Abstract
RNA structure regulates bacterial gene expression by several distinct mechanisms via environmental and cellular stimuli, one of which is temperature. While some genome-wide studies have focused on heat shock treatments and the subsequent transcriptomic changes, soil bacteria are less likely to experience such rapid and extreme temperature changes. Though RNA thermometers (RNATs) have been found in 5′ untranslated leader regions (5′ UTRs) of heat shock and virulence-associated genes, this RNA-controlled mechanism could regulate other genes as well. Using Structure-seq2 and the chemical probe dimethyl sulfate (DMS) at four growth temperatures ranging from 23°C to 42°C, we captured a dynamic response of the Bacillus subtilis transcriptome to temperature. Our transcriptome-wide results show RNA structural changes across all four temperatures and reveal nonmonotonic reactivity trends with increasing temperature. Then, focusing on subregions likely to contain regulatory RNAs, we examined 5′ UTRs to identify large, local reactivity changes. This approach led to the discovery of RNATs that control the expression of glpF (glycerol permease) and glpT (glycerol-3-phosphate permease); expression of both genes increased with increased temperature. Results with mutant RNATs indicate that both genes are controlled at the translational level. Increased import of glycerols at high temperatures could provide thermoprotection to proteins.
Keywords: RNA thermometers, RNA–protein, Structure-seq, glycerol permease
INTRODUCTION
RNA structure is known to regulate gene expression through a variety of conformational changes (Sharp 2009). These structural changes occur in response to both environmental and cellular stimuli by either exposing or sequestering key RNA regulatory elements (Dethoff et al. 2012; Ganser et al. 2019). In this way, RNA is involved in myriad cellular functions directly attributable to its ability to form secondary and tertiary structures (Mortimer et al. 2014).
In its simplest form, RNA secondary structure is comprised of hairpins with base-paired stems and unpaired loops (Tinoco and Bustamante 1999). While these structures can grow in complexity and involve interactions between themselves to form tertiary structures, secondary structural elements are sufficiently stable to influence a wide variety of biological processes. Moreover, many RNA elements can alter their structure based on ligand or protein binding; or in the case of a temperature increase, structured regions can melt to become unstructured (Ganser et al. 2019).
Common examples of bacterial RNA structural switches that control gene expression are transcription attenuators, riboswitches, and RNA thermometers (RNATs) (Roßmanith and Narberhaus 2016; Babitzke et al. 2019). These regulatory elements are typically found in 5′ untranslated regions (5′ UTRs) and control the expression of the downstream gene by altering their own structure. Transcription attenuators consist of overlapping and mutually exclusive antiterminator and intrinsic terminator structures, and binding of a regulatory molecule such as a protein or metabolite dictates which of the two structures form (Babitzke et al. 2019). Riboswitches bind a small molecule ligand, which in turn causes a conformational change in the RNA, leading to the regulation of transcription or translation. RNATs respond to a temperature change by altering the accessibility of the Shine–Dalgarno (SD) sequence and have been most commonly found upstream of heat shock proteins and virulence-associated genes (Roßmanith and Narberhaus 2016). Melting of the thermometer hairpin exposes the SD sequence, thereby allowing the ribosome to bind and initiate translation (Narberhaus 2010). In this way, RNATs, as well as other regulatory RNAs, are able to directly control expression in an RNA structure-dependent manner.
While many studies have examined RNA structure transcriptome-wide (Rouskin et al. 2014; Zubradt et al. 2017), we focused specifically on RNA structure in response to temperature changes transcriptome-wide using a technique that we developed called Structure-seq2 (Ritchey et al. 2017). This method takes advantage of in vivo dimethyl sulfate (DMS) probing and is readily amenable to different cellular conditions. We recently applied this method to tRNA modifications in Escherichia coli (Yamagami et al. 2022), amino acid starvation in Bacillus subtilis (Ritchey et al. 2020), as well as during heat shock (Su et al. 2018) and salt stress in plants (Tack et al. 2020).
Here, we used Structure-seq2 to probe RNAs in B. subtilis grown at four different temperatures ranging over almost 20°C. While previous studies have looked at changes to RNA structure and abundance between two temperatures varying by this much, these drastic and often rapid shifts in temperature are unlikely to occur frequently in nature (Budde et al. 2006; Twittenhoff et al. 2020). As a soil bacterium, B. subtilis experiences a wide range of temperatures, and is more likely to experience gradual changes in temperature such as during the day–night cycle (Budde et al. 2006).
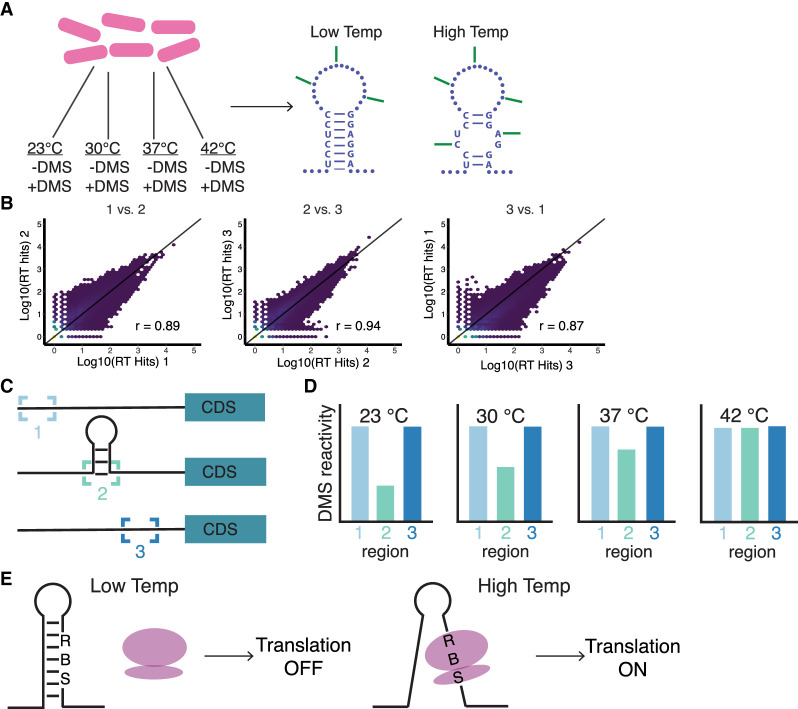
To more accurately capture the temperature changes experienced in the soil, B. subtilis cells were grown continuously (i.e., with no temperature shift) at 23°C, 30°C, 37°C, and 42°C and treated with DMS during exponential growth (Fig. 1A). Results from this study highlight the dynamic melting of RNA structure as the temperature increases. These results also demonstrate a nonmonotonic response to the temperature increases across the transcriptome and within subregions of the transcriptome (UTRs and coding sequences [CDS]). Our data accurately map to the well-characterized RNA kink-turn (k-turn) motif, validating the approach, and were used to identify large reactivity changes in 5′ UTRs over these temperatures (Fig. 1C,D). This analysis led to the discovery of two novel RNATs (Fig. 1E) that regulate the translation of the glycerol permease genes, glpF and glpT.
FIGURE 1.
Schematic for DMS treatment, library quality, data analysis, and reporter assays. (A) Generation of DMS-induced reverse transcription (RT) stops in Bacillus subtilis grown at one of four different temperatures. RNA samples are then prepped for Illumina sequencing using Structure-seq2. (B) Comparison of RT stops in three DMS-treated biological replicates grown at 37°C. Pearson correlation coefficients (r) are shown. (C) Schematic representing a 5′ untranslated region (5′ UTR) with unstructured (1 and 3) and structured (2) regions. (D) Reactivity profile of the 5′ UTR from (C) plotted for each temperature. (E) Schematic of RNAT-controlled translation initiation. At low temperatures, the ribosome binding site (RBS) is sequestered, and the ribosome (purple) does not bind. At high temperatures, the RBS is available for the ribosome to bind and initiate translation of the downstream gene.
