Significance
Dysregulation of the transcriptome-wide m6A methylome due to aberrant expression of the m6A writer complex has been widely observed in human cancers. But the mechanism is unknown. We report that histone acetyl-lysine reader BRD4 determines the m6A writer complex integrity via maintaining basal transcription of seven methyltransferase complex (MTC) components, thereby modulating global m6A levels. Meanwhile, METTL3 is critical for maintaining DNA damage response via m6A modification. We reveal that cotargeting BRD4 and PARP are highly synergistic in multiple PARPi-resistant human cancer PDX models, largely attributed to impaired MTC integrity and subsequently compromised m6A modification. We propose that the interplay between BRD4-dependent epigenetic and MTC-mediated epitranscriptomic networks may provide a biological rationale for both BET-targeted and combined DNA repair-targeted therapies.
Keywords: BET, m6A, DNA repair
Abstract
Aberrant transcripts expression of the m6A methyltransferase complex (MTC) is widely found across human cancers, suggesting a dysregulated signaling cascade which integrates m6A epitranscriptome to drive tumorigenesis. However, the responsible transcriptional machinery directing the expression of distinct MTC subunits remains unclear. Here, we identified an unappreciated interplay between the histone acetyl-lysine reader BRD4 and the m6A writer complex across human cancers. BRD4 directly stimulates transcripts expression of seven MTC subunits, allowing the maintenance of the nuclear writer complex integrity. Upon BET inhibition, this BRD4-MTC signaling cascade accounts for global m6A reduction and the subsequent dynamic alteration of BRD4-dependent transcriptome, resulting in impaired DNA damage response that involves activation of homologous recombination (HR) repair and repression of apoptosis. We further demonstrated that the combined synergy upon BET/PARP inhibition largely relies on disrupted m6A modification of HR and apoptotic genes, counteracting PARP inhibitor (PARPi) resistance in patient-derived xenograft models. Our study revealed a widespread active cross-talk between BRD4-dependent epigenetic and MTC-mediated epitranscriptomic networks, which provides a unique therapeutic vulnerability that can be leveraged in combined DNA repair-targeted therapy.
N 6-methyladenosine (m6A) is the most prevalent modification in eukaryotic mRNAs (1, 2). A multiprotein m6A writer complex catalyzes m6A methylation, a process that can be reversed by FTO and ALKBH5, two demethylases known as erasers (3–5). The writer complex includes a core component comprised of METTL3, METTL14, WTAP, and other regulatory subunits like VIRMA, ZC3H13, HAKAI, and RBM15 (6, 7). WTAP is required for optimal substrate recruitment and METTL3/14 localization (8). VIRMA is critical for deposition of m6A specifically to the 3′UTR (9). ZC3H13 promotes nuclear localization of the writer complex (10), whereas RBM15 facilitates the methylation of certain RNAs via binding to U-rich regions (11). Dynamic m6A modification has been linked to various biological processes and human diseases including cancer (12–14). However, the regulation of the writer complex itself remains elusive. We have recently reported an acetylation-dependent regulation of METTL3 localization that impacts on metastatic dissemination, which can be influenced by proinflammatory cytokines, allowing METTL3 to function at different cellular compartments through both m6A-dependent and -independent signaling pathways (15).
The major biological consequence of m6A on mRNAs is to maintain mRNA homeostasis, thereby modulating many RNA metabolism processes, including transcription, splicing, transport, and translation (16–18). m6A reader proteins orchestrate the dynamic turnover rate of m6A-modified transcripts via recruiting diverse regulatory machinery, resulting in either increased or decreased mRNA stability (19–24). Interestingly, the expression levels of the m6A writer complex are often dysregulated in various types of cancers, accompanied by globally increased m6A (25, 26). However, the mechanisms that govern aberrant expression of MTC subunits remain completely unknown.
Bromine domain and extra terminal domain (BET) family comprises bromodomain-containing protein BRD2, BRD3, BRD4, and testis-specific protein (BRDT). By recognizing acetylated lysine residues on histones or target proteins, BET family maintains and facilitates transcription via promoter binding or enhancer binding (27). Similar to the m6A writer complex, BET proteins play a pivotal role in regulating various cellular events and cancer development (27, 28). Small molecules that target BET proteins (BETi) have emerged as potential therapeutic agents (29, 30). The administration of BETi in cancer therapy is often based on cross-talks between BET proteins and essential oncogenic drivers in a context-dependent manner (31). However, the mechanisms underlying the wide range of sensitivity to BET inhibition remain largely unknown. Primary and acquired resistance to BET inhibition often occurs during cancer treatment (32, 33). Elucidating the mode of action of BET proteins, especially the identification of more generalized oncogenic pathways downstream of BET family, would optimize the development of new generation of BETi compounds to maximize its clinical benefit, as well as reducing cytotoxicity.
Here, we found that BET inhibition represses m6A writer complex components expression at the transcription level, followed by significantly reduced global m6A abundance. BRD4 is responsible for direct transactivation of MTC subunits and the subsequent nuclear retention of METTL3/METTL14. BETi treatment leads to reduced HR repair and increased apoptosis via dual regulation of m6A-dependent transcripts turnover. We further demonstrate that MTC subunit reconstitution counteracts the in vivo cytotoxicity caused by coadministration of BETi and PARPi, supportive of the contribution of the m6A writer complex to the combined BETi/PARPi synergistic effects in human cancers. In PARPi-resistant PDX models, BETi and PARPi cotreatment leads to profoundly impaired MTC expression and m6A modification, highlighting the in vivo relevance of the BRD4–MTC axis in directing DNA repair-targeted therapy.
Results
BRD4 Positively Correlates with the m6A Writer Complex Expression and m6A Abundance in Human Cancers.
We analyzed transcripts expression profiles of seven MTC subunits (METTL3, METTL14, WTAP, HAKAI, RBM15, VIRMA, and ZC3H13—MMWHRVZ in short) in a cohort of breast tumors (n = 50) obtained from The Cancer Genome Atlas (TCGA) database. Overall, these subunits were significantly higher in tumors compared to matched normal breast tissues (SI Appendix, Fig. S1A). To test whether this is associated with genetic aberrations, we assessed 2,173 primary breast cancer samples in TCGA using the cBioPortal visualizer (http://www.cbioportal.org/) and found that only VIRMA showed a 20% amplification rate (435/2,173), whereas the rest of the subunits displayed undetectable or very low frequency of mutations, amplifications, or homozygous deletions (SI Appendix, Fig. S1B). Interestingly, while analyzing our RNA-seq data derived from BETi-treated samples, we happened to find that BETi affected multiple aspects of RNA metabolism, accompanied by profoundly reduced transcripts expression of MMWHRVZ subunits (SI Appendix, Fig. S1 C and D). Analyzing published transcriptomic datasets from other groups also revealed significantly altered RNA biological pathways upon BET inhibition (34) (SI Appendix, Fig. S1E). These data suggest a potential role of BET family members in modulating m6A writer complex transcription and the subsequent impact on RNA metabolism.
