Abstract
Zucchini yellow mosaic virus (ZYMV) is a severe threat to cucurbit crops worldwide, including Pakistan. This study was pursued to evaluate the prevalence, geographic distribution, and molecular diversity of ZYMV isolates infecting cucurbits in Pakistan’s Pothwar region. Almost all the plant viruses act as a biotic stress on the host plants, which results in a yield loss. These viruses cause losses in single-infection or in mixed-infection cucurbit crops, and we have found a number of mixed-infected samples belonging to the Curubitaceae family. Serological detection of the tested potyviruses in the collected cucurbit samples revealed that ZYMV was the most prevalent virus, with a disease incidence (DI) at 35.2%, followed by Papaya ringspot virus (PRSV) with an incidence of 2.2%, and Watermelon mosaic virus (WMV) having an incidence as little as 0.5% in 2016. In the year 2017, a relatively higher disease incidence of 39.7%, 2.4%, and 0.3% for ZYMV, WMV, and PRSV, respectively, was recorded. ZYMV was the most prevalent virus with the highest incidence in Attock, Rawalpindi, and Islamabad, while PRSV was observed to be the highest in Islamabad and Jhelum. WMV infection was observed only in Rawalpindi and Chakwal. Newly detected Pakistani ZYMV isolates shared 95.8–97.0% nucleotide identities among themselves and 77.1–97.8% with other isolates retrieved from GenBank. Phylogenetic relationships obtained using different ZYMV isolates retrieved from GenBank and validated by in silico restriction analysis revealed that four Pakistani isolates clustered with other ZYMV isolates in group IIb with Chinese, Italian, Polish, and French isolates, while another isolate (MK848239) formed a separate minor clade within IIb. The isolate MK8482490, reported to infect bitter gourd in Pakistan, shared a minor clade with a Chinese isolate (KX884570). Recombination analysis revealed that the recently found ZYMV isolate (MK848239) is most likely a recombinant of Pakistani (MK848237) and Italian (MK956829) isolates, with a recombinant breakpoint between 266 and 814 nucleotide positions. Local isolate comparison and recombination detection may aid in the development of a breeding program that identifies resistant sources against recombinant isolates because the ZYMV is prevalent in a few cucurbit species grown in the surveyed areas and causes heavy losses and economic damage to the agricultural community.
Keywords: RNA, ZYMV, coat protein, in silico restriction, subgroup IIa and IIb, cucurbits, biotic stress
1. Introduction
Vegetables are high-value crops that frequently provide sustainable livelihoods for smallholders and large-scale commercial farmers. Cucurbits (Cucurbitaceae family) are major vegetable and fruit crops in Pakistan, grown throughout the country, including the Pothwar region [1]. This family includes around 95 extant genera with over 900 species [2]. The Pothwar region’s favorable climatic conditions facilitate the production of most cucurbits, cultivated as summer and winter crops throughout the year [3,4]. The main cultivated cucurbit species of the Pothwar region include melon (Cucumis melo L.), cucumber (C. sativus L.), squash (Cucurbita sp.), gourd (Luffa sp.), pumpkin (Cucurbita moschata), round gourd (Praecitrullus fistulosus (Stocks) Pangalo) and watermelon (Citrullus lanatus L.). Cucurbits play a vital role in the diet, as they contain vitamins and other dietary constituents (vitamins B and C, niacin, protein, lipid, carbohydrate, Ca, Fe, P) [5]. Several biotic and abiotic factors may limit cucurbit production. Among biotic factors, earlier studies reported approximately 90 different viruses infecting cucurbits from diverse regions of the world [4,6,7]. The cucurbit average yield in Pakistan is relatively lower than in other countries due to the region’s prevalence of diseases by RNA viruses. Among these RNA viruses, the most common are Zucchini yellow mosaic virus (ZYMV; Potyvirus), Cucurbit aphid-borne yellows virus (CABYV; Polerovirus), Papaya ringspot virus (PRSV; Potyvirus), Cucumber mosaic virus (CMV; Cucumovirus), Cucurbit chlorotic yellows virus (CCYV), and Watermelon mosaic virus (WMV; Potyvirus) [7,8,9,10,11,12].
ZYMV, belonging to genus Potyvirus in the family Potyviridae, is reported to incite severe diseases in crops grown as tropical and subtropical cucurbits, globally [13,14,15]. Virus particles are flexuous filaments of sizes ranging from 680 to 730 nm in length [13,16]. The potyviruses are reported to have been transmitted through numerous aphid species non-persistently and by seeds [17,18,19,20]. The aphid vectors play their role in spreading the virus within the cultivated fields and from cucurbit crops to weeds, acting as natural reservoirs of the virus during the offseason, and work as efficient sources of infection at the start of the growing season [14,21,22]. ZYMV causes typical symptoms of mosaic, leaf distortion, blisters, chlorosis on leaves, and general stunting in the affected cucurbit plants. In severe systemic infection, affected fruits develop malformation and surface discoloration, making them unmarketable [7,14,23]. Yield losses caused by ZYMV can range from mild to severe, with some reports suggesting reductions of up to 80% in affected crops [24]. The magnitude of yield losses depends on several factors, including the prevalence and virulence of the virus, susceptibility of cultivated varieties, agronomic practices, and environmental conditions [25]. Though a few earlier studies revealed the presence of ZYMV infection in bottle gourd [1], melon [26], ridge gourd [10], and round gourd [27] in other parts of the country, there is a critical need for an intensive study highlighting the comprehensive understanding of ZYMV prevalence in major cucurbits to devise proper management strategies. So, this study was conducted with the aim of providing an estimation of the incidence and molecular characterization of potyviruses, especially ZYMV, in the major cucurbit crops of the Pothwar region.
2. Results
2.1. Incidence and Distribution of Potyviruses
The result of general serological screening done by PTA–ELISA showed that, out of the 767 leaf samples collected from cucurbit plants exhibiting possible symptoms of viral diseases, 290 samples (37.8%) were infected with potyviruses in 2016. These positive samples were retested with specific DAS–ELISA and the incidence of ZYMV, PRSV, and WMV in the collected cucurbit samples was recorded to be 35.2%, 2.2%, and 0.5%, respectively (Table 1). In the year 2017, out of 795 samples, ZYMV was the most prevalent virus, with a relatively increased disease incidence of 39.7%, followed by WMV (2.4%) and PRSV (0.3%) (Table 1). In the Attock district, smooth gourd was found to be highly infected with potyviruses, followed by ridge gourd, round gourd, and cucumber, while bitter gourd was the lowest infected crop during 2016 and 2017. ZYMV was the most prevalent virus found heavily infecting ridge gourd, followed by smooth gourd, round gourd, and cucumber, while melon was the less-infecting crop in the Chakwal district during both sampling years. In the Jhelum and Islamabad districts, melon was observed without any potyvirus infection (Table 1).
Table 1.
