Abstract
Background:
Bacteriophage (phage) therapy has regained attention as an alternative to antimicrobial agents for eliminating bacteria; however, the emergence of phage-resistant bacteria during the therapy is a major concern. One method to control this emergence is to create a cocktail composed of multiple phages.
Materials and Methods:
In this study, we isolated 28 phages infecting Escherichia coli and evaluated their bacteriolysis (lysis) activity, lytic spectrum, adsorption rate constant, burst size, and titer of a 1-day incubation, followed by clustering of the phages based on these physiological characteristics.
Results:
The variation in lysis onset time and duration was more significant for cocktails of phages from different clusters than for phage cocktails from the same cluster.
Conclusions:
This suggests that a combination of phages with different physiological characteristics is necessary to create a cocktail that rapidly and continuously lyses bacteria over a prolonged duration while suppressing the emergence of resistant bacterial strains.
Keywords: bacteriophage, cocktail, Escherichia coli, phage therapy
Introduction
The development of drug resistance in bacteria rapidly eliminates treatment options available for bacterial infections.1–4 The World Health Organization has published a list of 12 antimicrobial-resistant microorganisms (ARMs), including Enterobacteriaceae and Acinetobacter baumannii, for which the development of new antimicrobial agents is urgently needed as they pose a huge threat to human health.5 Nevertheless, ARM infection, including the bacteria listed, leads to a reliance on natural healing process.4,6,7 Therefore, new approaches are required to control or prevent the emergence of drug-resistant bacteria.
Phages suppress bacteria differently than antibacterial drugs and have drawn attention because they do not cause an increase in ARMs.8,9 In general, bacteria acquire resistance to phages, and cocktailing phages is an effective means for avoiding this resistance. A cocktail consisting of phages that recognize different receptors delays the emergence of phage-resistant bacteria.10–14 However, several reports have not stated clear criteria for combining phages to create a cocktail.15,16
This is because identifying the receptor requires generally generating phage-resistant bacteria and then identifying the mutation location by whole genome sequencing of the wild type and resistant, so identifing receptor requires time-consuming, labor-intensive, and expensive.17,18 Therefore, a new method that is independent of receptor identification is required.
There are several considerations in phage therapy practice. First, phages can be lytic or lysogenic. Lysogenic phages do not lyse the host but rather enter lysogenic cycle, during which they are ineffective for bacteriostasis.16,19 Second, the infectious range of phages is generally narrow.19,20 Third, similar to drug-resistant bacteria, bacteria exposed to phages become resistant.10,21
Based on these factors, it is helpful to collect a wide variety of phages that infect the target bacteria and evaluate their characteristics when studying phage therapy. Since Escherichia coli, a member of the Enterobacteriaceae, has become increasingly drug resistant and is involved in the pathogenesis of inflammatory bowel disease, phage therapy for E. coli eradication is a relevant treatment option.19,22–26 Therefore, in this study, E. coli from a mouse with dextran sodium sulfate (DSS)-induced colitis was selected as the target bacterium for phage therapy.
In this study, we screened, characterized, and classified 13 phage strains that lyse E. coli derived from a colitis-induced mouse and summarized the lysis effects when combining two of these phage strains into cocktails. We then discussed whether phage classification based on physiological properties could be useful for preparing effective cocktails for long-term lysis. In addition, we conducted a genomic analysis of each phage and discussed whether phage classification by phenotype was consistent with phage classification by genotype.
Materials and Methods
Bacterial strains
E. coli strain, TK001, as the host bacteria, was isolated from the feces of a female C57BL/6JJcl mouse with induced colitis using DSS (MP Biomedicals, Soho, OH, USA). Unless otherwise stated, E. coli strain, TK001, was used for experiment. Luria–Bertani (LB) liquid medium (BD Difco, Franklin Lakes, NJ, USA) was used for bacterial culture.
Phage screening and isolations
Phage enrichment from sewage was performed as described previously.13 Mouse fecal supernatant was prepared from a female C57BL/6JJcl mouse with induced colitis using DSS. The fecal sample was homogenized, centrifuged, and the resulting supernatant was mixed with an equal volume of chloroform.
Phages were isolated from the phage concentrate or fecal supernatant, and purified more than three times using the double-layer agar plate method (Supplementary Table S1).13
Institutional review board approval
All animal experiments in this study were reviewed and approved by the ethical committee of Waseda University Academic Research Ethics Committee (approval number: 2020-A009).
Lysis activity
E. coli cultures in the logarithmic growth phase were diluted in LB medium to an optical density at 660 nm (OD) of 0.01. The diluted culture suspension (3.96 mL) was placed in an L-shaped test tube, and 40 μL of phage solution was added to achieve a multiplicity of infection (MOI) of 0.01. The test tubes were immediately placed in an Advantec TVS062CA biophotorecorder (Advantec Toyo Co. Ltd., Tokyo, Japan) and incubated with shaking. The “lysis onset time” was defined as the time at which the OD began to decline, and the period between this and the point when the OD started to increase again was termed the “lysis duration time.”
