Abstract
Objective
Schizophrenia is a serious mental disorder with complex clinical manifestations, while its pathophysiological mechanism is not fully understood. Accumulated evidence suggested the alteration in epigenetic pathway was associated with clinical features and brain dysfunctions in schizophrenia. DNA methyltransferases (DNMTs), a key enzyme for DNA methylation, are related to the development of schizophrenia, whereas the current research evidence is not sufficient. The aim of study was to explore the effects of gene polymorphisms of DNMTs on the susceptibility and symptoms of schizophrenia.
Methods
The study was case–control study that designed and employed the Diagnostic and Statistical Manual of Mental Disorders-Fifth Edition (DSM-5) as the diagnostic standard. 134 hospitalized patients with schizophrenia in the Third People's Hospital of Zhongshan City from January 2018 to April 2020 (Case group) as well as 64 healthy controls (Control group) from the same region were involved. Single nucleotide polymorphisms (SNPs) of DNMT1 genes (r s2114724 and rs 2228611) and DNMT3B genes (rs 2424932, rs 1569686, rs 6119954 and rs 2424908) were determined with massARRAY. Linkage disequilibrium analysis and haplotype analysis were performed, and genotype and allele frequencies were compared. The Hardy–Weinberg equilibrium was tested by the Chi-square test in SPSS software (version 20.0, SPSS Inc., USA). The severity of clinical symptoms was assessed by the Positive and Negative Syndrome Scale (PANSS). The correlation between DNMT1 genes (rs 2114724 and rs 2228611) and DNMT3B genes (rs2424932, rs1569686, rs6119954 and rs2424908) and clinical features was analyzed.
Results
There were no significant differences in genotype, allele frequency and haplotype of DNMT1 genes (rs 2114724 and rs 2228611) and DNMT3B genes (rs 2424932, rs 1569686, rs 6119954 and rs 2424908) between the case and healthy control group. There were significant differences in the PANSS total positive symptom scores, P3 (hallucinatory behavior), P6 (suspicious/persecution), G7 (motor retardation), and G15 (preoccupation) in patients with different DNMT1 gene rs 2114724 and rs 2228611 genotypes. The linkage disequilibrium analysis of gene polymorphic loci revealed that rs 2114724–rs 2228611 was complete linkage disequilibrium, and rs 1569686–rs 2424908, rs 2424932–rs 1569696 and rs 2424932–rs 2424908 were strongly linkage disequilibrium.
Conclusion
The polymorphisms alteration in genetic pathway may be associated with development of specific clinical features in schizophrenia.
Keywords: Schizophrenia, Single nucleotide polymorphism (SNP), DNA methyltransferases, Symptoms
Introduction
Schizophrenia is a chronic and debilitating mental disorder affecting more than 20 million individuals worldwide, with significant socioeconomic consequences [1, 2]. One of the main features of schizophrenia is the dissociation of thoughts, concepts, identities, and emotions [3–6], associated with dysregulated neural pathways in the brain that lead to positive and negative symptoms [4, 6, 7]. The etiology of schizophrenia is multifactorial, involving genetic predisposition, environmental factors, and epigenetic modifications [6]. It is estimated that schizophrenia has a heritability range of 79–81% [8]. Genetic risk loci have been identified in non-coding areas such as promoters and introns, indicating that gene regulation is crucial for the onset of psychotic disorders [9]. In the case of schizophrenia, research has identified a number of genetic risk factors that are associated with altered gene expression patterns [10–12], which may be influenced by epigenetic modifications [9]. Twin studies have also provided evidence that epigenetic factors may be involved in the development of schizophrenia [13].
Epigenetic modifications, mainly DNA methylation or histone modifications, are involved in gene regulation. Among these, DNA methylation is the most extensively investigated [14]. DNA methylation occurs through the covalent binding of methyl groups to cytosine-guanine dinucleotides (CpG) [15]. Several genes, including GAD1 [16], COMT [17], RELN [18], BDNF [19], CHRNA7 [20], SOX10 [21], EGR1 [22], have been identified as potential candidates for DNA methylation in schizophrenic patients. DNA methylation is established and maintained by methyltransferases, such as DNMT1, DNMT3A, and DNMT3B [23]. DNMTs were first recognized as stable repressive marks, which are important mechanisms for gene silencing during progression and the cycle of life [24]. It has been suggested that patients with schizophrenia may have DNMT gene dysfunction.
Studies have shown that DNMTs are essential for the generation of mature, functioning germ cells, and appropriate embryonic development [25, 26]. Abnormal DNA methylation patterns have been linked to an increased risk of certain human diseases in individuals with a hereditary predisposition [27]. Genetic variations, such as single nucleotide polymorphisms (SNPs), have been strongly associated with schizophrenia in several family studies [28, 29]. Recent case–control studies indicated a significant association between minor alleles of DNMT1 (rs2114724 and rs2228611), DNMT3B (rs2424932 and rs1569686) and susceptibility of schizophrenia, while it is rare [30].
Previous studies have focused on the three active DNMT enzymes: DNMT1, DNMT3A, and DNMT3B. Some investigations have linked schizophrenia to polymorphisms within these enzymes, but there is still considerable ambiguity regarding the relationship between these polymorphisms and the clinical features of schizophrenia. DNMT SNPs prevalence in southern Chinese populations has not been reported yet. Therefore, the aim of the present study was to explore the effects of DNMT gene polymorphisms on schizophrenia susceptibility and symptoms in this population.