Disease incidence of potyviruses infecting cucurbits in 2016–2017 from the Pothwar region of Pakistan.
| Location | Crop | D.I. % in 2016 | D.I. % in 2017 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Potyvirus a | ZYMV b | WMV c | PRSV d | Potyvirus | ZYMV | WMV | PRSV | ||||
| Samples * | D.I. % | Samples * | D.I. % | ||||||||
| Attock | Cucumber | 18 (40) | 45.0 | 40.0 | 0.0 | 5.0 | 17 (41) | 41.5 | 41.5 | 0.0 | 2.4 |
| Round gourd | 17 (32) | 53.1 | 50.0 | 0.0 | 3.13 | 18 (33) | 54.5 | 54.5 | 0.0 | 0.0 | |
| Watermelon | 8 (21) | 38.1 | 28.6 | 0.0 | 9.5 | 9 (21) | 42.9 | 33.3 | 0.0 | 9.5 | |
| Melon | 6 (21) | 28.6 | 19.0 | 0.0 | 9.5 | 8 (22) | 36.3 | 22.7 | 0.0 | 13.6 | |
| Pumpkin | 3 (10) | 30.0 | 30.0 | 0.0 | 0.0 | 5 (13) | 38.5 | 38.5 | 0.0 | 0.0 | |
| Bitter gourd | 2 (20) | 10.0 | 10.0 | 0.0 | 0.0 | 3 (21) | 14.3 | 14.3 | 0.0 | 0.0 | |
| Ridge gourd | 30 (38) | 78.9 | 78.9 | 0.0 | 0.0 | 32 (39) | 82.1 | 82.1 | 0.0 | 0.0 | |
| Smooth gourd | 5 (6) | 83.3 | 83.3 | 0.0 | 0.0 | 7 (8) | 87.5 | 87.5 | 0.0 | 0.0 | |
| Chakwal | Cucumber | 6 (25) | 24.0 | 24.0 | 0.0 | 0.0 | 7 (22) | 31.8 | 31.8 | 0.0 | 0.0 |
| Round gourd | 6 (17) | 35.3 | 35.3 | 0.0 | 0.0 | 4 (16) | 25.0 | 25.0 | 0.0 | 0.0 | |
| Watermelon | 4 (19) | 21.1 | 21.1 | 5.3 | 0.0 | 5 (18) | 27.8 | 27.8 | 0.0 | 5.6 | |
| Melon | 1 (11) | 9.1 | 0.0 | 0.0 | 9.1 | 2 (12) | 16.7 | 8.3 | 0.0 | 8.3 | |
| Pumpkin | 2 (13) | 15.4 | 15.4 | 0.0 | 0.0 | 3 (13) | 23.1 | 23.1 | 0.0 | 0.0 | |
| Bitter gourd | 1 (16) | 6.3 | 6.3 | 0.0 | 0.0 | 2 (16) | 12.5 | 12.5 | 0.0 | 0.0 | |
| Ridge gourd | 15 (25) | 60.0 | 60.0 | 0.0 | 0.0 | 19 (26) | 73.1 | 73.1 | 0.0 | 0.0 | |
| Smooth gourd | 4 (10) | 40.0 | 40.0 | 0.0 | 0.0 | 5 (8) | 62.5 | 62.5 | 0.0 | 0.0 | |
| Jhelum | Cucumber | 5 (21) | 23.8 | 23.8 | 0.0 | 0.0 | 6 (25) | 24.0 | 24.0 | 0.0 | 0.0 |
| Round gourd | 3 (7) | 42.9 | 42.9 | 0.0 | 0.0 | 4 (9) | 44.4 | 44.4 | 0.0 | 0.0 | |
| Watermelon | 4 (21) | 19.0 | 0.0 | 0.0 | 19.0 | 5 (21) | 23.8 | 9.5 | 0.0 | 14.3 | |
| Melon | 0 (5) | 0.0 | 0.0 | 0.0 | 0.0 | 0 (8) | 0.0 | 0.0 | 0.0 | 0.0 | |
| Pumpkin | 1 (14) | 7.1 | 7.1 | 0.0 | 0.0 | 2 (15) | 13.3 | 13.3 | 0.0 | 0.0 | |
| Bitter gourd | 1 (16) | 6.3 | 6.3 | 0.0 | 0.0 | 2 (16) | 6.3 | 6.3 | 0.0 | 0.0 | |
| Ridge gourd | 14 (26) | 53.8 | 53.8 | 0.0 | 0.0 | 15 (25) | 60.0 | 60.0 | 0.0 | 0.0 | |
| Smooth gourd | 3 (9) | 33.3 | 33.3 | 0.0 | 0.0 | 3 (9) | 33.3 | 33.3 | 0.0 | 0.0 | |
| Rawalpindi | Cucumber | 23 (68) | 33.8 | 32.4 | 1.5 | 0.0 | 25 (69) | 36.2 | 36.2 | 1.4 | 1.4 |
| Round gourd | 13 (26) | 50.0 | 50.0 | 0.0 | 0.0 | 15 (29) | 51.7 | 51.7 | 0.0 | 0.0 | |
| Watermelon | 11 (29) | 37.9 | 27.6 | 3.4 | 6.9 | 12 (30) | 40.0 | 30.0 | 3.3 | 10.0 | |
| Melon | 3 (18) | 16.7 | 5.6 | 5.6 | 5.6 | 4 (21) | 19.0 | 9.5 | 0.0 | 9.5 | |
| Pumpkin | 3 (15) | 20.0 | 20.0 | 0.0 | 0.0 | 4 (14) | 28.6 | 28.6 | 0.0 | 0.0 | |
| Bitter gourd | 3 (28) | 10.7 | 10.7 | 0.0 | 0.0 | 4 (29) | 13.8 | 13.8 | 0.0 | 0.0 | |
| Ridge gourd | 30 (37) | 81.1 | 81.1 | 0.0 | 0.0 | 33 (38) | 86.8 | 86.8 | 0.0 | 0.0 | |
| Smooth gourd | 8 (12) | 66.7 | 66.7 | 0.0 | 0.0 | 10 (15) | 66.7 | 66.7 | 0.0 | 0.0 | |
| Islamabad | Cucumber | 5 (13) | 38.5 | 38.5 | 0.0 | 0.0 | 2 (11) | 18.2 | 18.2 | 0.0 | 0.0 |
| Round gourd | 4 (9) | 44.4 | 44.4 | 0.0 | 0.0 | 6 (12) | 50.0 | 50.0 | 0.0 | 0.0 | |
| Watermelon | 2 (8) | 25.0 | 0.0 | 0.0 | 25.0 | 3 (8) | 37.5 | 12.5 | 0.0 | 25.0 | |
| Melon | 0 (7) | 0.0 | 0.0 | 0.0 | 0.0 | 0 (6) | 0.0 | 0.0 | 0.0 | 0.0 | |
| Pumpkin | 2 (7) | 28.6 | 28.6 | 0.0 | 0.0 | 3 (8) | 37.5 | 37.5 | 0.0 | 0.0 | |
| Bitter gourd | 2 (15) | 13.3 | 13.3 | 0.0 | 0.0 | 3 (15) | 20.0 | 20.0 | 0.0 | 0.0 | |
| Ridge gourd | 15 (23) | 65.2 | 65.2 | 0.0 | 0.0 | 18 (24) | 75.0 | 75.0 | 0.0 | 0.0 | |
| Smooth gourd | 7 (9) | 77.8 | 77.8 | 0.0 | 0.0 | 7 (9) | 77.8 | 77.8 | 0.0 | 0.0 | |
| Total % D. I. | 290 (767) | 37.8 | 35.2 | 0.5 | 2.2 | 331 (795) | 41.6 | 39.7 | 0.3 | 2.4 | |
* Samples: no. of +ive samples (Total tested samples); a Disease incidence of Potyvirus = (no. of Potyvirus +ive samples/Total tested samples) × 100; b Disease incidence of ZYMV = (no. of ZYMV +ive samples/Total tested samples) × 100; c Disease incidence of WMV = (no. of WMV +ive samples/Total tested samples) × 100; d Disease incidence of PRSV = (no. of PRSV +ive samples/Total tested samples) × 100.
The most common disease was ZYMV, which had the highest incidence in Attock, followed by Rawalpindi, Islamabad, and Chakwal, and the lowest incidence in Jhelum. The highest incidence of PRSV was detected in Islamabad, followed by Jhelum, Attock, and Chakwal, while in Rawalpindi, cucurbit crops were less infected with PRSV. WMV infection was observed only in Rawalpindi and Chakwal (Table 1).
