Abstract
Bacterial lineages acquire novel traits at diverse rates in part because the genetic background impacts the successful acquisition of novel genes by horizontal transfer. Yet, how horizontal transfer affects the subsequent evolution of core genes remains poorly understood. Here, we studied the evolution of resistance to quinolones in Escherichia coli accounting for population structure. We found 60 groups of genes whose gain or loss induced an increase in the probability of subsequently becoming resistant to quinolones by point mutations in the gyrase and topoisomerase genes. These groups include functions known to be associated with direct mitigation of the effect of quinolones, with metal uptake, cell growth inhibition, biofilm formation, and sugar metabolism. Many of them are encoded in phages or plasmids. Although some of the chronologies may reflect epidemiological trends, many of these groups encoded functions providing latent phenotypes of antibiotic low-level resistance, tolerance, or persistence under quinolone treatment. The mutations providing resistance were frequent and accumulated very quickly. Their emergence was found to increase the rate of acquisition of other antibiotic resistances setting the path for multidrug resistance. Hence, our findings show that horizontal gene transfer shapes the subsequent emergence of adaptive mutations in core genes. In turn, these mutations further affect the subsequent evolution of resistance by horizontal gene transfer. Given the substantial gene flow within bacterial genomes, interactions between horizontal transfer and point mutations in core genes may be a key to the success of adaptation processes.
Keywords: espistatic interactions, mutations, gene gain and loss, quinolones, mobile genetic elements
Introduction
Bacterial populations adapt rapidly to novel challenges such as bacteriophage predation, antibiotic therapy, or environmental perturbations. Adaptation is facilitated by point mutations and by genetic exchanges with other bacteria. Since the genome is made of thousands of genes linked in complex regulatory networks and encoding proteins with multiple physical interactions, these modifications may impact other processes than those directly involved in adaptation. The trade-off between the benefits and the cost of the novel variants shapes the outcome of their natural selection. Several studies have shown how the acquisition of novel functions by horizontal gene transfer (HGT) depends on the existing genetic background (Press et al. 2013; Szappanos et al. 2016). For example, metabolic pathways grow by transfer of genes encoding enzymes involved in reactions at the periphery of existing networks (Pal et al. 2005), whereas genome reduction involves an ordered loss of specific functions (Moran and Mira 2001; Pal et al. 2006; Tamames et al. 2007). Interactions between the genetic background and genes acquired horizontally may tune the probability of fixation of the latter. Such interactions may be a key to understand the adaptation of bacteria because their gene repertoires vary rapidly by HGT. For example, less than half of the average Escherichia coli genome corresponds to genes present in more than 99% of the strains (core genome). Consequently, the diversity of E. coli gene repertoires, its pan-genome, is more than an order of magnitude larger than the average genome (Touchon et al. 2009; Cummins et al. 2022). The large genomic variation caused by HGT and gene loss means that genetic backgrounds can be very different within a species. Hence, not only HGT may be affected by the genetic background, it is likely that the evolutionary trajectories of core genes are also affected by changes in gene repertoires (Batarseh et al. 2023). There are very few studies on the existence of the epistatic interactions between HGT and point mutations in core genes.
The evolution toward antibiotic resistance in bacterial pathogens (and commensals) is a recent example of the ability of bacteria to rapidly adapt to novel challenges (MacLean and San Millan 2019). Bacteria can counteract antibiotic therapies by diminishing the intracellular concentration of the antibiotic by reducing its influx or increasing its efflux. They can also protect the target of the antibiotic or modify it by mutation. Finally, they can inactivate the antibiotic using appropriate enzymes or evolve alternative pathways that make the target redundant (target bypass) (Munita and Arias 2016; Darby et al. 2023). These mechanisms may be acquired by mutation or by HGT (Davies and Davies 2010). While high environmental pressure imposed by antibiotics is central to the emergence of antibiotic resistance (Karve and Wagner 2022; Ghenu et al. 2023), epistatic interactions were shown to shape it (Weinreich et al. 2006; Salverda et al. 2011; Brandis et al. 2012; Brandis and Hughes 2013; San Millan et al. 2014). The evolutionary trajectories toward resistance are constrained by the existence of favorable mutational paths where intermediate steps have lower-than-average fitness costs and/or higher-than-average resistance (Rozen et al. 2007; Marcusson et al. 2009; Knopp and Andersson 2018; Patiño-Navarrete et al. 2020). Epistatic interactions may result in the fitness cost of a resistance being alleviated by the presence of another one, which may favor the evolution of multidrug resistance (Angst and Hall 2013; Borrell et al. 2013; Knopp and Andersson 2015; Moura de Sousa et al. 2017). For instance, the high transmission fitness of multiple drug resistance Mycobacterium tuberculosis strains of lineage 2 resulted from epistatic interactions between compensatory mutations in RNA polymerase and the rifampicin resistance-conferring mutation RpoB S450L (Loiseau et al. 2023). On the other hand, collateral sensitivity occurs when resistance to an antibiotic is linked with an increased susceptibility to another antibiotic (Hughes and Andersson 2017; Darby et al. 2023).
Differences in the genetic background influence the evolution of antibiotic resistance (Vogwill et al. 2016; Castro et al. 2020; Torres Ortiz et al. 2021), even under strong selection (Santos-Lopez et al. 2021). However, few studies identified chronologies between specific changes in the genetic background and the acquisition of the antibiotic resistance (Wong 2017). Here, we define chronologies as a set of events ordered in time. In a seminal study, the fitness cost of chromosomal resistance to several antibiotics acquired by point mutations was found to be in negative epistatic association with the presence of conjugative plasmids in more than half of the tested combinations (Silva et al. 2011). More recently, the presence of the efflux pump norA potentiated the subsequent evolution of point mutation conferring resistance to quinolones in Staphylococcus aureus (Papkou et al. 2020). Finally, the ability of E. coli to evolve high-level colistin resistance by point mutation in the core gene lpxC increased in the presence of a plasmid carrying the mcr-1 colistin resistance gene (Jangir et al. 2022). Epidemiological data seem to confirm these trends. For example, the E. coli strains from the clone ST131 comprise now the majority of extended spectrum beta-lactamase (ESBL)–producing isolates from the species (acquired by HGT), and they are almost invariably also resistant to fluoroquinolones (acquired by point mutations) (Nicolas-Chanoine et al. 2014). Hence, interactions between genes acquired by HGT and chromosomal mutations may be important. Given the differences in terms of genetic backgrounds across a bacterial species, caused by rampant HGT, epistatic interactions may contribute to explain why strains from certain lineages are more often antibiotic resistant than others (Leavis et al. 2006; Wyres et al. 2020).
We study the evolution of resistance to quinolones, a large group of broad-spectrum bactericidals widely used in human and veterinary medicine (Brown 1996; Redgrave et al. 2014; Bisacchi 2015). In 2017, they represented 9.5% of the antibiotic usage in the 30 EU/European Economic Area (EEA) countries (Bruyndonckx et al. 2021). Quinolones disrupt the function of the bacterial type II topoisomerases by inhibiting the catalytic activity of DNA gyrase (gyrA and gyrB) and topoisomerase IV (parC and parE). DNA gyrase introduces supercoils, and DNA topoisomerase IV prevents supercoiling from reaching unacceptably high levels. These proteins intervene during replication or transcription, and their mechanism involves the creation of a double strand break that is subsequently ligated. Quinolones stabilize the cleavage complex preventing the enzymes from performing the ligation step, thereby resulting in double strand breaks that may lead to replication stalling and cell death (Hopkins et al. 2005; Aldred et al. 2014; Pham et al. 2019). The primary target for quinolones is the gyrase in enterobacteria and the topoisomerase in Firmicutes (Hopkins et al. 2005). The mechanisms that provide high resistance to quinolones have been characterized in detail. They involve mutations in quinolone-resistance determining regions (QRDRs) of the target proteins. In E. coli, mutations appear most frequently at codons 83 and 87 of gyrA, near the active site of the gyrase, altering the binding of quinolones and reducing the cell's susceptibility to them (Willmott and Maxwell 1993). Mutations in gyrB are rarer, and some provide higher susceptibility to specific quinolones, a trait that can be masked by epistatic interactions with gyrA mutations (Herrera et al. 1993). Mutations in the topoisomerase subunits parC and parE of E. coli usually cooccur with mutations in gyrA (Everett et al. 1996; Heisig 1996), suggesting that the mutations in topoisomerase will not be fixed unless the sensitivity of the DNA gyrase has been reduced. Interestingly, a parC mutation was shown to strongly alleviate the cost of gyrA mutations. The relevance of epistatic interactions in the target protein mutations leading to quinolone resistance has been shown in many species, e.g. E. coli (Zhao et al. 1997; Marcusson et al. 2009), Streptococcus pneumoniae (Pan et al. 2001), Pseudomonas aeruginosa (Wong and Kassen 2011), and M. tuberculosis (Castro et al. 2020). A combination of sequence analysis of 195 resistant clinical isolates, experimental work, and mathematical modeling revealed the major trajectories of ciprofloxacin-resistance evolution in E. coli, explaining the prevalence of a few dominant genotypes and the order of accumulation of the mutations (Huseby et al. 2017). Epistatic interactions with other determinants of resistance for other antibiotics have also been reported (Trindade et al. 2009; Imamovic and Sommer 2013; Lázár et al. 2018) and shown to be sensitive to changes in the genetic background (Apjok et al. 2019). Of note, lower resistance level to quinolones can also be provided by plasmid-borne genes such as qnrA, qepA, AAC(6′)-Ib, and oqxAB (Martinez-Martinez et al. 1998; Robicsek et al. 2006; Hansen et al. 2007; Yamane et al. 2007).
Here, we wished to know if there is evidence in natural populations that gene gains and losses change the propensity of bacteria to adapt by specific point mutations that are known to result in antibiotic resistance. For this, we identified events of gene gain and loss in the E. coli phylogenetic tree that occurred systematically in branches preceding those where resistance to quinolones took place. This approach counts events of change in the phylogenetic tree and correlates them. It thus allows to explicitly account for population structure (if dozens of genomes with a specific mutation descend from one single ancestral event mutation, only one event is counted). We then computed the induction effect (termed here lambda) that one event has on the other, i.e. if a given gene gain or loss increased or decreased the frequency of subsequent acquisition of antibiotic resistance by point mutations. Of note, this approach is different from the ones used in bacterial GWAS where the goal is to identify frequent cooccurrences of events in the same branch or simultaneous presence of genes in extant taxa (Collins and Didelot 2018; Jaillard et al. 2018; Lees et al. 2018; Whelan et al. 2020). Here, the goal is to identify changes that were fixed in the lineage before the acquisition of mutations conferring resistance to quinolones. We focus on the latter because they are well known and occur in core essential genes. Also, it was shown that the same point mutation in gyrA conferring resistance to quinolones can have very different costs depending on the genetic background (Luo et al. 2005), a clear indication that preexisting variation in the latter may shape evolution in core genes.
Results
Frequency of the Mutations Providing Resistance to Quinolones
We analyzed 1 268 complete genomes of E. coli from RefSeq and 877 draft genomes of isolates sampled across multiple sources in Australia (named AUS [Touchon et al. 2020]). The first is our reference dataset, and the second is mentioned occasionally because it has much fewer clinical isolates (but the genomes are not fully assembled). The 2,145 genomes have 78,653 different gene families (pangenome) and 2,393 core gene families (genes present in a single copy in at least 99% of the genomes). The latter was used to construct a phylogenetic tree for each dataset (Fig. 1; supplementary fig. S1, Supplementary Material online). We built the trees using the same set of core genes to have comparable phylogenies and phylogeny-based analyses in the 2 datasets. The trees were well-resolved with more than 85% of the branches having more than 90% UltraFast Bootstrap.
Fig. 1.
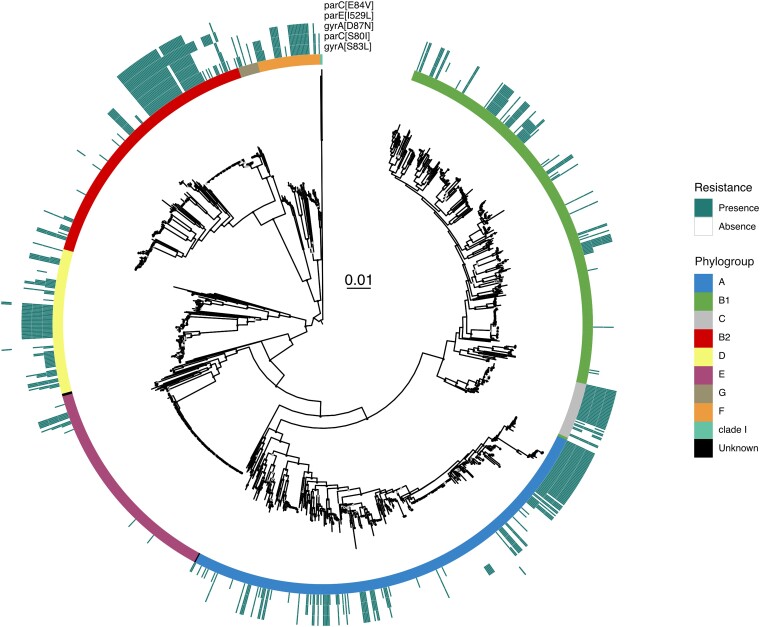
Phylogeny and distribution of quinolone resistance mutations in DNA gyrase (gyrA) and topoisomerase IV (parE and parC) genes in E. coli (RefSeq dataset). The maximum likelihood phylogenetic tree of the species was built with IQTree (model GTR + F + I + G4 and 1,000 ultrafast bootstraps), and its scale (in substitutions per site) is in the center. The tree was midpoint rooted. The E. coli phylogroups are represented by the colors in the inner center. The outer circles indicate the presence of the most frequent mutations across the species. Ultrafast bootstrap values superior to 75% are shown with a light gray circle and values superior to 90% with a black circle (see Supplementary material online).
