Abstract
Sulfonamide resistance in recent isolates of Streptococcus pyogenes was found to be associated with alterations of the chromosomally encoded dihydropteroate synthase (DHPS). There were 111 different nucleotides (13.8%) in the genes found in susceptible and resistant isolates, respectively, resulting in 30 amino acid changes (11.3%). These substantial changes suggested the possibility of a foreign origin of the resistance gene, in parallel to what has already been found for sulfonamide resistance in Neisseria meningitidis. The gene encoding DHPS was linked to at least three other genes encoding enzymes of the folate pathway. These genes were in the order GTP cyclohydrolase, dihydropteroate synthase, dihydroneopterin aldolase, and hydroxymethyldihydropterin pyrophosphokinase. The nucleotide differences in genes from resistant and susceptible strains extended from the beginning of the GTP cyclohydrolase gene to the end of the gene encoding DHPS, an additional indication for gene transfer in the development of resistance. Kinetic measurements established different affinities for sulfathiazole for DHPS enzymes isolated from resistant and susceptible strains.
Streptococcal infections were among the first infectious diseases that could be successfully treated when sulfonamides were introduced as chemotherapeutic agents (5). During the second World War, streptococcal infections in military training camps were controlled by prophylactic use of sulfonamides. However, in some cases, prophylaxis failed because sulfonamide-resistant streptococcal strains started to appear (4, 9). After the war, penicillins and other antibiotics replaced sulfonamides in the treatment of streptococcal infections, and sulfonamide resistance in Streptococcus pyogenes has received little attention since then.
Only a few earlier reports from larger surveys on antibiotic resistance in streptococci have included sulfonamide resistance. In one survey from 1965 at the Karolinska Hospital in Stockholm, Sweden, a resistance frequency of about 20% was found. Later, a similar study at the Boston City Hospital in 1972 showed 8.6% resistance (8).
The mechanism of sulfonamide resistance in S. pyogenes has not been studied before. The most thorough studies of sulfonamide resistance were in gram-negative bacteria, where the most common mechanism of resistance, e.g., in members of the family Enterobacteriaceae, was found to be the plasmid-mediated synthesis of drug-resistant variants of the target enzyme dihydropteroate synthase (DHPS) (11, 17, 18, 23). In Streptococcus pneumoniae, sulfonamide resistance can be mediated by mutations in the chromosomal gene coding for DHPS (15, 16). In Escherichia coli, mutants selected in the laboratory were found to contain point mutations in the folP gene, coding for DHPS (3, 6). In the protozoan parasite Plasmodium falciparum, sulfadoxine resistance has been associated with mutations in the DHPS-encoding gene (2, 25, 26). Sulfonamide resistance in Neisseria meningitidis was also shown to have a chromosomal location (13). In this case, however, the acquisition of resistance is not just by mutation but also by transformational recombination (6, 7, 19).
The present study was designed to determine the mechanism of resistance to sulfonamides in relatively recent isolates of S. pyogenes. The strains included in this study were isolated in 1985, and in these strains, the resistance was chromosomally located and was found to be associated with an altered DHPS. Large differences between resistant and susceptible variants of the enzyme suggested a recombinational mechanism rather than accumulated mutations for the acquisition of resistance.
MATERIALS AND METHODS
Collection of streptococcal strains.
Sulfonamide-resistant and -susceptible strains of S. pyogenes were collected at the Bacteriological Laboratory of Karolinska Hospital, Solna, Sweden. Resistance to sulfonamides is not routinely monitored, but collection of sulfonamide-resistant isolates was done by adding a disc containing sulfaisodimidine on streptococcal isolates analyzed by the laboratory. The sulfonamide-resistant isolates G52, G56, G71, G72, and G76 were all collected during 1985 and had MICs of >512 μg/ml. For reference, the three sulfonamide-susceptible isolates, G1, G2, and G68, which all showed MICs of <2 μg/ml, were collected at the same time. All isolates were kept frozen at −70°C until further laboratory analyses.
Cloning and nucleotide sequence determination of sulfonamide resistance determinants.
