Significance
Glucose-dependent insulinotropic polypeptide receptor (GIPR) plays an important role in energy metabolism in conjunction with glucagon and glucagon-like peptide-1 receptors. It also has different forms of splice variants (SVs). We found that N terminus–altered SV1 and SV2 of GIPR do not bind to the ligand and elicit signal transduction when expressed individually but are capable of differentially suppressing the wild-type (WT) GIPR signaling when coexpressed. Their cryoelectron microscopy structures show an unusual inward-folded transmembrane helix 6 and extracellular loop 3 that lock the receptor into a peptide-binding pocket-occupied conformation. It appears that SV1 and SV2 are incapacitated in terms of ligand-binding and signal transduction but could negatively affect that of the WT receptor in a ligand-independent and signaling-biased manner.
Keywords: glucose-dependent insulinotropic polypeptide receptor, splice variants, cryo-EM, biased signaling, ligand-independent modulation
Abstract
Glucose-dependent insulinotropic polypeptide receptor (GIPR) is a potential drug target for metabolic disorders. It works with glucagon-like peptide-1 receptor and glucagon receptor in humans to maintain glucose homeostasis. Unlike the other two receptors, GIPR has at least 13 reported splice variants (SVs), more than half of which have sequence variations at either C or N terminus. To explore their roles in endogenous peptide-mediated GIPR signaling, we determined the cryoelectron microscopy (cryo-EM) structures of the two N terminus–altered SVs (referred as GIPR-202 and GIPR-209 in the Ensembl database, SV1 and SV2 here, respectively) and investigated the outcome of coexpressing each of them in question with GIPR in HEK293T cells with respect to ligand binding, receptor expression, cAMP (adenosine 3,5-cyclic monophosphate) accumulation, β-arrestin recruitment, and cell surface localization. It was found that while both N terminus–altered SVs of GIPR neither bound to the hormone nor elicited signal transduction per se, they suppressed ligand binding and cAMP accumulation of GIPR. Meanwhile, SV1 reduced GIPR-mediated β-arrestin 2 responses. The cryo-EM structures of SV1 and SV2 showed that they reorganized the extracellular halves of transmembrane helices 1, 6, and 7 and extracellular loops 2 and 3 to adopt a ligand-binding pocket-occupied conformation, thereby losing binding ability to the peptide. The results suggest a form of signal bias that is constitutive and ligand-independent, thus expanding our knowledge of biased signaling beyond pharmacological manipulation (i.e., ligand specific) as well as constitutive and ligand-independent (e.g., SV1 of the growth hormone-releasing hormone receptor).
G protein–coupled receptors (GPCRs) represent the largest family of membrane proteins that are universally expressed in human tissues (1). They can recognize a diverse range of extracellular ligands and transduce signals to intracellular coupling partners, thereby governing crucial physiological functions (2). GPCR-mediated signaling and pharmacological activities could be profoundly affected by alternative splicing, leading to functional diversity (3, 4).
Splice variants (SVs) have been observed in many GPCRs and shown to be associated with a variety of important physiological functions and pathological processes (5). For instance, one of the growth hormone-releasing hormone receptor (GHRHR) SVs expressed in breast cancer cells is more potent than that of wild-type (WT) to promote cell proliferation, while SVs of corticotropin-releasing factor receptors 1 (CRF1R) and 2 (CRF2R) differentially express in reproductive tissues and act differently upon ligand stimulation, facilitating the transition from relaxed to contractile uterine states (6, 7). In addition, an SV of cholecystokinin B receptor is implicated in the development and progression of colonic and pancreatic carcinomas (8, 9).
Sequence variations of SVs include N terminus truncation or/and substitution, C terminus truncation or/and substitution, intracellular/extracellular loop (ICL/ECL) differences, severe truncation leading to variants with less than seven transmembrane helices (7TMs), or soluble variants (10). In general, N terminus variations impair ligand binding properties, as seen in CRF1R/CRF2R, calcitonin receptor (11), and parathyroid hormone 1 receptor (PTH1R) (12), while C terminus variations show altered signaling or protein interactions, like metabotropic glutamate receptors (13), μ-opioid receptor (14), and 5-hydroxytryptamine receptors (15). ICL differences affect G protein coupling preference and ECL differences change ligand specificity and binding kinetics, as illustrated by pituitary adenylate cyclase-activating polypeptide type 1 receptor (PAC1R) (16) and D3 dopamine receptor (17, 18), respectively. Variants with less than 7TMs caused by severe N terminus truncation exhibit a negative effect on WT receptor signaling (10).
Glucose-dependent insulinotropic polypeptide receptor (GIPR) belongs to the class B1 subfamily of GPCRs and is present in pancreatic cells, adipose tissues, and osteoblasts. Upon GIP stimulation, it regulates insulin secretion, fat accumulation, and bone formation by increasing intracellular adenosine 3,5-cyclic monophosphate (cAMP) levels (19–21). GIPR works with glucagon-like peptide-1 receptor (GLP-1R) and glucagon receptor (GCGR) to maintain glucose homeostasis. While either possessing none (GLP-1R) or only two SVs (GCGR), GIPR has at least 13 SVs with unknown functional properties (22–30). Two C terminus–truncated SVs of GIPR were investigated so far (3, 31). One has 419 residues with a truncated C terminus and 20-amino acid (AA) substitution (referred as GIPR-201 in the Ensembl database), which was shown to coexist with the WT and differentially affect its signaling profile (3). Another found in mouse β-cells only has 263 residues and the corresponding SV in human has 265 residues (referred as GIPR-203 in the Ensembl database). It not only reduces the intracellular trafficking but also decreases the expression of WT receptor in high-fat diet-fed mice, suggesting its involvement in obese-related hypersensitivity to GIP (31).
SVs of GIPR could be divided into several types according to their sequence variations (SI Appendix, Table S1) (32). Both C and N terminus alterations are present. Since SVs with N terminus modifications have not been studied to date, in this study, we constructed and expressed the two representative SVs with N terminus variation (referred as SV1 and SV2, respectively, hereafter) in HEK293T cells independently or jointly with GIPR to elaborate their signaling properties in GIPR-mediated pathways, mainly cAMP accumulation and β-arrestin recruitment. Furthermore, cryoelectron microscopy (cryo-EM) structures of both were obtained to depict the underlying molecular mechanism.
Results
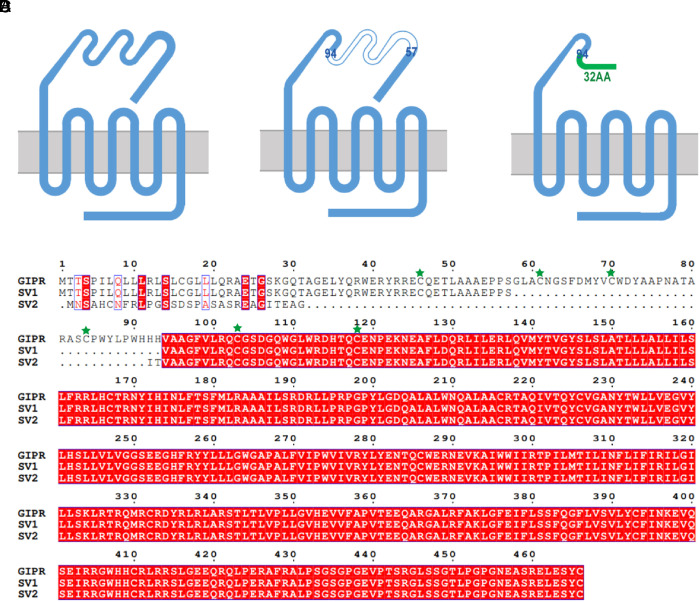
A schematic diagram of GIPR and its SV constructs is depicted in Fig. 1 A–C. SV1 (GIPR-202 in the Ensembl database) has 430 residues in total without one N-terminal segment (residues 58 to 93 in WT GIPR), while SV2 (GIPR-209 in the Ensembl database) possesses a 32-AA replaced N terminus in residues 1 to 93 of WT GIPR. Sequence alignment in Fig. 1D presents the detailed differences between GIPR and the two SVs in question.
Fig. 1.
Schematic diagram of GIPR and its SV constructs. (A) Construct of the WT GIPR. (B) Construct of SV1. Residues 58 to 93 are missing compared to that of WT. (C) Construct of SV2, whose ECD residues 1 to 93 are replaced by a 32-AA sequence (MNSAHCNFRLPGSSDSPASASREAGITEAGIT). (D) Sequence alignment of GIPR and the two SVs. The three conserved ECD disulfide bonds in class B1 GPCR (C46-C70, C61-C103, and C84-C118 for the WT GIPR) are highlighted by green stars.
SV1 and SV2 Neither Bind nor Affect GIP-Induced Signaling Pathways.
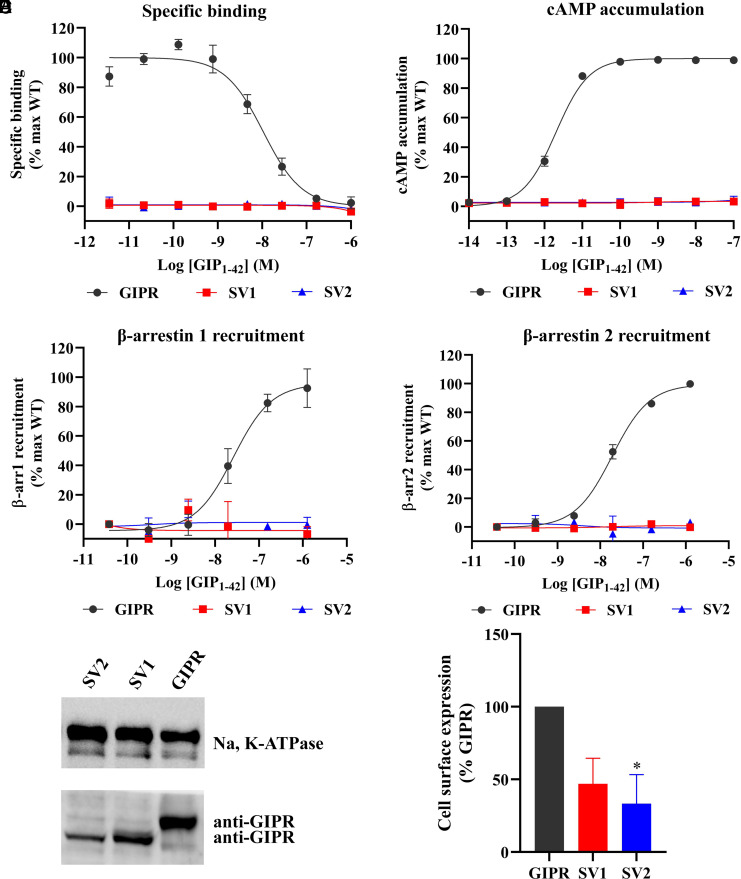
We first expressed GIPR and two SVs separately in HEK293T cells to investigate their abilities to bind GIP1-42 and elicit cAMP accumulation and β-arrestin recruitment. Fig. 2 A–D shows that neither of the SVs displayed any ligand-binding and signaling properties, whereas the WT receptor was highly active in each parameter analyzed. We further detected the membrane expression of each SV by western blot assay. Fig. 2E shows that both SVs were expressed on the membrane, although their expression levels were lower than that of the WT receptor.
