Abstract
In the course of clinical studies with the investigational streptogramin antimicrobial dalfopristin-quinupristin, isolates of vancomycin-resistant Enterococcus faecium were referred to our laboratory from across the United States. Seventy-two percent of the strains were of the VanA type, phenotypically and genotypically, while 28% were of the VanB type. High-level resistance to streptomycin or gentamicin was observed in 86 and 81%, respectively, of the VanA strains but in only 69 and 66%, respectively, of the VanB strains. These enterococci were resistant to ampicillin (MIC for 50% of the isolates tested [MIC50] and MIC90, 128 and 256 μg/ml, respectively) and to the other approved agents tested, with the exception of chloramphenicol (MIC90, 8 μg/ml) and novobiocin (MIC90, 1 μg/ml). Considering all of the isolates submitted, dalfopristin-quinupristin inhibited 86.4% of them at concentrations of ≤1 μg/ml and 95.1% of them at ≤2 μg/ml. However, for the data set comprised of only the first isolate submitted for each patient, 94.3% of the strains were inhibited at concentrations of ≤1 μg/ml and 98.9% were inhibited at concentrations of ≤2 μg/ml. Multiple drug resistance was very common among these isolates of vancomycin-resistant E. faecium, while dalfopristin-quinupristin inhibited the majority at concentrations that are likely to be clinically relevant.
The emergence of multiply antibiotic-resistant strains of Enterococcus faecium as increasingly common nosocomial pathogens has created a formidable challenge for both clinicians and hospital infection control officers (4, 12). Glycopeptide resistance has been especially noteworthy in this enterococcal species, in which resistance to penicillins had increased dramatically in recent years (18). Strains of E. faecium for which the MICs of ampicillin or penicillin are in excess of 100 μg/ml are now common (3, 34, 36). Data from the Centers for Disease Control and Prevention (7) or collected through a national surveillance program (23) point to a preponderance of the VanA phenotype among vancomycin-resistant strains of E. faecium and to high rates of resistance to other agents. Therapeutic regimens against infections caused by such multidrug-resistant strains have employed agents such as doxcycline or chloramphenicol (20, 26, 29) or unusual combinations like novobiocin-ciprofloxacin (24). Nevertheless, the mortality of patients infected with multiply antibiotic-resistant E. faecium remains high (11, 25, 33).
The streptogramin antimicrobial dalfopristin-quinupristin, designated RP 59500, has demonstrated activity in vitro against the majority of glycopeptide-resistant strains of E. faecium (2, 8, 15). As a result, this agent has been examined in the treatment of infections due to vancomycin-resistant E. faecium in humans. In the course of these investigations, enterococcal isolates were submitted to our laboratory for characterization and susceptibility testing. The present paper describes the glycopeptide resistance patterns and antimicrobial susceptibility profiles of these isolates of E. faecium from hospitals across the United States.
MATERIALS AND METHODS
Organisms.
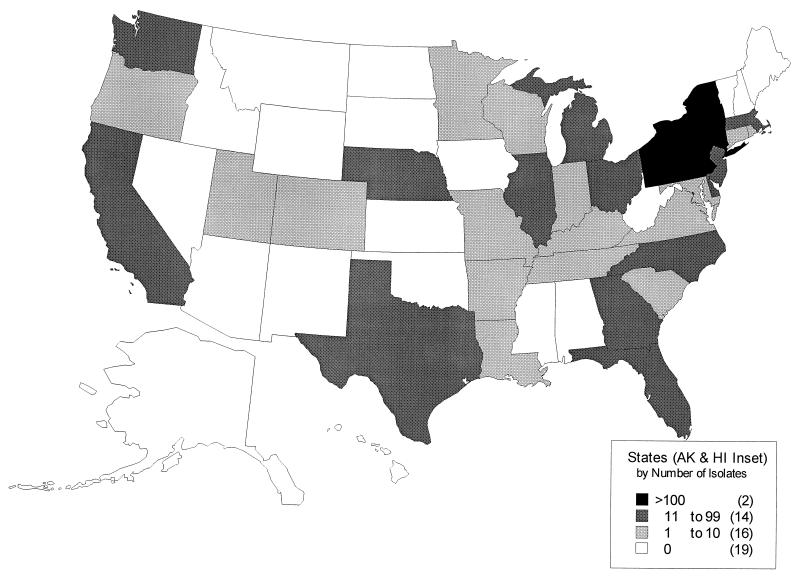
Enterococcal isolates were submitted by investigators in the United States (Fig. 1) who planned to use RP 59500 during 1994 to 1996 for treatment of E. faecium infections under an emergency use program of Rhône-Poulenc Rorer Pharmaceuticals, Inc., Collegeville, Pa. We did not attempt to distinguish actual pathogens from colonizing strains. Although other gram-positive bacteria were encountered among the specimens submitted, only strains identified in our laboratory as E. faecium and shown to be resistant to vancomycin were included in the present study.