2.2. Molecular Characterization of Zucchini Yellow Mosaic Virus (ZYMV)
PCR results revealed that ZYMV-specific primers [1] amplified a specific fragment of a size of 1.5 kb in all the ELISA-positive samples chosen from each surveyed location. Five sequences of ZYMV were acquired when amplified fragments were processed by Sanger sequencing in both directions. Five sequences of ZYMV were sequenced and subjected to BLASTn to confirm that all sequenced samples containing partial NIb, 3′UTR genomic regions, and complete CP showed nucleotide identities of 94.5–99.9% to the corresponding regions of other ZYMV isolates (same subgroup) from GenBank. After careful analysis, five ZYMV present-study isolates were submitted to GenBank (Table 2). Each sequence of ZYMV comprised of 211 nt of 3′UTR, 837 nt encoding 278 aa of CP, 363 nt encoding 121 aa of NIb, and followed by a poly-A tail of varying length. The proportions of guanine, uracil, adenine, and cytosine in the CP gene sequences of these isolates were 25.5%, 22.5%, 32.4%, and 19.5%, respectively (Table 2).
Table 2.
Features of Pakistani ZYMV isolates submitted in GenBank.
| Accession No. |
Isolate | Host | Viral cDNA (nt) |
NIb | Coat Protein Gene | 3′UTR (nt) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nt | aa | CP (nt) |
CP (aa) |
U (%) |
C (%) |
A (%) |
G (%) |
|||||
| MK848237 | AARC | Cucumber | 1426 | 363 | 121 | 837 | 278 | 23.42 | 18.64 | 32.97 | 24.97 | 226 |
| MK848238 | AAHSG | Smooth Gourd | 1426 | 363 | 121 | 837 | 278 | 23.42 | 19.12 | 32.74 | 24.73 | 226 |
| MK848239 | AAAP | Pumpkin | 1426 | 363 | 121 | 837 | 278 | 23.54 | 19.12 | 32.26 | 25.09 | 226 |
| MK848240 | AARBG | Bitter Gourd | 1426 | 363 | 121 | 837 | 278 | 23.78 | 18.76 | 31.78 | 25.69 | 226 |
| MK848241 | AAHWM | Watermelon | 1426 | 363 | 121 | 837 | 278 | 23.54 | 19.0 | 32.50 | 24.97 | 226 |
Moreover, the sequence identity matrix study showed that all the Pakistani ZYMV isolates shared a 95.8 to 97.0% similarity. Three recently identified isolates (MK848237, 97.6%; MK848238, 97.8%; MK848241, 97%) showed the highest similarity with the Pakistani ZYMV isolate AIRGPK (KR261952) reported in the year 2017 from round gourd, while MK848239 shared the highest similarity (96.7%) with the KX884565 isolate, reported to infect crayfish from China, and MK848240 shared maximum similarity (97.3%) with the French, Iranian, and South Korean isolates. All the newly characterized isolates showed the lowest similarity with sequence JF797206 isolated from cucumber in India (Table 3, Figure 1). We observed a 98.9–99.4% identity in the amino acid sequences of the CP genes of all isolates, while the formerly reported subgroup IIb ZYMV isolates shared 93.9–99.6% identities. Amino acid sequence identities of 88.1–92.4% and 88.5% were observed with the subgroup IIa and group I isolates, respectively, from other regions of the world.
Table 3.
Nucleotide identity of new Pakistani ZYMV isolates with isolates reported to infect several hosts in different countries around the world.
| Accession No. |
Location | Host | Year | NT Identity % Age | ||||
|---|---|---|---|---|---|---|---|---|
| MK848237 | MK848238 | MK848239 | MK848240 | MK848241 | ||||
| KR261952 | Pakistan | Round Gourd | 2014 | 97.0 | 96.8 | 96.1 | 95.2 | 97.0 |
| MK956829 | Italy | Pumpkin | 2019 | 97.6 | 97.8 | 96.1 | 97.2 | 97.6 |
| HM005312 | Mali | Watermelon | 2009 | 94.7 | 94.8 | 94.5 | 95.2 | 94.7 |
| HM005311 | Mali | Watermelon | 2009 | 95.8 | 95.5 | 95.3 | 96.8 | 95.8 |
| EU561044 | Poland | Cucumber | 2008 | 95.5 | 94.9 | 94.9 | 96.8 | 95.5 |
| EF122498 | China | Pumpkin | 2006 | 96.5 | 96.8 | 96.5 | 96.7 | 96.5 |
| MW345249 | Turkey | Pumpkin | 2011 | 97.2 | 96.8 | 96.5 | 96.9 | 97.2 |
| MH042025 | Republic of Korea | Pumpkin | 2016 | 96.9 | 97.0 | 96.3 | 97.2 | 96.9 |
| MH042026 | Republic of Korea | Cucumber | 2016 | 97.1 | 97.0 | 96.5 | 97.3 | 97.1 |
| KX884565 | China | Crayfish | 2014 | 97.0 | 96.9 | 96.5 | 97.3 | 97.0 |
| KX884570 | China | Spider | 2013 | 95.6 | 95.6 | 95.0 | 95.6 | 95.6 |
| AB188116 | Japan | Cucumber | 2004 | 97.3 | 97.0 | 96.7 | 97.2 | 97.3 |
| AB188115 | Japan | Cucumber | 2004 | 96.1 | 95.4 | 95.5 | 96.5 | 96.1 |
| MT383108 | Egypt | Zucchini | 2018 | 96.1 | 95.4 | 95.6 | 96.5 | 96.1 |
| MN598576 | Australia | Melon | 2019 | 94.8 | 94.3 | 94.4 | 95.3 | 94.8 |
| L31350 | USA | -- | 1995 | 95.6 | 94.8 | 95.3 | 96.3 | 95.6 |
| KU366270 | Iran | Pumpkin | 2013 | 95.8 | 95.4 | 95.5 | 96.3 | 95.8 |
| MW449260 | France | Zucchini | 2022 | 96.2 | 96.3 | 96.0 | 97.3 | 96.2 |
| MW449262 | France | Melon | 2022 | 94.3 | 94.1 | 94.0 | 94.5 | 94.3 |
| MW449263 | France | Melon | 2022 | 93.8 | 93.6 | 93.1 | 94.6 | 93.8 |
| MK033873 | China | Pumpkin | 2017 | 97.3 | 97.5 | 96.5 | 97.3 | 97.3 |
| AY995216 | New Zealand | Zucchini | 2016 | 96.2 | 96.5 | 95.8 | 96.5 | 96.2 |
| OQ335839 | Italy | Pumpkin | 2023 | 93.2 | 92.9 | 92.9 | 92.9 | 93.2 |
| KJ614229 | India | Bitter Gourd | 2013 | 96.1 | 95.8 | 95.6 | 96.9 | 96.1 |
| JF797206 | India | Cucumber | 2011 | 78.5 | 78.5 | 77.7 | 79.0 | 78.5 |
| MW345250 | Turkey | Cucumber | 2009 | 93.7 | 93.7 | 93.3 | 94.4 | 93.7 |
| OK558793 | Canada | - | 2001 | 93.9 | 93.9 | 93.3 | 94.6 | 93.9 |
| AY188994 | Israel | - | 2004 | 77.6 | 77.6 | 77.1 | 78.3 | 77.6 |
| D00692 | USA | Zucchini | 1990 | 93.7 | 93.9 | 93.1 | 94.6 | 93.7 |
| OM471983 | UK | Pumpkin | 2022 | 95.9 | 95.3 | 95.6 | 96.5 | 95.9 |
| LC726785 | China | Vegetables | 2019 | 95.7 | 95.0 | 95.2 | 96.2 | 95.7 |
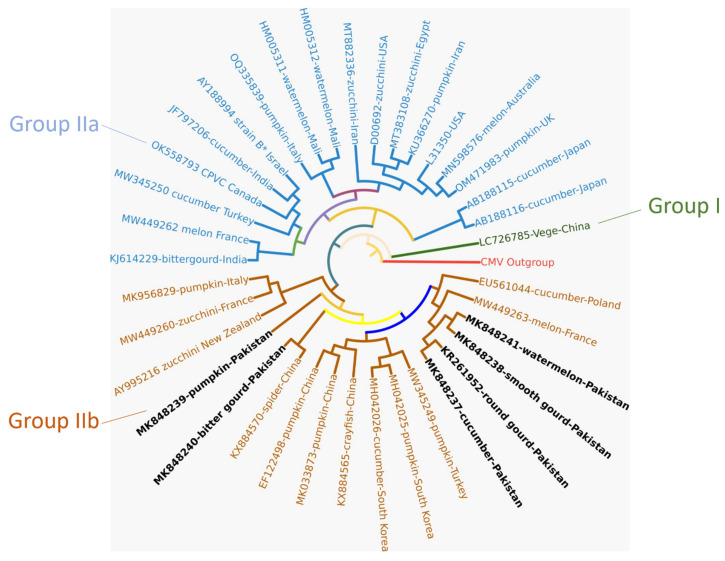
Figure 1.