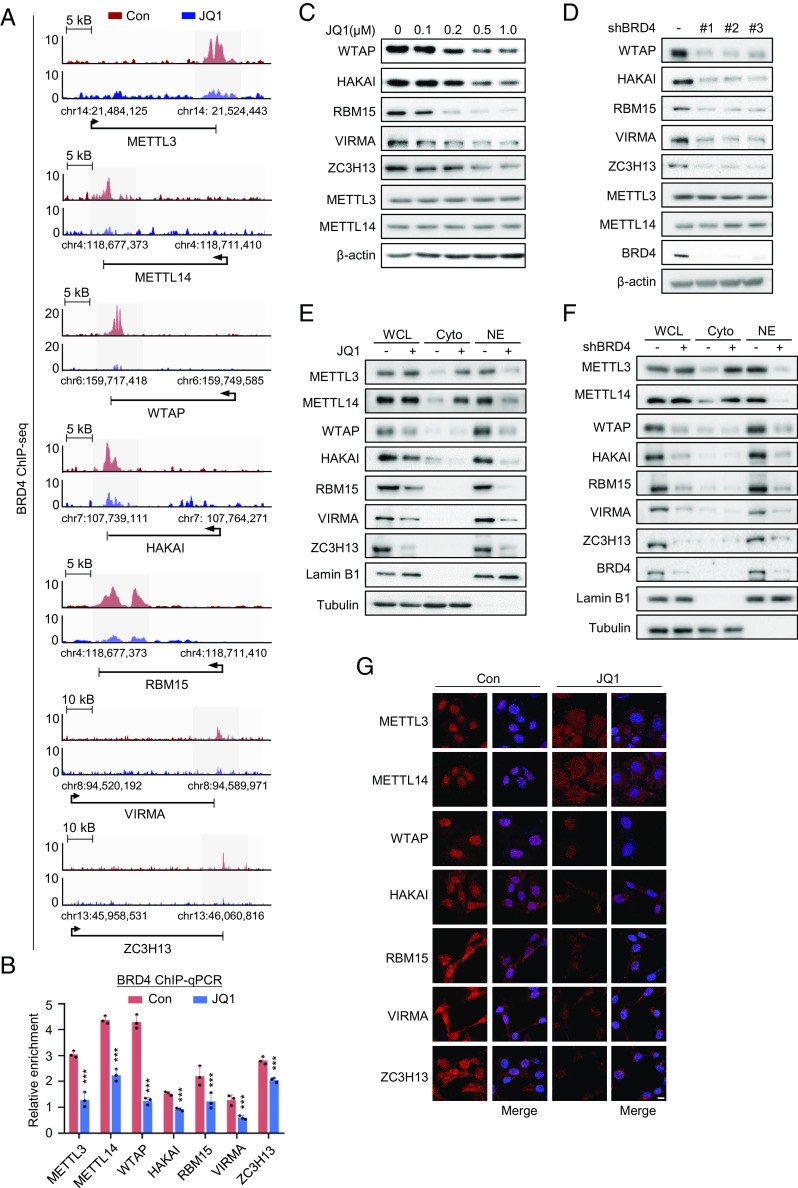
We next examined the correlation between MTC subunits and BET family members in human cancers. A positive correlation between BRD4 and MMWHRZ subunits (except VIRMA) was found in breast (n = 1,085), ovarian (n = 426), prostate (n = 492), and pancreatic cancers (n = 179) from the TCGA database (SI Appendix, Fig. S1 F–I). We assessed the correlation among m6A levels and transcripts levels of BET and MTC subunits in 36 pairs of frozen human breast cancers and their adjacent normal tissues (Fig. 1A and SI Appendix, Fig. S1J). Except ZC3H13, the average expression levels of MMWHRV subunits were all significantly up-regulated in tumor regions, accompanied by a remarkable increase of m6A. By contrast, the mRNA transcript of FTO was largely down-regulated in tumor samples, whereas ALKBH5 showed no difference. Among 3 BRD family members tested, only BRD4 mRNA was profoundly increased in tumor areas. Meanwhile, we stratified these tumor samples as BRD2/3/4-low and -high according to their transcripts abundance. A significant positive correlation between m6A levels and BRD4 expression was found (Fig. 1B), whereas BRD2 and BRD3 showed no correlation with m6A at all (SI Appendix, Fig. S1K). Meanwhile, the absence of BRD2 or BRD3 failed to reduce total m6A abundance (SI Appendix, Fig. S1 L and M). We also performed a Pearson correlation test (R) between BETs and the MTC subunits among these samples based on transcripts expression (SI Appendix, Fig. S1N). While BRD2 or BRD3 showed no detectable correlation with MTC components, BRD4 displayed a significant positive correlation with six subunits, except VIRMA. We also assessed protein levels of BRD4 in correlation with the MTC in tumor samples (n = 23). Except METTL3 and METTLE14, the rest of the subunits exhibited significant correlation with BRD4 protein expression (SI Appendix, Fig. S1O). We further confirmed that BRD4 inhibition (treatment with additional BETis including Molibresib and IBET-151, or KD BRD4) decreased transcript levels of MMWHRVZ subunits and global m6A modification in MDA-MB-231 cells (Fig. 1 C–F). Similar results were obtained in a variety of human cancer cells (SI Appendix, Fig. S1 P–R), indicative of a widespread and specific regulation of MTC-dependent m6A signaling by BRD4.
Fig. 1.

BRD4 positively correlates with m6A writer complex expression and m6A abundance in human cancers. (A) LC-MS/MS quantification of the m6A/A ratio in polyadenylated RNA in paired normal and breast tumor samples (n = 36 per group, pairs connected with lines, P = 0.0002). (B) LC-MS/MS quantification of the m6A/A ratio in polyadenylated RNA in human breast cancer biopsies with low (n = 14) or high (n = 22) BRD4 expression. Samples were divided into “low” and “high” groups based on BRD4 levels measured by QRT-PCR. The cutoff was determined by the best-performing threshold in terms of the P values between both groups (regardless of the direction of the change in m6A), as determined by the t test (P = 0.0064). (C and D) QRT-PCR quantification of the indicated mRNAs in MDA-MB-231 cells treated with BETi for 24 h (C) or infected with the indicated shBRD4 lentiviruses (D). (E and F) LC-MS/MS quantification of the m6A/A ratio in polyadenylated RNA isolated from MDA-MB-231 cells treated with BETi for 24 h (E), or infected with the indicated shBRD4 lentiviruses (F). (A and B) Data are represented as mean ± SEM, and P values were calculated by the two-tailed paired t test. *P < 0.05, **P < 0.01, ***P < 0.001. (C–F) Data are represented as mean ± SD of three biologically independent experiments, and P values were calculated by Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001.
BRD4 Transactivates MTC Subunits and Modulates Nuclear Retention of METTL3/METTL14.
We next performed anti-BRD4 chromatin immunoprecipitation sequencing (ChIP-seq) using MDA-MB-231 cells treated with BETi. In untreated cells, BRD4 was enriched at promoter regions of MMWHRVZ subunits (Fig. 2A). As expected, BETi caused a remarkable decrease of BRD4 deposition at these chromatin loci (Fig. 2B). Meanwhile, BETi also profoundly reduced promoter-associated marks including H3K4me3 and H3K4me2 as well as promoter-bound RNA Pol II (SI Appendix, Fig. S2 A–C). These data indicate a direct transactivation of MTC subunits by BRD4 and its cofactors at promoters. Interestingly, only WHRVZ protein expression was largely diminished in response to BRD4 inhibition in MDA-MB-231 cells, whereas METTL3 and METTL14 remained unchanged (Fig. 2 C and D and SI Appendix, Fig. S2 D and E). These results were also observed in a number of additional human cancer cell lines (SI Appendix, Fig. S2F). Of note, METTL3 and METTL14 are long-lived proteins, as supported by their remarkably long protein half-life (SI Appendix, Fig. S2G), providing a potential explanation to the constant METTL3/METTL14 protein levels despite BETi-caused transcriptional repression. Meanwhile, expression levels of m6A erasers (FTO and ALKBH5) or m6A reader proteins [including YT521-B homology (YTH) domain family proteins YTHDF1-3 and insulin-like growth factor 2 mRNA-binding proteins IGF2BPs] were not affected upon BET inhibition (SI Appendix, Fig. S2 H and I).
Fig. 2.
BRD4 transactivates MTC subunits and modulates nuclear retention of METTL3/METTL14. (A) ChIP-seq analysis of BRD4-binding profiles at promoters of the indicated MTC genes. JQ1(0.5 μM, 24 h) triggered a strong decrease in BRD4 binding at the indicated gene loci in MDA-MB-231. (B) MDA-MB-231 cells were treated with DMSO or JQ1(0.5 μM, 24 h) and then subjected to ChIP with anti-BRD4 antibody. ChIP samples were analyzed by QRT-PCR. (C) Immunoblotting of the indicated proteins in MDA-MB-231 cells treated with the indicated doses of JQ1 for 48 h. (D) Immunoblotting of the indicated proteins in MDA-MB-231 cells infected with indicated shBRD4 lentiviruses. (E and F) Western blotting analysis of whole cell lysate (WCL), cytoplasmic (Cyto), and nuclear (NE) fractions of MDA-MB-231 cells treated with 0.5 μM JQ1 for 48 h (E), or infected with the shBRD4 lentivirus (F). (G) Representative immunofluorescence for METTL3 (red) and DAPI (blue, cell nuclei) in MDA-MB-231 cells treated with JQ1 (0.5 μM, 48 h). (Scale bars, 10 μm.) All data are represented as mean ± SD of three biologically independent experiments. All P values were calculated by Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001.