Materials and methods
Patient and control samples
Patients or their legal guardians provided informed consent after receiving authorization from the ethics committee of Zhongshan Third People’s Hospital for this study. The investigation focused on patients with schizophrenia who were treated at the hospital between 2018 and 2020. Participants were included if they met the following criteria: a diagnosis of schizophrenia as per the Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition, made by two attending physicians with consistent training, age between 16 and 60 years, and Han ethnicity. The study excluded patients with diabetes, hypertension, cancer, and other nervous system or mental diseases. However, other mental disorders were not excluded.
The control group at Zhongshan City’s Third People’s Hospital consisted of physical exam mentally healthy patients, employees, nurses, and volunteers. To be included in the control group, subjects had to meet the following criteria: no history of mental illness, no family history of mental illness, pass a physical exam, Han ethnicity, and between 18 and 60 years of age after psychological screening. The control group excluded adopted or single-parent children with unknown family histories, as well as individuals with serious somatic disorders such as diabetes, hypertension, or cancer.
Methods
The experimental methods used in this research included the collection of general data and clinical information, evaluation of psychotic symptoms, genomic DNA extraction, primer creation, PCR amplification, alkaline phosphatase treatment, single-base extension reaction, resin purification, chip sampling, mass spectrometric detection, and data analysis, which were based on past studies [31, 32].
The collected data were sorted and evaluated using the statistical program SPSS 20.0 (IBM, Armonk, NY, USA). The analysis of variance was performed, and results were presented as mean and standard deviation since the data exhibited normal distribution. Nonparametric statistical approaches were applied to non-normal data, and the results were expressed numerically as the median value (upper quartile, lower quartile). The genotype frequency was analyzed using the Chi-square test. regarding as categorical data. Hardy–Weinberg equilibrium was examined using SPSS 20.0 Chi-square test, and genotype and allele frequencies were compared between the schizophrenia and control groups. The linkage disequilibrium (LD) and pairwise LD coefficients in DNMT1 and DNMT3B were implemented with Haploview 4.2. ORs and confidence intervals (CIs) were computed for 95% of the data, and G*Power 3.1 was used for power analysis.
Results
Test of Hardy–Weinberg equilibrium for DNMT1 and DNMT3B
We performed a Hardy–Weinberg equilibrium analysis on the genotype data of 134 individuals with schizophrenia, focusing on six DNMT gene polymorphisms (rs2114724, rs2228611, rs2424932, rs1569686, rs6119954, and rs2424908). The results, presented in Tables 1 and 2, indicate that the genotype frequencies at these loci did not deviate significantly from the predicted population frequencies (p > 0.05). No outliers were identified as the cause of these results.
Table 1.
Hardy–Weinberg equilibrium test for DNMT1 genotypes
| Locus | Group (n) | Genotype | Frequency (%) | X2 | P |
|---|---|---|---|---|---|
| rs2114724 | Schizophrenia group (134) | CC | 57 (42.5) | 0.84 | 0.66 |
| CT | 57 (42.5) | ||||
| TT | 20 (15.0) | ||||
| Control group (64) | CC | 30 (46.9) | 0.06 | 0.97 | |
| CT | 27 (42.2) | ||||
| TT | 7 (10.9) | ||||
| rs2228611 | Schizophrenia group (134) | CC | 57 (42.5) | 0.84 | 0.66 |
| CT | 57 (42.5) | ||||
| TT | 20 (15.0) | ||||
| Control group (64) | CC | 30 (46.9) | 0.06 | 0.97 | |
| CT | 27 (42.2) | ||||
| TT | 7 (10.9) |
Table 2.
Hardy–Weinberg equilibrium test for DNMT3B genotypes
| Locus | Group (n) | Genotype | Frequency (%) | X2 | P |
|---|---|---|---|---|---|
| rs1569686 | Schizophrenia group (134) | GG | 3 (2.2) | 17.8 | 0.00 |
| GT | 9 (6.7) | ||||
| TT | 122 (91.1) | ||||
| Control group (64) | GG | 1 (1.6) | 1.71 | 0.43 | |
| GT | 7 (10.9) | ||||
| TT | 56 (87.5) | ||||
| rs2424908 | Schizophrenia group (134) | CC | 23 (17.2) | 0.78 | 0.68 |
| CT | 59 (44.0) | ||||
| TT | 52 (38.8) | ||||
| Control group (64) | CC | 9 (14.1) | 0.16 | 0.92 | |
| CT | 32 (50.0) | ||||
| TT | 23 (35.9) | ||||
| rs2424932 | Schizophrenia group (134) | AA | 0 (0) | 0.29 | 0.86 |
| AG | 12 (8.9) | ||||
| GG | 122 (91.0) | ||||
| Control group (64) | AA | 0 (0) | 0.28 | 0.87 | |
| AG | 8 (12.5) | ||||
| GG | 56 (87.5) | ||||
| rs6119954 | Schizophrenia group (134) | AA | 8 (6.0) | 0.54 | 0.76 |
| AG | 56 (41.8) | ||||
| GG | 70 (52.2) | ||||
| Control group (64) | AA | 3 (4.7) | 0.94 | 0.62 | |
| AG | 28 (43.8) | ||||
| GG | 33 (51.5) |
Results of genotype and allelic analysis for DNMT1 and DNMT3B
Tables 3 and 4 demonstrate that there were no significant differences in genotype frequencies between the two groups at the DNMT1 gene loci rs2114724 and rs2228611 (p > 0.05). Likewise, no significant differences were observed in the genotype frequencies between the two groups at the DNMT3B gene loci rs2424932, rs1569686, rs6119954, and rs2424908 (p > 0.05).
