Summary
The needs fulfilled by sleep are unknown, though the effects of insufficient sleep are manifold. To better understand how the need to sleep is sensed and discharged, much effort has gone into identifying the neural circuits involved in regulating arousal, especially those that promote sleep. In prevailing models the dorsal fan-shaped body (dFB) plays a central role in this process in the fly brain. In the present study we manipulated various properties of the dFB including its electrical activity, synaptic output, and endogenous gene expression. In each of these experimental contexts we were unable to identify any effect on sleep that could be unambiguously mapped to the dFB. Furthermore, we found evidence that sleep phenotypes previously attributed to the dFB were caused by genetic manipulations that inadvertently targeted the ventral nerve cord. We also examined expression of two genes whose purported effects have been attributed to functions within a specific subpopulation of dFB neurons. In both cases we found little to no expression in expected cells. Collectively our results cast doubt on the prevailing hypothesis that the dFB plays a central role in promoting sleep.
eTOC Blurb
De et al. find that activation of the dorsal fan-shaped body (dFB) is not necessary or sufficient to promote sleep in Drosophila. The authors attribute previously reported sleep-promoting effects of the dFB partly to off-target effects of drivers and to mislocalization of molecules to the dFB.
Introduction
Sleep has wide-ranging effects on organismal fitness, and its disruption is thought to contribute to dysfunction, including metabolic disorders, neurodegenerative diseases, and other health problems (reviewed in1–4). Therefore, much effort has been devoted to determining how sleep is regulated with an eye to ultimately unraveling the biological needs it fulfills. The fruit fly Drosophila melanogaster has been widely adopted to study both the mechanisms underlying regulation of sleep and the impact of sleep on organismal function. Fruit flies share many of the neurotransmitter systems that have been implicated in mammalian arousal5, and studies using flies have predicted subsequent discoveries of genes involved in control of mammalian sleep6,7. While much is known in both flies and mammals about the function of the circadian clock in regulating the timing of sleep, very little is known about homeostatic control of sleep duration. An exception to this generalization is the growing consensus in flies that a brain structure called the dorsal fan-shaped body (dFB) is a major regulator of sleep need.
Evidence from various studies suggests that the dFB is both necessary and sufficient to promote daily sleep and homeostatic regulation of sleep need8–13, and that the dFB is the locus at which genes such as Shaker (Sh), Hyperkinetic (Hk), Sandman (Sand) and Allatostatin A (AstA) function to regulate sleep9,14,15. However, it is unclear whether neurons outside the dFB were inadvertently targeted in previous studies and thus might be responsible for sleep-promoting effects that have been attributed to the dFB. Because of the centrality of this structure to how sleep regulation is discussed and studied in flies, we tested the main underpinnings of the hypothesis that the dFB promotes sleep.
Results
Activation of the dFB is not sufficient to promote sleep
One of the earliest studies to suggest that the dFB promotes sleep involved exciting neurons targeted by drivers that express in the dFB. Regardless of the driver/transgene combination, experimental animals manipulated in this way moved less during the day, and this result was interpreted as an increase in sleep10. We repeated this experiment using two reagents from the original study: a transgene encoding the depolarizing ion channel, TrpA1, and a driver named 104y. We focused on TrpA1 because its activity can be transiently and reversibly induced16, unlike other ion channels used in the original study whose effects on sleep might have been due to developmental defects in targeted neurons10. We selected 104y among drivers used in the original study because it was more commonly used to interrogate dFB-driven sleep in subsequent studies17–19.
We reproduced conditions from the original study by activating dFB neurons in 104y>TrpA1 animals at 31° C during the day. However, we used a milder six hour heat pulse from zeitgeber time (ZT) 0–6 instead of the twelve hour heat pulse from ZT0–12 that was used in the original study so that we could avoid misconstruing locomotor impairment for sleep. Specifically, we reasoned that normal waking activity should be observable at the end of a short induction period. As previously reported10, we found that experimental flies moved less upon TrpA1 activation, which translated into an apparent increase in sleep during the heat pulse. However, the same behavior unexpectedly persisted for at least six additional hours beyond the end of dFB stimulation (Figure 1A). To assess whether induced immobility was reversible, as expected for sleep, we delivered pulses of mechanical agitation to animals during the heat pulse. We found that control animals lacking either the 104y driver or the TrpA1 transgene responded to the additional stimulus with locomotion as fast as we were able to make a measurement (~1 min), whereas experimental 104y>TrpA1 animals were unresponsive until ~3 hrs later on average (Figure 1B). Thus, transient activation of neurons targeted by the 104y driver caused a persistent increase in immobility for hours beyond the duration of stimulation, and the induced immobility was not rapidly reversible. These results suggest that activating 104y neurons impairs locomotion rather than induces sleep.
Figure 1. Activation of broadly expressing 104y neurons promotes seizures and paralysis.
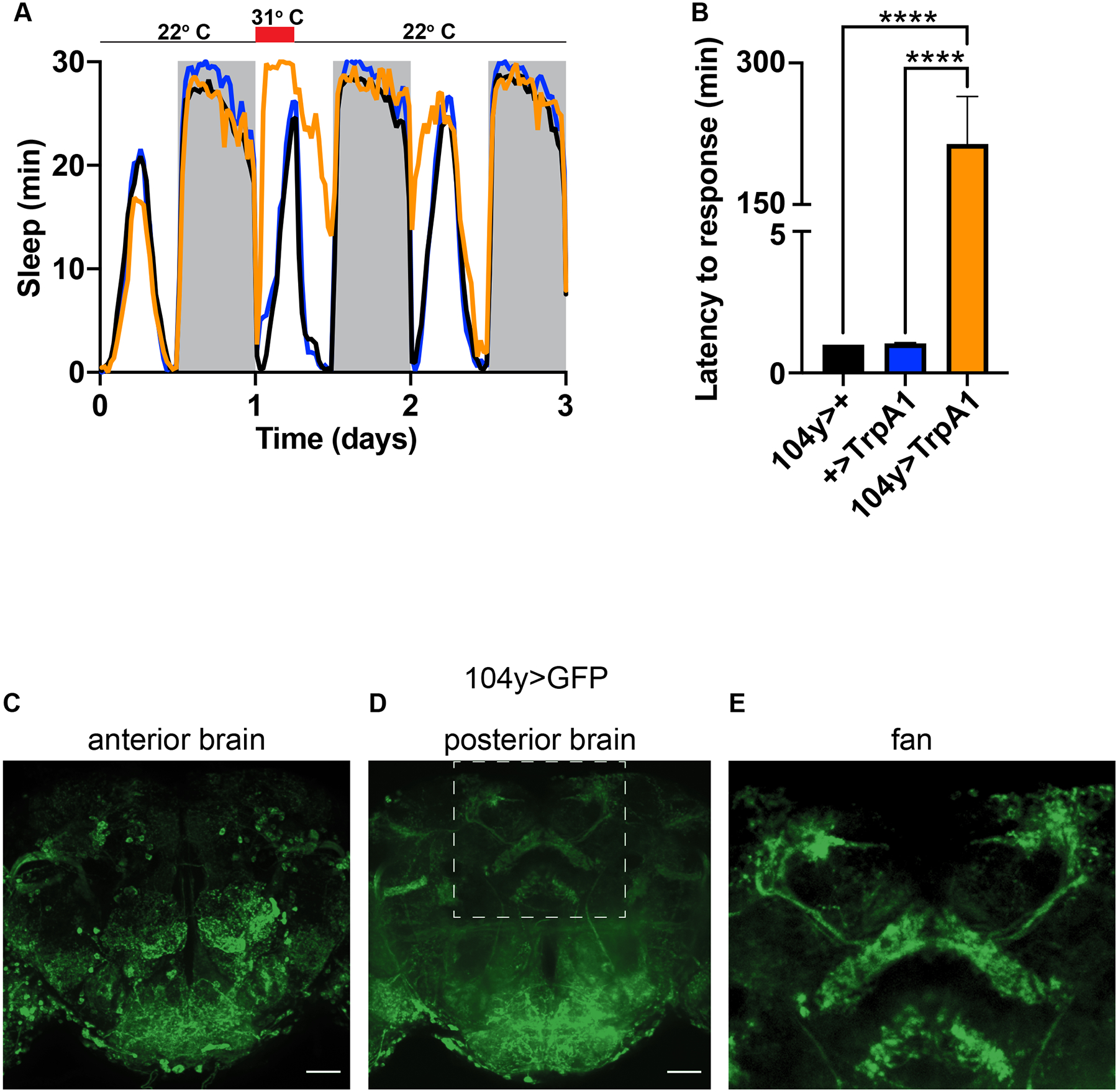
(A) Thermogenetic activation of 104y neurons from ZT0–6 causes persistent immobility for hours beyond the duration of the heat pulse. N≥26 for each group. (B) Thermogenetic activation of 104y neurons prevents animals from responding to strong mechanical agitation at ZT1. N≥38 for each group. (C-E) Expression of GFP in (C) anterior, (D) posterior, and (E) an inset of posterior brain depicting the dFB. C-D are max projections of whole mounts. E shows zoomed-in dFB from D. ****, p<.0001 by one-way ANOVA. Scale bars = 40 um. See also Videos S1 and S2.