Previous studies have reported that WTAP is required for anchoring METTL3 and METTL14 in nuclear speckles (8). Likewise, VIRMA, HAKAI, RBM15, and ZC3H13 have also been found to facilitate the nuclear accumulation of METTL3 and METTL14 (9–11). Indeed, we observed a significant nuclear to cytosolic translocation of METTL3/METTL14 upon BETi treatment or silencing BRD4 (Fig. 2 E–G), implying an indirect negative impact of BETi on the nuclear retention of METTL3/METTL14, via direct suppression of the WHRVZ subunits. We further confirmed that acetylation of METTL3 did not change upon BRD4 inhibition (SI Appendix, Fig. S2J), excluding acetylation-mediated translocation of METTL3. By contrast, no translocation of FTO, ALKBH5, or m6A reader proteins was observed (SI Appendix, Fig. S2 K and L). Taken all together, BRD4 modulates the cellular abundance of the m6A writer complex, via both direct transactivation and the maintaining of the nuclear METTL3/METTL14 abundance.
BET Inhibition Modulates Gene Transcription by Repressing the m6A Writer Complex.
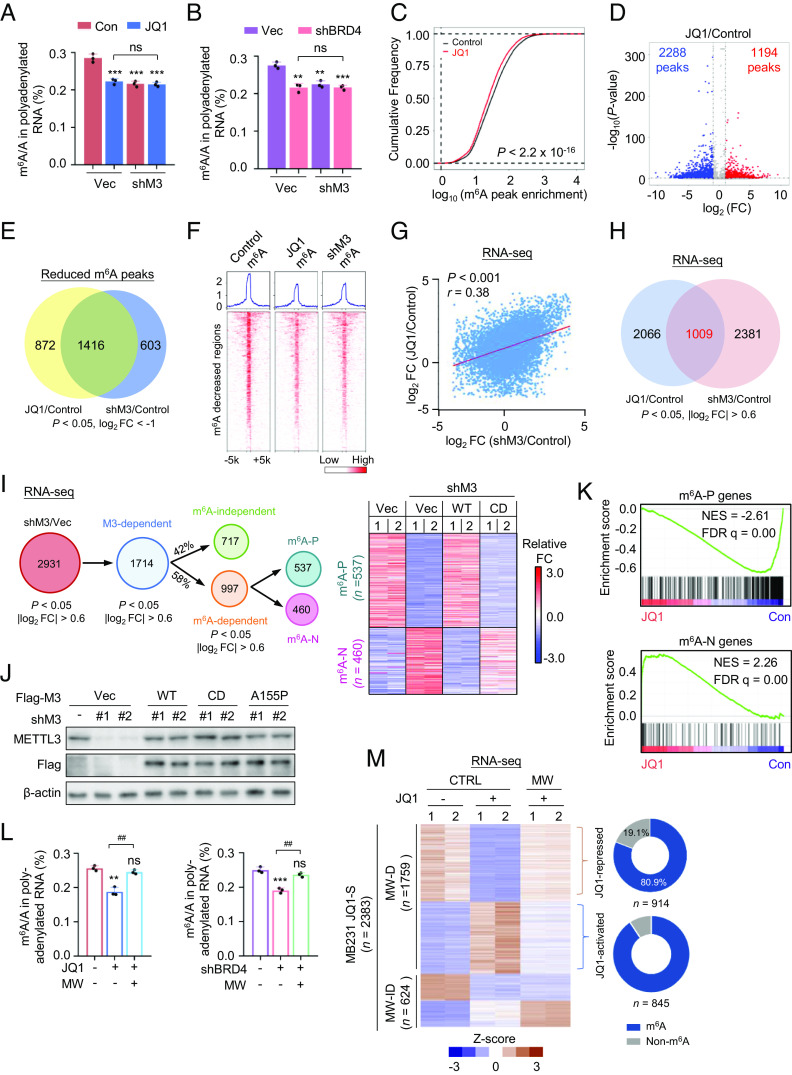
To elucidate whether BETi may regulate m6A modification via the MTC, we measured m6A in METTL3-depleted cells bearing BRD4 silencing or BETi treatment. In the absence of METTL3, BRD4 inhibition failed to further decrease m6A abundance (Fig. 3 A and B and SI Appendix, Fig. S3A), suggesting METTL3 dependency. To gain molecular insights into how BRD4 may affect the m6A-modified transcripts and the subsequent impact on global transcriptome, we mapped m6A methylome in BETi-treated cells as well as in METTL3-deficient cells. Differential m6A peaks were consistent between two biological replicates, indicating good reproducibility (SI Appendix, Fig. S3B). Comparison of m6A methylome between control and BETi-treated cells revealed the following findings: 1) Significant global alteration of methylation sites due to BETi-treatment (herein defined as the BETi-m6A signature) (Fig. 3 C and D). Our m6A sites were enriched for the known m6A consensus motifs (SI Appendix, Fig. S3C). 2) Gene Ontology (GO) analysis of the differential MeRIP-seq candidates enriched pathways involved in chromatin modification, RNA splicing, and pathways regulating cell fate (SI Appendix, Fig. S3D), reflecting concordance between m6A-mediated diverse biological processes and BRD4-directed cellular events, thereby highlighting a previously unrecognized interplay between epigenetic and epitranscriptomic pathways driven by BRD4.
Fig. 3.
BET inhibition modulates gene transcription by repressing the m6A writer complex. (A) LC-MS/MS quantification of the m6A/A ratio in polyadenylated RNA isolated MDA-MB-231 cells either infected with shMETTL3 lentivirus or treated with 0.5 μM JQ1. (B) LC-MS/MS quantification of the m6A/A ratio in polyadenylated RNA isolated from MDA-MB-231 cells infected with the indicated lentiviruses. (C) Cumulative distribution of differential m6A peak intensity between JQ1-treated and DMSO-treated MDA-MB-231 cells. (D) Volcano plot for JQ1-treated and DMSO-treated MDA-MB-231 cells, showing peaks with differential m6A intensity. Fold change (FC) is the ratio of IP over input for JQ1-treated and DMSO-treated cells. (E) Venn diagram showing the overlap between differential down-regulated m6A peaks in JQ1-treated and METTL3-deficient MDA-MB-231 cells. MeRIP-seq data of METTL3 knockdown and control MDA-MB-231 cells were obtained from GEO database (GSE183017). (F) Heatmap analysis of MeRIP-seq reads density in m6A-marked regions with significant difference in JQ1-treated vs. control MDA-MB-231 cells, and shMETTL3 vs. control MDA-MB-231 cells. m6A modified regions were sorted according to m6A reads density level. (G) Correlation analysis of differentially expressed genes in JQ1-treated or METTL3-depleted MDA-MB-231 cells normalized to control cells. P values were calculated by Fisher’s exact test. Gene expression profiles of METTL3-depleted and control MDA-MB-231 cells were obtained from GEO database (GSE183017). (H) Venn diagram showing the overlap between JQ1-dependent and METTL3-dependent transcriptional profiles. (I) Schematic for identification of METTL3-dependent and m6A-dependent genes determined by RNA-seq analysis of MDA-MB-231 cells reconstituted with METTL3WT or METTL3CD mutant constructs (Left). Heatmap of RNA-seq data showing an m6A-dependent signature (Right). (J) Western blotting analysis of the METTL3 expression level in METTL3WT, METTL3CD, or METTL3A155P reconstituting MDA-MB-231 cells. (K) GSEA of m6A positively regulated (Upper) and m6A negatively regulated gene sets (Lower) from Fig. 3I compared with transcriptional profiles of JQ1-treated MDA-MB-231 cells. (L) LC-MS/MS quantification of the m6A/A ratio in polyadenylated RNA isolated from MW reconstituting MDA-MB-231 cells subjected to the indicated treatment. (M) Heatmap of differentially expressed genes (P < 0.05, |log2FC| > 0.6) between MW reconstituting vs. control (CTRL) MDA-MB-231 cells following JQ1 treatment for 24 h. All data are represented as mean ± SD of three biologically independent experiments. All P values were calculated by Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001. #P < 0.05, ##P < 0.01, ###P < 0.001.