Fig. 2.
Ligand-binding, signaling profiles and cell surface expression of GIPR and its SVs. (A) Competitive inhibition of 125I-GIP1-42 binding to GIPR and SVs by unlabeled GIP1-42. Binding affinity is quantified by reduction of radioactivity (counts per minute, CPM). (B) Concentration–response curves of cAMP accumulation elicited by GIP1-42 at GIPR and SVs. (C and D) β-arrestins 1 (β-arr1) and 2 (β-arr2) recruitment by GIPR and SVs. Concentration–response characteristics are shown as the area-under-the-curve (AUC) across the time course–response curve (0 to 10 min) for each concentration. Data shown are means ± SEM of at least three independent experiments (n = 3 to 5) performed in quadruplicate (cAMP accumulation) or duplicate (specific binding and β-arrestin recruitment). Signals were normalized to the maximum (max) response of the WT GIPR, and concentration–response curves were analyzed using a three-parameter logistic equation. (E) Cell surface expression of GIPR and each SV by western blot using antibodies against GIPR and Na, K-ATPase (internal reference), Left panel. The Right panel shows the quantification. Data shown are means ± SEM of three independent experiments (n = 3). One-way ANOVA was used to determine statistical difference (*P < 0.05).
Structure Determination of GIPR SV1 and SV2.
In order to elucidate the molecular basis underlying the pharmacological properties of independently expressed SV1 and SV2, we determined their cryo-EM structures. To prepare high-quality human GIPR SV1–Gs complexes and SV2–Gs complexes, the 45 AAs at the C terminus of the receptor were truncated, and the NanoBiT tethering strategy was applied as previously described (33). To enhance the expression level, a BRIL fusion protein and an optimized maltose binding protein–maltose binding protein (OMBP-MBP) tag were added to the N and C termini of the LgBiT subunit, respectively. Additionally, we introduced one mutation (T6.44bF, class B1 GPCR numbering in superscript) to help stabilize the SV1–Gs/SV2–Gs complexes as previously described (33). This mutation did not affect ligand binding and signaling properties (33). Large-scale purification was followed, and the SV1/SV2–Gs complexes were collected by size-exclusion chromatography (SEC) for cryo-EM studies, with all components of the complex identified in the sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (SI Appendix, Fig. S1).
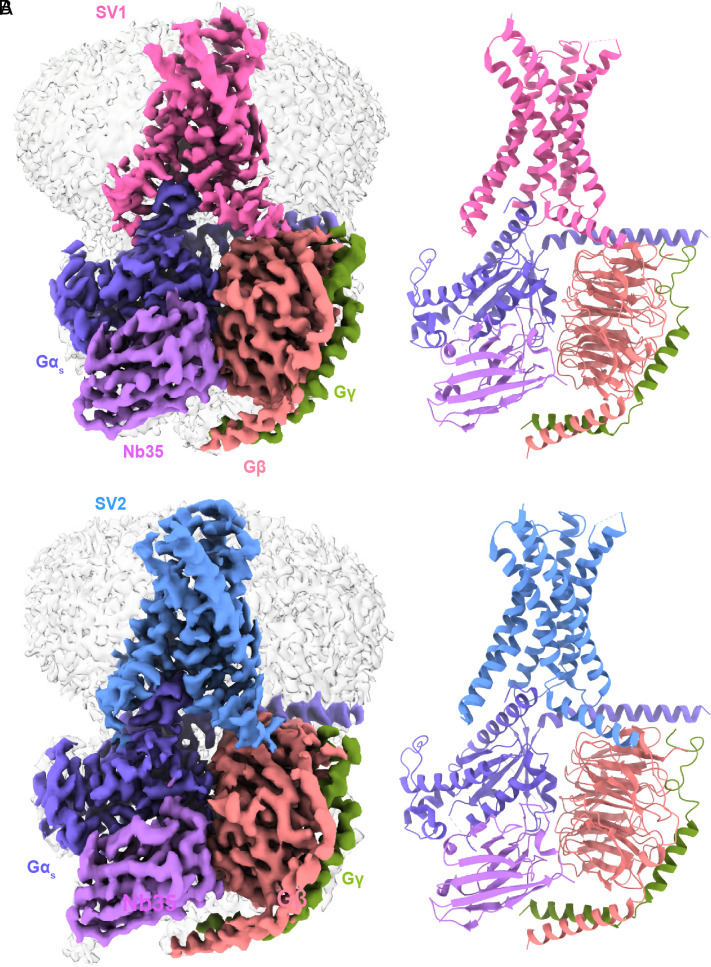
The SV1–Gs and SV2–Gs complexes were imaged using a Titan Krios equipped with a Gatan K3 Summit direct electron detector. Two-dimensional (2D) classification showed a clear secondary structure feature. Different directions of the particles enabled high-resolution cryo-EM maps of SV1–Gs complexes (3.23 Å) and SV2–Gs complexes (3.13 Å) (SI Appendix, Fig. S2). Apart from the α-helical domain of the Gαs, the extracellular domain (ECD) and the ECL1 of the receptor and the absence of GIP, individual receptors (SV1 and SV2) and heterotrimeric Gαs protein in respective complexes were clearly visible. The cryo-EM density of the ECL3 is relatively poor owing to its intrinsic flexibility. Generally, these maps allowed unambiguous modeling of the secondary structure and side chain orientation of the major components of the complexes (Fig. 3 and SI Appendix, Fig. S3). The absence of the GIP indicates that the final structures of SVs are likely ligand-free. This is in line with our pharmacology data that neither SV1 nor SV2 displayed any signaling function (Fig. 2).
Fig. 3.
Cryo-EM structures of SV1 and SV2 of GIPR in complex with Gs. (A) Cryo-EM map (Left) and structural model (Right) of SV1 in complex with Gs. (B) Cryo-EM map (Left) and structural model (Right) of SV2 in complex with Gs. SV1 is shown in hot pink, SV2 in cornflower blue, Gαs in medium slate blue, Gβ subunit in light salmon, Gγ subunit in olive, and Nb35 in medium purple.
Structure Comparison between SVs and GIPR.
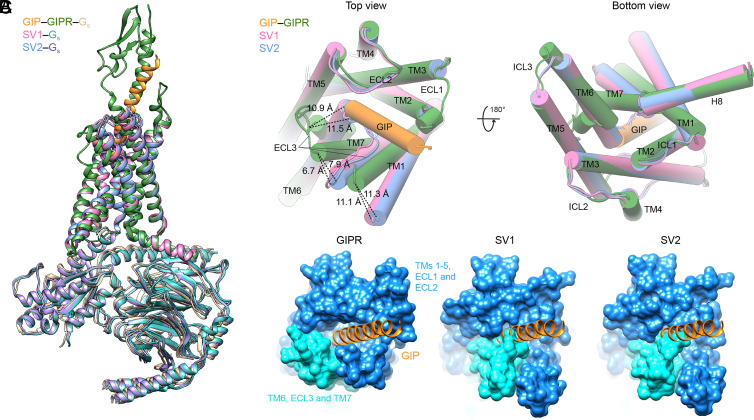
Compared to the GIP-bound GIPR structure (33), the unique ECD sequences of SV1 and SV2 rendered the receptor undergo significant conformational changes, including ECD dynamics, inward movements of the extracellular half of TM6 and ECL3, and outward movements of the extracellular halves of TM1 and TM7, thereby revealing a peptide-binding pocket-occupied conformation unseen before (Fig. 4).
Fig. 4.
Structural comparison of GIPR, SV1, and SV2. (A) Structural comparison of GIP–GIPR–Gs (PDB code: 7DTY), SV1–Gs, and SV2–Gs. The structures of SV1 and SV2 are superimposed on the GIP-bound GIPR using the Cα carbons of the TMD residues. (B) Extracellular view (Left) and intracellular view (Right) of the receptor TMD. The ECD of GIPR and all the G proteins are omitted for clarity. The receptors and peptides are colored as labeled. (C) Surface representation of the TMD peptide-binding pocket among GIPR, SV1, and SV2 structures. The GIP is shown in orange, the N-terminal half of TMD (TMs 1-5, ECL1, and ECL2) in dodger blue, and the C-terminal half of TMD (TM5, ECL3, and TM6) in cyan.
The ECD of the class B1 GPCRs is considered to play a crucial role in ligand recognition. Upon ligand binding, it can initiate a series of events resulting in subsequent signal transduction (34). Because of the absence of one segment (residues 58 to 93 in WT GIPR) that abolished the three conserved ECD disulfide bonds (C46-C70, C61-C103, and C84-C118 for GIPR) (35), the ECD regions of SV1 and SV2 were not resolvable, reflecting their high conformational flexibility (Figs. 1D, 3, and 4A).