Lytic spectrum of phage isolates
Phage was spotted onto soft agar (0.5%) containing overnight bacterial culture.27 Transparent and translucent spots were confirmed as successful infections. The bacteria, their sources, and purpose of use are summarized in Supplementary Table S2.
Adsorption test of phage isolates
Adsorption tests were performed under OD 0.1 and MOI 0.01 conditions (technical replicates ≥2, biological replicates ≥2). Phages were added to the bacterial sample, and after 0, 2, 5, and 10 min (OD = 0.1), 10 μL of the culture was added to 990 μL of sodium chloride-magnesium sulfate (SM) buffer and 100 μL of chloroform, after which the solution was vortexed and stored on ice. The bacterial debris were subsequently precipitated by centrifugation (7,607 g, 5 min, 23°C), and 100 μL of the supernatant was used for titer measurement.
The plaque-forming unit (PFU) value (P) was divided by the PFU value at time 0 (P0) to calculate the reduction ratio. The adsorption rate constants were determined by plotting the reduction ratio (P/P0) as a function of time and fitting the data with the following equation28:
where N0 is the bacterial concentration (OD) [−] at 0 min, t is the time [min], and k is the adsorption rate constant [/min].
Burst size measurement
One step test was performed under OD 0.1 and MOI 0.01 conditions (technical replicates ≥2, biological replicates ≥2). Ten minutes after the addition of the phage, the sample was centrifuged (21,130 g, 1 min, 23°C), and the supernatant was replaced with LB medium to remove the free phages. This procedure was repeated three times. At 20, 30, 40, 50, and 60 min (up to 70 min depending on the phage) after the addition of phage to the sample of bacteria (OD = 0.1), 10 μL of the culture was added to 990 μL of SM buffer, and the sample was vortexed and stored on ice.
For all samples except for those at the initial sampling time of 20 min, 100 μL of chloroform was predosed into the SM buffer to prevent phage growth during sampling. The sample collected after 20 min was used for titer measurement within 30 min of phage administration. The burst size was calculated by dividing the maximum titer value [PFU/plate] during the sampling time by the initial titer value [PFU/plate].
Yield assessment of phage isolates
Phages (3.0 × 106 PFU/mL, 10 μL) were added to 990 μL of TK001 (OD 0.1) in the log growth phase and cultured by shaking (120 rpm, 24 h, 37°C). The culture medium was centrifuged (21,130 g, 5 min, 23°C), the supernatant was mixed with 100 μL of chloroform, and the solution was vortexed followed by centrifugation (21,130 g, 5 min, 23°C). Phage titer (yield) was determined as described above.
Cocktail of phages
Cocktails were prepared by mixing equal amounts of different phages with the same titer [PFU/mL] (MOI = 0.01, technical replicates ≥2, biological replicates ≥1). Onset extension ratio was calculated by dividing the lysis onset time of the cocktail by the earlier onset time of the individual phages in the cocktail. The duration extension ratio was calculated by dividing the lysis duration time of the cocktail by the longer lysis duration time of the individual phages.
Principal component analysis and clustering
For the lytic spectrum, the lysis spectrum data were normalized by allocating values of 1 to the presence of lysis and 0 to the absence of lysis for all the bacteria, excluding TK001–TK005, in which lysis occurred with all phages. The yield was converted to logarithm base 10, and principal component analysis (PCA) was performed on the normalized data. A vector indicating the degree of contribution of each feature was plotted based on the factor loadings of the feature. The Euclidean distance between the phages was calculated using the principal component values of each phage. Hierarchical clustering was performed using the minimum variance method29 based on the calculated distances, to provide a total of three clusters.
Phage DNA extraction and analysis
Phage DNA was extracted using a phage DNA isolation kit (Norgen Biotech, Canada), according to the manufacturer's protocol. DNA was sequenced using contract analysis (BGI) with DNBSEQ G400 (150 bp read length), and the data were filtered with SOAPnuke software (BGI, Shenzhen, Guangdong, China) using the following parameters: “ -n 0.01 -l 20 -q 0.4 –adaMis 3 –outQualSys 1.”30
Genome analysis of phages
Velvet software (ver. 1.2.10) was used for de novo assembly.31 The genome annotation was performed by DFAST with default options.32,33 Phage nucleotide differences were scanned using GenomeMatcher software (ver. 3.06, options: blast+, parameters: defaults).34
Data availability
The phage genomes were deposited in the DNA Data Bank of Japan with the following accession numbers: ΦWec172 (LC739530), ΦWec174 (LC739531), ΦWec177 (LC739532), ΦWec179 (LC739533), ΦWec181 (LC739534), ΦWec186 (LC739535), ΦWec187 (LC739536), ΦWec188 (LC739537), ΦWec189 (LC739538), ΦWec190 (LC739539), ΦWec191 (LC739540), ΦWec193 (LC739541), and ΦWec196 (LC739542).