FIG. 1.
Sources of vancomycin-resistant E. faecium isolates by region of the United States. AK, Alaska; HI, Hawaii.
Identification.
Bacterial colonies that had the morphological appearance of enterococci on horse blood agar plates were identified by biochemical properties (API 20 Strep system; bioMérieux Vitek, Inc., Hazelwood, Mo.), by an aac6′-Ii gene probe specific for E. faecium (9, 10), or by both. Pigment production was tested on cotton-tipped applicators. Motility was assessed in motility agar tubes (Motility B Medium; Remel, Lenexa, Kans.) which were incubated at 30°C for up to 72 h. Isolates were tested for β-lactamase production by spotting growth from plates onto nitrocefin disks (BBL, Becton Dickinson Microbiology Systems, Cockeysville, Md.) which were examined for a color change for up to 30 min. E. faecalis CH-19 was used as a positive control (31).
Dilution susceptibility studies.
Susceptibility to the antimicrobial dalfopristin-quinupristin (70:30) and comparative antibiotics was determined by the agar dilution method (27). Bacteria were suspended to a density of ca. 107 CFU/ml in Mueller-Hinton broth and applied to the surface of Mueller-Hinton II agar (BBL) plates with an inoculator yielding approximately 104 CFU per spot. Plates were incubated in room air at 35°C for 18 to 20 h. Resistance to streptomycin at 2,000 μg/ml, gentamicin at 500 μg/ml, and vancomycin at 6 μg/ml was tested on Synergy Quad plates (Remel) in accordance with the manufacturer’s recommendations. Plates were read at 24 and 48 h of incubation at 35°C.
Disk testing.
Inhibition zones around 15-μg dalfopristin-quinupristin disks were determined by National Committee for Clinical Laboratory Standards methods against selected control strains (28). Plates containing 25 ml of Mueller-Hinton II agar were inoculated over the entire surface with a cotton-tipped applicator moistened in a saline suspension of the test organism at a density of ca. 108 CFU/ml (0.5 McFarland standard). The disks were applied, and the plates were incubated overnight at 35°C.
Antimicrobials.
Dalfopristin-quinupristin (RP 59500) susceptibility test powder and disks were provided by Rhône-Poulenc Rorer Pharmaceuticals, Inc. Teicoplanin and ciprofloxacin susceptibility test powders were, respectively, the generous gifts of Hoechst Marion Roussel Research Institute; Hoechst Marion Roussel, Inc., Cincinnati, Ohio, and Bayer Corp., West Haven, Conn. Other agents were purchased from Sigma Chemical Co., St. Louis, Mo.
DNA hybridization methods.
Bacteria were lifted from agar plates inoculated as growth controls in susceptibility tests by using dry nylon transfer membranes (MSI, Westboro, Mass.). Colony lysis was accomplished by using a technique adapted from Sambrook et al. (32). Filters were placed on filter paper saturated with the following solutions and incubated at room temperature unless otherwise stated: TE (10 mM Tris [pH 8], 1 mM EDTA) with lysozyme at 5 mg/ml, 30 min at 37°C; 10% sodium dodecyl sulfate, 3 min; 0.5 N sodium hydroxide with 1.5 M sodium chloride, 5 min; 1.5 M sodium chloride with 1 M Tris (pH 8), 5 min; 2× SSC (20× SSC is 3 M sodium chloride plus 0.3 M sodium citrate, pH 7), 5 min. Bacterial DNA was bound to the filter by UV cross-linking. A 489-bp intragenic fragment of aac6′-Ii was amplified by PCR of genomic DNA from E. faecium ATCC 19434 by using primers 5′-GAT TTA CTG AGA CTG ACT TGG-3′ and 5′-GAG AAT CTG GTC GAG GAA TAA-3′, which correspond to bp 223 to 243 and 692 to 712, respectively, of the sequence published by Costa et al. (10). The PCR product was cloned into pCRII (Invitrogen, San Diego, Calif.). Probes for vanA and vanB2 were prepared by PCR amplification of ca. 630-bp intragenic fragments from E. faecium 228 (19) and E. faecalis SF300, respectively, by using primers described previously (17). The fragments were cloned into pCR II and pBluescript II (Stratagene, La Jolla, Calif.), respectively. All probe fragments were restricted with EcoRI, purified, and labeled with digoxigenin-dUTP by using a kit from Boehringer Mannheim Corp., Indianapolis, Ind. Hybridization was performed under stringent conditions.