RESULTS
Transcriptome-wide changes in reactivity across temperatures
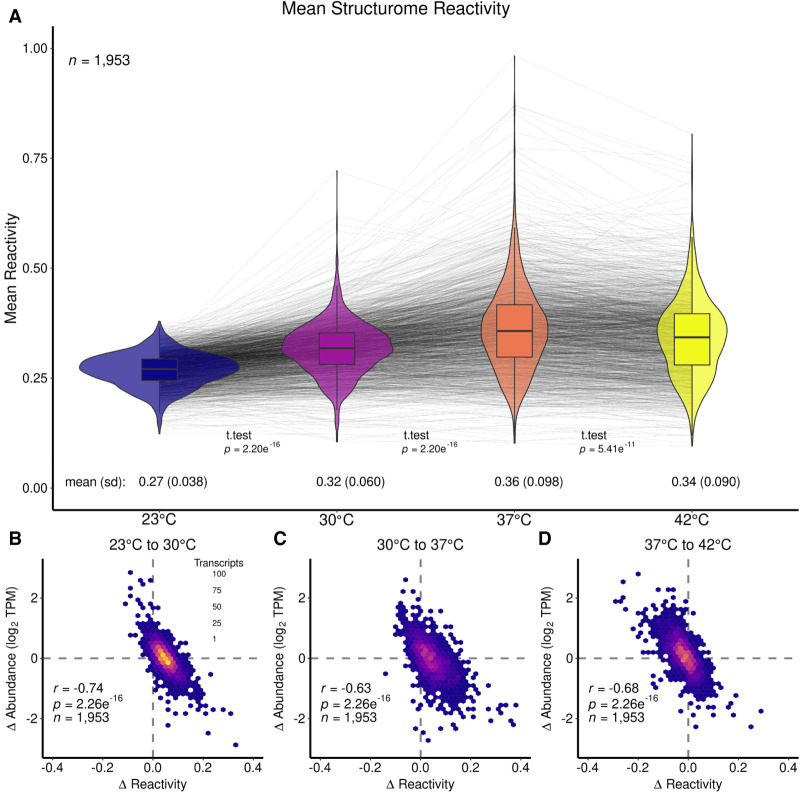
To examine global changes in the reactivity of the B. subtilis transcriptome in response to temperature, cells were grown at four temperatures 23°C, 30°C, 37°C, and 42°C. Once the cultures reached the mid-exponential phase, they were treated with DMS (Fig. 1A). The resulting RNA samples were then prepared as Structure-seq2 libraries (Ritchey et al. 2020) and sequenced, which generated RT stops that were highly correlated (Fig. 1B). The sequencing reads were processed using StructureFold2 (Tack et al. 2018). This procedure resulted in 1953 genes that met threshold criteria and with sufficient coverage in all four temperatures (described in Materials and Methods). Figure 2A provides violin plots with overlaid box plots for the mean reactivity per transcript across all four temperatures of 23°C, 30°C, 37°C, and 42°C. The spider lines of the plot mark the reactivity change path of a single transcript from temperature to temperature. This approach highlights that it is not just the transcripts with high reactivity that become more accessible to DMS reactivity with temperature, but rather the transcriptome undergoes myriad structural changes in response to temperature.
FIGURE 2.
Transcriptome-wide changes in reactivity across four growth temperatures in B. subtilis. (A) Mean structurome changes in reactivity for all transcripts at 23°C (blue), 30°C (purple), 37°C (orange), and 42°C (yellow) shown as violin plots with box plots in the center. Gray spider lines link individual transcript reactivity in each condition. Global mean values are given below the violin plot for each temperature, as well as standard deviation (sd) values. Additionally, t-test P-values are given between the compared data sets. Mean reactivity increased from 23°C to 30°C, and further increased at 37°C, then slightly decreased at 42°C. (B–D) Scatter plots showing Δ reactivity (from higher temperature to lower temperature) against Δ abundance (from higher temperature to lower temperature) for (B) 23°C–30°C, (C) 30°C–37°C, and (D) 37°C–42°C. All plots show an inverse correlation with the strongest correlation shown in (B).
At the lowest temperature tested (23°C), the transcriptome is the least reactive with a mean reactivity of 0.27 (Fig. 2A). At intermediate temperatures of 30°C and 37°C (Fig. 2A), there is an increase in the reactivity of the transcriptome at each temperature (mean reactivity at 30°C increases to 0.32, mean reactivity at 37°C increases further to 0.36). The 30°C temperature is in the middle of the mesophilic range, of which B. subtilis is a member, and 37°C is typically used as the optimal growth temperature in laboratory settings (Droffner and Yamamoto 1985; Kobayashi et al. 2003). Changes in the distribution of reactivity of the transcriptome with temperature are also evident. In particular, the sd of the reactivity broadens markedly from 0.038 to 0.060 to 0.098 as the temperature changes from 23°C to 30°C to 37°C (Fig. 2A). The interquartile range (iqr) and median absolute deviation (mad) follow a similar trend (Supplemental Table T.4). Both the largest mean reactivity and sd in reactivity occur at 37°C; however, it is not necessarily the transcripts with the most reactivity at 30°C that exhibited the most reactivity at 37°C, as revealed by crossing of many of the spider lines. Overall, these results reflect the structural shifts of the transcriptome from less reactive and less different (i.e., more protected and more similar) at the lower temperature to more reactive and more different (i.e., less protected and less similar) at 37°C.
Intriguingly, at the highest temperature of 42°C there was a slight decrease in mean reactivity (Fig. 2A; mean reactivity = 0.34) compared to at 37°C (mean reactivity = 0.36). At 42°C, there was also a slight decrease in sd (Fig. 2A; sd reactivity = 0.090) compared to 37°C (sd reactivity = 0.098). The mean and sd values across the set of four temperatures show that the connection between reactivity and temperature was nonmonotonic. The increase in temperature from 37°C to 42°C could begin to unfold RNA structures, yet here the mean reactivity decreased indicating that RNA structure accessibility was under more biological regulation than pure thermodynamic control. While the mean reactivity of the transcriptome decreased from 37°C to 42°C, when comparing the 42°C data to that from either of the lower temperatures (23°C or 30°C) it still showed an increase in reactivity.
These changes in the reactivity of the transcriptome with temperature can be compared to changes in the relative abundance of transcripts with temperature. Previously, it was shown that there is an inverse correlation between changes in transcript abundance and changes in transcript reactivity (Su et al. 2018; Ritchey et al. 2020; Tack et al. 2020). This trend was seen for this study as well (Fig. 2B–D). When focusing on changes between 23°C and 30°C, the reactivity changes and abundance changes resulted in an inverse correlation and an r-value of −0.74 (Fig. 2B). Thus, those transcripts that were more reactive at 30°C compared to 23°C were also less abundant, whereas those transcripts that were more abundant were less reactive at 30°C (Fig. 2B). The same trend held true when comparing 30°C and 37°C (Fig. 2C), which has an r-value of −0.63, and when comparing 37°C and 42°C (Fig. 2D), which has an r-value of −0.68. All inverse correlation plots had P-values of 2.3 × 10−16.
Reactivity changes with temperature across subregions of the transcriptome
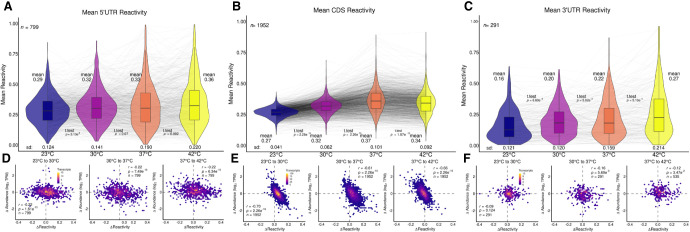
While the reactivity across the entire transcript is informative, it was estimated that at least 4% of the B. subtilis genome is controlled by cis-acting RNAs with the majority located in 5′ UTRs (Irnov et al. 2006, 2010). Thus, the transcriptome was divided into the three subregions of 5′ UTR, CDS, and 3′ UTR, and the mean reactivity at all temperatures for each of these subregions was plotted (Fig. 3A–C). The reactivity pattern for each transcript CDS region (Fig. 3B) mimicked the reactivity pattern found for each transcript in its entirety across the four temperatures (Fig. 2A), which is unsurprising since each transcript is mostly CDS. Hence, the mean CDS reactivity for 23°C, 30°C, 37°C, and 42°C (Fig. 3B; 0.27, 0.32, 0.37, and 0.34, respectively) was nearly identical to the means for the entire transcriptome, both in values and in the shape of the nonmonotonic trend (Fig. 2A; 0.27, 0.32, 0.36, and 0.34, respectively). However, the reactivity for the UTRs showed entirely different patterns. Both the 5′ UTR (Fig. 3A) and 3′ UTR (Fig. 3C) regions showed a wider distribution of reactivity values as compared to the entire transcript (e.g., sd of 0.124 for 5′ UTR and 0.121 for 3′ UTR vs. 0.041 for CDS at 23°C). Furthermore, the reactivity in both the 5′ UTR and 3′ UTR regions showed a monotonic trend between increases in reactivity with each increase in temperature, including the increase from 37°C to 42°C (Fig. 3A: 0.29, 0.32, 0.33, and 0.36, respectively; Fig. 3C: 0.16, 0.20, 0.22, and 0.27, respectively).
FIGURE 3.
