J et al. [43] | 1005 | 0.94 |
| Serum samples | |||
| Sensitivity | Ramírez AM et al. [96] | 265 | 73.63 |
| Specificity | Ramírez AM et al. [96] | 265 | 97.6 |
| PPV | Ramírez AM et al. [96] | 265 | 96.73 |
| NPV | Ramírez AM et al. [96] | 265 | 75 |
| Kappa Coefficient | Ramírez AM et al. [96] | 265 | 0.69 |
| Mixed samples | |||
| Sensitivity | Silva ferreira B I et al. [42] | 65 | 98.04 |
| Omar S et al. [85] | 319 | 95 | |
| Yip CCY et al. [87] | 106 | 99.1 | |
| Yang M et al. [97] | 35 | 100 | |
| Specificity | Silva ferreira B I et al. [42] | 65 | 100 |
| Omar S et al. [85] | 319 | 97 | |
| Yip CCY et al. [87] | 106 | 100 | |
| Yang M et al. [97] | 28 | 100 | |
| PPV | Silva ferreira B I et al. [42] | 65 | 100 |
| Omar S et al. [85] | 319 | 82.4 | |
| Yang M et al. [97] | 63 | 100 | |
| NPV | Silva ferreira B I et al. [42] | 65 | 93.3 |
| Omar S et al. [85] | 319 | 99.9 | |
| Yang M et al. [97] | 63 | 100 | |
| Kappa Coefficient | Silva ferreira B I et al. [42] | 65 | 0.96 |
PPV: positive predictive value; NPV: negative predictive value.
3.6. Sputum Samples
The study by Torres A et al. [33] indicated a sensitivity, specificity, PPV, NPV, and kappa coefficient of 86%, 37%, 38%, 85%, and 0.73, respectively. The study by Villota S D et al. [22] reported a sensitivity and specificity of 86% and 37%, respectively. The details are provided in Table 3.
3.7. Broncho Aspirate Samples
Only a single study by Pace V D et al. [23] used broncho aspirate samples for the detection of COVID-19 using RT-PCR. The sensitivity, specificity, and kappa coefficient were 96%, 100%, and 0.94, respectively. The details are provided in Table 3.
3.8. Throat Swab Samples
Two studies [30,95] used throat samples for the detection of COVID-19 by RT-PCR. The study by Wang B et al. [30] reported a sensitivity, specificity, PPV, NPV, and kappa coefficient of 97.62%, 100%, 100%, 98.52%, and 0.985, respectively. The study by Lu Y et al. [95] reported a sensitivity, specificity, PPV, NPV, and kappa coefficient of 94.4%, 100%, 100%, 99%, and 0.996, respectively. The details are provided in Table 3.
3.9. Gargle Samples
The study by Dumaresq J et al. [43] reported a sensitivity and kappa coefficient of 95.3% and 0.94, respectively, in gargle samples for the detection of COVID-19 using the RT-PCR technique. The details are provided in Table 3.
3.10. Serum Samples
Ramírez AM et al. [96] reported a sensitivity, specificity, PPV, NPV, and kappa coefficient of 73.63%, 97.6%, 96.73%, 75%, and 0.69, respectively. The details are provided in Table 3.
3.11. Mixed Samples
The sensitivity and specificity were reported by four studies using mixed samples for the detection of COVID-19. The studies by Ferreira BLS et al. [42], Omar S et al. [85], Yip CCY et al. [87], and Yang M et al. [97] reported a sensitivity of 98.04%, 95%, 99.1%, and 100%, respectively. Similarly, the specificity was observed to be 100%, 97%, 100%, and 100%, respectively. The PPV and NPV were reported by three studies [42,85,97], and they were 100% and 93.3%, 82.4, and 99.9%, and 100% for studies by Ferreira BLS et al. [42], Omar S et al. [85], and Yang M et al. [97], respectively. The kappa coefficient was reported by only one study, Ferreira BLS et al. [42], and it was 0.96. The details are provided in Table 3.
3.12. Publication Bias
A visual inspection of the funnel plot reveals an obvious asymmetry, which represents the chances of publication bias. This was confirmed statistically by Egger’s test (p = 0.00003) but not Begg’s test (p = 0.0982). The funnel plot is provided in Supplementary File S3.
3.13. Sensitivity Analysis
The sensitivity analysis was performed by altering the analysis model from the random effect model to the fixed effect model on NPS sensitivity analysis (Figure 2A). This made a small change in the overall effect measure, which is 91.06% (95%CI: 88.91 to 93.21) in the random effect model and 94.53% (95%CI: 94.53 to 94.54) in the fixed effect model. The sensitivity analysis result is provided in Supplementary File S4.
4. Discussion
COVID-19 can manifest in a variety of forms ranging from simple flu-like illness to death [103]. Various samples are used for the diagnosis of COVID-19 using many techniques, including RT-PCR. The diagnostic performance of various sampling approaches needs to be investigated to gain a better picture of all these aspects [104].
Our review provided evidence that pharyngeal samples (combined nasopharyngeal/oropharyngeal) have an equivalent sensitivity to nasopharyngeal samples, whereas saliva samples have a lesser sensitivity compared to the two other types of samples. A previous systematic review reported a comparable diagnostic performance with pooled nasal and throat swabs in comparison with nasopharyngeal swabs, which is considered to be the gold standard technique. Moreover, the self-collection of samples has influenced diagnostic accuracy [104].
As indicated in our review, respiratory samples, combined nasopharyngeal/oropharyngeal samples, broncho aspirate samples, throat swab samples, gargle samples, and mixed samples had better sensitivity than other samples, like serum and saliva, compared to nasopharyngeal swabs. Similarly, the study by Becker et al. recorded that saliva had approximately 30% lesser sensitivity than NPS, and it was 50% less sensitive in those cohorts of samples taken less than 21 days from the first symptom occurrence [105]. Similar findings were observed in the previous meta-analysis by Lee et al. [106].
Combined NPS/OPS, saliva, respiratory, broncho aspirate, throat, and mixed samples had better specificity than NPS. The current review indicates lower specificity with NPS than other specimens except for the sputum sample, which had reduced specificity compared to NPS. A community study by Torres et al. reported that saliva had 99.1% relative specificity to NPS [107]. Better diagnostic accuracy and specificity with saliva samples have been reported by many other studies [108,109,110]. Moreover, another study by Sasikala et al. reported that there was no difference between the diagnostic performance of saliva samples collected by healthcare workers and the patients themselves [111].