We next compared our BETi-m6A signature to METTL3 knockdown MeRIP-seq data, which has been previously reported by our group (M3-KD-m6A signature) (SI Appendix, Fig. S3E). The general m6A distribution showed no major differences among the indicated samples (SI Appendix, Fig. S3F). However, when we compared these two signatures, we found a significant overlap of reduced m6A peaks, which included about 62% (1,416/2,288) BETi-dependent peaks and 70% (1,416/2,019) METTL3-dependent peaks, respectively (Fig. 3E). Heatmap analysis revealed similar reduction patterns of m6A peaks in BETi-treated and METTL3-depleted samples (Fig. 3F). Global gene expression analysis displayed a positive correlation (r = 0.38) between JQ1-dependent and METTL3-dependent transcriptional profiles (Fig. 3G). In addition, 1,009 genes were further identified as common targets between these two datasets (Fig. 3H), among which 967 genes harbored m6A modification (SI Appendix, Fig. S3G). These data indicate a potential connection between BRD4-mediated gene transcription and m6A modification.
BRD4 directs gene transcription through diverse mechanisms. Here, we specifically focused on BETi-dependent transcriptome orchestrated by m6A-mediated regulation. To this end, we first need to identify bona fide m6A-dependent transcripts via rescue experiments in METTL3-knockdown cells (Fig. 3 I and J). Among the altered 2,931 transcripts (shM3 vs. Con, P < 0.05, |log2FC| > 0.6) upon METTL3 depletion, 1,714 mRNAs can be rescued by METTL3WT reconstitution (WT vs. shM3, P < 0.05, |log2FC| > 0.6), which we defined as the METTL3-dependent signature. These genes were further filtered according to RNA-seq data from METTL3CD (a catalytically inactive form of METTL3) reconstituting cells. Of note, 997 targets (CD vs. WT, P < 0.05, |log2FC| > 0.6) were only rescued by METTL3WT, but not by METTL3 CD add-back, which we named the m6A-dependent signature. These m6A-dependent transcripts were next stratified as m6A positively regulated (m6A-P, mRNA levels decreased upon METTL3 depletion, n = 537) and m6A negatively regulated targets (m6A-N, mRNA levels increased upon METTL3 depletion, n = 460). Gene set enrichment analysis (GSEA) demonstrated that expression levels of m6A-P genes were remarkably down-regulated by BETi or BRD4 loss, whereas m6A-N targets were profoundly up-regulated upon BETis exposure or silencing BRD4, respectively (Fig. 3K and SI Appendix, Fig. S3 H and I). These results highlight a general reliance of a BETi-mediated transcriptional program on METTL3-dependent m6A modification.
To systematically dissect the direct contribution of m6A modification to BETi-induced transcriptomic alteration, we performed MTC subunit reconstitution. A series of reconstituting cells, including adding back METTL3 alone, WTAP alone, or combined METTL3/WTAP (MW), respectively, were generated in MDA-MB-231 cells depleted of endogenous METTL3 and/or WTAP (SI Appendix, Fig. S3 J–L). The reason to combine METTL3 and WTAP for reconstitution lies in the fact that these two subunits, together with METTL14, are considered catalytic core components of the MTC. In the absence of HRVZ subunits caused by BETi treatment, WTAP add-back alone is insufficient to restore nuclear METTL3/METTL14 abundance. Coexpression of ectopic METTL3 and WTAP is expected to restore a functional m6A writer complex under BETi treatment. Indeed, compared to their individual adding back, MW reconstitution almost completely restored nuclear METTL3/METTL14 abundance in JQ1-treated cells, yielding a significant restoration of the reduced global m6A abundance upon JQ1 exposure or BRD4 loss (Fig. 3L and SI Appendix, Fig. S3M). RNA-seq analysis revealed 2,383 altered mRNA transcripts in JQ1-treated MDA-MB-231 (CTRL group, JQ1-treated vs. untreated, named as MB231 JQ1-S, P < 0.05, |log2FC| > 0.6) (Fig. 3M). Furthermore, MW reconstitution restored 74% (1,759/2,383) of JQ1-S transcripts to an extent close to their expression levels in DMSO-treated cells. Among them, 914 JQ1-repressed and 845 JQ1-activated mRNAs were identified as MW-dependent transcripts (named MW-D in short), which contained 739 (80.9%, JQ1-repressed) and 765 (90.5%, JQ1-activated) genes that harbored annotated m6A sites, respectively. These data suggest that the majority of BETi-dependent transcriptome is orchestrated by MTC-mediated regulation, including both m6A-dependent and -independent mechanisms.
It has been suggested that m6A sites are relatively conserved across tissues (35). Meanwhile, m6A-marked mRNAs are coordinately regulated via control of m6A writing and reading processes, which may generate distinct outcomes regarding the ultimate mRNA fate. We were curious whether the BRD4–MTC axis may represent a widespread regulatory mode modulating a common subset of transcripts across cell types. To this end, we compared published JQ1-specific transcriptomic signatures obtained from the indicated datasets to our MB231 JQ1-signature (JQ1-S, Fig. 3M) (SI Appendix, Fig. S3N). The overlapping gene hits were called common#1 cluster, which was used for further overlapping with MW-dependent genes (MW-D, Fig. 3M), resulting in common#2 cluster which was potentially related to MTC-dependent regulation. Common#2 cluster was then sorted as JQ1-up-regulated and -down-regulated groups, followed by calculating the percentage of m6A-marked transcripts. Although the percentage of common#1 varied from 23 to 31.5% among different tumor cell lines, likely reflecting tissue-specific transcriptomic regulation, the percentage of common#2 among independent groups was between 54% and 67% (calculated over common#1 total gene numbers), suggesting a probability that a proportion of common JQ1-dependent signature genes across human cancer cell lines might be MW-dependent. Similar to data obtained from MDA-MB-231 cells in Fig. 3M, 82.9 to 95.3% of transcripts in common#2 among different tumor cell lines were m6A-modified. We also identified a common#1 feature (n = 538) among four different breast cancer cell lines exposed to JQ1, which contained 378 (70%) genes overlapped with a MW-dependent signature with a broad range of basic cellular activities like cell cycle regulation (SI Appendix, Fig. S3 O–Q). Meanwhile, we took a close examination of expression levels of MTC subunits from published RNA-seq datasets, along with the known BRD4 targets including Myc, BRCA1, RAD51, and CtIP (31, 36, 37). BETi treatment reduced these transcripts expression to various extents in a context-dependent manner (SI Appendix, Fig. S3R), highlighting that BETi-dependent modulation of MMWHRVZ subunits occurs as a general regulation across tumor cell lines.
Altogether, our results indicate that BETi suppresses global m6A modification via MTC subunits, which in turn influences gene transcription across human cancers, underscoring an essential and widespread role of the m6A epitranscriptome as a downstream signaling mediating BRD4-dependent transcriptional program.
BETi Suppresses HR Repair and Induces Apoptosis via MTC-Mediated m6A Modification.
To characterize the role of m6A modification in mediating BRD4-dependent biological functions, we attempted to identify both BETi-activated and BETi-repressed transcripts harboring m6A modifications. According to GO analysis of JQ1-dependent transcripts with m6A annotations in MDA-MB-231 cells, we noticed inactivation of DNA double-strand break (DSB) repair pathway in down-regulated genes (Fig. 4A), as well as activation of an apoptotic pathway in up-regulated genes (SI Appendix, Fig. S4A). HR and NHEJ reporter assays further revealed that BETis-treated cells exhibited HR deficiency (SI Appendix, Fig. S4 B and C). Based on the published Peng-HR signature (38) and Sun-apoptotic gene signature (39), respectively, we performed GSEA to analyze m6A-modified transcriptional profiles of BETi-treated MDA-MB-231 cells. JQ1 administration significantly down-regulated HR genes and up-regulated apoptotic hits (Fig. 4B and SI Appendix, Fig. S4D). Similar results were obtained in Molibresib-treated or BRD4-depleted cells (SI Appendix, Fig. S4 E–H). To specifically verify m6A dependency in BRD4-mediated transcription, we next selected our candidate transcripts with reduced m6A peaks from the BETi-m6A signature and further filtered them based on RNA-seq data. Given that m6A modification has been reported to both positively and negatively impact transcripts’ stability, we picked up four HR genes (UHRF1, MCM3, EXO1, and ASF1B) which were down-regulated upon JQ1 treatment (FC < 1.5), and four proapoptotic targets (FC > 1.5) (PIDD, MST1, Caspase-9, and BNIP3) that were induced by JQ1 (Fig. 4C and SI Appendix, Figs. S3R and S4I).