To test this possibility more thoroughly, we examined the behavior of 104y>TrpA1 animals using a video camera before and during stimulation from the heat pulse. At 22° C experimental and control animals similarly roamed the length of the glass tubes in which they were placed (Video 1). However, after shifting to 31° C for one hour, 104y>TrpA1 animals fell on their sides and persistently shook their legs (Video 2). These effects are reminiscent of seizures, which are not commonly attributed to activation of the dFB, and likely went unnoticed in prior studies because video recording was not used to monitor behavior of freely moving animals10,17.
Early studies that used the 104y driver to study sleep interpreted phenotypes as having arisen from effects on the dFB10,18,19. However, the specificity of the driver has been a lingering caveat since those studies did not address its complete expression pattern in the brain. To determine the extent to which 104y might in fact target neurons outside the dFB and thus confound earlier interpretations, we examined expression of GFP under control of the driver in dissected fly brains. We found that in addition to the dFB, 104y labeled many other central neurons (Figure 1C–E). These results suggest that conclusions based on selective targeting of the dFB by 104y may need to be re-evaluated due to the driver’s apparent lack of specificity.
More recent studies have largely switched from using the 104y driver to the 23E10 driver, presumably because the latter more selectively targets the dFB. Despite questions about behavioral changes in 104y>TrpA1 animals, inducible activation of targeted 23E10 neurons has also been reported to increase sleep in flies11,13. This finding has reinforced the hypothesis that the dFB is a sleep-promoting structure in the fly brain. We reproduced these results using thermogenetics: activation of neurons in 23E10>TrpA1 animals with a 32° C pulse from ZT0–6 indeed induced immobility. Furthermore, unlike in 104y>TrpA1 animals, this phenotype was restricted to the duration of the heat pulse and was rapidly reversible with brief mechanical agitation, consistent with it representing sleep (Figure 2A,B; Figure S1A).
Figure 2. Sleep-promoting effects of 23E10 neurons map to the VNC.
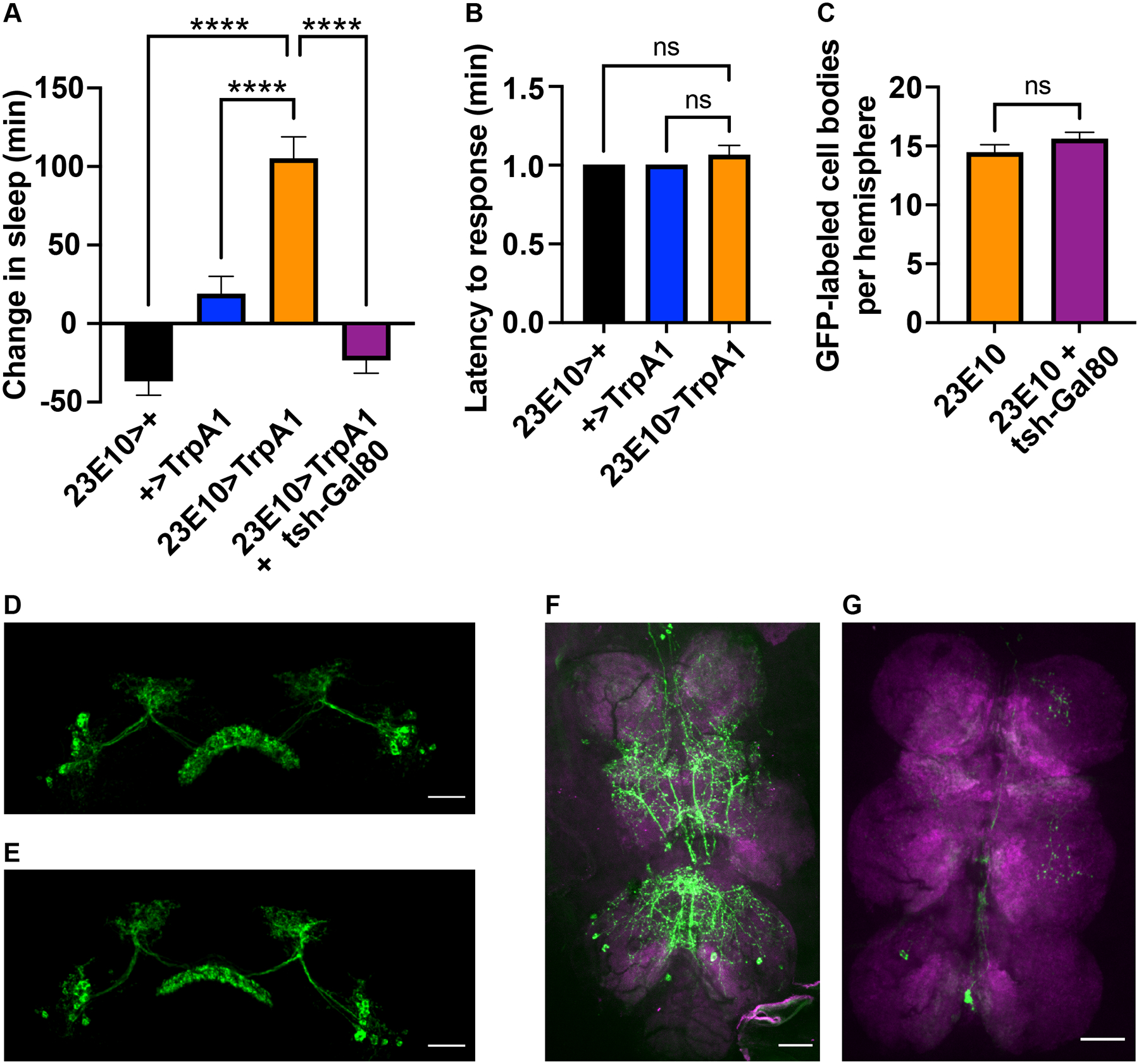
(A) Thermogenetic activation of 23E10 neurons at 32° C from ZT0–6 promotes sleep and is blocked by tsh-Gal80. N≥42 for each genotype. (B) Sleep in A is rapidly reversible in the presence of strong mechanical agitation at ZT1. N≥42 for each group. (C) Tsh-Gal80 does not reduce the number of dFB cell bodies in the brain that are labeled by 23E10>GFP. N≥24 for each group. (D-E) Representative images of dFBs in 23E10>GFP brains in the absence (D) vs presence (E) of tsh-Gal80. (F-G) Representative images of 23E10>GFP VNCs in the absence (F) vs presence (G) of tsh-Gal80. All images are max projections of whole mount preparations. Data are presented as mean ± SEM. **** p<.0001 and ns = not significant by one-way ANOVA. Scale bars = 40 um. See also Figure S1.
However, the origins of 23E10-driven sleep has not been conclusively mapped to the dFB. This issue has largely been ignored because within the brain 23E10 more selectively labels the dFB than other drivers. However, 23E10 also expresses sparsely in the ventral nerve cord (VNC)20. To address whether the sleep-like behavior of 23E10>TrpA1 animals originates in the brain or VNC, we made use of tsh-Gal80. This genetic element represses Gal4 activity under the control of the tsh promoter, which has been reported to express broadly throughout the VNC but only to a limited extent in the brain21. We verified that a driver derived from the tsh promoter does not express in the dFB (Figure S1C–E). Nevertheless, we found that tsh-Gal80 was able to suppress induction of sleep in 23E10>TrpA1 animals (Figure 2A; Figure S1B). These results suggest that neurons in the VNC rather than in the dFB are responsible for 23E10-driven sleep.
To determine if tsh-Gal80 truly selectively targets sleep-promoting neurons in the VNC rather than in the dFB, we expressed GFP under control of the 23E10 driver in the presence or absence of tsh-Gal80. Then we dissected labeled brains and VNCs and examined them by confocal microscopy. As expected, in the absence of tsh-Gal80 we observed strong 23E10 expression in both locations. However, in the presence of tsh-Gal80, 23E10 expression in the VNC was nearly abolished, whereas the number of 23E10-labeled dFB-projecting cell bodies in the brain was unaffected (Figure 2C–G). Thus, 23E10 sleep-promoting neurons appear to coincide with tsh-Gal80-targeted neurons in the VNC rather than to reside in the brain. Put another way, activation of the dFB appears to be insufficient to promote sleep.
The dFB is not necessary for daily sleep or for sleep homeostasis
Several studies have also reported that inhibiting neuronal activity or knocking down certain genes in the dFB reduces daily sleep or recovery sleep following sleep deprivation8,9,12. If confirmed, these results would suggest that the dFB is required for daily sleep or for sleep homeostasis. To test the first possibility we targeted the hyperpolarizing potassium channels EKO22 and Kir2.123 to 104y and 23E10 neurons and measured the amount of sleep in experimental animals vs controls. 104y>Kir2.1 animals did not survive to adulthood, supporting our hypothesis that the 104y driver expresses broadly, possibly also during development. However, other driver>K channel combinations produced seemingly healthy adults in abundance. In each case, though, we found that hyperpolarizing dFB neurons had no significant effect on total daily sleep (Figure 3A–C; Figure S2A–C). Similarly we found no significant change in total daily sleep when synaptic transmission was blocked in dFB neurons selectively in adulthood using a temperature-sensitive dominant-negative dynamin transgene called shibirets (shits)24 (Figure 3D).
Figure 3. Reducing excitability or synaptic transmission in the dFB does not affect sleep.