The phylogenetic analysis of 37 isolates of ZYMV using the maximum-likelihood method with the Jukes–Cantor model, supported by robustness assessments, keeping the bootstrap value at 1000.
2.3. Phylogenetic Analysis and In Silico RFLP-Based Phylogeny-Based Comparison
Phylogenetic analysis revealed that all the sequences were outlined into two major groups, i.e., group I and group II. Group I consisted of a single isolate reported from China, while group IIa included 18 isolates reported from Japan, the United Kingdom, Australia, the United States, Iran, Egypt, Mali, Italy, Israel, India, Canada, Turkey, and France. Group IIb also contained 18 isolates, including 5 new and 1 previously reported from Pakistan, which were scattered among several minor clades. One of the isolates (MK848239), reported to infect pumpkin in Pakistan, was found to be relatively divergent, as it did not share its position in a minor clade within group IIb with any other isolate. Another isolate (MK8482490), reported to infect bitter gourd in Pakistan, shared a minor clade with a Chinese isolate (KX884570). Four other newly reported isolates from Pakistan infecting watermelon, cucumber, smooth gourd, and round gourd shared a separate minor clade with isolates of ZYMV reported from Poland and France.
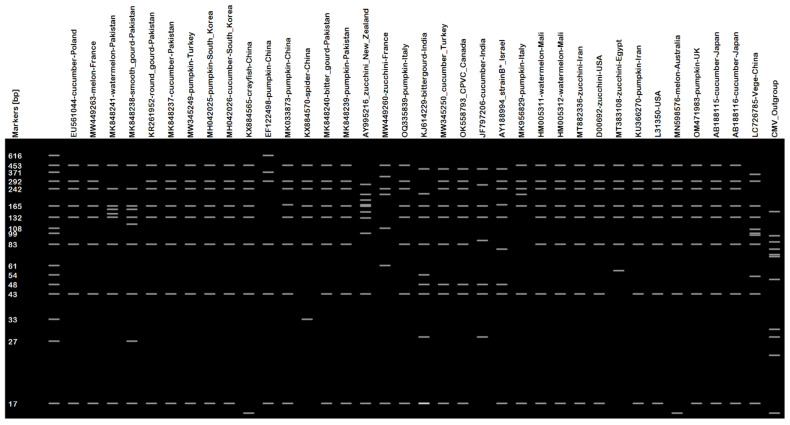
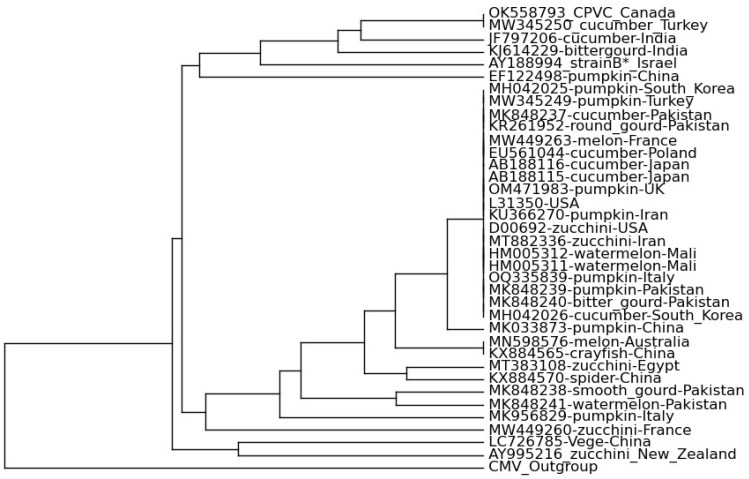
In silico RFLP simulation of ZYMV nucleotide sequences showed the presence of seven to nine cut positions of each enzyme. Virtual gel resulting from RFLP simulation revealed that nearly all the ZYMV isolates contained a distinct 43 bp band, while the CMV outgroup isolates lacked it. Similarly, group I exhibited a unique 99 bp fragment, whereas a 48 bp fragment distinguished group IIa isolates from the others (Figure 2). Phylograms (Figure 3) obtained after the virtual gel analysis in PyElph v1.4 affirmed the findings of the phylogeny analysis, i.e., the clustering of isolates in two main clades, except the shuffling of group IIa isolates to IIb isolates in minor clades. The AY995216 isolate from New Zealand also shifted from IIb to group I, clustering with the Chinese Vege isolate. This shift of isolate clustering may be due to silent mutation in the sequences of these isolates at restriction points [28]. Amino acid analysis confirms the occurrence of specific potyvirus motifs: QMKAAA in all the Pakistani ZYMV isolates, the CP region’s MVWCIEN, and the NIb area’s GNNS are the main criteria for potyvirus identification [29,30].
Figure 2.
Virtual gel after the in silico RFLP simulation of CP genes of the ZYMV isolates.
Figure 3.
In silico RFLP simulation and genotyping of 37 isolates of ZYMV based on the CP gene.
2.4. Recombination Analysis
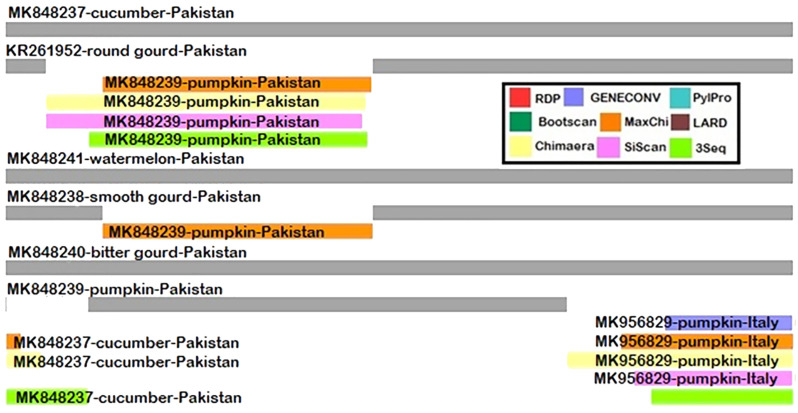
Recombination analysis showed that our isolate AAAP (MK848239) is likely to be recombinant with Pakistani (MK848237) and Italian (MK956829) ZYMV pumpkin isolates (Figure 4). Five statistical methods, i.e., Chimaera, GENECONV, MaxChi, 3SEQ, and SiScan, confirmed this recombination event, with a recombination breakpoint between nucleotides 266 and 814. A second recombination event was also detected between nucleotide 74–650 through four statistical methods in RDP4, which showed that the Pakistani KR261952 isolate reported in our previous study [27] is likely a recombinant, originating from the Australian (MN598576) and Pakistani (MK848239) ZYMV pumpkin isolate (Table 4).
Figure 4.
Recombinant events detected in ZYMV isolates using Standard Methods in RDP4.
Table 4.