Fig. 4.
BETi suppresses HR repair and induces apoptosis via MTC-mediated m6A modification. (A) Gene ontology functional enrichment analysis of JQ1-repressed genes bearing m6A sites in MDA-MB-231 cells. (B) GSEA of the transcriptional profile of JQ1-treated MDA-MB-231 cells compared with Peng-HR gene sets. (C) Genomic visualization of the m6A MeRIP-seq normalized signal in MDA-MB-231 cells treated with JQ1 or METTL3 depletion for the indicated METTL3-dependent m6A substrates. (D) MeRIP-qPCR analysis of the indicated m6A substrates normalized to input in MDA-MB-231 cells subjected to the indicated treatment. (E) QRT-PCR quantification of the indicated mRNAs in MDA-MB-231 cells subjected to the indicated treatment. (F) Western blotting analysis of the indicated proteins in MDA-MB-231 cells subjected to the indicated treatment. (G) Western blotting analysis of the indicated proteins in MDA-MB-231 cells reconstituted with the indicated METTL3 constructs. (H) QRT-PCR quantification of the indicated mRNAs in MW reconstituting MDA-MB-231 cells treated with JQ1 (0.5 μM, 24 h). For P values, * compared to CTRL. $ compared to MW. # compared to JQ1-treated CTRL cells. (I) Immunoblotting of the indicated proteins in MW reconstituting MDA-MB-231 cells treated with JQ1 (0.5 μM, 48 h). All data are represented as mean ± SD. All P values were calculated by Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001. $P < 0.05, $$P < 0.01, $$$P < 0.001. #P < 0.05, ##P < 0.01, ###P < 0.001.
MeRIP-qPCR analysis was utilized to validate the significant decreases in m6A deposition of the selected transcripts in cells treated with JQ1 or depleted of METTL3 (Fig. 4D). QRT-PCR results verified downregulation of HR genes and upregulation of proapoptotic genes upon BRD4 inhibition or METTL3 depletion (Fig. 4E and SI Appendix, Fig. S4J). BRD4 inhibition showed altered protein expression of the candidate genes that was consistent with their mRNA expression patterns (Fig. 4F and SI Appendix, Fig. S4 K–M). Rescue experiments were next conducted to examine METTL3 dependency. As shown in Fig. 4G, METTL3WT or METTL3A155P (a mutant METTL3 with impaired mRNA translation function) reconstitution almost completely rescued candidate protein expression in METTL3 deficient cells, whereas METTL3CD failed to do so, indicating that the m6A catalytic activity of METTL3 is indispensable for modulating expression levels of these candidate genes. Moreover, MW reconstitution conferred partial yet significant resistance to JQ1-mediated changes of these candidate molecules at both transcription and translation levels (Fig. 4 H and I). These results support a pivotal role of METTL3-dependent m6A modification in mediating BRD4-driven transcriptional regulation of HR and apoptotic genes.
We next investigated the exact mechanism by which m6A modification modulates the expression of our HR/apoptotic genes. Upon JQ1 treatment or METTL3 depletion, their altered mRNA abundance was largely attributed to changes in their mRNA stability (SI Appendix, Fig. S4N). Since m6A can be recognized by different types of reader proteins, we therefore sought to elucidate the responsible readers by individual gene silencing. IGF2BP2/3 depletion displayed robust suppression of UHRF1, MCM3, EXO1, and ASF1B, whereas silencing YTHDF2 induced MST1 expression. YTHDF3 knockdown up-regulated PIDD1, Caspase-9, and BNIP3 expression (SI Appendix, Fig. S4 O and P). Further evaluation of their mRNA stability revealed consistent patterns (SI Appendix, Fig. S4 Q–R). These results demonstrate an essential role of the m6A machinery in orchestrating BRD4-dependent transcription.
METTL3 Deficiency Impairs HR Repair and Confers Sensitivity to PARPi.
Given our selected candidate genes are critical for DDR, we hypothesized that loss of core components of the m6A writer complex should result in impaired DNA repair and the activation of apoptosis. To test this, we first investigated whether METTL3 deficiency may have an impact on DDR. Upon ionizing radiation (IR), loss of METTL3 resulted in increased γH2AX foci at late time points (8 h and 24 h, respectively), indicative of impaired DNA repair (SI Appendix, Fig. S5A). Using HR and NHEJ reporter assays, we further determined that METTL3 silencing led to attenuated HR repair, without affecting NHEJ activity (SI Appendix, Fig. S5 B and C). Of note, METTL3 depletion in MDA-MB-231 cells did not influence cell cycle distribution profiles (SI Appendix, Fig. S5D), excluding possible secondary effects arising from METTL3’s involvement in controlling cell cycle progression under this context. Importantly, the reduced HR repair efficiency in METTL3-depleted cells was significantly rescued by METTL3WT and METTL3A155P reconstitution (SI Appendix, Fig. S5 E and F), but not by METTL3CD adding back, indicating that the catalytic activity of METTL3 is indispensable for the HR repair process. We noticed that our selected m6A-methylated transcripts are involved in different stages of HR repair, reflecting a prominent role of METTL3 in DDR. HR repair is initiated by resection of DSBs to generate single-stranded DNA (ssDNA). We therefore set out to identify whether METTL3 may modulate DNA end resection, and the subsequent chromatin loading of RPA and RAD51, two ssDNA-binding proteins involved in HR repair at DSBs and stalled forks. Native BrdU staining showed significantly reduced ssDNA in METTL3 deficient cells (SI Appendix, Fig. S5G). Consistently, we observed markedly impaired IR-induced RPA and RAD51 foci formation upon METTL3 loss (SI Appendix, Fig. S5 H and I), indicative of compromised HR repair function. Similar results were obtained in METTL3-depleted cells exposed to camptothecin (CPT) (SI Appendix, Fig. S5 J–L). Meanwhile, METTL3 deficiency drastically exacerbated IR-induced cell death (SI Appendix, Fig. S5M). These results demonstrate a crucial survival role of METTL3 in promoting HR repair and blocking apoptosis under DNA damage conditions. HR defect is a prerequisite for clinical response to PARP inhibitors when treating patients with various cancer types, including breast, ovarian, prostate, and pancreatic cancers (40). The above results prompted us to explore potential synergistic cytotoxicity between METTL3 silencing and PARPi administration. Indeed, we observed greatly enhanced DNA damage in PARPi-treated HR-competent TNBC cell lines (MDA-MB-231, SUM159, Hs578T, and BT549 cells) upon METTL3 depletion, as determined by positive γH2AX foci (SI Appendix, Fig. S5N) and comet assay (SI Appendix, Fig. S5O). These results were further supported by the colony formation assay, where decreased cell viability was detected in METTL3-deficient cells treated with different PARPis (Olaparib and Talazoparib) (SI Appendix, Fig. S5 P and Q). Again, the attenuated long-term survival phenotype due to METTL3 loss was rescued by METTL3WT and METTL3A155P, but not by METTL3CD (SI Appendix, Fig. S5 R and S). Collectively, these data support our hypothesis that METTL3 deficiency results in synergistic responses to PARPi.
UHRF1 Is the Critical Downstream Effector Mediating the HR Function of METTL3.
To determine the functional contribution of our selected HR genes in mediating HR repair downstream of METTL3, we generated METTL3-deficient MDA-MB-231 cells stably expressing UHRF1, MCM3, EXO1, and ASF1B, respectively (SI Appendix, Fig. S6 A–D). Ectopic expression of individual HR genes can partially restore BrdU/RPA32/RAD51 positive foci and reduce γH2AX foci in METTL3-depleted cells upon IR (SI Appendix, Fig. S6 E–H), indicating that METTL3-dependent HR repair involves multiple downstream effectors acting at different steps. Given DNA damage-induced apoptosis often occurs as a result of failed DDR process, which involves complex mechanisms, we therefore skipped the rescue experiments to test the contribution of those apoptotic candidates downstream of METTL3.