The transmembrane domains (TMDs) of SV1 and SV2 are highly similar with a Cα rmsd (root mean square deviation)value of 0.52 Å but significantly different from that of WT GIPR bound by GIP (Cα rmsd values = 1.42 and 1.43 Å, respectively; Fig. 4A). In the GIP–GIPR–Gs complex structure, GIP deeply inserts into the TMD core and makes polar and nonpolar contacts with the extracellular halves of TMs 1-3 and 5-7, ECLs 1-3, and ECD (33). The TMs 2-5 of SV1/SV2 overlapped well with these of WT GIPR, while notable conformational differences were observed in the positions of TMs 1, 6, and 7 and ECL3 (Fig. 4B). Specifically, the extracellular halves of TM1 and TM7 of SV1 moved away from the peptide-binding pocket by 11.1 and 6.7 Å (measured by the Cα of A1261.28b and A3687.33b in GIPR), respectively. Similar outward movements (11.3 and 7.9 Å) were also observed in SV2. Meanwhile, the extracellular halves of TM6 of both SV1 and SV2 moved inward (by 11.5 and 10.9 Å as measured by the Cα of V3566.55b in GIPR, respectively) to occupy the position of the N terminus of GIP. Consequently, the ECL3 moved inward and stood upward in line with TM2 and TM3, largely overlapping with the positions occupied by GIP residues A2 to A13 (Fig. 4B). These TM-level movements of SV1 and SV2 closed the TMD peptide-binding pocket and caused a ligand-binding pocket-occupied conformation, thereby abolishing peptide binding (Fig. 4C). Consistently, molecular dynamics (MD) simulations found that the extracellular half of TM6 and ECL3 intimately interacted with TMs 1-5 and ECL2, i.e., GIPR SV1 adopted a peptide-binding pocket-occupied conformation, as supported by both the interface area and representative minimum distances (SI Appendix, Fig. S4).
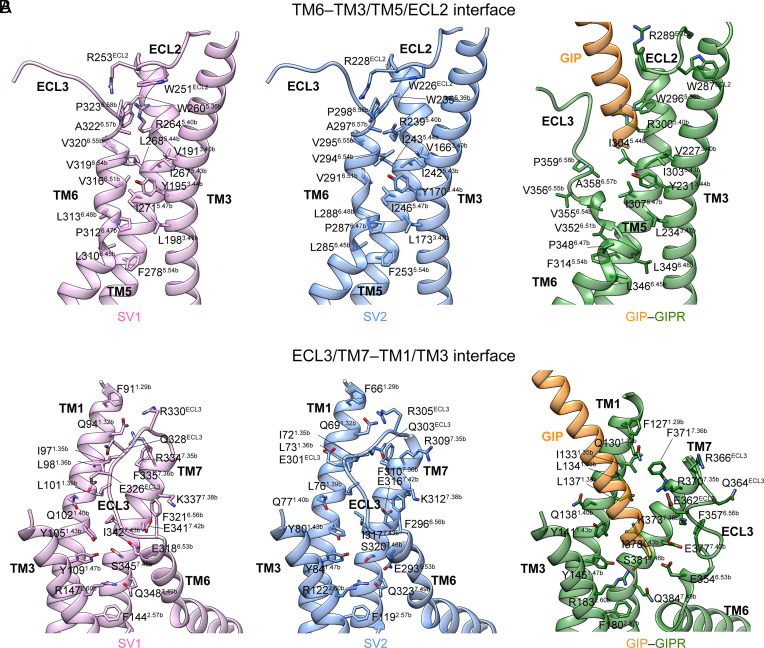
The unusual conformations of SV1 and SV2 were stabilized by massive contacts formed within the interface between TM6/ECL3/TM7 and TM1/TM3/ECL2/TM5 (Fig. 5). Specifically, the extracellular half of SV1 TM6 formed massive hydrophobic contacts with the TM3 residues V1913.40b, Y1953.44b, and L1983.47b (via P3126.47b, V3166.51b, V3196.54b, and A3226.57b) and TM5 residues W2605.36b, R2645.40b, I2675.43b, L2685.44b, I2715.47b, and F2785.54b (via L3106.45b, L3136.48b, V3166.51b, and V3206.55b) (Fig. 5A). Similar hydrophobic interactions were also found in SV2 (Fig. 5B). Meanwhile, two potent hydrogen bonds (G2906.50b–Y1703.44b and V2946.54b–R2395.40b) and one weak hydrogen bond (V2956.55b–R228ECL2) were observed in SV2, while only one hydrogen bond between the side chain of Y1953.44b and the main chain oxygen atom of G3156.50b was seen in SV1. These interactions are absent in WT GIPR whose TM6 is separated from TM3 due to the insertion of bound peptide N terminus (Fig. 5A). The ECL3 of SV1 and SV2 were clasped by TM1 and ECL2 with the formation of several polar contacts including hydrogen bonds (E326ECL3–Q1021.40b and R330ECL3–Q941.32b for SV1; R305ECL3–Q691.32b for SV2). However, ECL3 of WT GIPR is too distant from TM1/ECL2 to contribute notable interaction (Fig. 5B). Of note is the distinct side chain orientation of W287ECL2 in WT GIPR (corresponding to W251ECL2 in SV1 and W226ECL2 in SV2): It inserted to the TM3-TM4 crevice in the GIP–GIPR–Gs structure but occupied the TMD bundle in the SV1–Gs and SV2–Gs structures, reflecting the different conformational dynamics of ECL2 between GIPR and SV1/SV2. Consistently, mutating residues of ECL2 (E288, R289, and N290) in GIPR to alanine significantly impaired the ligand binding, cAMP accumulation, pERK1/2 signaling, and β-arrestin 2 recruitment (36). The TM7 of SV1 and SV2 stood upward in line with TM1 and approached to the latter compared to the WT GIPR, thereby providing additional contacts (Fig. 5B). For example, the side chain of S3457.46b in SV1 rotated outward and pointed to Y1091.47b with the formation of one hydrogen bond. Meanwhile, Q3487.49b rotated inward to connect R1472.60b via two hydrogen bonds. These polar interactions were also observed in SV2 but not WT GIPR, indicating that these structural features of SV1 and SV2 are responsible for their altered behavior in signal transduction.
Fig. 5.
Molecular interactions within the TM6/ECL3/TM7-TM1/TM3/ECL2/TM5 interfaces of GIPR, SV1, and SV2. (A) The extracellular halves of TM6 of both SV1 and SV2 fold inward to form massive polar and nonpolar interactions with TM3, ECL2, and TM5. The structures of SV1 and SV2 are superimposed on the GIP-bound GIPR (PDB code: 7DTY) using the Cα carbons of the TMD residues. (B) The ECL3 and extracellular half of TM7 of SV1 and SV2 are clasped by TM1, TM3 and ECL2 with the formation of multiple contacts.
In the G protein coupling interface, there was a negligible difference between each SV and GIPR. Compared to the WT GIPR, the polar contacts between SV1/SV2 and C terminus of Gαs were reduced (SI Appendix, Fig. S5A). Meanwhile, the five hydrogen bonds between R380GαH5 (GαH5 means α5 helix of Gα) and ICL2 with the backbone atoms of L2453.58b, V2463.59b, L2473.60b and V248ICL2 in the GIP–GIPR–Gs structure were reduced to one (L2113.60b) in SV1 and none in SV2 (SI Appendix, Fig. S5B).
Structure Comparison between GIPR SVs and GHRHR SV1.
We have previously reported the cryo-EM structure of a SV of GHRHR (GHRHR SV1) (37). Similar to the SV1 and SV2 of GIPR, GHRHR SV1 also lacks a portion of the ECD, whose first 89 AAs were replaced by a distinct 25-AA sequence (38). In spite of this change that also destroyed three disulfide bonds, GHRHR SV1 could still engage GHRH and activate the downstream signaling pathways in favor of β-arrestin recruitment (37). Structural comparison shows that the receptor backbone of peptide-free GHRHR SV1 (PDB code: 7V9L) is almost identical to that of GHRH–bound (PDB code: 7V9M) (SI Appendix, Fig. S6A) but significantly different from that of GIPR SV1 or SV2 (SI Appendix, Fig. S6 B and C), especially in the extracellular regions. The sharp kink of TM6 was found in both peptide-free and GHRH-bound GHRHR SV1 structures but absent in that of GIPR SV1–Gs and SV2–Gs, indicating that SVs of GHRHR and GIPR adopt distinct conformations in TM6. As a comparison, the ECL2, ECL3, TM1, and TM7 displayed more profound movements in the cases of GIPR SV1 and SV2. The TMs 2-5 aligned well among the above four structures (SI Appendix, Fig. S6), consistent with their rigidities in maintaining 7TMs architecture.
GIPR and the Two SVs are Colocalized in the Cell.
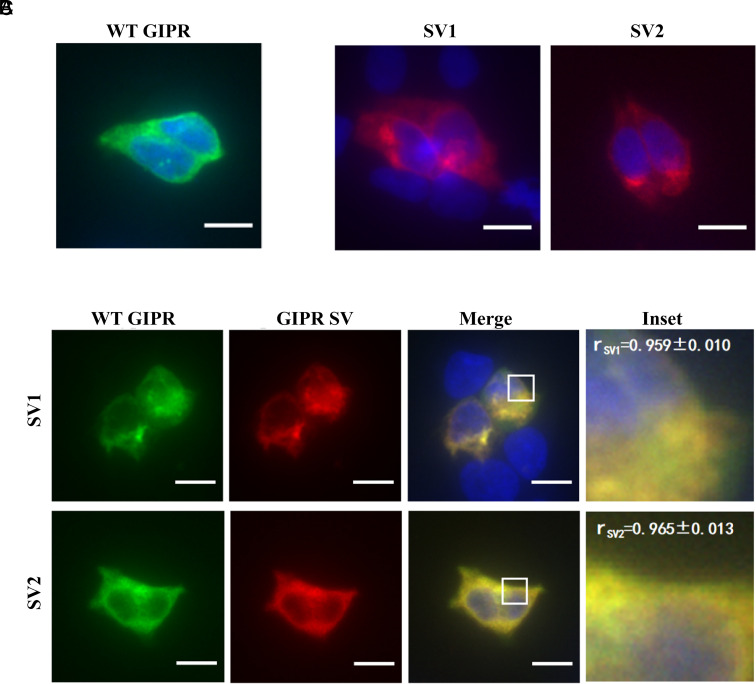
Since SVs are usually expressed in cells and tissues where GIPRs are present (31, 39), we cotransfected GIPR with each SV to assess their location. GIPR and SV1 or SV2 were colocalized either on the membrane or the cytoplasm of transfected HEK293T cells with Pearson’s correlation coefficients (r) above 0.94 (Fig. 6C). Fig. 6 A and B show immunofluorescence staining of HEK293T cells expressing GIPR, SV1 or SV2 individually.