Table 3.
Comparison of DNMT1 rs2114724 and rs2228611 genotype frequencies between the two groups
| Group | Cases | rs2114724 | rs2228611 | ||||
|---|---|---|---|---|---|---|---|
| CC | CT | TT | CC | CT | TT | ||
| Schizophrenia group | 134 | 57 | 57 | 20 | 57 | 57 | 20 |
| Control group | 64 | 30 | 27 | 7 | 30 | 27 | 7 |
| p | / | 0.708 | 0.692 | ||||
Table 4.
Comparison of DNMT3B rs1569686, rs2424908, rs2424932 and rs6119954 genotype frequencies between the two groups
| Group | Cases | rs1569686 | rs2424908 | rs2424932 | rs6119954 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GG | GT | TT | CC | CT | TT | AA | AG | GG | AA | AG | GG | ||
| Schizophrenia group | 134 | 3 | 9 | 122 | 23 | 59 | 52 | 0 | 12 | 122 | 8 | 56 | 70 |
| Control group | 64 | 1 | 7 | 56 | 9 | 32 | 23 | 0 | 8 | 56 | 3 | 28 | 33 |
| p | / | 0.573 | 0.709 | 0.436 | 0.918 | ||||||||
Table 5 displays no significant differences in the frequency of the rs2114724, rs2228611, rs1569686, rs2424908, rs2424932, and rs6119954 alleles between the two groups (p > 0.05).
Table 5.
Comparison of rs2114724, rs2228611, rs1569686, rs2424908, rs2424932 and rs6119954 allele frequencies between the two groups
| Group | Cases | rs2114724 | rs2228611 | rs1569686 | rs2424908 | rs2424932 | rs6119954 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | T | C | T | G | T | C | T | A | G | A | G | ||
| Schizophrenia group | 134 | 171 | 97 | 171 | 97 | 15 | 253 | 105 | 163 | 12 | 256 | 72 | 196 |
| Control group | 64 | 87 | 41 | 87 | 41 | 9 | 119 | 50 | 78 | 8 | 120 | 34 | 94 |
| p | / | 0.416 | 0.416 | 0.576 | 0.902 | 0.451 | 0.949 | ||||||
Linkage disequilibrium analysis for DNMT1 and DNMT3B
Genotyping for two SNPs in DNMT1 and four SNPs in DNMT3B was successful, and Fig. 1 illustrates their linkage disequilibrium (LD). The two SNPs in DNMT1 formed a block within 25 kb (chr19: 10154572-10156401), contrasting the four SNPs in DNMT3B. Due to DNMT1's strong linkage, tagged SNPs were run in Haploview.
Fig. 1.
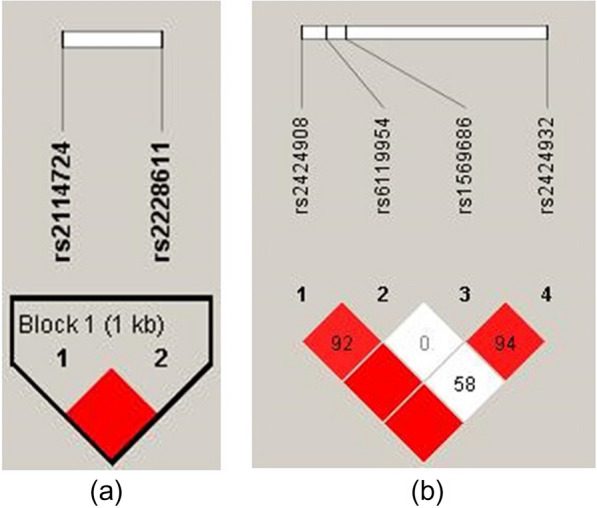
Linkage disequilibrium (LD) pattern of DNMT1 (a) and DNMT3B (b) SNPs from Haploview analysis. The D′ values for LD are represented by the numbers inside the boxes; the miss numbers were 100. The strength of the LD between pairwise pairings of SNPs is shown by the areas that are shaded in the LD map (white represent low LD; red represent high LD)
Haplotypes of the SNPs in DNMT1 and DNMT3B
We examined the haplotype distribution of DNMT1 (rs2114724 and rs2228611) and DNMT3B (rs1569686, rs2424908, rs2424932, and rs6119954) in both the schizophrenia and control groups. Haplotypes with a frequency of at least 3% were selected for analysis to investigate their association with schizophrenia. Two distinct DNMT1 haplotypes were identified, with the CT haplotype being the most common in both controls (68.0%) and patients (63.8%). The most common DNMT3B haplotype in both groups was GTGT (59.3% and 60.9%, respectively). The haplotype frequencies at gene loci rs2114724–rs2228611, rs2424932–rs1569686–rs6119954–rs2424908, and overall did not differ significantly between patients and controls (p > 0.05, Table 6).
Table 6.