To elucidate the crucial downstream effector mediating METTL3-dependent PARPi response, we carefully evaluated our HR gene candidates because HR is the immediate responsive mechanism determining PARPi sensitivity, and activation of proapoptotic program often occurs when HR repair is compromised. We first characterized the clinical implications of our selected HR genes utilizing TCGA database containing approximately 10,000 samples across 20 cancer types. UHRF1, MCM3, EXO1, and ASF1B mRNAs were aberrantly expressed in all tumor samples, compared to normal tissues (SI Appendix, Fig. S6 I–L). Importantly, the Kaplan–Meier survival analysis showed that high UHRF1 expression in patients with breast, ovarian, prostate, and pancreatic cancers had significantly shorter overall survival (SI Appendix, Fig. S6M). Based on the relatively strong prognostic potential, we focused on UHRF1 to evaluate its contribution in mediating METTL3-dependent HR repair and prosurvival functions under PARPi treatment. The colony formation assay confirmed that ectopic UHRF1 can partially yet significantly blunt PARPi sensitivity in METTL3 knockdown cells (SI Appendix, Fig. S6N). Using mice xenografted with MDA-MB-231 cells, we found that the enhanced PARPi sensitivity upon METTL3 depletion was remarkably disrupted when adding back ectopic UHRF1 (SI Appendix, Fig. S6 O–Q). Likewise, IHC analysis of γH2AX in xenografted tumors revealed a greater percentage of γH2AX-positive cells upon PARPi treatment and METTL3 loss, which can be largely reversed by UHRF1 expression (SI Appendix, Fig. S6R). Meanwhile, in the absence of METTL3, UHRF1 depletion failed to further impair HR efficiency (SI Appendix, Fig. S6S), indicating that these two proteins act in a linear pathway. Collectively, these data demonstrate a vital role of METTL3 in regulating HR repair via epitranscriptional modulation of an array of essential HR factors like UHRF1, further highlighting that targeting METTL3 may serve as a potential therapeutic option to maximize PARPi sensitivity.
BETi Synergizes with PARPi Mainly through an Impaired MTC–HR Axis.
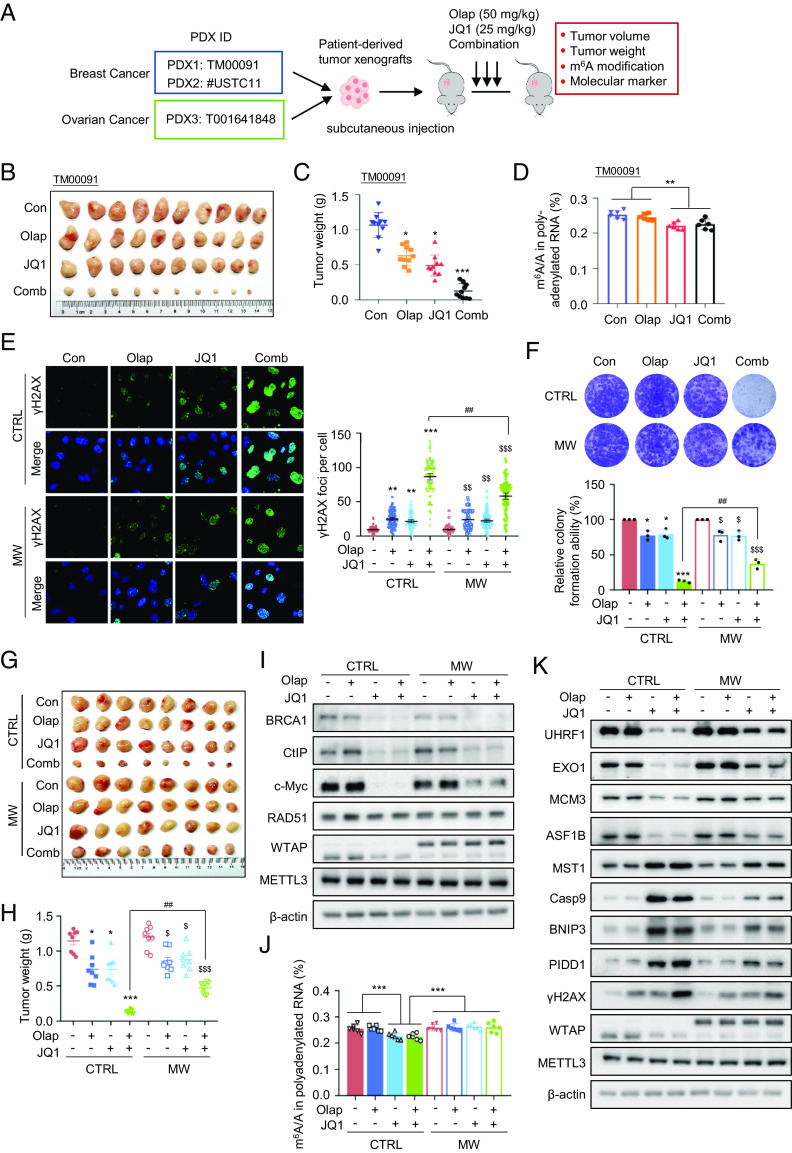
Emerging studies have shown that combination of BETi with PARPi is synergistic in ablating tumor cell survival. Given a causal role of the METTL3–HR axis in DDR and PARPi response, we explored the biological significance of the m6A writer complex in mediating the synthetic lethality upon cotargeting BRD4 and PARPi. We first tested whether BETi-mediated synergistic cytotoxicity with PARPi is associated with impaired m6A machinery in vivo. Three patient-derived xenografts PDX models were utilized, including TM00091 and #USTC11, which are TNBC PDX tissues, and T001641848, which is an ovarian cancer PDX derived from an HGSOC patient who developed acquired resistance to PARPi (Fig. 5A and SI Appendix, Fig. S7A). In agreement with previous findings, JQ1 displayed profound synergy with PARPi in blocking tumor growth (Fig. 5 B and C and SI Appendix, Fig. S7 B and E). Administration of JQ1 alone or JQ1/PARPi showed significantly decreased m6A levels in xenografted tumors (Fig. 5D and SI Appendix, Fig. S7 F and G). Furthermore, expression levels of WHRVZ subunits were down-regulated upon JQ1 treatment and displayed a similar alteration pattern as observed in cell lines (SI Appendix, Fig. S7 H–M). Meanwhile, downregulation of HR regulators and upregulation of proapoptotic factors were observed in JQ1-treated samples (SI Appendix, Fig. S7 N–S). Collectively, these data support that inhibition of BRD4 profoundly suppresses MTC signaling, which in turn diminishes global m6A methylation in vivo.
Fig. 5.
BETi synergizes with PARPi mainly through an impaired MTC–HR axis. (A) Schematic showing experimental processing of the indicated PDX models. (B and C) Images of xenografted tumors in the indicated groups (B) and quantification of tumor weight (C). (D) LC-MS/MS quantification of the m6A/A ratio in polyadenylated RNA isolated from xenografted tumors. (E) Representative images of γH2AX foci in the indicated MDA-MB-231 cells treated with Olaparib (2 μM), JQ1 (0.5 μM), or a combination for 72 h (Left). (Scale bars, 15 µm.) Data are represented as mean ± 95% CI. Statistical analysis was performed using the two-tailed unpaired t-test. Each point represents a cell. A total of 100 cells quantified in each group were obtained from one experiment (Right). Data are representative of three independent experiments. (F) Clonogenic assay of the indicated MDA-MB-231 cells subjected to the indicated treatment (Upper). Lower panel showing the quantification result. Data are represented as mean ± SD of three biologically independent experiments. P values were calculated by Student’s t test. (G and H) Images of xenografted tumors (G) in the indicated groups and quantification (H) of tumor weight. (I) Western blotting analysis of the indicated proteins in xenografted tumors. (J) LC-MS/MS quantification of the m6A/A ratio in polyadenylated RNA isolated from xenografted tumors. (K) Western blotting analysis of the indicated proteins in xenografted tumors. (C, D, H, and J) Data are represented as mean ± SEM. All P values were calculated by Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001. #P < 0.05, ##P < 0.01, ###P < 0.001. $P < 0.05, $$P < 0.01, $$$P < 0.001.