Statistical analysis
The data were analyzed using R (ver. 4.0.3) and its packages.35–46
Results
Lysis activity
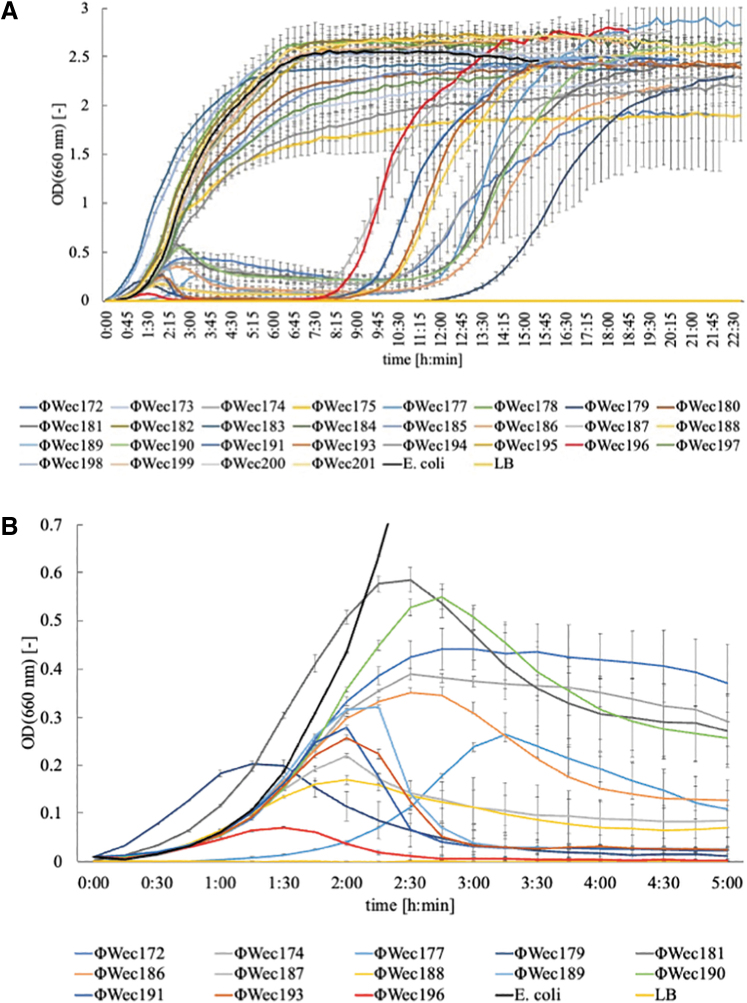
E. coli TK001, in the logarithmic growth phase, was diluted in LB medium to an OD of 0.01. Screened phages were added to the medium to achieve an MOI of 0.01. The OD was measured every 15 min (Fig. 1A).
FIG. 1.
Lysis kinetics of TK001 infected with phages. (A) The absorbance of TK001 (initial OD = 0.01) cultures infected with each phage incubated in LB medium in L-shaped test tubes by shaking at 40 rpm at 37°C. OD660 was measured every 15 min. An MOI of 0.01 was used for all phages. The horizontal axis is time [h: min], and the vertical axis is OD660 [−]. Plots (line) and error bars are expressed as the mean ± SD of three technically independent experiments. (B) Magnified view of absorbance of the culture medium showing the lysis curve. LB, Luria–Bertani; MOI, multiplicity of infection; OD, optical density; SD, standard deviation.
Of the 28 phage strains tested, 15 showed increasing ODs that remained constant at a specific value, similar to the positive control (only TK001 added to the LB medium). In contrast, 13 isolates showed a lysis curve in which the OD decreased to ∼0 (Fig. 1B). The 13 phage strains that showed this lysis curve differed in terms of lysis onset time and duration (Table 1).
Table 1.
Lysis Onset Time and Lysis Duration of TK001 Lytic Phage
| Phage | Lysis onset time | Lysis duration time |
|---|---|---|
| ΦWec172 | 2:50 | 7:30 |
| ΦWec174 | 2:50 | 6:25 |
| ΦWec177 | 3:15 | 6:50 |
| ΦWec179 | 1:20 | 9:15 |
| ΦWec181 | 2:25 | 8:25 |
| ΦWec186 | 2:30 | 7:10 |
| ΦWec187 | 2:00 | 4:15 |
| ΦWec188 | 2:00 | 6:15 |
| ΦWec189 | 2:10 | 4:40 |
| ΦWec190 | 2:45 | 7:35 |
| ΦWec191 | 1:55 | 4:35 |
| ΦWec193 | 2:00 | 4:55 |
| ΦWec196 | 1:30 | 4:30 |
Lytic spectrum
The lytic spectra of the phage were determined using a spot test. The lysis spectra of the 13 phages against 17 bacteria that showed discoloration in the spot area for at least one phage are summarized in Figure 2. The spectra for all phages and bacteria tested in this study are summarized in Supplementary Figure S2.