RESULTS
Organisms.
A total of 875 isolates were identified as vancomycin-resistant E. faecium and included in this study. Although this data set contained multiple isolates collected at different times or from separate culture sites for many patients, analysis of this entire collection served to detect potentially infrequent events. All of the strains grew on screening plates containing vancomycin at 6 μg/ml. The aac6′-Ii gene was detected in all of the 829 strains examined. No isolate (0 of 875) produced pigment or β-lactamase. None of the 194 isolates tested for motility was motile. Three hundred seven isolates (35%) were recovered from blood or catheter tip cultures, 51 were from urine (5.8%), 185 (21%) were from stool samples or rectal swabs, and the remainder were from various other sites.
To minimize the bias of multiple isolates submitted per patient, we also examined the subgroup of 423 isolates which remained after likely duplicate strains were excluded. Specifically, for any patient, strains were considered different if the MICs of vancomycin, teicoplanin, or dalfopristin-quinupristin differed by fourfold or more; otherwise, they were considered to be possible duplicates. Finally, we considered a third data set of 352 strains, which represented the first isolate submitted for each patient.
Glycopeptide resistance profiles.
From the entire collection, 631 isolates (72%) were classified as having the VanA phenotype on the basis of resistance to both vancomycin (MICs, 64 to >512 μg/ml) and teicoplanin (MICs, 8 to >256 μg/ml). The remaining 244 strains (28%) were classified as VanB. For these, the MICs of vancomycin ranged from 16 to >512 μg/ml and those of teicoplanin ranged from 0.25 to 2 μg/ml. From the restricted collection of 423 strains, 70% were VanA and 30% were VanB.
The genotypic glycopeptide resistance profiles of 797 isolates were evaluated with vanA and vanB gene probes (78 strains were not tested). Of these, 573 (72%) were vanA and 224 (28%) were vanB. Genotyping of 392 of the 423 isolates in the second data set gave similar results: 70% vanA and 30% vanB. Results based on the first-isolate data set were 73% vanA and 27% vanB. Discordance between the phenotype and genotype was encountered with only three strains. Each was genotypically vanB, but phenotypically VanA (MICs of teicoplanin, ≥64 μg/ml). The strains were recovered in Illinois, Pennsylvania, and New York.
Antimicrobial susceptibility.
Susceptibility results for the entire collection, for the 423-strain subgroup, and for the first-isolate subgroup are shown in Tables 1, 2, and 3, respectively. Results for the three groups were virtually identical. With the exception of chloramphenicol and novobiocin, resistance to other agents was common. Only two strains (one VanA and one VanB) were inhibited by ampicillin at concentrations below 32 μg/ml. There was little difference in susceptibility to the agents shown in Table 1 between the VanA and VanB isolates. Aside from teicoplanin (which was the basis for the grouping of the isolates), only rifampin appeared to be more active against the VanB strains.
TABLE 1.