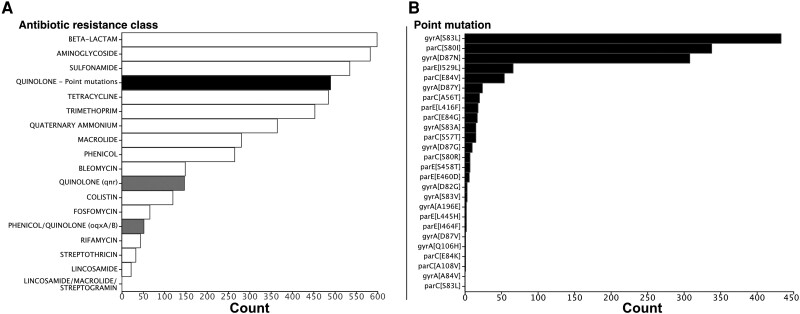
We extracted from the proteome of the RefSeq dataset the sequences of the gyrA, gyrB, parC, and parE genes. We screened these sequences for 39 different quinolone resistance mutations retrieved from the literature (supplementary table S1, Supplementary Material online). The mutations were remarkably frequent in the RefSeq dataset, where 39% of the strains had at least one mutation (supplementary table S2, Supplementary Material online). As a comparison, the AUS dataset had fewer resistance mutations (11% of the strains, supplementary table S3, Supplementary Material online), and these were concentrated in the human and poultry isolates (86.4%, supplementary table S4, Supplementary Material online). Hence, the absolute frequency of these mutations in the RefSeq dataset should be interpreted in the light of the presumed high frequency of clinical strains in the set. The search for antibiotic resistance genes (ARGs) in the RefSeq dataset showed that resistance to quinolones (including the point mutations, qnr/qep, and the plasmid-encoded multidrug efflux pump oqx) is the most frequent in the species, since 637 strains of 1,268 have at least one of the 3 types of resistance (Fig. 2a; supplementary table S5, Supplementary Material online). Among these mechanisms of resistance to quinolones, the ones based on mutations were by far the most frequent.
Fig. 2.
Distribution of the antibiotic resistance classes detected in the RefSeq dataset. a) Antibiotic resistance classes are ordered according to their frequency (total number in the dataset). Resistances to quinolone are separated into 3 distinct classes: resistance conferred by point mutations of core genes (black), by acquisition of qnr genes or oqx genes (gray), b) Mutations are ordered according to their frequency. The first letter in the name in the square brackets correspond to the ancestral amino acid, the number to the position of the transition, and the last letter to the amino acid conferring the resistance, e.g. gyrA[S83L] means that the mutation is in gyrA at position 83 and involves a substitution Ser->Leu.
The mutations conferring resistance to quinolones had very different prevalence in the species (Fig. 2b). The single mutation gyrA[S83L], which confers a significant increase in resistance at a low cost (Marcusson et al. 2009), is present in 34% of the strains. The 2 other most frequent mutations (parC[S80I] and gyrA[D87N]) were both present in more than 26% and 24% of the strains, respectively. The list of mutations in order of frequency then includes a mutation in parE and another in parC. Of note, mutations in parE and gyrB are much less frequent than those in gyrA and parC. Apart from the sheer abundance of mutations, higher in the RefSeq than in the genomes of the Australian (AUS) dataset (see Methods section), both datasets showed similar patterns in terms of the most frequent types of mutations (supplementary fig. S2, Supplementary Material online). Hence, and in agreement with the literature (Saenz et al. 2003; Johnning et al. 2015), a group of 3 mutations in 2 (of the 4) key genes targeted by quinolones (gyrA[S83L], gyrA[D87N], and parC[S80I]) are by far the most frequent in E. coli.
We then mapped the resistant isolates on the phylogenetic tree. We found them scattered across the species. They are present in all phylogroups, but their prevalence can be quite different. In phylogroups E and B1, these mutations are rare, whereas in phylogroup F, they are very frequent. Hence, there is a nonuniform distribution of mutations for resistance across the species, with certain clades containing many more mutations than others (Fig. 1). The AUS dataset shows similar results, with high frequency of resistance mutations in phylogroup F and low in E and B1 (supplementary fig. S1, Supplementary Material online). Overall, the clustered distribution of resistances and the frequent presence of multiple mutations suggest that they accumulate in nonrandom ways.
Order of Acquisition of the Mutations Conferring Resistance to Quinolone
The phylogenetic analysis shows a strong cooccurrence of the different mutations conferring quinolone resistance. It is well known that the combination of gyrA[S83L], parC[S80I], and gyrA[D87N] confers high level of resistance with limited fitness cost (Bagel et al. 1999). These 3 mutations cooccurred much more frequently together (182) than separately (90). The other low-frequency mutations are often associated with them (supplementary fig. S3, Supplementary Material online). Only one relatively rare mutation (gyrA[S83A]) was more typically found alone than in combination with the 3 major ones (supplementary table S2 and fig. S3, Supplementary Material online). The systematic identification of large combinations of mutations raises the possibility that there exist typical chronologies for their accumulation. Indeed, it was shown using a combination of experimental biology and modeling that given the resistance they provide, the most likely path to adaptation was gyrA[S83L] -> parC[S80I] -> gyrA[D87N] (Marcusson et al. 2009; Huseby et al. 2017).
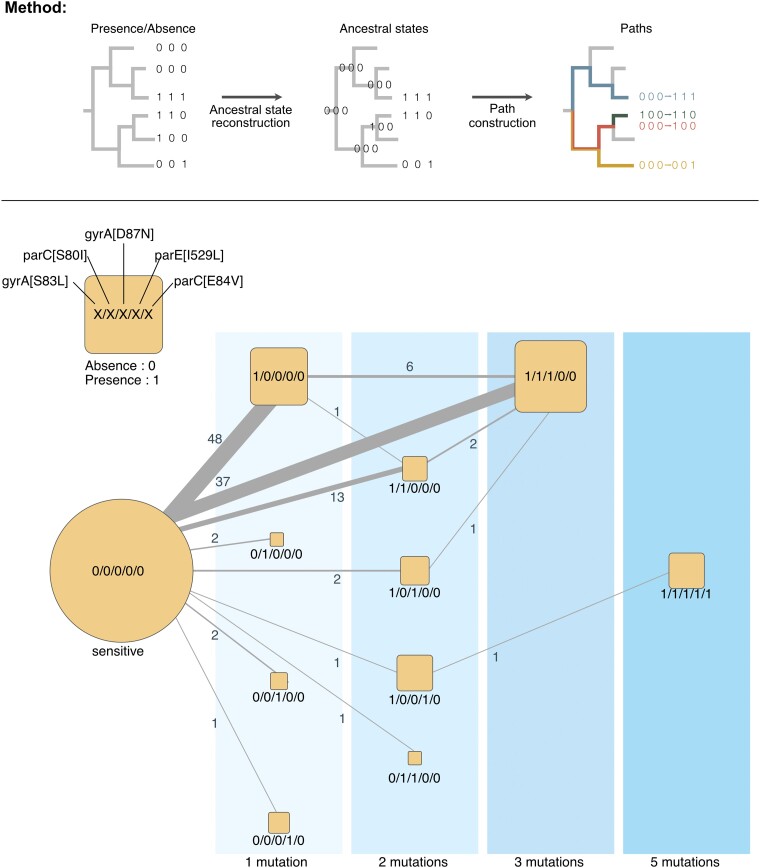
To test this hypothesis, we searched for the preferential chronologies of the 5 main mutations on the RefSeq data using the species phylogenetic tree (see Methods section; Fig. 3). We did not consider the reverse mutations because these are very rare (1.7% of all paths across the tree) and because reversions are sometimes associated with poorly resolved regions of the tree and may thus be spurious. We observed similar trajectories toward the acquisition of the 3 main mutations in AUS dataset (Figs. 3; supplementary fig. S4 and tables S6 and S7, Supplementary Material online). In the RefSeq data, we identified 48 occurrences of lineages acquiring only the gyrA[S83L] mutation. This suggests that the mutation gyrA[S83L] is the first one fixed in most lineages. Some of the clones with this mutation then gave rise to the triple mutated strain. The other most frequent trajectory was the one going from the fully sensitive combination to the triple mutant (37 occurrences). Hence, our analysis failed to reveal unequivocally the substitution paths from the ancestral sensitive state to the triple mutant (gyrA[S83L], parC[S80I] and gyrA[D87N]). This suggests that the other 2 major mutations are acquired and fixed very quickly, such that they are inferred to arrive jointly in a single branch of the phylogenetic tree.
Fig. 3.
Chronologies of acquisition of the main resistance mutations in E. coli of the RefSeq dataset. Graphical representation of the method: Starting from the phylogenetic tree and the presence/absence matrix of the mutation conferring the resistance to quinolone, we inferred the ancestral state of each mutation at every node. These states were used to reconstruct the chronologies of acquisition of the mutation conferring resistance to quinolone. Reversions were very rare (<1% of all transitions) and were not plotted for clarity. The chronologies identified in this figure represent successions of events in the tree and thus account explicitly for the population structure. The vertical areas represent the number of distinct mutations in genomes. The size of the square scales with the number of genomes observed (leaves of the tree). The edges represent the chronologies of acquisition of one or several mutations as inferred from the reconstruction of ancestral states on the tree. The edge size is proportional to the frequency of the respective transition, with the labels showing this exact number.
To identify the paths toward the triple mutant, we used 3 genomic datasets already available in our laboratory consisting of very closely related genomes (one single ST): 757 of ST410, 85 ST131, and 579 for ST167. In ST410, of the 757 strains, most (656) contain mutations conferring resistance to quinolone. This trait is not ancestral, as shown by the phylogenetic reconstruction of the sequences (supplementary fig. S5, Supplementary Material online). This is consistent with previous works, which dated the emergence of quinolone resistance within ST131 or ST410 (some basal ST131/ST410 lineages still missing it) in the early 1990s (Stoesser et al. 2016; Roer et al. 2018). Yet, almost all the resistant clones (653/656) harbor the gyrA[D87N]/gyrA[S83L]/parC[S80I] combination of mutations, and only 2 among the basal isolates contain the gyrA[S83L] alone. We found similarly inconclusive results for the other STs. Hence, even at this fine level of granularity, one cannot reconstruct the precise order of fixation of the 3 mutations (supplementary figs. S6–S10 and table S8, Supplementary Material online).
The quick accumulation of the 3 mutations could be due to homologous recombination replacing in one single event the ancestral sequences by the triple mutant. This seems unlikely because parC and gyrA are 1.39 Mb apart in the strain MG1655, and even parC and parE are more than 8 kb part (while observed average recombination tracts in E. coli are less than 1 kb [Didelot et al. 2012]). Still, we searched for evidence of recombination in the genomes using Gubbins. This analysis showed that in 7 of the 39 events of acquisition of the 2 mutations in gyrA, there was a recombination tract that was acquired overlapping the position of the mutations at the same time (same branch in the tree). This suggests that recombination may occasionally contribute to the emergence of the double mutant in the lineages. Yet, of the 37 events of acquisition of the 3 mutations in a branch of the tree, only one coincided with the acquisition of recombination tracts at both genes (different tracts). Overall, these results suggest that gyrA[S83L] is the main initial driver of the evolution of quinolone resistance in E. coli and that the subsequent mutations are fixed almost simultaneously by rapid accumulation of novel point mutations, although recombination may occasionally accelerate the process.
Clusters of Gene Gains and Losses Shaping the Emergence of Resistance Mutations
To investigate how the dynamics of gene repertoires favor the acquisition of resistance to quinolones by point mutations in the core genes targeted by the antibiotics, we searched for genes that were frequently gained or lost before the emergence of these mutations. Since most resistance mutations cooccurred, and they were all shown to have significant individual effects on resistance, we defined taxa as resistant when they had at least one resistance mutation. We then reconstructed the ancestral states of resistance (as defined above) and of each gene family of the pan-genome to infer the moments of gains and losses of the gene families and of the resistance. Finally, we used Evo-Scope to compare the chronology of gains and losses of every family in the pangenome with the acquisition of the resistance to the antibiotic. Of note, this software searches for successions of events in different branches of the tree. The 2 events are thus well-separated in time. This allowed to identify the significant chronologies, i.e. when one event of gene gain or loss was followed by the acquisition of resistance more frequently than expected given the frequency of events and their distribution on the phylogenetic tree (Behdenna et al. 2016). We identified 183 gene gains and 26 gene losses occurring frequently before the acquisition of the resistance (P < 10−5 after correction for multiple tests, Fig. 4, supplementary table S9, Supplementary Material online, P-value distributions in supplementary figs. S11 and S12, Supplementary Material online).
Fig. 4.