FIG. 2.
Lytic spectrum of TK001 infected with phages. The measurement was performed by spotting the phages. Black indicates complete lysis (Supplementary Fig. S1A), gray indicates incomplete lysis (Supplementary Fig. S1B), and white indicates no lysis (Supplementary Fig. S1C); TK002–TK004, Escherichia coli isolated from the upper, middle, and lower small intestine of DSS-induced colitis mice; TK005, Escherichia coli isolated from feces of healthy mice; TWCC 59747, Escherichia coli isolated from blood cultures of patients with suspected bacterial translocation; Ef, Escherichia fergusonii NBRC102419; Sp, Salmonella enterica subsp. Enterica serovar Pullorum NBRC3163; St, Salmonella enterica subsp. Enterica serovar Typhimurium NBRC13245; Se, Salmonella enterica subsp. enterica serovar Enteritidis NBRC3313; Sm, Salmonella enterica subsp. enterica serovar Minnesota NBRC15335; Ca, Citrobacter amalonaticus isolated from feces of DSS-induced colitis mice; DSS, dextran sodium sulfate.
All phages showed bacteriolytic activity against all E. coli strains (TK001–TK005) isolated from mice, regardless of the location of these isolates or the condition of the isolated mice. In contrast, bacteriolytic activity against human E. coli and laboratory strains differed significantly among the phages. The bacteriolytic activity against Enterobacteriaceae other than E. coli also differed among phages.
Other physiological characteristics
The adsorption rate constant, burst size, and yield were measured using the logarithmic growth phase cultures. Each phage had a unique set of physiological characteristics (Supplementary Fig. S3–S7).
PCA and clustering
PCA results based on the lysis onset time, lysis duration time, lytic spectrum, adsorption rate constant, burst size, and yield data showed that the proportion of variance was 48.4% for the first principal component and 22.1% for the second principal component (Supplementary Table S3). The factor loadings for lysis duration time and certain lytic spectra were significant and contributed considerably to the characterization of each phage (Supplementary Table S4 and Supplementary Fig. S8).
Grouping the phages into three clusters resulted in Cluster No. 1 containing six phages, Cluster No. 2 containing three phages, and Cluster No. 3 containing four phages (Fig. 3).
FIG. 3.
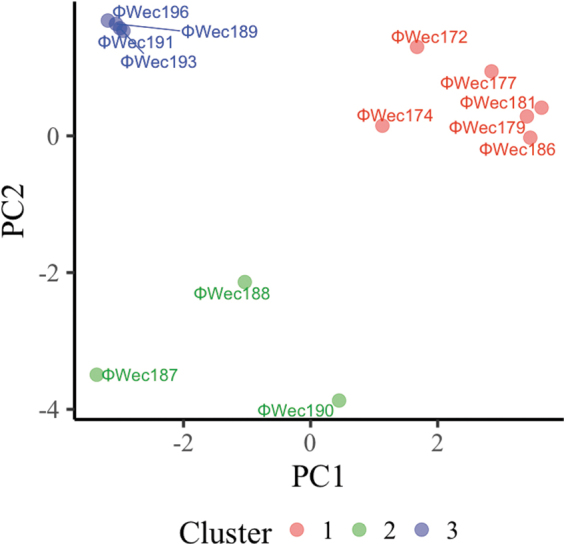
Clustering of normalized phage phenotypes. Each phage was plotted with the first principal component (PC1) on the horizontal axis and the second principal component (PC2) on the vertical axis. Cluster 1 is red, Cluster 2 is green, and Cluster 3 is blue. The Euclidean distance between the phages was calculated using the values of each principal component of each phage, and hierarchical clustering was performed using the minimum variance method (Ward's method) to produce three clusters.