This review suggests that throat and respiratory samples had a similar positive predictive value (PPV) compared to NPS, while all other specimens had a lower PPV than NPS. Wang H et al. also found that NPS had better performance than other samples and recommended it as the best specimen for detecting COVID-19 through RT-PCR techniques [112]. The findings from this study can be used to develop protocols and guidelines for diagnosing COVID-19 and similar infections. Although NPS is considered the gold standard for diagnosing COVID-19, other samples have also been found to be equally helpful. Head-to-head analysis between different sampling strategies and specimens needs to be studied to develop the best alternative, cost-effective, and accurate diagnostic techniques.
This review had some limitations. First, there was a significant level of heterogeneity in all the meta-analyses performed, so caution should be taken when interpreting the findings. Second, English language restriction might have contributed to the exclusion of studies. However, comprehensive literature searches in all the available databases helped to collate the maximum possible information. Third, the variation in the RT-PCR techniques used and their processes might have contributed to the findings. Fourth, there was a lack of information with respect to sampling techniques and time of sampling. Hence, further research studies should focus on this. Future meta-analyses that emphasize subgroup analysis based on COVID-19 status, severity, and other important parameters should be planned.
5. Conclusions
The current meta-analysis suggests that NPS has a better or similar sensitivity than other samples, especially the specimens collected from any parts of the respiratory system, while the relative specificity of NPS was lower compared to other samples. Caution should be taken while interpreting the results due to the high heterogeneity in the analyses.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/diagnostics13193057/s1, Supplementary File S1: Detailed search strategy in various databases; Supplementary File S2: Characteristics of the RT-PCR techniques; Supplementary File S3: The funnel plot for publication bias; Supplementary File S4: Sensitivity analysis.
Author Contributions
Conceptualization, K.B. and K.S.A.A.-S.; methodology, K.B.; literature search: K.B.; software, K.B.; validation, K.S.A.A.-S., H.A.-L.M. and M.A.A.A.; formal analysis, K.B; investigation, K.A.A.; data curation, K.B.; writing—original draft preparation, K.B., K.S.A.A.-S. and A.M.; writing-review and editing, K.B., K.S.A.A.-S. and A.M.; visualization, K.A.A.; supervision, K.B.; project administration, K.B. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Ethical approval was not required for this study as it is a systematic review and does not include direct human data.
Informed Consent Statement
Patient consent was waived as this is a secondary research (meta-analysis) of published literature.
Data Availability Statement
All the data related to this paper is provided in the text or as a Supplementary Material along with this paper. Any additional information can be made available from the corresponding author upon request.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Khan S., Gionfriddo M.R., Cortes-Penfield N., Thunga G., Rashid M. The trade-off dilemma in pharmacotherapy of COVID-19: Systematic review, meta-analysis, and implications. Expert Opin. Pharmacother. 2020;21:1821–1849. doi: 10.1080/14656566.2020.1792884. [DOI] [PubMed] [Google Scholar]
- 2.Tahamtan A., Ardebili A. Real-time RT-PCR in COVID-19 detection: Issues affecting the results. Expert Rev. Mol. Diagn. 2020;20:453–454. doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foundation of Innovative New Diagnostics Sars-Cov-2 Molecular Assay Evaluation: Results. Mar 18, 2022. [(accessed on 2 February 2023)]. Available online: https://www.finddx.org/covid-19/sarscov2-eval-molecular/molecular-eval-results/
- 4.Lippi G., Simundic A.M., Plebani M. Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (COVID-19) Clin. Chem. Lab. Med. 2020;58:1070–1076. doi: 10.1515/cclm-2020-0285. [DOI] [PubMed] [Google Scholar]
- 5.Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for Typical Coronavirus Disease 2019 (COVID-19) Pneumonia: Relationship to Negative RT-PCR Testing. Radiology. 2020;296:E41–E45. doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of Chest CT and RT-PCR Testing for Coronavirus Disease 2019 (COVID-19) in China: A Report of 1014 Cases. Radiology. 2020;296:E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao A.T., Tong Y.X., Zhang S. False negative of RT-PCR and prolonged nucleic acid conversion in COVID-19: Rather than recurrence. J. Med. Virol. 2020;92:1755–1756. doi: 10.1002/jmv.25855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wyllie A.L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P., Warren J.L., Geng B., Muenker M.C., Moore A.J., et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N. Engl. J. Med. 2020;383:1283–1286. doi: 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wyllie A.L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P., Geng B., Muenker M.C., Moore A.J., Vogels C.B., et al. Saliva is more sensitive for SARS-CoV-2 detection in COVID-19 patients than nasopharyngeal swabs. MedRxiv. 2020;22:2020-04. doi: 10.1056/NEJMc2016359. [DOI] [Google Scholar]