Activity of dalfopristin-quinupristin against vanomycin-resistant E. faeciuma
| Strains and antimicrobial | No. of strains | MIC50 | MIC90 | MIC range |
|---|---|---|---|---|
| All isolates (875 strains) | ||||
| RP 59500 | 875 | 1 | 2 | 0.25–32 |
| Vancomycin | 875 | 512 | 512 | 16–>512 |
| Teicoplanin | 875 | 32 | 64 | 0.12–>256 |
| Ampicillin | 875 | 128 | 256 | 4–>256 |
| Doxycycline | 868 | 8 | 32 | 0.12–>64 |
| Rifampin | 868 | 8 | >128 | ≤0.06–>128 |
| Chloramphenicol | 868 | 8 | 8 | 4.0–64 |
| Novobiocin | 868 | 0.5 | 1 | 0.25–16 |
| Erythromycin | 871 | >128 | >128 | 1–>128 |
| Ciprofloxacin | 813 | >128 | >128 | 1–>128 |
| VanA+ strains | ||||
| RP 59500 | 631 | 1 | 2 | 0.25–32 |
| Vancomycin | 631 | 512 | 512 | 64–>512 |
| Teicoplanin | 631 | 64 | 128 | 8–>256 |
| Ampicillin | 631 | 128 | 256 | 8–>256 |
| Doxycycline | 625 | 4 | 32 | 0.12–64 |
| Rifampin | 625 | 8 | >128 | ≤0.06–>128 |
| Chloramphenicol | 625 | 8 | 8 | 4.0–32 |
| Novobiocin | 625 | 0.5 | 1 | 0.25–16 |
| Erythromycin | 627 | >128 | >128 | 1–>128 |
| Ciprofloxacin | 581 | >128 | >128 | 1–>128 |
| VanB+ strains | ||||
| RP59500 | 244 | 1 | 2 | 0.25–16 |
| Vancomycin | 244 | 256 | 512 | 16–>512 |
| Teicoplanin | 244 | 0.5 | 1 | 0.25–2 |
| Ampicillin | 244 | 128 | 256 | 4–>256 |
| Doxycycline | 243 | 16 | 32 | 0.12–>64 |
| Rifampin | 243 | ≤0.06 | 16 | ≤0.06–>128 |
| Chloramphenicol | 243 | 8 | 16 | 4.0–64 |
| Novobiocin | 243 | 0.5 | 1 | 0.25–4 |
| Erythromycin | 244 | >128 | >128 | 1–>128 |
| Ciprofloxacin | 232 | >128 | >128 | 4–>128 |
MICs are reported in micrograms per milliliter. MIC50 and MIC90, MICs for 50 and 90% of the isolates tested, respectively.
TABLE 2.
In vitro susceptibility of 423 strains of E. faecium selected to minimize inclusion of duplicate isolatesa
| Antimicrobial | No. of strains | MIC50 | MIC90 | MIC range |
|---|---|---|---|---|
| RP 59500 | 423 | 1 | 2 | 0.25–32 |
| Vancomycin | 423 | 512 | 512 | 16–>512 |
| Teicoplanin | 423 | 32 | 128 | 0.25–>256 |
| Ampicillin | 423 | 128 | 256 | 4–>256 |
| Doxycycline | 423 | 8 | 32 | 0.12–64 |
| Rifampin | 423 | 8 | >128 | ≤0.06–>128 |
| Chloramphenicol | 423 | 8 | 8 | 4.0–64 |
| Novobiocin | 423 | 0.5 | 1 | 0.25–16 |
| Erythromycin | 423 | >128 | >128 | 1–>128 |
| Ciprofloxacin | 381 | >128 | >128 | 1–>128 |
See Table 1, footnote a.
TABLE 3.
In vitro susceptibilities of 352 first-isolate E. faecium strainsa
| Antimicrobial | No. of strains | MIC50 | MIC90 | MIC range |
|---|---|---|---|---|
| RP 59500 | 352 | 1 | 1 | 0.25–8 |
| Vancomycin | 352 | 512 | 512 | 16–>512 |
| Teicoplanin | 352 | 32 | 128 | 0.25–>256 |
| Ampicillin | 352 | 128 | 256 | 32–>256 |
| Doxycycline | 349 | 8 | 32 | 0.12–64 |
| Rifampin | 349 | 8 | >128 | ≤0.06–>128 |
| Chloramphenicol | 349 | 8 | 8 | 4–64 |
| Novobiocin | 349 | 0.5 | 1 | 0.5–1 |
| Erythromycin | 351 | >128 | >128 | 1–>128 |
| Ciprofloxacin | 298 | >128 | >128 | 1–>128 |
See Table 1, footnote a.
