Abstract
An uncoupler of oxidative phosphorylation, 2,4-dinitrophenol, and an aconitase inhibitor, fluoroacetic acid, both of which are known to lower the cellular ATP pool, protected Escherichia coli cells from the bactericidal actions of gyrase poisons including quinolone antibiotics, nalidixic acid and ciprofloxacin, and the epipodophyllotoxins VP-16 and VM-26. Using purified E. coli DNA gyrase, we examined the effect of ATP on gyrase-mediated DNA cleavage in the presence of these gyrase poisons. ATP was shown to stimulate gyrase-mediated DNA cleavage from 10- to more than 100-fold in the presence of these gyrase poisons. ADP antagonized the stimulatory effect of ATP. Consequently, gyrase-mediated DNA cleavage induced by gyrase poisons is modulated by the ATP concentration/ADP concentration ([ATP]/[ADP]) ratio. Coumermycin A1, an inhibitor of the ATPase subunit of DNA gyrase, like ADP, also effectively antagonized the stimulatory effect of ATP on gyrase-mediated DNA cleavage induced by gyrase poisons. Furthermore, coumermycin A1, like DNP and fluoroacetic acid, also protected cells from the bactericidal action of gyrase poisons. In the aggregate, our results are consistent with the notion that the [ATP]/[ADP] ratio, through its modulatory effect on the gyrase-mediated DNA cleavage, is an important determinant of cellular susceptibility to gyrase poisons.
Type II DNA topoisomerases have been demonstrated to be very effective molecular targets for therapeutic agents ranging from antibiotics to antitumor drugs (for reviews, see references 9, 13, and 23). Many drugs have been classified as topoisomerase II poisons on the basis of their effectiveness in converting cellular topoisomerase II into DNA-breaking nuclease (9, 23). However, the molecular mechanism(s) by which these drugs kill cells remains unclear (4, 7, 13, 23).
Most type II DNA topoisomerases require ATP as an energy cofactor in enzyme catalysis (26, 35, 36, 38). Binding of ATP is apparently sufficient to trigger one round of strand passage (26, 37, 38). Upon binding to ATP, yeast DNA topoisomerase II was shown to undergo a conformational change to form a circular protein clamp (20–22, 31, 32, 34, 38), a result consistent with earlier studies with Drosophila DNA topoisomerase II (30) and crystallographic studies with yeast DNA topoisomerase II (5). However, the role of this circular clamp conformation in enzyme catalysis and drug action has not been established.
In addition to ATP, ADP also appears to play an important role in modulating the activity of topoisomerase II. ADP is an effective inhibitor of both the ATPase and the strand-passing activity of DNA topoisomerase II and it can also compete with the binding of the nonhydrolyzable analog of ATP, ADPNP, to gyrase (1, 2, 25, 30, 35, 37). Studies with bacteria have demonstrated that cellular supercoiling is affected by the ATP concentration/ADP concentration ([ATP]/[ADP]) ratio (15, 16). This effect was attributed to the effect of the [ATP]/[ADP] ratio on the supercoiling activity of DNA gyrase on the basis of the results of in vitro studies (39). Studies with Drosophila DNA topoisomerase II have similarly demonstrated that ADP can effectively compete with ATP in enzyme catalysis (30).
A potential role of ATP in cellular susceptibility to topoisomerase II poisons has been suggested from a number of studies. The cytotoxicity of VM-26 was greatly reduced in L1210 cells cotreated with a number of ATP inhibitors such as 2,4-dinitrophenol (DNP), sodium cyanide, and 2-deoxyglucose (18). Hypoxic tumor cells, which have a lowered ATP level, have also been demonstrated to be more resistant to VP-16 (17, 41). Two lines of evidence from studies with bacteria have suggested a potential role of ATP in the bactericidal action of nalidixic acid. First, mutations conferring resistance to nalidixic acid have been mapped to enzymes involved in the tricarboxylic acid (TCA) cycle (12, 19). Second, DNP has been shown to protect cells from the bactericidal action of nalidixic acid, presumably due to the lowered cellular ATP pool (7). These studies suggest a potential role of the ATP pool in modulating cellular susceptibility to topoisomerase II poisons in both bacteria and mammalian cells. However, the precise role(s) of ATP in cell killing by topoisomerase II poisons remains unclear due to the multiple effects of ATP on cellular functions.
In order to evaluate the role of ATP as a determinant of cellular susceptibility to topoisomerase II poisons, we have studied the mechanisms of action of quinolones and epipodophyllotoxins using bacteria as a model system. Our results indicate that the gyrase-mediated DNA damage induced by quinolones and epipodophyllotoxins is greatly stimulated by ATP. In addition, ADP antagonizes the stimulatory effect of ATP. Consequently, the gyrase-mediated DNA cleavage in vitro is affected by the [ATP]/[ADP] ratio. In vivo, inhibitors of the ATP pool such as DNP and fluoroacetic acid and the ATP-antagonizing gyrase poison coumermycin A1 protect cells from the bactericidal action of quinolones and epipodophyllotoxins. Together, these results suggest that the [ATP]/[ADP] ratio modulates the cellular susceptibility to topoisomerase II poisons in part through its modulation of topoisomerase II-mediated DNA damage at the level of the topoisomerase II-drug-DNA ternary cleavable complex.