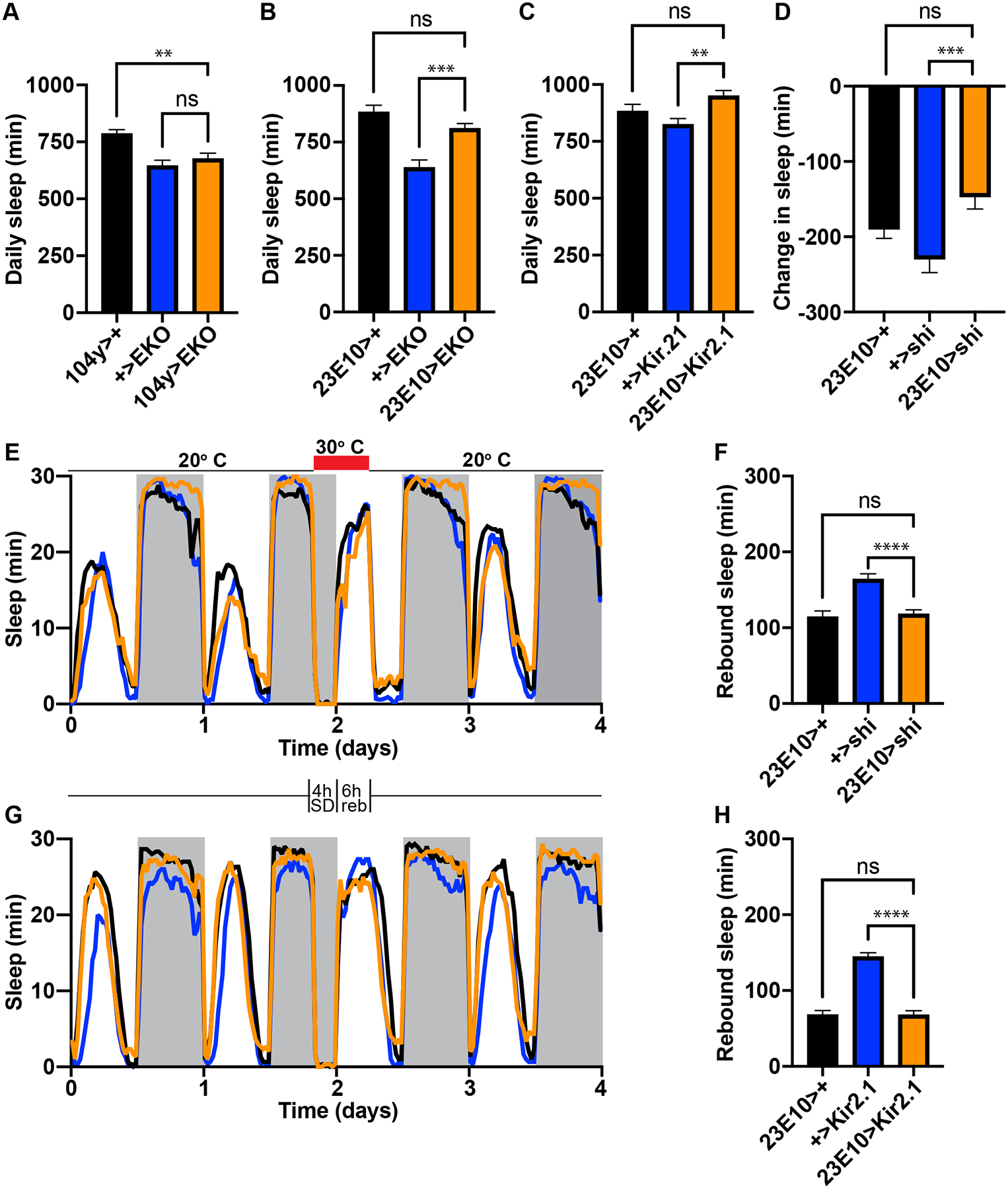
(A-C) Daily sleep in controls vs experimental animals expressing the K channels EKO or Kir2.1 under control of the dFB drivers 104y or 23E10. N≥35 for A; N=48 for B; N≥47 for each group in C. (D) A 24 hr heat pulse to 30° C does not significantly reduce sleep in 23E10>shits animals relative to controls. N≥83 for each group. (E, G) Sleep profiles of controls vs experimental animals during baseline conditions, sleep deprivation, the rebound period and a subsequent day of recovery. Animals were mechanically sleep-deprived from ZT20–24 at the end of the second day, and rebound was measured from ZT0–6 on the third day, as indicated by the schematic between panels E and G. Shits was activated by a 30° C heat pulse throughout both periods in E (red bar) to block synaptic transmission from 23E10 neurons. (F, H) Quantification of rebound sleep in E and G. N=79 for each group in E and F; N≥88 for each group for each group in G and H. Data are presented as mean ± SEM. ** p<.01; **** p<.0001; ns = not significant by one-way ANOVA. See also Figure S2.
Next we asked whether the dFB might be required to sense or discharge sleep need. We asked this question because several reports have suggested that sleep homeostasis can be impaired by manipulating gene expression in 23E10 neurons8,9. However, these reports did not address whether impairment of neurons outside the dFB or development of sleep circuits might be responsible. To address the potential requirement of the dFB in sleep homeostasis specifically in adult animals, we deprived animals of sleep for four hours at the end of the night and then measured rebound sleep immediately afterward while blocking synaptic transmission in the dFB using 104y>shits and 23E10>shits. In each case we found that rebound sleep was not significantly reduced in experimental animals relative to controls (Figure 3E,F; Figure S2D,E). In fact, 104y>shits animals exhibited enhanced recovery sleep (Figure S2D,E). However, this effect was not similarly observed in 23E10>shits animals, thus suggesting that neurons outside the dFB are responsible for enhanced rebound sleep in 104y>shits (Figure 3E,F). Failure to reduce rebound in 104y>shits and 23E10>shits animals was not caused by inadequate sleep deprivation since sleep loss in both genotypes was equivalent to or greater than in controls (Figure S2G,H). Collectively our data suggest that synaptic transmission in the dFB is not required for sleep homeostasis.
We also asked whether electrical transmission in the dFB might be important for regulating sleep need. To address this question we mechanically sleep-deprived and measured rebound sleep from animals with constitutively hyperpolarized dFB neurons due to targeted expression of EKO or Kir2.1 in 23E10 neurons. We found that experimental 23E10>EKO and 23E10>Kir2.1 animals recovered approximately the same amount of lost sleep as their respective controls (Figure S2F; Figure 3G,H), despite the fact that each set of genotypes was equivalently sleep-deprived (Figure S2I,J). These data therefore suggest that like chemical transmission, electrical transmission in the dFB is also dispensable for sleep homeostasis.
Genetic perturbations of the dFB do not significantly alter sleep
Loss-of-function mutations or RNAi knockdown of various genes have been reported to reduce sleep in flies. In some cases the 23E10 driver was used to map these phenotypes to the dFB. Genes that fall into this category encode the voltage-gated K channel Sh and its modulatory subunit Hk14,15, the leak K channel Sand15 and the neuropeptide AstA9. Since our data cast doubt on the sleep-promoting role of the dFB, we repeated key experiments in the reports cited above using the 23E10 driver and the same RNAi transgenes against Sh, Hk and Sand. We first confirmed the authenticity of parental stocks by amplifying their transgenes by PCR of genomic DNA and sequencing the resulting products. Then we expressed RNAi transgenes against Sh, Hk, and Sand in 23E10 neurons, and we measured sleep in experimental animals and in controls. In contrast to previous reports, we found that RNAi targeting of each gene had no significant effect on total daily sleep (Figure 4A–C; Figure S3A–C).
Figure 4. Knockdown of Sh, Hk and Sand in 23E10 neurons has no effect on daily sleep.