Subregions of the transcriptome show changes in reactivity across four temperatures. Violin plots overlayed with box plots (A–C) and scatter plots (D–F) are formatted as in Figure 2. Mean reactivity across four temperatures for (A) 5′ UTR, (B) CDS, and (C) 3′ UTR. Global trends in mean reactivity exhibited by the entire transcriptome (Fig. 2) are similar to those displayed in the CDS plot (B). (D–F) Scatter plots showing Δ reactivity against Δ abundance of 23°C–30°C (left), 30°C–37°C (middle), and 37°C–42°C (right) for (D) 5′ UTR, (E) CDS, and (F) 3′ UTR. Trends exhibited by the entire transcriptome are likewise displayed in the CDS plots (E).
Parallel to the comparison between changes in reactivity and changes in abundance described above for entire transcripts (Fig. 2B–D), the relationship between these two parameters is shown for each of the subregions (Fig. 3D–F). While the inverse correlation between change in reactivity and change in abundance observed for the entire transcriptome (Fig. 2B–D) was maintained for the CDS with an average r-value of −0.66 and a P-value of 2.3 × 10−16 (Fig. 3E), this relationship was modest for the 5′ UTR with an average r-value of −0.22 and a P-value ≤ 7.5 × 10−10 (Fig. 3D) and absent in the 3′ UTR with a P-value range of 0.0057–0.12 (Fig. 3F). Notably, while the change in reactivity with temperature remained nonmonotonic for the CDS, increasing from 23°C to 37°C and then decreasing between 37°C and 42°C, it became monotonic for the 5′ UTR and 3′ UTR, as mentioned above. The sd followed a similar trend of being nonmonotonic for the CDS, which is similar to the entire transcriptome, and increasing monotonically for the UTRs (Supplemental Table T.4). Thus, the UTRs appear to be under thermodynamic control over the entire temperature range.
Structure-seq2 identified known RNA motifs
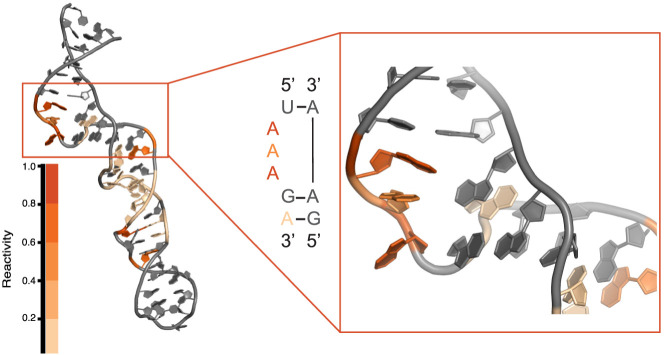
The wide distribution of reactivity seen for 5′ UTRs implies that this portion of the transcriptome is structurally dynamic under the four temperature conditions. To provide confidence that unpaired and available residues of an unknown structure could be identified by Structure-seq2, as provided in the next section, the reactivity of a portion of the 5′ UTR for tyrS was examined. This 5′ UTR contains a T box riboswitch, an RNA structural element that changes conformation to regulate transcription of the downstream gene (Grundy et al. 2002). A solution structure for a truncated version of the tyrS 5′ UTR from B. subtilis (PDB ID: 2KZL) shows a k-turn structural motif (Wang and Nikonowicz 2011). This well-known motif serves as an ideal candidate for accessing the accuracy of the in vivo probing method specifically used here.
K-turns (Fig. 4) are classified by a 3 nt bulge flanked on the 5′ end by Watson–Crick pairings and on the 3′ end by highly conserved G•A and A•G mismatches, although the identity can vary (Schroeder et al. 2010). This arrangement produces a strong kink in the helix (Schroeder et al. 2010). Specifically for the 5′ UTR for tyrS, the bulge consists of three A residues. Because of the unique geometry formed by this motif, the bulged A residues and potentially the A residues of the 3′ mismatches (since the base pairs involve only the Hoogsteen edge of the A residues) could be probed using Structure-seq2. Mapping the 37°C reactivity data from the 5′ UTR of tyrS (Fig. 4) showed strong reactivity for the bulged A residues (orange and dark orange) and slight reactivity for the second A•G base pair (pale orange). The reactivity data from the other temperatures showed a similar pattern (Supplemental Fig. 1). This result confirmed that the Structure-seq2 data collected here can accurately report on in vivo RNA structures, and further motivated the search for unique motifs or elements that change structure at different temperatures.
FIGURE 4.
Mapping in vivo reactivity onto a known RNA motif. The tyrS T box riboswitch from B. subtilis contains a k-turn in the lower portion of the structure (PDB ID: 2KZL). This particular k-turn is characterized by three bulged A residues followed by two GA base pairs. The bulged A residues show moderate (orange) to strong (dark orange) DMS reactivity at 37°C.
Identification of glpF and glpT as RNA thermometers
Given that Structure-seq2 accurately probes accessible RNA residues and the wide distribution of reactivity changes across the temperature range in the 5′ UTR, this region of the transcriptome was chosen to search for thermo-responsive RNAs. The reactivity trends of the transcriptome and subregions presented above represent reactivity averaged over many transcripts; however, the reactivity pattern across temperatures of a single transcript can help to identify structural changes that occur in particular regions of a specific RNA. For example, it is known that RNATs are RNA structures that change conformation in response to changes in temperature (Narberhaus 2010). Often found upstream of heat shock proteins and virulence genes, this type of RNA element could be used more generally to regulate gene expression in response to temperature changes. To this end, reactivity plots per nucleotide for the 5′ UTR of all individual transcripts with overlapping coverage thresholds (see Materials and Methods) across all four temperatures were analyzed, focusing on nucleotides with large changes in reactivity.
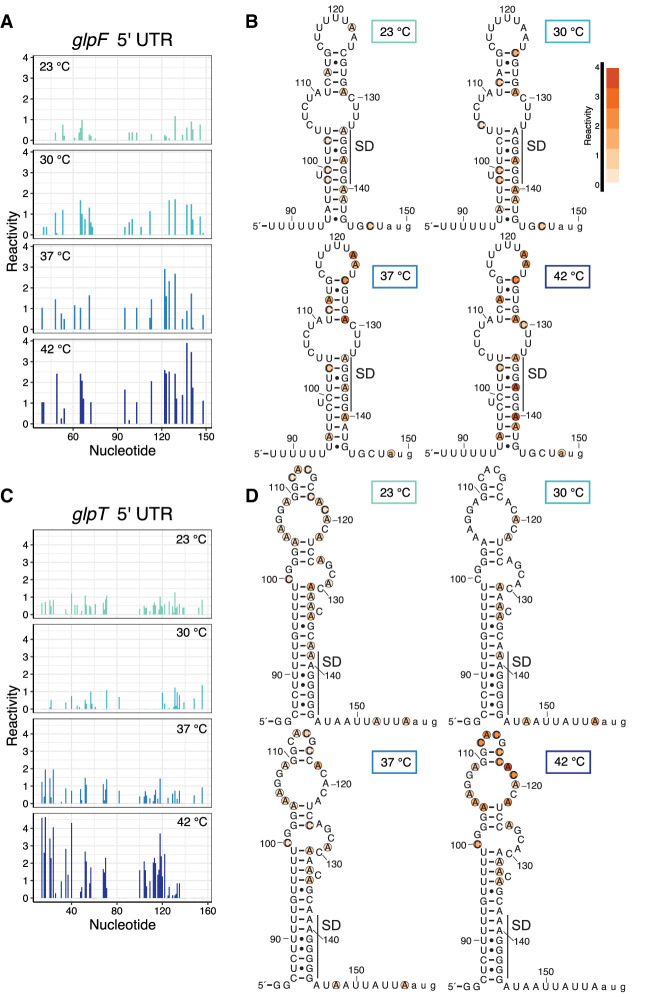
From this analysis, the 5′ UTRs of glpF and glpT emerged as potential candidates for RNATs. These genes are part of the glycerol regulon, of which there are four operons: glpP, glpFK, glpD, and glpTQ (Glatz et al. 1996; Darbon et al. 2002). Specifically involved in glycerol (glpF) and glycerol-3-phosphate (glpT) transport, the 5′ UTRs for both of these operons contain inverted repeats similar to that found in glpD (Rutberg 1997). The transcriptionally controlled termination of glpD is regulated by binding of GlpP, with the expectation that similar control exists for all of the glp regulon (Rutberg 1997). Consistent with that hypothesis, intrinsic terminators were identified in the 5′ UTRs of glpF and glpT by Term-seq (Mondal et al. 2016). Strikingly, both glpF and glpT have nucleotides in the 5′ UTR immediately downstream from these terminators with large reactivity increases with increasing temperature (Fig. 5A,C, respectively). These reactivity increases can be compared to minimal changes in the 5′ UTRs of glpD (Supplemental Fig. 2A), which is encoded downstream from glpF (glpK is the second gene of the glpF operon but did not meet the coverage criterion), and of glpQ (Supplemental Fig. 2B), the second gene in the glpT operon. For glpF, this includes nucleotide 90 and extends to the start codon at position 150 (Fig. 5A) and shows gradual increases in reactivity at each temperature throughout the predicted secondary structure in this region. Mapping the reactivity onto this predicted structure highlights the A and C residues that became more reactive with increasing temperature (Fig. 5B). Notably, A137 and A140, which are part of the glpF SD sequence, increased in reactivity, and therefore gave this region of the UTR increased accessibility at the highest temperature (Fig. 5B). Similarly, glpT had two regions of nucleotides that showed increased reactivity at each temperature (Fig. 5C). Focusing on the downstream region (nucleotide 90 and extending to the start codon at position 158), increased reactivity (and therefore increased accessibility) was observed (Fig. 5D; nucleotides 104–123) for nucleotides in a large apical hairpin structure that includes a 5 × 5 internal loop, GG/CC stem, and CACG hairpin loop regions just upstream of the glpT SD sequence. Notably, the CACG hairpin loop is a member of the stable YNMG RNA tetraloop family (Proctor et al. 2002). These changes in reactivity suggested that the opening of the entire apical structure (internal loop, Watson–Crick pairing, and stable tetraloop) might stimulate translation (see below). In summary, for both glpF and glpT, regions of increased reactivity with temperature map onto a secondary structure, on or near the SD sequence. This type of secondary structure is a hallmark of all known RNATs (Narberhaus 2010; Kortmann and Narberhaus 2012; Nshogozabahizi et al. 2019).