Fig. 6.
Colocalization of GIPR and its SVs. Immunofluorescence staining of HEK293T cells transfected with GIPR-HA (A) or each SV-FLAG (B) alone. To estimate their colocalization, cotransfection of GIPR and individual SVs (C) was performed at a ratio of 1:3 (green, GIPR-HA; red, SV-FLAG; yellow, merge). Data show representative results from three independent experiments. The inset demonstrates the overlapping positions of GIPR and SVs in the cells. Cells were observed by DeltaVision™ Ultra. (Scale bar, 10 μm.) Pearson’s correlation coefficients (r) were determined using the Image J colocalization threshold plugin. Values are means ± SD.
SV1 and SV2 Differentially Modulate GIPR-Mediated Signaling.
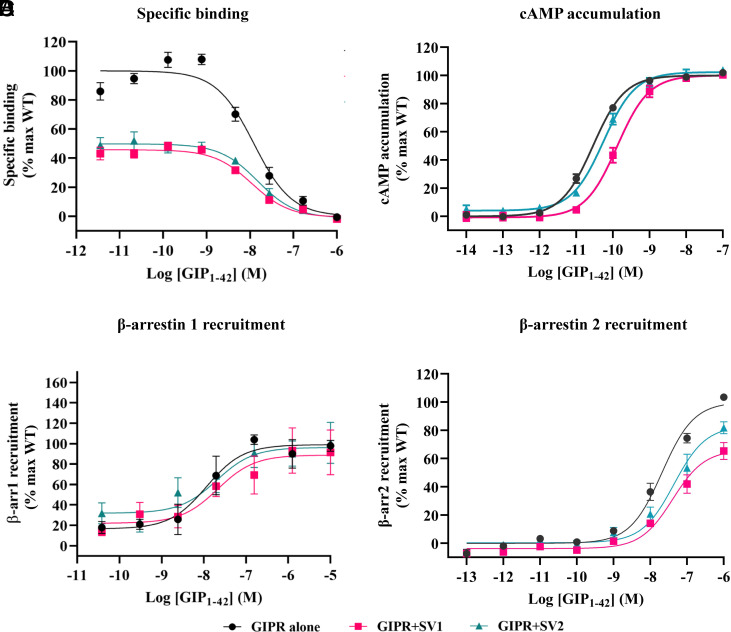
We then cotransfected GIPR with each SV to study whether they influence the signaling profile of the WT receptor. While the binding affinity (Span) of GIP1-42 to the cognate receptor was significantly reduced by 0.53 and 0.49-fold following cotransfection of SV1 or SV2 (Fig. 7A and Table 1), both of them increased the EC50 values of cAMP signaling of the WT receptor (by 3.47 and 0.91-fold, respectively; Fig. 7B and Table 1). Although SV1 and SV2 slightly decreased the Emax values of β-arrestin 2 recruitment (by 0.34 and 0.17-fold, respectively; Fig. 7D and Table 1), neither of them influenced on β-arrestin 1 recruitment (Fig. 7C).
Fig. 7.
Effects of GIPR SVs on ligand binding and the WT GIPR-mediated signal transduction in HEK293T cells coexpressing GIPR and individual SVs. (A) Effects of SVs on competitive binding of 125I-GIP1-42 to GIPR. (B) Effects of SVs on GIP1-42 induced cAMP accumulation at GIPR. (C and D) Effects of SVs and GIPR on β-arrestins 1 (β-arr1) and 2 (β-arr2) recruitment by GIPR. Cells were cotransfected with GIPR and each SV at a ratio of 1:3. Data shown are means ± SEM of at least three independent experiments (n = 3 to 6) performed in quadruplicate (cAMP accumulation) or duplicate (specific binding and β-arrestin recruitment). Signals were normalized to the maximum (max) response of GIPR, and concentration–response curves were analyzed using a three-parameter logistic equation.
Table 1.
Effects of SVs on ligand binding and GIPR-mediated signal transduction in HEK293T cells coexpressing GIPR and individual SVs
| Ligand binding | cAMP accumulation | β-arr1 recruitment | β-arr2 recruitment | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| pIC50 ± SEM | Span (%) ± SEM | pEC50 ± SEM | Emax (%) ± SEM | Δlog (τ/KA) | pEC50 ± SEM | Emax (%) ± SEM | Δlog (τ/KA) | pEC50 ± SEM | Emax (%) ± SEM | Δlog (τ/KA) | |
| WT GIPR | 7.91 ± 0.10 | 100.00 ± 4.87 | 10.54 ± 0.03 | 100.00 ± 0.88 | 0 | 7.94 ± 0.26 | 99.03 ± 6.83 | 0 | 7.68 ± 0.09 | 100.00 ± 3.83 | 0 |
| GIPR+SV1 | 7.96 ± 0.11 | 46.52 ± 2.30*** | 9.89 ± 0.05*** | 100.01 ± 1.87 | −0.33 ± 0.14 | 7.67 ± 0.44 | 88.87 ± 9.90 | −0.37 ± 0.44 | 7.39 ± 0.12 | 66.15 ± 4.09** | −0.67 ± 0.17* |
| GIPR+SV2 | 7.82 ± 0.13 | 51.01 ± 3.26*** | 10.26 ± 0.05** | 102.39 ± 1.44 | 0.12 ± 0.11 | 7.70 ± 0.39 | 96.31 ± 8.45 | 0.18 ± 0.43 | 7.36 ± 0.14 | 83.14 ± 5.53 | −0.25 ± 0.12 |
cAMP accumulation, ligand binding and β-arrestin 1/2 recruitment assays were performed in HEK293T cells. Whole-cell binding assay was performed in CHO-K1 cells. All the measures were fitted to nonlinear regression three-parameter logistic curves. pEC50 is the negative logarithm of the concentration of an agonist that gives a half of the maximum response. pIC50 is the negative logarithm of the 50% inhibitory concentration in competitive radiolabeled ligand binding assay. Emax and span values are the percentage (%) of the maximum response in cells expressing GIPR only. LogR value (log τ/KA, i.e., logarithm of the transduction ratio) is determined by nonlinear regression using the operational model equation. τ is the efficacy value of the agonist and was corrected by cell surface expression of the receptor. KA is dissociation constant. Changes in transduction ratio (Δlog R) were calculated to determine the relative effectiveness of the SVs. Data shown are means ± SEM of at least three independent experiments. One-way ANOVA was used to determine statistical difference (*P < 0.05, **P < 0.01 and ***P < 0.001). β-arr1, β-arrestin 1; β-arr2, β-arrestin 2; WT, wild-type.
SV1 and SV2 Modulate WT GIPR Cell Surface Expression.
When GIPR was cotransfected with each of the SVs, its cell surface expression was significantly decreased (SI Appendix, Fig. S7). Briefly, SV1 and SV2 decreased WT GIPR cell surface expression by 54.5% and 53.1%, respectively.
Effects of RAMPs (Receptor Activity-Modulating Proteins) on GIPR SV1 and SV2 Signaling.
RAMPs were reported to modulate signaling profiles of class B1 GPCRs in an isoform-specific (RAMP1, RAMP2 and RAMP3) and receptor-dependent manner (40–42). To investigate their effects on GIPR SV1 and SV2 signaling, we coexpressed WT GIPR, SV1 or SV2 with RAMPs. In the presence of RAMP2, the ability of SV1 and SV2 to activate G protein was markedly increased, as seen from the improved EC50 values (29.64 nM for SV1 and 26.57 nM for SV2) in the cAMP accumulation assay (SI Appendix, Fig. S8A). Coexpression of RAMP1 only had a nominal effect on SV1/SV2-mediated cAMP accumulation. Meanwhile, no recruitment of β-arrestin 1/2 was observed when GIPR SVs were coexpressed with RAMPs (SI Appendix, Fig. S8 B and C and Table S4).
Discussion
GIP, GLP-1 and glucagon together play a pivotal role in glucose homeostasis mediated via their respective receptors (43, 44). Glucagon increases the release of glucose, while GIP and GLP-1 work synergistically to cause postprandial insulin secretion, regulate glucagon secretion, stimulate β cell proliferation, and protect it from apoptosis (45–49). Of note is that GIP promotes the release of both insulin and glucagon (50), thereby modulating the action of GLP-1 and glucagon on sugar metabolism, probably involving some SVs of GIPR. In order to examine this proposition, we determined the cryo-EM structures of two N terminus–altered SVs of GIPR and investigated their signaling profiles in HEK293T cells expressing GIPR and SVs independently or jointly.
A common feature of the two GIPR SVs is that they neither bind to the endogenous agonist (GIP) nor elicit signal transduction. This was confirmed by the cryo-EM structures of SV1 and SV2, as they adopted a peptide-free conformation state that differs from the GIP–GIPR–Gs structure. Considering that both SVs lack the key ECD residues important for peptide recognition [three disulfide bonds (C46–C70, C61–C103 and C84–C118) and multiple peptide-binding groove residues (M67, Y68, Y87, L88, P89 and W90)] (35), they likely have more dynamic ECDs to weaken the binding of peptide C terminus (35), consistent with their high flexibilities observed in our cryo-EM studies. Moreover, their TM6 and ECL3 showed profound inward movements toward the peptide-binding pocket to exhibit an occupied conformation, thereby preventing ligand interaction. These differences in ECD and TMD explain the reason why SV1 and SV2 do not bind GIP and elicit downstream signaling.