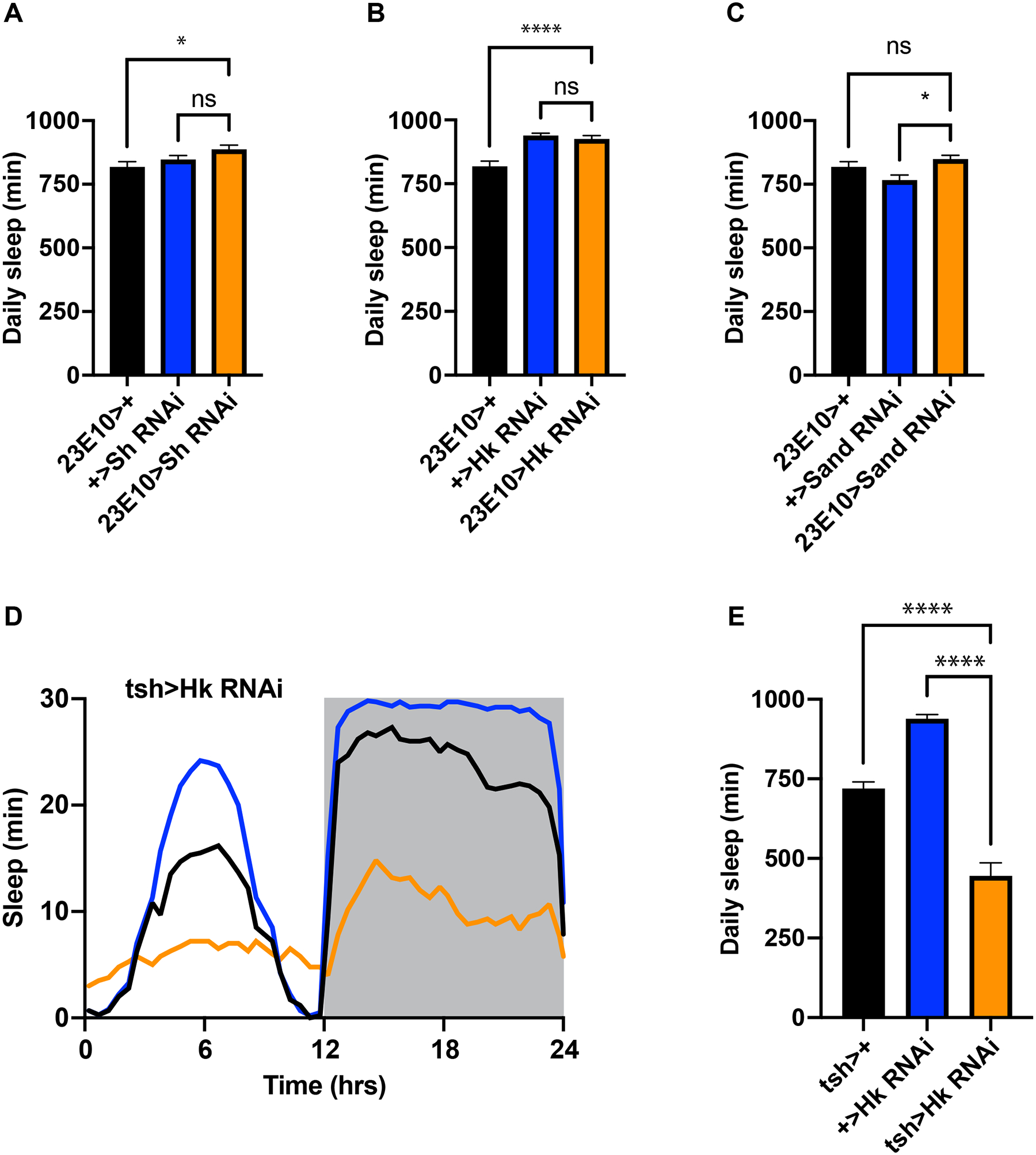
(A-C) Daily sleep in controls and experimental animals in which RNAi transgenes against Sh (A), Hk (B) or Sand (C) are expressed under the control of the 23E10 driver. N≥42 in A; N≥58 for B; N≥42 for each group in C; (D) Sleep profile of controls and experimental animals in which an RNAi against Hk is expressed under the control of tsh-Gal4. (E) Quantification of results in (E). N≥42 for each group in D and E. Data are presented as mean ± SEM. ** p<.01; **** p<.0001; ns = not significant by one-way ANOVA. See also Figure S3.
Loss-of-function mutations in Sh and Hk strongly reduce sleep25,26. Since we were unable to identify related phenotypes by RNAi targeting of the same genes in 23E10 neurons, we hypothesized that their effects might map to neurons outside the dFB. Since the data we reported in the previous section suggested that one important locus for sleep regulation is the VNC, we crossed the VNC-selective driver tsh-Gal4 to our RNAi against Hk and measured sleep in experimental animals and in their genetic controls. We found that tsh>Hk RNAi animals exhibited a significant reduction in total daily sleep (Figure 4D,E). Collectively these results strongly suggest that the VNC is required for some sleep-promoting effects that have been previously attributed to the dFB.
Some molecules reported to function in the dFB to promote sleep are undetectable or nearly undetectable there using sensitive quantitative methods
Since we were unable to measure a reduction in sleep when we knocked down Sand with the 23E10 driver, we asked whether this molecule is expressed in dFB neurons. No antibody is available to detect Sand protein by immunohistochemistry, and the expression pattern of Sand transcript has not previously been assessed. Therefore, we used RNAscope to examine the distribution of Sand mRNA in whole mount fly brains. We found Sand mRNA sparsely and diffusely spread throughout the central brain, with no obvious concentration in any particular region (Figure 5A). At high magnification, RNAscope has been reported to be sufficiently sensitive to detect individual transcripts27. Therefore, we used this technique in combination with confocal microscopy at high magnification to quantify Sand transcript in 23E10-expresssing dFB neurons. Specifically, we counted puncta encoding Sand mRNA in volumetric reconstructions of 23E10 cell bodies at 60x magnification (Figure 5B–D). We found at least one molecule of Sand transcript in only 7.6% of all volumetrically reconstructed 23E10 cell bodies we examined. If we instead set our threshold for a Sand-positive cell at two molecules of Sand transcript, then overlap with 23E10 labeling dropped to 2.1% of cells (Figure 5J).
Figure 5. Sand and AstA transcripts exhibit little to no expression in 23E10 neurons.
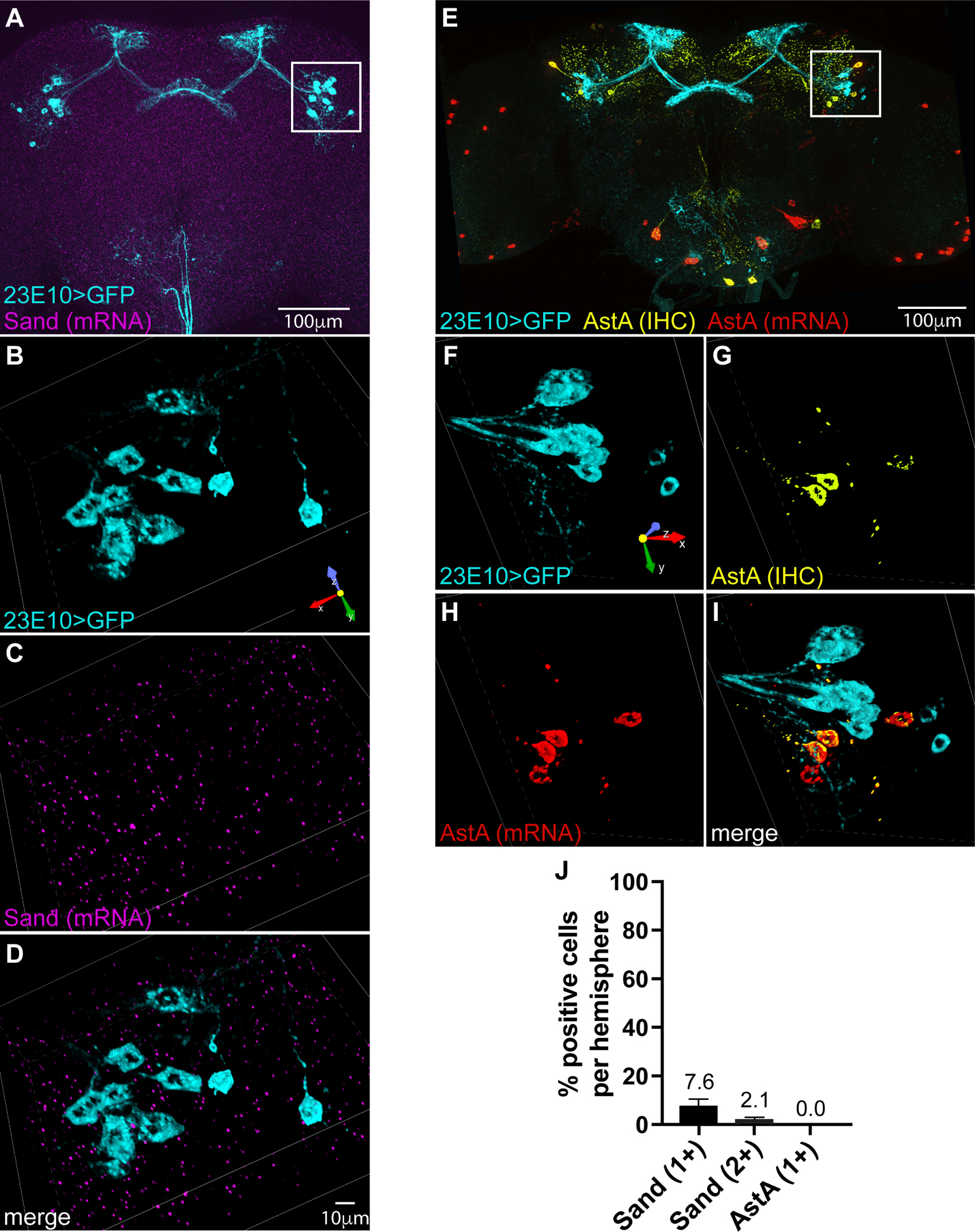
(A) Whole-brain image with multiplexed labeling of 23E10-driven GFP by IHC and Sand transcript by RNAscope. (B-D) Rotated volumetric reconstruction (.25 um slices) of inset in A showing 23E10-labeled cell bodies by IHC (B), labeling of Sand mRNA by RNAscope (C), and a merge of both labels (D). (E) Volume projection of whole mount brain with multiplexed labeling of 23E10-driven GFP by IHC, AstA protein by IHC and AstA transcript by RNAscope. (F-I) Rotated volumetric reconstruction (.25 um slices) of inset in E showing 23E10-labeled cell bodies by IHC (F), labeling of AstA protein by IHC (G), labeling of AstA mRNA by RNAscope (H), and a merge of all three labels (I). (J) Average percentage of 23E10 cell bodies per hemisphere that show one or two puncta of each mRNA. N=36 (AstA) and N=71 (Sand) total GFP-positive cells analyzed per group. See also Figure S4.