MATERIALS AND METHODS
Chemicals, drugs, and enzymes.
Nalidixic acid, coumermycin A1, DNP, and fluoroacetic acid were purchased from Sigma Chemical Co. The epipodophyllotoxins VP-16 (etoposide) and VM-26 (teniposide) were gifts from Bristol-Myers and Squibb Co. Ciprofloxacin was a gift from Tom Rowe (University of Florida, Gainsville). All drugs were dissolved in dimethyl sulfoxide (Sigma Chemical Co.) and were kept in aliquots frozen at −20°C. Purified Escherichia coli DNA gyrase was kindly provided by Roger L. McMacken (Johns Hopkins University) and Martin Gellert (National Institutes of Health, Bethesda, Md.).
Gyrase-mediated DNA cleavage assays.
Gyrase-mediated DNA cleavage assays were performed under the conditions described previously for the mammalian topoisomerase II cleavage assay (28), with slight modification. Briefly, BamHI-digested YEpG DNA was labeled at the 3′ end with [α-32P]dATP and the large fragment of E. coli DNA polymerase I. Reaction mixtures (20 μl each) contained 50 mM Tris (pH 8.0), 100 mM KCl, 8 mM MgCl2, 0.1 mM dithiothreitol, 0.5 mM EDTA, 30 μg of bovine serum albumin per ml, approximately 10 ng of E. coli DNA gyrase, and 10 ng of end-labeled [32P]YEpG DNA. Gyrase poisons, ATP, ADP, and/or coumermycin A1 were added to the reaction mixtures at the indicated concentrations. The reaction mixtures were incubated at 37°C for 30 min, and the reactions were then terminated by adding 5 μl of a stop buffer (5% sodium dodecyl sulfate, 2.5 mg of proteinase K per ml). After another hour of protease digestion at 37°C, the reactions were then analyzed on a 0.8% agarose gel in 0.5× TPE buffer (45 mM Tris-phosphate [pH 8.0], 1 mM EDTA). The gels were then dried onto 3MM chromatographic paper and were autoradiographed at −80°C by using Kodak XAR-5 films and Dupont lightening plus intensifying screens.
Determination of bactericidal effects of gyrase drugs.
The strain E. coli AS19 (40) was chosen for this study because of the higher level of permeability of its membrane to certain drugs. Briefly, logarithmically growing cells were diluted to 106 to 107 cells/ml in Luria broth (LB) with 5 g of NaCl per liter. After the addition of drug, the cells were incubated at 37°C for another 3 h. Subsequently, the cells were serially diluted and plated onto LB plates. The colonies were counted after 16 to 24 h of incubation.
RESULTS
ATP stimulates gyrase-mediated DNA cleavage in the presence of the quinolone antibiotics nalidixic acid and ciprofloxacin and the antitumor epipodophyllotoxins VP-16 and VM-26.
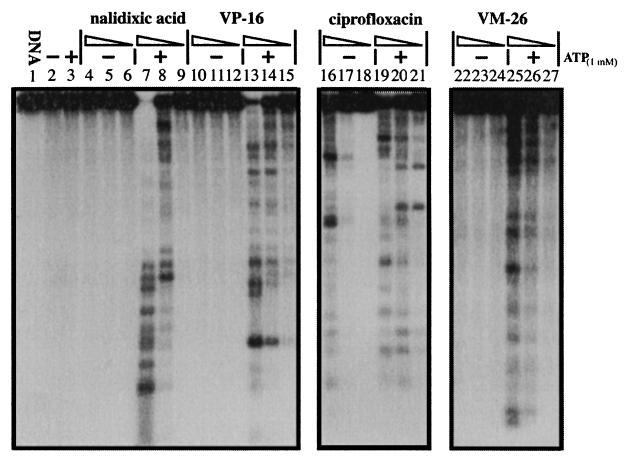
On the basis of their poisoning effects on DNA gyrase, ciprofloxacin is about 100- to 1,000-fold more potent than nalidixic acid (13) (Fig. 1). As indicated in Fig. 1 (compare lanes 7 to 9 with lanes 4 to 6, respectively), the gyrase-mediated DNA cleavage induced by nalidixic acid was strongly (more than 100-fold) stimulated by ATP (1 mM). The effect of ATP on the gyrase-mediated DNA cleavage induced by ciprofloxacin was less dramatic (over 10-fold) (Fig. 1; compare lanes 19 to 21 with lanes 16 to 18, respectively). It was also evident that ATP altered the cleavage efficiency at some of the sites where cleavage was induced by ciprofloxacin (Fig. 1; compare lanes 16 and 19), consistent with results from previous studies with oxolinic acid (27, 36).
FIG. 1.