Interestingly, GHRHR SV1 can independently induce signal transduction (37). The different pharmacological profiles between GHRHR SV1 and GIPR SV1/SV2 may reflect the distinct modulating role of ECD and conformational flexibility of TMD among members of class B1 GPCRs. For instance, ECD-truncated GHRHR, CRF1R, PTH1R, and PAC1R could be activated at high concentrations of agonists, whereas ECD-truncated GIPR (SI Appendix, Fig. S9 and Table S5), GCGR and GLP-1R could not (37, 51). Another notable difference comes from the TMD sequences/structures between GHRHR and GIPR. The peptide-free GHRHR SV1 is able to maintain the peptide-binding pocket, induce the sharp kink of TM6, and trigger the outward movement of TM7 in a manner similar to that bound by GHRH. Clearly, GIPR SV1 and SV2 fail to maintain the opening of peptide-binding pocket, thereby acting as a silent receptor when expressed individually.
When coexpressed with WT GIPR, SV1 and SV2 reduced peptide binding that is consistent with decreased receptor expression, but different from their impact on cAMP accumulation and β-arrestin 2 recruitment. It appears that partial loss of the N terminus (SV1, missing 36 residues) led to signal bias toward β-arrestin 2 recruitment and cAMP accumulation, whereas N terminus truncation (missing 93 AAs plus insertion of 32 residues, SV2) had no impact on β-arrestin 2 recruitment but slightly affected cAMP accumulation. Both of them had no effect on β-arrestin 1 responses. In line with previous findings showing that SVs are capable of altering signaling profiles compared to WT receptors (52, 53), our data suggest a constitutive biased mechanism different from signal bias caused by various ligands. For example, SVs of the C-X-C chemokine receptor 3 could activate different signaling pathways through biased agonism (54), and SV1 of GHRHR preferentially transduces signals via β-arrestins, whereas WT GHRHR predominantly activates Gs proteins (37). However, unlike SVs of other GPCRs, that of GIPR are incapacitated in terms of ligand-binding and signal transduction per se, but negatively affect that of the WT receptor in a ligand-independent and signaling-biased manner.
Bidirectional regulation of carbohydrate levels by GIP is essential to the maintenance of glucose homeostasis, although this hinders the development of therapeutic agents targeting GIPR (55). It seems that such a unique modulation of gut hormone actions is finely tuned by SVs with differentiated functionalities. It remains elusive if the above-described phenomenon constitutes a “doubly insured” mechanism for signal modulation in order to fine-tune the action of GIP. Such a complex modulation could be also observed in the regulation of GIPR or SV signaling by RAMPs: GIPR-mediated cAMP accumulation is negatively modulated by SV1, SV2, and RAMP3, while that of SV1 and SV2 is positively modulated by RAMP2 and RAMP3 (SI Appendix, Fig. S8 and Table S4) (40). Clearly, in-depth structural and biochemical studies in both nonengineered cells and in a physiologically relevant system are required to solve the puzzle.
Materials and Methods
Construct.
cDNAs were inserted into the pcDNA3.1 vector by one-step cloning. Addition of Flag- and HA (hemagglutinin)-tags to WT GIPR or SVs was carried out by site-directed mutagenesis. WT GIPR or SVs were cloned to the backbone of Rluc8 at the C terminus. All constructs were confirmed by DNA sequencing (GENEWIZ, Suzhou, China). To optimize the cotransfection assays, three GIPR vs. SV ratios (1:1, 1:3 and 1:6) were tried. Since the impact of 1:1 on GIPR activity was hard to observe and that of 1:3 and 1:6 was similar, we selected 1:3 for the entire study.
The pFastBac vector (Invitrogen) was used to clone the human GIPR SV1 DNA (Genewiz) carrying one mutation (T3096.44bF) (33, 56, 57) or the human GIPR SV2 DNA (Genewiz) with one mutation (T2846.44bF), and the native signal peptide of both was replaced by that of HA. A tobacco etch virus (TEV) protease site and 2GSA linker were present between the N terminus of the extracellular domain (ECD) and an added BRIL fusion protein. Forty-five C-terminal residues (Q386-C430 for SV1; Q361-C405 for SV2) were truncated from the receptor. Before a TEV protease cleavage site and an OMBP-MBP tag, the addition of LgBiT was made to the end of helix 8 with a 15AA polypeptide linker in between (SI Appendix, Fig. S1) (56). A mutated (S54N, G226A, E268A, N271K, K274D, R280K, T284D and I285T) dominant-negative bovine Gαs (DNGαs) construct was employed to stabilize the GIP1-42–SV1–Gs and GIP1-42–SV2–Gs complex (33, 58). A C-terminal SmBiT34 (peptide 86, Promega) connected with a 15AA polypeptide linker was cloned with rat Gβ1. The pFastBac vector was used to clone the modified rat Gβ1 and bovine Gγ2 (33, 56).
Cell Culture.
HEK293T cells were maintained in Dulbecco's Modified Eagle's Medium (DMEM) (Gibco) supplemented with 10% fetal bovine serum (FBS) (Gibco) and 100 mM sodium pyruvate (Gibco). CHO-K1 cells were maintained in F12 (Gibco) supplemented with 10% FBS. All cells were incubated in a humidified environment at 37 °C in 5% CO2. Spodoptera frugiperda 9 (Sf9) (Invitrogen) and High Five™ insect cells (ThermoFisher Scientific) were cultured in ESF 921 serum-free medium (Expression Systems) at 27 °C and 120 rpm (33).
Western Blot Assay.
HEK293T cells (3.5 × 105 cells/mL) were grown for 24 h before transfection with 13 μg plasmid. Cell surface expression of SVs were then determined by western blot 24 h posttransfection. Membrane protein was isolated using the Minute™ plasma membrane protein isolation kit (Invent Biotechnologies) according to the manufacturer’s protocol. After membrane protein isolation, samples were mixed with 5 × SDS loading buffer, loaded and run on SDS-PAGE gel, and then transferred to polyvinylidene difluoride (PVDF) membrane (Merck Millipore) for western blot analysis. The PVDF membrane was blocked by protein-free rapid blocking buffer (EpiZyme Biotec) and incubated with rabbit polyclonal antibody against GIPR (Abcam) and rabbit antibody against Na, K-ATPase (Cell Signaling Technology) followed by three washes with Tris-buffered saline with 0.1% Tween 20 (TBST) (EpiZyme). The membrane was then reacted with horseradish peroxidase-conjugated anti-rabbit antibody (Signalway antibody). Protein bands were detected with enhanced chemiluminescence western blotting detection reagent (Share-bio), and images were collected using ChemiDocTM XRS+ imager (Bio-Rad) and quantified by Image Lab software.
cAMP Accumulation Assay.
GIP1-42 stimulated cAMP accumulation was measured by a LANCE Ultra cAMP kit (PerkinElmer) (37, 56). Cells were seeded onto 6-well cell culture plates and transiently transfected with 4 μg plasmid using Lipofectamine 2000 transfection reagent (Invitrogen). When SVs coexpressed with RAMPs, cells were transfected with 4 μg plasmid containing GIPR or SV1 or SV2:RAMP1 or RAMP2 or RAMP3 or pcDNA3.1 at a ratio of 1:1. After 24 h culture, the transfected cells were seeded onto 384-well microtiter plates at a density of 3,000 cells per well in HBSS (Gibco) supplemented with 5 mM N-2-hydroxyethylpiperazine-N-2-ethane sulfonic acid (HEPES) (Gibco), 0.1% (w/v) bovine serum albumin (BSA) and 0.5 mM 3-isobutyl-1-methylxanthine (Sigma-Aldrich). The cells were stimulated with different concentrations of GIP1-42 for 40 min at room temperature (RT). Eu and Ulight were then diluted by cAMP detection buffer and added to the plates separately to terminate the reaction (32, 33, 56). Plates were incubated at RT for 40 min, and the fluorescence intensity was measured at 620 nm and 650 nm by an EnVision multilabel plate reader (PerkinElmer) (33, 56).
Whole-Cell Binding Assay.
CHO-K1 cells were seeded at a density of 30,000 cells/well in Isoplate-96 plates (PerkinElmer). The WT GIPR or SVs were transiently transfected with 0.2 μg plasmid using Lipofectamine 2000 transfection reagent (32, 33, 56). Twenty-four hours after transfection, cells were washed twice and incubated with blocking buffer (F12 supplemented with 33 mM HEPES and 0.1% BSA, pH 7.4) for 2 h at 37 °C. For homogeneous binding, cells were incubated in binding buffer with a constant concentration of 125I-GIP (40 pM, PerkinElmer) and increasing concentrations of unlabeled GIP1-42 (3.57 pM to 1 μM) at RT for 3 h. Following incubation, cells were washed three times with ice-cold PBS and lysed by addition of 50 μL lysis buffer (PBS supplemented with 20 mM Tris-HCl and 1% Triton X-100, pH 7.4). Fifty µL of scintillation cocktail (OptiPhase SuperMix, PerkinElmer) was added, and the plates were subsequently counted for radioactivity (counts per minute, CPM) in a scintillation counter (MicroBeta2 Plate Counter, PerkinElmer) (32, 33, 56).
β-arrestin 1/2 Recruitment.
HEK293T cells (3.5 × 105 cells/mL) were grown in 6-well plates for 24 h before transfection with 4 μg plasmid containing a GIPR/SV-Rluc8:Venus-β-arrestin1/2 at ratio of 1:9, or a GIPR-Rluc8:SV:Venus-β-arrestin1/2 at a ratio of 1:3:9 (32, 33). When SVs coexpression with RAMPs, cells were transfected with 4 μg plasmid containing GIPR-Rluc8 or SV1-Rluc8 or SV2-Rluc8:Venus-β-arrestin 1 or Venus-β-arrestin 2:GRK5:RAMP1 or RAMP2 or RAMP3 or pcDNA3.1, respectively, at a ratio of 1:5:4:1. Transiently transfected cells were then seeded onto poly-D-lysine coated 96-well culture plates (50,000 cells/well) in DMEM with 10% FBS. Cells were grown overnight before incubation in assay buffer (HBSS supplemented with 10 mM HEPES and 0.1% BSA, pH 7.4) for 30 min at 37 °C. Coelenterazine-h (Yeasen Biotech) was added to a final concentration of 5 μM for 5 min before bioluminescence resonance energy transfer (BRET) readings were made using an EnVision plate reader (32, 33). BRET baseline measurements were collected for 15 cycles prior to ligand addition. Following peptide addition, BRET was measured for 55 cycles. The BRET signal (ratio of 535 nm over 470 nm emission) was corrected to the baseline and then vehicle-treated condition to determine ligand-induced changes in BRET response. Concentration–response values were obtained from the area-under-the-curve of the responses elicited by GIP1-42 (32, 33).