- 10.Beyene G.T., Alemu F., Kebede E.S., Alemayehu D.H., Seyoum T., Tefera D.A., Assefa G., Tesfaye A., Habte A., Bedada G., et al. Saliva is superior over nasopharyngeal swab for detecting SARS-CoV2 in COVID-19 patients. Sci. Rep. 2021;11:22640. doi: 10.1038/s41598-021-02097-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X., Tan L., Wang X., Liu W., Lu Y., Cheng L., Sun Z. Comparison of nasopharyngeal and oropharyngeal swabs for SARS-CoV-2 detection in 353 patients received tests with both specimens simultaneously. Int. J. Infect. Dis. 2020;94:107–109. doi: 10.1016/j.ijid.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manzoor S. Comparison of oropharyngeal and nasopharyngeal swabs for detection of SARS-COV-2 in patients with COVID-19. Chest. 2020;158:A2473. doi: 10.1016/j.chest.2020.09.050. [DOI] [Google Scholar]
- 13.Page M.J., McKenzie J.E., Bossuyt P.M., Boutron I., Hoffmann T.C., Mulrow C.D., Shamseer L., Tetzlaff J.M., Akl E.A., Brennan S.E., et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ. 2020;372:n71. doi: 10.1016/j.ijsu.2021.105906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.RevMan . Review Manager (RevMan). [Computer Program] 5.4 ed. The Nordic Cochrane Centre, The Cochrane Collaboration; Copenhagen, Denmark: 2020. [Google Scholar]
- 15.Deeks J.J., Higgins J.P., Altman D.G., Group C.S.M. Cochrane Handbook for Systematic Reviews of Interventions. John Wiley & Sons; Hoboken, NJ, USA: 2019. Analysing data and undertaking meta-analyses; pp. 241–284. [Google Scholar]
- 16.Sterne J., Egger M., Moher D.J. Chapter 10: Addressing reporting biases. In: Higgins J.P.T., Green S., editors. Cochrane Handbook for Systematic Reviews of Interventions. John Wiley & Sons; Hoboken, NJ, USA: 2008. [Google Scholar]
- 17.Higgins J.P., Thomas J., Chandler J., Cumpston M., Li T., Page M.J., Welch V.A. Cochrane Handbook for Systematic Reviews of Interventions. John Wiley & Sons; Hoboken, NJ, USA: 2019. [Google Scholar]
- 18.Escobar D.F., Díaz P., Díaz-Dinamarca D., Puentes R., Alarcón P., Alarcón B., Rodríguez I., Manzo R.A., Soto D.A., Lamperti L., et al. Validation of a Methodology for the Detection of Severe Acute Respiratory Syndrome Coronavirus 2 in Saliva by Real-Time Reverse Transcriptase-PCR. Front. Public Health. 2021;9:743300. doi: 10.3389/fpubh.2021.743300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh J., Yadav A.K., Pakhare A., Kulkarni P., Lokhande L., Soni P., Dadheech M., Gupta P., Masarkar N., Maurya A.K. Comparative analysis of the diagnostic performance of five commercial COVID-19 qRT PCR kits used in India. Sci. Rep. 2021;11:22013. doi: 10.1038/s41598-021-00852-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Figueroa S., Freire-Paspuel B., Vega-Mariño P., Velez A., Cruz M., Cardenas W.B., Garcia-Bereguiain M.A. High sensitivity-low cost detection of SARS-CoV-2 by two steps end point RT-PCR with agarose gel electrophoresis visualization. Sci. Rep. 2021;11:21658. doi: 10.1038/s41598-021-00900-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LeGoff J., Kernéis S., Elie C., Mercier-Delarue S., Gastli N., Choupeaux L., Fourgeaud J., Alby M.L., Quentin P., Pavie J., et al. Evaluation of a saliva molecular point of care for the detection of SARS-CoV-2 in ambulatory care. Sci. Rep. 2021;11:21126. doi: 10.1038/s41598-021-00560-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Villota S.D., Nipaz V.E., Carrazco-Montalvo A., Hernandez S., Waggoner J.J., Ponce P., Coloma J., Orlando A., Cevallos V. Alternative RNA extraction-free techniques for the real-time RT-PCR detection of SARS-CoV-2 in nasopharyngeal swab and sputum samples. J. Virol. Methods. 2021;298:114302. doi: 10.1016/j.jviromet.2021.114302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Pace V., Caligiuri P., Ricucci V., Nigro N., Galano B., Visconti V., Da Rin G., Bruzzone B. Rapid diagnosis of SARS-CoV-2 pneumonia on lower respiratory tract specimens. BMC Infect. Dis. 2021;21:926. doi: 10.1186/s12879-021-06591-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanwar N., Banerjee D., Sasidharan A., Abdulhamid A., Larson M., Lee B., Selvarangan R., Liesman R.M. Comparison of diagnostic performance of five molecular assays for detection of SARS-CoV-2. Diagn. Microbiol. Infect. Dis. 2021;101:115518. doi: 10.1016/j.diagmicrobio.2021.115518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michel J., Neumann M., Krause E., Rinner T., Muzeniek T., Grossegesse M., Hille G., Schwarz F., Puyskens A., Förster S., et al. Resource-efficient internally controlled in-house real-time PCR detection of SARS-CoV-2. Virol. J. 2021;18:110. doi: 10.1186/s12985-021-01559-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu S., Shi X., Chen Q., Jiang Y., Zuo L., Wang L., Jiang M., Lin Y., Fang S., Peng B., et al. Comparative evaluation of six nucleic acid amplification kits for SARS-CoV-2 RNA detection. Ann. Clin. Microbiol. Antimicrob. 2021;20:38. doi: 10.1186/s12941-021-00443-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee K.K., Doudesis D., Ross D.A., Bularga A., MacKintosh C.L., Koch O., Johannessen I., Templeton K., Jenks S., Chapman A.R., et al. Diagnostic performance of the combined nasal and throat swab in patients admitted to hospital with suspected COVID-19. BMC Infect. Dis. 2021;21:318. doi: 10.1186/s12879-021-05976-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borkakoty B., Bali N.K. TSP-based PCR for rapid identification of L and S type strains of SARS-CoV-2. Indian J. Med. Microbiol. 2021;39:73–80. doi: 10.1016/j.ijmmb.2021.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hata D.J., White E.L., Jr., Bridgeman M.M., Gasson S.L., Jones D.S., Vicari B.R., Van Siclen C.P., Palmucci C., Marquez C.P., Parkulo M.A., et al. Performance Accuracy of a Laboratory-Developed Real-Time RT-PCR Method for Detection of SARS-CoV-2 in Self-Collected Saliva Specimens. Ann. Clin. Lab. Sci. 2021;51:741–749. [PubMed] [Google Scholar]