Analysis of haplotype distribution between the schizophrenia and control groups
| Genes | Haplotype | Cases, n (frequency) | Controls, n (frequency) | p | OR (95% CI) |
|---|---|---|---|---|---|
| DNMT1 | CT | 171 (0.638) | 87 (0.680) | 0.416 | 0.831 (0.531, 1.299) |
| TT | 97 (0.362) | 41 (0.320) | 0.416 | 1.204 (0.770, 1.882) | |
| DNMT3B | AGGC | 11 (0.041) | 8 (0.062) | 0.382 | 0.660 (0.259, 1.684) |
| GTAC | 65 (0.245) | 33 (0.258) | 0.830 | 0.948 (0.583, 1.542) | |
| GTGC | 25 (0.095) | 8 (0.062) | 0.296 | 1.551 (0.678, 3.552) | |
| GTGT | 159 (0.593) | 78 (0.609) | 0.987 | 0.997 (0.644, 1.541) |
Analysis of the association between genotypes and clinical symptoms of schizophrenia
Table 7 displays the associations between DNMT1 (rs2114724 and rs2228611) and DNMT3B gene (rs1569686, rs2424908, rs2424932, and rs6119954) and clinical mental symptoms of schizophrenia. Statistically significant differences in PANSS overall positive symptom scores, P3 (hallucinatory behavior), P6 (suspicious/persecution), G7 (motor retardation), and G15 (preoccupation) were observed when comparing patients with different DNMT1 gene rs2114724 and rs2228611 genotypes. Moreover, significant differences were found in PANSS general symptom scores G1 (somatic concern), G5 (mannerisms and posturing), and G8 (uncooperativeness) among patients with different DNMT3B gene rs6119954 genotypes (p < 0.05). Additionally, significant differences were noted in the PANSS general symptom score G10 (disorientation) among individuals with different DNMT3B gene rs2424908 genotypes (p < 0.05).
Table 7.
Relationships between different DNMT1 and DNMT3B genotypes and PANSS scores in schizophrenia group
| SNP | rs2114724 | rs2228611 | rs1569686 | rs2424908 | rs2424932 | rs6119954 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype (n) | CC (57) | CT (57) | TT (20) | CC (57) | CT (57) | TT (20) | GG (3) | GT (9) | TT (122) | CC (23) | CT (59) | TT (52) | AG (12) | GG (122) | AA (8) | AG (56) | GG (70) | |
| P1 (delusions) | X2 (P) | 6.201 (0.045) | 6.201 (0.045) | 3.054 (0.217) | 2.183 (0.336) | 1.078 (0.281) | 1.664 (0.435) | |||||||||||
| P2 (conceptual disorganization) | X2 (P) | 0.715 (0.699) | 0.715 (0.699) | 0.158 (0.924) | 0.542 (0.763) | 0.044 (0.965) | 1.555 (0.460) | |||||||||||
| P3 (hallucinatory behavior) | X2 (P) | 7.851 (0.020) | 7.851 (0.020) | 5.685 (0.058) | 0.052 (0.975) | 1.878 (0.060) | 0.020 (0.990) | |||||||||||
| P4 (excitement) | X2 (P) | 0.300 (0.861) | 0.300 (0.861) | 0.888 (0.641) | 4.028 (0.133) | 0.894 (0.371) | 3.662 (0.160) | |||||||||||
| P5 (grandiosity) | X2 (P) | 1.264 (0.532) | 1.264 (0.532) | 0.918 (0.632) | 0.219 (0.896) | 0.315 (0.753) | 5.937 (0.051) | |||||||||||
| P6 (suspiciousness/persecution) | X2 (P) | 10.387 (0.006) | 10.387 (0.006) | 2.659 (0.265) | 0.209 (0.901) | 1.292 (0.196) | 0.431 (0.806) | |||||||||||
| P7 (hostility) | X2 (P) | 1.954 (0.376) | 1.954 (0.376) | 2.142 (0.343) | 1.062 (0.588) | 0.203 (0.839) | 1.474 (0.478) | |||||||||||
| Total of positive symptoms | X2 (P) | 8.471 (0.014) | 8.471 (0.014) | 1.995 (0.369) | 0.310 (0.856) | 0.967 (0.334) | 0.610 (0.737) | |||||||||||
| N1 (blunted affect) | X2 (P) | 0.877 (0.645) | 0.877 (0.645) | 1.579 (0.454) | 2.963 (0.227) | 0.723 (0.470) | 2.963 (0.227) | |||||||||||
| N2 (emotional withdrawal) | X2 (P) | 2.692 (0.260) | 2.692 (0.260) | 2.830 (0.243) | 3.709 (0.157) | 0.208 (0.837) | 2.899 (0.235) | |||||||||||
| N3 (poor rapport) | X2 (P) | 1.376 (0.502) | 1.376 (0.502) | 0.863 (0.649) | 1.382 (0.501) | 0.790 (0.430) | 1.337 (0.512) | |||||||||||
| N4 (passive/apathetic social withdrawal) | X2 (P) | 1.886 (0.389) | 1.886 (0.389) | 3.987 (0.136) | 1.640 (0.440) | 0.487 (0.627) | 0.085 (0.958) | |||||||||||
| N5 (difficulty in abstract thinking) | X2 (P) | 1.329 (0.515) | 1.329 (0.515) | 0.736 (0.692) | 2.147 (0.342) | 0.520 (0.603) | 1.537 (0.464) | |||||||||||