Next we examined expression of AstA in the fly brain. The study that reported a sleep-promoting function for AstA used immunohistochemistry (IHC) to indicate that indeed this molecule is expressed in the dFB9. We obtained the same monoclonal antibody and compared the distribution of AstA protein to expression of GFP targeted to 23E10 neurons. We confirmed that AstA is indeed expressed in the dFB. However, we found that the protein is localized to dFB neurons other than those targeted by the 23E10 driver (Figure S4A–F). We independently confirmed the distinction between AstA-positive/23E10-negative and AstA-negative/23E10-positive dFB neurons using RNAscope. Consistent with our immunohistochemical data, we found that AstA transcript co-localizes to dFB neurons that stain positive for AstA protein but not to GFP-labeled 23E10 neurons (Figure 5E–I). This result is consistent with another report that the 23E10 driver labels only a subset of neurons that project to the dFB28. Our results indicate that these neurons lack AstA.
To rule out the possibility that AstA message might not be visible at the low magnification we used to visualize 23E10 neurons in whole brains, we also used confocal microscopy at 60x magnification to count puncta encoding AstA mRNAs in volumetric reconstructions of 23E10 cell bodies. Consistent with our immunohistochemical findings, we found no AstA transcript in any of the 23E10 cells that we examined (Figure 5J).
Discussion
Over the last twelve years a near consensus has emerged that the central complex functions as a major sleep-regulating locus within the fly brain. However, many aspects of this hypothesis have remained untested. For example, the expression of key effector molecules in relevant neurons has been assumed in most cases and has never been directly examined. Similarly, the impact of central complex neuronal subpopulations on sleep has been inferred using genetic tools whose specificity has not been rigorously assessed. Despite these important caveats, the hypothesis that the central complex regulates sleep is sufficiently entrenched that is a focus of reviews and is discussed extensively throughout a recent paper describing the connectome of the Drosophila central complex29,30. Within this overall structure, the dFB and the ellipsoid body (EB) have received the most attention. The dFB has consistently been proposed to promote sleep, whereas the EB has been proposed to have mixed sleep- and wake-promoting functions31–33. Recently, however, the role of the EB in regulating sleep has been challenged. For example, drivers used to study functions of the EB in regulating arousal have been shown to also target peripheral ppk neurons. Furthermore, activation of ppk neurons phenocopies at least some of the sleep-regulating behaviors ascribed to the EB. Lastly, blocking expression of some EB drivers in ppk neurons reduces major sleep-regulating effects that have been attributed to the EB34,35.
The dFB has not yet undergone such scrutiny. Therefore, we set out to independently confirm the proposed role of this region of the central complex in promoting sleep. Our results suggest that seizure activity or sleep-inducing effects outside the dFB account for phenotypes caused by activating neurons with the broadly expressing 104y driver, which have previously been described as sleep10,36. Our results also suggest that sleep phenotypes elicited by the more selective 23E10 driver should be attributed to expression in sleep-regulating neurons in the VNC, rather than in the dFB11. Notably another recent study came to a similar conclusion37. However, we cannot rule out the possibility that dFB neurons promote sleep when co-activated with VNC neurons. We also cannot rule out the possibility that the dFB utilizes neurons other than those marked by the 23E10 driver to promote sleep.
Other discrepancies between our results and those in previous reports are more difficult to reconcile. For example, we found that blocking synaptic output or constitutively hyperpolarizing 23E10 neurons does not reduce recovery sleep after sleep deprivation. Thus, in contrast to previous reports8,9, our results suggest that 23E10 neurons are not required for sleep homeostasis. However, we cannot rule out the possibility that these manipulations were not fully effective in blocking synaptic output or in silencing 23E10 neurons. Additionally, it is worth noting that for these particular experiments our assay conditions deviated from those in previous reports that came to the opposite conclusion. For example, we sleep-deprived animals by mechanically agitating them for 2s every minute from ZT20–24, whereas at least one previous report mechanically agitated animals for 5s every hour from ZT12–248. We also measured rebound sleep from ZT0–6, whereas other studies measured it from ZT0–24. After testing a variety of conditions, we chose ours because they allowed us to achieve maximal rebound with minimal sleep deprivation, and animals deprived under our conditions returned to baseline behavior 24 hrs later. Thus, our assay conditions are sufficiently sensitive to measure rebound sleep, and they are also sufficiently mild that our measurements do not appear to be contaminated by injury-induced locomotor deficits. It is unclear if the same criteria apply to prior studies on which ours was based. Those studies deprived animals of sleep for three times longer than we did, measured rebound sleep over 24 hrs, and did not describe behavior beyond the initial rebound period8,9. We also measured rebound sleep as an absolute change over baseline, whereas previous studies measured it as a relative change. We chose our method because it is less dependent on uniform levels of baseline sleep between groups, which can vary considerably between genotypes, particularly in the daytime. Thus, at least for measuring rebound sleep, it is possible that methodological differences between our study and prior studies could account for the different results we describe here. To avoid such a possibility in the future, we suggest the field should establish standards for measuring sleep homeostasis. We note that other studies have adopted criteria similar to ours to ensure greater rigor and reproducibility of results34,35,38.
We were also unable to reproduce previous reports that Sh, Hk, and Sand function in 23E10-positive dFB neurons to promote sleep8,9,14. Our results were obtained with identical drivers and RNAi lines to those used in previous studies. We even amplified RNAi elements from genomic DNA of parental stocks by PCR and verified their identities by DNA sequencing. Furthermore, we confirmed the functionality of our RNAi line for Hk by knocking down expression in the VNC with tsh-Gal4, which resulted in a significant reduction in sleep, much like the phenotype reported for loss-of-function mutants of Hk26. While this result does not preclude a role for Hk in the dFB per se, it certainly suggests an alternative locus for the gene’s sleep-regulating function. It is also difficult to reconcile previous reports of sleep-promoting functions of Sand and AstA with the extremely low levels of transcripts we measured for these genes as well as with the undetectable levels of AstA protein we found in cell bodies of 23E10-positive dFB neurons.
There are caveats to our imaging of mRNA expression though. For example, it is possible that a low expression of Sand in only a few 23E10 neurons is sufficient to promote sleep. However, such a scenario would not apply to AstA since it was undetectable in 23E10 neurons. It is also unsupported by our behavioral data. A second possibility is that some of the signal for Sand and AstA fell below our detection limit. Although we cannot rule out such a possibility, we note that RNAscope has been reported to be sufficiently sensitive to allow detection of single molecules of mRNA27. Lastly, Sand and AstA could be required during development but taper in expression by adulthood. However, such a scenario would contradict existing models in which Sand and AstA function in adult dFB neurons to regulate daily sleep9.
In summary, our findings call into question important aspects of the prevailing hypothesis that the dFB functions as a sleep-promoting locus in the fly brain. While we do not suggest that the hypothesis is necessarily wrong, we believe it should be re-examined, including in the context of other supportive data. For example, elegant studies have shown that 23E10 dFB neurons change their firing properties in response to the wake-promoting neurotransmitter dopamine, to prior sleep deprivation and to oxidative stress8,14,15. While these correlated changes may be causally related to each other and to 23E10-driven changes in sleep, they may also reflect correlated changes found broadly throughout the nervous system in response to stress or sleep deprivation with little impact on sleep itself.
Sleep-promoting neurons have also been identified outside the central complex in the fly brain. These consist of DN1 and DN3 neurons in the circadian clock network as well as proposed clock outputs such as TuBu and claw neurons39–43. We also previously demonstrated that peripheral ppk neurons seem to function as rare inputs to the sleep homeostat34,35,44. These sensory neurons terminate in the adjacent subesophogeal zone (SEZ) and antennal mechanosensory and motor center (AMMC). The latter region is known for responding to vibratory stimuli, which are among the rare sensory signals that have been shown to trigger sleep homeostasis in flies. The SEZ and AMMC have also been proposed to be responsible for the sleep-promoting effects of other sensory pathways that terminate there45–47. Thus, we suggest that these regions may represent alternative locations to look for neurons that convert wake-promoting signals into the need to sleep.
STAR Methods
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, William Joiner (wjoiner@ucsd.edu).
Materials availability
This study did not generate new unique reagents.
Data and Code Availability
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report new, original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Drosophila melanogaster were grown at 20–22° C on standard cornmeal-molasses medium prepared by the Fly Kitchen in the Department of Biology at UCSD. The background of all flies was w1118.
Method Details
Fly stocks
Fly lines obtained from the Bloomington Stock Center included: 104y-Gal4 (81014), 23E10-Gal4 (49032), and UAS-smGFP-HA,LexAop-smGFP-V5 (64092). The following RNAi lines were obtained from the Vienna Drosophila Resource Center: Sh (104474), Hk (101402), Sand (47977), UAS-TrpA1, UAS-EKO and UAS-Kir2.1 were from previous studies35,49. UAS-shits was kindly provided by Gerry Rubin. Tsh-Gal4 and tsh-Gal80 were kindly provided by Julie Simpson. Identities of RNAi lines were confirmed by PCR amplification of genomic insertions followed by DNA sequencing.