Considering only the collection of first isolates, dalfopristin-quinupristin inhibited 98.9% of the strains at ≤2 μg/ml and 94.3% of them at ≤1 μg/ml. Analysis of the duplicate-minimized 423-strain data set revealed that 92.2 and 84.9% of the strains were susceptible to these concentrations, respectively, consistent with the recovery of strains with increased resistance to the agent in subsequent cultures from some patients. For the collection as a whole, dalfopristin-quinupristin inhibited 832 (95.1%) of the 875 isolates at ≤2 μg/ml and 756 strains (86.4%) at ≤1 μg/ml. Strains for which the dalfopristin-quinupristin MICs were ≥4 μg/ml were all verified to be aac6′-Ii positive. These were recovered from patients treated with this agent and were found more frequently among enteric cultures (16 [8.7%] of 185) than among blood and catheter tip cultures (11 [3.6%] of 307) or urine cultures (2 [3.9%] of 51). Comparison of the enteric cultures with those from these other sites showed that the difference in resistance to the streptogramin (8.7 versus 3.9%) was statistically significant (χ2 = 6.99, P = 0.008).
High-level resistance to gentamicin was common, whether based on the entire collection (604 [77%] of 784 isolates tested), on the duplicate-minimized data (298 [77%] of 387 isolates), or on the first-isolate data set (239 [77%] of 311 isolates). High-level resistance to streptomycin was seen in 80, 82, and 82% of the strains in these three groups, respectively. High-level aminoglycoside resistance was significantly more common among VanA isolates. High-level resistance to gentamicin was found in 463 of 571 VanA and 141 of 213 VanB strains (81 versus 66%; χ2 = 19.44, P = 10−5), and high-level resistance to streptomycin was noted in 86% of VanA and 69% of VanB isolates (χ2 = 29.89, P < 10−7).
Activity of dalfopristin-quinupristin against control stains.
Shown in Table 4 are the activities of dalfopristin-quinupristin against American Type Culture Collection control strains. Results were consistent over multiple runs extending over several months.
TABLE 4.
Activity of dalfopristin-quinupristin against control strains
| Strain | MIC (μg/ml) observeda | Inhibition zone size (mm) observed |
|---|---|---|
| Enterococcus faecium ATCC 19434 | 0.543 | 241, 251, 262 |
| Enterococcus faecalis ATCC 29212 | 420, 823 | 124, 135, 151 |
| Enterococcus faecalis ATCC 51299 | 1626, 3216 | 81, 91 |
| Staphylococcus aureus ATCC 29213 | 0.534, 1.09 | NTb |
| Staphylococcus aureus ATCC 25923 | NT | 239, 2411 |
Each subscript indicates the number of runs for which the MIC was observed.
NT, not tested.
DISCUSSION
The large number of vancomycin-resistant E. faecium isolates collected in the course of these clinical studies provided an opportunity to examine the glycopeptide resistance profiles and general antibiotic susceptibility patterns of isolates prevailing across the United States during 1994 to 1996. Vancomycin-resistant E. faecium isolates were widespread geographically. The proportion of VanA (72%) to VanB (28%) isolates was slightly lower than, but in general agreement with, that reported for E. faecium isolates submitted to the Centers for Disease Control and Prevention from 1988 to 1992 (83% VanA) or described in a 1992 survey of 97 U.S. laboratories (79% VanA) (7, 23).
When resistance phenotypes were assigned, strains for which the teicoplanin MIC was 8 μg/ml were considered resistant and classified VanA, despite the fact that MICs of ≤8 μg/ml fall into the susceptible range based on National Committee for Clinical Laboratory Standards breakpoints (27). Our approach seemed appropriate for E. faecium because of the bimodal distribution of teicoplanin MICs (0.25 to 2 μg/ml or ≥8 μg/ml). Furthermore, for glycopeptide-susceptible E. faecium isolates collected before 1989, teicoplanin MICs ranged from 0.25 to 2 μg/ml as well (18). Finally, all three isolates for which the teicoplanin MICs were = 8 μg/ml in our study were genotypically vanA. The only discordance between the phenotypic and genotypic classifications occurred with three phenotype VanA (teicoplanin MICs ≥64 μg/ml), genotype vanB strains, almost certainly representing teicoplanin-resistant mutants of vanB isolates, as have been previously described (21).
High levels of ampicillin or penicillin are required to inhibit many contemporary isolates of E. faecium, and this was confirmed here (3, 36). In addition, we had previously noted that 61% of the vancomycin-susceptible isolates of E. faecium collected in Boston during 1989 and 1990 exhibited high-level resistance to gentamicin, a remarkable fact given that the first such strain was detected here in 1986 (13, 18). The rate of high-level gentamicin resistance in the current collection of VanB isolates (66%) was similar to that for the glycopeptide-susceptible isolates just described, but the frequency of resistance among the VanA strains was significantly higher at 81%. Although we have not studied the genetics of resistance in these isolates, linkage between the vanA gene and gentamicin resistance genes on single plasmids has been described in other enterococcal species (5, 35). This observation might offer the most plausible explanation for the higher rate of gentamicin resistance among VanA E. faecium strains.