| N6 (lack of spontaneity and flow of conversation) | X2 (P) | 0.720 (0.698) | 0.720 (0.698) | 0.019 (0.990) | 0.500 (0.779) | 0.138 (0.890) | 0.037 (0.982) | |||||||||||
| N7 (stereotyped thinking) | X2 (P) | 4.471 (0.107) | 4.471 (0.107) | 3.009 (0.222) | 0.607 (0.738) | 0.554 (0.580) | 2.333 (0.311) | |||||||||||
| Total of negative symptoms | X2 (P) | 2.359 (0.307) | 2.359 (0.307) | 1.404 (0.496) | 2.412 (0.299) | 0.077 (0.938) | 2.287 (0.319) | |||||||||||
| G1 (somatic concern) | X2 (P) | 1.253 (0.534) | 1.253 (0.534) | 3.377 (0.185) | 3.283 (0.194) | 0.331 (0.740) | 7.148 (0.028) | |||||||||||
| G2 (anxiety) | X2 (P) | 5.256 (0.072) | 5.256 (0.072) | 1.602 (0.449) | 1.266 (0.531) | 0.662 (0.508) | 0.155 (0.925) | |||||||||||
| G3 (guilt feeling) | X2 (P) | 5.584 (0.061) | 5.584 (0.061) | 1.275 (0.529) | 0.008 (0.996) | 0.626 (0.531) | 2.286 (0.319) | |||||||||||
| G4 (tension) | X2 (P) | 2.819 (0.244) | 2.819 (0.244) | 2.708 (0.258) | 0.810 (0.667) | 0.184 (0.854) | 0.004 (0.998) | |||||||||||
| G5 (mannerisms and posturing) | X2 (P) | 0.300 (0.861) | 0.300 (0.861) | 0.539 (0.764) | 1.022 (0.600) | 0.583 (0.560) | 6.077 (0.048) | |||||||||||
| G6 (depression) | X2 (P) | 4.176 (0.124) | 4.176 (0.124) | 0.276 (0.871) | 1.539 (0.463) | 0.217 (0.828) | 1.688 (0.430) | |||||||||||
| G7 (motor retardation) | X2 (P) | 6.656 (0.036) | 6.656 (0.036) | 1.690 (0.430) | 0.307 (0.857) | 0.276 (0.782) | 1.384 (0.500) | |||||||||||
| G8 (uncooperativeness) | X2 (P) | 3.692 (0.158) | 3.692 (0.158) | 1.840 (0.399) | 0.127 (0.938) | 0.068 (0.946) | 6.923 (0.031) | |||||||||||
| G9 (unusual thought content) | X2 (P) | 5.050 (0.080) | 5.050 (0.080) | 0.839 (0.657) | 1.691 (0.429) | 0.283 (0.777) | 1.464 (0.481) | |||||||||||
| G10 (disorientation) | X2 (P) | 0.371 (0.831) | 0.371 (0.831) | 2.825 (0.243) | 6.927 (0.031) | 0.433 (0.665) | 3.325 (0.190) | |||||||||||
| G11 (poor attention) | X2 (P) | 6.003 (0.050) | 6.003 (0.050) | 2.197 (0.333) | 0.232 (0.890) | 0.222 (0.825) | 1.853 (0.396) | |||||||||||
| G12 (lack of judgment and insight) | X2 (P) | 2.399 (0.301) | 2.399 (0.301) | 2.151 (0.341) | 1.135 (0.567) | 0.463 (0.643) | 3.279 (0.194) | |||||||||||
| G13 (disturbance of volition) | X2 (P) | 3.959 (0.138) | 3.959 (0.138) | 5.174 (0.075) | 1.434 (0.488) | 0.634 (0.526) | 0.117 (0.943) | |||||||||||
| G14 (poor impulse control) | X2 (P) | 1.365 (0.505) | 1.365 (0.505) | 0.354 (0.838) | 3.256 (0.196) | 0.036 (0.971) | 0.210 (0.900) | |||||||||||
| G15 (preoccupation) | X2 (P) | 10.222 (0.006) | 10.222 (0.006) | 1.372 (0.504) | 0.236 (0.888) | 1.065 (0.287) | 0.649 (0.723) | |||||||||||
| G16 (active social avoidance) | X2 (P) | 1.724 (0.422) | 1.724 (0.422) | 1.199 (0.549) | 1.340 (0.512) | 0.420 (0.675) | 1.087 (0.581) | |||||||||||
| Total of general symptom | X2 (P) | 5.710 (0.058) | 5.710 (0.058) | 1.283 (0.527) | 0.027 (0.986) | 0.191 (0.849) | 1.415 (0.493) | |||||||||||
| Positive symptoms | X2 (P) | 7.938 (0.019) | 7.938 (0.019) | 2.579 (0.275) | 2.389 (0.303) | 1.245 (0.213) | 1.617 (0.446) | |||||||||||
| Negative symptoms | X2 (P) | 1.493 (0.474) | 1.493 (0.474) | 1.924 (0.382) | 3.379 (0.185) | 0.066 (0.948) | 1.888 (0.389) | |||||||||||
| Cognitive impairment | X2 (P) | 0.951 (0.621) | 0.951 (0.621) | 0.525 (0.769) | 0.959 (0.619) | 0.180 (0.857) | 2.162 (0.339) | |||||||||||
| Excited/hostile | X2 (P) | 2.261 (0.322) | 2.261 (0.322) | 0.937 (0.626) | 1.646 (0.439) | 0.525 (0.600) | 3.353 (0.187) | |||||||||||
| Anxiety/depression | X2 (P) | 4.999 (0.082) | 4.999 (0.082) | 0.779 (0.677) | 1.293 (0.524) | 0.303 (0.762) | 0.891 (0.640) | |||||||||||
| PANSS (total) | X2 (P) | 5.368 (0.068) | 5.368 (0.068) | 0.683 (0.711) | 0.953 (0.621) | 0.030 (0.976) | 2.158 (0.340) | |||||||||||
The bold values mean the positive association between loci frequence of specific SNP with the type of clinical symptoms
Evaluation of statistical power
G*Power calculated power. This sample had 92.372% power to identify a meaningful relationship (α < 0.05) with an effect size index of 0.5.