Behavioral assays
Sleep measurements were performed as previously described using 5 min of inactivity as a proxy for sleep based on its correlation with elevated arousal threshold50,51. Briefly, one- to five-day old female flies were loaded into 5×65 mm glass tubes containing 5% sucrose and 2% agarose. Animals were entrained for 2–3 days on a 12 hr:12 hr light:dark cycle prior to measuring sleep/wake cycles using the Drosophila Activity Monitoring System (DAMS; Trikinetics) and custom software written in Matlab (Mathworks). Temperatures were maintained at 25° C except for during thermogenetic experiments (see below for details).
Thermogenetic activation of targeted neurons was performed using a baseline temperature of 22° C for at least 48 hrs prior to delivery of a heat pulse from ZT0–6 at either 31° C (for experiments involving 104y>TrpA1) or 32° C (for experiments involving 23E10>TrpA1). Following the heat pulse, ambient temperature was returned to 22° C. The resulting change in sleep was measured as sleep during the 6 hr heat pulse minus sleep during the equivalent baseline period 24 hrs earlier.
To determine the necessity of chemical synaptic transmission in dFB neurons for baseline sleep, 23E10>shits animals and their genetic controls were maintained at 22° C for at least 48 hrs prior to delivery of a heat pulse to 30° C from ZT0–24. The resulting change in sleep was measured as sleep during the 24 hr heat pulse minus sleep during the baseline period 24 hrs immediately beforehand.
To measure sleep homeostasis, flies in DAMS monitors were loaded into a VX-2500 multi-tube vortexer (VWR) fitted with a custom base. Monitors were maintained at 20° C for at least 48 hrs prior to elevating the ambient temperature to 30° C to block synaptic transmission in the dFB of animals expressing shits. Animals were then mechanically agitated at the lowest intensity setting for 2 sec/min from ZT20–24. After 6 hrs of recovery, the ambient temperature was returned to 20° C. Sleep deprivation was calculated as the total amount of sleep from ZT20–24 during mechanical agitation minus sleep during the same period the previous day. Rebound was calculated as the difference in sleep between the subsequent two 6 hr periods.
To measure response latencies in 104y>TrpA1, 23E10>TrpA1 and controls, animals in DAMS monitors were loaded into a vortexer and entrained in 12:12 L:D for at least two days at 22° C. A heat pulse to 31° C (104y>TrpA1) or 32° C (23E10>TrpA1) was then delivered from ZT0–6, and the vortexer was activated at its lowest setting for 2 sec every min from ZT1–1.5. Sleep latency was calculated as the time it took for each animal to cross the middle of its tube for the first time after initiating agitation.
For video analysis flies were loaded into glass tubes and entrained as described above, then placed under a video camera (IPEVO; model VZ-R) at ZT0 on the day of the experiment. At ZT2 movement was recorded for 5 min before raising the temperature and repeating the recording at ZT3.
Immunohistochemistry
For immunostaining of whole-mount brains and ventral nerve cord (VNC), 3–5 days old female brains (N = 8–10 for each genotype) or VNCs (N = 8–10 for each genotype) were dissected in ice-cold PBS and fixed in 4% PFA for 1 h at room temperature or overnight at 4°C. After 6 brief washes (10 mins each) in PBS + 0.3% Triton X-100 (PBST), brains were blocked in 5% normal donkey serum (The Jackson Laboratory) in PBST for 2 h at room temperature or overnight at 4°C. Brains were incubated for 2 nights at 4°C with 1:1000 rabbit anti-V5 (Thermo Fisher Scientific), and/or 1:100 rabbit anti-HA (Rockland), and/or 1:100 rat anti-HA (Roche), and/or 1:1000 rabbit anti-GFP (Invitrogen), and/or 1:500 mouse anti-AstA (Developmental Studies Hybridoma Bank), and/or 1:100 mouse anti-nc82 (Developmental Studies Hybridoma Bank). After 6 brief washes in PBST, brains were incubated overnight at 4°C in 1:1000 Alexa Fluor 488 anti-rabbit (Thermo Fisher Scientific), and/or 1:1000 Alexa Fluor 568 anti-rat (Thermo Fisher Scientific), and/or 3:1000 Alexa Fluor 633 anti-mouse (Thermo Fisher Scientific) before a final 6 brief washes in PBS at room temperature. Brains were mounted in ‘Vectashield’ (Vector Laboratories) mounting reagent and imaged at 20x or 60x magnification on a Nikon Eclipse Ti2-E confocal microscope at 1 micron intervals and reassembled for maximum projection using Fiji (SciJava ecosystems).
Cell counting
Brains of 3–5 day old female flies expressing GFP under control of the 23E10 driver in the presence or absence of tsh-Gal80 were dissected and immunostained as described above. Total number of cells projecting to the dorsal fan-shaped body was counted per brain and averaged for each genotype.
RNAscope
Adult Drosophila brains were labeled with RNAscope probes using the Multiplex Fluorescent Detection Kit v2 (Advanced Cell Diagnostics, ACD) according to the manufacturer’s protocol with several modifications. Day 1: Dissected brains were fixed overnight at room temperature (RT) in 4% formaldehyde/PBS on a rocker set at the lowest speed. Day 2: Brains were washed for 10 min at RT in PBST [PBS + 0.3% Triton X-100]. Brains were then dehydrated stepwise in PBST-methanol solutions (25%, 50%, 75%, and 100% methanol/PBST) for 10 min for each step at RT on a slow rocker and then stored at −20° C.
Day 3: Brains were restored to RT in 100% methanol and rehydrated stepwise from 100% methanol to PBST + 1% BSA in the reverse of the dehydration procedure. For all washes and incubations care was taken to ensure that brains were freely floating and not attached to the wall of the tube or clumped with other brains. Rehydrated brains were then treated with 500 ul of pre-warmed 1X Target Retrieval solution (ACD) and incubated for 5.5 minutes in a 100°C heat block. Samples were immediately washed at RT for 1 min in 650 ul PBST + 1% BSA, then for 1 min in 100% methanol, then for 10 min in 650 ul PBST + 1% BSA. Brains were then incubated at RT for 25 min in 750 ul 4% formaldehyde/PBS to post-fix, followed by a 10 min wash in 650 ul PBST + 1% BSA. Wash solution was removed and replaced with two drops of Protease Plus (ACD) and incubated for 10 min in a 40°C heat block, followed by another RT wash for 10 min in PBST + 1% BSA. Finally, brains were rinsed in two drops of pre-warmed probe diluent (ACD) prior to addition of pre-mixed probes (prepared according to manufacturer’s instructions). Brains were incubated with ~ 50–100 ul of pre-mixed probe overnight in a 40° C heat block.
Day 4: Samples were removed from the heat block, and manufacturer’s instructions for labeling procedures were followed with the following modifications. All RT washes were performed in 650 ul volumes, and 40° C incubations were performed in a heat block. For AMPs 1–3, HRP C1-C3 and HRP-blocker (ACD), 2 −3 drops of each solution was added to the brains after removing as much of the previous solution as possible. RNAscope® Probes used in this study were: Dm-Sand-C2 (573821-C2) and Dm-AstA-C3 (ACD 573861-C3). C2 and C3 probes were diluted 1:50 in 50–100 ul of probe diluent (ACD). Opal dyes (Akoya Biosciences) for HRP probe conjugation (Akoya Biosciences) were diluted 1:1000 in TSA buffer (ACD) and stored up to 1 month at 4° C. Dm-Sand-C2 was conjugated to Opal-690 and Dm-AstA-C3 probe was conjugated to Opal-620.
For multiplexed co-labeling with IHC, the final HRP-blocker step of RNAscope probe conjugation on day 4 was followed by washing brains at RT 3x for 10 min in PBST, then by blocking brains for 1 hr in block solution [10% normal goat serum (Life Technologies), 1% BSA, 0.3% Triton X-100]. Primary antibody [rabbit anti-GFP (Life Technologies, 1:600) or mouse anti-AstA (Developmental Studies Hybridoma Bank, 1:100)] was diluted in block solution, and brains were incubated in primary antibody at 4° C for 24–48 hrs on a slow rocker.
Day 5 (multiplexed co-labeling continued): Samples were washed at RT 4x for 15 min each in PBST and then incubated with secondary antibody [goat-anti-mouse-HRP or goat-anti-rabbit-HRP (VWR)] diluted 1:600 in block solution at 4°C overnight on a slow rocker.
Day 6: Opal dyes (Opal 480 for anti-GFP and Opal 540 for anti-AstA) were conjugated to the HRP secondary by a 30 min incubation in a 40° C heat block, followed by two 5 min washes at RT in RNAscope wash buffer (ACD), then a 15 min incubation in two drops of HRP-blocker (ACD) in a 40°C heat block. Brains were washed at RT for a final 2x for 5 min each in RNAscope wash buffer (ACD) before mounting as described above. For samples that required labeling with more than one antibody (ie. anti-GFP and anti-AstA), the entire labeling procedure was performed sequentially over days 4–6.