Recombination events detected in Pakistan isolates of ZYMV sequences.
| Recombinant Isolate | Breaking Point | Major Parent |
Minor Parent |
p-Value | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Starting | Ending | RDP | GENECONV | BootScan | MaxChi | Chimera | SiScan | 3SEQ | |||
| MK848239 | 266 | 814 | MK848237 | MK956829 | NS | 1.257 × 10−2 | NS | 3.121 × 10−3 | 3.925 × 10−2 | 2.045 × 10−8 | 5.253 × 10−4 |
| KR261952 | 74 | 650 | MN598576 | MK848239 | NS | NS | NS | 1.295 × 10−1 | 2.269 × 10−3 | 1.143 × 10−2 | 9.417 × 10−4 |
3. Discussion
As with all viruses, plant viruses implement their mechanism of highjacking the nuclear machinery of the host cells and dedicate the plant resources for the replication and movement of its particles to new cells and tissues for the sake of invasion. Apart from the direct effects, a number of genes present in potyviruses are reported to interact with Ca2+ sensors and activate the related transcriptional procedures. Few genes which show altered transcribed abundance are related to the process of photosynthesis, signaling, and defense responses [31]. As a consequence of potyvirus infection, the photosynthetic and defense machinery is under stress, leading to yield loss in the longer run. Among plant viruses, the genus Potyvirus is considered one of the largest groups of plant viruses, with a large number of definite and tentative candidates. Among the members of potyviruses, ZYMV, WMV, and PRSV have been reported to be devastating to several important cucurbit crops, particularly from the economic point of view [32,33]. ZYMV may cause up to 80% yield losses in different crops, depending on the crop germplasm, age, viral load, vector population, and favorable environmental conditions [24,25,34]. The classification of potyviruses has sometimes led to confusion because of their striking variations in symptomatology, host range, and genetic makeup, with markable changes among the strains and the tentative candidates assigned to this genus [35]. Considering the diverse nature of this genus, the use of molecular approaches has been a successful tool for characterizing the local potyvirus isolates infecting cucurbits. This study helped in estimating the status of the prevalence of three species of potyviruses infecting cucurbits grown in the Pothwar region using serological identification and molecular characterization, which aided in understanding the evolutionary and recombination histories of the identified isolates in recent years.
The results of surveys in the Pothwar region revealed that ZYMV was the most prevalent virus, with a disease incidence of 37.52%, followed by PRSV (2.3%) and WMV (0.38%) in 2016–17, which showed that ZYMV is the most widespread among the tested viruses in the Pothwar region infecting cucurbits, and in line with the reports of other scientists working on the identification of this virus in the country [1,10,27,34]. Considering the disease-incidence data of the potyviruses, all crops other than cucumber have a substantially lower proportion of disease, implying that cucumber crop cultivation is vulnerable to these diseases, although other cucurbit crops may hold a desirable position in the region. Sequence identity matrix findings indicate that Pakistani ZYMV isolates exhibited 94.6–97.9% identities with other ZYMV isolates retrieved from GenBank, while they shared 95.8–97.0% nucleotide identities with one another. Interestingly, the similarity data reveal that one of the closely related isolates was reported as early as 1990 from the USA, which had a 95.8% (on average) similarity with all the newly reported isolates from Pakistan. It can be concluded that the virus has a good level of adaptation to several cucurbit hosts cultivated in different countries over the last three decades; however, the similarity percentage did not go below 94.6%. It has also been reported that variation in the genomic regions, viz. the CP gene, helper component–protease (HC-Pro), or P3 protein genes of plant viruses, especially ZYMV, leads to varying expression of symptoms, transmission, and host range [14,23].
In the nucleotide-based phylogenetic analysis, it was revealed that the five new Pakistani isolates of ZYMV, along with thirty-two other isolates retrieved from GenBank, NCBI, produced two distinct groups, including group I and group II. Group I includes a single isolate reported from China, while group II has two subgroups, group IIa and b. Group IIa comprises of 18 isolates reported from several countries and which are relatively different from the isolates reported from Pakistan, regardless of the host. Whereas, in group IIb, all isolates were reported to infect cucumber, watermelon, pumpkin, bitter gourd, round gourd, and smooth gourd Pakistan. It was observed that the isolates reported from Pakistan are phylogenetically related to each other, and apparently the host has no impact on the development of new strains. The phylogenetic tree depicts a low genetic diversity among the isolates of ZYMV reported to infect several hosts from different countries. The overall picture of the phylogenetic tree shows that isolates of ZYMV characterized from several hosts in different countries over the years are different, but the level of variance is not very high. These phylogeny results do not highlight any fact about the route of the virus entry into the country, because almost all the isolates share similar levels of identity.
The clustering pattern of isolates were validated using the in silico RFLP simulation of the obtained sequences, which revealed the similar pattern, except for the isolates of the IIa group, most of which clustered in group IIb, along with divergent patterns of isolate AY995216, which clustered with group I from IIb. This shift may be due to a silent mutation in the sequences of these isolates at restriction points. Silent mutations can occur at restriction-enzyme-recognition sequences, allowing the introduction or removal of restriction sites without affecting the protein translation [28]. Integration of information from phylogeny and virtual restriction simulations helped researchers to identify clustering patterns and variations, including the clustering of isolates in different groups and the impact of silent mutations on the clustering pattern [28,36].
Maina, et al. [37,38] recorded recombination events in the CP, as well as in the NIb, CI, P1, and P3 regions of the ZYMV isolates, and they also confirmed the formation of new groups due to these recombinant isolates. Variation may appear in the genome due to the mutation, recombination, and reassortment of viral genome elements [39]. The quarantine department must take serious testing and legislation measures to control the invasion of new viruses to the country.
Recombination analysis aids in the study of host adaptation and virus evolution in the abolition or emergence of variation in the viral genome. In this work, CP-gene-based recombination analysis revealed the detection of two recombination events, indicating that ZYMV pumpkin (MK848239) and ZYMV round gourd (KR261952) isolates are likely to be recombinant. Similar results of recombination in CP gene sequences have been elaborated in other potyviruses, i.e., PRSV [40,41], Plum pox virus (PPV) [42], Sugarcane mosaic virus (SCMV) [43,44], Yam mosaic virus (YMV) [45], Bean yellow mosaic virus (BYMV) [46], Potato virus Y (PVY) [47], and Chilli veinal mottle virus (ChiVMV) [48]. The fitness of potyvirus populations in changing conditions confirmed the importance of recombination studies [49]. The presence of recombinant isolates and various groups of ZYMV isolates with widespread distribution creates an alarming situation for productive crop yields that must be remedied through suitable viral detection and management strategies. Moreover, the quarantine department must implement rigorous testing and regulatory measures to prevent the introduction of new viruses into the country.
4. Materials and Methods
4.1. Surveys and Collection of Important Vegetable Crop Samples
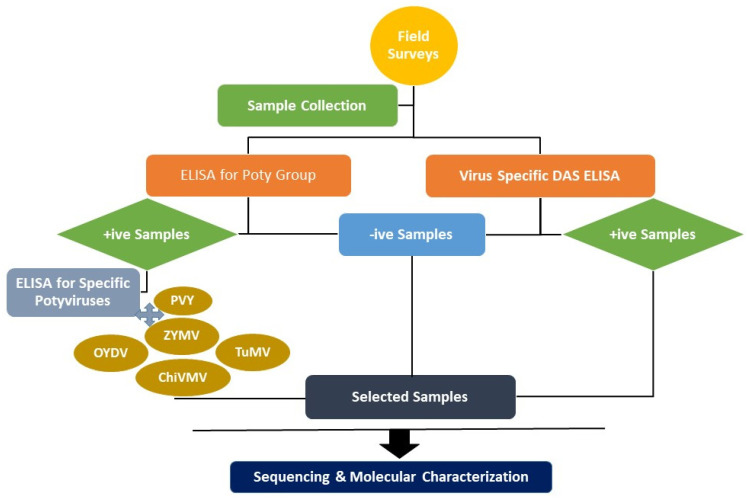
Vegetable-growing areas of the Pothwar region (Figure 5) were surveyed using a random stratified design during 2016–2017 [50] to record the incidence and distribution of potyviruses, as per the scheme given in Figure 6.
Figure 5.
Map of the Pothwar region (colored) in the Punjab province of Pakistan.
Figure 6.