Discussion
Accumulating evidence suggests that the development of schizophrenia is influenced by the interaction of heredity and environmental factors [33]. Epigenetic mechanisms, such as DNA methylation with DNA methyltransferases (DNMTs), are crucial for the effect of environmental factors on genetic susceptible. In the dynamic procedure methylation, the DNMT1 is responsible majorly for maintaining methylation during cell division [34, 35].
This study investigated the genotype and allele distribution of DNMT1 (rs2114724 and rs2228611) and DNMT3B (rs1569686, rs2424908, rs2424932, and rs6119954) in 134 patients with schizophrenia and 64 healthy controls. The results showed no statistical difference between the case and control groups, which contradicted to the few previous study in India population [36]. The study found that the TT genotype and T allele in DNMT1 rs2114724 and the AA genotype and A allele in DNMT1 rs2228611 were associated with schizophrenia. However, there was no significant difference in genotype and allele distribution in DNMT3B gene (rs2424932, rs1569686, rs6119954, and rs2424908) between the case group and the control group. In line with study from population with shared ethical background [37], indicated that the genotype frequency and allelic frequency of DNMT1 rs2114724 and rs2228611 were also correlated with schizophrenia. Few studies employing Chinese Han population reported similar findings with the current study. For instance, Zhai et al. [38] investigated the genotype and allele distribution of DNMT1 rs2114724 and rs2228611 in a Chinese population and found no significant difference between the schizophrenic patients and healthy controls. Furthermore, Zhang Chen et al. [39] revealed that there was no statistical difference in genotype and allele distribution of DNMT3B rs2424908 between the schizophrenic case and control groups. Although the significant difference in genotype and allele distribution of DNMT3B rs6119954 was identified in the schizophrenic patients, which indicated dysfunction of DNMR3B. There are couple of reasons contribute to the discrepancies. Firstly, racial genetic background will affect on the difference in allelic frequency, according to the previous findings [38, 39]. Secondly, technology employed for identifying the genetic variation attribute partly to difference of observational results. Previous study indicated that low effective values for identifying genetic variation in promoter upstream regions with Methylated DNA Immunoprecipitation (MeDIP) comparing to microarray [40]. Finally, sample size in the literatures may contribute to variation in the findings [41].
This study used linkage disequilibrium analysis to identify an absolute linkage block between rs2114724 and rs2228611 in DNMT1 and the four loci in DNMT3B. These loci were significantly correlated with positive symptoms of schizophrenia, including hallucinatory behavior, suspicion/persecution, motor retardation, and preoccupation. This is the first report linking rs2114724 and rs2228611 in DNMT1 with positive symptoms of schizophrenia. DNMT1 gene may affect the clinical symptoms of schizophrenia by regulating the expression of genes involved in the dopaminergic and GABAergic systems [41]. Cumulating evidence has suggested that DNMT1 as well as DNMT3 established and maintained dynamically the DNA methylation in dopaminergic, GABAergic, glutamatergic, serotonergic pathways of neurotransmission, which response for regulating the clinical phenotypes [42–46].
Although this study provides new insights into the role of DNMTs in schizophrenia, limitations exist, and further research is necessary to validate the findings in larger samples and different populations. Future studies should also explore the specific regulatory mechanisms of DNA methylation in the development of schizophrenia.
Author contributions
JZ and TYJ contributed to the conception and design of the study and provided approval for publication of the content. JJP designed the study and organized the database, performed statistical analysis, and were responsible for manuscript writing and editing. BGD and XGL conducted quality supervision and guidance for the project execution process. JL was responsible for the testing of experimental samples. JW, CYH, JMY and ZQX were responsible for clinical data collection and clinical evaluation. All authors reviewed and approved the final version of the manuscript.
Funding
Medical Science and Technology Research Fund Project of Guangdong (No. A2021205); Social Welfare Science and Technology Research Project in Zhongshan (No. 2019B1107, No. 2018B1033, No. 2022B1061); Social Welfare Science and Technology Research Major Project in Zhongshan (No. 2018B1006, 2021B3019).
Availability of data and materials
The original data presented in the study are available upon to enquiry from the corresponding authors.
Declarations
Ethics approval and consent to participate
Study protocol carried out in the research is followed in accordance with the ethical guidelines laid down as per the Declaration of Helsinki. The study was approved by the Ethics Committee of the Third People’s Hospital of Zhongshan, and written informed consent was obtained from either the patients or their guardians.
Competing interests
The authors declare that this research was conducted in the absence of any commercial or financial relationships that may be construed as a potential competing interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Tingyun Jiang, Email: jzshantou@126.com.
Jie Zhang, Email: dr.zhangjie@gmail.com.