- 30.Wang B., Hu M., Ren Y., Xu X., Wang Z., Lyu X., Wu W., Li Z., Gong X., Xiang Z., et al. Evaluation of seven commercial SARS-CoV-2 RNA detection kits based on real-time polymerase chain reaction (PCR) in China. Clin. Chem. Lab. Med. 2020;58:e149–e153. doi: 10.1515/cclm-2020-0271. [DOI] [PubMed] [Google Scholar]
- 31.Mollaei H.R., Afshar A.A., Kalantar-Neyestanaki D., Fazlalipour M., Aflatoonian B. Comparison five primer sets from different genome region of COVID-19 for detection of virus infection by conventional RT-PCR. Iran. J. Microbiol. 2020;12:185–193. doi: 10.18502/ijm.v12i3.3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pierri B., Mancusi A., Proroga Y.T.R., Capuano F., Cerino P., Girardi S., Vassallo L., Lo Conte G., Tafuro M., Cuomo M.C., et al. SARS-CoV-2 detection in nasopharyngeal swabs: Performance characteristics of a real-time RT-qPCR and a droplet digital RT-PCR assay based on the exonuclease region (ORF1b, nsp 14) J. Virol. Methods. 2020;300:114420. doi: 10.1016/j.jviromet.2021.114420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Torres A., Fors M., Rivero T., Pantoja K., Ballaz S. Comparison between RT-qPCR for SARS-CoV-2 and expanded triage in sputum of symptomatic and asymptomatic COVID-19 subjects in Ecuador. BMC Infect. Dis. 2021;21:558. doi: 10.1186/s12879-021-06272-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pearson J.D., Trcka D., Lu S., Hyduk S.J., Jen M., Aynaud M.M., Hernández J.J., Peidis P., Barrios-Rodiles M., Chan K., et al. Comparison of SARS-CoV-2 indirect and direct RT-qPCR detection methods. Virol J. 2021;18:99. doi: 10.1186/s12985-021-01574-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kriegova E., Fillerova R., Raska M., Manakova J., Dihel M., Janca O., Sauer P., Klimkova M., Strakova P., Kvapil P. Excellent option for mass testing during the SARS-CoV-2 pandemic: Painless self-collection and direct RT-qPCR. Virol. J. 2021;18:95. doi: 10.1186/s12985-021-01567-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Onyilagha C., Mistry H., Marszal P., Pinette M., Kobasa D., Tailor N., Berhane Y., Nfon C., Pickering B., Mubareka S., et al. Evaluation of mobile real-time polymerase chain reaction tests for the detection of severe acute respiratory syndrome coronavirus 2. Sci. Rep. 2021;11:9387. doi: 10.1038/s41598-021-88625-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Desmet T., Paepe P., Boelens J., Coorevits L., Padalko E., Vandendriessche S., Leroux-Roels I., Aerssens A., Callens S., Braeckel E.V., et al. Combined oropharyngeal/nasal swab is equivalent to nasopharyngeal sampling for SARS-CoV-2 diagnostic PCR. BMC Microbiol. 2021;21:31. doi: 10.1186/s12866-021-02087-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanji J.N., Zelyas N., MacDonald C., Pabbaraju K., Khan M.N., Prasad A., Hu J., Diggle M., Berenger B.M., Tipples G. False negative rate of COVID-19 PCR testing: A discordant testing analysis. Virol. J. 2021;18:13. doi: 10.1186/s12985-021-01489-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gómez-Romero L., Tovar H., Moreno-Contreras J., Espinoza M.A., de-Anda-Jáuregui G. Automated Reverse Transcription Polymerase Chain Reaction Data Analysis for Sars-CoV-2 Detection. Rev. Investig. Clin. Organo Hosp. Enfermedades Nutr. 2021;73:339–346. doi: 10.24875/RIC.21000189. [DOI] [PubMed] [Google Scholar]
- 40.Milosevic J., Lu M., Greene W., He H.Z., Zheng S.Y. An Ultrafast One-Step Quantitative Reverse Transcription-Polymerase Chain Reaction Assay for Detection of SARS-CoV-2. Front. Microbiol. 2021;12:749783. doi: 10.3389/fmicb.2021.749783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pekosz A., Parvu V., Li M., Andrews J.C., Manabe Y.C., Kodsi S., Gary D.S., Roger-Dalbert C., Leitch J., Cooper C.K. Antigen-Based Testing but Not Real-Time Polymerase Chain Reaction Correlates with Severe Acute Respiratory Syndrome Coronavirus 2 Viral Culture. Clin. Infect. Dis. 2021;73:e2861–e2866. doi: 10.1093/cid/ciaa1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferreira B.I.D.S., da Silva-Gomes N.L., Coelho W.L.D.C.N.P., da Costa V.D., Carneiro V.C.S., Kader R.L., Amaro M.P., Villar L.M., Miyajima F., Alves-Leon S.V., et al. Validation of a novel molecular assay to the diagnostic of COVID-19 based on real time PCR with high resolution melting. PLoS ONE. 2021;16:e0260087. doi: 10.1371/journal.pone.0260087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dumaresq J., Coutlée F., Dufresne P.J., Longtin J., Fafard J., Bestman-Smith J., Bergevin M., Vallières E., Desforges M., Labbé A.C. Natural spring water gargle and direct RT-PCR for the diagnosis of COVID-19 (COVID-SPRING study) J. Clin. Virol. 2021;144:104995. doi: 10.1016/j.jcv.2021.104995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morecchiato F., Coppi M., Baccani I., Maggini N., Ciccone N., Antonelli A., Rossolini G.M. Evaluation of extraction-free RT-PCR methods for faster and cheaper detection of SARS-CoV-2 using two commercial systems. Int. J. Infect. Dis. 2021;112:264–268. doi: 10.1016/j.ijid.2021.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olearo F., Nörz D., Hoffman A., Grunwald M., Gatzemeyer K., Christner M., Both A., Campos C.E.B., Braun P., Andersen G., et al. Clinical performance and accuracy of a qPCR-based SARS-CoV-2 mass-screening workflow for healthcare-worker surveillance using pooled self-sampled gargling solutions: A cross-sectional study. J. Infect. 2021;83:589–593. doi: 10.1016/j.jinf.2021.