Scheme used to detect plant RNA viruses that infect vegetable samples from the region of Pothwar in Pakistan.
A total of 767 and 795 leaf samples were collected in years 2016 and 2017, respectively, from eight to ten randomly selected vegetable fields in five districts of Pothwar region of Punjab, including Islamabad, Rawalpindi, Chakwal, Attock, and Jhelum (Table 1). The samples were collected based on typical virus-like symptoms, such as ring spots, mosaic, chlorotic, mottling, stunting, and puckering. All the collected samples were stored in iceboxes and brought to the Plant Virology lab, Department of Plant Pathology, PMAS Arid Agriculture University, Rawalpindi, Pakistan, for further processing. The samples were kept at −20 °C until they were processed for viral agent detection and characterization.
4.2. Serological Diagnosis of RNA Viruses
All the collected plant samples were initially tested serologically using a group-specific ELISA kit, i.e., potyvirus group ELISA kit (art. no. 163072, Bioreba, Reinach, Switzerland), and later by virus-specific ELISA using commercially available kits for ZYMV (art. no. 161272, Bioreba), PRSV (art. no. 151972, Bioreba), and WMV (art. no. 161172, Bioreba), with slight modification in the method of Clark and Adams [51]. The optical density (OD 405 nm) of the yellow color was determined using the ELISA reader with positive and negative controls (Bioreba, Switzerland) as a reference [52]. The samples with a 2X or greater OD 405 nm value than the healthy (control) samples were referred to as positive. ELISA-positive samples were used to determine the disease incidence percentage using the proportionate formula as given by Ahsan et al. [53]:
| (1) |
4.3. RNA Extraction and Molecular Characterization of ZYMV
After the serological screening, five representative samples infected with ZYMV were selected for molecular characterization based on coat protein (CP) gene sequences to ascertain genetic diversity. For this purpose, total RNA was extracted from ELISA-positive ZYMV samples by using TRIzol Reagent (Thermo Scientific, Waltham, MA, USA) according to the manufacturer’s protocol. First-strand complementary DNA (cDNA) was synthesized using the RevertAid RT Reverse Transcription Kit (Thermo Scientific, USA) and ZYMV CP downstream primer (ZYMV R Not 1-25, 5′- AAC TGG AAG AAT TCG CGG CCG CAC GAAT) [1] by following instructions of the manufacturer. The CP gene was amplified by the primer pair (ZYMV F-8193-22; 5′-CACCAAGCAATGYTRGTTG-3′ and ZYMV R Not 1-25) [1] in a thermal cycler using GreenTaq Green PCR Master Mix (2X) (Thermo Scientific, USA) following the reaction conditions; 94 °C for 3 min followed by 35 cycles of 94 °C for 30 s, 52 °C for 45 s, and 72 °C for 90 s, and a final extension at 72 °C for 10 min. RT-PCR products were run through 1% agarose gel added with ethidium bromide (100 μg mL−1). The amplicons of ~1.50 kb was purified using the GeneJET PCR Purification Kit (Thermo Scientific, USA) and cloned in the pTZ57R/T vector (InsTAcloneTM PCR cloning kit, Thermo Scientific, USA) with E. coli strain XL1-Blue competent cells. The manufacturer’s instructions were followed to isolate the recombinant plasmid DNA using the GeneJET Plasmid Miniprep Kit (Thermo Scientific, USA). ECoR1 and HindIII enzymes were used for the restriction digestion and validation of transformants, while M13 forward and reverse primers were employed for the sequencing of positive clones in both directions from Macrogen (Seoul, Republic of Korea).
The ExPASy translate tool [54] was employed to obtained amino acid sequences from nucleotides and subjected to BLASTn [55] and BLASTp [56] analysis for identification and to calculate sequence identity percentages. A total of 32 resembling ZYMV CP gene sequences from the NCBI database were downloaded and aligned with 6 isolates that were newly identified from Pakistan, along with an isolate of CMV (outgroup) using the CLUSTAL W program embedded in the MEGA7 software [57]. The aligned sequences along with a CMV isolate as an outgroup were subjected to phylogenetic reconstructions using the Maximum Likelihood with the Jukes–Cantor model, supported by robustness assessments, through 1000 bootstrap replicates. The identities of nucleotide and amino acid sequences were determined with the Sequence Identity Matrix option in BioEdit 7.2 [58].
4.4. In Silico RFLP and Recombination Analysis
The CP-based in silico restriction fragment length polymorphism (RFLP) of new Pakistani isolates and 32 retrieved sequences of ZYMV isolates from GenBank were executed using the DdeI, HinfI, and HpaII restriction enzymes in CLC Main Workbench 20 (QIAGEN, Aarhus, Denmark). The resulting gel (Figure 3) showed the presence of 3-3-5 bands of different sizes for each isolate analyzed in PyElph v1.4 [36] for the construction of phylograms and serotyping of isolates. Recombinant events in the 5 new Pakistani and 32 other ZYMV isolates were analyzed with RDP4 [59] with default settings, viz. RDP, GENECONV, BootScan, MaxChi, Siscan, Chimaera, and 3SEQ.
5. Conclusions
ZYMV is the most prevalent virus infecting all the major cucurbits grown in the Pothwar region of Pakistan. In silico analysis of ZYMV sequences revealed the presence of five new group A and B isolates closely related to isolates from Pakistan, China, Japan Poland, Italy, and Australia. Recombinant strain detection and higher incidence of the virus across the Pothwar region underscore the disease’s magnitude, forcing us to formulate new breeding programs to identify resistant sources and apprise our farmers about the disease.
Author Contributions
Conceptualization, M.A. (Muhammad Ashfaq); investigation, M.A. (Muhammad Ahsan) and M.T.S.; methodology, M.A.A., M.A. (Muhammad Ahsan) and M.A. (Muhammad Ashfaq); software, M.U. and M.A. (Muhammad Ashfaq); validation, M.A. (Muhammad Ashfaq); formal analysis, M.A. (Muhammad Ashfaq), M.A.M., M.A.A., M.T.S. and M.U.; resources, M.A. (Muhammad Ashfaq); M.A.A.-S.; writing—original draft preparation, M.A. (Muhammad Ahsan) and M.T.S.; writing—review and editing, M.A. (Muhammad Ahsan), M.A.M, M.A.A. and M.T.S.; supervision, M.A. (Muhammad Ashfaq); project administration, M.A. (Muhammad Ashfaq) and M.A.A.-S.; funding acquisition, M.A. (Muhammad Ashfaq) and M.A.A.-S. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
All the information and data have been added to the manuscript. However, the details of genetic sequences have been uploaded to GenBank, NCBI, and can be accessed using the accession numbers which are publicly available.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
The authors extend their appreciation to the Deputyship for Research and Innovation, ‘Ministry of Education’ in Saudi Arabia for funding this research (IFKSUOR3-418-1).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Ali A., Natsuaki T., Okuda S. Identification and molecular characterization of viruses infecting cucurbits in Pakistan. J. Phytopathol. 2004;152:677–682. doi: 10.1111/j.1439-0434.2004.00915.x. [DOI] [Google Scholar]