FIGURE 5.
Reactivity changes with temperature identify potential regulatory RNA elements. (A) Reactivity per nucleotide for the glpF 5′ UTR at 23°C, 30°C, 37°C, and 42°C. Numbering extends from the start of transcription to the start codon. (B) Reactivity for nucleotides 88–150 of the glpF 5′ UTR mapped onto the predicted secondary structure. Normalized reactivity ranges from low (pale orange) to high (dark orange). The position of the SD sequence is shown. (C) Reactivity per nucleotide for the glpT 5′ UTR at 23°C, 30°C, 37°C, and 42°C. Numbering extends from the start of transcription to the start codon. (D) Reactivity for nucleotides 84–158 of the glpT 5′ UTR mapped onto the predicted secondary structure. Normalized reactivity ranges from low (pale orange) to high (dark orange). The position of the SD sequence is shown. The schematics in panels B and D begin immediately downstream from experimentally identified intrinsic terminators and extend to the AUG start codon (shown in lowercase letters).
After identifying specific regions on the glpF and glpT 5′ UTRs containing reactivity changes with parallel temperature changes, it was imperative to validate the behavior by an additional method in vivo. Since RNATs function by melting of an SD-sequestering hairpin at elevated temperatures to regulate gene expression, in vivo expression assays were performed to test whether the changes detected using Structure-seq2 could lead to changes in gene expression. A similar approach was recently undertaken in the pathogenic bacterium, Yersinia pseudotuberculosis, where in vivo probing scores led to the discovery of new RNATs that were verified by translational fusions to bgaB (a thermostable β-galactosidase) and compared to a known RNAT from Y. pseudotuberculosis (5′ UTR of katA) (Twittenhoff et al. 2020). To validate the candidates identified by this study (glpF and glpT), similar translational fusions were generated and tested.
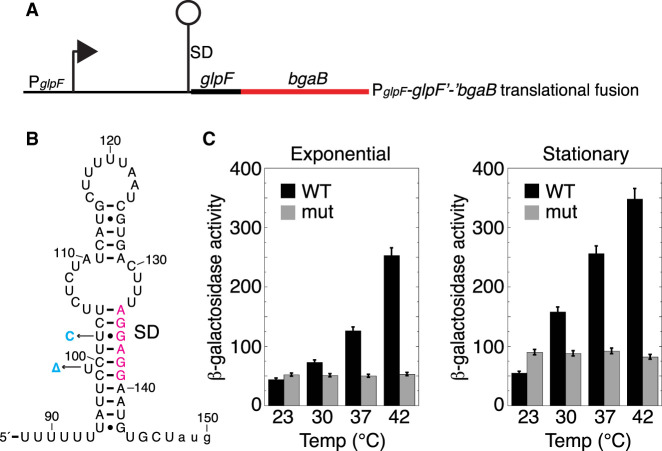
glpF expression was examined using a glpF′-′bgaB translational fusion containing the natural promoter, the 5′ UTR (including the SD sequence and potential thermometer hairpin), and the start codon fused in frame to the bgaB CDS (Fig. 6A,B). Expression of the fusion was measured by growing cultures at each of the four temperatures used for the in vivo probing studies (Fig. 6C). Expression steadily increased with increasing temperature, culminating in a fivefold increase during the exponential phase and a sevenfold increase during early stationary phase growth (Fig. 6C). These results are consistent with the SD-sequestering hairpin functioning as an RNAT. Since the transcript levels for glpF were about twofold lower at 42°C compared to 23°C (Supplemental Table D.5), the increased expression for glpF underrepresents the impact of the RNAT. To confirm that the increased expression was the result of changes to the RNA structure, the SD-sequestering hairpin was mutated to increase the stability of the hairpin. By mutating U102 to C (a U•G to CG transition) and deleting the bulged U99, the predicted stability of the glpF UTR according to Mfold (Zuker 2003) increased by nearly 7 kcal/mol from −23.80 kcal/mol to −30.50 kcal/mol (Fig. 6B; mutations shown in cyan). Expression from this mutant fusion did not increase with increasing temperature in either growth phase (Fig. 6C). Taken together, these results demonstrate that glpF expression responds to temperature changes and that the SD-sequestering hairpin functions as an RNAT.
FIGURE 6.
Effect of temperature on glpF expression. (A) Schematic representation of the glpF′-′bgaB translational fusion showing the promoter (P) and SD sequence. The glpF RNAT is depicted as a lollipop. (B) glpF RNAT. SD sequence is in magenta and two point mutations that stabilize the thermometer are in cyan. (C) β-Galactosidase activity of the glpF′-′bgaB translational fusion grown at the indicated temperatures during exponential and stationary phase growth. Values are averages of at least three independent experiments ± sd. WT, wild type fusion; mut, mutant fusion with the two mutations shown in (B).
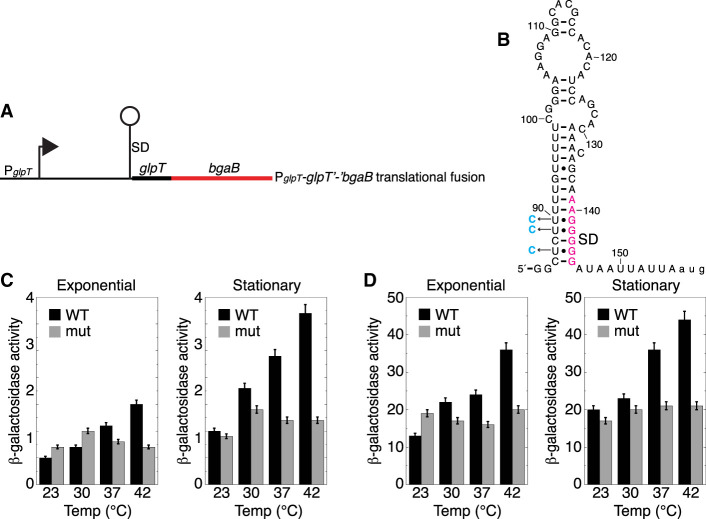
Likewise, glpT expression was tested using a similar translational fusion, including the natural promoter, 5′ UTR (including the SD sequence and potential thermometer hairpin), and start codon fused in frame to the bgaB CDS (Fig. 7A,B). Unlike glpF, the reactive nucleotides for glpT do not overlap with the SD sequence (Fig. 5D), but instead involve the nucleotides in the loop of the SD-containing hairpin. However, a stretch of mostly U residues (including four consecutive U residues) is predicted to pair with the SD sequence, which is another hallmark feature of a specific class of known RNATs (Waldminghaus et al. 2007; Narberhaus 2010). Similarly, expression of the fusion was measured from cultures grown at each of the four in vivo probing temperatures. Expression of the fusion steadily increased with increasing temperature, culminating in a threefold increase during the exponential phase and a fourfold increase during stationary phase growth (Fig. 7C). Furthermore, this temperature-dependent increase in expression was also observed for the glpT fusion in which the intrinsic terminator was deleted, indicating that the increase in expression was associated with the SD-sequestering hairpin and not from transcription attenuation (Fig. 7D). As for glpT, the transcript levels were about twofold lower at 42°C compared to 23°C (Supplemental Table D.5), indicating that the increased expression underrepresents the impact of the RNAT. Mutations were made to the stretch of U residues opposite the SD sequence (Fig. 7B; mutations shown in cyan in three U•G to CG transitions) to strengthen base-pairing and increase the predicted stability of the hairpin according to Mfold (Zuker 2003) by over 8 kcal/mol (similar to our change in the glpF hairpin) from −13.30 kcal/mol to −21.80 kcal/mol. As was expected due to the increased hairpin stability, increased expression was not observed at the higher temperatures in either the growth phase in the presence or absence of the intrinsic terminator (Fig. 7C,D). Taken together, these results demonstrate that glpT expression responds to temperature changes and that the SD-sequestering hairpin functions as an RNAT.