Lysis activity of the two-phage cocktails
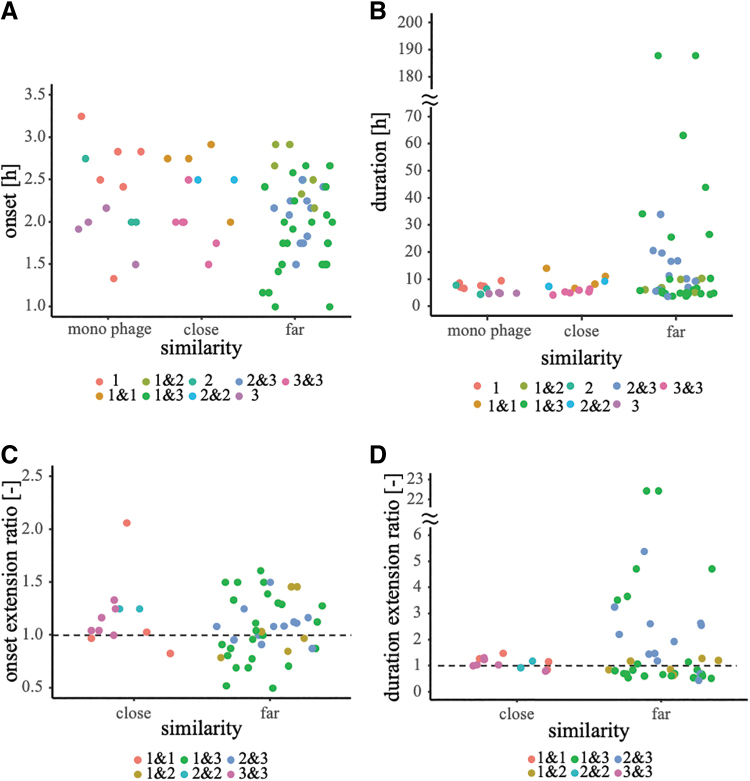
The lysis activities of 78 combinations were examined. Certain phage cocktails showed earlier or later lysis onset time than the single phage, and in some cases, a longer or shorter lysis duration than the original (Fig. 4A, B), Supplementary Table S5. Excluding the 24 cocktail pairs that did not show lysis, the distantly related cocktail pairs tended to show greater variation in both onset extension and duration extension ratios than the closely related pairs (Fig. 4C, D).
FIG. 4.
Lysis onset time or duration of cocktails with same or different cluster phages. Similarity on the horizontal axis indicates the similarity of the phages; “close” is a combination of phages in the same cluster, “far” is a combination of phages belonging to different clusters, and “monophage” is a phage that does not constitute the cocktail (as in Supplementary Table S5). The plots are color coded according to the combination of the phage clusters that make up the cocktail. (A) Perspective relationship and lysis onset time of phages constituting the cocktail. The vertical axis shows the lysis onset time (h). (B) Perspective relationship of phages in the cocktail and the lysis duration. The vertical axis indicates the lysis duration (h). (C) Onset extension ratio and perspective relationship of cocktail-constitutive phages. The onset extension ratio was calculated by dividing the lysis onset time of the cocktail by the earlier onset time of the phages constituting the cocktail. (D) Duration extension ratio and perspective relationship of cocktail-constitutive phages. The duration extension ratio was calculated by dividing the lysis duration time of the cocktail by the longer of the lysis durations of the phages constituting the cocktail. The duration of bacteriolysis was set to 0 h for those combinations in which bacteriolysis did not begin.
Phage genome
Whole genomes of the 13 phage strains were sequenced using shotgun sequencing. The complete genome of each phage was obtained (Supplementary Table S6). Differences in genome size were detected by GenomeMatcher for two pairs of phages with the same genome size (Supplementary Fig. S9).
Analysis of genome size and guanine and cytosine content (%) indicated that 9 of the 13 phages were distinct from each other, except for 2 pairs (ΦWec181 and ΦWec186, and ΦWec193 and ΦWec196) that shared the same genome size (Supplementary Table S6). Despite the genomes being the same size, there were regions of differences between the phage (Supplementary Fig. S9). For these pairs, Wec193 and ΦWec196 did not differ significantly in physiological characteristics. In contrast, ΦWec181 and ΦWec186 showed a 10-fold difference in yield.
Discussion
There have been limited studies on tailoring phage cocktails with high bacteriolytic efficacy (which are less likely to produce resistant bacteria) without phage receptor identification; therefore, this study proposed conditions for the preparation of high-efficiency phage cocktails. In this study, we isolated phages that lyse E. coli from colitis-induced mouse and classified these phages based on their physiological characteristics. We then evaluated the relationship between the cocktail lysis duration and the phage characteristics that composed the cocktail. The results suggest that phage clustering based on physiological characteristics is a prerequisite for extending the lysis duration of the phage cocktail.
Characterization of lytic phage cocktails
Greater than half of all combinations had an onset extension ratio of >1, which indicated a delay in the lysis onset time for more than half of the cocktails compared with the single phage (Fig. 4C; Supplementary Table S5; Supplementary Fig. S10). However, some cocktails had an onset extension ratio of <1, signifying a shorter lysis onset time than that of the single phage (Fig. 4C; Supplementary Table S5; Supplementary Fig. S10). In addition, some of the cocktails showed increased lysis durations (duration extension ratio >1) compared with the single phage, whereas others show decreased durations.