References
- 1.Stuchlik A, Sumiyoshi T. Cognitive deficits in schizophrenia and other neuropsychiatric disorders: convergence of preclinical and clinical evidence. Front Behav Neurosci. 2014;8:444. doi: 10.3389/fnbeh.2014.00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.James SL, Abate D, Abate KH, Abay SM, Abbafati C, Abbasi N, Abbastabar H, Abd-Allah F, Abdela J, Abdelalim A, et al. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: A systematic analysis for the global burden of disease study 2017. Lancet. 2018;392:1789–1858. doi: 10.1016/S0140-6736(18)32279-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birnbaum R, Weinberger DR. Genetic insights into the neurodevelopmental origins of schizophrenia. Nat Rev Neurosci. 2017;18:727–740. doi: 10.1038/nrn.2017.125. [DOI] [PubMed] [Google Scholar]
- 4.Kim JS, Shin KS, Jung WH, Kim SN, Kwon JS, Chung CK. Power spectral aspects of the default mode network in schizophrenia: an MEG study. BMC Neurosci. 2014;15:104. doi: 10.1186/1471-2202-15-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moskowitz A, Heim G. Eugen Bleuler’s dementia praecox or the group of schizophrenias (1911): a centenary appreciation and reconsideration. Schizophr Bull. 2011;37:471–479. doi: 10.1093/schbul/sbr016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Owen MJ, Sawa A, Mortensen PB. Schizophrenia. Lancet. 2016;388:86–97. doi: 10.1016/S0140-6736(15)01121-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Howes OD, Murray RM. Schizophrenia: an integrated sociodevelopmental-cognitive model. Lancet. 2014;383:1677–1687. doi: 10.1016/S0140-6736(13)62036-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hilker R, Helenius D, Fagerlund B, Skytthe A, Christensen K, Werge TM, Nordentoft M, Glenthøj B. Heritability of schizophrenia and schizophrenia spectrum based on the nationwide Danish twin register. Biol Psychiatry. 2018;83:492–498. doi: 10.1016/j.biopsych.2017.08.017. [DOI] [PubMed] [Google Scholar]
- 9.Mendizabal I, Berto S, Usui N, Toriumi K, Chatterjee P, Douglas C, Huh I, Jeong H, Layman T, Tamminga CA, et al. Cell type-specific epigenetic links to schizophrenia risk in the brain. Genome Biol. 2019;20:135. doi: 10.1186/s13059-019-1747-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gusev A, Mancuso N, Won H, Kousi M, Finucane HK, Reshef Y, Song L, Safifi A, McCarroll S, Neale BM. Transcriptome wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat Genet. 2018;50:538–548. doi: 10.1038/s41588-018-0092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, Ruderfer DM, Oh EC, Topol A, Shah HR. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19:1442–1453. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaffe AE, Straub RE, Shin JH, Tao R, Gao Y, Collado-Torres L, Kam-Thong T, Xi HS, Quan J, Chen Q. Developmental and genetic regulation of the human cortex transcriptome illuminate schizophrenia pathogenesis. Nat Neurosci. 2018;21:1117–1125. doi: 10.1038/s41593-018-0197-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cardno AG, Gottesman II. Twin studies of schizophrenia: from bow-and-arrow concordances to star wars Mx and functional genomics. Am J Med Genet. 2000;97:12–17. [PubMed] [Google Scholar]
- 14.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 15.Zeng Y, Chen T. DNA methylation reprogramming during mammalian development. Genes. 2019;10:257. doi: 10.3390/genes10040257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishioka M, Bundo M, Kasai K, Iwamoto K. DNA methylation in schizophrenia: Progress and challenges of epigenetic studies. Genome Med. 2012;4:96. doi: 10.1186/gm397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez-Jauand M, Sitges C, Rodriguez V, Picornell A, Ramon M, Buskila D, Montoya P. Pain sensitivity in fibromyalgia is associated with catechol-O-methyltransferase (COMT) gene. Eur J Pain. 2013;17:16–27. doi: 10.1002/j.1532-2149.2012.00153.x. [DOI] [PubMed] [Google Scholar]
- 18.Sozuguzel MD, Sazci A, Yildiz M. Female gender specific association of the Reelin (RELN) gene rs7341475 variant with schizophrenia. Mol Biol Rep. 2019;46:3411–3416. doi: 10.1007/s11033-019-04803-w. [DOI] [PubMed] [Google Scholar]
- 19.Janicijevic SM, Dejanovic SD, Borovcanin M. Interplay of brain-derived neurotrophic factor and cytokines in schizophrenia. Serb J Exp Clin Res. 2018;108:110–117. [Google Scholar]
- 20.Leonard S, Freedman R. Genetics of chromosome 15q13–q14 in schizophrenia. Biol Psychiatry. 2006;60:115–122. doi: 10.1016/j.biopsych.2006.03.054. [DOI] [PubMed] [Google Scholar]
- 21.Iwamoto K, Bundo M, Yamada K, Takao H, Iwayama-Shigeno Y, Yoshikawa T, Kato T. DNA methylation status of SOX10 correlates with its downregulation and oligodendrocyte dysfunction in schizophrenia. J Neurosci. 2005;25:5376–5381. doi: 10.1523/JNEUROSCI.0766-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu T-M, Chen S-J, Hsu S-H, Cheng M-C. Functional analyses and effect of DNA methylation on the EGR1 gene in patients with schizophrenia. Psychiatry Res. 2019;275:276–282. doi: 10.1016/j.psychres.2019.03.044. [DOI] [PubMed] [Google Scholar]
- 23.Gowher H, Liebert K, Hermann A, Xu G, Jeltsch A. Mechanism of stimulation of catalytic activity of Dnmt3A and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L. J Biol Chem. 2005;280(14):13341–13348. doi: 10.1074/jbc.M413412200. [DOI] [PubMed] [Google Scholar]