FIGURE 7.
Effect of temperature on glpT expression. (A) Schematic representation of the glpT′-′bgaB translational fusion showing the P and SD sequence. The glpT RNAT is depicted as a lollipop. (B) glpT RNAT. SD sequence is in magenta and three point mutations that stabilize the thermometer are in cyan. (C) β-Galactosidase activity of the glpT′-′bgaB translational fusion grown at the indicated temperatures during exponential and stationary phase growth. Values are averages of at least three independent experiments ± sd. WT, wild type fusion; mut, mutant fusion with the three mutations shown in (B). (D) Same as (C) except the intrinsic terminator was deleted from the glpT leader region.
DISCUSSION
Exploring RNA structure in vivo has expanded from the initial work of studying one RNA at a time to the current abilities of studying many RNAs in a cell simultaneously (Mortimer et al. 2014; Mitchell et al. 2019). The role RNA plays in cellular functions and processes has not been fully enumerated, but it is clear that the structure of RNA is a critical regulator (Mortimer et al. 2014; Mitchell et al. 2019). In this study, we set out to survey changes to the B. subtilis transcriptome across four temperatures. The reactivity changes with the temperature of the transcriptome, subregions, and of individual RNAs represent the dynamic nature of RNA structure. These changes led to the discovery of two RNAs that function as thermo-responsive elements known as RNATs.
Previous studies that probed RNA in an analogous way, resulted in a decrease in reactivity upon amino acid starvation in B. subtilis, and to an increase in reactivity during both heat and salt stress in plants (Su et al. 2018; Ritchey et al. 2020; Tack et al. 2020). In the current study, the lowest mean reactivity was observed for 23°C. As the temperature increased to 30°C and 37°C, the mean reactivity of the transcriptome also increased. Then, at the highest temperature of 42°C, the mean reactivity of the transcriptome decreased slightly giving an overall nonmonotonic character. Changes in the distribution of reactivity followed a similar trend, with the sd of the reactivity increasing from 23°C to 37°C and then decreasing slightly to 42°C. These changes are consistent with the transcriptome becoming less protected and less similar as temperature increases from 23°C to 37°C, and then reversing slightly as the temperature rises to 42°C. For the lower temperature range, these changes reflect expected trends in thermodynamics as RNA structure is known to melt with increasing temperature, with some transcripts being more sensitive to temperature than others. For the higher temperature range, the changes may reflect increased binding of proteins, whose identity and function can be probed in future studies. The greater range in reactivity at 30°C and 37°C could reflect higher expression at these optimal temperatures. Finally, the 5′ UTR and 3′ UTR showed monotonic behavior for both the mean reactivity and the sd of the reactivity over the entire temperature range of 23°C–42°C, suggesting these regions may remain under thermodynamic control over the entire temperature range. Additionally, the sd of the reactivity of the UTRs was two to three times greater than of the CDS, which is suggestive of greater unevenness in reactivity and therefore of more structure of the UTRs. Perhaps the thermodynamic control for the UTRs is due to their having more structure than the CDS and being less dominated by protein binding. These observations are consistent with the known importance of RNA structure in the 5′ UTRs in B. subtilis (Babitzke et al. 2019).
Another probing technique called Lead-seq was used to look at transcriptome changes at two temperatures in the human pathogen Y. pseudotuberculosis, and it was similarly found that the transcriptome was less structured at 37°C compared to 25°C (Twittenhoff et al. 2020). Increased temperature also led to an increase in transcriptome reactivity in rice, as well as for tRNA in E. coli (Su et al. 2018; Yamagami et al. 2022). However, what makes our present study unique is the probing of RNA structural reactivity across four temperatures rather than just two, thereby revealing nonmonotonic relationships between temperature and reactivity, which was not possible to observe in the other studies.
Distinct regions of the transcriptome can participate in cellular mechanisms differently. For instance, in B. subtilis a much higher density of NusA-stimulated and NusG-dependent RNA polymerase pauses were located in 5′ UTRs compared to the CDS, which is consistent with pausing playing a role in a variety of transcription attenuation mechanisms (Yakhnin et al. 2020; Jayasinghe et al. 2022). Likewise, the same propensity could exist for conferring translational control by elements in 5′ UTRs compared to the CDS. Indeed, the mean reactivity trends for the UTRs and CDS differ. Whereas the nonmonotonic trend between temperature and reactivity seen in the transcriptome was observed in the CDS, a monotonic trend existed for both the 5′ UTR and 3′ UTR (Fig. 3). Since regulatory RNAs, which could include unique temperature-responsive elements, tend to be in the 5′ UTRs, it was encouraging to see a continual increase in reactivity in the 5′ UTR with increasing temperature given that RNA structure is continuously destabilized with temperature. In fact, the mean reactivity for the 5′ UTR was higher than the mean reactivity for the CDS at the lowest and highest temperatures (0.29 vs. 0.27 at 23°C and 0.36 vs. 0.34 at 42°C). The higher reactivity in the 5′ UTR at the low temperature could be the result of less translation, and therefore less ribosome-dependent protection of the transcript in the translation initiation region. Additionally, the higher reactivity in the 5′ UTR at the high temperature is likely due to the melting of RNA structures making the RNA more accessible.
Further differences in reactivity between the subregions of the transcriptome are exemplified by comparing the difference in abundance versus the difference in reactivity. The strong inverse correlation for these differences seen in the transcriptome (Fig. 2B) is similarly seen in the CDS (Fig. 3E), yet that correlation is not present in either UTR (Fig. 3D,F). A strong correlation between abundance and reactivity in the CDS of the E. coli transcriptome was also observed in a similar study (Del Campo et al. 2015). Comparing mRNA abundance with parallel analysis of RNA structure (PARS) scores (a measure of in vivo structural propensity) for the CDS revealed the same correlation of higher abundant transcripts having more structure (in the case of Structure-seq, more structure results in less reactivity; Del Campo et al. 2015).
In addition to global changes to the transcriptome in response to temperature, individual transcripts can respond in a structure-dependent manner. In fact, RNATs function in this way to regulate gene expression. To find such elements, the reactivity profiles per nucleotide across the four temperatures were compared for all 5′ UTRs, and two candidates were chosen for further testing. glpF and glpT had regions with large reactivity changes that paralleled the temperature changes. This phenomenon could be explained by structural changes of a thermo-responsive RNA element, like an RNAT. In vivo expression assays confirmed this hypothesis.
Unsurprisingly, the RNATs discovered here are not associated with heat shock genes, where they have been found previously, as the B. subtilis cells were not exposed to a heat shock treatment. Instead, the cells were grown continuously at 23°C, 30°C, 37°C, and 42°C, which was not expected to elicit a heat shock response but would still be capable of altering cell physiology. The two newly identified RNAs that act as thermometers control the expression of glpF and glpT, which encode permeases responsible for transporting glycerol or glycerol-3-phosphate into the cell, respectively (Darbon et al. 2002). The precise role for the increase in these solutes is not known; however, one hypothesis is explored here. Many bacteria and archaea accumulate similar solutes with the distinction that the archaeal solute often carries a negative charge compared to the bacterial counterpart (Roeßler and Müller 2001). For example, glucosylglycerol is accumulated by cyanobacteria in response to osmolality while the companion solute, diglycerol phosphate, which carries a negative charge, accumulates in Archaeoglobus fulgidus (Reed et al. 1986). The accumulation of diglycerol phosphate in A. fulgidus was shown to increase in response to salt stress and also to provide thermoprotective properties to certain proteins typically inactivated by heat (Lamosa et al. 2000).