The delay in the lysis onset time for greater than half of the cocktails may relate to the reduction in the absolute amount of phage in the earlier onset time by half compared with that of the single phage. The shortening in the lysis onset time may be due to noncompetitive infection, proliferation, and lysis by phages in the cocktail. Bacterial resistance may have been effective for both the phages in the cocktail, thereby causing a shorter lysis duration, whereas longer lysis durations may indicate that bacteria resistant to one phage were susceptible to the other phage.
Variation in the extension ratios of the onset and duration of lysis tended to be more significant in the cocktails of phages with different clusters than in those with the same clusters. The cocktails of phages with different clusters showed no bias toward larger or smaller extension ratios, which suggests the presence of other phage physiological characteristics that contributed significantly to the performance of the phage cocktail, but were not measured in this study. However, as discussed above, cocktails composed of phages from different clusters contained more cocktails with smaller onset extension ratios or larger duration extension ratios compared with cocktails composed of the same cluster phages.
In other words, distantly related phages appear to differ from each other physiologically, and have different mechanisms of infection and lysis than closely related phages. Therefore, even if a bacterium acquires resistance to one phage, the other phage can multiply through a different pathway. This is supported by the fact that all 13 combinations with a lysis duration ratio >2, having increased persistence, were distantly related phage combinations.
It is also noteworthy that among the 13 pairs of this distantly related phage cocktails, the cocktail of ΦWec181 and ΦWec191, a combination of Clusters 1 and 3, had a lysis duration ratio >6, and the cocktail of Wec179 and ΦWec193, or that of ΦWec181 and ΦWec193, had a lysis duration ratio of >22 (Supplementary Table S5).
Conclusion
In this study, we isolated and characterized 13 phages lysing E. coli, followed by classifying them into three groups based on their physiological properties. Then, we assessed the lysis activity of 78 phage cocktails comprising two phage strains by analyzing lysis curves. As a result, we observed that the combinations involving distinct cluster phages had a greater tendency to exhibit altered lysis activity, particularly those with prolonged lysis durations, indicating a delay in the development of resistant bacteria.
The findings suggest that phages must be combined with different characteristics to obtain an effective cocktail that is less prone to the emergence of resistant bacteria. This information is essential for the preparation of phage cocktails to design phage therapy in the future.
Supplementary Material
Acknowledgments
We thank Yasunori Tanji and Kazuhiko Miyanaga for the helpful discussions. We thank Ken Kikuchi (Tokyo Women's Medical University, Tokyo, Japan) for providing the clinical isolates. We thank Editage for English language editing.
Authors' Contributions
T.K. contributed to conceptualization, data curation, formal analysis, writing—original draft, and investigation; O.T. and S.T. were involved in conceptualization, supervision, and writing—review and editing.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
This research was supported by a Waseda University Grant for Special Research Projects (Project number: 2022C-160).
Supplementary Material
References
- 1. Papanicolaou GA, Medeiros AA, Jacoby GA. Novel plasmid-mediated β-lactamase (MIR-1) conferring resistance to oxyimino- and α-methoxy β-lactams in clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother 1990;34(11):2200–2209; doi: 10.1128/AAC.34.11.2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Horii T, Arakawa Y, Ohta M, et al. Plasmid-mediated AmpC-type β-lactamase isolated from Klebsiella pneumoniae confers resistance to broad-spectrum β-lactams, including moxalactam. Antimicrob Agents Chemother 1993;37(5):984–990; doi: 10.1128/AAC.37.5.984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Doi Y, Shibata N, Shibayama K, et al. Characterization of a novel plasmid-mediated cephalosporinase (CMY-9) and its genetic environment in an Escherichia coli clinical isolate. Antimicrob Agents Chemother 2002;46(8):2427–2434; doi: 10.1128/AAC.46.8.2427-2434.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tzouvelekis LS, Markogiannakis A, Piperaki E, et al. Treating infections caused by carbapenemase-producing Enterobacteriaceae. Clin Microbiol Infect 2014;20(9):862–872; doi: 10.1111/1469-0691.12697 [DOI] [PubMed] [Google Scholar]
- 5. Olivia Lawe Davies. WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed; 2017. Available from: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed [Last accessed: September 2, 2022].