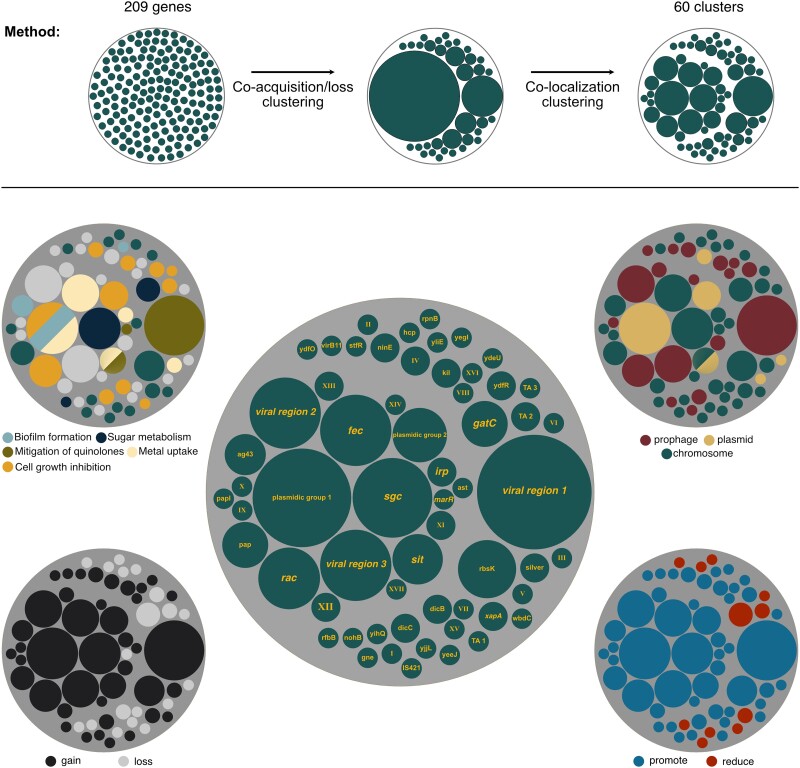
Cluster of genes consistently acquired or lost prior the acquisition of the quinolone resistance. Graphical representation of the method: the 209 genes were clustered in 2 steps using hierarchical clustering. Genes were initially clustered if they were often coacquired or colost in the branches of the tree and then clustered if they were colocalized in the genome. The center figure represents the different clusters according to their size. Clusters with known functions or encoded by a specific mobile genetic element type were named accordingly. Others were given roman numbers. Clusters are represented according to their location on prophage, plasmid, or chromosome (top right), to their influence on the acquisition of the quinolone resistance (bottom right), to the nature of the event (bottom left), and to the functions they encode (top left).
Our method searches for preferential chronologies between a type of event that occurs in a set of branches of the phylogenetic tree and resistance mutations that emerge later (on higher branches). It is not aimed at identifying events that are associated, such as genes directly involved in the phenotype. Yet, it may also reveal associations in the sense usually employed in genome-wide association studies (GWAS). To assess if chronologies differ significantly from GWAS, we performed a GWAS analysis to identify genes whose presence or absence is associated with quinolone resistance. We then identified those positively or negatively associated with the trait (quinolone resistance) while accounting for the phylogenetic structure of the data using a linear mixed model (LMM) (see Methods section). This analysis yielded no significant associations at the same statistical threshold (correction for multiple tests and P < 10−5). Lowering the significance threshold revealed 53 genes (P < 0.01). Most of the 20 genes positively associated with quinolone resistance encode transporters, regulators, transposases, or enzymes associated with antibiotic resistance (e.g. beta-lactams or tetracycline, supplementary table S10, Supplementary material online). None of the 209 genes involved in the chronologies identified by the Evo-Scope analysis as occurring more often than expected before the resistance mutations was identified by the GWAS analysis. This may seem surprising, but while requiring frequent ordered acquisition of the 2 traits, our method does not require a very significant overlap of the presence of the genes with the resistance, contrary to the GWAS. For instance, the gene encoding the toxin RelE/ParE (not to be confounded with the topoisomerase ParE) was yielded by the Evo-Scope analysis as significantly acquired before the acquisition of the resistance to quinolone (e-value = 8.6 × 10−11). However, this gene was not detected as significantly associated with the resistance by the GWAS analysis (e-value = 0.275). The gene was acquired 130 times and lost 21 times in the tree and is present in 192 genomes (of which 60 have the resistance mutations, i.e. a fraction close to the one observed across the species). This suggests that our chronologies identify events that favor the acquisition of resistance without contributing very strongly to it. This is not very surprising, considering that strong resistance is well-known to be due to mutations in a few core genes (and that was how it was defined in the GWAS).
Many of the events identified in the chronologies were acquisitions (see below, Fig. 4). Genes acquired by HGT are often initially grouped in large mobile genetic elements (MGEs) or at least in fragments of DNA with more than one gene. If one of these genes is biologically associated with the emergence of resistance, other genes under strong genetic linkage, e.g. entering the genome in the same event of transfer, may also be identified as significantly associated with the emergence of resistance. Indeed, an analysis of the presence of the 209 genes revealed cooccurrence of many of them (supplementary fig. S13, Supplementary Material online). To control for the effect of genetic linkage, we clustered the genes in function of their tendency to be acquired or lost in the same branches of the species tree (supplementary tables S11–S13, Supplementary Material online). We then subdivided these clusters using information on genomic colocalization to group genes that tend to be neighbors (see Methods section; Fig. 4; supplementary table S14, Supplementary Material online). This led to the identification of 26 clusters of genes and 34 isolated genes (singletons) (Fig. 4; supplementary table S15, Supplementary Material online). Hence, our analysis suggests that there is a minimal number of 60 genes whose gain/loss affect the probability of acquisition of resistance mutations.
The Evo-Scope method provides a parameter (λ), which is a measure of the impact of an event (here gene gain or loss) in increasing the likelihood of another (acquisition of resistance mutations) in a higher branch of the tree. These values were not used for the clustering but are expected to be similar among genes under strong genetic linkage. Indeed, genes in clusters had more homogeneous lambda values relative to the others (levene Test: P-value = 5.71 × 10−06, mean square of the error = 0.591, one-way analysis of variance ***), meaning that genes within the same cluster have approximately similar impacts on the acquisition rate of the quinolone resistance from the statistical point of view. Of the 60 clusters, the majority (49) promotes the acquisition of the resistance mutations (λ>1), while only 11 clusters reduce it (λ<1) (Fig. 4, bottom right). Most of the latter are close to the significance threshold. This means that most of the significant events of gene gain and loss associated with mutations tend to increase the likelihood of the resistance mutations.
The clustering procedure aimed at identifying blocks of genes that were acquired at the same time (along the same branch). As expected, all the clusters were exclusively made of groups of either lost or gained genes, not both. Of the 60 clusters, the majority (41) correspond to gene gains and 19 correspond to gene losses. One might expect that the first correspond to gain of functions and the latter to losses. Bacteria need to acquire all the genes involved in a function to express it. However, the loss of only few genes is sufficient to disrupt the activity of entire pathways. This may explain why our analysis reveals that groups of gains are larger than groups of losses (Mann–Whitney U test: P-value = 0.0108) (Fig. 4, bottom left). The acquisition of functions in large MGEs may also contribute to explain these results. Except for one cluster that was either part of a plasmid or of a chromosome (depending on the genome), clusters were made exclusively of plasmid, prophage genes, or neither of the 2. Eighteen clusters, including the largest, were in prophages. All the prophage clusters are associated with promotion of resistance mutations. Five clusters were on plasmids including the second largest (Fig. 4, top right). In conclusion, our analysis identified 60 groups of genes, most of them corresponding to genetic acquisitions. Many of these are in MGEs and they tend to increase the likelihood of acquisition of resistance mutations.
Acquired Functions That Induced Subsequent Resistance Acquisition
Among the 60 clusters identified above, 17 lack genes with predicted functions. The other clusters tend to have many genes involved in closely related functions, organized in loci with the characteristics of operons (cooriented closely spaced genes). The analysis of the AUS dataset revealed much fewer genes (62) significantly associated with the emergence of quinolones (supplementary table S16, Supplementary Material online), which is probably caused by the much lower frequency of resistance in that dataset. Of these, 27 were also identified in the RefSeq analysis (supplementary table S17, Supplementary Material online). Most of the other are of unknown function or related to MGEs. Among all the genes of known or putative function that were identified as affecting the emergence of the resistance mutations, we found a few recurrent broad functions involved in mitigation of the effect of quinolones, growth arrest, metal metabolism, biofilm formation, and sugar metabolism.
Mitigation of the Effect of Quinolones
The loss of the marR gene is associated with an increase in the probability of subsequent acquisition of resistance in the RefSeq dataset. Inactivating mutations in marR were previously found to be associated with decrease of sensitivity to different antibiotics including quinolones (Maneewannakul and Levy 1996). marR code for a repressor of marA, which code for a transcriptional activator of acrAB and tolC. Increase in the activity of the AcrAB-TolC multidrug efflux pump has been shown to increase resistance to quinolones (Wang et al. 2001). Inactivating mutations in marR are frequent in E. coli clinical isolates resistant to fluoroquinolones (Komp Lindgren et al. 2003). In viral region 1, we found a gene encoding DinI-like protein. DinI turns off the SOS responses through inhibition of the RecA coprotease activity (Yasuda et al. 1998). E. coli mutants deficient in SOS induction were previously shown to survive longer in the presence of several quinolones, suggesting that induction of the SOS response by quinolones is harmful to wild-type cells (Piddock and Walters 1992). Finally, the cluster named sit encodes CrcB, a plasmid-encoded protein whose overexpression not only increases the supercoiling level of plasmids but also reduces the sensitivity of gyrase and topoisomerase IV temperature-sensitive E. coli mutants to nalidixic acid (1st quinolone) (Sand et al. 2003).
Cell Growth Inhibition
Functions associated with bacterial growth arrest are amongst the most represented functions in the 60 groups of genes. Slow growth decreases the bactericidal efficacy of antibiotics, whether because it increases tolerance or persistence (Schumacher et al. 2009). Of note, evolution of tolerance was found to systematically precede the experimental evolution of resistance to ampicillin (Levin-Reisman et al. 2017). We found 8 clusters encoding toxin-antitoxin (TA) systems (NinE, rac, plasmidic group 1, plasmidic group 2, ydfR, TA1, TA2, and TA3) whose acquisition was inferred to increase the acquisition rate of resistance mutations. TAs have been describe to induce persister phenotypes that are highly tolerant to antibiotics, including quinolones (Dorr et al. 2010), although recent data have questioned this (Goormaghtigh et al. 2018). Three of the identified TA hits are from the ParDE family, which was described to contribute to more than a 1,000-fold increase in survival in the presence of supra-minimum inhibitory concentration (MIC) concentrations of quinolones (Kamruzzaman and Iredell 2019). Four groups encode cell division inhibitors (dicC, dicB, kill, and rac). Single-cell imaging showed that ofloxacin persisters formed polynucleoid filamentous cells. This phenotype was independent of the conserved filamentation inducer genes sulA or damX, suggesting that it was controlled by other cell division inhibitors (Goormaghtigh and Van Melderen 2019). Interestingly, it was previously found that cryptic prophages of E. coli contribute significantly to resistance to sublethal concentrations of quinolone through proteins that inhibit cell division, notably KilR of Rac and DicB of Qin (Wang et al. 2010), both of which were identified in our analysis. Hence, the presence of TA and cell division inhibitors seems to provide a favorable ground to the acquisition of resistance mutations, presumably because they provide more time for the mutations to emerge under antibiotic therapy.
Metal Uptake
Five clusters encode genes encoding proteins involved in the uptake of metals, such as iron, zinc, manganese, or copper. In all cases, the acquisition of these genes is associated with a subsequent increase in the rate of acquisition of quinolone resistance. Notably, 4 clusters (fec, sit, silver, and irp) encode almost exclusively genes involved in metals intake, showing that the effect is not related with genetic linkage to other functions. In viral region 2, there is a gene encoding a protein with a domain TonB/TolA, C-terminal. This domain is also involved in iron transport (Braun and Hantke 2011). The case of iron uptake could be regarded as surprising, because iron is associated with the creation of reactive oxygen species that were suggested to add to the toxicity of antibiotics. Yet, iron acquisition was shown to increase the MIC of quinolones in E. coli (Smith and Lewin 1988) and to promote the acquisition of quinolone resistance (Méhi et al. 2014). Moreover, several projects are considering therapies that combine ciprofloxacin and iron chelators that reduce the iron availability to bacteria to suppress the growth rate of drug-resistant subpopulations (Allan and Holbein 2022). Of note, 1 of the 2 groups of genes detected only in the AUS dataset consists of yet another iron siderophore systems of the aerobactin pathway of iron uptake (iucABD and iutA; supplementary table S16, Supplementary material online) (Challis 2005). Other metals, especially copper and zinc, were associated with elevated resistance rates against several antibiotics (Poole 2017). For example, minimal selective concentration for ciprofloxacin resistance increased up to 5-fold in the presence of zinc (Vos et al. 2020).
Biofilm Formation
We found 3 clusters with genes involved in biofilm formation. Of these, the acquisition of 2 clusters, ag43 and plasmidic group 1, is associated with increased probability of acquisition of resistance mutations. These clusters contain the Ag43 gene and the epsB/macA/macB genes, respectively, which enhance biofilm formation (Kjaergaard et al. 2000; van der Woude and Henderson 2008; Baugh et al. 2012; Gerwig et al. 2014). The third cluster contains only the chromosomal gene yliE/pdeI, whose overexpression reduces biofilm formation (Boehm et al. 2009) and whose mutant formed more biofilm in the mouse gut (Da Re et al. 2013). In our analysis, the loss of this cluster increases the acquisition rate of the quinolone resistance across the tree. Taken together, this suggests that a higher capacity to form biofilms increases the acquisition rate of quinolone resistance. Quinolones are known to be efficient at diffusing through biofilms. (Brooun et al. 2000). However, when compared with planktonic lifestyle, bacteria in biofilms developed more mutants with low-level resistance to quinolones. This is because the biofilm growth mode promotes the upregulation of efflux pumps (Ciofu et al. 2022). Even if these low-level resistances do not reach the clinical resistance level, biofilms might increase the time of survival of bacteria, giving them more opportunities for the emergence of mutations responsible for high-resistance levels to quinolone (Costerton et al. 1999; Lewis 2001).