For cloning of the sulfonamide resistance determinant, the plasmid vector pUC19 (27) was used. It was propagated in E. coli JM83 (28). For selection of clones with sulfonamide resistance genes, Iso-Sensitest agar (Oxoid) supplemented with ampicillin (50 μg/ml) and sulfathiazole (12.5 μg/ml) was used. The E. coli C-167ts20 strain (21) and a knockout derivative of strain C600 (C600ΔfolP::Kmr) (7) were used for detection of DHPS activity by complementation. For measurement of DHPS activity, E. coli C600ΔfolP::Kmr was used as the host. Nucleotide sequence determinations were done by the dideoxy-chain termination method (20) after cloning in the phage vectors M13mp18 and M13mp19, which were propagated in E. coli JM105 (28).
DNA preparations.
DNA from S. pyogenes strains was prepared by a method obtained from F. Walter (27a), Jena, Germany. Bacteria were grown in 100 ml of Todd-Hewitt broth (Difco) and harvested by centrifugation. The pellet was resuspended in 5 ml of lysis buffer (50 mM Tris-HCl [pH 8.0], 50 mM EDTA, 5% sucrose, 1% Triton X-100) and incubated at 37°C for 2 to 4 h. Mutanolysin was added to a final concentration of 2 mg/ml, and digestion was continued for another 1 to 2 h. Sodium dodecyl sulfate was added to a final concentration of 1%, and proteinase K was added to a final concentration of 50 μg/ml. Incubation was continued for another hour at 37°C. The lysate was extracted twice with an equal volume of phenol and twice with chloroform. The DNA was precipitated by the addition of ethanol, washed several times in 70% ethanol, and finally dissolved in TE buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA).
Other methods.
Digestions with restriction endonucleases, agarose gel electrophoresis, ligation, and transformation were done by standard procedures.
PCR.
DNA amplifications were done in a Perkin-Elmer Thermocycler in a buffer of 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 0.01% gelatin, and 0.2 mM of each deoxynucleoside triphosphate. Twenty picomoles of each primer and approximately 1 μg of template DNA was used. Taq DNA polymerase was purchased from Promega and Pharmacia, and 2.5 units were used in each reaction mixture. Primers were designed according to the sequence of the folP gene of the resistant strain G56. The primer 5′-GGGATCCAGGAGAGGACTATGAAGATT-3′ specified the 5′ end of the gene, and the primer 5′-AATGCTTTCCTCACATCAACTGACTCA-3′ specified the 3′ end of the gene.
Determination of DHPS activity.
For measurement of DHPS activity, E. coli C600ΔfolP::Kmr was used as the host. Genes encoding DHPS from the streptococcal strains were cloned in pUC19 and introduced into the host by transformation. Cell extracts were prepared as described earlier (24) and partially purified by gel filtration on Sephacryl S-200 followed by ion-exchange chromatography on DEAE-Sephacel. Determination of DHPS activity was done as described previously by the incorporation of 14C-labelled p-aminobenzoic acid (PABA) into dihydropteroic acid (24).
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this report have been deposited at the EMBL with the following accession numbers: AJ000685 (S. pyogenes G56 folE, folP, folQ, and folK genes), AJ000686 (S. pyogenes G1 folP gene), and AJ000687 (S. pyogenes G72 folP gene).
RESULTS
Collection of sulfonamide-resistant isolates of S. pyogenes.
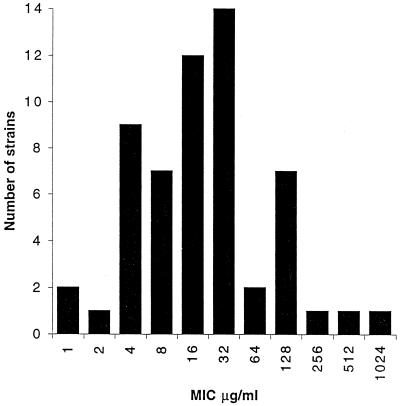
Sulfonamide susceptibility testing of clinical isolates of S. pyogenes is not routinely performed today. However, sulfonamide-resistant strains were looked for and found at the bacteriological laboratory of the Karolinska Hospital in 1985 and 1987. Five resistant strains (MIC ≥ 512 μg/ml) collected in 1985 were used in the present investigation of the mechanism of sulfonamide resistance. A more thorough survey was done in December 1987, when all laboratory cultures of S. pyogenes were tested for resistance to sulfaisodimidine and their MICs were determined. The most interesting result from this survey was the large range of MICs from a few very low MICs (<2 μg/ml) to a few very high MICs (>512 μg/ml), while the majority of strains showed intermediate levels of resistance, with a peak at 32 μg/ml (Fig. 1).