- 6. Gutiérrez-Gutiérrez B, Salamanca E, de Cueto M, et al. Effect of appropriate combination therapy on mortality of patients with bloodstream infections due to carbapenemase-producing Enterobacteriaceae (INCREMENT): A retrospective cohort study. Lancet Infect Dis 2017;17(7):726–734; doi: 10.1016/S1473-3099(17)30228-1 [DOI] [PubMed] [Google Scholar]
- 7. Tamma PD, Goodman KE, Harris AD, et al. Comparing the outcomes of patients with carbapenemase-producing and non-carbapenemase-producing carbapenem-resistant Enterobacteriaceae bacteremia. Clin Infect Dis 2017;64(3):257–264; doi: 10.1093/cid/ciw741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schooley RT, Biswas B, Gill JJ, et al. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob Agents Chemother 2017;61(10):e00954-17; doi: 10.1128/AAC.00954-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jault P, Leclerc T, Jennes S, et al. Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): A randomised, controlled, double-blind phase 1/2 trial. Lancet Infect Dis 2019;19(1):35–45; doi: 10.1016/S1473-3099(18)30482-1 [DOI] [PubMed] [Google Scholar]
- 10. Tanji Y, Shimada T, Yoichi M, et al. Toward rational control of Escherichia coli O157:H7 by a phage cocktail. Appl Microbiol Biotechnol 2004;64(2):270–274; doi: 10.1007/s00253-003-1438-9 [DOI] [PubMed] [Google Scholar]
- 11. Gu J, Liu X, Li Y, et al. A method for generation phage cocktail with great therapeutic potential. PLoS One 2012;7(3):e31698; doi: 10.1371/journal.pone.0031698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang Y, Shen W, Zhong Q, et al. Development of a bacteriophage cocktail to constrain the emergence of phage-resistant Pseudomonas aeruginosa. Front Microbiol 2020;11(March):1–12; doi: 10.3389/fmicb.2020.00327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tanji Y, Shimada T, Fukudomi H, et al. Therapeutic use of phage cocktail for controlling Escherichia coli O157:H7 in gastrointestinal tract of mice. J Biosci Bioeng 2005;100(3):280–287; doi: 10.1263/jbb.100.280 [DOI] [PubMed] [Google Scholar]
- 14. Bai J, Jeon B, Ryu S. Effective inhibition of Salmonella Typhimurium in fresh produce by a phage cocktail targeting multiple host receptors. Food Microbiol 2019;77(November 2017):52–60; doi: 10.1016/j.fm.2018.08.011 [DOI] [PubMed] [Google Scholar]
- 15. Sultana S, Reuteler G, Moine D, et al. Oral phage therapy of acute bacterial diarrhea with two coliphage preparations: A randomized trial in children from Bangladesh. EBioMedicine 2016;4:124–137; doi: 10.1016/j.ebiom.2015.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang Y, Barr JJ. Phage cocktail targeting STEC O157: H7 has comparable efficacy and superior recovery compared with enrofloxacin in an Enteric Murine Model. Microbiol Spectr 2022;10(3):e0023222; doi: 10.1128/spectrum.00232-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nakamura K, Fujiki J, Nakamura T, et al. Fluctuating bacteriophage-induced galU deficiency region is involved in trade-off effects on the phage and fluoroquinolone sensitivity in Pseudomonas aeruginosa. Virus Res 2021;306(July):198596; doi: 10.1016/j.virusres.2021.198596 [DOI] [PubMed] [Google Scholar]
- 18. Liang J, Li X, Zha T, et al. DTDP-rhamnosyl transferase RfbF, is a newfound receptor-related regulatory protein for phage phiYe-F10 specific for Yersinia enterocolitica serotype O:3. Sci Rep 2016;6(15):1–11; doi: 10.1038/srep22905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sausset R, Petit MA, Paepe M De. New insights into intestinal phages. Mucosal Immunol 2020;13:205–215; doi: 10.1038/s41385-019-0250-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hsu BB, Gibson TE, Yeliseyev V, et al. Dynamic modulation of the gut microbiota and metabolome by bacteriophages in a Mouse Model. Cell Host Microbe 2019;25(6):803–814.e5; doi: 10.1016/j.chom.2019.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Barrangou R, Moineau S, Romero DA, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science (80-) 2007;315(March):1709–1712. [DOI] [PubMed] [Google Scholar]
- 22. Lupp C, Robertson ML, Wickham ME, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2007;2(2):119–129; doi: 10.1016/j.chom.2007.06.010 [DOI] [PubMed] [Google Scholar]