Sugar Metabolism
Three clusters contain genes involved in sugar metabolism. Of these, 2 clusters, sgc and gatC, contain genes involved in the galactitol metabolism. The gain of sgc increases the rate of acquisition of the quinolone resistance, while gatC reduces it when lost. A recent screen showed that mutants in the galactitol pathway repressor have a reduced susceptibly to fluoroquinolone in Salmonella, suggesting that this pathway is associated with low-level resistance to the antibiotic (Turner et al. 2020). The 3rd cluster contains the rfbB gene that is involved in the dTDP-rhamnose biosynthesis (Marolda and Valvano 1995). Intracellular dTDP-rhamnose concentration correlates with the MIC of nalidixic acid and norfloxacin (Zampieri et al. 2017). This occurs because dTDP-rhamnose upregulates gyrA transcription in E. coli in the presence of norfloxacin and nalidixic acid. This helps cells to cope with the quinolones by sequestering the antibiotic and thereby reducing its effect. Taken together, these results suggest that the metabolism of certain sugars may facilitate the acquisition of quinolone resistance by contributing to reduced susceptibility.
Cooccurrence of the Quinolone Resistance with Other Antibiotic Resistance Classes
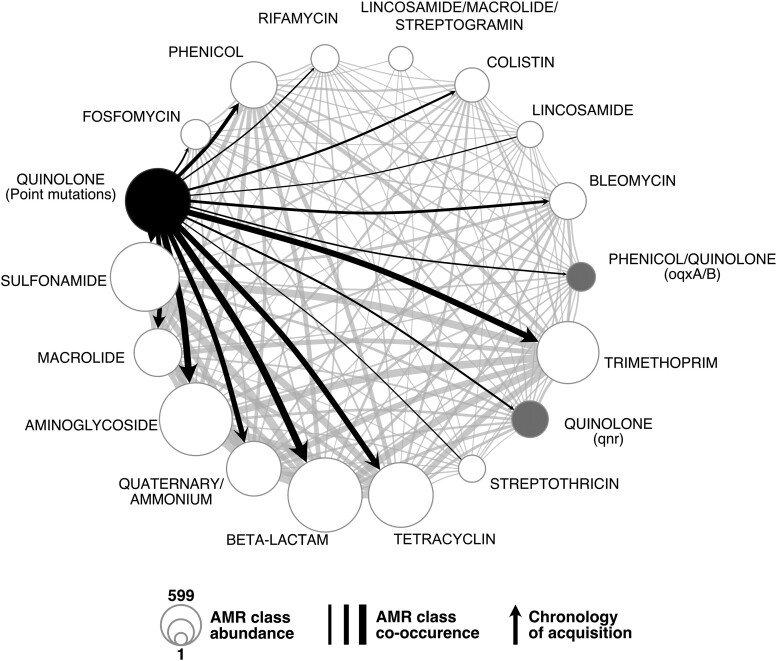
We have shown above that E. coli strains encode numerous ARGs (Fig. 2A; supplementary table S5, Supplementary Material online). Among the 490 RefSeq genomes with point mutations conferring resistance to quinolone, 400 also encode well-known determinants of resistance to beta-lactams, 372 to aminoglycosides, and 352 to sulfonamides (Fig. 5; supplementary fig. S14, Supplementary Material online). This raises the question of how the presence of one antibiotic resistance influences the acquisition of another. We screened our chronologies for ARGs systematically acquired before the resistance mutations and found none that was below the statistical significance level. Hence, it seems that in lineages resistant to quinolones, the acquisition of resistance to quinolones occurs early on, often being among the first resistance mechanisms acquired.
Fig. 5.
Cooccurrence of the different AMR classes (as defined by AMRfinder+), and the point mutations conferring the resistance to quinolone in the RefSeq dataset. The node size represents the number of times the resistance was found across the genomes. The resistances to quinolone conferred by point mutations and by qnr or oqx genes are represented in black and gray, respectively. The edge width represents the number of cooccurrences in a genome. Edges were converted into arrows when the chronologies between acquisition of mutations and the AMR classes were significant (mutations emerging first). When the resistance is conferred by other mechanisms, genes involved are annotated between brackets.
We then checked if other classes of ARGs were systematically acquired after the acquisition of the quinolone resistance mutations. We found 43 ARGs that were significantly more likely to be acquired after the emergence of quinolone resistance (Fig. 5; supplementary table S18 and fig. S15, Supplementary Material online). For these families, the acquisition of resistance to quinolone increased the rate of subsequent acquisition of the ARG (λ > 1). These families correspond to various antibiotic resistance classes among which the most diverse are for resistance to aminoglycosides (12/43), phenicol (6/43), beta-lactamases (5/43), and trimethoprim (5/43). Hence, resistance to quinolones seems to precede resistance to most other antibiotics. Alternatively, quinolone resistance may be less frequently lost and thus remain for longer periods in lineages than resistances that are very costly and/or encoded in MGEs that are easily lost.
As mentioned above, low-level resistance to quinolones can also be acquired through the acquisition of certain plasmid-associated genes. Interestingly, accumulation of these resistance mechanisms is frequent. Three of the families identified as systematically being acquired after the point mutations conferring resistance to quinolone correspond to genes that provide low-level resistance to quinolones (either qnr or oqx). Hence, we observe an initial emergence of point mutations conferring fluoroquinolone resistance and then the acquisition of specific resistance genes by HGT. This is in line with historical data: resistance to quinolones due to point mutations is almost as old as the quinolone usage. In contrast, resistance due to acquired genes is more recent. This accumulation is clinically relevant because the resistance effect provided by these novel genes affects resistance to all quinolones (at least for qnr), and it is additive to the one provided by the point mutation (resulting in increased resistance) (Ruiz 2019).
Discussion
In this study, we have searched to identify chronologies shedding light on the evolution of resistance to quinolones in E. coli. We have searched for them before, during, and after the acquisition of resistance. We found many genes that were preferentially gained/lost before the acquisition of the resistance mutations. During the process of acquisition of the mutations, there is a preferred initial one and then a very quick succession of mutations whose order we could not trace. Once resistance to quinolones was acquired, other resistances quickly accumulated in the lineages. It should be emphasized that these chronologies correspond to pairs of events that are far apart in time (they are found in different branches of the phylogenetic tree). Resistance mutations are usually very recent, whereas the preceding acquisitions or losses of genes can be recent or very ancient (supplementary fig. S16, Supplementary Material online). Simultaneous dating of recent and ancient events in the same tree is very difficult because the observed density of polymorphism per unit of time increases from the root to the tip (Rolland et al. 2023). This effect and the lack of information on the dates of isolation of the RefSeq strains precluded a reliable dating (in years from present) of the chronologies. In any case, they should not be interpreted in the framework of hysteresis, which is a transgenerational change in cellular physiology (Roemhild et al. 2018). Instead, we interpret the chronologies for gene gains/losses as changes that predispose a lineage to the acquisition of resistance, but that were selected for other reasons. We think many of the genes identified by this approach fall into 3 categories: functions in epistatic interaction with quinolone resistance, functions coselected in lineages for ecological/epidemiological reasons, and genes in genetic linkage with the former.
Many of the functions frequently acquired before the resistance mutations might make the bacterium more tolerant, resistant, or able to persist in the presence of quinolones. This is the case of functions that are known to arrest growth (TAs), those that diminish the availability of quinolones as a side-effect (transporters, metal chelators), or those that increase the expression of the gyrase. These functions could allow the bacterium to cope with the presence of quinolones for a longer period and thus provide time for resistance mutations to arise. This does not require any kind of foresight for natural selection. These functions might have their own adaptive role and may have been fixed independently of the possible advantage they may subsequently give under quinolone treatment. In some cases, the phenotype under selection before antibiotics is also adaptive under antibiotic therapy. In other cases, the phenotypes under selection with or without antibiotics may be very different, as observed for the acquisition of low-dose resistance to quinolones by overproduction of gyrase by the dTDP-rhamnose (Zampieri et al. 2017). Such latent phenotypes may be frequent across microbial systems, as previously suggested for gene regulatory circuits (Payne and Wagner 2014), moonlighting proteins (Henderson and Martin 2011), and biofilm formation (Nucci et al. 2023). The constant gain and loss of genes in bacterial genomes may produce a wide diversity of such latent phenotypes in natural populations rendering certain lineages better prepared than others to acquire specific novel traits (like antibiotic resistance). Such predisposition does not preclude the acquisition of resistance in other lineages that lack these genes, because the trait is accessible by mutation and is under strong selection. What we propose is that acquisition of certain genes might increase the probability of subsequent emergence of resistance to quinolones either because they increase the likelihood of appearance of resistance mutations or because they provide less costly genetic backgrounds.
Epistatic interactions in the process of accumulation of the mutations conferring resistance to quinolones have been described before (Silva et al. 2011; Hughes and Andersson 2017). The chronology usually starts by the mutation gyrA[S83L] (which was shown to have the largest effect and low cost) followed by quick fixation of the parC[S80I] and gyrA[D87N] mutations, which further increase resistance at low cost (Komp Lindgren et al. 2005; Rozen et al. 2007; Marcusson et al. 2009; Hughes and Andersson 2017; Huseby et al. 2017). The second most frequent evolutionary trajectory is the simultaneous acquisition of the 3 mutations. We thought this could be due to recombination, since the simultaneous acquisition of 4 mutations in gyrA, gyrB, parC, and parE after one single event of chromosomal conjugation was described in mycoplasma (Faucher et al. 2019). Yet, we detected only one case with clear evidence that the mutations in gyrA and parC were acquired in the same branch of the tree by recombination. Alternatively, the lack of time-resolution at the scale of the species trees could explain the apparent simultaneous emergence of all mutations. To test this idea, we performed the same analysis on 3 STs for which many genomes were available, which should improve the resolution of our study. Even in this case, we failed to untangle the order of acquisition of the mutations. While this matter remains to be completely resolved, we propose that natural selection for resistance by multiple mutations may be so strong in the clinical environment (Santos-Lopez et al. 2021; Karve and Wagner 2022) that the multiple point mutations are quickly acquired in succession and outcompete the other mutants. If so, then these lineages have frequently endured 3 successive (possibly nested) selective sweeps.
Epidemiological factors may explain some of the identified chronologies. The lineages of pathogens are more likely to encounter antibiotic pressure. Mutations conferring the resistance to quinolone are then more likely to be selected in such lineages. Hence, the acquisition of virulence factors may be associated with an increase in the rate of emergence of quinolone resistance once it started to be used in the clinic. Of note, the most obvious virulence factors (such as toxins, protein secretion systems, or their effectors) were not revealed by our analysis. But, we did find functions that while being very frequent in environmental bacteria are also associated with host colonization, like biofilm formation or iron uptake. Although there are data associating these traits with the acquisition of quinolone resistance outside the context of pathogenesis (Méhi et al. 2014; Liang et al. 2023), one cannot exclude the possibility that some of our results are due to epidemiological factors.
Epidemiological factors seem particularly pertinent to explain the chronologies of ARG acquisitions after the emergence of quinolone resistance. None of the genes systematically acquired before the quinolone resistance was annotated as ARG. However, genes encoding resistance to many antibiotic classes such as beta-lactams and aminoglycosides are consistently acquired after quinolone resistance. This suggests that quinolone resistances were usually acquired before the bacteria became multidrug resistant. Historically, resistance to quinolone was not the first one acquired by E. coli where resistance to penicillin was described in the 40s while quinolone resistance only in the 70s (Abraham and Chain 1940; Gellert et al. 1977). However, quinolone resistance is the one that best correlates with its antibiotic usage in E. coli across European countries (Redgrave et al. 2014), in agreement with this and other works showing that it can emerge very rapidly (Komp Lindgren et al. 2003). The accumulation of the full set of resistance mutations makes the clones fitter when compared with other antibiotic resistances, so that the resistance is less likely to be lost (Rozen et al. 2007; Marcusson et al. 2009). It may also be difficult to lose the resistance to quinolones by multiple mutations since it requires the reversion of all of them, whereas many other resistances may be lost simply by plasmid loss, gene deletion, or inactivation. Several studies have reported that mutations in the core genome often do not reverse even when antibiotic resistance pressure was removed (Pallecchi et al. 2012; Baker et al. 2013; Johnning et al. 2015). This combination of high acquisition/low reversion rates could be the reason why the quinolone resistance appears very early in lineages of bacteria becoming multidrug resistant. Nevertheless, some studies have shown that resistance to quinolones does promote resistance to other antibiotics. For example, substitution at gyrA[87] in Salmonella influences sensitivity to other types of antibiotics and results in increased expression of stress response pathways (Webber et al. 2013). Hence, even the chronologies of gene gains/loss after acquisition of resistance may result from epistatic interactions.
A specificity of our approach is that functions acquired as the result of the same event of transfer will have similar statistical signal. Yet, this does not mean they are all biologically pertinent. One single gene under strong selection and strong linkage with others may lead to the entire set of genes coming out as significant in our analysis. Our clustering approach allowed to regroup such genes together and provides a more parsimonious analysis of the functions that may explain the chronologies. Several of these groups only have genes of unknown function. It is beyond the scope of this work to elucidate these functions or to disentangle the most important in each group of linkage. Yet, our analysis reveals many functions that were previously shown to be involved in the acquisition of quinolone resistance (or associated with it). This validates our approach and provides a rich list of genes for future experimental analysis.