FIG. 1.
Distribution of MICs of sulfaisodimidine for 65 isolates of S. pyogenes collected at the bacteriological laboratory of Karolinska Hospital, Solna, Sweden, during December 1987.
Cloning and sequencing of the sulfonamide resistance determinant.
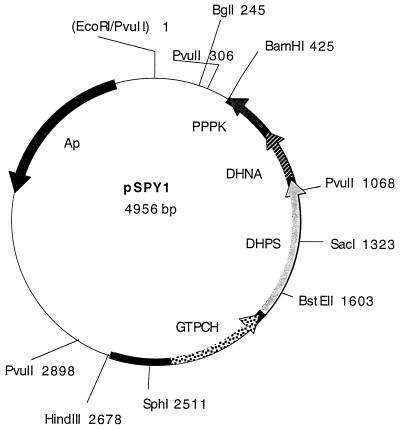
DNA preparations from the strains isolated in 1985 failed to show plasmid bands by agarose gel electrophoresis, and we therefore decided to clone the resistance determinant from a resistant strain of S. pyogenes. This was done by partial digestion of DNA from the streptococcal isolate G56 with Sau3A and inserting the resulting fragments in the plasmid vector pUC19 that had been digested with BamHI. The ligation mixture was used to transform the E. coli JM83 strain with selection on Iso-Sensitest agar plates containing 50 μg of ampicillin per ml and 12.5 μg of sulfathiazole per ml. In this way, six colonies were obtained; the plasmids from these colonies contained inserts of 5 kb, which showed similar restriction enzyme digestion patterns. After subcloning in pUC19, a 2.3-kb HindIII-BamHI fragment present in all six transformants was shown to confer sulfonamide resistance to strain JM83. A restriction enzyme digestion map of the resulting recombinant plasmid pSPY1 made with enzymes used to make subclones for sequence determinations is shown in Fig. 2.
FIG. 2.
Restriction map and localization of potential open reading frames in the recombinant plasmid pSPY1, carrying a 2.3-kb fragment from the sulfonamide-resistant strain G56. The fragment was cloned between the HindIII and BamHI sites of the vector pUC19. Potential open reading frames were identified after sequencing and comparison with published sequences. Abbreviations: GTPCH, GTP cyclohydrolase; DHNA, dihydroneopterin aldolase; PPPK, hydroxymethyldihydropterin pyrophosphokinase.
In order to show that the cloned DNA expressed a DHPS enzyme, the recombinant plasmid pSPY1 was used to transform the thermosensitive E. coli C-167ts20 strain (21). The results of this experiment showed that pSPY1 was able to complement the thermosensitivity of the mutant and support growth at 42°C. The transformation also allowed the mutant to grow in 12.5 μg of sulfathiazole per ml, which is normally sufficient to inhibit the growth of C-167ts20 at the permissive temperature of 30°C. These results were taken as evidence for the production of thermostable DHPS in C-167ts20 carrying pSPY1 and thus that pSPY1 encodes a DHPS.