ATP stimulates gyrase-mediated DNA cleavage in the presence of quinolone antibiotics and epipodophyllotoxins. Gyrase-mediated DNA cleavage in the presence (+) or absence (−) of 1 mM ATP was performed as described in Materials and Methods. Gyrase poisons were serially diluted with 10-fold differences in concentrations. The concentrations of nalidixic acid (1,000 to 10 μM), ciprofloxacin (10 to 0.1 μM), VP-16 (300 to 3 μM), and VM-26 (100 to 1 μM) are indicated above the corresponding lanes (with decreasing concentrations for each set of lanes indicated by the triangles).
The epipodophyllotoxins VP-16 (etoposide) and VM-26 (teniposide) are potent antitumor drugs which poison mammalian DNA topoisomerases IIα and IIβ (3, 8, 23). As shown in Fig. 1 (lanes 10 to 15 and 22 to 27), they also poisoned E. coli DNA gyrase, albeit much less potently than ciprofloxacin. Again, ATP (1 mM) strongly stimulated (more than 100-fold) gyrase-mediated DNA cleavage in the presence of either VP-16 or VM-26 (Fig. 1).
ADP antagonizes the stimulatory effect of ATP on gyrase-mediated DNA cleavage.
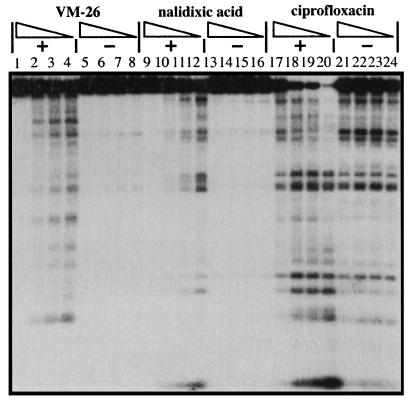
The known inhibitory effect of ADP on the ATPase and supercoiling activities of E. coli DNA gyrase prompted us to examine the effect of ADP on gyrase-mediated DNA cleavage. As indicated in Fig. 2, in the absence of ATP, ADP (from 0.9 to 0 mM) had a minimal effect on gyrase-mediated DNA cleavage in the presence of VM-26, nalidixic acid, or ciprofloxacin (Fig. 2, lanes 5 to 8, 13 to 16, and 21 to 24, respectively). However, in the presence of 0.1 mM ATP, ADP effectively inhibited gyrase-mediated DNA cleavage by ciprofloxacin, nalidixic acid, and VM-26 in a dose-dependent manner (Fig. 2). At the highest concentration of ADP (0.9 mM), the stimulatory effect of ATP (0.1 mM) was nearly completely abolished (Fig. 2, lanes 1 to 4, 9 to 12, and 17 to 20).
FIG. 2.
ADP effectively antagonizes the stimulatory effect of ATP on gyrase-mediated DNA cleavage. Gyrase-mediated DNA cleavage was performed as described in Materials and Methods, with the concentrations of ADP (0.9, 0.3, 0.1, and 0 mM [with decreasing concentrations indicated by the triangles]) and ATP (0.1 mM), which was present (+) or absent (−), indicated above each lane. The concentrations of nalidixic acid, ciprofloxacin, and VM-26 were 800, 1.5, and 100 μM, respectively.
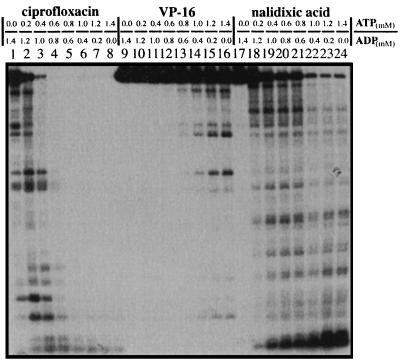
In a separate experiment, we varied the [ATP]/[ADP] ratio by maintaining a constant amount of [ATP] and [ADP] (Fig. 3). Again, the [ATP]/[ADP] ratio strongly affected gyrase-mediated DNA cleavage in the presence of ciprofloxacin, VP-16, or nalidixic acid (Fig. 3, lanes 1 to 8, 9 to 16, and 17 to 24, respectively). Gyrase-mediated DNA cleavage increased with increasing [ATP]/[ADP] ratios in the presence of all three compounds tested.
FIG. 3.
The [ATP]/[ADP] ratio modulates gyrase-mediated DNA cleavage. Gyrase-mediated DNA cleavage was performed as described in Materials and Methods. The combined concentration of ATP and ADP was 1.4 mM. The [ATP]/[ADP] ratios varied from 0/1.4 to 1.4/0, as indicated. The concentrations of nalidixic acid, ciprofloxacin, and VP-16 were 200, 1.5, and 300 μM, respectively.
Coumermycin A1 antagonizes ATP in gyrase-mediated DNA cleavage.