The clustering of genes in linkage also allowed to study the vehicles of gene acquisition, i.e. the MGEs that promoted the gene transfer. Several of these groups of genes are in plasmids or prophages. If the acquisition of one gene carried by an MGE influences the acquisition of the mutation conferring resistance due to epistatic interactions, other genes carried by the same MGE will hitchhike with it. This has often been theorized as being one of the potential reasons why costly and autonomous MGEs, such as prophages and conjugative elements, encode traits adaptive for bacteria (Rankin et al. 2011). Hence, we do not think that the entire MGEs are favoring the acquisition of quinolones, only a few of their genes. MGEs, especially plasmids, are known to carry ARGs to the bacteria they infect. But, our results suggest they are also responsible for facilitating the acquisition of antibiotic resistance by carrying functions that make the bacterium better prepared to acquire the resistance itself. In this context, the identification of prophages having such an effect is intriguing (even though it fits previous works (Wang et al. 2010). Quinolones are known to induce the SOS response, which induces the lytic cycle of many prophages (Baharoglu and Mazel 2014). It is tempting to speculate that prophages might carry traits favoring resistance or tolerance to quinolones to avoid induction at a moment where most potential hosts in the near environment are not promising hosts for infection. These genes may not have evolved to tackle quinolone resistance, presumably a recent threat to E. coli but may have become advantageous under it. Anti-SOS genes have been found in both conjugative plasmids and prophages where they may serve to manipulate the host responses or those of other mobile elements (Bagdasarian et al. 1992; Azulay et al. 2022). This fits our observation that the prophages identified in this work encode functions resulting in growth arrest or mitigation of the effect of quinolones.
Bacteria colonize diverse environments that vary in terms of selective pressures. Epistatic interactions create complex evolutionary patterns of adaptation. Our results suggest that this complexity further increases when a mix of HGT and mutations produce bacteria with very different genetic backgrounds within lineages. While these interactions can facilitate the emergence of antibiotic resistance, they may also render evolutionary processes less predictable (Palmer et al. 2015). They may contribute to explain why within many microbial species some lineages tend to accumulate traits like virulence factors and antibiotic resistance. Interestingly, recent studies show that the decline of resistance to a cephalosporin in Pseudomonas is also contingent on the genetic background (Hernando-Amado et al. 2022). Understanding how frequent chronologies of events result from genetic, functional, and epidemiological interactions will shed further light on the evolution of antibiotic resistance and how one can forecast it.
Methods
Genome Data and Pangenome Construction
We analyzed 3 datasets in this study.
The 1st dataset is the one used across the study unless otherwise specified. It includes 1,585 genomes of E. coli identified from 21,086 complete bacterial genomes retrieved from NCBI RefSeq representing 6,124 species of Bacteria (http://ftp.ncbi.nih.gov/genomes/refseq/bacteria/), in March 2021. This dataset has the advantage of being very large and diverse, and we know the structure of the genome (chromosome, plasmids). This facilitates the study of the MGEs allowing gene acquisition, such as plasmids and phages, and provides high-quality data for the identification of mutations. It is usually presumed that it contains many clinical isolates.
The 2nd dataset was used to control for the presumed overrepresentation of clinical isolates in the RefSeq dataset. It includes a collection of 1,294 E. coli draft genomes from isolates recovered in Australia between 1993 and 2015 chosen to represent the phylogenetic diversity of the species (Touchon et al. 2020). It includes less than 25% of clinical isolates and allows to control for sampling biases commonly encountered in public databases.
The 2,879 E. coli genomes were analyzed for assembly quality using their L90 value and for genetic distance using Mash v2.2 (Ondov et al. 2016). We removed from further analysis 369 strains from the Australian dataset because they had a L90 superior to 100 (i.e. the sum of the 100 longest contigs does not cover at least 90% of the genome size, suggesting that they are highly fragmented). Additionally, 365 were removed because they were very similar (MASH distance <10−4). This resulted in a dataset of 2,145 (877 from the Australian dataset and 1,268 RefSeq sequences) completely sequenced and assembled E. coli genomes.
The 3rd dataset was only used to pinpoint the chronology of mutations for quinolone resistance at a short evolutionary time scale. We used all E. coli sequence type (ST) 131 (ST131) genomes from RefSeq (n = 85), and we retrieved all available E. coli ST 410 and 167 (ST410 and ST167) genomes from Enterobase (n = 1,000 and n = 801) (last accessed in December 2021) (Zhou et al. 2020). We removed from further analysis 45 ST410 and 29 ST167 genomes because they had a L90 superior to 100. Additionally, 198 ST410 and 193 ST167 strains were removed because they were too similar (MASH distance <0.0001). This resulted in a dataset of 757 E. coli ST410, 85 E. coli ST131, and 579 E. coli ST167 genomes.
Identification of the Pangenome and of the Core Genome
The pangenome and core genome of the 2,145 E. coli from the RefSeq and Australian datasets were computed using PanACoTA v.1.3.1 (Perrin and Rocha 2021). Briefly, the pangenome was constructed by clustering all protein sequences in the set of genomes using MMseqs2 (protein identity >80%). We retrieved the core genome from this pangenome. It consists of 2393 gene families present in exactly 1 copy in at least 99.0% of all genomes. In the remaining 1% genomes, we accepted the presence of 0 or several members of the gene family (in which case the sequence of the gene is not used for the strain). The pangenome (27,556 gene families) and core genome (3,933 gene families) for the 757 ST410 E. coli were built the same way.
Phylogenetic Reconstruction
The multiple sequence alignments of the families of the core genes were computed using the “align” PanACoTA v1.3.1 module. Briefly, the protein sequences of the core genes were aligned with MAFFT v7.467 (-auto parameters) (Katoh and Standley 2014) and then back-translated to nucleotide alignments (i.e. each amino acid was replaced by the original codon) and concatenated. The large multiple sequence alignment including the RefSeq and Australian dataset was then separated in an alignment with the 1,268 RefSeq genomes and another with the 877 genomes from the Australian dataset. The phylogenetic inference was done from the resulting multiple alignments using IQ-TREE 2.0.6 (Nguyen et al. 2015) with the ultrafast bootstrap option (-bb 1,000 bootstraps) and with the best fitting model estimated using ModelFinder Plus (-MPF) (Kalyaanamoorthy et al. 2017). The best model for the 3 datasets was GTR + F + I + G4 according to the bayesian information criterion (BIC). Trees were rooted using the midpoint function from the phangorn packages (v.2.5.5) for R (Schliep 2011). The root obtained is in agreement with the literature, as strains belonging to clade 1 (outgroup closest to the E. coli species) are indeed the most external.
Mutation Profile
Point mutations leading to quinolone resistance in E. coli are found in the DNA gyrase and the DNA topoisomerase IV genes, i.e. in gyrA, gyrB, parC, and parE (Hopkins et al. 2005). We retrieved the sequences of these proteins using blastp v2.12.0 (default parameters, identity threshold of 90%) (Camacho et al. 2009). We then built a multiple alignment of the proteins using MAFFT v7.429 (with -auto parameters). The alignments were parsed using Biopython (Cock and Whitworth 2010). We looked for point mutations leading to quinolone resistance, at each expected position (accounting for gaps) using the comprehensive list provided by Hopkins et al. (Hopkins et al. 2005) for E. coli (Table S1). For each dataset, the distribution of every combination of resistance mutation was summarized in an upset plot computed using the R package UpsetR v1.4.0 (Conway et al. 2017).
Inference of Ancestral Gene Repertoires
We counted the number of occurrences of each family of the pangenome in all genomes. This was used to build a gene presence (1 or more copies)/absence matrix in all leaves of the phylogenetic trees. From this occurrence matrix, we inferred the ancestral state (presence or absence) of each gene family at every internal node of the phylogenetic trees with PastML v1.9.33 (Ishikawa et al. 2019). We used the JOINT method with default parameters, and this method reconstructs the states of the scenario with the highest likelihood. From the ancestral state matrix computed by PastML, we inferred the gene gains and losses at all the branches of the species tree by subtracting the gene content of the child node to the gene content of the parent node.
Trajectories of Acquisition of Mutations
To infer the history of the quinolone resistance mutations, we constructed a matrix with the presence or absence of each type of mutation in gyrA, gyrB, parC, and parE in each strain. The ancestral states for each mutation were inferred the same way as described in the previous section for the genes in the pangenome. To identify the acquisition chronologies of the multiple mutations, every path starting from the root and leading to a leaf containing at least one quinolone resistance mutation was extracted using the ape package v5.3 for R (Paradis and Schliep 2019). Paths were then traversed from the root to the first acquisition of a mutation conferring quinolone resistance (as inferred by PastML). All the paths arising from the point in the tree where this first mutation occurred were then traversed from this event of acquisition to the next event on the path, and this recursively until the last event of acquisition on each path (for the scripts used to perform this analysis, see Supplementary material online). This way, we only consider the number of events in the tree, not the number of taxa affected by them. This is important because events of gain and loss within a gene family can be regarded as independent, whereas resistant taxa are not (they may result from the same ancestral mutation event). Counting resistant taxa would inflate and bias the statistics, overrepresenting some chronologies. Counting events allows to identify independent events across the tree. This procedure results in a collection of paths corresponding to every single chronology of mutation acquisition.
Detection of Preferential Chronologies
Preferential chronologies of events leading to the acquisition of resistance to quinolones were identified in a 2-step procedure using the program Evo-Scope v1.0.0 (Godfroid et al. 2022).
In the first step, we analyze all possible chronologies to identify events often preceding the acquisition of resistance to quinolones. This is done with the Epics module of Evo-Scope (with the -S parameter). The procedure identifies the number of occurrences of an event E1 (e.g. acquisition of resistance) following an event E0 (e.g. acquisition of a certain gene) on a phylogenetic tree. The observed values of all pairs of events are compared with the expected numbers under a null model of uniform rates of distribution of events on the tree (Behdenna et al. 2016). P-values were adjusted for multiple tests using the “fdr_bh” method (P < 10−5) (Benjamini–Hochberg correction) from SciPy v.1.10.1 (Benjamini and Hochberg 1995; Virtanen et al. 2020).
In the 2nd step, we retrieved the significant chronologies identified above and inferred the interaction type and strength of correlated evolution between these pairs using the Epocs module of Evo-Scope (Behdenna et al. 2022). This module classifies interactions, by maximum likelihood, in 3 different categories depending on the influence of a trait on the occurrence of the other one.
Scenario of independence (-Si): The occurrence of event E0 does not change the occurrence rate of E1.
Scenario of asymmetric induction (-Sa, -Sb): The occurrence of event E0 changes the occurrence rate of event E1, or the occurrence of event E1 changes the occurrence rate of event E0.
Scenario of reciprocal induction (-Sl): Event E0 enhances the occurrence rate of event E1, and reciprocally, event E1 enhances the occurrence rate of event E0.
These scenarios are described by models containing from 2 to 4 parameters, which are associated to each trait. The parameters are divided into natural occurrence rates (i.e. rates at which the trait mutates from present to absent or from absent to present) and modified occurrence rates (i.e. rates at which the trait mutates from present to absent or from absent to present after a change of state of the other trait). The ratio λ between the modified occurrence rates and the natural occurrence rates can be interpreted as an induction factor. If λ > 1, the induction between the 2 traits is positive (i.e. E0 increases the occurrence rate of the subsequent event E1), whereas it is negative when λ < 1 (i.e. E0 decreases the occurrence rate of the subsequent event E1). The model best describing the data under study is selected following a significant likelihood ratio test (Neyman and Pearson 1933)
Detection of Recombination
The alignment of the core genome that was used to build the phylogeny was scanned with Gubbins v2.4.1 to find recombination tracts (Croucher et al. 2015). The maximum likelihood tree previously built was provided for the first iteration using the “—starting-tree” command. The position of the recombination tracks was compared with the position of mutations in gyrA (gyrA[S83L] and gyrA[D87N]) and in parC (parC[S80I]) in the alignment of the core genome to identify overlaps that could indicate that mutations arrived by recombination. We then compared the nodes at which recombination occurred with nodes where the mutations gyrA[S83L], gyrA[D87N], and parC[S80I] were acquired. When overlapping recombination tracks and acquisition of mutations occurred at the same nodes, we considered the mutations to be acquired by events of recombination.
Genome-Wide Association Study
We performed a GWAS for the presence of fluoroquinolone resistance using 1,268 E. coli genomes with pyseer v.1.3.9 (Lees et al. 2018). The association between the gene presence/absence and the resistance phenotype (defined by the presence of a known resistance mutation) was assessed with an LMM in which the STs were considered as covariates. We used the recombination-free phylogenetic tree produced by Gubbins. This tree allowed us to generate a distance and a kinship matrix with the scripts coming with pyseer. The LMM used the multidimensional scaling (MDS) of these matrices to control for population structure. Ten dimensions were included in the MDS. To address the problem of multiple comparisons in our analysis, we used a Benjamini–Hochberg procedure (Benjamini and Hochberg 1995) on the P-value of the association already adjusted for population structure using the p.adjust function with the “BH” method in R v4.2.2. For a corrected P-value inferior to 0.05, we deemed the association between the gene presence/absence and the resistance as significant.
Identification of Groups of Genes in Genetic Linkage
Gene families consistently acquired before the resistance to quinolone can be in genetic linkage, e.g. if they are systematically coacquired within a plasmid, a phage, or recombination. In such cases, there may be only one gene that effectively changes the likelihood of acquisition of the resistance, but the method will also highlight genes in strong linkage with this one. To control for this effect, we took all the genes highlighted by the analysis of chronologies and clustered them using 2 key information: first coacquisition or coloss in the phylogenetic tree and then colocalization in the genome.