The nucleotide sequences for both strands of the entire 2.3-kb fragment were determined and are shown in Fig. 3 together with the predicted amino acid sequence for three complete and two partial open reading frames. The first complete open reading frame was identified as coding for a GTP cyclohydrolase by comparison with database sequences. The highest similarity was found with GTP cyclohydrolase from Bacillus subtilis (accession no. P19465; 64.9% amino acid identity). The corresponding gene in E. coli is designated folE. Immediately following the stop codon of the folE gene, another reading frame starts and it was identified as coding for the DHPS enzyme by comparison with previously known DHPS enzymes. Here the highest degree of similarity was scored for the DHPS from Staphylococcus aureus (45.1% amino acid identity) (10). The DHPS from S. pneumoniae showed only 41.3% amino acid identity (15). The considerable difference in the sequences of S. pyogenes and S. pneumoniae is remarkable, and the similarity between these two DHPS enzymes is not higher than that for most other DHPSs. The open reading frame starting 6 bp after the stop codon of the folP analog (coding for DHPS) shows a reasonable similarity to identified and proposed genes coding for dihydroneopterin aldolase from Staphylococcus haemolyticus (51.2% amino acid identity) (12), B. subtilis (41.2%) (22), and to a lesser degree from S. pneumoniae (26.9%) (14). The gene coding for DHNA has been designated folQ. Downstream the folQ gene, a partial open reading frame that could be identified as a hydroxymethyldihydropterin pyrophosphokinase (folK) was found. The closest similarity with other organisms was in this case with E. coli (40.7%) (accession no. P26281), while the corresponding protein in S. pneumoniae (accession no. P22291) showed 36.8% amino acid identity. Preceding the folE gene, another potential open reading frame could be identified as similar to the sulB gene of S. pneumoniae (14). This gene codes for a dihydrofolate synthetase, and the gene designation is folC. Thus, five genes involved in the biosyntheses of folates could be identified in the cloned fragment. Although similar arrangements of corresponding genes have been found in B. subtilis and S. pneumoniae, the order of the individual genes is different in these cases.
FIG. 3.
Nucleotide sequence and deduced amino acid sequences for potential open reading frames of the cloned 2.3-kb HindIII-BamHI fragment from S. pyogenes G56, which is resistant to sulfonamides. Nucleotides that are different from those in the sequence published by the Streptococcal Genome Sequencing Project are indicated above the G56 sequence.
Comparison between nucleotide sequences for the folP gene from sulfonamide-susceptible and -resistant isolates.
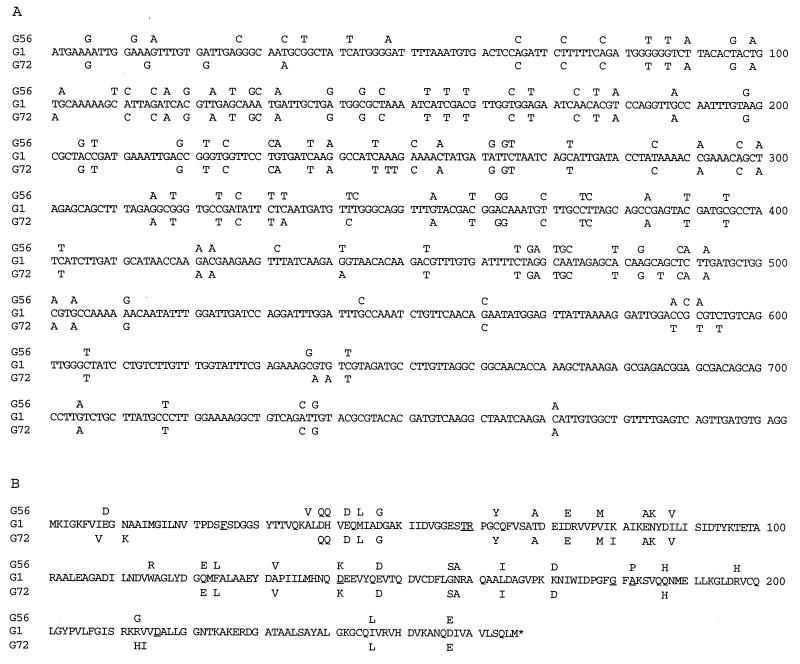
The nucleotide sequence of the folP gene from G56 cloned in plasmid pSPY1 was used to design oligonucleotide primers for PCR amplification of the corresponding sequence from other strains of S. pyogenes, both resistant and susceptible to sulfonamides. DNA from the resistant strains G52, G71, and G76 all gave distinct bands of the expected size, whereas DNA from the susceptible strains G1, G2, G68, and from the resistant strain G72 gave several bands in most experiments, one of which was of the expected size. Samples of DNA from PCR amplifications were purified by agarose gel electrophoresis and electroeluted from the gels. The purified DNA samples were cloned in M13 phages for nucleotide sequence determinations. The complete nucleotide sequences from the susceptible strain G1 and from the resistant strain G72 were determined, and a comparison of these sequences to that from G56 are shown in Fig. 4A. The number of differences between the folP genes from these three strains is remarkable, but they are not evenly distributed. In the 5′ part, all three sequences differ in many positions. From nucleotide position 65, G56 and G72 are almost identical and differ from G1. Toward the 3′ end, the three sequences again become more similar but still exhibit some differences. The same pattern appears when the deduced amino acid sequences are compared in Fig. 4B, although it is evident that many differences in nucleotide sequence do not influence the amino acid sequence.