Given that the BRD4–MTC axis can regulate HR and proapoptotic factors, we next investigated whether BET inhibition may sensitize PARPi response via MTC-dependent modulation of these processes. We performed the HR reporter assay and found that BETis markedly decreased HR repair, which can be largely rescued by MW reconstitution (SI Appendix, Fig. S7T), suggesting that MTC signaling serves as a crucial downstream effector mediating BRD4-dependent HR repair efficiency. Likewise, the enhanced DNA damage upon BETi and PARPi cotreatment, evaluated by γH2AX and comet assays, was significantly ablated in MW reconstituting cells (Fig. 5E and SI Appendix, Fig. S7 U and V). MW reconstitution also drastically reduced the proportion of apoptotic cells upon BETi/PARPi coadministration (SI Appendix, Fig. S7W). Consistent with these observations, MW reconstitution significantly restored long-term survival of BETi/PARPi-treated MDA-MB-231 cells (Fig. 5F) as well as rescued the in vivo tumor growth rate in BETi/PARPi-treated xenograft animal models (Fig. 5 G and H). Notably, CtIP and BRCA1, which have been previously reported as BRD4 downstream targets mediating HR repair activity, remained decreased in MW reconstituting cells and xenografted tumor samples exposed to BETi (Fig. 5I and SI Appendix, Fig. S7 X and Y), whereas RAD51 protein expression was unchanged. We noticed partial restoration of the c-Myc level upon MW reconstitution, consistent with its mRNA, is subjected to m6A modification (41). Furthermore, we found that in xenografted tumor samples, the level of m6A was significantly reduced upon BETi, which can be markedly rescued by MW reconstitution (Fig. 5J). Meanwhile, restoration of altered expression of HR and apoptotic genes was achieved by MW reconstitution as well (Fig. 5K and SI Appendix, Fig. S7Z). Our data indicate that MW reconstitution partially yet significantly rescued DNA damage, apoptosis rate, and the in vivo anticancer cytotoxicity triggered by cotreatment with BETi/PARPi. All these results indicate a pivotal role of m6A signaling in mediating the enhanced synergy between BETi and PARPi.
Discussion
Accumulating evidence has reported aberrant transcripts expression of the m6A writer complex in human cancers, accompanied by elevated m6A abundance, thereby endowing cells with oncogenic properties (25, 26). However, molecular mechanisms underlying the transcriptional regulation of MTC subunits remain completely unknown. We found that BET inhibitors can drastically repress transcriptome-wide m6A deposition by suppressing seven MTC subunits. Further elucidation of the mechanistic details revealed a prominent role of BRD4 in guiding the basal transcription machinery required for homeostatic maintenance of the m6A writer complex under pathophysiological conditions.
Given a pivotal role of MTC in RNA biology and protein translation, it is plausible that BRD4 may control diverse cellular activities through these MTC-related biological processes. Since the primary function of m6A methylation is to regulate mRNA homeostasis, we therefore specifically focused on investigating the functional contribution of m6A-dependent posttranscriptional output downstream of BRD4. It is conceivable that BRD4-mediated transcriptome might be indirectly influenced by MTC-dependent transcription/translation as well, highlighting an essential role of BRD4 via residing at the top of the hierarchy of the transcriptional network.
It is intriguing that a significant proportion of BRD4-dependent transcripts harbors annotated m6A sites. Given that the majority of m6A sites are found in the same transcripts in the same location across different tissues (35), this highlights a prominent contribution of the m6A writer complex in mediating a wide range of cellular activities driven by BRD4. We specifically characterized BRD4-dependent dual regulation of HR and proapoptotic factors. Our ChIP-seq data reveal genome-wide localization of BRD4, which supports a direct transcriptional regulation of HR/apoptotic genes by BRD4 via promoter binding. Unexpectedly, these transcripts are subsequently modified by m6A methylation that is indirectly controlled by BRD4, which acts as an additional essential layer of regulation to ensure a prompt stress response governed by these factors. BETi, in general, leads to transcriptional repression. However, a substantial amount of genes are up-regulated in BETi-treated cells for unknown reasons. Our data revealed that most of these BETi negatively regulated genes are in fact MTC-dependent m6A substrates, providing mechanistic insights underlying BETi-mediated induction of a certain set of transcripts.
By analyzing published BETi-dependent transcriptome datasets, we unraveled BRD4-dependent modulation of MTC subunits across human cancer cell lines. It is intriguing that a significant proportion of common#1 targets is overlapped with a MW-dependent signature in MDA-MB-231 cells, and the majority of common#2 mRNAs harbor annotated m6A sites. We speculate that this unappreciated BRD4–MTC axis may evolve to modulate dynamic expression of a subset of common transcripts across cell/tissue types, enabling the prompt activation of fundamental cellular events like HR and apoptosis. This is in fact in agreement with a recent finding demonstrating that ubiquitously expressed genes are more likely to be m6A regulated (42). The contribution of MTC as essential downstream effectors mediating BRD4-directed gene transcription was evaluated using METTL3/WTAP reconstitution, where altered expression of HR/apoptotic factors upon BET inhibition can be partially yet significantly rescued, supportive of MTC subunits dependency. One plausible explanation to the partial rescue effects is that METTL3/WTAP reconstitution probably can only restore partial m6A regulatory machinery since the additional MTC components still remain repressed by BETi. Nevertheless, these modes of action provide a strong scientific rationale for predicting sensitivity to BETi administration, either by itself or in combination therapies, in tumors harboring aberrantly expressed MTC subunits.
Emerging evidence has demonstrated the synergy between PARPi and BETi, which occurs via distinct signaling pathways. Here, we elucidated the functional contribution of compromised MTC signaling in mediating cytotoxicity triggered by cotargeting BET and PARP. It turns out that m6A methylation accounts for both METTL3-dependent and BRD4-specific DDR process, which involves induction of essential HR factors and repression of classical proapoptotic transcripts. Consistent with our in vitro data, xenografted tumors reconstituted with METTL3/WTAP confer significant resistance to combined BETi/PARPi treatment, regardless of the presence of compromised HR signaling (CtIP and BRCA1) reported by other groups, suggesting a central role of MTC in mediating the survival advantage upon BETi/PARPi cotreatment. METTL3 has been found to modulate the DDR process via different mechanisms (43, 44). In our study, we identified a cluster of m6A-modified HR transcripts acting in different stages of HR repair, along with a group of apoptotic transcripts, indicative of a pivotal role of METTL3-dependent m6A substrates in the orchestrating DDR process. We noticed that a previous study reported that enforced expression of CtIP was sufficient to reverse the effects of BRD4i on DNA end resection, HR function, and PARPi sensitivity, even in the absence of RAD51 or BRCA1, through mechanisms including both HR repair and recovery of replication fork (36). Indeed, DNA replication fork reversal and fork stability have been found to contribute to PARPi resistance independent of HR repair (45). We have identified several m6A-marked genes required for replication fork progression. ASF1B has been shown to coordinate histone supply with replicative unwinding of DNA at replication sites (46). Replication helicase MCM3 has been found to associate with STAG2, which is essential for DNA replication fork progression (47). Meanwhile, it is plausible that additional gene targets regulated by m6A-dependent mechanism may contribute to BETi/PARPi responsiveness, independently of loss of CtIP and BRCA1. Based on our findings, we propose that high levels of MTC subunits may serve as potential biomarkers of decreased sensitivity to PARPi monotherapy. However, patients with an aberrant BRD4–MTC axis may benefit from BETi/PARPi combination therapy. Given dysregulated BRD4 expression is often found in human cancers, our findings suggest that BETi/PARPi therapy strategies can be deployed for the treatment of human cancers bearing overpresented MTC-m6A signaling, regardless of BRCA status, thus expanding the therapeutic utility of PARP inhibitors. Of note, pharmacological inhibitors targeting METTL3 have been reported, which displayed selective reduction of m6A levels as well as impaired protein translation when treating acute myeloid leukemia (AML) (48). It is worth pointing out that m6A deposition on RNA substrates might be context-specific (49). For instance, increasing evidence supports a critical and unique role for individual MTC components in site-specific and transcript-specific m6A methylation (50, 51). Moreover, METTL3/METTL14 can regulate m6A methylation in a threshold-dependent manner. These lines of evidence suggest that the overall transcriptome-wide m6A modification is influenced by multifaceted mechanisms, raising the possibility that blocking METTL3 alone might be insufficient to fully disrupt aberrant MTC-driven oncogenic events. Our findings demonstrate a prominent role for BRD4 in maintaining the abundance of individual MTC subunits and the nuclear m6A writer complex integrity, highlighting the utility of BET inhibitors as anticancer therapeutic intervention by disrupting the MTC-dependent epitranscriptome.