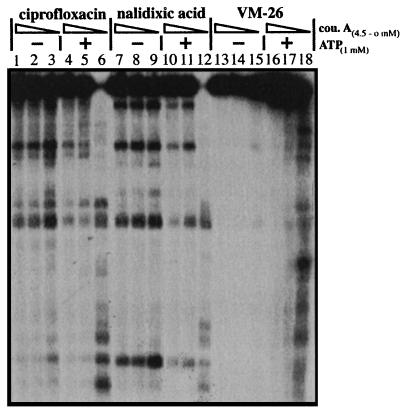
Coumermycin A1, like novobiocin, is an inhibitor of DNA gyrase (9). Resistance to coumermycin A1 mapped at the gyrB gene which encodes the ATPase subunit of gyrase (9, 10, 13). In vitro studies have suggested that coumermycin A1 inhibits DNA gyrase by competing with ATP (35, 36). To test whether coumermycin A1, like ADP, can also antagonize ATP in gyrase-mediated DNA cleavage, we tested the effect of coumermycin A1 on gyrase-mediated DNA cleavage in the presence and absence of 1 mM ATP (Fig. 4). Like ADP, coumermycin A1 had a minimal effect on gyrase-mediated DNA cleavage in the absence of ATP (Fig. 4). However, in the presence of ATP, coumermycin A1 strongly inhibited gyrase-mediated DNA cleavage (Fig. 4). In the presence of 0.45 μM coumermycin A1, the stimulatory effect of ATP (1 mM) on gyrase-mediated DNA cleavage was completely abolished (Fig. 4). However, different from ADP, coumermycin A1 at concentrations higher than 4.5 μM inhibited gyrase-mediated DNA cleavage even in the absence of ATP (data not shown), suggesting that the mechanism of coumermycin A1 inhibition is not strictly competitive with ATP, consistent with results from recent studies with an N-terminal fragment of the GyrB subunit (1, 2, 24).
FIG. 4.
Coumermycin A1 antagonizes the stimulatory effect of ATP on gyrase-mediated DNA cleavage. Gyrase-mediated DNA cleavage was performed as described in Materials and Methods. The concentrations of coumermycin A1 (cou. A; 4.5, 0.45, and 0 μM) with (+) or without (−) 1 mM ATP are indicated. The concentrations of nalidixic acid, ciprofloxacin, and VM-26 were 2 mM, 1.5 μM, and 100 μM, respectively.
Modulation of the bactericidal activities of gyrase inhibitors by DNP and fluoroacetic acid.
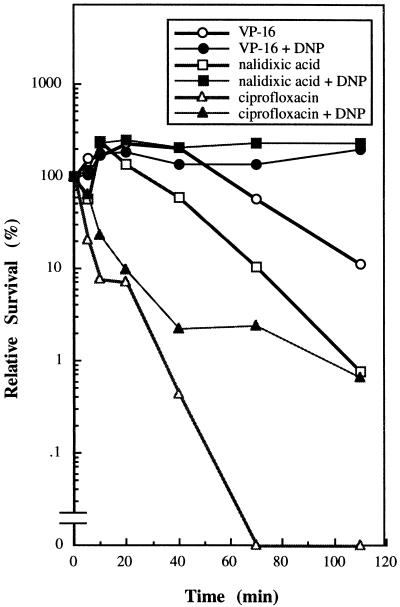
Previous studies have shown that DNP, an uncoupler of oxidative phosphorylation which lowers the cellular ATP pool, can protect cells from the bactericidal action of nalidixic acid (7). Consistent with this observation, we showed that DNP (1 mM) can protect cells from the bactericidal effect of a number of gyrase poisons, including nalidixic acid, VP-16, and ciprofloxacin (Fig. 5). The potential role of the cellular ATP pool in modulating the cellular susceptibility to gyrase poisons also has been suggested from studies showing that mutations in the genes encoding enzymes involved in the TCA cycle can confer resistance to nalidixic acid (12, 19). In order to strengthen the notion that the ATP pool may be important in modulating the bactericidal actions of gyrase poisons, we have tested the protective effect of fluoroacetic acid, an aconitase inhibitor. Aconitase, which catalyzes the conversion of citrate to isocitrate, is one of the key enzymes involved in the TCA cycle. Simultaneous treatment of cells with 40 mM fluoroacetic acid and quinolones or epipodophyllotoxins greatly reduced the bactericidal activities of these compounds. The relative survivals, defined as the ratio of viable numbers of drug-treated cells in the presence of 40 mM of fluoroacetic acid over that in its absence, of cells treated with nalidixic acid (400 μM), ciprofloxacin (0.4 μM), and VM-26 (100 μM) were 120, 190, and 204, respectively (the values are the averages of two independent determinations). The bactericidal effects of these drugs were determined as described in Materials and Methods. This result, which complements the results of earlier studies of drug resistance (4, 7, 12, 18), suggests an important role of the cellular ATP pool in determining cellular susceptibility to quinolones and epipodophyllotoxins.
FIG. 5.