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ghoshal U., Garg A., Vasanth S., Arya A.K., Pandey A., Tejan N., Patel V., Singh V.P. Assessing a chip-based rapid RTPCR test for SARS CoV-2 detection (TrueNat assay): A diagnostic accuracy study. PLoS ONE. 2021;16:e0257834. doi: 10.1371/journal.pone.0257834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Balaska S., Pilalas D., Takardaki A., Koutra P., Parasidou E., Gkeka I., Tychala A., Meletis G., Fyntanidou B., Metallidis S., et al. Evaluation of the Advanta Dx SARS-CoV-2 RT-PCR Assay, a High-Throughput Extraction-Free Diagnostic Test for the Detection of SARS-CoV-2 in Saliva: A Diagnostic Accuracy Study. Diagnostics. 2021;11:1766. doi: 10.3390/diagnostics11101766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watanabe Y., Oikawa R., Suzuki T., Funabashi H., Asai D., Hatori Y., Takemura H., Yamamoto H., Itoh F. Evaluation of a new point-of-care quantitative reverse transcription polymerase chain test for detecting severe acute respiratory syndrome coronavirus 2. J. Clin. Lab. Anal. 2021;35:e23992. doi: 10.1002/jcla.23992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Domnich A., De Pace V., Pennati B.M., Caligiuri P., Varesano S., Bruzzone B., Orsi A. Evaluation of extraction-free RT-qPCR methods for SARS-CoV-2 diagnostics. Arch. Virol. 2021;166:2825–2828. doi: 10.1007/s00705-021-05165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim Y.K., Chang S.H. Clinical usefulness of extraction-free PCR assay to detect SARS-CoV-2. J. Virol. Methods. 2021;296:114217. doi: 10.1016/j.jviromet.2021.114217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carvalho R.F., Oliveira M.D.S., Ribeiro J., Dos Santos I.G.C., Almeida K.S., Conti A.C.M., Alexandrino B., Campos F.S., Soares C.M.A., Ribeiro Júnior J.C. Validation of conventional PCR-like alternative to SARS-CoV-2 detection with target nucleocapsid protein gene in naso-oropharyngeal samples. PLoS ONE. 2021;16:e0257350. doi: 10.1371/journal.pone.0257350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kritikos A., Caruana G., Brouillet R., Miroz J.P., Abed-Maillard S., Stieger G., Opota O., Croxatto A., Vollenweider P., Bart P.A., et al. Sensitivity of Rapid Antigen Testing and RT-PCR Performed on Nasopharyngeal Swabs versus Saliva Samples in COVID-19 Hospitalized Patients: Results of a Prospective Comparative Trial (RESTART) Microorganisms. 2021;9:1910. doi: 10.3390/microorganisms9091910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brotons P., Perez-Argüello A., Launes C., Torrents F., Subirats M.P., Saucedo J., Claverol J., Garcia-Garcia J.J., Rodas G., Fumado V., et al. Validation and implementation of a direct RT-qPCR method for rapid screening of SARS-CoV-2 infection by using non-invasive saliva samples. Int. J. Infect. Dis. 2021;110:363–370. doi: 10.1016/j.ijid.2021.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laverack M., Tallmadge R.L., Venugopalan R., Cronk B., Zhang X., Rauh R., Saunders A., Nelson W.M., Plocharczyk E., Diel D.G. Clinical evaluation of a multiplex real-time RT-PCR assay for detection of SARS-CoV-2 in individual and pooled upper respiratory tract samples. Arch. Virol. 2021;166:2551–2561. doi: 10.1007/s00705-021-05148-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Avetyan D., Chavushyan A., Ghazaryan H., Melkonyan A., Stepanyan A., Zakharyan R., Hayrapetyan V., Atshemyan S., Khachatryan G., Sirunyan T., et al. SARS-CoV-2 detection by extraction-free qRT-PCR for massive and rapid COVID-19 diagnosis during a pandemic in Armenia. J. Virol. Methods. 2021;295:114199. doi: 10.1016/j.jviromet.2021.114199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hernandez M.M., Banu R., Shrestha P., Patel A., Chen F., Cao L., Fabre S., Tan J., Lopez H., Chiu N., et al. RT-PCR/MALDI-TOF mass spectrometry-based detection of SARS-CoV-2 in saliva specimens. J. Med. Virol. 2021;93:5481–5486. doi: 10.1002/jmv.27069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hernández C., Florez C., Castañeda S., Ballesteros N., Martínez D., Castillo A., Muñoz M., Gomez S., Rico A., Pardo L., et al. Evaluation of the diagnostic performance of nine commercial RT-PCR kits for the detection of SARS-CoV-2 in Colombia. J. Med. Virol. 2021;93:5618–5622. doi: 10.1002/jmv.27051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leber W., Lammel O., Redlberger-Fritz M., Mustafa-Korninger M.E., Glehr R.C., Camp J., Agerer B., Lercher A., Popa A., Genger J.W., et al. Rapid, early and accurate SARS-CoV-2 detection using RT-qPCR in primary care: A prospective cohort study (REAP-1) BMJ Open. 2021;11:e045225. doi: 10.1136/bmjopen-2020-045225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gadkar V.J., Goldfarb D.M., Young V., Watson N., Al-Rawahi G.N., Srigley J.A., Tilley P. Development and validation of a new triplex real-time quantitative reverse Transcriptase-PCR assay for the clinical detection of SARS-CoV-2. Mol. Cell. Probes. 2021;58:101744. doi: 10.1016/j.mcp.2021.101744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bruno A., de Mora D., Freire-Paspuel B., Rodriguez A.S., Paredes-Espinosa M.B., Olmedo M., Sanchez M., Romero J., Paez M., Gonzalez M., et al. Analytical and clinical evaluation of a heat shock SARS-CoV-2 detection method without RNA extraction for N and E genes RT-qPCR. Int. J. Infect. Dis. 2021;109:315–320. doi: 10.1016/j.ijid.2021.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun R., Achkar S., Ammari S., Bockel S., Gallois E., Bayle A., Battistella E., Salviat F., Merad M., Laville A., et al. Systematic Screening of COVID-19 Disease Based on Chest CT and RT-PCR for Cancer Patients Undergoing Radiation Therapy in a Coronavirus French Hotspot. Int. J. Radiat. Oncol. Biol. Phys. 2021;110:947–956. doi: 10.1016/j.ijrobp.2021.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rigo S., Durigon S., Basaglia G., Avolio M., De Rosa R. Molecular diagnosis of SARS-Cov-2, performances and throughput by Direct RT-PCR. New Microbiol. 2021;44:173–176. [PubMed] [Google Scholar]