This project confirmed the results of previous studies demonstrating activity of dalfopristin-quinupristin against the great majority of glycopeptide-resistant E. faecium isolates (16, 22). Virtually all (98.9%) of the first isolates recovered from the patients were inhibited by the agent at ≤2 μg/ml. Even from the complete 875-strain collection, which included isolates collected after the start of treatment, the streptogramin inhibited 95% at ≤2 μg/ml and 86% at ≤1 μg/ml. Provisional breakpoints of ≤1 μg/ml for the susceptible and 2 μg/ml for the intermediately susceptible designations have been proposed for this antimicrobial (1). Of the 43 resistant isolates encountered, all of which were confirmed to be E. faecium by gene probe, 35 were inhibited at 4 μg/ml, a concentration which may still be transiently achievable in serum (14).
Plasmid-mediated resistance to the streptogramin has been described (30) but appears to be rare in the United States. However, Chow et al. (6) have reported the development of decreased susceptibility to the drug in a strain of E. faecium following therapy. The MIC for a pretreatment bloodstream isolate was 0.25 μg/ml, while that for a clonally indistinguishable blood isolate recovered after initially successful treatment was 2 μg/ml. While probably uncommon, minor increases in the MIC for sequential isolates may not be rare. Of the patients whose isolates were studied here, 31 of 33 individuals from whom organisms for which the dalfopristin-quinupristin MICs were >2 μg/ml were recovered also, at some point, harbored at least one strain for which the MIC was ≤2 μg/ml. The clinical significance of possible small, stepwise increases in resistance is unknown and must await correlation of microbiologic data with treatment outcomes in these investigations.
ACKNOWLEDGMENT
This study was supported by Rhône-Poulenc Rorer Pharmaceuticals, Inc.
REFERENCES
- 1.Barry, A. L., P. C. Fuchs, and S. D. Brown. 1997. Provisional interpretive criteria for quinupristin/dalfopristin susceptibility tests. J. Antimicrob. Chemother. 39(Suppl. A):87–92. [DOI] [PubMed]
- 2.Bonilla H F, Perri M B, Kauffman C A, Zervos M J. Comparative in vitro activity of quinupristin/dalfopristin against multidrug resistant Enterococcus faecium. Diagn Microbiol Infect Dis. 1996;25:127–131. doi: 10.1016/s0732-8893(96)00123-x. [DOI] [PubMed] [Google Scholar]
- 3.Bush L M, Calmon J, Cherney C L, Wendeler M, Pitsakis P, Poupard J, Levison M E, Johnson C C. High-level penicillin resistance among isolates of enterococci. Implications for treatment of enterococcal infections. Ann Intern Med. 1989;110:515–520. doi: 10.7326/0003-4819-110-7-515. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Nosocomial enterococci resistant to vancomycin—United States, 1989–1993. Morbid Mortal Weekly Rep. 1993;42:597–599. [PubMed] [Google Scholar]
- 5.Cercenado E, Ünal S, Eliopoulos C T, Rubin L G, Isenberg H D, Moellering R C, Jr, Eliopoulos G M. Characterization of vancomycin-resistance in Enterococcus durans. J Antimicrob Chemother. 1995;36:821–825. doi: 10.1093/jac/36.5.821. [DOI] [PubMed] [Google Scholar]
- 6.Chow J W, Donabedian S M, Zervos M J. Emergence of increased resistance to quinupristin/dalfopristin during therapy for Enterococcus faecium bacteremia. Clin Infect Dis. 1997;24:90–91. doi: 10.1093/clinids/24.1.90. [DOI] [PubMed] [Google Scholar]