FIG. 4.
Comparison of the folP nucleotide sequences (A) and DHPS amino acid sequences (B) from S. pyogenes G56 (Sur), G1 (Sus), and G72 (Sur). For the sulfonamide-resistant isolates, only those nucleotides that differ from the G1 sequence are indicated. In panel B, amino acids that have been associated with sulfonamide resistance in DHPS from other organisms are underlined.
The complete nucleotide sequence of S. pyogenes is being determined at the University of Oklahoma (http://www.genome.ou.edu/strep.html). One contig (contig 264, release of 25 September 1997) covers the sequences contained in pSPY1. Comparison between contig 264 and pSPY1 revealed that the region of divergence in the resistant variant and the susceptible variant extends from the beginning of the folE gene to the end of the folP gene, totalling ca. 1,330 bp. In Fig. 3, all nucleotides in contig 264 that differ from those in the sequence of G56 are indicated above the sequence of G56. The start of the divergent region is easily detectable at nucleotide 360 of the cloned segment in pSPY1, but the end of the divergent region is somewhat uncertain because of differences outside this highly divergent region. These small divergences are most likely due to natural strain variation. The overall sequence differences between strain G56 and contig 264 in the divergent part was 16.8%. In the latter part of the sequence covering the folQ and folK genes, the sequence difference was only 0.8%. The amino acid sequence differences for the different genes were also different: 7.3% for folE, 7.9% for folP, 1.7% for folQ, and 0% for the partially sequenced folK gene. Thus, both for nucleotide differences and for amino acid differences, a divergent part covering the two genes folE and folP was evident. Comparison between the nucleotide sequence of the folP gene from the susceptible strain G1 and contig 264 revealed a few differences (the identity between these two sequences is 98%), which could reflect natural strain variation.
Determination of DHPS enzyme kinetics.
For the determination of DHPS enzyme characteristics in S. pyogenes, the plasmids pSPY1 (carrying the sulfonamide resistance gene from G56) and pSPY2 (carrying the corresponding gene from the sulfonamide-susceptible strain G1) were introduced into an E. coli C600 knockout mutant lacking endogenous DHPS activity (7). Cell extracts were prepared from these plasmid-carrying cells, and the DHPS activity was measured in a partially purified extract. Enzyme kinetics were analysed by varying the concentration of the substrate PABA with the other substrate 2-hydroxy-4-amino-6-hydroxymethyl-pteridine-pyrophosphate in excess. The Km for PABA was determined in the absence of inhibitor by Lineweaver-Burk diagrams, and Ki was determined by measuring the apparent Km in the presence of inhibitor. Ki determinations showed a distinct difference between the two enzymes (27.4 μM for the resistant enzyme and 0.2 μM for the susceptible variant). A difference in Km for PABA was also apparent from these determinations (2.5 μM for the resistant enzyme and 0.7 μM for the susceptible enzyme).
