Plexiform neurofibromas (PNF) most commonly occur in patients with neurofibromatosis type 1 (NF1), who have a germline heterozygous mutation in NF1. The NF1 gene encodes for neurofibromin, a GTPase activating protein that functions to negatively regulate RAS and its effector signaling pathways. Loss of heterozygosity (LOH) of NF1, therefore, leads to tumorigenesis via RAS activation, and biallelic NF1 inactivation is essential for the development of PNF.1 Approximately 15–30% of patients with NF1 will develop a PNF in their lifetime, most frequently in the head and neck region, extremities, or pelvis,2 but PNF is exceedingly rare outside this population of genetically predisposed individuals. Therefore, for patients who present with a histologically verified PNF, regardless of other clinical features, it is reasonable to proceed to genetic testing for NF1.
Very little is known about sporadic PNF, and published literature includes a small number of single-patient case reports with sporadic, non-NF1-associated PNF.3–6 Further, molecular characterization of these tumors has not been reported to date in the published literature. Herein, we report a unique case of a patient without an identifiable constitutional NF1 mutation, who developed a benign PNF in the liver, that subsequently underwent malignant transformation to MPNST with genomic characteristics resembling a NF1-associated malignancy. Additional germline and molecular tumor testing performed in the context of suspected mosaicism was unable to confirm a diagnosis of NF1.
Case Description
A previously healthy 9-year-old male first presented with a 3-year history of intermittent low back pain. Imaging revealed a T2-hyperintense, T1-hypointense, mildly enhancing polylobulated mass in the region of porta hepatis that was subsequently confirmed by biopsy as a liver plexiform neurofibroma.7 Genetic evaluation at the time of initial diagnosis revealed a family history that was not suggestive of NF1, and the patient did not meet the clinical criteria for the diagnosis of NF1, with only 1 café-au-lait-macule (CALM) on his right lateral ankle, and no other remarkable clinical findings. Genetic testing was therefore not performed. Recommendations included close clinical and imaging follow-up. Four years later, the patient was seen again by the genetics consultation team, and NF1 testing was performed on blood, but did not detect a sequence alteration in NF1. No additional clinical findings suggestive of NF1 had emerged at that time.
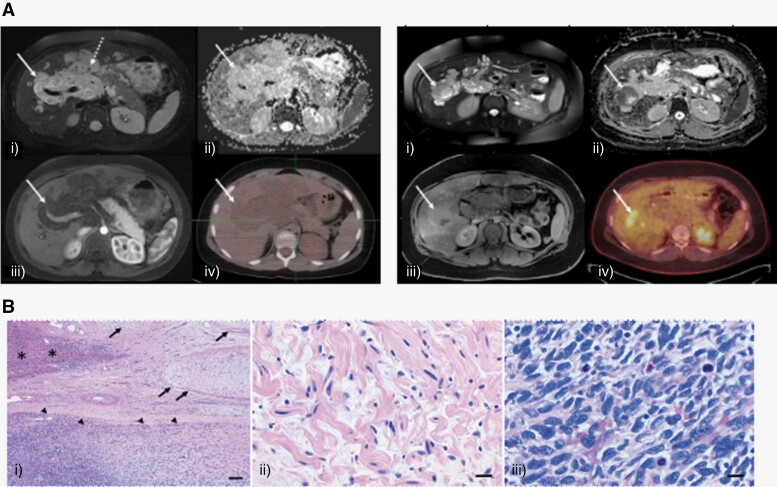
Eight years after his initial presentation, surveillance MRI of the abdomen revealed an interval increase in tumor size of the PNF, with changes in the appearance and enhancement pattern of a new, well-defined, hyperintense, 4.8-cm area centered in the right hepatic lobe, within the known plexiform neurofibroma, with MRI characteristics (suspiciously low apparent diffusion coefficient value) suggestive of malignancy (Figure 1A). A PET/CT scan revealed a focal region of FDG activity corresponding to the new mass in the right lobe of liver (SUV 8.2) and no evidence of other distant metastatic lesions. A biopsy of the mass was performed, and pathology revealed a malignant spindle cell neoplasm with regions of pleomorphism, with apoptotic bodies and necrosis, with loss of S100 protein expression, SOX10 and p53 positivity in a subset of cells, and loss of expression of p16 and H3K27me3, consistent with MPNST (Figure 1B). Diagnosis of MPNST arising from PNF again prompted further investigation into a possible diagnosis of germline NF1 and, at this time, testing for NF1 alterations in blood was pursued again. The single CALM was also biopsied and tested for NF1 mutations, but again, did not reveal any pathogenic variants.
Figure 1.