- 7.Clark N C, Cooksey R C, Hill B C, Swenson J M, Tenover F C. Characterization of glycopeptide-resistant enterococci from U.S. hospitals. Antimicrob Agents Chemother. 1993;37:2311–2317. doi: 10.1128/aac.37.11.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins L A, Malanoski G J, Eliopoulos G M, Wennersten C B, Ferraro M J, Moellering R C., Jr In vitro activity of RP59500, an injectable streptogramin antibiotic, against vancomycin-resistant gram-positive organisms. Antimicrob Agents Chemother. 1993;37:598–601. doi: 10.1128/aac.37.3.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coque T M, Murray B E. Identification of Enterococcus faecalis strains by DNA hybridization and pulsed-field gel electrophoresis. J Clin Microbiol. 1995;33:3368–3369. doi: 10.1128/jcm.33.12.3368-3369.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costa Y, Galimand M, Leclercq R, Duval J, Courvalin P. Characterization of the chromosomal aac(6′)-Ii gene specific for Enterococcus faecium. Antimicrob Agents Chemother. 1993;37:1896–1903. doi: 10.1128/aac.37.9.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edmond M B, Ober J F, Dawson J D, Weinbaum D L, Wenzel R P. Vancomycin-resistant enterococcal bacteremia: natural history and attributable mortality. Clin Infect Dis. 1996;23:1234–1239. doi: 10.1093/clinids/23.6.1234. [DOI] [PubMed] [Google Scholar]
- 12.Eliopoulos G M. Antibiotic resistance in Enterococcus species: an update. Curr Clin Top Infect Dis. 1996;16:21–51. [PubMed] [Google Scholar]
- 13.Eliopoulos G M, Wennersten C, Zighelboim-Daum S, Reiszner E, Goldmann D, Moellering R C., Jr High-level resistance to gentamicin in clinical isolates of Streptococcus (Enterococcus) faecium. Antimicrob Agents Chemother. 1988;32:1528–1532. doi: 10.1128/aac.32.10.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Etienne, S. D., G. Montay, A. Le Liboux, A. Frydman, and J. J. Garaud. 1992. A phase I, double-blind, placebo-controlled study of the tolerability and pharmacokinetic behavior of RP 59500. J. Antimicrob. Chemother. 30:(Suppl. A):123–131. [DOI] [PubMed]
- 15.Fass R J. In vitro activity of RP 59500, a semisynthetic injectable pristinamycin, against staphylococci, streptococci, and enterococci. Antimicrob Agents Chemother. 1991;35:553–559. doi: 10.1128/aac.35.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freeman C, Robinson A, Cooper B, Mazens-Sullivan M, Quintiliani R, Nightingale C. In vitro antimicrobial susceptibility of glycopeptide-resistant enterococci. Diagn Microbiol Infect Dis. 1995;21:47–50. doi: 10.1016/0732-8893(94)00113-b. [DOI] [PubMed] [Google Scholar]
- 17.Gold H S, Ünal S, Cercenado E, Thauvin-Eliopoulos C, Eliopoulos G M, Wennersten C B, Moellering R C., Jr A gene conferring resistance to vancomycin but not teicoplanin in isolates of Enterococcus faecalis and Enterococcus faecium demonstrates homology with vanB, vanA, and vanC genes of enterococci. Antimicrob Agents Chemother. 1993;37:1604–1609. doi: 10.1128/aac.37.8.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grayson M L, Eliopoulos G M, Wennersten C B, Ruoff K L, De Girolami P C, Ferraro M J, Moellering R C., Jr Increasing resistance to β-lactam antibiotics among clinical isolates of Enterococcus faecium: a 22-year review at one institution. Antimicrob Agents Chemother. 1991;35:2180–2184. doi: 10.1128/aac.35.11.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Handwerger S, Pucci M J, Kolokathis A. Vancomycin resistance is encoded on a pheromone response plasmid in Enterococcus faecium 228. Antimicrob Agents Chemother. 1990;34:358–360. doi: 10.1128/aac.34.2.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hardalo C, Rigsby M, Chen L, Dykewicz C, Elmore J, Nagy-Agren S, Wetherill P, Edberg S, Hierholzer W. Program and abstracts of the Infectious Diseases Society of America Annual Meeting. 1993. Chloramphenicol treatment of resistant Enterococcus faecium infections, abstr. 110. [Google Scholar]
- 21.Hayden M K, Trenholme G M, Schultz J E, Sahm D F. In vivo development of teicoplanin resistance in a VanB Enterococcus faecium isolate. J Infect Dis. 1993;167:1224–1227. doi: 10.1093/infdis/167.5.1224. [DOI] [PubMed] [Google Scholar]