WT GIPR Surface Expression.
HEK293T cells (3.5 × 105 cells/mL) were grown in 6-well plates for 24 h before transfection with 4 μg plasmid containing GIPR:pcDNA3.1 (GIPR alone), or GIPR:GIPR SV1 or GIPR:GIPR SV2 at a ratio of 1:3. The GIPR construct has its native signal peptide replaced by the HA signal peptide, between which and the ECD, a Flag tag with 3GSA linker was inserted [HA-Flag-3GSA-GIPR(22-466)]. No modification was made to the SV constructs. Flow cytometry was used to assess cell surface GIPR expression via detecting the Flag tag 24 h after transfection in HEK293T cells (56, 59). Briefly, PBS containing 5% BSA (w/v) was applied at RT for 15 min to block approximately 2 × 105 cells. The GIPR-expressing cells were then reacted with an anti-Flag primary antibody (1:300 diluted with PBS containing 5% BSA, Sigma-Aldrich) at RT for 1 h. After three times washing with PBS containing 1% BSA (w/v), the cells were incubated for 1 h at RT in the dark with an anti-mouse Alexa Fluor 488 conjugated secondary antibody (1:1,000 diluted with PBS containing 5% BSA, Invitrogen). Following another three times washing, PBS containing 1% BSA (200 μL) was utilized to resuspend the cells, and fluorescent signals were detected by the BD Accuri C6 flow cytometer system (BD Biosciences) at excitation 488 nm and emission 530 nm. Ten thousand (10,000) cellular events were recorded for each sample, and the BD Accuri C6 software v1.0.264.21 was employed to calculate the total fluorescence intensity of cell population that shows positive expression. Data were normalized to the GIPR alone group.
Immunofluorescence Staining.
HEK293T cells were seeded in 6-well plates and transfected with 4 μg plasmid containing GIPR-HA or/and SV-FLAG (32, 40). After 24 h, cells were collected and reseeded in 96-well plates until they reached 50~70% confluence. Cells were washed with PBS before fixation with 4% paraformaldehyde for 15 min. Then, they were washed three more times and blocked with 5% BSA plus 0.1% Triton X-100 for 1 h at RT. Rabbit anti-HA primary antibody (diluted 1:500, Cell Signaling Technology) or/and mouse anti-FLAG primary antibody (diluted 1:300, Sigma-Aldrich) were diluted with incubation buffer (PBS supplemented with 5% BSA) for 1 h followed by 3-time wash. Cells were reacted with 200 μL interaction buffer containing donkey anti-rabbit Alexa 488-conjugated secondary antibody or/and donkey anti-mouse Alexa 647-conjugated secondary antibody (diluted 1:1,000, Invitrogen) at RT for 1 h in the dark. After final washing, nuclei were stained with Hoechst 33258 for 5 min. Cells were imaged using a high-resolution microscope DeltaVision™ Ultra (GE Healthcare, Boston, USA) (32, 40).
SV Protein Expression.
Baculoviruses containing the above complex constructs were prepared by the Bac-to-Bac system (Invitrogen). SV1 or SV2 and DNGαs heterotrimer were coexpressed in High Five™ cells (32, 33). Briefly, insect cells were grown in ESF 921 culture medium to a density of 3.0 × 106 cells/mL. The cells were then infected with BRIL-TEV-2GSA-SV1(22-385)(T309F)-15AA-LgBiT-TEV-OMBP-MBP or BRIL-TEV-2GSA-SV2(2-360)(T284F)-15AA-LgBiT-TEV-OMBP-MBP, DNGαs, Gβ1-peptide 86 and Gγ2, respectively, at a ratio of 1:3:3:3. After 48-h incubation at 27 °C, the cells were collected by centrifugation and stored at –80 °C until use.
Nb35 (Nanobody-35) Expression and Purification.
Nb35 with a 6× his tag at the C terminus was expressed in the periplasm of E. coli BL21 (DE3) cells (33, 56, 59). Briefly, the Nb35 target gene was transformed in the bacterium and amplified in TB culture medium with 100 μg/mL ampicillin, 2 mM MgCl2, 0.1% (w/v) glucose at 37 °C, 180 rpm. When OD600 reached 0.7 to 1.2, 1 mM isopropylthio-β-galactoside was added to induce expression followed by overnight incubation at 28 °C. The cell pellet was then collected under 4 °C and stored at –80 °C. Nb35 was purified by SEC using a HiLoad 16/600 Superdex 75 column (GE Healthcare) with running buffer containing 20 mM HEPES and 100 mM NaCl, pH 7.4. Fractions of Nb35 were concentrated to ~3 mg/mL and quickly frozen in the liquid nitrogen with 10% glycerol and stored in –80 °C.
Complex Formation and Purification.
For the GIP1-42–SV1–Gs or GIP1-42–SV2–Gs complex, cell pellets were lysed in a buffer containing 20 mM HEPES, 100 mM NaCl, pH 7.4, 10 mM MgCl2, 1 mM MnCl2 and 10% glycerol supplemented with EDTA-free protease inhibitor cocktail (TragetMol) (33, 56, 59). Cell membranes were then collected by ultracentrifugation at 4 °C, 90,000 g for 35 min. A buffer consisting of 20 mM HEPES, 100 mM NaCl, pH 7.4, 10 mM MgCl2, 1 mM MnCl2 and 10% glycerol was used to resuspend the collected membranes. To assemble the GIP1-42–SV1–Gs or GIP1-42–SV2–Gs complex, 15 μM GIP1-42 (GL Biochem) was added to the preparation accompanied by 100 μM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), 25 mU/mL apyrase (Sigma-Aldrich), 15 μg/mL Nb35 and 100 U salt active nuclease (Sigma-Aldrich) supplemented with protease inhibitor cocktail for 1.5 h incubation at RT (33, 56). The membrane was then solubilized with 0.5% (w/v) lauryl maltose neopentylglycol (LMNG, Anatrace) and 0.1% (w/v) cholesterol hemisuccinate (CHS, Anatrace) with additional 2 μM GIP1-42 for 2 h at 4 °C. The supernatant was isolated by centrifugation at 90,000 g for 35 min, and the solubilized complex was incubated with amylose resin (New England Biolabs) for 2 h at 4 °C. The resin was collected by centrifugation at 550 g and loaded onto a gravity flow column. The resin in the column was washed with 30 column volumes of buffer containing 20 mM HEPES, pH 7.4, 100 mM NaCl, 10% (v/v) glycerol, 5 mM MgCl2, 1 mM MnCl2, 25 μM TCEP, 5 μM GIP1-42, 0.03% (w/v) LMNG, 0.01% (w/v) glyco-diosgenin (GDN, Anatrace) and 0.008% (w/v) CHS. The protein was then incubated with a buffer consisting of 20 mM HEPES, pH 7.4, 100 mM NaCl, 10% (v/v) glycerol, 5 mM MgCl2, 1 mM MnCl2, 25 μM TCEP, 50 μM GIP1-42, 0.03% (w/v) LMNG, 0.01% (w/v) GDN, 0.008% (w/v) CHS and 30 μg/mL His-tagged TEV protease on the column overnight at 4 °C (33, 56). The flow through was collected and concentrated to 500 μL using a 100-kDa filter (Merck Millipore). SEC was performed by loading the protein onto Superose 6 Increase 10/300GL (GE Healthcare) column with running buffer containing 20 mM HEPES, pH 7.4, 100 mM NaCl, 10 mM MgCl2, 100 μM TCEP, 5 μM GIP1-42, 0.00075% (w/v) LMNG, 0.00025% (w/v) GDN and 0.0002% (w/v) CHS. The GIP1-42–SV1–Gs or GIP1-42–SV2–Gs complexes were collected and concentrated for cryo-EM analysis (33, 56).
Data Acquisition and Image Processing.
The purified GIP1-42–SV1–Gs or GIP1-42–SV2–Gs complexes at a concentration of 18 to 20 mg/mL were applied to glow-discharged holey carbon grids (Quantifoil R1.2/1.3, Au 300 mesh) that were subsequently vitrified by plunging into liquid ethane using a Vitrobot Mark IV (ThermoFisher Scientific) (33, 56). A Titan Krios equipped with a Gatan K3 Summit direct electron detector and serial EM3.7 was used to acquire cryo-EM images. The microscope was operated at 300 kV accelerating voltage, at a nominal magnification of 46,685× in counting mode, corresponding to a pixel size of 1.071 Å. Totally, 5,429 (SV1) and 5,422 movies (SV2) were obtained with a defocus range of −1.2 to −2.2 μm. An accumulated dose of 80 electrons per Å2 was fractionated into a movie stack of 36 frames (56).
Dose-fractionated image stacks were subjected to beam-induced motion correction using MotionCor2.1 (60). A sum of all frames, filtered according to the exposure dose, in each image stack was used for further processing. Contrast transfer function parameters for each micrograph were determined by Gctf v1.06 (61). Automated particle selection and data processing were performed using cryoSPARC v3.2.0+211012 and RELION-3.0 beta2 (62).
For the dataset of the GIP1-42–SV1–Gs–Nb35 complex, automated particle selection yielded 6,613,994 particles, which were subjected to reference-free 2D classification, producing 1,684,203 particles with well-defined averages. This subset of particle projections was subjected to a round of three-dimensional (3D) classification resulting in one well-defined subset with 834,757 projections. This subset of particle projections was further subjected to two rounds of 3D classification resulting in one well-defined subset with 596,712 projections. These particles were subsequently subjected to CTF refinement and Bayesian polishing, which generated a map with an indicated global resolution of 3.23 Å at an FSC of 0.143 (56).