First, 2 gene families were clustered together if they were consistently coacquired or colost in the same branch or node of the tree. To assess the statistical significance of these simultaneous events, we used the Epics module of the program Evo-Scope with the parameter -I (i.e, Identity matrix). Thus, the program compares the number of cooccurrences of 2 events E1 and E0 in a branch of the tree with their expected cooccurrence under a null model of uniform distribution of events on the tree. Pairs of events that frequently cooccurred in time were then clustered by single-linkage using the agglomerative clustering algorithm from scikit-learn v1.2.2 (parameters: affinity=“precomputed,” distance_threshold = 1, linkage=“single,” n_clusters = None) (Pedregosa et al. 2011). We used single-linkage to obtain large clusters that could then be further split.
Second, we split the clusters of coacquired or colost genes using information on the distance between the genes in the genomes. For every pair of gene families in a cluster of cooccurrences, we computed the median number of genes between them in the genomes (in which they cooccur). As all pairs of genes need a distance, when genes were present on different DNA molecules (e.g. one in the chromosome and another in a plasmid), the number of genes in the largest replicon in both bacteria was set as the distance between them. This way, the distance between genes on different DNA molecules will be above the clustering threshold (they will not be clustered together). These values were then used to cluster the gene families by average-linkage using the agglomerative clustering algorithm from scikit-learn v1.2.2 (parameters: affinity=“precomputed,” distance_threshold = 30, linkage=“average,” n_clusters = None). At the end of these procedures, all clusters were checked for homogeneity of induction values (λ) and event type (loss or gain).
Functional Annotation of the Pangenome Families
We picked at random a representative sequence of each pangenome family. These sequences were annotated using eggNOG-mapper v2.1.9. In order to be exhaustive, we also fetched the gene name and product functions from the RefSeq annotations. When both eggNOG and RefSeq yield different gene names, we used the RefSeq gene name.
Detection of the Antibiotic Resistance Pangenome Families
We picked at random a representative sequence of each pangenome family. These sequences were screened for ARGs using AMRfinderPlus v3.10.18 with default parameters (Feldgarden et al. 2021). If the representative sequence was identified as an AMR gene, the pangenome family was considered as an antibiotic resistance one.
Statistics
Unless mentioned otherwise, all statistics were performed within R (v3.6.3).
Supplementary Material
Supplementary material is available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We acknowledge Eugen Pfeifer for providing the viral regions data and Manuel Ares-Arroyo for scientific discussions. This project was funded by the INCEPTION project Path2Resistance (PIA/ANR-16-CONV-0005), Equipe FRM (Equipe FRM/EQU201903007835), Laboratoire d’Excellence IBEID (ANR-10-LABX-62-IBEID), and SEQ2DIAG (ANR-20-PAMR-0010). This work used the computational and storage services (MAESTRO cluster) provided by the IT department at Institut Pasteur, Paris. We thank Frédéric Barras, Jessica El Khoury, and Laurence Van Melderen for help on the interpretation of our results.
Contributor Information
Charles Coluzzi, Institut Pasteur, Université Paris Cité, CNRS, UMR3525, Microbial Evolutionary Genomics, Paris, France.
Martin Guillemet, Institut Pasteur, Université Paris Cité, CNRS, UMR3525, Microbial Evolutionary Genomics, Paris, France.
Fanny Mazzamurro, Institut Pasteur, Université Paris Cité, CNRS, UMR3525, Microbial Evolutionary Genomics, Paris, France; Collège Doctoral, Sorbonne Université, Paris, France.
Marie Touchon, Institut Pasteur, Université Paris Cité, CNRS, UMR3525, Microbial Evolutionary Genomics, Paris, France.
Maxime Godfroid, SMILE Group, Center for Interdisciplinary Research in Biology (CIRB), Collège de France, CNRS, INSERM, Université PSL, Paris, France.
Guillaume Achaz, SMILE Group, Center for Interdisciplinary Research in Biology (CIRB), Collège de France, CNRS, INSERM, Université PSL, Paris, France.
Philippe Glaser, Institut Pasteur, Université de Paris Cité, CNRS, UMR6047, Unité EERA, Paris, France.
Eduardo P C Rocha, Institut Pasteur, Université Paris Cité, CNRS, UMR3525, Microbial Evolutionary Genomics, Paris, France.
Author Contributions
C.C., M.G., P.G., and E.P.C.R. contributed to design of the study. C.C., M.G., and F.M. performed the analyses. M.T., M.Go., and G.A. contributed with data or methods. C.C., M.G., and E.P.C.R. interpreted the analyses). C.C. and E.P.C.R. wrote the draft). All authors participated during writing of the manuscript and approved the submitted version.
Data Availability
All major data, or identifiers to public repositories with the data, are incorporated into the article and its online Supplementary material. Other data, e.g. intermediary results files, scripts underlying this article will be shared on reasonable request to the corresponding author.
References
- Abraham EP, Chain E. An enzyme from bacteria able to destroy penicillin. Nature 1940:146(3713):837. 10.1038/146837a0. [DOI] [PubMed] [Google Scholar]
- Aldred KJ, Kerns RJ, Osheroff N. Mechanism of quinolone action and resistance. Biochemistry 2014:53(10):1565–1574. 10.1021/bi5000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan DS, Holbein BE. Iron chelator DIBI suppresses formation of ciprofloxacin-induced antibiotic resistance in Staphylococcus aureus. Antibiotics-Basel. 2022:11(11). 10.3390/antibiotics11111642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angst DC, Hall AR. The cost of antibiotic resistance depends on evolutionary history in Escherichia coli. BMC Evol Biol. 2013:13(1):163. 10.1186/1471-2148-13-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apjok G, Boross G, Nyerges A, Fekete G, Lazar V, Papp B, Pal C, Csorgo B. Limited evolutionary conservation of the phenotypic effects of antibiotic resistance mutations. Mol Biol Evol. 2019:36(8):1601–1611. 10.1093/molbev/msz109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azulay G, Pasechnek A, Stadnyuk O, Ran-Sapir S, Fleisacher AM, Borovok I, Sigal N, Herskovits AA. A dual-function phage regulator controls the response of cohabiting phage elements via regulation of the bacterial SOS response. Cell Rep. 2022:39(3):110723. 10.1016/j.celrep.2022.110723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagdasarian M, Bailone A, Angulo JF, Scholz P, Bagdasarian M, Devoret R. Psib, an anti-SOS protein, is transiently expressed by the F sex factor during its transmission to an Escherichia coli K-12 recipient. Mol Microbiol. 1992:6(7):885–893. 10.1111/j.1365-2958.1992.tb01539.x. [DOI] [PubMed] [Google Scholar]
- Bagel S, Hüllen V, Wiedemann B, Heisig P. Impact of gyrA and parC mutations on quinolone resistance, doubling time, and supercoiling degree of Escherichia coli. Antimicrob Agents Chemother. 1999:43(4):868–875. 10.1128/AAC.43.4.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baharoglu Z, Mazel D. SOS, the formidable strategy of bacteria against aggressions. FEMS Microbiol Rev. 2014:38(6):1126–1145. 10.1111/1574-6976.12077. [DOI] [PubMed] [Google Scholar]
- Baker S, Duy PT, Nga TVT, Dung TTN, Phat VV, Chau TT, Turner AK, Farrar J, Boni MF. Fitness benefits in fluoroquinolone-resistant Salmonella Typhi in the absence of antimicrobial pressure. Elife 2013:2:e01229. 10.7554/eLife.01229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batarseh TN, Batarseh SN, Rodriguez-Verdugo A, Gaut BS. Phenotypic and genotypic adaptation of Escherichia coli to thermal stress is contingent on genetic background. Mol Biol Evol. 2023:40(5):5. 10.1093/molbev/msad108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh S, Ekanayaka AS, Piddock LJV, Webber MA. Loss of or inhibition of all multidrug resistance efflux pumps of Salmonella enterica serovar Typhimurium results in impaired ability to form a biofilm. J Antimicrob Chemother. 2012:67(10):2409–2417. 10.1093/jac/dks228. [DOI] [PubMed] [Google Scholar]
- Behdenna A, Godfroid M, Petot P, Pothier J, Lambert A, Achaz G. A minimal yet flexible likelihood framework to assess correlated evolution. Syst Biol. 2022:71(4):823–838. 10.1093/sysbio/syab092. [DOI] [PubMed] [Google Scholar]
- Behdenna A, Pothier J, Abby SS, Lambert A, Achaz G. Testing for independence between evolutionary processes. Syst Biol. 2016:65(5):812–823. 10.1093/sysbio/syw004. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc B: Stat Methodol. 1995:57:289–300. 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- Bisacchi GS. Origins of the quinolone class of antibacterials: an expanded “discovery story”. J Med Chem. 2015:58(12):4874–4882. 10.1021/jm501881c. [DOI] [PubMed] [Google Scholar]
- Boehm A, Steiner S, Zaehringer F, Casanova A, Hamburger F, Ritz D, Keck W, Ackermann M, Schirmer T, Jenal U. Second messenger signalling governs Escherichia coli biofilm induction upon ribosomal stress. Mol Microbiol. 2009:72(6):1500–1516. 10.1111/j.1365-2958.2009.06739.x. [DOI] [PubMed] [Google Scholar]
- Borrell S, Teo Y, Giardina F, Streicher EM, Klopper M, Feldmann J, Muller B, Victor TC, Gagneux S. Epistasis between antibiotic resistance mutations drives the evolution of extensively drug-resistant tuberculosis. Evol Med Public Health. 2013:2013(1):65–74. 10.1093/emph/eot003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandis G, Hughes D. Genetic characterization of compensatory evolution in strains carrying rpoB Ser531Leu, the rifampicin resistance mutation most frequently found in clinical isolates. J Antimicrob Chemother. 2013:68(11):2493–2497. 10.1093/jac/dkt224. [DOI] [PubMed] [Google Scholar]
- Brandis G, Wrande M, Liljas L, Hughes D. Fitness-compensatory mutations in rifampicin-resistant RNA polymerase. Mol Microbiol. 2012:85(1):142–151. 10.1111/j.1365-2958.2012.08099.x. [DOI] [PubMed] [Google Scholar]
- Braun V, Hantke K. Recent insights into iron import by bacteria. Curr Opin Chem Biol. 2011:15(2):328–334. 10.1016/j.cbpa.2011.01.005. [DOI] [PubMed] [Google Scholar]
- Brooun A, Liu S, Lewis K. A dose-response study of antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 2000:44(3):640–646. 10.1128/AAC.44.3.640-646.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SA. Fluoroquinolones in animal health. J Vet Pharmacol Ther. 1996:19(1):1–14. 10.1111/j.1365-2885.1996.tb00001.x. [DOI] [PubMed] [Google Scholar]
- Bruyndonckx R, Adriaenssens N, Versporten A, Hens N, Monnet DL, Molenberghs G, Goossens H, Weist K, Coenen S, ESAC-Net Study Group . Consumption of antibiotics in the community, European Union/European Economic Area, 1997-2017. J Antimicrob Chemother. 2021:76(Supplement_2):ii7–ii13. 10.1093/jac/dkab172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinform. 2009:10(1):421. 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro RAD, Ross A, Kamwela L, Reinhard M, Loiseau C, Feldmann J, Borrell S, Trauner A, Gagneux S. The genetic background modulates the evolution of fluoroquinolone-resistance in Mycobacterium tuberculosis. Mol Biol Evol. 2020:37(1):195–207. 10.1093/molbev/msz214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis GL. A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases. Chembiochem. 2005:6(4):601–611. 10.1002/cbic.200400283. [DOI] [PubMed] [Google Scholar]
- Ciofu O, Moser C, Jensen PØ, Høiby N. Tolerance and resistance of microbial biofilms. Nat Rev Microbiol. 2022:20(10):621–635. 10.1038/s41579-022-00682-4. [DOI] [PubMed] [Google Scholar]
- Cock PJA, Whitworth DE. Evolution of relative reading frame bias in unidirectional prokaryotic gene overlaps. Mol Biol Evol. 2010:27(4):753–756. 10.1093/molbev/msp302. [DOI] [PubMed] [Google Scholar]
- Collins C, Didelot X. A phylogenetic method to perform genome-wide association studies in microbes that accounts for population structure and recombination. PLoS Comput Biol. 2018:14(2):e1005958. 10.1371/journal.pcbi.1005958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway JR, Lex A, Gehlenborg N. Upsetr: an R package for the visualization of intersecting sets and their properties. Bioinformatics 2017:33(18):2938–2940. 10.1093/bioinformatics/btx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science 1999:284(5418):1318–1322. 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Parkhill J, Harris SR. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015:43(3):e15. 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins EA, Hall RJ, Connor C, McInerney JO, McNally A. Pangenome evolution in Escherichia coli is sequence type, not phylogroup, specific. Cold Spring Harbor Laboratory; 2022. 10.1099/mgen.0.000903. [DOI] [Google Scholar]
- Darby EM, Trampari E, Siasat P, Gaya MS, Alav I, Webber MA, Blair JMA. Molecular mechanisms of antibiotic resistance revisited. Nat Rev Microbiol. 2023:21(5):280–295. 10.1038/s41579-022-00820-y. [DOI] [PubMed] [Google Scholar]