- 2.Ebert A.W., Drummond E., Giovannini P., van Zonneveld M. A Global Conservation Strategy for Crops in the Cucurbitaceae Family. Global Crop Diversity Trust; Bonn, Germany: 2021. [Google Scholar]
- 3.Ali A., Hussain A., Ahmad M. Occurrence and molecular characterization of Cucumber green mottle mosaic virus in cucurbit crops of KPK, Pakistan. Braz. J. Microbiol. 2014;45:1247–1253. doi: 10.1590/S1517-83822014000400015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashfaq M., Waqas M., Ahmed N., Raheel M., Abbas H.T., Masroor A., Ikram R.M., Riaz H., Ishtiaq M., Khan Z. Molecular characterization and identification of economically important Potyviruses in Cucurbitaceae family from Gujranwala division of Punjab, Pakistan. J. King Saud Univ.-Sci. 2021;33:101642. doi: 10.1016/j.jksus.2021.101642. [DOI] [Google Scholar]
- 5.Rolnik A., Olas B. Vegetables from the Cucurbitaceae family and their products: Positive effect on human health. Nutrition. 2020;78:110788. doi: 10.1016/j.nut.2020.110788. [DOI] [PubMed] [Google Scholar]
- 6.Ali A. Chapter 28—Epidemiology and evolution of poytviruses infecting cucurbits. In: Awasthi L.P., editor. Applied Plant Virology. Academic Press; Cambridge, MA, USA: 2020. pp. 405–417. [Google Scholar]
- 7.Lecoq H., Desbiez C. Advances in Virus Research. Volume 84. Elsevier; Amsterdam, The Netherlands: 2012. Viruses of cucurbit crops in the Mediterranean region: An ever-changing picture; pp. 67–126. [DOI] [PubMed] [Google Scholar]
- 8.Caciagli P. Vegetable viruses. In: Mahy B.W., Van-Regenmortel M.H., editors. Desk Encyclopedia of Plant and Fungal Virology. 1st ed. Academic Press; Cambridge, MA, USA: 2009. [Google Scholar]
- 9.Romay G., Lecoq H., Geraud-Pouey F., Chirinos D., Desbiez C. Current status of cucurbit viruses in Venezuela and characterization of Venezuelan isolates of Zucchini yellow mosaic virus. Plant Pathol. 2014;63:78–87. doi: 10.1111/ppa.12072. [DOI] [Google Scholar]
- 10.Ashfaq M., Saeed U., Mukhtar T., Haq M. First report of Zucchini yellow mosaic virus in ridge gourd in Pakistan. Plant Dis. 2015;99:1870. doi: 10.1094/PDIS-05-15-0553-PDN. [DOI] [Google Scholar]
- 11.Ahsan M., Ashfaq M., Mukhtar T., Abbasi N.A., Asad Z. First report of Cucurbit aphid borne yellows virus (CABYV) infecting melon in Pakistan. J. Plant Pathol. 2020;102:563–564. doi: 10.1007/s42161-019-00450-z. [DOI] [Google Scholar]
- 12.Nouman M., Farooq T., Parveen R., ud-Din Umar U., Aslam M.N., Moustafa M., Shakeel M.T. First report of cucumber-infecting cucurbit chlorotic yellows virus (CCYV) associated with cucurbit yellows disease in Pakistan. J. Plant Pathol. 2023;105:323–324. doi: 10.1007/s42161-022-01240-w. [DOI] [Google Scholar]
- 13.Desbiez C., Lecoq H. Zucchini yellow mosaic virus. Plant Pathol. 1997;46:809–829. doi: 10.1046/j.1365-3059.1997.d01-87.x. [DOI] [Google Scholar]
- 14.Coutts B., Kehoe M., Webster C., Wylie S., Jones R. Zucchini yellow mosaic virus: Biological properties, detection procedures and comparison of coat protein gene sequences. Arch. Virol. 2011;156:2119–2131. doi: 10.1007/s00705-011-1102-0. [DOI] [PubMed] [Google Scholar]
- 15.Lecoq H., Katis N. Advances in Virus Research. Volume 90. Elsevier; Amsterdam, The Netherlands: 2014. Control of cucurbit viruses; pp. 255–296. [DOI] [PubMed] [Google Scholar]
- 16.Lisa V., Boccardo G., D’Agostino G., Dellavalle G., d’Aquilio M. Characterization of a potyvirus that causes zucchini yellow mosaic. Phytopathology. 1981;71:667–672. doi: 10.1094/Phyto-71-667. [DOI] [Google Scholar]
- 17.Coutts B., Kehoe M., Jones R. Minimising losses caused by Zucchini yellow mosaic virus in vegetable cucurbit crops in tropical, sub-tropical and Mediterranean environments through cultural methods and host resistance. Virus Res. 2011;159:141–160. doi: 10.1016/j.virusres.2011.04.015. [DOI] [PubMed] [Google Scholar]
- 18.Yuan C., Ullman D.E. Comparison of efficiency and propensity as measures of vector importance in zucchini yellow mosaic potyvirus transmission by Aphis gossypii and A. craccivora. Phytopathology. 1996;86:698–703. doi: 10.1094/Phyto-86-698. [DOI] [Google Scholar]
- 19.Katis N., Tsitsipis J., Lykouressis D., Papapanayotou A., Margaritopoulos J., Kokinis G., Perdikis D.C., Manoussopoulos I. Transmission of Zucchini yellow mosaic virus by colonizing and non-colonizing aphids in Greece and new aphid species vectors of the virus. J. Phytopathol. 2006;154:293–302. doi: 10.1111/j.1439-0434.2006.01096.x. [DOI] [Google Scholar]
- 20.Coutts B., Kehoe M., Jones R. Zucchini yellow mosaic virus: Contact transmission, stability on surfaces, and inactivation with disinfectants. Plant Dis. 2013;97:765–771. doi: 10.1094/PDIS-08-12-0769-RE. [DOI] [PubMed] [Google Scholar]
- 21.Coutts B., Jones R. Incidence and distribution of viruses infecting cucurbit crops in the Northern Territory and Western Australia. Aust. J. Agric. Res. 2005;56:847–858. doi: 10.1071/AR04311. [DOI] [Google Scholar]
- 22.Lecoq H., Desbiez C. Watermelon Mosaic Virus and Zucchini Yellow Mosaic Virus. Academic Press; Cambridge, MA, USA: 2008. [Google Scholar]
- 23.Gal-On A. Zucchini yellow mosaic virus: Insect transmission and pathogenicity—The tails of two proteins. Mol. Plant Pathol. 2007;8:139–150. doi: 10.1111/j.1364-3703.2007.00381.x. [DOI] [PubMed] [Google Scholar]
- 24.Rao G.P., Reddy M.G. Chapter 38—Overview of yield losses due to plant viruses. In: Awasthi L.P., editor. Applied Plant Virology. Academic Press; Cambridge, MA, USA: 2020. pp. 531–562. [Google Scholar]
- 25.Lobell D.B., Cassman K.G., Field C.B. Crop yield gaps: Their importance, magnitudes, and causes. Annu. Rev. Environ. Resour. 2009;34:179–204. doi: 10.1146/annurev.environ.041008.093740. [DOI] [Google Scholar]
- 26.Malik A.H., Mansoor S., Iram S., Briddon R.W., Zafar Y. Severe disease of melon in North West frontier province is associated with simultaneous infection of two RNA viruses. Pak. J. Bot. 2010;42:361–367. [Google Scholar]
- 27.Ashfaq M., Ahsan M. First Report of Zucchini yellow mosaic virus in Round Gourd (Praecitrullus fistulosus) in Pakistan. Plant Dis. 2017;101:265. doi: 10.1094/PDIS-08-16-1161-PDN. [DOI] [Google Scholar]
- 28.Shankarappa R. Introduction of restriction enzyme recognition sequences by silent mutation. Curr. Protoc. Mol. Biol. 2001;35:A.3E.1–A.3E.3. doi: 10.1002/0471142727.mba03es35. [DOI] [PubMed] [Google Scholar]
- 29.Ha C., Revill P., Harding R.M., Vu M., Dale J.L. Identification and sequence analysis of potyviruses infecting crops in Vietnam. Arch. Virol. 2008;153:45–60. doi: 10.1007/s00705-007-1067-1. [DOI] [PubMed] [Google Scholar]