2, DNP protects cells against gyrase poisons. The cells were exposed to various gyrase poisons in the presence or absence of 1 mM DNP as described in Materials and Methods. At various times the numbers of viable cells were scored by determining the number of colonies that formed on LB plates. The concentrations of nalidixic acid, ciprofloxacin, and VP-16 were 200, 1, and 500 μM, respectively.
Coumermycin A1 protects cells from the bactericidal actions of quinolones and epipodophyllotoxins.
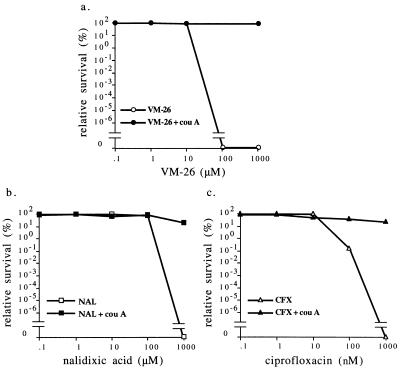
Reduction of the cellular ATP pool can affect many cellular processes. It is unclear whether the protective effect of the reduced ATP pool is due to reduced gyrase-mediated DNA cleavage as observed in vitro or some other event (e.g., the processing of the cleavable complexes). As indicated in Fig. 4, coumermycin A1 can specifically abolish the stimulatory effect of ATP on gyrase-mediated DNA cleavage, presumably by competing with ATP binding to gyrase (1, 10, 24, 35, 36). Coumermycin A1 probably does not interact with other important proteins in cells since the molecular target of coumermycin A1 has been determined to be GyrB on the basis of genetic and biochemical studies (1, 9–11, 13, 23, 24). We therefore tested whether coumermycin A1 also could protect cells from the bactericidal actions of quinolones and epipodophyllotoxins. As indicated in Fig. 6, cotreatment of cells with coumermycin A1 (20 μM) and the gyrase poisons nalidixic acid, ciprofloxacin, or VM-26 almost completely eliminated the cytotoxicities of these gyrase poisons. This result strongly suggests that the protective effect of coumermycin A1 is related to its effect on DNA gyrase and is presumably due to its antagonizing effect on ATP binding to gyrase.
FIG. 6.
Coumermycin A1 protects cells against gyrase poisons. Cell viability was measured as described in Materials and Methods. The cells were exposed to increasing concentrations of the gyrase poisons VM-26 (a), nalidixic acid (NAL) (b), and ciprofloxacin (CFX) (c) in the presence or absence of 20 μM coumermycin A1 (cou A). Relative survival (in percent) was defined as the ratio of viable cell numbers in the presence of the gyrase poison over the viable cell numbers in its absence.
DISCUSSION
The [ATP]/[ADP] ratio was initially shown to affect cellular supercoiling through its effect on DNA gyrase (15, 16). At a higher [ATP]/[ADP] ratio, DNA gyrase is more active in catalysis. The effect of ATP is understandable since gyrase requires ATP hydrolysis for its DNA supercoiling reaction. However, the effect of ADP is less clear. Studies of Drosophila DNA topoisomerase II have suggested that ADP is competitive with ATP in enzyme catalysis. In the case of Drosophila DNA topoisomerase II, the apparent Km for ATP is 280 μM and the Ki for ADP is 120 μM (30). A simple explanation is that either ATP or ADP can bind to Drosophila DNA topoisomerase II and DNA gyrase at the same binding site with a high affinity. This is in agreement with the recent observations showing that ADP can efficiently inhibit ADPNP binding to DNA gyrase (37).
Our in vitro results indicate that the [ATP]/[ADP] ratio also significantly affects the actions of topoisomerase II poisons. ATP stimulates (more than 100-fold) the formation of the cleavable complexes induced by DNA gyrase poisons including quinolones and epipodophyllotoxins. This stimulation is specifically antagonized by ADP. One possible explanation for this result is that the major drug target for these type II topoisomerase poisons is in fact the ATP-bound form of DNA gyrase. In the presence of ATP, DNA gyrase forms an ATP-gyrase complex. When this complex is bound to DNA it can effect the strand-passing reaction. ADP can competitively inhibit the formation of the ATP-gyrase complex and thereby inhibit ATPase and the catalytic activity of DNA gyrase. In the presence of gyrase poisons, the ATP-gyrase complex is effectively trapped as a cleavage complex by quinolones and epipodophyllotoxins. The formation of the cleavage complex in the presence of quinolones and epipodophyllotoxins is inhibited by ADP, since ADP competitively inhibits the formation of the ATP-gyrase complex. Why the ATP-bound gyrase is a more effective target for quinolones and epipodophyllotoxins is still unclear. Studies with yeast topoisomerase II have clearly established that the ATP-bound topoisomerase II undergoes a conformational change to form a circular protein clamp (21–23, 31–33). It is possible that certain topoisomerase II poisons (e.g., etoposide and adriamycin) preferentially interact with topoisomerase II in this ATP-bound conformation to alter the cleavage-religation equilibrium. In the case of DNA gyrase, studies by the hydroxyl radical footprint method have suggested that ATP binding may also change the conformation of gyrase (29). Moreover, the pattern of conformational change induced by the quinolones is the same as that induced by the ATP (29). It seems reasonable to assume that the conformational change of the ATP-gyrase complex is associated with the increased affinity for quinolones and epipodophyllotoxins. More detailed studies are necessary to establish the precise role of ATP in the catalytic cycle of gyrase and gyrase poisoning by quinolones and epipodophyllotoxins.