Many compatible solutes have been shown to confer osmoprotective abilities to the cell, and some have further been studied to see if the same solutes also offer thermoprotection (Holtmann and Bremer 2004, Hoffmann and Bremer 2011). Specifically, glycine betaine is a solute previously shown to function as an osmoprotectant in B. subtilis (Holtmann and Bremer 2004, Hoffmann and Bremer 2011). Additionally, when tested at 37°C and 52°C in a minimal medium with and without glycine betaine, there was a noticeable thermoprotection to the growth of B. subtilis with glycine betaine at the higher temperature (Holtmann and Bremer 2004). Furthermore, a glycine betaine metabolism intermediate, dimethylglycine, exhibited similar thermoprotection for B. subtilis after both a heat stress and a cold stress (Bashir et al. 2014).
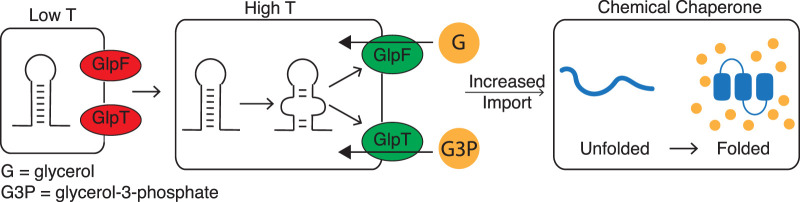
Previous studies showed that salt-stressed E. coli accumulates glycerol via GlpF-mediated diffusion (Richey and Lin 1972; Heller et al. 1980). In another study, glycerol and other solutes were tested for thermoprotection during heat stress in vitro (Diamant et al. 2001). Increased levels of glycerol were shown to behave as a chemical chaperone. For example, the model protein mitochondrial malate dehydrogenase (MDH) was protected from heat inactivation through stabilization of the native fold at 44°C by glycerol (Diamant et al. 2001). Additionally, when heat-denatured MDH was prebound to GroEL (a known protein chaperone) it exhibited a refolding rate 12× faster in the presence of glycerol than with no osmolyte present (Diamant et al. 2001). The ability of glycerol to chaperone proteins by stabilizing the folded structure has been further examined, and it was proposed that these characteristics extend from glycerol preventing partial unfolding of the native protein, thereby inhibiting the formation of aggregation-prone intermediates (Vagenende et al. 2009). In this same way, B. subtilis may accumulate glycerols to promote the thermoprotection of proteins (Fig. 8) at high temperatures through RNAT-controlled glycerol uptake.
FIGURE 8.
Schematic representing possible model of regulation. At low temperatures, the 5′ UTR for both glpF and glpT showed little reactivity and are therefore depicted as folded and import is low (red). As temperature increases, the 5′ UTR reactivity increased and is depicted as melting from a structured RNA to a more unstructured RNA. This in turn increased expression and could result in increased import (green) of glycerol (G) and glycerol-3-phosphate (G3P) by GlpF and GlpT, respectively. These solutes could function as chemical chaperones to protect proteins during this temperature increase.
MATERIALS AND METHODS
Bacterial strains, plasmids, and oligonucleotides
All B. subtilis strains, plasmids, and oligonucleotides can be found in Supplemental Tables S1–S3, respectively.
Bacterial growth and in vivo treatment
Single colonies of B. subtilis strain PLBS338 (Yakhnin et al. 2004) were inoculated in a 2 mL standing overnight culture in Luria–Bertani (LB) medium for three biological replicates. Next, 0.2 mL of the overnight culture was diluted 100-fold into 20 mL of LB medium and then grown in a shaking water bath to mid-exponential phase (75 Klett), for each of the three replicates. Culture and growth temperatures were as follows: 23°C, 30°C, and 37°C samples were inoculated and grown at 23°C, 30°C, and 37°C, respectively, while 42°C samples were inoculated from overnight cultures grown at 37°C.
Once growth had reached the mid-exponential phase for each of the four growth temperatures, control samples (−DMS) were removed prior to DMS treatment and then the remaining cultures were treated with DMS in a safety hood while wearing appropriate PPE. For the control samples, 6 mL of culture was transferred to a tube containing 0.45 g dithiothreitol (DTT) and slightly agitated for 2 min for a mock quench at the growth culture temperature in the shaking water bath. After 2 min, the solution was poured into an equal volume of a partially frozen killing buffer containing 10 mM Tris-HCl (pH 7.2), 5 mM MgCl2, 25 mM NaN3, 1.5 mM chloramphenicol, and 12.5% ethanol. For the treated samples (+DMS), DMS was added to the remaining 14 mL culture to a final concentration of 50 mM, and incubation was continued at the growth temperature. After 5 min, 1.05 g DTT was added to the culture and slightly agitated at the growth temperature in the shaking water bath for 2 min to quench the reaction. Immediately, 6 mL of the quenched culture was added to an equal volume of partially frozen killing buffer. All samples were centrifuged at 10,000 rpm for 10 min at 4°C to pellet cells, which were subsequently resuspended into 1 mL ice-cold killing buffer. Resuspended cells were transferred to a microfuge tube and the pellet/wash procedure was repeated. Final cell pellets were stored at −20°C.
Library preparation
Cell pellets for both control and treated samples from each temperature were lysed with lysozyme (10 mg/mL), and total RNA was extracted using the RNeasy Mini Kit (Qiagen). RNA quality was checked using a Prokaryotic Total RNA Nano Bioanalyzer (Agilent) scan and examining accompanying RIN scores, as well as using a 1% agarose gel. rRNA was depleted (∼70% depletion efficiency) using a RiboZero rRNA Removal Kit (Illumina), and RNA quality was again checked using an mRNA Pico Bioanalyzer scan. Next, Structure-seq2 library preparation followed previously published protocols (Ritchey et al. 2017, 2019). Specifically, 500 ng of the rRNA-depleted RNA was reverse transcribed using a random hexamer, 1 U/µL Superscript III (Invitrogen), 500 µM each dNTP, 62.5 µM of biotinylated-dCTP (Tri-Link Biotechnologies), and 62.5 µM biotinylated-dUTP (Thermo Fisher). Reactions were purified using an RNA Clean & Concentrator-5 (C&C) Kit (Zymo Research). Biotin-incorporated cDNAs were pulled down using magnetic streptavidin beads, and then purified with C&C. A DNA hairpin adaptor was ligated to the 3′ end of the cDNAs and again reactions were purified with C&C, samples pulled down, and purified with C&C. PCR was performed for 25 cycles with primers having extended regions complementary to the Illumina sequencing primers. Library samples were size selected between 200 and 600 nt on a denaturing 8.3 M urea 10% polyacrylamide gel, gel-purified, ethanol precipitated, and then resuspended in water. Sequencing was performed using Illumina NextSeq High Output 150 nt single-end runs.
Data processing and analysis
Reads were checked for quality with FastQC before using StructureFold2 (Tack et al. 2018) for processing and analysis, as done previously (Ritchey et al. 2020). Briefly, sequencing resulted in approximately 756 million reads spread across the 24 libraries (four temperatures, ±DMS, and triplicate). Next, reads were trimmed of the end adapters, low-quality bases at the 3′ end, and short reads (20 bases or less) using CutAdapt v1.16 (Supplemental Table W.1; Martin 2011). Reads were mapped to the genome (Supplemental Table W.2) with a 93% mapping rate and then mapped to a custom-built B. subtilis transcriptome, following a previous procedure (Ritchey et al. 2020) using Bowtie2-2.3.4.1 (Langmead and Salzberg 2012) and filtered (Supplemental Table W.3) using SAMtools v0.1.19 (Li et al. 2009). Following these steps, filtered files were used to generate RT stop count (RTSC) files via StructureFold2 (Tack et al. 2018). These files were checked for coverage, specificity, and correlation between repeats (Supplemental Tables W.4, T.1, T.2; Supplemental Fig. 3). These files were merged if the correlation was good and transcript coverage was at or above the threshold of one RT stop per A or C residue. The RTSC files were then used to generate React files by a natural logarithm transformation, length normalization, and a final 2%–8% normalization, as part of the StructureFold2 module (Supplemental Tables D.1–D.4; Tang et al. 2015; Tack et al. 2018). Transcript abundance was estimated by calculating the sum of all RT stop counts, including run-off stops, per transcript resulting in transcripts per million (TPM) reads, according to the StructureFold2 module (Supplemental Tables T.3, D.5; Tang et al. 2015; Tack et al. 2018). All analyses were performed using R statistical Software (v4.1.2; R Core Team 2021). Reactivity structure mapping was done using the R2easyR R package (https://github.com/JPSieg/R2easyR) and R2R (Weinberg and Breaker 2011).