DISCUSSION
Sulfonamide resistance in S. pyogenes has been reported since the second World War, but the mechanism of resistance has not been investigated. In this report, we see that in isolates of resistant strains, the resistance determinant is chromosomally located and encodes a drug-resistant variant of the enzyme DHPS, the target of sulfonamide action. The simplest explanation would then be one or several mutational changes in the folP gene of S. pyogenes, as has been shown for sulfadoxine resistance in P. falciparum, where defined mutations in the gene coding for DHPS have been found in some cases of drug-resistant isolates (2, 25, 26). The pronounced differences in sequences from resistant and susceptible strains of S. pyogenes led to the conclusion that in this organism sulfonamide resistance has not developed merely by the accumulation of point mutations. Instead, it is most likely that the resistance gene has been introduced by transformational recombination. A parallel case is found in N. meningitidis, where sulfonamide resistance could be explained by the recombinational spread of resistance genes or gene fragments (19). Although we cannot detect plasmids in the recent isolates, we cannot exclude the possibility that the early sulfonamide-resistant S. pyogenes isolates actually carried plasmids that were later incorporated into the chromosome. There are other possibilities for recombination. The results presented here with a limited area of divergent sequences are most likely caused by transformation or possibly transduction. The same pattern with defined limits for an area of divergent sequence has been defined for sulfonamide resistance in N. meningitidis (19).
That the difference in phenotype can be attributed to these differences in the DHPS amino acid sequence was borne out by preliminary measurements of enzyme kinetics, which showed that the DHPS enzyme specified by the G56 (Sur) strain is indeed less susceptible to sulfathiazole inhibition and that its Km for PABA is increased in comparison with the DHPS present in the susceptible G1 strain. This parallels the situation in N. meningitidis where similar differences in Km for PABA in susceptible and resistant variants of the enzyme appear (7).
Two questions still remain. One concerns the amino acid changes that are really crucial for development of resistance. The many differences in DHPS amino acid sequences in the resistant and susceptible strains make it difficult to determine those changes that are important in changing the enzyme to a different response to the inhibitor. Comparison with mutants isolated in the laboratory offers little help. An E. coli ts mutant had a Phe-to-Ile change at position 25 (numbered according to the G56 sequence) (3). Another E. coli mutant and two clinical isolates of N. meningitidis with an Phe-to-Leu change at position 25 have been reported (6, 7). In the S. pyogenes resistant strains, there is a Phe at this position, however. Site-directed mutagenesis in a N. meningitidis isolate showed that the Phe-to-Leu change was absolutely necessary for the resistance phenotype (6). Also, another change, Cys to Gly at position 194, was shown to be involved but not necessary for resistance (6). In both of these identified positions, the sulfonamide-resistant S. pyogenes isolates have the same amino acids as the susceptible isolates. Any point mutations involved in resistance of S. pyogenes thus must be found in some other location in the sequence. Some clinical isolates of N. meningitidis were found to be resistant to sulfonamides due to an Ser-Gly insertion next to a conserved part of the enzyme (6). However, other sulfonamide-resistant strains of N. meningitidis lack the Ser-Gly insertion, and in S. pyogenes, there is no obvious difference between the sulfonamide-resistant and -susceptible strains in the corresponding part of the sequence (6). Figure 4 highlights the mutations that have been associated with sulfonamide resistance in other species and compares the differences in resistant and susceptible isolates of S. pyogenes. In most cases, the differences do not coincide with the earlier described mutations in folP genes from other organisms. There are only two possible exceptions. The first is at position 141 where a change from Lys to Glu is found in the corresponding position of DHPS from some isolates of P. falciparum (26). The second is that the Pro found at position 182 in G56 corresponds to an Ala in the susceptible isolate G1 (and in the second resistant isolate, G72). In the corresponding position in the pppk-dhps gene of P. falciparum, a mutation resulting in the change of an Ala in the wild type to a Gly in the sulfonamide-resistant strain K1 is the only difference in this strain compared to susceptible strains (2). The recent publication of the three-dimensional structure of DHPS from S. aureus (10) and E. coli (1) where substrate binding regions in the protein are defined offers help in locating the most likely part of the sequence to look for important changes in relation to resistance development. These differences can now be investigated by site-directed mutagenesis according to the procedure described by Fermér et al. (6) for the meningococcal folP gene.
The second question concerns the origin of the resistance gene. The large number of silent changes and the fact that the preceding gene coding for GTP cyclohydrolase is different from the reference strain at least suggest that this gene is not originally from S. pyogenes but introduced by transformation from a different bacterial species. This possibility can now be tested by using the sequence derived from the resistant strains as a probe in hybridizations or PCR screenings against different types of streptococci and other related bacteria.