For the dataset of the GIP1-42–SV2–Gs–Nb35 complex, automated particle selection yielded 5,853,096 particles, which were subjected to reference-free 2D classification, producing 1,512,371 particles with well-defined averages. This subset of particle projections was subjected to a round of 3D classification resulting in one well-defined subset with 663,512 projections. This subset of particle projections was further subjected to two rounds of 3D classification resulting in one well-defined subset with 463,406 projections. These particles were subsequently subjected to CTF refinement and Bayesian polishing, which generated a map with an indicated global resolution of 3.13 Å at an FSC of 0.143 (56).
Model Building and Refinement.
The models of the SV1–Gs complex and SV2–Gs complex were built using the cryo-EM structure of the GIP1-42–GIPR–Gs complex (PDB code: 7DTY) (33) as the starting point. The models were docked into the EM density maps using UCSF Chimera (63), followed by iterative manual adjustment and rebuilding in COOT (64). Real space refinement was performed using Phenix (65). The model statistics were validated with comprehensive validation (cryo-EM) in Phenix (66). The final refinement statistics are provided in SI Appendix, Table S2. Structural figures were prepared in UCSF Chimera 1.16 and Chimera X 1.2.4.
MD Simulation.
Molecular dynamic simulations were performed by Gromacs 2021.4. The cryo-EM structure of GIPR SV1 was prepared by the Protein Preparation Wizard (Schrodinger 2021-2) to add missing backbone and side chain atoms. The receptor chain termini were capped with acetyl and methylamide. All the titratable residues were left in their dominant states at pH 7.0 (33, 56). To build MD simulation systems, the receptor was embedded in a bilayer composed of 256 POPC lipids and solvated with 0.15 M NaCl in explicit TIP3P waters using CHARMM-GUI Membrane Builder v3.7 (67). The CHARMM36-CAMP force filed (68) was adopted for protein, lipids, and salt ions. The Particle Mesh Ewald method was used to treat all electrostatic interactions beyond a cutoff of 12 Å, and the bonds involving hydrogen atoms were constrained using the LINCS algorithm (69). The complex system was first relaxed using the steepest descent energy minimization, followed by slow heating of the system to 310 K with restraints. The restraints were reduced gradually over 50 ns. Finally, 1,000 ns production run was carried out for each simulation, with a time step of 2 fs in the NPT ensemble at 310 K and 1 bar using the V-rescale thermostat (70) and the semi-isotropic Parrinello-Rahman barostat (71), respectively. To restrain the GIPR SV1 in its G protein complex conformation, harmonic restraints were placed on all Cα atoms within 5 Å of the G protein binding interface during the MD simulation. The buried interface areas were calculated with FreeSASA using the Shrake–Rupley algorithm with a probe radius of 1.2 Å (72).
Statistical Analysis.
Statistical analysis was performed using GraphPad Prism 8.4 (GraphPad Software). For signaling assays, data of individual experiments were normalized to the maximum responses in cells expressing only the WT GIPR. Nonlinear curve fit was performed using a three-parameter logistic equation [log (agonist vs. response)]. All data are presented as means ± SEM. Significant differences were determined by one-way ANOVA with Dunnett’s test. For colocalization analysis, Pearson’s correlation coefficients (r) were performed using the colocalization threshold plugin of ImageJ. Five separate regions of interest were selected, and means ± SD were determined. Transduction ratio (Log τ/KA) was determined by operational modeling of the concentration–response data, and differences in relative transduction ratio across two individual groups were determined relative to GIPR alone [ΔLog (τ/KA)] (36).
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We are grateful to Zhaotong Cong, Yan Chen, and Chao Zhang for technical assistance. The cryo-EM data were collected at Cryo-Electron Microscopy Research Center, Shanghai Institute of Materia Medica, Chinese Academy of Sciences. This work was partially supported by National Natural Science Foundation of China 82273961 (M.-W.W.), 81872915 (M.-W.W.), 82073904 (M.-W.W.), 82273985 (D.Y.), 82121005 (D.Y.), 81973373 (D.Y.), 21704064 (Q.Z.), and 82204474 (F.Z.); National Science & Technology Major Project of China—Key New Drug Creation and Manufacturing Program 2018ZX09735–001 (M.-W.W.) and 2018ZX09711002–002–005 (D.Y.); STI2030-Major Project 2021ZD0203400 (Q.Z.) and 2022ZD0213000 (F.Z.); the National Key Basic Research Program of China 2018YFA0507000 (M.-W.W.); Novo Nordisk-CAS Research Fund grant NNCAS-2017–1-CC (D.Y.); SA-SIBS Scholarship Program (D.Y.); China Postdoctoral Science Foundation grant 2022M713266 (F.Z.); and Shanghai Sailing Program 22YF1457200 (F.Z.).
Author contributions
D.Y. and M.-W.W. designed research; F.Z., K.H., Q.Z., and A.D. performed research; L.S., H.L., W.L., S.L., X.C., Y.L., Y.X., and W.F. contributed new reagents/analytic tools; F.Z., K.H., Q.Z., D.Y., and M.-W.W. analyzed data; and F.Z., K.H., Q.Z., and M.-W.W. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Dehua Yang, Email: dhyang@simm.ac.cn.
Ming-Wei Wang, Email: mwwang@simm.ac.cn.
Data, Materials, and Software Availability
The atomic coordinates and the electron microscopy maps have been deposited in the Protein Data Bank (PDB) under accession codes 8ITL (SV1–Gs complex) (73) and 8ITM (SV2–Gs complex) (74) and Electron Microscopy Data Bank (EMDB) accession codes EMD-35706 (SV1–Gs complex) (75) and EMD-35707 (SV2–Gs complex) (76), respectively. All relevant data are available from the authors and/or included in the manuscript or SI Appendix.
Supporting Information
References
- 1.Pavlos N. J., Friedman P. A., GPCR signaling and trafficking: The long and short of it. Trends Endocrinol. Metab. 28, 213–226 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strange P. G., Signaling mechanisms of GPCR ligands. Curr. Opin. Drug. Discov. Dev. 11, 196–202 (2008). [PubMed] [Google Scholar]
- 3.Marti-Solano M., et al. , Combinatorial expression of GPCR isoforms affects signalling and drug responses. Nature 587, 650–656 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Furness S. G., Wootten D., Christopoulos A., Sexton P. M., Consequences of splice variation on Secretin family G protein-coupled receptor function. Br. J. Pharmacol. 166, 98–109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding W. Q., Kuntz S. M., Miller L. J., A misspliced form of the cholecystokinin-B/gastrin receptor in pancreatic carcinoma: Role of reduced sellular U2AF35 and a suboptimal 3’-splicing site leading to retention of the fourth intron. Cancer Res. 62, 947–952 (2002). [PubMed] [Google Scholar]
- 6.Grammatopoulos D. K., et al. , A novel spliced variant of the type 1 corticotropin-releasing hormone receptor with a deletion in the seventh transmembrane domain present in the human pregnant term myometrium and fetal membranes. Mol. Endocrinol. 13, 2189–2202 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Grammatopoulos D., et al. , Human corticotropin-releasing hormone receptor: Differences in subtype expression between pregnant and nonpregnant myometria. J. Clin. Endocrinol. Metab. 83, 2539–2544 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Nelson C. P., Challiss R. A., “Phenotypic” pharmacology: The influence of cellular environment on G protein-coupled receptor antagonist and inverse agonist pharmacology. Biochem. Pharmacol. 73, 737–751 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Hellmich M. R., et al. , Human colorectal cancers express a constitutively active cholecystokinin-B/gastrin receptor that stimulates cell growth. J. Biol. Chem. 275, 32122–32128 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Markovic D., Challiss R. A., Alternative splicing of G protein-coupled receptors: Physiology and pathophysiology. Cell Mol. Life Sci. 66, 3337–3352 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nag K., Sultana N., Kato A., Hirose S., Headless splice variant acting as dominant negative calcitonin receptor. Biochem. Biophys. Res. Commun. 362, 1037–1043 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Joun H., et al. , Tissue-specific transcription start sites and alternative splicing of the parathyroid hormone (PTH)/PTH-related peptide (PTHrP) receptor gene: A new PTH/PTHrP receptor splice variant that lacks the signal peptide. Endocrinology 138, 1742–1749 (1997). [DOI] [PubMed] [Google Scholar]
- 13.Cai Z., Schools G. P., Kimelberg H. K., Metabotropic glutamate receptors in acutely isolated hippocampal astrocytes: Developmental changes of mGluR5 mRNA and functional expression. Glia 29, 70–80 (2000). [DOI] [PubMed] [Google Scholar]
- 14.Lu Z., et al. , Mediation of opioid analgesia by a truncated 6-transmembrane GPCR. J. Clin. Invest. 125, 2626–2630 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coupar I. M., Desmond P. V., Irving H. R., Human 5-HT(4) and 5-HT(7) receptor splice variants: Are they important? Curr. Neuropharmacol. 5, 224–231 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCulloch D. A., et al. , ADP-ribosylation factor-dependent phospholipase D activation by VPAC receptors and a PAC(1) receptor splice variant. Mol. Pharmacol. 59, 1523–1532 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Richtand N. M., Behavioral sensitization, alternative splicing, and d3 dopamine receptor-mediated inhibitory function. Neuropsychopharmacology 31, 2368–2375 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karpa K. D., Lin R., Kabbani N., Levenson R., The dopamine D3 receptor interacts with itself and the truncated D3 splice variant d3nf: D3–D3nf interaction causes mislocalization of D3 receptors. Mol. Pharmacol. 58, 677–683 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Campbell J. E., Targeting the GIPR for obesity: To agonize or antagonize? Potential mechanisms. Mol. Metab. 46, 101139 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yue J. T. Y., Lam T. K. T., Antiobesogenic effects of central GIPR antagonism. J. Clin. Invest. 129, 3532–3535 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yabe D., Seino Y., Two incretin hormones GLP-1 and GIP: Comparison of their actions in insulin secretion and beta cell preservation. Prog. Biophys. Mol. Biol. 107, 248–256 (2011). [DOI] [PubMed] [Google Scholar]
- 22.EMBL-EBI, Gene: GIPR ENSG00000010310 structural variants. https://asia.ensembl.org/Homo_sapiens/Gene/StructuralVariation_Gene?db=core;g=ENSG00000010310;r=19:45668221-45683722 (Accessed 13 January 2022).