- 24.Symmank J, Bayer C, Schmidt C, et al. DNMT1 modulates interneuron morphology by regulating Pak6 expression through crosstalk with histone modifications. Epigenetics. 2018;13(5):536–556. doi: 10.1080/15592294.2018.1475980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases DNMT3A and DNMT3B are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 26.Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking DNMT3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 27.Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 28.Ripke S, Neale BM, Corvin A, et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malhotra D, Sebat J. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell. 2012;148(6):1223–1241. doi: 10.1016/j.cell.2012.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saradalekshmi KR, Neetha NV, Sathyan S, Nair IV, Nair CM, Banerjee M. DNA methyl transferase (DNMT) gene polymorphisms could be a primary event in epigenetic susceptibility to schizophrenia. PLoS ONE. 2014;9(5):e98182–e98188. doi: 10.1371/journal.pone.0098182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ping J, Zhang J, Wan J, Banerjee A, Huang C, Yu J, Jiang T, Du B. Correlation of four single nucleotide polymorphisms of the RELN gene with Schizophrenia. East Asian Arch Psychiatry. 2021;31(4):112–118. doi: 10.12809/eaap2168. [DOI] [PubMed] [Google Scholar]
- 32.Ping J, Zhang J, Wan J, Huang C, Luo J, Du B, Jiang T. A polymorphism in the BDNF gene (rs11030101) is associated with negative symptoms in Chinese Han patients with schizophrenia. Front Genet. 2022;16(13):849227. doi: 10.3389/fgene.2022.849227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palha JA, Santos NC, Marques F, et al. Do genes and environment meet to regulate cerebrospinal fluid dynamics? Relevance for schizophrenia. Front Cell Neurosci. 2012;6(8):31. doi: 10.3389/fncel.2012.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pogribny IP, Beland FA. DNA hypomethylation in the origin and pathogenesis of human diseases. Cell Mol Life Sci. 2009;66(14):2249–2261. doi: 10.1007/s00018-009-0015-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grayson DR, Guidotti A. The dynamics of DNA methylation in schizophrenia and related psychiatric disorders. Neuropsychopharmaeology. 2013;38(1):138–166. doi: 10.1038/npp.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saradalekshmi KR, Neetha NV, Satyan S, et al. DNA methyltransferase (DNMT) gene polymorphisms could be a primary event in epigenetic susceptibility to schizophrenia. PLoS ONE. 2014;9(5):e98182. doi: 10.1371/journal.pone.0098182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saxena S, Maroju PA, Choudhury S, Voina VC, Naik P, Gowdhaman K, Kkani P, Chennoju K, Ganesh Kumar S, Ramasubramanian C, Prasad Rao G, Jamma T, Narayan KP, Mohan KN. Functional analysis of DNMT1 SNPs (rs2228611 and rs2114724) associated with schizophrenia. Genet Res. 2021;31(2021):6698979. doi: 10.1155/2021/6698979.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhai J, Miao Q, Gao Y, Chen M, Zhou YN, Wei XL, Li J. The relationship between DNA methyltransferase 1 gene polymorphism and cognitive function in the first episode, drug-naïve schizophrenia. Chin J Nerv Ment Dis. 2018;44:166–170. [Google Scholar]
- 39.Zhang C, Fang Y, Xie B, Cheng W, Du Y, Wang D, Yu S. DNA methyltransferase 3B gene increases risk of early onset schizophrenia. Neurosci Lett. 2009;462(3):308–311. doi: 10.1016/j.neulet.2009.06.085. [DOI] [PubMed] [Google Scholar]
- 40.Mill J, Tang T, Kaminsky Z, Khare T, Yazdanpanah S, Bouchard L, Jia P, Assadzadeh A, Flanagan J, Schumacher A, Wang SC, Petronis A. Epigenomic profiling reveals DNA methylation changes associated with major psychosis. Am J Hum Genet. 2008;82(3):696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu X, Ye J, Wang Z, Zhao C. Epigenetic age acceleration was delayed in schizophrenia. Schizophr Bull. 2021;47(3):803–811. doi: 10.1093/schbul/sbaa164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Magwai T, Shangase KB, Oginga FO, Chiliza B, Mpofana T, Xulu KR. DNA methylation and schizophrenia: current literature and future perspective. Cells. 2021;10(11):2890. doi: 10.3390/cells10112890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshino Y, Kawabe K, Mori T, Mori Y, Yamazaki K, Numata S, Nakata S, Yoshida T, Iga J, Ohmori T, Ueno S. Low methylation rates of dopamine receptor D2 gene promoter sites in Japanese schizophrenia subjects. World J Biol Psychiatry. 2016;17(6):449–456. doi: 10.1080/15622975.2016.1197424. [DOI] [PubMed] [Google Scholar]
- 44.Huang HS, Akbarian S. GAD1 mRNA expression and DNA methylation in prefrontal cortex of subjects with schizophrenia. PLoS ONE. 2007;2(8):e809. doi: 10.1371/journal.pone.0000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kordi-Tamandani DM, Dahmardeh N, Torkamanzehi A. Evaluation of hypermethylation and expression pattern of GMR2, GMR5, GMR8, and GRIA3 in patients with schizophrenia. Gene. 2013;515(1):163–166. doi: 10.1016/j.gene.2012.10.075. [DOI] [PubMed] [Google Scholar]
- 46.Carrard A, Salzmann A, Malafosse A, Karege F. Increased DNA methylation status of the serotonin receptor 5HTR1A gene promoter in schizophrenia and bipolar disorder. J Affect Disord. 2011;132(3):450–453. doi: 10.1016/j.jad.2011.03.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original data presented in the study are available upon to enquiry from the corresponding authors.