- 63.Banko A., Petrovic G., Miljanovic D., Loncar A., Vukcevic M., Despot D., Cirkovic A. Comparison and Sensitivity Evaluation of Three Different Commercial Real-Time Quantitative PCR Kits for SARS-CoV-2 Detection. Viruses. 2021;13:1321. doi: 10.3390/v13071321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tastanova A., Stoffel C.I., Dzung A., Cheng P.F., Bellini E., Johansen P., Duda A., Nobbe S., Lienhard R., Bosshard P.P., et al. A Comparative Study of Real-Time RT-PCR-Based SARS-CoV-2 Detection Methods and Its Application to Human-Derived and Surface Swabbed Material. J. Mol. Diagn. 2021;23:796–804. doi: 10.1016/j.jmoldx.2021.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Noor F.A., Safain K.S., Hossain M.W., Arafath K., Mannoor K., Kabir M. Development and performance evaluation of the first in-house multiplex rRT-PCR assay in Bangladesh for highly sensitive detection of SARS-CoV-2. J. Virol. Methods. 2021;293:114147. doi: 10.1016/j.jviromet.2021.114147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fitoussi F., Dupont R., Tonen-Wolyec S., Bélec L. Performances of the VitaPCR™ SARS-CoV-2 Assay during the second wave of the COVID-19 epidemic in France. J. Med. Virol. 2021;93:4351–4357. doi: 10.1002/jmv.26950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Freire-Paspuel B., Garcia-Bereguiain M.A. Analytical and Clinical Evaluation of “AccuPower SARS-CoV-2 Multiplex RT-PCR kit (Bioneer, South Korea)” and “Allplex 2019-nCoV Assay (Seegene, South Korea)” for SARS-CoV-2 RT-PCR Diagnosis: Korean CDC EUA as a Quality Control Proxy for Developing Countries. Front. Cell. Infect. Microbiol. 2021;11:630552. doi: 10.3389/fcimb.2021.630552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dierks S., Bader O., Schwanbeck J., Groß U., Weig M.S., Mese K., Lugert R., Bohne W., Hahn A., Feltgen N., et al. Diagnosing SARS-CoV-2 with Antigen Testing, Transcription-Mediated Amplification and Real-Time PCR. J. Clin. Med. 2021;10:2404. doi: 10.3390/jcm10112404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nakura Y., Wu H.N., Okamoto Y., Takeuchi M., Suzuki K., Tamura Y., Oba Y., Nishiumi F., Hatori N., Fujiwara S., et al. Development of an efficient one-step real-time reverse transcription polymerase chain reaction method for severe acute respiratory syndrome-coronavirus-2 detection. PLoS ONE. 2021;16:e0252789. doi: 10.1371/journal.pone.0252789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stockdale A.J., Fyles F., Farrell C., Lewis J., Barr D., Haigh K., Abouyannis M., Hankinson B., Penha D., Fernando R., et al. Sensitivity of SARS-CoV-2 RNA polymerase chain reaction using a clinical and radiological reference standard. J. Infect. 2021;82:260–268. doi: 10.1016/j.jinf.2021.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kortela E., Kirjavainen V., Ahava M.J., Jokiranta S.T., But A., Lindahl A., Jääskeläinen A.E., Jääskeläinen A.J., Järvinen A., Jokela P., et al. Real-life clinical sensitivity of SARS-CoV-2 RT-PCR test in symptomatic patients. PLoS ONE. 2021;16:e0251661. doi: 10.1371/journal.pone.0251661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Altamimi A.M., Obeid D.A., Alaifan T.A., Taha M.T., Alhothali M.T., Alzahrani F.A., Albarrag A.M. Assessment of 12 qualitative RT-PCR commercial kits for the detection of SARS-CoV-2. J. Med. Virol. 2021;93:3219–3226. doi: 10.1002/jmv.26900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Visseaux B., Collin G., Houhou-Fidouh N., Le Hingrat Q., Ferré V.M., Damond F., Ichou H., Descamps D., Charpentier C. Evaluation of three extraction-free SARS-CoV-2 RT-PCR assays: A feasible alternative approach with low technical requirements. J. Virol. Methods. 2021;291:114086. doi: 10.1016/j.jviromet.2021.114086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cassinari K., Alessandri-Gradt E., Chambon P., Charbonnier F., Gracias S., Beaussire L., Alexandre K., Sarafan-Vasseur N., Houdayer C., Etienne M., et al. Assessment of Multiplex Digital Droplet RT-PCR as a Diagnostic Tool for SARS-CoV-2 Detection in Nasopharyngeal Swabs and Saliva Samples. Clin. Chem. 2021;67:736–741. doi: 10.1093/clinchem/hvaa323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Carrillo R.J., Sarmiento A.D., Ang M.A., Diwa M.H., Dungog C.C., Tan D.I., Lacuata J.A., Salud J.E., Lopa R.A., Velasco J.M., et al. Validation of snort-spit saliva in detecting COVID-19 using RT-PCR and rapid antigen detection test. Acta Medica Philipp. 2021:55. doi: 10.47895/amp.v55i2.2779. [DOI] [Google Scholar]
- 76.Girish P., Jayasankar P., Abhishek P., Sumeeta S., Gunvant P., Shalin P. Comparative analysis of the naso/oropharyngeal swab and oral bio-fluid (whole saliva) samples for the detection of SARS-CoV-2 using RT-qPCR. Indian J. Dent. Res. 2021;32:206–210. doi: 10.4103/ijdr.ijdr_483_21. [DOI] [PubMed] [Google Scholar]