ACKNOWLEDGMENTS
This work was supported by grants from the Swedish Medical Research Council to Ola Sköld and Göte Swedberg.
The technical support of Katarina Johansson, Peter Blomgren, and Kristine Flick-Fries in PCR analysis and nucleotide sequence determinations has been most valuable. We also thank the pharmacy students Sarah Hood, Mojgan Saboonchi, and Katrin Ström for help with sequence determinations and enzymatic assays. We are grateful for the release of data from the Streptococcal Genome Sequencing Project and acknowledge the efforts of B. A. Roe, S. Clifton, Mike McShan, and Joseph Ferreti, University of Oklahoma.
REFERENCES
- 1.Achari A, Somers D O, Champness J N, Bryant P K, Rosemond J, Stammers D K. Crystal structure of the anti-bacterial sulfonamide drug target dihydropteroate synthase. Nat Struct Biol. 1997;4:490–497. doi: 10.1038/nsb0697-490. [DOI] [PubMed] [Google Scholar]
- 2.Brooks D R, Wang P, Read M, Watkins W M, Sims P F G, Hyde J E. Sequence variation of the hydroxymethyldihydropterin pyrophosphokinase: dihydropteroate synthase gene in lines of the human malaria parasite, Plasmodium falciparum, with differing resistance to sulfa. Eur J Biochem. 1994;224:397–405. doi: 10.1111/j.1432-1033.1994.00397.x. [DOI] [PubMed] [Google Scholar]
- 3.Dallas W S, Gowen J E, Ray P H, Cox M J, Dev I K. Cloning, sequencing, and enhanced expression of the dihydropteroate synthase gene of Escherichia coli MC4100. J Bacteriol. 1992;174:5961–5970. doi: 10.1128/jb.174.18.5961-5970.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Damrosch D S. Chemoprophylaxis and sulfonamide resistant streptococci. JAMA. 1946;130:124–128. doi: 10.1001/jama.1946.02870030004002. [DOI] [PubMed] [Google Scholar]
- 5.Domagk G. Ein Beitrag zur Chemotherapie der Bakteriellen Infektionen. Dtsch Med Wochenschr. 1935;61:250–253. [Google Scholar]
- 6.Fermér C, Kristiansen B-E, Sköld O, Swedberg G. Sulfonamide resistance in Neisseria meningitidis as defined by site-directed mutagenesis could have its origin in other species. J Bacteriol. 1995;177:4669–4675. doi: 10.1128/jb.177.16.4669-4675.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fermér C, Swedberg G. Adaptation to sulfonamide resistance in Neisseria meningitidis may have required compensatory changes to retain enzyme function: kinetic analysis of dihydropteroate synthases from N. meningitidis expressed in a knockout mutant of Escherichia coli. J Bacteriol. 1997;179:831–837. doi: 10.1128/jb.179.3.831-837.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finland M, Garner C, Wilcox C, Sabath L D. Susceptibility of beta-hemolytic streptococci to 65 antibacterial agents. Antimicrob Agents Chemother. 1976;9:11–19. doi: 10.1128/aac.9.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamburger M, Mattman L H, Grosch D S, Hurst V. Susceptibility to sulfadiazine of hemolytic streptococci recovered in army camps. Am J Med. 1946;1946:23–27. doi: 10.1016/0002-9343(46)90018-6. [DOI] [PubMed] [Google Scholar]
- 10.Hampele I C, D’Arcy A, Dale G E, Kostrewa D, Nielsen J, Oefner C, Page M G P, Schönfeld H-J, Stüber D, Then R L. Structure and function of the dihydropteroate synthase from Staphylococcus aureus. J Mol Biol. 1997;268:21–30. doi: 10.1006/jmbi.1997.0944. [DOI] [PubMed] [Google Scholar]
- 11.Huovinen P, Sundström L, Swedberg G, Sköld O. Trimethoprim and sulfonamide resistance. Antimicrob Agents Chemother. 1995;39:279–289. doi: 10.1128/aac.39.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kellam P, Dallas W S, Ballantine S P, Delves C J. Functional cloning of the dihydropteroate synthase gene of Staphylococcus haemolyticus. FEMS Microbiol Lett. 1995;134:165–169. doi: 10.1111/j.1574-6968.1995.tb07932.x. [DOI] [PubMed] [Google Scholar]