Materials and Methods
Detailed materials and methods are available in SI Appendix. This includes the following: plasmids and cloning procedures, siRNAs, antibodies, clinical specimens, immunoblotting, subcellular fractionation, quantitative RT-PCR, RNA m6A quantification by LC-MS/MS, clonogenic survival assay, X-ray irradiation, immunofluorescence, comet assay, HR and NHEJ reporter assays, MeRIP-seq and MeRIP-qPCR, MeRIP-seq data processing, computational analysis of RNA-seq data, chromatin immunoprecipitation (ChIP) and ChIP-seq, computational analysis of ChIP-seq data, immunohistochemistry, flow cytometry-based cell cycle and cell death analysis, animal studies, statistics, and reproducibility.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Dr. Cixiong Zhang (School of Life Sciences, Xiamen University) for LC-MS/MS analysis, Dr. Jialiang Huang (School of Life Sciences, Xiamen University) for helping with MeRIP-seq analysis, and Dr. Jian Zhang (Fourth Military Medical University), Dr. Yuntao Xie (Peking University Cancer Hospital & Institute), and Hongxia Wang (Shanghai Jiao Tong University School of Medicine) for the collection of breast cancer samples. We apologize to colleagues whose work could not be cited due to space constraints. This work was supported by the National Natural Science Foundation of China Distinguished Young Scholars Program (81725012), the National Natural Science Foundation of China project (U2005203), the National Key R&D Program of China (2017YFA0504502 to H.Y.), and the Natural Science Foundation of China (82021003).
Author contributions
W.L. and H.Y. designed research; X.L., L.P., and M.R. performed research; T.S. and J.Y. contributed new reagents/analytic tools; J.D., Y.L., Q.L., X.H., and C.L. analyzed data; and H.Y. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Jing Ye, Email: yejing@fmmu.edu.cn.
Wen Liu, Email: w2liu@xmu.edu.cn.
Han You, Email: hyou@xmu.edu.cn.
Data, Materials, and Software Availability
The MeRIP-seq, RNA-seq, and ChIP-seq data generated in this study have been deposited in the Gene Expression Omnibus database under accession codes GSE219190 (52), GSE219191 (53), and GSE219192 (54). All other data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Wang X., et al. , N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang X., et al. , N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi H., Wei J., He C., Where, when, and how: Context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell 74, 640–650 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jia G., et al. , N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng G., et al. , ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X., et al. , Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 534, 575–578 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Huang H., Weng H., Chen J., m(6)A modification in coding and non-coding RNAs: Roles and therapeutic implications in cancer. Cancer Cell 37, 270–288 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ping X. L., et al. , Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24, 177–189 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yue Y., et al. , VIRMA mediates preferential m(6)A mRNA methylation in 3’UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 4, 10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wen J., et al. , Zc3h13 regulates nuclear RNA m(6)A methylation and mouse embryonic stem cell self-renewal. Mol. Cell 69, 1028–1038.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patil D. P., et al. , m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Batista P. J., et al. , m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem. Cell 15, 707–719 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frye M., Harada B. T., Behm M., He C., RNA modifications modulate gene expression during development. Science 361, 1346–1349 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geula S., et al. , Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347, 1002–1006 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Li Y., et al. , METTL3 acetylation impedes cancer metastasis via fine-tuning its nuclear and cytosolic functions. Nat. Commun. 13, 6350 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barbieri I., et al. , Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature 552, 126–131 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng X., et al. , RNA N(6)-methyladenosine modification in cancers: current status and perspectives. Cell Res. 28, 507–517 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyer K. D., et al. , 5’ UTR m(6)A promotes cap-independent translation. Cell 163, 999–1010 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du H., et al. , YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 7, 12626 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edupuganti R. R., et al. , N(6)-methyladenosine (m(6)A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol. 24, 870–878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu P. J., et al. , Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 27, 1115–1127 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang H., et al. , Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20, 285–295 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang F., et al. , Fragile X mental retardation protein modulates the stability of its m6A-marked messenger RNA targets. Hum. Mol. Genet. 27, 3936–3950 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao W., et al. , Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61, 507–519 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Lin S., Choe J., Du P., Triboulet R., Gregory R. I., The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell 62, 335–345 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Q., et al. , METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut 69, 1193–1205 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Doroshow D. B., Eder J. P., LoRusso P. M., BET inhibitors: A novel epigenetic approach. Ann. Oncol. 28, 1776–1787 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Stathis A., Bertoni F., BET proteins as targets for anticancer treatment. Cancer Discov. 8, 24–36 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Loven J., et al. , Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Filippakopoulos P., et al. , Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delmore J. E., et al. , BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–917 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewin J., et al. , Phase Ib trial with birabresib, a small-molecule inhibitor of bromodomain and extraterminal proteins, in patients with selected advanced solid tumors. J. Clin. Oncol. 36, 3007–3014 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Kurimchak A. M., et al. , Resistance to BET bromodomain inhibitors is mediated by kinome reprogramming in ovarian cancer. Cell Rep. 16, 1273–1286 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shu S., et al. , Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature 529, 413–417 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Campos M. A., et al. , Deciphering the “m(6)A Code” via antibody-independent quantitative profiling. Cell 178, 731–747.e16 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Sun C., et al. , BRD4 inhibition is synthetic lethal with PARP inhibitors through the induction of homologous recombination deficiency. Cancer Cell 33, 401–416.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang L., et al. , Repression of BET activity sensitizes homologous recombination-proficient cancers to PARP inhibition. Sci. Transl. Med. 9, eaal1645 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peng G., et al. , Genome-wide transcriptome profiling of homologous recombination DNA repair. Nat. Commun. 5, 3361 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun Y., et al. , Context-dependent tumor-suppressive BMP signaling in diffuse intrinsic pontine glioma regulates stemness through epigenetic regulation of CXXC5. Nat. Cancer 3, 1105–1122 (2022). [DOI] [PubMed] [Google Scholar]
- 40.Quigley D., et al. , Analysis of circulating cell-free DNA identifies multiclonal heterogeneity of BRCA2 reversion mutations associated with resistance to PARP inhibitors. Cancer Discov. 7, 999–1005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Achour C., Bhattarai D. P., Groza P., Roman A. C., Aguilo F., METTL3 regulates breast cancer-associated alternative splicing switches. Oncogene 42, 911–925 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu J., et al. , Landscape and regulation of m(6)A and m(6)Am methylome across human and mouse tissues. Mol. Cell 77, 426–440 e426 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Zhang C., et al. , METTL3 and N6-methyladenosine promote homologous recombination-mediated repair of DSBs by modulating DNA-RNA hybrid accumulation. Mol. Cell 79, 425–442.e7 (2020). [DOI] [PubMed] [Google Scholar]
- 44.Xiang Y., et al. , RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature 543, 573–576 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ray Chaudhuri A., et al. , Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature 535, 382–387 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Groth A., et al. , Regulation of replication fork progression through histone supply and demand. Science 318, 1928–1931 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Mondal G., Stevers M., Goode B., Ashworth A., Solomon D. A., A requirement for STAG2 in replication fork progression creates a targetable synthetic lethality in cohesin-mutant cancers. Nat. Commun. 10, 1686 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yankova E., et al. , Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 593, 597–601 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang H., et al. , Histone H3 trimethylation at lysine 36 guides m(6)A RNA modification co-transcriptionally. Nature 567, 414–419 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Su S., et al. , Cryo-EM structures of human m(6)A writer complexes. Cell Res. 32, 982–994 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Knuckles P., et al. , Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes. Dev. 32, 415–429 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu X., et al. , m6A methylome in JQ1 treatment and control in MDA-MB-231 cells. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE219190. Deposited 1 December 2022.
- 53.Lu X., et al. , BET inhibition abrogates global transcription via repressing m6A writer complex. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE219191. Deposited 1 December 2022.
- 54.Lu X., et al. , BRD4 ChIP-seq in JQ1 treatment and control in MDA-MB-231 cells. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE219192. Deposited 1 December 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
The MeRIP-seq, RNA-seq, and ChIP-seq data generated in this study have been deposited in the Gene Expression Omnibus database under accession codes GSE219190 (52), GSE219191 (53), and GSE219192 (54). All other data are included in the article and/or SI Appendix.