- 77.Freire-Paspuel B., Garcia-Bereguiain M.A. Clinical Performance and Analytical Sensitivity of Three SARS-CoV-2 Nucleic Acid Diagnostic Tests. Am. J. Trop. Med. Hyg. 2021;104:1516–1518. doi: 10.4269/ajtmh.20-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dong L., Zhou J., Niu C., Wang Q., Pan Y., Sheng S., Wang X., Zhang Y., Yang J., Liu M., et al. Highly accurate and sensitive diagnostic detection of SARS-CoV-2 by digital PCR. Talanta. 2021;224:121726. doi: 10.1016/j.talanta.2020.121726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gupta-Wright A., Macleod C.K., Barrett J., Filson S.A., Corrah T., Parris V., Sandhu G., Harris M., Tennant R., Vaid N., et al. False-negative RT-PCR for COVID-19 and a diagnostic risk score: A retrospective cohort study among patients admitted to hospital. BMJ Open. 2021;11:e047110. doi: 10.1136/bmjopen-2020-047110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dimke H., Larsen S.L., Skov M.N., Larsen H., Hartmeyer G.N., Moeller J.B. Phenol-chloroform-based RNA purification for detection of SARS-CoV-2 by RT-qPCR: Comparison with automated systems. PLoS ONE. 2021;16:e0247524. doi: 10.1371/journal.pone.0247524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Alaifan T., Altamimi A., Obeid D., Alshehri T., Almatrrouk S., Albarrag A. SARS-CoV-2 direct real-time polymerase chain reaction testing in laboratories with shortage challenges. Future Virol. 2021;16:133–139. doi: 10.2217/fvl-2020-0187. [DOI] [Google Scholar]
- 82.Onwuamah C.K., Okwuraiwe A.P., Salu O.B., Shaibu J.O., Ndodo N., Amoo S.O., Okoli L.C., Ige F.A., Ahmed R.A., Bankole M.A., et al. Comparative performance of SARS-CoV-2 real-time PCR diagnostic assays on samples from Lagos, Nigeria. PLoS ONE. 2021;16:e0246637. doi: 10.1371/journal.pone.0246637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Price T.K., Bowland B.C., Chandrasekaran S., Garner O.B., Yang S. Performance Characteristics of Severe Acute Respiratory Syndrome Coronavirus 2 RT-PCR Tests in a Single Health System: Analysis of >10,000 Results from Three Different Assays. J. Mol. Diagn. 2021;23:159–163. doi: 10.1016/j.jmoldx.2020.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Trobajo-Sanmartín C., Adelantado M., Navascués A., Guembe M.J., Rodrigo-Rincón I., Castilla J., Ezpeleta C. Self-Collection of Saliva Specimens as a Suitable Alternative to Nasopharyngeal Swabs for the Diagnosis of SARS-CoV-2 by RT-qPCR. J. Clin. Med. 2021;10:299. doi: 10.3390/jcm10020299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Omar S., Brown J.M., Mathivha R.L., Bahemia I., Nabeemeeah F., Martinson N. The impact of a mobile COVID-19 polymerase chain reaction laboratory at a large tertiary hospital during the first wave of the pandemic: A retrospective analysis. S. Afr. Med. J. 2021;111:957–960. doi: 10.7196/SAMJ.2021.v111i10.15690. [DOI] [PubMed] [Google Scholar]
- 86.Bergevin M.A., Freppel W., Robert G., Ambaraghassi G., Aubry D., Haeck O., Saint-Jean M., Carignan A. Validation of saliva sampling as an alternative to oro-nasopharyngeal swab for detection of SARS-CoV-2 using unextracted rRT-PCR with the Allplex 2019-nCoV assay. J. Med. Microbiol. 2021;70:001404. doi: 10.1099/jmm.0.001404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yip C.C.Y., Leung K.H., Ng A.C.K., Chan K.H., To K.K.W., Chan J.F.W., Hung I.F.N., Cheng V.C.C., Sridhar S. Comparative evaluation of a dual-target real-time RT-PCR assay for COVID-19 diagnosis and assessment of performance in pooled saliva and nasopharyngeal swab samples. Expert Rev. Mol. Diagn. 2021;21:741–747. doi: 10.1080/14737159.2021.1933445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Renzoni A., Perez F., Ngo Nsoga M.T., Yerly S., Boehm E., Gayet-Ageron A., Kaiser L., Schibler M. Analytical Evaluation of Visby Medical RT-PCR Portable Device for Rapid Detection of SARS-CoV-2. Diagnostics. 2021;11:813. doi: 10.3390/diagnostics11050813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tsujimoto Y., Terada J., Kimura M., Moriya A., Motohashi A., Izumi S., Kawajiri K., Hakkaku K., Morishita M., Saito S., et al. Diagnostic accuracy of nasopharyngeal swab, nasal swab and saliva swab samples for the detection of SARS-CoV-2 using RT-PCR. Infect. Dis. 2021;53:581–589. doi: 10.1080/23744235.2021.1903550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mio C., Cifù A., Marzinotto S., Marcon B., Pipan C., Damante G., Curcio F. Validation of a One-Step Reverse Transcription-Droplet Digital PCR (RT-ddPCR) Approach to Detect and Quantify SARS-CoV-2 RNA in Nasopharyngeal Swabs. Dis. Markers. 2021;2021:8890221. doi: 10.1155/2021/8890221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lau Y.L., Ismail I.B., Mustapa N.I.B., Lai M.Y., Tuan Soh T.S., Haji Hassan A., Peariasamy K.M., Lee Y.L., Abdul Kahar M.K.B., Chong J., et al. Development of a reverse transcription recombinase polymerase amplification assay for rapid and direct visual detection of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) PLoS ONE. 2021;16:e0245164. doi: 10.1371/journal.pone.0245164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shen L., Cui S., Zhang D., Lin C., Chen L., Wang Q. Comparison of four commercial RT-PCR diagnostic kits for COVID-19 in China. J. Clin. Lab. Anal. 2021;35:e23605. doi: 10.1002/jcla.23605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Freire-Paspuel B., Garcia-Bereguiain M.A. Poor sensitivity of “AccuPower SARS-CoV-2 real time RT-PCR kit (Bioneer, South Korea)”. Virol. J. 2020;17:178. doi: 10.1186/s12985-020-01445-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Guo J.J., Yu Y.H., Ma X.Y., Liu Y.N., Fang Q., Qu P., Guo J., Lou J.L., Wang Y.J. A multiple-center clinical evaluation of a new real-time reverse transcriptase PCR diagnostic kit for SARS-CoV-2. Future Virol. 