- 13.Kristiansen B-E, Rådström P, Jenkins A, Ask E, Facinelli B, Sköld O. Cloning and characterization of a DNA fragment that confers sulfonamide resistance in a serogroup B, serotype 15 strain of Neisseria meningitidis. Antimicrob Agents Chemother. 1990;34:2277–2279. doi: 10.1128/aac.34.11.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lacks S A, Greenberg B, Lopez P. A cluster of four genes encoding enzymes for five steps in the folate biosynthetic pathway of Streptococcus pneumoniae. J Bacteriol. 1995;177:66–74. doi: 10.1128/jb.177.1.66-74.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopez P, Espinosa M, Greenberg B, Lacks S A. Sulfonamide resistance in Streptococcus pneumoniae: DNA sequence of the gene encoding dihydropteroate synthase and characterization of the enzyme. J Bacteriol. 1987;169:4320–4326. doi: 10.1128/jb.169.9.4320-4326.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maskell J P, Sefton A M, Hall L M C. Mechanism of sulfonamide resistance in clinical isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:2121–2126. doi: 10.1128/aac.41.10.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rådström P, Swedberg G. Evolution of antibiotic resistance plasmids: both RSF1010 and a self-transmissible plasmid contain sulII, one of two known genes for plasmid-borne sulfonamide-resistant dihydropteroate synthase. Antimicrob Agents Chemother. 1988;32:1684–1692. doi: 10.1128/aac.32.11.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rådström P, Swedberg G, Sköld O. Genetic analysis of sulfonamide resistance and its dissemination in gram-negative bacteria illustrate new aspects of R plasmid evolution. Antimicrob Agents Chemother. 1991;35:1840–1848. doi: 10.1128/aac.35.9.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rådström P, Fermér C, Kristiansen B-E, Jenkins A, Sköld O, Swedberg G. Transformational exchanges in the dihydropteroate synthase gene of Neisseria meningitidis, a novel mechanism for the acquisition of sulfonamide resistance. J Bacteriol. 1992;174:6386–6393. doi: 10.1128/jb.174.20.6386-6393.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sköld O. R-factor-mediated resistance to sulfonamides by a plasmid-borne, drug-resistant dihydropteroate synthase. Antimicrob Agents Chemother. 1976;9:49–54. doi: 10.1128/aac.9.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slock J, Stahly D P, Han C-Y, Six E W, Crawford I P. An apparent Bacillus subtilis folic acid biosynthetic operon containing pab, an amphibolic trpG gene, a third gene required for synthesis of para-aminobenzoic acid, and the dihydropteroate synthase gene. J Bacteriol. 1990;172:7211–7226. doi: 10.1128/jb.172.12.7211-7226.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sundström L, Rådström P, Swedberg G, Sköld O. Site-specific recombination promotes linkage between trimethoprim- and sulfonamide resistance genes. Sequence characterization of dhfrV and sulI and a recombination active locus of Tn21. Mol Gen Genet. 1988;213:191–201. doi: 10.1007/BF00339581. [DOI] [PubMed] [Google Scholar]
- 24.Swedberg G, Castensson S, Sköld O. Characterization of mutationally altered dihydropteroate synthase and its ability to form a sulfonamide-containing dihydrofolate analog. J Bacteriol. 1979;137:129–136. doi: 10.1128/jb.137.1.129-136.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Triglia T, Cowman A F. Primary structure and expression of the dihydropteroate synthase gene of Plasmodium falciparum. Proc Natl Acad Sci USA. 1994;91:7141–7153. doi: 10.1073/pnas.91.15.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Triglia T, Menting J G T, Wilson C, Cowman A F. Mutations in dihydropteroate synthase are responsible for sulfone and sulfonamide resistance in Plasmodium falciparum. Proc Natl Acad Sci USA. 1997;94:13944–13949. doi: 10.1073/pnas.94.25.13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 27a.Walter, J. Personal communication.
- 28.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]