- Da Re S, Valle J, Charbonnel N, Beloin C, Latour-Lambert P, Faure P, Turlin E, Le Bouguénec C, Renauld-Mongénie G, Forestier C, et al. Identification of commensal Escherichia coli genes involved in biofilm resistance to pathogen colonization. PLoS One. 2013:8(5):e61628. 10.1371/journal.pone.0061628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010:74(3):417–433. 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X, Meric G, Falush D, Darling AE. Impact of homologous and non-homologous recombination in the genomic evolution of Escherichia coli. Bmc Genomics. 2012:13(1):256. 10.1186/1471-2164-13-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorr T, Vulic M, Lewis K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 2010:8(2):e1000317. 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett MJ, Jin YF, Ricci V, Piddock LJ. Contributions of individual mechanisms to fluoroquinolone resistance in 36 Escherichia coli strains isolated from humans and animals. Antimicrob Agents Chemother. 1996:40(10):2380–2386. 10.1128/AAC.40.10.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucher M, Nouvel L-X, Dordet-Frisoni E, Sagné E, Baranowski E, Hygonenq M-C, Marenda M-S, Tardy F, Citti C. Mycoplasmas under experimental antimicrobial selection: the unpredicted contribution of horizontal chromosomal transfer. PLoS Genet. 2019:15(1):e1007910. 10.1371/journal.pgen.1007910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldgarden M, Brover V, Gonzalez-Escalona N, Frye JG, Haendiges J, Haft DH, Hoffmann M, Pettengill JB, Prasad AB, Tillman GE, et al. AMRFinderplus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci Rep. 2021:11(1):12728. 10.1038/s41598-021-91456-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M, Mizuuchi K, O'Dea MH, Itoh T, Tomizawa J-I. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc Natl Acad Sci USA. 1977:74(11):4772–4776. 10.1073/pnas.74.11.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerwig J, Kiley TB, Gunka K, Stanley-Wall N, Stülke J. The protein tyrosine kinases EpsB and PtkA differentially affect biofilm formation in Bacillus subtilis. Microbiology 2014:160(4):682–691. 10.1099/mic.0.074971-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghenu A-H, Amado A, Gordo I, Bank C. Epistasis decreases with increasing antibiotic pressure but not temperature. Philos Trans R Soc B Biol Sci. 2023:378(1877):1877. 10.1098/rstb.2022.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfroid M, Coluzzi C, Lambert A, Glaser P, Rocha EPC, Achaz G. Evo-Scope: fully automated assessment of correlated evolution on phylogenetic trees. Cold Spring Harbor Laboratory; 2022. 10.1101/2022.12.08.519595. [DOI]
- Goormaghtigh F, Fraikin N, Putrins M, Hallaert T, Hauryliuk V, Garcia-Pino A, Sjodin A, Kasvandik S, Udekwu K, Tenson T, et al. Reassessing the role of type II toxin-antitoxin systems in formation of Escherichia coli type II persister cells. MBio. 2018:9(3):e00640–e00618. 10.1128/mBio.00640-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goormaghtigh F, Van Melderen L. Single-cell imaging and characterization of Escherichia coli persister cells to ofloxacin in exponential cultures. Sci Adv. 2019:5(6):eaav9462. 10.1126/sciadv.aav9462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen LH, Jensen LB, Sorensen HI, Sorensen SJ. Substrate specificity of the OqxAB multidrug resistance pump in Escherichia coli and selected enteric bacteria. J Antimicrob Chemother. 2007:60(1):145–147. 10.1093/jac/dkm167. [DOI] [PubMed] [Google Scholar]
- Heisig P. Genetic evidence for a role of parC mutations in development of high-level fluoroquinolone resistance in Escherichia coli. Antimicrob Agents Chemother. 1996:40(4):879–885. 10.1128/AAC.40.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson B, Martin A. Bacterial virulence in the moonlight: multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect Immun. 2011:79(9):3476–3491. 10.1128/IAI.00179-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernando-Amado S, Laborda P, Valverde JR, Martinez JL. Rapid decline of ceftazidime resistance in antibiotic-free and sublethal environments is contingent on genetic background. Mol Biol Evol. 2022:39(3):3. 10.1093/molbev/msac049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera G, Aleixandra V, Urios A, Blanco M. Quinolone action in Escherichia coli cells carrying gyrA and gyrB mutations. FEMS Microbiol Lett. 1993:106(2):187–191. 10.1111/j.1574-6968.1993.tb05957.x. [DOI] [PubMed] [Google Scholar]
- Hopkins KL, Davies RH, Threlfall EJ. Mechanisms of quinolone resistance in Escherichia coli and Salmonella: recent developments. Int J Antimicrob Agents. 2005:25(5):358–373. 10.1016/j.ijantimicag.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Hughes D, Andersson DI. Evolutionary trajectories to antibiotic resistance. Annu Rev Microbiol. 2017:71(1):579–596. 10.1146/annurev-micro-090816-093813. [DOI] [PubMed] [Google Scholar]
- Huseby DL, Pietsch F, Brandis G, Garoff L, Tegehall A, Hughes D. Mutation supply and relative fitness shape the genotypes of ciprofloxacin-resistant Escherichia coli. Mol Biol Evol. 2017:34(5):1029–1039. 10.1093/molbev/msx052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamovic L, Sommer MOA. Use of collateral sensitivity networks to design drug cycling protocols that avoid resistance development. Sci Transl Med. 2013:5(204):204ra132–204ra132. 10.1126/scitranslmed.3006609. [DOI] [PubMed] [Google Scholar]
- Ishikawa SA, Zhukova A, Iwasaki W, Gascuel O. A fast likelihood method to reconstruct and visualize ancestral scenarios. Mol Biol Evol. 2019:36(9):2069–2085. 10.1093/molbev/msz131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillard M, Lima L, Tournoud M, Mahé P, Van Belkum A, Lacroix V, Jacob L. A fast and agnostic method for bacterial genome-wide association studies: bridging the gap between k-mers and genetic events. PLoS Genet. 2018:14(11):e1007758. 10.1371/journal.pgen.1007758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangir PK, Yang Q, Shaw LP, Caballero JD, Ogunlana L, Wheatley R, Walsh T, Maclean RC. Pre-existing chromosomal polymorphisms in pathogenic E. coli potentiate the evolution of resistance to a last-resort antibiotic. Elife 2022:11:e78834. 10.7554/eLife.78834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnning A, Kristiansson E, Fick J, Weijdegard B, Larsson DGJ. Resistance mutations in gyrA and parC are common in Escherichia communities of both fluoroquinolone-polluted and uncontaminated aquatic environments. Front Microbiol. 2015:6:1355. 10.3389/fmicb.2015.01355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. Modelfinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017:14(6):587. 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamruzzaman M, Iredell J. A ParDE-family toxin antitoxin system in major resistance plasmids of Enterobacteriaceae confers antibiotic and heat tolerance. Sci Rep. 2019:9(1):1–12. 10.1038/s41598-019-46318-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karve S, Wagner A. Environmental complexity is more important than mutation in driving the evolution of latent novel traits in E. coli. Nat Commun. 2022:13(1):5904. 10.1038/s41467-022-33634-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT: iterative refinement and additional methods. Methods Mol Biol. 2014:1079:131–146. 10.1007/978-1-62703-646-7_8. [DOI] [PubMed] [Google Scholar]
- Kjaergaard K, Schembri MA, Ramos C, Molin S, Klemm P. Antigen 43 facilitates formation of multispecies biofilms. Environ Microbiol. 2000:2(6):695–702. 10.1046/j.1462-2920.2000.00152.x. [DOI] [PubMed] [Google Scholar]
- Knopp M, Andersson DI. Amelioration of the fitness costs of antibiotic resistance due to reduced outer membrane permeability by upregulation of alternative porins. Mol Biol Evol. 2015:32(12):3252–3263. 10.1093/molbev/msv195. [DOI] [PubMed] [Google Scholar]
- Knopp M, Andersson DI. Predictable phenotypes of antibiotic resistance mutations. MBio. 2018:9(3):3. 10.1128/mBio.00770-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komp Lindgren P, Karlsson A, Hughes D. Mutation rate and evolution of fluoroquinolone resistance in Escherichia coli isolates from patients with urinary tract infections. Antimicrob Agents Chemother. 2003:47(10):3222–3232. 10.1128/AAC.47.10.3222-3232.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komp Lindgren P, Marcusson LL, Sandvang D, Frimodt-Moller N, Hughes D. Biological cost of single and multiple norfloxacin resistance mutations in Escherichia coli implicated in urinary tract infections. Antimicrob Agents Chemother. 2005:49(6):2343–2351. 10.1128/AAC.49.6.2343-2351.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lázár V, Martins A, Spohn R, Daruka L, Grézal G, Fekete G, Számel M, Jangir PK, Kintses B, Csörgő B, et al. Antibiotic-resistant bacteria show widespread collateral sensitivity to antimicrobial peptides. Nat Microbiol. 2018:3(6):718–731. 10.1038/s41564-018-0164-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavis HL, Bonten MJ, Willems RJL. Identification of high-risk enterococcal clonal complexes: global dispersion and antibiotic resistance. Curr Opin Microbiol. 2006:9(5):454–460. 10.1016/j.mib.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Lees JA, Galardini M, Bentley SD, Weiser JN, Corander J. Pyseer: a comprehensive tool for microbial pangenome-wide association studies. Bioinformatics 2018:34(24):4310–4312. 10.1093/bioinformatics/bty539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin-Reisman I, Ronin I, Gefen O, Braniss I, Shoresh N, Balaban NQ. Antibiotic tolerance facilitates the evolution of resistance. Science 2017:355(6327):826–830. 10.1126/science.aaj2191. [DOI] [PubMed] [Google Scholar]
- Lewis K. Riddle of biofilm resistance. Antimicrob Agents Chemother. 2001:45(4):999–1007. 10.1128/AAC.45.4.999-1007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Zhang J, Hu J, Li X, Li B. Fluoroquinolone residues in the environment rapidly induce heritable fluoroquinolone resistance in Escherichia coli. Environ Sci Technol. 2023:57(12):4784–4795. 10.1021/acs.est.2c04999. [DOI] [PubMed] [Google Scholar]
- Loiseau C, Windels EM, Gygli SM, Jugheli L, Maghradze N, Brites D, Ross A, Goig G, Reinhard M, Borrell S, et al. The relative transmission fitness of multidrug-resistant Mycobacterium tuberculosis in a drug resistance hotspot. Nat Commun. 2023:14(1):1988. 10.1038/s41467-023-37719-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo N, Pereira S, Sahin O, Lin J, Huang S, Michel L, Zhang Q. Enhanced in vivo fitness of fluoroquinolone-resistant Campylobacter jejuni in the absence of antibiotic selection pressure. Proc Natl Acad Sci USA. 2005:102(3):541–546. 10.1073/pnas.0408966102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean RC, San Millan A. The evolution of antibiotic resistance. Science 2019:365(6458):1082–1083. 10.1126/science.aax3879. [DOI] [PubMed] [Google Scholar]
- Maneewannakul K, Levy SB. Identification for mar mutants among quinolone-resistant clinical isolates of Escherichia coli. Antimicrob Agents Chemother. 1996:40(7):1695–1698. 10.1128/AAC.40.7.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcusson LL, Frimodt-Møller N, Hughes D. Interplay in the selection of fluoroquinolone resistance and bacterial fitness. PLoS Pathog. 2009:5(8):e1000541. 10.1371/journal.ppat.1000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marolda CL, Valvano MA. Genetic analysis of the dTDP-rhamnose biosynthesis region of the Escherichia coli VW187 (O7:K1) rfb gene cluster: identification of functional homologs of rfbB and rfbA in the rff cluster and correct location of the rffE gene. J Bacteriol. 1995:177(19):5539–5546. 10.1128/jb.177.19.5539-5546.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Martinez L, Pascual A, Jacoby GA. Quinolone resistance from a transferable plasmid. Lancet 1998:351(9105):797–799. 10.1016/S0140-6736(97)07322-4. [DOI] [PubMed] [Google Scholar]
- Méhi O, Bogos B, Csörgő B, Pál F, Nyerges Á, Papp B, Pál C. Perturbation of iron homeostasis promotes the evolution of antibiotic resistance. Mol Biol Evol. 2014:31(10):2793–2804. 10.1093/molbev/msu223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Mira A. The process of genome shrinkage in the obligate symbiont Buchnera aphidicola. Genome Biol. 2001:2(12):1–12. 10.1186/gb-2001-2-12-research0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura de Sousa J, Balbontin R, Durao P, Gordo I. Multidrug-resistant bacteria compensate for the epistasis between resistances. PLoS Biol. 2017:15(4):e2001741. 10.1371/journal.pbio.2001741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munita JM, Arias CA. Mechanisms of antibiotic resistance. Microbiol Spectr. 2016:4(2):2. 10.1128/microbiolspec.VMBF-0016-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyman J, Pearson ES. IX. On the problem of the most efficient tests of statistical hypotheses. Philos Trans A Math Phys Eng Sci. 1933:231:289–337. 10.1098/rsta.1933.0009. [DOI] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015:32(1):268–274. 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas-Chanoine M-H, Bertrand X, Madec J-Y. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev. 2014:27(3):543–574. 10.1128/CMR.00125-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nucci A, Rocha EPC, Rendueles O. 2023. Latent evolution of biofilm formation depends on life-history and genetic background. npj Biofilms Microbiomes. 2023:9:53. 10.1038/s41522-023-00422-3. [DOI]
- Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, Phillippy AM. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016:17(1):132. 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal C, Papp B, Lercher MJ. Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nat Genet. 2005:37(12):1372–1375. 10.1038/ng1686. [DOI] [PubMed] [Google Scholar]
- Pal C, Papp B, Lercher MJ, Csermely P, Oliver SG, Hurst LD. Chance and necessity in the evolution of minimal metabolic networks. Nature 2006:440(7084):667–670. 10.1038/nature04568. [DOI] [PubMed] [Google Scholar]