2020;15:673–681. doi: 10.2217/fvl-2020-0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lu Y., Li L., Ren S., Liu X., Zhang L., Li W., Yu H. Comparison of the diagnostic efficacy between two PCR test kits for SARS-CoV-2 nucleic acid detection. J. Clin. Lab. Anal. 2020;34:e23554. doi: 10.1002/jcla.23554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Martín Ramírez A., Zurita Cruz N.D., Gutiérrez-Cobos A., Rodríguez Serrano D.A., González Álvaro I., Roy Vallejo E., Gómez de Frutos S., Fontán García-Rodrigo L., Cardeñoso Domingo L. Evaluation of two RT-PCR techniques for SARS-CoV-2 RNA detection in serum for microbiological diagnosis. J. Virol. Methods. 2022;300:114411. doi: 10.1016/j.jviromet.2021.114411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang M., Cao S., Liu Y., Zhang Z., Zheng R., Li Y., Zhou J., Zong C., Cao D., Qin X. Performance verification of five commercial RT-qPCR diagnostic kits for SARS-CoV-2. Clin. Chim. Acta Int. J. Clin. Chem. 2022;525:46–53. doi: 10.1016/j.cca.2021.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sahoo A., Shulania A., Chhabra M., Kansra S., Achra A., Nirmal K., Katiyar S., Duggal N. Performance of Chip Based Real Time RTPCR (TrueNat) and Conventional Real Time RT-PCR for Detection of SARS-CoV-2. J. Clin. Diagn. Res. 2021;15:DC25–DC28. doi: 10.7860/JCDR/2021/50628.15680. [DOI] [Google Scholar]
- 99.Hofman P., Boutros J., Benchetrit D., Benzaquen J., Leroy S., Tanga V., Bordone O., Allégra M., Lespinet V., Fayada J., et al. A rapid near-patient RT-PCR test for suspected COVID-19: A study of the diagnostic accuracy. Ann. Transl. Med. 2021;9:921. doi: 10.21037/atm-21-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jamal A.J., Mozafarihashjin M., Coomes E., Powis J., Li A.X., Paterson A., Anceva-Sami S., Barati S., Crowl G., Faheem A., et al. Sensitivity of Nasopharyngeal Swabs and Saliva for the Detection of Severe Acute Respiratory Syndrome Coronavirus 2. Clin. Infect. Dis. 2021;72:1064–1066. doi: 10.1093/cid/ciaa848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ngaba G.P., Kalla G.C.M., Assob J.C.N., Njouendou A.J., Jembe C.N., Mboudou E.T., Mbopi-Keou F.X. Comparative analysis of two molecular tests for the detection of COVID-19 in Cameroon. Pan. Afr. Med. J. 2021;39:214. doi: 10.11604/pamj.2021.39.214.30718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Procop G.W., Brock J.E., Reineks E.Z., Shrestha N.K., Demkowicz R., Cook E., Ababneh E., Harrington S.M. A Comparison of Five SARS-CoV-2 Molecular Assays with Clinical Correlations. Am. J. Clin. Pathol. 2021;155:69–78. doi: 10.1093/ajcp/aqaa181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rashid M., Rajan A.K., Thunga G., Shanbhag V., Nair S. Impact of Diabetes in COVID-19 Associated Mucormycosis and its Management: A Non-Systematic Literature Review. Curr. Diabetes Rev. 2023;19:e240222201411. doi: 10.2174/1573399818666220224123525. [DOI] [PubMed] [Google Scholar]
- 104.Tsang N.N.Y., So H.C., Ng K.Y., Cowling B.J., Leung G.M., Ip D.K.M. Diagnostic performance of different sampling approaches for SARS-CoV-2 RT-PCR testing: A systematic review and meta-analysis. Lancet Infect. Dis. 2021;21:1233–1245. doi: 10.1016/S1473-3099(21)00146-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Becker D., Sandoval E., Amin A., De Hoff P., Diets A., Leonetti N., Lim Y.W., Elliott C., Laurent L., Grzymski J., et al. Saliva is less sensitive than nasopharyngeal swabs for COVID-19 detection in the community setting. MedRxiv. 2020 doi: 10.1101/2020.05.11.20092338. [DOI] [Google Scholar]
- 106.Lee R.A., Herigon J.C., Benedetti A., Pollock N.R., Denkinger C.M. Performance of Saliva, Oropharyngeal Swabs, and Nasal Swabs for SARS-CoV-2 Molecular Detection: A Systematic Review and Meta-analysis. J. Clin. Microbiol. 2021;59:e02881-20. doi: 10.1128/JCM.02881-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Torres M., Collins K., Corbit M., Ramirez M., Winters C.R., Katz L., Ross M., Relkin N., Zhou W. Comparison of saliva and nasopharyngeal swab SARS-CoV-2 RT-qPCR testing in a community setting. J. Infect. 2021;82:84–123. doi: 10.1016/j.jinf.2020.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Altawalah H., AlHuraish F., Alkandari W.A., Ezzikouri S. Saliva specimens for detection of severe acute respiratory syndrome coronavirus 2 in Kuwait: A cross-sectional study. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2020;132:104652. doi: 10.1016/j.jcv.2020.104652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Atieh M.A., Guirguis M., Alsabeeha N.H.M., Cannon R.D. The diagnostic accuracy of saliva testing for SARS-CoV-2: A systematic review and meta-analysis. Oral Dis. 2022;28((Suppl. S2)):2347–2361. doi: 10.1111/odi.13934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Huber M., Schreiber P.W., Scheier T., Audigé A., Buonomano R., Rudiger A., Braun D.L., Eich G., Keller D.I., Hasse B., et al. High Efficacy of Saliva in Detecting SARS-CoV-2 by RT-PCR in Adults and Children. Microorganisms. 2021;9:642. doi: 10.3390/microorganisms9030642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang H., Liu Q., Hu J., Zhou M., Yu M.Q., Li K.Y., Xu D., Xiao Y., Yang J.Y., Lu Y.J., et al. Nasopharyngeal Swabs Are More Sensitive than Oropharyngeal Swabs for COVID-19 Diagnosis and Monitoring the SARS-CoV-2 Load. Front. Med. 2020;7:334. doi: 10.3389/fmed.2020.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sasikala M., Sadhana Y., Vijayasarathy K., Gupta A., Daram S.K., Podduturi N.C.R., Reddy D.N. Comparison of saliva with healthcare workers- and patient-collected swabs in the diagnosis of COVID-19 in a large cohort. BMC Infect. Dis. 2021;21:648. doi: 10.1186/s12879-021-06343-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data related to this paper is provided in the text or as a Supplementary Material along with this paper. Any additional information can be made available from the corresponding author upon request.