- 30.Zheng L., Wayper P.J., Gibbs A.J., Fourment M., Rodoni B.C., Gibbs M.J. Accumulating variation at conserved sites in potyvirus genomes is driven by species discovery and affects degenerate primer design. PLoS ONE. 2008;3:e1586. doi: 10.1371/journal.pone.0001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baebler S., Krecic-Stres H., Rotter A., Kogovsek P., Cankar K., Kok E.J., Gruden K., Kovac M., Zel J., Pompe-Novak M. and Ravnikar, M. PVYNTN elicits a diverse gene expression response in different potato genotypes in the first 12 h after inoculation. Mol. Plant Pathol. 2009;10:263–275. doi: 10.1111/j.1364-3703.2008.00530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lecoq H., Dafalla G., Desbiez C., Wipf-Scheibel C., Delécolle B., Lanina T., Ullah Z., Grumet R. Biological and molecular characterization of Moroccan watermelon mosaic virus and a potyvirus isolate from Eastern Sudan. Plant Dis. 2001;85:547–552. doi: 10.1094/PDIS.2001.85.5.547. [DOI] [PubMed] [Google Scholar]
- 33.Fauquet C.M., Mayo M.A., Maniloff J., Desselberger U., Ball L.A. Virus Taxonomy: VIIIth Report of the International Committee on Taxonomy of Viruses. Academic Press; Cambridge, MA, USA: 2005. [Google Scholar]
- 34.Asad Z., Ashfaq M., Mukhtar T., Tariq M. Incidence and distribution of Zucchini yellow mosaic virus (ZYMV) infecting Cucumber (Cucumis sativus L) crop in Pothowar, Pakistan. Pure Appl. Biol. 2019;8:2036–2043. doi: 10.19045/bspab.2019.80148. [DOI] [Google Scholar]
- 35.Ali A., Natsuaki T., Okuda S. The complete nucleotide sequence of a Pakistani isolate of Watermelon mosaic virus provides further insights into the taxonomic status in the Bean common mosaic virus subgroup. Virus Genes. 2006;32:307–311. doi: 10.1007/s11262-005-6915-z. [DOI] [PubMed] [Google Scholar]
- 36.Pavel A.B., Vasile C.I. PyElph-a software tool for gel images analysis and phylogenetics. BMC Bioinform. 2012;13:1–6. doi: 10.1186/1471-2105-13-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maina S., Coutts B.A., Edwards O.R., de Almeida L., Kehoe M.A., Ximenes A., Jones R.A. Zucchini yellow mosaic virus populations from east Timorese and northern Australian cucurbit crops: Molecular properties, genetic connectivity, and biosecurity implications. Plant Dis. 2017;101:1236–1245. doi: 10.1094/PDIS-11-16-1672-RE. [DOI] [PubMed] [Google Scholar]
- 38.Maina S., Barbetti M.J., Edwards O.R., Minemba D., Areke M.W., Jones R.A. Zucchini yellow mosaic virus genomic sequences from Papua New Guinea: Lack of genetic connectivity with Northern Australian or East Timorese genomes, and new recombination findings. Plant Dis. 2019;103:1326–1336. doi: 10.1094/PDIS-09-18-1666-RE. [DOI] [PubMed] [Google Scholar]
- 39.Holmes E.C. The evolution of viral emergence. Proc. Natl. Acad. Sci. USA. 2006;103:4803–4804. doi: 10.1073/pnas.0601166103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu Z., Mo C., Zhang S., Li H. Characterization of papaya ringspot virus isolates infecting transgenic papaya ‘Huanong No. 1’in South China. Sci. Rep. 2018;8:8206. doi: 10.1038/s41598-018-26596-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gorane A., Verma R., Naik A., Nikam T., Ade A., Tripathi S. Genetic variability and recombination analysis of papaya infecting isolates of Papaya ringspot virus based on CP gene from Western India. Arch. Phytopathol. Plant Prot. 2021;54:2294–2305. doi: 10.1080/03235408.2021.1975463. [DOI] [Google Scholar]
- 42.Mavrodieva V., James D., Williams K., Negi S., Varga A., Mock R., Levy L. Molecular analysis of a Plum pox virus W isolate in plum germplasm hand carried into the USA from the Ukraine shows a close relationship to a Latvian isolate. Plant Dis. 2013;97:44–52. doi: 10.1094/PDIS-01-12-0104-RE. [DOI] [PubMed] [Google Scholar]
- 43.Li Y., Liu R., Zhou T., Fan Z. Genetic diversity and population structure of Sugarcane mosaic virus. Virus Res. 2013;171:242–246. doi: 10.1016/j.virusres.2012.10.024. [DOI] [PubMed] [Google Scholar]
- 44.Xie X., Chen W., Fu Q., Zhang P., An T., Cui A., An D. Molecular variability and distribution of Sugarcane mosaic virus in Shanxi, China. PLoS ONE. 2016;11:e0151549. doi: 10.1371/journal.pone.0151549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bousalem M., Douzery E., Fargette D. High genetic diversity, distant phylogenetic relationships and intraspecies recombination events among natural populations of Yam mosaic virus: A contribution to understanding potyvirus evolution. J. Gen. Virol. 2000;81:243–255. doi: 10.1099/0022-1317-81-1-243. [DOI] [PubMed] [Google Scholar]
- 46.Wylie S., Jones R. Role of recombination in the evolution of host specialization within Bean yellow mosaic virus. Phytopathology. 2009;99:512–518. doi: 10.1094/PHYTO-99-5-0512. [DOI] [PubMed] [Google Scholar]
- 47.Cuevas J.M., Delaunay A., Visser J.C., Bellstedt D.U., Jacquot E., Elena S.F. Phylogeography and molecular evolution of Potato virus Y. PLoS ONE. 2012;7:e37853. doi: 10.1371/journal.pone.0037853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahmad A., Ashfaq M. Genetic diversity and recombination analysis based on capsid protein gene of Chilli veinal mottle virus isolates from Pakistan. Eur. J. Plant Pathol. 2018;151:891–900. doi: 10.1007/s10658-018-1423-x. [DOI] [Google Scholar]
- 49.Sztuba-Solińska J., Urbanowicz A., Figlerowicz M., Bujarski J.J. RNA-RNA recombination in plant virus replication and evolution. Annu. Rev. Phytopathol. 2011;49:415–443. doi: 10.1146/annurev-phyto-072910-095351. [DOI] [PubMed] [Google Scholar]
- 50.Khan M.G., Reddy K.G., Rao D.K. Designing stratified sampling in economic and business surveys. J. Appl. Stat. 2015;42:2080–2099. doi: 10.1080/02664763.2015.1018674. [DOI] [Google Scholar]
- 51.Clark M.F., Adams A. Characteristics of the microplate method of enzyme-linked immunosorbent assay for the detection of plant viruses. J. Gen. Virol. 1977;34:475–483. doi: 10.1099/0022-1317-34-3-475. [DOI] [PubMed] [Google Scholar]
- 52.Ashfaq M., Iqbal S., Mukhtar T., Shah H. Screening for resistance to cucumber mosaic cucumovirus in chilli pepper. J. Anim. Plant Sci. 2014;24:791–795. [Google Scholar]
- 53.Ahsan M., Ashfaq M., Riaz H., Khan Z., Hamza M., Asad Z. Genetic diversity and molecular characterization of Cucumber mosaic cucumovirus (CMV) subgroup II infecting Spinach (Spinacia oleracea) and Pea (Pisum sativum) in Pothwar region of Pakistan. Braz. J. Biol. 2021;83:e245865. doi: 10.1590/1519-6984.245865. [DOI] [PubMed] [Google Scholar]
- 54.Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R.D., Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Z., Schwartz S., Wagner L., Miller W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 56.Stephen F.A. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumar S., Stecher G., Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hall T.A. Proceedings of the Nucleic Acids Symposium Series. Oxford University Press; Oxford, UK: 1999. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT; pp. 95–98. [Google Scholar]
- 59.Martin D.P., Murrell B., Khoosal A., Muhire B. Bioinformatics. Springer; Berlin/Heidelberg, Germany: 2017. Detecting and analyzing genetic recombination using RDP4; pp. 433–460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the information and data have been added to the manuscript. However, the details of genetic sequences have been uploaded to GenBank, NCBI, and can be accessed using the accession numbers which are publicly available.