- Pallecchi L, Bartoloni A, Riccobono E, Fernandez C, Mantella A, Magnelli D, Mannini D, Strohmeyer M, Bartalesi F, Rodriguez H, et al. Quinolone resistance in absence of selective pressure: the experience of a very remote community in the Amazon forest. PLoS Negl Trop Dis. 2012:6(8):e1790. 10.1371/journal.pntd.0001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer AC, Toprak E, Baym M, Kim S, Veres A, Bershtein S, Kishony R. Delayed commitment to evolutionary fate in antibiotic resistance fitness landscapes. Nat Commun. 2015:6(1):7385. 10.1038/ncomms8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X-S, Yague G, Fisher LM. Quinolone resistance mutations in Streptococcus pneumoniae GyrA and ParC proteins: mechanistic insights into quinolone action from enzymatic analysis, intracellular levels, and phenotypes of wild-type and mutant proteins. Antimicrob Agents Chemother. 2001:45(11):3140–3147. 10.1128/AAC.45.11.3140-3147.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papkou A, Hedge J, Kapel N, Young B, Maclean RC. Efflux pump activity potentiates the evolution of antibiotic resistance across S. aureus isolates. Nat Commun. 2020:11(1):3970. 10.1038/s41467-020-17735-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E, Schliep K. Ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2019:35(3):526–528. 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- Patiño-Navarrete R, Rosinski-Chupin I, Cabanel N, Gauthier L, Takissian J, Madec J-Y, Hamze M, Bonnin RA, Naas T, Glaser P. Stepwise evolution and convergent recombination underlie the global dissemination of carbapenemase-producing Escherichia coli. Genome Med. 2020:12(1):10. 10.1186/s13073-019-0699-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne JL, Wagner A. Latent phenotypes pervade gene regulatory circuits. BMC Syst Biol. 2014:8(1):64. 10.1186/1752-0509-8-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V. et al. Scikit-learn: machine learning in python. J Mach Learn Res. 2011:12:2825–2830. https://jmlr.csail.mit.edu/papers/v12/pedregosa11a.html. [Google Scholar]
- Perrin A, Rocha EPC. PanACoTA: a modular tool for massive microbial comparative genomics. NAR Genom Bioinform. 2021:3(1):lqaa106. 10.1093/nargab/lqaa106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham TDM, Ziora ZM, Blaskovich MAT. Quinolone antibiotics. MedChemComm. 2019:10(10):1719–1739. 10.1039/C9MD00120D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piddock LJ, Walters RN. Bactericidal activities of five quinolones for Escherichia coli strains with mutations in genes encoding the SOS response or cell division. Antimicrob Agents Chemother. 1992:36(4):819–825. 10.1128/AAC.36.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole K. At the nexus of antibiotics and metals: the impact of Cu and Zn on antibiotic activity and resistance. Trends Microbiol. 2017:25(10):820–832. 10.1016/j.tim.2017.04.010. [DOI] [PubMed] [Google Scholar]
- Press MO, Li H, Creanza N, Kramer G, Queitsch C, Sourjik V, Borenstein E. Genome-scale co-evolutionary inference identifies functions and clients of bacterial Hsp90. PLoS Genet. 2013:9(7):e1003631. 10.1371/journal.pgen.1003631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin DJ, Rocha EPC, Brown SP. What traits are carried on mobile genetic elements, and why? Heredity (Edinb). 2011:106(1):1–10. 10.1038/hdy.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redgrave LS, Sutton SB, Webber MA, Piddock LJV. Fluoroquinolone resistance: mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol. 2014:22(8):438–445. 10.1016/j.tim.2014.04.007. [DOI] [PubMed] [Google Scholar]
- Robicsek A, Strahilevitz J, Jacoby GA, Macielag M, Abbanat D, Hye Park C, Bush K, Hooper DC. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat Med. 2006:12(1):83–88. 10.1038/nm1347. [DOI] [PubMed] [Google Scholar]
- Roemhild R, Gokhale CS, Dirksen P, Blake C, Rosenstiel P, Traulsen A, Andersson DI, Schulenburg H. Cellular hysteresis as a principle to maximize the efficacy of antibiotic therapy. Proc Natl Acad Sci USA. 2018:115(39):9767–9772. 10.1073/pnas.1810004115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roer L, Overballe-Petersen S, Hansen F, Schonning K, Wang M, Roder BL, Hansen DS, Justesen US, Andersen LP, Fulgsang-Damgaard D, et al. Escherichia coli sequence type 410 is causing new international high-risk clones. mSphere. 2018:3(4):e00337–18. 10.1128/mSphere.00337-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland J, Henao-Diaz LF, Doebeli M, Germain R, Harmon LJ, Knowles LL, Liow LH, Mank JE, Machac A, Otto SP, et al. Conceptual and empirical bridges between micro- and macroevolution. Nat Ecol Evol. 2023:7(8):1181–1193. 10.1038/s41559-023-02116-7. [DOI] [PubMed] [Google Scholar]
- Rozen DE, McGee L, Levin BR, Klugman KP. Fitness costs of fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother. 2007:51(2):412–416. 10.1128/AAC.01161-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz J. Transferable mechanisms of quinolone resistance from 1998 onward. Clin Microbiol Rev. 2019:32(4):4. 10.1128/CMR.00007-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz Y, Zarazaga M, Brinas L, Ruiz-Larrea F, Torres C. Mutations in gyrA and parC genes in nalidixic acid-resistant Escherichia coli strains from food products, humans and animals. J Antimicrob Chemother. 2003:51(4):1001–1005. 10.1093/jac/dkg168. [DOI] [PubMed] [Google Scholar]
- Salverda MLM, Dellus E, Gorter FA, Debets AJM, van der Oost J, Hoekstra RF, Tawfik DS, de Visser JAGM. Initial mutations direct alternative pathways of protein evolution. PLoS Genet. 2011:7(3):e1001321. 10.1371/journal.pgen.1001321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sand O, Gingras M, Beck N, Hall C, Trun N. Phenotypic characterization of overexpression or deletion of the Escherichia coli crcA, cspE and crcB genes. Microbiology 2003:149(8):2107–2117. 10.1099/mic.0.26363-0. [DOI] [PubMed] [Google Scholar]
- San Millan A, Peña-Miller R, Toll-Riera M, Halbert Z, McLean A, Cooper B, MacLean R. Positive selection and compensatory adaptation interact to stabilize non-transmissible plasmids. Nat Commun. 2014:5:1–11. 10.1038/ncomms6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Lopez A, Marshall CW, Haas AL, Turner C, Rasero J, Cooper VS. The roles of history, chance, and natural selection in the evolution of antibiotic resistance. Elife 2021:10:e70676. 10.7554/eLife.70676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliep KP. Phangorn: phylogenetic analysis in R. Bioinformatics 2011:27(4):592–593. 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher MA, Piro KM, Xu W, Hansen S, Lewis K, Brennan RG. Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science 2009:323(5912):396–401. 10.1126/science.1163806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva RF, Mendonca SCM, Carvalho LM, Reis AM, Gordo I, Trindade S, Dionisio F. Pervasive sign epistasis between conjugative plasmids and drug-resistance chromosomal mutations. PLoS Genet. 2011:7(7):e1002181. 10.1371/journal.pgen.1002181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JT, Lewin CS. Chemistry and mechanisms of action of the quinolone antibacterials. In: Andriole VT, editor. The quinolones. San Diego (CA): Academic Press; 1988. p. 22–82. [Google Scholar]
- Stoesser N, Sheppard AE, Pankhurst L, De Maio N, Moore CE, Sebra R, Turner P, Anson LW, Kasarskis A, Batty EM, et al. Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. MBio. 2016:7(2):e02162. 10.1128/mBio.02162-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szappanos B, Fritzemeier J, Csorgo B, Lazar V, Lu X, Fekete G, Balint B, Herczeg R, Nagy I, Notebaart RA, et al. Adaptive evolution of complex innovations through stepwise metabolic niche expansion. Nat Commun. 2016:7(1):11607. 10.1038/ncomms11607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamames J, Moya A, Valencia A. Modular organization in the reductive evolution of protein-protein interaction networks. Genome Biol. 2007:8(5):R94. 10.1186/gb-2007-8-5-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres Ortiz A, Coronel J, Vidal JR, Bonilla C, Moore DAJ, Gilman RH, Balloux F, Kon OM, Didelot X, Grandjean L. Genomic signatures of pre-resistance in Mycobacterium tuberculosis. Nat Commun. 2021:12(1):7312. 10.1038/s41467-021-27616-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Bingen E, Bonacorsi S, Bouchier C, Bouvet O, et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009:5(1):e1000344. 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon M, Perrin A, De Sousa JAM, Vangchhia B, Burn S, O’Brien CL, Denamur E, Gordon D, Rocha EPC. Phylogenetic background and habitat drive the genetic diversification of Escherichia coli. PLoS Genet. 2020:16(6):e1008866. 10.1371/journal.pgen.1008866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trindade S, Sousa A, Xavier KB, Dionisio F, Ferreira MG, Gordo I. Positive epistasis drives the acquisition of multidrug resistance. PLoS Genet. 2009:5(7):e1000578. 10.1371/journal.pgen.1000578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner AK, Eckert SE, Turner DJ, Yasir M, Webber MA, Charles IG, Parkhill J, Wain J. A whole-genome screen identifies Salmonella enterica serovar Typhi genes involved in fluoroquinolone susceptibility. J Antimicrob Chemother. 2020:75(9):2516–2525. 10.1093/jac/dkaa204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Woude MW, Henderson IR. Regulation and function of Ag43 (flu). Annu Rev Microbiol. 2008:62(1):153–169. 10.1146/annurev.micro.62.081307.162938. [DOI] [PubMed] [Google Scholar]
- Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, Burovski E, Peterson P, Weckesser W, Bright J, et al. Scipy 1.0: fundamental algorithms for scientific computing in python. Nat Methods. 2020:17(3):261–272. 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogwill T, Kojadinovic M, Maclean RC. Epistasis between antibiotic resistance mutations and genetic background shape the fitness effect of resistance across species of Pseudomonas. Proc R Soc B: Biol Sci. 2016:283(1830):20160151. 10.1098/rspb.2016.0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos M, Sibleyras L, Lo LK, Hesse E, Gaze W, Klumper U. Zinc can counteract selection for ciprofloxacin resistance. FEMS Microbiol Lett. 2020:3:367. 10.1093/femsle/fnaa038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Dzink-Fox JL, Chen M, Levy SB. Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: role of acrR mutations. Antimicrob Agents Chemother. 2001:45(5):1515–1521. 10.1128/AAC.45.5.1515-1521.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kim Y, Ma Q, Hong SH, Pokusaeva K, Sturino JM, Wood TK. Cryptic prophages help bacteria cope with adverse environments. Nat Commun. 2010:1(1):147. 10.1038/ncomms1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber MA, Ricci V, Whitehead R, Patel M, Fookes M, Ivens A, Piddock LJV. Clinically relevant mutant DNA gyrase alters supercoiling, changes the transcriptome, and confers multidrug resistance. MBio. 2013:4(4):4. 10.1128/mBio.00273-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich DM, Delaney NF, Depristo MA, Hartl DL. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 2006:312(5770):111–114. 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- Whelan FJ, Rusilowicz M, McInerney JO. Coinfinder: detecting significant associations and dissociations in pangenomes. Microbial Genomics. 2020:6(3):3. 10.1099/mgen.0.000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmott CJ, Maxwell A. A single point mutation in the DNA gyrase: a protein greatly reduces binding of fluoroquinolones to the gyrase-DNA complex. Antimicrob Agents Chemother. 1993:37(1):126–127. 10.1128/AAC.37.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A. Epistasis and the evolution of antimicrobial resistance. Front Microbiol. 2017:8:246. 10.3389/fmicb.2017.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A, Kassen R. Parallel evolution and local differentiation in quinolone resistance in Pseudomonas aeruginosa. Microbiology 2011:157(4):937–944. 10.1099/mic.0.046870-0. [DOI] [PubMed] [Google Scholar]
- Wyres KL, Lam MM, Holt KE. Population genomics of Klebsiella pneumoniae. Nat Rev Microbiol. 2020:18(6):344–359. 10.1038/s41579-019-0315-1. [DOI] [PubMed] [Google Scholar]
- Yamane K, Wachino J-i, Suzuki S, Kimura K, Shibata N, Kato H, Shibayama K, Konda T, Arakawa Y. New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. AntAllanimicrob Agents Chemother. 2007:51(9):3354–3360. 10.1128/AAC.00339-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda T, Morimatsu K, Horii T, Nagata T, Ohmori H. Inhibition of Escherichia coli RecA coprotease activities by DinI. EMBO J. 1998;17(11):3207–3216. 10.1093/emboj/17.11.3207. [DOI] [PMC free article] [PubMed]
- Zampieri M, Zimmermann M, Claassen M, Sauer U. Nontargeted metabolomics reveals the multilevel response to antibiotic perturbations. Cell Rep. 2017:19(6):1214–1228. 10.1016/j.celrep.2017.04.002. [DOI] [PubMed] [Google Scholar]
- Zhao X, Xu C, Domagala J, Drlica K. DNA topoisomerase targets of the fluoroquinolones: a strategy for avoiding bacterial resistance. Proc Natl Acad Sci USA. 1997:94(25):13991–13996. 10.1073/pnas.94.25.13991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Alikhan N-F, Mohamed K, Fan Y, Achtman M. The EnteroBase user's Guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020:30(1):138–152. 10.1101/gr.251678.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All major data, or identifiers to public repositories with the data, are incorporated into the article and its online Supplementary material. Other data, e.g. intermediary results files, scripts underlying this article will be shared on reasonable request to the corresponding author.