β-Galactosidase assay
B. subtilis cultures containing translational fusions with bgaB were grown at 23°C, 30°C, 37°C, and 42°C in LB supplemented with 0.5% glycerol and 5 µg/mL chloramphenicol. Cells were grown until the mid-exponential or early stationary phase. Four milliliters of each culture was collected, pelleted by centrifugation, washed with 10 mM Tris-HCl (pH 7.5), and frozen at −20°C. For β-galactosidase assays, cells were resuspended in 4 mL of Z buffer (Miller 1972) and OD600 values were measured. Samples (0.1 mL) were either used undiluted or diluted 10-fold with Z buffer (pH 6.4) depending on β-galactosidase activity (determined empirically). Ten microliters of fresh lysozyme solution (10 mg/mL) was added, and the mixtures were incubated for 5 min at 37°C prior to the addition of 10 µL of 10% Triton X-100 and 200 µL o-nitrophenyl-β-galactoside (ONPG). Reaction mixtures were incubated at 65°C until a yellow color appeared (Jensen et al. 2017). Reactions were stopped by adding 600 µL of 1 M sodium carbonate solution. Samples were centrifuged for 5 min, the clear supernatants were transferred into cuvettes and absorbance was measured at 420 nm and 550 nm. β-Galactosidase activity was calculated as previously described (Miller 1972).
DATA DEPOSITION
Processed data metrics can be found in the Supplemental Tables. Raw sequencing reads are available as FASTQ files, and processed data files are available as RTSC files on the Gene Expression Omnibus (GEO) database at the National Center for Biotechnology Information (NCBI) with the series entry GSE224126.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
The authors would like to thank the Penn State Genomics Core Facility at University Park, Pennsylvania for assistance with sequencing. This work was supported by the National Institutes of Health (GM127064 to P.C.B., GM128311 to E.A.J., and GM098399 to P.B.). Funding for open access charge was provided by the National Institutes of Health.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079652.123.
MEET THE FIRST AUTHOR
Elizabeth A. Jolley.

Meet the First Author(s) is an editorial feature within RNA, in which the first author(s) of research-based papers in each issue have the opportunity to introduce themselves and their work to readers of RNA and the RNA research community. Elizabeth (Liz) Jolley is the first author of this paper, “Transcriptome-wide probing reveals RNA thermometers that regulate translation of glycerol permease genes in Bacillus subtilis.” Liz is an Assistant Research Professor in the laboratory of Dr. Philip Bevilacqua at Pennsylvania State University. Her primary research focus is on RNA structure and function and how they relate to bacterial gene regulation.
What are the major results described in your paper and how do they impact this branch of the field?
There are two major results in this paper. The first is the nonmonotonic trend of in vivo DMS reactivity with increasing temperature across the Bacillus subtilis transcriptome. And the second is the discovery of RNATs regulating genes involved with glycerol transport. Both of these findings were somewhat unexpected and therefore really exciting!
What led you to study RNA or this aspect of RNA science?
As an Assistant Research Professor, I have now been studying RNA for a little over 10 years. When I first started my PhD, RNA was a little less widely known than it is today. I instantly had an interest in the stepsister of DNA that seemed to be doing a lot in the cell. That initial RNA interest has never left me. There is still more to learn about how RNA functions!
What are some of the landmark moments that provoked your interest in science or your development as a scientist?
A line from the classic 1992 film, Ferngully: The Last Rainforest, has always stuck with me: There are worlds within worlds. The longer I study science, and RNA in particular, the more and more I realize the truth of this statement. Whether on the small scale, like compartments within cells, or on the large scale, with exploration and discoveries across the universe, science is helping to explain the magic of the world(s) around us.
If you were able to give one piece of advice to your younger self, what would that be?
When I first started graduate school, I was a single mom of three young kids. I was unsure of myself and my future. I thought I can't be a scientist; my experiments usually don't work. My advice to my younger self would be to be confident! My experiments don't have to work every time for me to like what I do or to qualify me as a scientist.
REFERENCES
- Babitzke P, Lai Y-J, Renda AJ, Romeo T. 2019. Posttranscriptional initiation control of gene expression mediated by bacterial RNA-binding proteins. Ann Rev Microbiol 73: 43–67. 10.1146/annurev-micro-020518-115907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashir A, Hoffmann T, Smits SHJ, Bremer E. 2014. Dimethylglycine provides salt and temperature stress protection to Bacillus subtilis. Appl Environ Microbiol 80: 2773–2785. 10.1128/AEM.00078-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde I, Steil L, Scharf C, Völker U, Bremer E. 2006. Adaptation of Bacillus subtilis to growth at low temperature: a combined transcriptomic and proteomic appraisal. Microbiology 152: 831–853. 10.1099/mic.0.28530-0 [DOI] [PubMed] [Google Scholar]
- Darbon E, Servant P, Poncet S, Deutscher J. 2002. Antitermination by GlpP, catabolite repression via CcpA and inducer exclusion triggered by P-GlpK dephosphorylation control Bacillus subtilis glpfk expression. Mol Microbiol 43: 1039–1052. 10.1046/j.1365-2958.2002.02800.x [DOI] [PubMed] [Google Scholar]
- Del Campo C, Bartholomäus A, Fedyunin I, Ignatova Z. 2015. Secondary structure across the bacterial transcriptome reveals versatile roles in mRNA regulation and function. PLoS Genet 11: e1005613. 10.1371/journal.pgen.1005613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethoff EA, Chugh J, Mustoe AM, Al-Hashimi HM. 2012. Functional complexity and regulation through RNA dynamics. Nature 482: 322–330. 10.1038/nature10885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamant S, Eliahu N, Rosenthal D, Goloubinoff P. 2001. Chemical chaperones regulate molecular chaperones in vitro and in cells under combined salt and heat stress. J Biol Chem 276: 39586–39591. 10.1074/jbc.M103081200 [DOI] [PubMed] [Google Scholar]
- Droffner ML, Yamamoto N. 1985. Isolation of thermophilic mutants of Bacillus subtilis and Bacillus pumilus and transformation of the thermophilic trait to mesophilic strains. J Gen Microbiol 131: 2789–2794. 10.1099/00221287-131-10-2789 [DOI] [PubMed] [Google Scholar]
- Ganser LR, Kelly ML, Herschlag D, Al-Hashimi HM. 2019. The roles of structural dynamics in the cellular functions of RNA. Nat Rev Mol Cell Biol 20: 474–489. 10.1038/s41580-019-0136-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatz E, Nilsson R, Rutberg L, Rutber B. 1996. A dual role for the Bacillus subtilis glpD leader and the GlpP protein in the regulated expression of glpD: antitermination and control of mRNA stability. Mol Microbiol 19: 319–328. 10.1046/j.1365-2958.1996.376903.x [DOI] [PubMed] [Google Scholar]
- Grundy FJ, Moir TR, Haldeman MT, Henkin TM. 2002. Sequence requirements for terminators and antiterminators in the T box transcription antitermination system: disparity between conservation and functional requirements. Nucleic Acids Res 30: 1646–1655. 10.1093/nar/30.7.1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller KB, Lin EC, Wilson TH. 1980. Substrate specificity and transport properties of the glycerol facilitator in Escherichia coli. J Bacteriol 144: 274–278. 10.1128/jb.144.1.274-278.1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann T, Bremer E. 2011. Protection of Bacillus subtilis against cold stress via compatible-solute acquisition. J Bacteriol 193: 1552–1562. 10.1128/JB.01319-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtmann G, Bremer E. 2004. Thermoprotection of Bacillus subtilis by exogenously provided glycine betaine and structurally related compatible solutes: involvement of Opu transporters. J Bacteriol 186: 1683–1693. 10.1128/JB.186.6.1683-1693.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irnov I, Kertsburg A, Winkler WC. 2006. Genetic control by cis-acting regulatory RNAs in Bacillus subtilis: general principles and prospects for discovery. Cold Spring Harb Symp Quant Biol 71: 239–249. 10.1101/sqb.2006.71.021 [DOI] [PubMed] [Google Scholar]