Our in vitro studies with DNA gyrase poisons also have suggested that the [ATP]/[ADP] ratio may play an important role in determining the cellular susceptibility to gyrase poisons including quinolones and epipodophyllotoxins. Indeed, earlier studies with nalidixic acid have demonstrated that mutations in enzymes involved in the TCA cycle can modulate cellular resistance to nalidixic acid (12, 19). Furthermore, DNP, an uncoupler of oxidative phosphorylation, was shown to protect cells from the bactericidal action of nalidixic acid (7). DNP (1 mM) has been shown to reduce the ATP level from 4.2 to 0.67 mM and the [ATP]/[ADP] ratio from 12 to 0.44, a 27-fold change (33). In our current studies, we have confirmed that DNP can indeed protect cells from the bactericidal actions of both quinolone antibiotics and epipodophyllotoxins (Fig. 5). In addition, fluoroacetic acid, an aconitase inhibitor, was shown to protect cells from the bactericidal actions of quinolones and epipodophyllotoxins (see Results). These results are consistent with a potential role of ATP in determining cellular susceptibility to gyrase poisons. However, whether the ATP effect is exclusively through DNA gyrase is unclear since ATP affects many cellular processes including those which may be involved in the processing of gyrase cleavage complexes (e.g., DNA replication, RNA transcription, or the actions of DNA helicases) (6, 14, 23).
In order to clarify the role of ATP in the bactericidal actions of gyrase poisons, we have tested coumermycin A1 for its protective effect on cell killing by gyrase poisons. Coumermycin A1 specifically antagonizes the effect of ATP on DNA gyrase without significantly affecting other cellular ATP-requiring enzymes (1, 9–11, 13, 23, 24, 35, 36). As expected, coumermycin A1 antagonized the effect of ATP on gyrase-mediated DNA cleavage in vitro and protected cells from the bactericidal actions of gyrase poisons in vivo. These results strongly suggest that the protective effect of coumermycin A1 on the bactericidal actions of gyrase poisons in vivo is mediated through DNA gyrase rather than other cellular enzymes which may indirectly affect the bactericidal actions of gyrase poisons. In the aggregate, these results are consistent with the notion that the [ATP]/[ADP] ratio is an important determinant of cellular susceptibility to gyrase poisons.
Whether the ATP antagonizing effect of coumermycin A1 on gyrase-mediated DNA cleavage is primarily responsible for the protective effect of coumermycin A1 on the bactericidal actions of gyrase poisons is not certain. Our in vitro results suggest such a possibility. However, coumermycin A1 is also known to inhibit DNA replication through its inhibitory effect on the catalytic activity of DNA gyrase (9). If the collision between the replication fork and the cleavage complex induced by gyrase poisons is primarily responsible for cell killing, inhibition of DNA replication by coumermycin A1 is sufficient to account for the protective effect of coumermycin A1. However, studies with mammalian cells have suggested that replication is only partially responsible for the cytotoxic actions of epipodophyllotoxins (6). RNA transcription and other unidentified processes also have been suggested to be involved (for a review, see reference 23). If the mechanism of cell killing by quinolones and epipodophyllotoxins also involves multiple mechanisms, the large protective effect of coumermycin A1 on the bactericidal actions of gyrase poisons cannot be explained solely by the inhibitory effect of coumermycin A1 on DNA replication. We therefore suggest that the antagonistic effect of coumermycin A1 on ATP-stimulated, gyrase-mediated DNA cleavage is primarily responsible for the protective effect of coumermycin A1 on the bactericidal actions of gyrase poisons. Taken together, our results support the notion that the [ATP]/[ADP] ratio, through its modulatory effect on the gyrase-mediated DNA cleavage, is an important determinant of cellular susceptibility to gyrase poisons.
ACKNOWLEDGMENT
This work was supported by an NIH grant (grant CA-39662) to Leroy F. Liu.