- 23.NCBI, Gastric inhibitory polypeptide receptor isoform X1 [Homo sapiens]. Available at https://www.ncbi.nlm.nih.gov/protein/XP_011525012.1. (Accessed 13 January 2022).
- 24.NCBI, Gastric inhibitory polypeptide receptor isoform X2 [Homo sapiens]. Available at https://www.ncbi.nlm.nih.gov/protein/XP_047294555.1. (Accessed 13 January 2022).
- 25.NCBI, Gastric inhibitory polypeptide receptor isoform X4 [Homo sapiens]. Available at https://www.ncbi.nlm.nih.gov/protein/XP_011525015.1. (Accessed 13 January 2022).
- 26.NCBI, Gastric inhibitory polypeptide receptor isoform X5 [Homo sapiens]. Available at https://www.ncbi.nlm.nih.gov/protein/XP_047294557.1. (Accessed 13 January 2022).
- 27.NCBI, Gastric inhibitory polypeptide receptor isoform X7 [Homo sapiens]. Available at https://www.ncbi.nlm.nih.gov/protein/XP_011525018.1. (Accessed 13 January 2022).
- 28.NCBI, Gastric inhibitory polypeptide receptor isoform X6 [Homo sapiens]. Available at https://www.ncbi.nlm.nih.gov/protein/XP_011525017.1. (Accessed 13 January 2022).
- 29.NCBI, Gastric inhibitory polypeptide receptor isoform X8 [Homo sapiens]. Available at https://www.ncbi.nlm.nih.gov/protein/XP_047294558.1. (Accessed 13 January 2022).
- 30.NCBI, Gastric inhibitory polypeptide receptor isoform 2 precursor [Homo sapiens]. Available at https://www.ncbi.nlm.nih.gov/protein/NP_001295347.1. (Accessed 13 January 2022).
- 31.Harada N., et al. , A novel GIP receptor splice variant influences GIP sensitivity of pancreatic beta-cells in obese mice. Am. J. Physiol. Endocrinol. Metab 294, E61–68 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Hang K., et al. , Ligand-independent modulation of GIPR signaling by splice variants. bioRxiv [Preprint] (2022). https://www.biorxiv.org/content/10.1101/2022.01.24.477496v2 (Accessed 20 July 2023).
- 33.Zhao F. H., et al. , Structural insights into hormone recognition by the human glucose-dependent insulinotropic polypeptide receptor. Elife 10, e68719 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang D., et al. , G protein-coupled receptors: Structure- and function-based drug discovery. Signal Transduct. Target Ther. 6, 7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parthier C., et al. , Crystal structure of the incretin-bound extracellular domain of a G protein-coupled receptor. Proc. Natl. Acad. Sci. U.S.A. 104, 13942–13947 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuliantie E., et al. , Insights into agonist-elicited activation of the human glucose-dependent insulinotropic polypeptide receptor. Biochem. Pharmacol. 192, 114715 (2021). [DOI] [PubMed] [Google Scholar]
- 37.Cong Z., et al. , Constitutive signal bias mediated by the human GHRHR splice variant 1. Proc. Natl. Acad. Sci. U.S.A. 118, e2106606118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rekasi Z., Czompoly T., Schally A. V., Halmos G., Isolation and sequencing of cDNAs for splice variants of growth hormone-releasing hormone receptors from human cancers. Proc. Natl. Acad. Sci. U.S.A. 97, 10561–10566 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.GTEx, Isoform expression of GIPR: ENSG00000010310.8 gastric inhibitory polypeptide receptor [Source:HGNC Symbol;Acc:HGNC:4271]. https://gtexportal.org/home/transcriptPage. (2022).
- 40.Shao L., et al. , Modulating effects of RAMPs on signaling profiles of the glucagon receptor family. Acta Pharm. Sin. B 12, 637–650 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kotliar I. B., Lorenzen E., Schwenk J. M., Hay D. L., Sakmar T. P., Elucidating the interactome of G protein-coupled receptors and receptor activity-modifying proteins. Pharmacol. Rev. 75, 1–34 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harris M., et al. , RAMPs regulate signalling bias and internalisation of the GIPR. bioRxiv [Preprint] (2021). https://www.biorxiv.org/content/10.1101/2021.04.08.436756v2 (Accessed 09 June 2023).
- 43.Cho Y. M., Merchant C. E., Kieffer T. J., Targeting the glucagon receptor family for diabetes and obesity therapy. Pharmacol. Ther 135, 247–278 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Sekar R., Singh K., Arokiaraj A. W., Chow B. K., Pharmacological actions of glucagon-like peptide-1, gastric inhibitory polypeptide, and glucagon. Int. Rev. Cell Mol. Biol. 326, 279–341 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Seino Y., Fukushima M., Yabe D., GIP and GLP-1, the two incretin hormones: Similarities and differences. J. Diabetes Investig. 1, 8–23 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hansotia T., Drucker D. J., GIP and GLP-1 as incretin hormones: Lessons from single and double incretin receptor knockout mice. Regul. Pept. 128, 125–134 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Skow M. A., Bergmann N. C., Knop F. K., Diabetes and obesity treatment based on dual incretin receptor activation: “Twincretins”. Diabetes Obes. Metab. 18, 847–854 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Alexiadou K., Anyiam O., Tan T., Cracking the combination: Gut hormones for the treatment of obesity and diabetes. J. Neuroendocrinol. 31, e12664 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Estall J. L., Drucker D. J., Glucagon and glucagon-like peptide receptors as drug targets. Curr. Pharm. Des. 12, 1731–1750 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Gasbjerg L. S., et al. , Glucose-dependent insulinotropic polypeptide (GIP) receptor antagonists as anti-diabetic agents. Peptides 100, 173–181 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Zhao L. H., et al. , Differential requirement of the extracellular domain in activation of class B G protein-coupled receptors. J. Biol. Chem. 291, 15119–15130 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maggio R., et al. , Variants of G protein-coupled receptors: A reappraisal of their role in receptor regulation. Biochem. Soc. Trans. 44, 589–594 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Kochman K., Superfamily of G-protein coupled receptors (GPCRs)–extraordinary and outstanding success of evolution. Postepy Hig. Med. Dosw (Online) 68, 1225–1237 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Berchiche Y. A., Sakmar T. P., CXC chemokine receptor 3 alternative splice variants selectively activate different signaling pathways. Mol. Pharmacol. 90, 483–495 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Killion E. A., et al. , Anti-obesity effects of GIPR antagonists alone and in combination with GLP-1R agonists in preclinical models. Sci. Transl. Med. 10, eaat3392 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Zhao F. H., et al. , Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors. Nat. Commun. 13, 1057 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wootten D., Simms J., Miller L. J., Christopoulos A., Sexton P. M., Polar transmembrane interactions drive formation of ligand-specific and signal pathway-biased family B G protein-coupled receptor conformations. Proc. Natl. Acad. Sci. U.S.A. 110, 5211–5216 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liang Y. L., et al. , Dominant negative G proteins enhance formation and purification of agonist-GPCR-G protein complexes for structure determination. ACS Pharmacol. Transl. Sci. 1, 12–20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu Y. N., et al. , A distinctive ligand recognition mechanism by the human vasoactive intestinal polypeptide receptor 2. Nat. Commun. 13, 2272 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scheres S. H., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pettersen E. F., et al. , UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 65.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen V. B., et al. , MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu E. L., et al. , CHARMM-GUI membrane builder toward realistic biological membrane simulations. J. Comput. Chem. 35, 1997–2004 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guvench O., et al. , CHARMM additive all-atom force field for carbohydrate derivatives and its utility in polysaccharide and carbohydrate-protein modeling. J. Chem. Theory Comput. 7, 3162–3180 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hess B., P-LINCS: A parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 4, 116–122 (2008). [DOI] [PubMed] [Google Scholar]
- 70.Bussi G., Donadio D., Parrinello M., Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 014101 (2007). [DOI] [PubMed] [Google Scholar]
- 71.Aoki K. M., Yonezawa F., Constant-pressure molecular-dynamics simulations of the crystal-smectic transition in systems of soft parallel spherocylinders. Phys. Rev. A 46, 6541–6549 (1992). [DOI] [PubMed] [Google Scholar]
- 72.Mitternacht S., FreeSASA: An open source C library for solvent accessible surface area calculations. F1000Res. 5, 189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao F., et al. , Cryo-EM structure of GIPR splice variant 1 (SV1) in complex with Gs protein. Protein Data Bank. https://www.rcsb.org/structure/8ITL. Deposited 22 March 2023. [Google Scholar]
- 74.Zhao F., et al. , Cryo-EM structure of GIPR splice variant 2 (SV2) in complex with Gs protein. Protein Data Bank. https://www.rcsb.org/structure/8ITM. Deposited 22 March 2023. [Google Scholar]
- 75.Zhao F., et al. , Cryo-EM structure of GIPR splice variant 1 (SV1) in complex with Gs protein. Electron Microscopy Data Bank. https://www.emdataresource.org/EMD-35706. Deposited 22 March 2023. [Google Scholar]
- 76.Zhao F., et al. , Cryo-EM structure of GIPR splice variant 2 (SV2) in complex with Gs protein. Electron Microscopy Data Bank. https://www.emdataresource.org/EMD-35707. Deposited 22 March 2023. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
The atomic coordinates and the electron microscopy maps have been deposited in the Protein Data Bank (PDB) under accession codes 8ITL (SV1–Gs complex) (73) and 8ITM (SV2–Gs complex) (74) and Electron Microscopy Data Bank (EMDB) accession codes EMD-35706 (SV1–Gs complex) (75) and EMD-35707 (SV2–Gs complex) (76), respectively. All relevant data are available from the authors and/or included in the manuscript or SI Appendix.