- Irnov I, Sharma CM, Vogel J, Winkler WC. 2010. Identification of regulatory RNAs in Bacillus subtilis. Nucleic Acids Res 38: 6637–6651. 10.1093/nar/gkq454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasinghe OT, Mandell ZF, Yakhnin AV, Kashlev M, Babitzke P. 2022. Transcriptome-wide effects of NusA on RNA polymerase pausing in Bacillus subtilis. J Bacteriol 204: e0053421. 10.1128/jb.00534-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen TØ, Pogrebnyakov I, Falkenberg KB, Redl S, Nielsen AT. 2017. Application of the thermostable β-galactosidase, BgaB, from Geobacillus stearothermophilus as a versatile reporter under anaerobic and aerobic conditions. AMB Express 7: 169–179. 10.1186/s13568-017-0469-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, et al. 2003. Essential Bacillus subtilis genes. Proc Natl Acad Sci 100: 4678–4683. 10.1073/pnas.0730515100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortmann J, Narberhaus F. 2012. Bacterial RNA thermometers: molecular zippers and switches. Nat Rev Microbiol 10: 255–265. 10.1038/nrmicro2730 [DOI] [PubMed] [Google Scholar]
- Lamosa P, Burke A, Peist R, Huber R, Liu M-Y, Silva G, Rodrigues-Pousada C, LeGall J, Maycock C, Santos H. 2000. Thermostabilization of proteins by digylcerol phosphate, a new compatible solute from the hyperthermophile Archaeoglobus fulgidus. Appl Environ Microbiol 66: 1974–1979. 10.1128/AEM.66.5.1974-1979.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Subgroup. 2009. The sequence alignment/map format and samtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17: 10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Miller JH. 1972. Formulas and recipes. Experiments in molecular genetics, pp. 352–355. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- Mitchell D III, Assmann SA, Bevilacqua PC. 2019. Probing RNA structure in vivo. Curr Opin Struct Biol 59: 151–158. 10.1016/j.sbi.2019.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal S, Yakhnin AV, Sebastian A, Albert I, Babitzke P. 2016. NusA-dependent transcription termination prevents misregulation of global gene expression. Nat Microbiol 1: 15007. 10.1038/nmicrobiol.2015.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer SA, Kidwell MA, Doudna JA. 2014. Insights into RNA structure and function from genome-wide studies. Nat Rev Genet 15: 469–479. 10.1038/nrg3681 [DOI] [PubMed] [Google Scholar]
- Narberhaus F. 2010. Translational control of bacterial heat shock and virulence genes by temperature-sensing mRNAs. RNA Biol 7: 84–89. 10.4161/rna.7.1.10501 [DOI] [PubMed] [Google Scholar]
- Nshogozabahizi JC, Aubrey KL, Ross JA, Thakor N. 2019. Applications and limitations of regulatory RNA elements in synthetic biology and biotechnology. J Appl Microbiol 127: 968–984. 10.1111/jam.14270 [DOI] [PubMed] [Google Scholar]
- Proctor DJ, Schaak JE, Bevilacqua JM, Falzone CJ, Bevilacqua PC. 2002. Isolation and characterization of a family of stable RNA tetraloops with the motif YNMG that participate in tertiary interactions. Biochemistry 41: 12062–12075. 10.1021/bi026201s [DOI] [PubMed] [Google Scholar]
- R Core Team. 2021. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
- Reed RH, Borowitzka LJ, Mackay MA, Chudek JA, Foster R, Warr SRC, Moore DJ, Stewart WDP. 1986. Organic solute accumulation in osmotically stressed cyanobacteria. FEMS Microbiol Rev 2: 51–56. 10.1111/j.1574-6968.1986.tb01842.x [DOI] [Google Scholar]
- Richey DP, Lin ECC. 1972. Importance of facilitated diffusion for effective utilization of glycerol by Escherichia coli. J Bacteriol 112: 784–790. 10.1128/jb.112.2.784-790.1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchey LE, Su Z, Tack DC, Assmann SA, Bevilacqua PC. 2017. Structure-seq2: sensitive and accurate genome-wide profiling of RNA structure in vivo. Nucleic Acids Res 45: e135. 10.1093/nar/gkx533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchey LE, Su Z, Assmann SA, Bevilacqua PC. 2019. In vivo genome-wide RNA structure probing with Structure-seq. Methods Mol Biol 1933: 305–341. 10.1007/978-1-4939-9045-0_20 [DOI] [PubMed] [Google Scholar]
- Ritchey LE, Tack DC, Yakhnin H, Jolley EA, Assmann SM, Bevilacqua PC, Babitzke P. 2020. Structure-seq2 probing of RNA structure upon amino acid starvation reveals both known and novel RNA switches in Bacillus subtilis. RNA 26: 1431–1447. 10.1261/rna.075986.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeßler M, Müller V. 2001. Osmoadaptation in bacteria and archaea: common principles and differences. Environ Microbiol 3: 743–754. 10.1046/j.1462-2920.2001.00252.x [DOI] [PubMed] [Google Scholar]
- Roßmanith J, Narberhaus F. 2016. Exploring the modular nature of riboswitches and RNA thermometers. Nucleic Acids Res 44: 5410–5423. 10.1093/nar/gkw232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouskin S, Zubradt M, Washietl S, Kellis M, Weismann JS. 2014. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 505: 701–705. 10.1038/nature12894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutberg B. 1997. Antitermination of transcription of catabolic operons. Mol Microbiol 23: 413–421. 10.1046/j.1365-2958.1997.d01-1867.x [DOI] [PubMed] [Google Scholar]
- Schroeder KT, McPhee SA, Ouellet J, Lilley DM. 2010. A structural database for k-turn motifs in RNA. RNA 16: 1463–1468. 10.1261/rna.2207910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PA. 2009. The centrality of RNA. Cell 136: 577–580. 10.1016/j.cell.2009.02.007 [DOI] [PubMed] [Google Scholar]
- Su Z, Tang Y, Ritchey LE, Tack DC, Zhu M, Bevilacqua PC, Assmann SM. 2018. Genome-wide RNA structurome reprogramming by acute heat shock globally regulates mRNA abundance. Proc Natl Acad Sci 115: 12170–12175. 10.1073/pnas.1807988115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tack DC, Tang Y, Ritchey LE, Assmann SM, Bevilacqua PC. 2018. StructureFold2: bringing chemical probing data into the computational fold of RNA structural analysis. Methods 143: 12–15. 10.1016/j.ymeth.2018.01.018 [DOI] [PubMed] [Google Scholar]
- Tack DC, Su Z, Yu Y, Bevilacqua PC, Assmann SM. 2020. Tissue-specific changes in the RNA structurome mediate salinity response in Arabidopsis. RNA 26: 492–511. 10.1261/rna.072850.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Bouvier E, Kwok CK, Ding Y, Nekrutenko A, Bevilacqua PC, Assmann SM. 2015. StructureFold: genome-wide RNA secondary structure mapping and reconstruction in vivo. Bioinformatics 31: 2668–2675. 10.1093/bioinformatics/btv213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I, Bustamante C. 1999. How RNA folds. J Mol Biol 293: 271–281. 10.1006/jmbi.1999.3001 [DOI] [PubMed] [Google Scholar]
- Twittenhoff C, Brandenburg VB, Righetti F, Nuss AM, Mosig A, Dersch P, Narberhaus F. 2020. Lead-seq: transcriptome-wide structure probing in vivo using lead(II) ions. Nucleic Acids Res 48: e71. 10.1093/nar/gkaa404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagenende V, Yap MGS, Trout BL. 2009. Mechanisms of protein stablilization and prevention of protein aggregation by glycerol. Biochemistry 48: 11084–11096. 10.1021/bi900649t [DOI] [PubMed] [Google Scholar]
- Waldminghaus T, Heidrich N, Brantl S, Narberhaus F. 2007. FourU: a novel type of RNA thermometer in Salmonella. Mol Microbiol 65: 413–424. 10.1111/j.1365-2958.2007.05794.x [DOI] [PubMed] [Google Scholar]
- Wang J, Nikonowicz EP. 2011. Solution structure of the k-turn and specifier loop domains from the Bacillus subtilis tyrS T-box leader RNA. J Mol Biol 408: 99–117. 10.1016/j.jmb.2011.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Breaker RR. 2011. R2R—software to speed the depiction of aesthetic consensus RNA secondary structures. BMC Bioinform 12: 3. 10.1186/1471-2105-12-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakhnin H, Zhang H, Yakhnin AV, Babitzke P. 2004. The trp RNA-binding attenuation protein of Bacillus subtilis regulates translation of the tryptophan transport gene trpP (yhaG) by blocking ribosome binding. J Bacteriol 186: 278–286. 10.1128/JB.186.2.278-286.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakhnin AV, Fitzgerald PC, McIntosh C, Yakhnin H, Kireeva M, Turek-Herman J, Mandell ZF, Kashlev M, Babitzke P. 2020. NusG controls transcription pausing and RNA polymerase translocation throughout the Bacillus subtilis genome. Proc Natl Acad Sci 117: 21628–21636. 10.1073/pnas.2006873117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagami R, Sieg JP, Assmann SM, Bevilacqua PC. 2022. Genome-wide analysis of the in vivo tRNA structurome reveals RNA structural and modification dynamics under heat stress. Proc Natl Acad Sci 119: e2201237119. 10.1073/pnas.2201237119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubradt M, Gupta P, Persad S, Lambowitz AM, Weissman JS, Rouskin S. 2017. DMS-MaPseq for genome-wide or targeted RNA structure probing in vivo. Nat Methods 14: 75–82. 10.1038/nmeth.4057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415. 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]