REFERENCES
- 1.Ali J A, Jackson A P, Howell A J, Maxwell A. The 43-kilodalton N-terminal fragment of the DNA gyrase B protein hydrolyzes ATP and binds coumarin drugs. Biochemistry. 1993;32:2717–2724. doi: 10.1021/bi00061a033. [DOI] [PubMed] [Google Scholar]
- 2.Ali J A, Orphanides G, Maxwell A. Nucleotide binding to the 43-kilodalton N-terminal fragment of the DNA gyrase B protein. Biochemistry. 1995;34:9801–9808. doi: 10.1021/bi00030a018. [DOI] [PubMed] [Google Scholar]
- 3.Austin C A, Marsh K L, Wasserman R A, Willmore E, Sayer P J, Wang J C, Fisher L M. Expression, domain structure, and enzymatic properties of an active recombinant human DNA topoisomerase II beta. J Biol Chem. 1995;270:15739–15746. doi: 10.1074/jbc.270.26.15739. [DOI] [PubMed] [Google Scholar]
- 4.Beck W T, Danks M K. Mechanisms of resistance to drugs that inhibit DNA topoisomerases. Semin Cancer Biol. 1991;2:235–244. [PubMed] [Google Scholar]
- 5.Berger J M, Gamblin S J, Harrison S C, Wang J C. Structure and mechanism of DNA topoisomerase II. Nature. 1996;379:225–232. doi: 10.1038/379225a0. [DOI] [PubMed] [Google Scholar]
- 6.D’Arpa P, Beardmore C, Liu L F. Involvement of nucleic acid synthesis in cell killing mechanisms of topoisomerase poisons. Cancer Res. 1990;50:6919–6924. [PubMed] [Google Scholar]
- 7.Deitz W H, Cook T M, Goss W A. Mechanism of action of nalidixic acid on Escherichia coli. III. Conditions required for lethality. J Bacteriol. 1966;91:768–773. doi: 10.1128/jb.91.2.768-773.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drake F H, Hofmann G A, Bartus H F, Mattern M R, Crooke S T, Mirabelli C K. Biochemical and pharmacological properties of p170 and p180 forms of topoisomerase II. Biochemistry. 1989;28:8154–8160. doi: 10.1021/bi00446a029. [DOI] [PubMed] [Google Scholar]
- 9.Drlica K, Franco R J. Inhibitors of DNA topoisomerases. Biochemistry. 1988;27:2253–2259. doi: 10.1021/bi00407a001. [DOI] [PubMed] [Google Scholar]
- 10.Gilbert E J, Maxwell A. The 24 kDa N-terminal sub-domain of the DNA gyrase B protein binds coumarin drugs. Mol Microbiol. 1994;12:365–373. doi: 10.1111/j.1365-2958.1994.tb01026.x. [DOI] [PubMed] [Google Scholar]
- 11.Gormley N A, Orphanides G, Meyer A, Cullis P M, Maxwell A. The interaction of coumarin antibiotics with fragments of DNA gyrase B protein. Biochemistry. 1996;35:5083–5092. doi: 10.1021/bi952888n. [DOI] [PubMed] [Google Scholar]
- 12.Helling R B, Kukora J S. Nalidixic acid-resistant mutants of Escherichia coli deficient in isocitrate dehydrogenase. J Bacteriol. 1971;105:1224–1226. doi: 10.1128/jb.105.3.1224-1226.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hooper D C, Wolfson J S. Mode of action of the quinolone antimicrobial agents. Rev Infect Dis. 1988;10:S14–S21. doi: 10.1093/clinids/10.supplement_1.s14. [DOI] [PubMed] [Google Scholar]
- 14.Howard M T, Neece S H, Matson S W, Kreuzer K J. Disruption of a topoisomerase-DNA cleavage complex by a helicase. Proc Natl Acad Sci USA. 1994;91:12031–12035. doi: 10.1073/pnas.91.25.12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsieh L-S, Burger R M, Drlica K. Bacterial DNA supercoiling and [ATP]/[ADP]: change associated with a transition to anaerobic growth. J Mol Biol. 1991;219:443–450. doi: 10.1016/0022-2836(91)90185-9. [DOI] [PubMed] [Google Scholar]
- 16.Hsieh L-S, Rouviere-Yaniv J, Drlica K. Bacterial DNA supercoiling and [ATP]/[ADP]: changes associated with salt shock. J Bacteriol. 1991;173:3914–3917. doi: 10.1128/jb.173.12.3914-3917.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hughes C S, Shen J W, Subjeck J R. Resistance to etoposide induced by three glucose-regulated stresses in Chinese ovary cells. Cancer Res. 1989;49:4452–4454. [PubMed] [Google Scholar]
- 18.Kupfer G, Bodley A L, Liu L F. Involvement of intracellular ATP in cytotoxicity of topoisomerase II-targeting antitumor drugs. NCI Monogr. 1987;4:37–40. [PubMed] [Google Scholar]
- 19.Lakshmi T M, Helling R B. Selection of citrate synthase deficient in icd mutants of Escherichia coli. J Bacteriol. 1976;127:76–83. doi: 10.1128/jb.127.1.76-83.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindsley J E, Wang J C. Proteolysis patterns of epitopically labeled yeast DNA topoisomerase II suggest an allosteric transition in the enzyme induced by ATP binding. Proc Natl Acad Sci USA. 1991;88:10485–10489. doi: 10.1073/pnas.88.23.10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindsley J E, Wang J C. On the coupling between ATP usage and DNA transport by yeast DNA topoisomerase II. J Biol Chem. 1993;268:8096–8104. [PubMed] [Google Scholar]