RNAscope-labeled samples were imaged at the UCSD Nikon Imaging Core on a CREST X-Light V2 spinning disc confocal scanhead mounted on a Nikon Ti2-E microscope. A 60X 1.27NA water immersion objective and Hamamatsu Orca Fusion sCMOS camera were used for acquiring images. Z-stacks were acquired with 0.25 um optical steps at 0.11um/pixel resolution. Acquisition at 16-bit depth allows for a maximum intensity of 65,000 shades in each channel, ensuring that signal saturation is nearly impossible (for example, typical signal intensity range from 1000–35,000, well below the saturation limit). Image were processed using NIS Elements analysis software first by 3 dimensional deconvolution to remove out of focus fluorescence and noise, and then a difference of Gaussian processing step was used to sharpen bright spots into resolvable puncta. Bright spots were thresholded for average diameter, intensity and contrast and automatically labeled and counted using the NIS Elements Analysis software.
For spatial labeling of cell volumes, NIS Elements segment.ai software was trained for multiple sessions of 1000x iterations on several hand-curated cell volume binaries and then the trained model was used to rapidly segment cells volumes in additional data sets. All cell volumes generated using the trained model were visually verified and manually edited for accuracy.
Quantification and Statistical Analysis
Statistical comparisons were performed using GraphPad Prism version 8 (GraphPad Software). One-way ANOVA with Tukey’s multiple comparison post-test were used to calculate p-values for experimental vs. control groups. Bar graphs are presented as mean ± standard error of the mean. Each behavioral experiment was repeated independently at least three times. Each immunohistochemistry experiment was repeated independently at least twice.
Supplementary Material
Video S1. Normal locomotor activity of 104y>TrpA1 animals at 22°C, Related to Figure 1.
Video S2. Seizure-like activity of 104y>TrpA1 animals at 31°C, Related to Figure 1.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-V5 | Thermo Fisher | PA1993 |
| Rabbit anti-HA | Rockland | RL600-401-384 |
| Rat anti-HA | Roche | 11867423001 |
| Rabbit anti-GFP | Invitrogen, Life Technologies | A11122 |
| Mouse anti-AstA | DSHB | 5F10-c |
| Mouse anti-nc-82 | DSHB | Nc-82-c |
| Goat anti-rabbit Alexa Fluor 488 | Thermo Fisher | A11008 |
| Goat anti-mouse Alexa Fluor 633 | Thermo Fisher | A11009 |
| Goat anti-mouse-HRP | VWR | 95959–740 |
| Goat anti-rabbit-HRP | VWR | 95959–088 |
| Bacterial and virus strains | ||
| Biological samples | ||
| Chemicals, peptides, and recombinant proteins | ||
| Vectashield | Vector labs | H-1000 |
| Critical commercial assays | ||
| Multiplex Fluorescent Detection Kit v2 | Advanced Cell Diagnostics | 323110 |
| RNAscope probe Dm-Sand-C2 | Advanced Cell Diagnostics | 573821-C2 |
| RNAscope probe Dm-AstA-C3 | Advanced Cell Diagnostics | 573861-C3 |
| Opal dye 540 | Akoya Bioscience | FP1494001KT |
| Opal dye 690 | Akoya Bioscience | FP1497001KT |
| Opal dye 620 | Akoya Bioscience | FP1495001KT |
| Opal dye Polaris 480 | Akoya Bioscience | FP1500001KT |
| Deposited data | ||
| Experimental models: Cell lines | ||
| Experimental models: Organisms/strains | ||
| 104y-Gal4 | Bloomington Drosophila Stock Center | BDSC:81014; FlyBase: FBti0072312 |
| 23E10-Gal4 | Bloomington Drosophila Stock Center | BDSC:49032; FlyBase: FBti0134066 |
| UAS-smGFP-HA,LexAop-smGFP-V5 | Bloomington Drosophila Stock Center | BDSC:64092; FlyBase: FBti0169244, FBti0169271 |
| Sh RNAi | Vienna Drosophila Resource Center | VDRC: 104474 |
| Hk RNAi | Vienna Drosophila Resource Center | VDRC: 101402 |
| Sand RNAi | Vienna Drosophila Resource Center | VDRC: 47977 |
| UAS-TrpA1 | Seidner et al.35 | Flybase: FBtp0040248 |
| UAS-EKO | White et al.22 | Flybase: FBtp0014962 |
| UAS-Kir2.1 | Baines et al.23 | Flybase: FBto0000566 |
| UAS-shits | Gerry Rubin | Flybase: FBtp0013545 |
| tsh-Gal4 | Julie Simpson | N/A |
| tsh-Gal80 | Julie Simpson | N/A |
| Oligonucleotides | ||
| Recombinant DNA | ||
| Software and algorithms | ||
| NIS Elements Analysis AR 5.41.01 | Nikon | |
| ImageJ | Schneider et al.48 | https://imagej.nih.gov/ij/ |
| Prism 8 | Graphpad | |
| Other | ||
Highlights.
Off-target effects of dFB drivers account for previously reported effects on sleep.
Silencing or selectively activating neurons in the dFB has no effect on sleep.
23E10 dFB neurons lack AstA, which was reported to function there to promote sleep.
Hk functions in the VNC rather than its reported locus, the dFB, to promote sleep.
Acknowledgements
This work was supported by National Institutes of Health Grant R01 GM125080 and R21 NS123690 to WJJ. The authors thank Eric Griffis, Peng Guo and the UCSD Nikon Imaging Center for assistance with imaging. The authors also thank Nick Stavropoulos for helpful comments on the manuscript.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- 1.Duan D, Kim LJ, Jun JC, and Polotsky VY (2023). Connecting insufficient sleep and insomnia with metabolic dysfunction. Ann N Y Acad Sci 1519, 94–117. 10.1111/nyas.14926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Joiner WJ (2018). The Neurobiological Basis of Sleep and Sleep Disorders. Physiology (Bethesda) 33, 317–327. 10.1152/physiol.00013.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mander BA, Winer JR, Jagust WJ, and Walker MP (2016). Sleep: A Novel Mechanistic Pathway, Biomarker, and Treatment Target in the Pathology of Alzheimer’s Disease? Trends Neurosci 39, 552–566. 10.1016/j.tins.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang C, and Holtzman DM (2020). Bidirectional relationship between sleep and Alzheimer’s disease: role of amyloid, tau, and other factors. Neuropsychopharmacology 45, 104–120. 10.1038/s41386-019-0478-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joiner WJ (2016). Unraveling the Evolutionary Determinants of Sleep. Curr Biol 26, R1073–R1087. 10.1016/j.cub.2016.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joiner WJ, Friedman EB, Hung HT, Koh K, Sowcik M, Sehgal A, and Kelz MB (2013). Genetic and anatomical basis of the barrier separating wakefulness and anesthetic-induced unresponsiveness. PLoS Genet 9, e1003605. 10.1371/journal.pgen.1003605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Funato H, Miyoshi C, Fujiyama T, Kanda T, Sato M, Wang Z, Ma J, Nakane S, Tomita J, Ikkyu A, et al. (2016). Forward-genetics analysis of sleep in randomly mutagenized mice. Nature. 10.1038/nature20142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donlea JM, Pimentel D, and Miesenbock G (2014). Neuronal machinery of sleep homeostasis in Drosophila. Neuron 81, 860–872. 10.1016/j.neuron.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donlea JM, Pimentel D, Talbot CB, Kempf A, Omoto JJ, Hartenstein V, and Miesenbock G (2018). Recurrent Circuitry for Balancing Sleep Need and Sleep. Neuron 97, 378–389 e374. 10.1016/j.neuron.2017.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donlea JM, Thimgan MS, Suzuki Y, Gottschalk L, and Shaw PJ (2011). Inducing sleep by remote control facilitates memory consolidation in Drosophila. Science 332, 1571–1576. 10.1126/science.1202249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ni JD, Gurav AS, Liu W, Ogunmowo TH, Hackbart H, Elsheikh A, Verdegaal AA, and Montell C (2019). Differential regulation of the Drosophila sleep homeostat by circadian and arousal inputs. Elife 8. 10.7554/eLife.40487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kottler B, Bao H, Zalucki O, Imlach W, Troup M, van Alphen B, Paulk A, Zhang B, and van Swinderen B (2013). A sleep/wake circuit controls isoflurane sensitivity in Drosophila. Curr Biol 23, 594–598. 10.1016/j.cub.2013.02.021. [DOI] [PubMed] [Google Scholar]