- 22.Johnson C C, Slavoski L, Schwartz M, May P, Pitsakis P G, Shur A L, Levison M E. In vitro activity of RP 59500 (quinupristin/dalfopristin) against antibiotic-resistant strains of Streptococcus pneumoniae and enterococci. Diagn Microbiol Infect Dis. 1995;21:169–173. doi: 10.1016/0732-8893(95)00068-l. [DOI] [PubMed] [Google Scholar]
- 23.Jones R N, Sader H S, Erwin M E, Anderson S A the Enterococcus Study Group. Emerging multiply resistant enterococci among clinical isolates. I. Prevalence data from 97 medical center surveillance study in the United States. Diagn Microbiol Infect Dis. 1995;21:85–93. doi: 10.1016/0732-8893(94)00147-o. [DOI] [PubMed] [Google Scholar]
- 24.Linden P, Pasculle A W, Manez R, Silverman D, Kramer D J, Kusne S. Program and abstracts of the 33rd Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1993. Utilization of novobiocin and ciprofloxacin for the treatment of serious infection due to vancomycin resistant Enterococcus faecium, abstr. 1027; p. 307. [Google Scholar]
- 25.Linden P K, Pasculle A W, Manez R, Kramer D J, Fung J J, Pinna A D, Kusne S. Differences in outcomes for patients with bacteremia due to vancomycin-resistant Enterococcus faecium or vancomycin-susceptible E. faecium. Clin Infect Dis. 1996;22:663–670. doi: 10.1093/clinids/22.4.663. [DOI] [PubMed] [Google Scholar]
- 26.Montecalvo M A, Horowitz H, Wormser G P, Seiter K, Carbonaro C A. Effect of novobiocin-containing antimicrobial regimens on infection and colonization with vancomycin-resistant Enterococcus faecium. Antimicrob Agents Chemother. 1995;39:794. doi: 10.1128/AAC.39.3.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically–third edition; approved standard. NCCLS document M7-A3. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1993. [Google Scholar]
- 28.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility tests–fifth edition; approved standard. NCCLS document M2-A5. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1993. [Google Scholar]
- 29.Norris A H, Reilly J P, Edelstein P H, Brennan P J, Schuster M G. Chloramphenicol for the treatment of vancomycin-resistant enterococcal infections. Clin Infect Dis. 1995;20:1137–1144. doi: 10.1093/clinids/20.5.1137. [DOI] [PubMed] [Google Scholar]
- 30.Rende-Fournier R, Leclercq R, Galimand M, Duval J, Courvalin P. Identification of the satA gene encoding a streptogramin A acetyltransferase in Enterococcus faecium BM4145. Antimicrob Agents Chemother. 1993;37:2119–2125. doi: 10.1128/aac.37.10.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rice L B, Eliopoulos G M, Wennersten C, Goldmann D, Jacoby G A, Moellering R C., Jr Chromosomally mediated β-lactamase production and gentamicin resistance in Enterococcus faecalis. Antimicrob Agents Chemother. 1991;35:272–276. doi: 10.1128/aac.35.2.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. pp. 1.98–1.99. [Google Scholar]
- 33.Shay D K, Maloney S A, Montecalvo M, Banerjee S, Wormser G P, Arduino M J, Bland L A, Jarvis W R. Epidemiology and mortality risk of vancomycin-resistant enterococcal bloodstream infections. J Infect Dis. 1995;172:993–1000. doi: 10.1093/infdis/172.4.993. [DOI] [PubMed] [Google Scholar]
- 34.Venditti M, Tarasi A, Gelfusa V, Nicastri E, Penni A, Martino P. Antimicrobial susceptibilities of enterococci isolated from hospitalized patients. Antimicrob Agents Chemother. 1993;37:1190–1192. doi: 10.1128/aac.37.5.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woodford N, Jones B L, Baccus Z, Ludlam H A, Brown D F. Linkage of vancomycin and high-level gentamicin resistance genes on the same plasmid in a clinical isolate of Enterococcus faecalis. J Antimicrob Chemother. 1995;35:179–184. doi: 10.1093/jac/35.1.179. [DOI] [PubMed] [Google Scholar]
- 36.Woodford N, Morrison D, Johnson A P, George R C. Antimicrobial resistance amongst enterococci isolated in the United Kingdom: a reference laboratory perspective. J Antimicrob Chemother. 1993;32:344–346. doi: 10.1093/jac/32.2.344. [DOI] [PubMed] [Google Scholar]