- 22.Lindsley J E, Wang J C. Study of allosteric communication between promoters by immunotagging. Nature. 1993;361:749–750. doi: 10.1038/361749a0. [DOI] [PubMed] [Google Scholar]
- 23.Liu L F. DNA topoisomerase poisons as antitumor drugs. Annu Rev Biochem. 1989;58:351–375. doi: 10.1146/annurev.bi.58.070189.002031. [DOI] [PubMed] [Google Scholar]
- 24.Maxwell A. The interaction between coumarin drugs and DNA gyrase. Mol Microbiol. 1993;9:681–668. doi: 10.1111/j.1365-2958.1993.tb01728.x. [DOI] [PubMed] [Google Scholar]
- 25.Maxwell A, Rau D C, Gellert M. Proceedings of the fourth conversation in the discipline of biomolecular stereodynamics. In: Sarma R H, Sarma M H, editors. Biomolecular stereodynamics III. Albany, N.Y: Adenine Press; 1986. pp. 137–146. [Google Scholar]
- 26.Menzel R, Gellert M. The biochemistry and biology of DNA gyrase. In: Liu L F, editor. Advances in pharmacology-DNA topoisomerases: biochemistry and molecular biology. San Diego, Calif: Academic Press, Inc.; 1994. pp. 39–61. [DOI] [PubMed] [Google Scholar]
- 27.Morrison A, Higgins N P, Cozzarelli N R. Interaction between DNA gyrase and its cleavage sites on DNA. J Biol Chem. 1980;255:2211–2219. [PubMed] [Google Scholar]
- 28.Nelson E M, Tewey K M, Liu L F. Mechanism of anti-tumor drugs: poisoning of mammalian DNA topoisomerase II on DNA by anti-tumor drug m-MASA. Proc Natl Acad Sci USA. 1984;81:1361–1365. doi: 10.1073/pnas.81.5.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orphanides D, Maxwell A. Evidence for a conformational change in the DNA gyrase-DNA complex from hydroxyl radical footprint. Nucleic Acids Res. 1994;22:1567–1575. doi: 10.1093/nar/22.9.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osheroff N. Eucaryotic topoisomerase II: characterization of enzyme turnover. J Biol Chem. 1986;261:9944–9950. [PubMed] [Google Scholar]
- 31.Roca J, Wang J C. The capture of a DNA duplex helix by an ATP-dependent protein clamp: a key step in DNA transport by type II DNA topoisomerase. Cell. 1992;71:833–840. doi: 10.1016/0092-8674(92)90558-t. [DOI] [PubMed] [Google Scholar]
- 32.Roca J, Ishida R, Berger J M, Andoh T, Wang J C. Antitumor bisdioxopiperazines inhibit yeast DNA topoisomerase II by trapping the enzyme in the form of a closed protein clamp. Proc Natl Acad Sci USA. 1994;91:1781–1785. doi: 10.1073/pnas.91.5.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rohweer J M, Jensen P R, Shinohara Y, Postma P W, Westerhoff H V. Changes in the cellular energy state affect the activity of the bacterial phosphotransferase system. Eur J Biochem. 1996;235:225–230. doi: 10.1111/j.1432-1033.1996.00225.x. [DOI] [PubMed] [Google Scholar]
- 34.Schultz P, Olland S, Oudet P, Hancock R. Structure and conformational changes of DNA topoisomerase II visualized by electron microscopy. Proc Natl Acad Sci USA. 1996;93:5936–5940. doi: 10.1073/pnas.93.12.5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sugino A, Cozzarelli N R. The intrinsic ATPase of DNA gyrase. J Biol Chem. 1980;255:6299–6306. [PubMed] [Google Scholar]
- 36.Sugino A, Higgins N P, Brown P K, Peebles C L, Cozzarelli N R. Energy coupling in the DNA gyrase and the mechanism of action of novobiocin. Proc Natl Acad Sci USA. 1978;75:4838–4842. doi: 10.1073/pnas.75.10.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamura J K, Bate A D, Gellert M. Slow interaction of 5′-adenylyl-β,γ-imidodiphosphate with Escherichia coli gyrase. J Biol Chem. 1992;267:9214–9222. [PubMed] [Google Scholar]
- 38.Wang J C. DNA topoisomerases. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 39.Westerhoff H V, O’Dea M H, Maxwell A, Gellert M. DNA supercoiling by gyrase: a static head analysis. Cell Biophys. 1988;12:157–181. doi: 10.1007/BF02918357. [DOI] [PubMed] [Google Scholar]
- 40.Wu H-Y, Liu L F. DNA looping alters DNA conformation during transcription. J Mol Biol. 1991;219:615–622. doi: 10.1016/0022-2836(91)90658-s. [DOI] [PubMed] [Google Scholar]
- 41.Yamauchi T, Raffin T A, Yang P, Sikic B I. Differential protective effects of varying degrees of hypoxia on the cytotoxicities of etoposide and bleomycin. Cancer Chemother Pharmacol. 1987;19:282–286. doi: 10.1007/BF00261473. [DOI] [PubMed] [Google Scholar]