- 13.Troup M, Yap MH, Rohrscheib C, Grabowska MJ, Ertekin D, Randeniya R, Kottler B, Larkin A, Munro K, Shaw PJ, and van Swinderen B (2018). Acute control of the sleep switch in Drosophila reveals a role for gap junctions in regulating behavioral responsiveness. Elife 7. 10.7554/eLife.37105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kempf A, Song SM, Talbot CB, and Miesenbock G (2019). A potassium channel beta-subunit couples mitochondrial electron transport to sleep. Nature 568, 230–234. 10.1038/s41586-019-1034-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pimentel D, Donlea JM, Talbot CB, Song SM, Thurston AJ, and Miesenbock G (2016). Operation of a homeostatic sleep switch. Nature 536, 333–337. 10.1038/nature19055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamada FN, Rosenzweig M, Kang K, Pulver SR, Ghezzi A, Jegla TJ, and Garrity PA (2008). An internal thermal sensor controlling temperature preference in Drosophila. Nature 454, 217–220. 10.1038/nature07001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yap MHW, Grabowska MJ, Rohrscheib C, Jeans R, Troup M, Paulk AC, van Alphen B, Shaw PJ, and van Swinderen B (2017). Oscillatory brain activity in spontaneous and induced sleep stages in flies. Nat Commun 8, 1815. 10.1038/s41467-017-02024-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Q, Liu S, Kodama L, Driscoll MR, and Wu MN (2012). Two dopaminergic neurons signal to the dorsal fan-shaped body to promote wakefulness in Drosophila. Curr Biol 22, 2114–2123. 10.1016/j.cub.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ueno T, Tomita J, Tanimoto H, Endo K, Ito K, Kume S, and Kume K (2012). Identification of a dopamine pathway that regulates sleep and arousal in Drosophila. Nat Neurosci 15, 1516–1523. 10.1038/nn.3238. [DOI] [PubMed] [Google Scholar]
- 20.Jenett A, Rubin GM, Ngo TT, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, et al. (2012). A GAL4-driver line resource for Drosophila neurobiology. Cell Rep 2, 991–1001. 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clyne JD, and Miesenbock G (2008). Sex-specific control and tuning of the pattern generator for courtship song in Drosophila. Cell 133, 354–363. 10.1016/j.cell.2008.01.050. [DOI] [PubMed] [Google Scholar]
- 22.White BH, Osterwalder TP, Yoon KS, Joiner WJ, Whim MD, Kaczmarek LK, and Keshishian H (2001). Targeted attenuation of electrical activity in Drosophila using a genetically modified K(+) channel. Neuron 31, 699–711. 10.1016/s0896-6273(01)00415-9. [DOI] [PubMed] [Google Scholar]
- 23.Baines RA, Uhler JP, Thompson A, Sweeney ST, and Bate M (2001). Altered electrical properties in Drosophila neurons developing without synaptic transmission. J Neurosci 21, 1523–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poodry CA, and Edgar L (1979). Reversible alteration in the neuromuscular junctions of Drosophila melanogaster bearing a temperature-sensitive mutation, shibire. J Cell Biol 81, 520–527. 10.1083/jcb.81.3.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cirelli C, Bushey D, Hill S, Huber R, Kreber R, Ganetzky B, and Tononi G (2005). Reduced sleep in Drosophila Shaker mutants. Nature 434, 1087–1092. 10.1038/nature03486. [DOI] [PubMed] [Google Scholar]
- 26.Bushey D, Huber R, Tononi G, and Cirelli C (2007). Drosophila Hyperkinetic mutants have reduced sleep and impaired memory. J Neurosci 27, 5384–5393. 10.1523/JNEUROSCI.0108-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie F, Timme KA, and Wood JR (2018). Using Single Molecule mRNA Fluorescent in Situ Hybridization (RNA-FISH) to Quantify mRNAs in Individual Murine Oocytes and Embryos. Sci Rep 8, 7930. 10.1038/s41598-018-26345-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davie K, Janssens J, Koldere D, De Waegeneer M, Pech U, Kreft L, Aibar S, Makhzami S, Christiaens V, Bravo Gonzalez-Blas C, et al. (2018). A Single-Cell Transcriptome Atlas of the Aging Drosophila Brain. Cell 174, 982–998 e920. 10.1016/j.cell.2018.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hulse BK, Haberkern H, Franconville R, Turner-Evans D, Takemura SY, Wolff T, Noorman M, Dreher M, Dan C, Parekh R, et al. (2021). A connectome of the Drosophila central complex reveals network motifs suitable for flexible navigation and context-dependent action selection. Elife 10. 10.7554/eLife.66039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shafer OT, and Keene AC (2021). The Regulation of Drosophila Sleep. Curr Biol 31, R38–R49. 10.1016/j.cub.2020.10.082. [DOI] [PubMed] [Google Scholar]
- 31.Yan W, Lin H, Yu J, Wiggin TD, Wu L, Meng Z, Liu C, and Griffith LC (2023). Subtype-Specific Roles of Ellipsoid Body Ring Neurons in Sleep Regulation in Drosophila. J Neurosci 43, 764–786. 10.1523/JNEUROSCI.1350-22.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu S, Liu Q, Tabuchi M, and Wu MN (2016). Sleep Drive Is Encoded by Neural Plastic Changes in a Dedicated Circuit. Cell 165, 1347–1360. 10.1016/j.cell.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ho MCW, Tabuchi M, Xie X, Brown MP, Luu S, Wang S, Kolodkin AL, Liu S, and Wu MN (2022). Sleep need-dependent changes in functional connectivity facilitate transmission of homeostatic sleep drive. Curr Biol 32, 4957–4966 e4955. 10.1016/j.cub.2022.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Satterfield LK, De J, Wu M, Qiu T, and Joiner WJ (2022). Inputs to the sleep homeostat originate outside the brain. J Neurosci 42, 5695–5704. 10.1523/JNEUROSCI.2113-21.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seidner G, Robinson JE, Wu M, Worden K, Masek P, Roberts SW, Keene AC, and Joiner WJ (2015). Identification of Neurons with a Privileged Role in Sleep Homeostasis in Drosophila melanogaster. Curr Biol 25, 2928–2938. 10.1016/j.cub.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dag U, Lei Z, Le JQ, Wong A, Bushey D, and Keleman K (2019). Neuronal reactivation during post-learning sleep consolidates long-term memory in Drosophila. Elife 8. 10.7554/eLife.42786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones JD, Holder BL, Eiken KR, Vogt A, Velarde AI, Elder AJ, McEllin JA, and Dissel S (2023). Regulation of sleep by cholinergic neurons located outside the central brain in Drosophila. PLoS Biol 21, e3002012. 10.1371/journal.pbio.3002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andreani T, Rosensweig C, Sisobhan S, Ogunlana E, Kath W, and Allada R (2022). Circadian programming of the ellipsoid body sleep homeostat in Drosophila. Elife 11. 10.7554/eLife.74327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo F, Chen X, and Rosbash M (2017). Temporal calcium profiling of specific circadian neurons in freely moving flies. Proc Natl Acad Sci U S A 114, E8780–E8787. 10.1073/pnas.1706608114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo F, Holla M, Diaz MM, and Rosbash M (2018). A Circadian Output Circuit Controls Sleep-Wake Arousal in Drosophila. Neuron 100, 624–635 e624. 10.1016/j.neuron.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 41.Guo F, Yu J, Jung HJ, Abruzzi KC, Luo W, Griffith LC, and Rosbash M (2016). Circadian neuron feedback controls the Drosophila sleep--activity profile. Nature 536, 292–297. 10.1038/nature19097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lamaze A, Kratschmer P, Chen KF, Lowe S, and Jepson JEC (2018). A Wake-Promoting Circadian Output Circuit in Drosophila. Curr Biol 28, 3098–3105 e3093. 10.1016/j.cub.2018.07.024. [DOI] [PubMed] [Google Scholar]
- 43.Sun L, Jiang RH, Ye WJ, Rosbash M, and Guo F (2022). Recurrent circadian circuitry regulates central brain activity to maintain sleep. Neuron 110, 2139–2154 e2135. 10.1016/j.neuron.2022.04.010. [DOI] [PubMed] [Google Scholar]
- 44.Grace KP (2023). Leveraging Simplicity to Generate Fundamental Insights into the Complex Nature of Sleep-Drives. Sleep. 10.1093/sleep/zsad026. [DOI] [PubMed] [Google Scholar]
- 45.Lone SR, Potdar S, Venkataraman A, Sharma N, Kulkarni R, Rao S, Mishra S, Sheeba V, and Sharma VK (2021). Mechanosensory Stimulation via Nanchung Expressing Neurons Can Induce Daytime Sleep in Drosophila. J Neurosci 41, 9403–9418. 10.1523/JNEUROSCI.0400-21.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ozturk-Colak A, Inami S, Buchler JR, McClanahan PD, Cruz A, Fang-Yen C, and Koh K (2020). Sleep Induction by Mechanosensory Stimulation in Drosophila. Cell Rep 33, 108462. 10.1016/j.celrep.2020.108462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Melnattur K, Zhang B, and Shaw PJ (2020). Disrupting flight increases sleep and identifies a novel sleep-promoting pathway in Drosophila. Sci Adv 6, eaaz2166. 10.1126/sciadv.aaz2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Joiner WJ, Crocker A, White BH, and Sehgal A (2006). Sleep in Drosophila is regulated by adult mushroom bodies. Nature 441, 757–760. 10.1038/nature04811. [DOI] [PubMed] [Google Scholar]
- 50.Huber R, Hill SL, Holladay C, Biesiadecki M, Tononi G, and Cirelli C (2004). Sleep homeostasis in Drosophila melanogaster. Sleep 27, 628–639. 10.1093/sleep/27.4.628. [DOI] [PubMed] [Google Scholar]
- 51.Robinson JE, Paluch J, Dickman DK, and Joiner WJ (2016). ADAR-mediated RNA editing suppresses sleep by acting as a brake on glutamatergic synaptic plasticity. Nat Commun 7, 10512. 10.1038/ncomms10512. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. Normal locomotor activity of 104y>TrpA1 animals at 22°C, Related to Figure 1.
Video S2. Seizure-like activity of 104y>TrpA1 animals at 31°C, Related to Figure 1.
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report new, original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