- 23. Zhilu X, Xiangqian D, Keli Y, et al. Association of adherent-invasive Escherichia coli with severe gut mucosal dysbiosis in Hong Kong Chinese population with Crohn's disease. Gut Microbes 2021;13(1):1994833; doi: 10.1080/19490976.2021.1994833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Osaka T, Moriyama E, Arai S, et al. Meta-analysis of fecal microbiota and metabolites in experimental colitic mice during the inflammatory and healing phases. Nutrients 2017;9(12):1–13; doi: 10.3390/nu9121329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Håkansson, Tormo-Badia N, Baridi A, et al. Immunological alteration and changes of gut microbiota after dextran sulfate sodium (DSS) administration in mice. Clin Exp Med 2015;15(1):107–120; doi: 10.1007/s10238-013-0270-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rath HC, Schultz M, Freitag R, et al. Different subsets of enteric bacteria induce and perpetuate experimental colitis in rats and mice. Infect Immun 2001;69(4):2277–2285; doi: 10.1128/IAI.69.4.2277-2285.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mattila S, Ruotsalainen P, Jalasvuori M. On-demand isolation of bacteriophages against drug- resistant bacteria for personalized phage therapy bacteria strains and culturing. Front Microbiol 2015;6(November):1–7; doi: 10.3389/fmicb.2015.01271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shao Y, Wang IN. Bacteriophage adsorption rate and optimal lysis time. Genetics 2008;180(1):471–482; doi: 10.1534/genetics.108.090100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ward JH. Hierarchical grouping to optimize an objective function. J Am Stat Assoc 1963;58(301):236–244; doi: 10.1080/01621459.1963.10500845 [DOI] [Google Scholar]
- 30. Chen Y, Chen Y, Shi C, et al. SOAPnuke: A MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience 2018;7(1):1–6; doi: 10.1093/gigascience/gix120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 2008;18(5):821–829; doi: 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tanizawa Y, Fujisawa T, Kaminuma E, et al. DFAST and DAGA: Web-based integrated genome annotation tools and resources. Biosci Microbiota Food Health 2016;35(4):173–184; doi: 10.12938/bmfh.16-003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tanizawa Y, Fujisawa T, Nakamura Y. DFAST: A flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics 2018;34(6):1037–1039; doi: 10.1093/bioinformatics/btx713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ohtsubo Y, Ikeda-Ohtsubo W, Nagata Y, et al. GenomeMatcher: A graphical user interface for DNA sequence comparison. BMC Bioinformatics 2008;9(vi):1–9; doi: 10.1186/1471-2105-9-376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria; 2017. [Google Scholar]
- 36. Maechler M, Rousseeuw P, Struyf A, et al. Cluster: Cluster Analysis Basics and Extensions. Comprehensive R Archive Network (CRAN); 2019. Available from: https://cran.r-project.org/package=cluster [Last accessed: September 2, 2022].
- 37. Wickham H, Averick M, Bryan J, et al. Welcome to the Tidyverse. J Open Source Softw 2019;4(43):1686; doi: 10.21105/JOSS.01686 [DOI] [Google Scholar]
- 38. Barret Schloerke, Di Cook, Joseph Larmarange, et al. GGally: Extension to “Ggplot2.” Comprehensive R Archive Network (CRAN); 2021. Available from: https://cran.r-project.org/package=GGally [Last accessed: September 2, 2022].
- 39. Vincent QV. Ggbiplot: A Ggplot2 Based Biplot; 2011. Available from: https://github.com/vqv/ggbiplot [Last accessed: September 2, 2022].
- 40. Wickham H. Ggplot2: Elegant Graphics for Data Analysis; 2016. Available from: https://ggplot2.tidyverse.org/ [Last accessed: September 2, 2022].
- 41. Slowikowski K. Ggrepel: Automatically Position Non-Overlapping Text Labels with “Ggplot2.” Comprehensive R Archive Network (CRAN); 2021. Available from: https://cran.r-project.org/package=ggrepel [Last accessed: September 2, 2022].
- 42. Alboukadel K, Fabian M. Extract and Visualize the Results of Multivariate Data Analyses [R Package Factoextra Version 1.0.7]. Comprehensive R Archive Network (CRAN); 2020. Available from: https://cran.r-project.org/package=factoextra [Last accessed: September 2, 2022].
- 43. Schauberger P, Walker A. Openxlsx: Read, Write and Edit Xlsx Files. Comprehensive R Archive Network (CRAN); 2021. Available from: https://cran.r-project.org/package=openxlsx [Last accessed: September 2, 2022].
- 44. Wickham H. The split-apply-combine strategy for data analysis. J Stat Softw 2011;40(1):1–29; doi: 10.18637/jss.v040.i01. [DOI] [Google Scholar]
- 45. Wickham H. Reshaping data with the reshape Package. J Stat Softw 2007;21(12):1–20; doi: 10.18637/JSS.V021.I12 [DOI] [Google Scholar]
- 46. Bache MS, Wickham H. Magrittr: A Forward-Pipe Operator for R. R Package Version 2.0.1. Comprehensive R Archive Network (CRAN); 2022. Available from: https://cran.r-project.org/package=magrittr [Last accessed: September 2, 2022].
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The phage genomes were deposited in the DNA Data Bank of Japan with the following accession numbers: ΦWec172 (LC739530), ΦWec174 (LC739531), ΦWec177 (LC739532), ΦWec179 (LC739533), ΦWec181 (LC739534), ΦWec186 (LC739535), ΦWec187 (LC739536), ΦWec188 (LC739537), ΦWec189 (LC739538), ΦWec190 (LC739539), ΦWec191 (LC739540), ΦWec193 (LC739541), and ΦWec196 (LC739542).