(A) PNF: (left) axial T2-fat suppressed (FS) (i), apparent diffusion coefficient (ADC) map (ii), T1-FS postcontrast (iii) and F18-fluorodeoxyglucose positron emission tomography-computed tomography (18F-FDG PET-CT) (iv) through the upper abdomen shows a T2-hyperintense periportal soft tissue mass (arrow) without associated restricted diffusion (minimum ADC value (ADCmin) of 1.7 × 10−3 mm2/s and maximum ADC value (ADCmax) of 2.6 × 10−3 mm2/s), enhancement or radiotracer uptake (maximum Standard Uptake Value (SUVmax) of 2.2). Note, subtle target sign (dotted arrow, central T2 hypointensity and peripheral T2 hyperintensity) indicative of plexiform neurofibroma is visible on fluid-sensitive images. MPNST: (right) Axial T2-FS (i), ADC map (ii), T1-FS postcontrast (iii) and 18F-FDG PET-CT (iv) through the upper abdomen shows a discrete nodular lesion (arrow) in the background of periportal plexiform neurofibroma with associated restricted diffusion (ADCmin of 0.7 × 10−3 mm2/s, ADCmax of 2.8 × 10−3 mm2/s and hypointense signal on ADC map), enhancement and hypermetabolic activity (SUVmax of 8.2) compatible with malignant peripheral nerve sheath tumor. (B) Histologic examination demonstrated a plexiform neurofibroma (arrows) involving the liver parenchyma (asterisks) with development of MPNST (arrowheads). Scale bars = 200 microns (i). Bland Schwann cells with shredded collaged and absent mitotic activity in the neurofibroma component. Scale bar = 20 microns (ii). The MPNST demonstrated a cellular spindle cell neoplasm with brisk mitotic activity compatible with high grade. Scale bars = 20 microns (iii).
Due to high speculation of mosaicism, we performed next-generation sequencing (NGS) of adjacent normal liver tissue, PNF, and MPNST in parallel. NGS assays were conducted by the Johns Hopkins Molecular Diagnostics Laboratory and used human reference sequence genome assembly hg19 to analyze the coding region of cancer-related genes. Tumor purity for the PN and MPNST were 40% and 95%, respectively, as reviewed by the pathologist. This testing identified copy number loss of NF1, homozygous deletion of CDKN2A/B, and a likely pathogenic variant in SUZ12 in the malignant tumor tissue (Table 1), but not in the PNF or the normal liver tissue (Table 2). Of note, while the mutation identified in SUZ12 (p.N126fs) is not classified definitively as pathogenic, we suspect that this variant may be pathogenic as frameshift loss of function mutations in SUZ12 are commonly seen in NF1-associated MPNST and H3K27me3 staining was absent in the tumor specimen.8 All samples harbored a missense variant in PMS2 which was classified as a possible germline variant as it was present in normal liver tissue and not a known pathogenic variant. There was not a family history suggestive of Lynch syndrome, which is associated with pathogenic variants of PMS2, and therefore further testing was not recommended. Additional genomic variants identified, including BLM, DMN2, ETV6, NLRP1, and others, were found in all tested samples and also thought to be germline polymorphisms.
Table 1.
Molecular characterization of MPNST, PNF, and adjacent normal liver
| Pathogenicity | MPNST | PNF* | Normal liver*** | CALM * | Blood** | |
|---|---|---|---|---|---|---|
| NF1 copy loss | Pathogenic | + | − | − | − | − |
| CDKN2A/B loss | Pathogenic | + | − | − | NT | NT |
| SUZ12 p.N126fs | VUS, possibly pathogenic | + | − | − | NT | NT |
| TYK2 p.P117S | VUS | + | + | + | NT | NT |
| PMS2 p.V717M | VUS | + | + | + | NT | NT |
NT, not tested; VUS, variant of unknown significance.
*Sequencing for NF1 variants in CALM and PNF was performed at University of Alabama at Birmingham (UAB). No reportable NF1 variants were identified in the CALM but for the PNF, sequencing failed due to DNA quality.
**Peripheral blood was tested in 2016 at UAB and did not reveal any NF1 sequence alteration.
***Next-generation sequencing (NGS) was performed using the in house JHU solid tumor panel (v3.0), for the normal adjacent liver, PNF and MPNST.
Table 2.
Other coding variants present in normal liver, PNF, and MPNST.
| Variant allele frequency (VAF) | |||||||
|---|---|---|---|---|---|---|---|
| Chr:Pos | Base change | Gene | AA-change | Pathogenicity | Normal liver | PNF | MPNST |
| Chr7:6022480 | C>T | PMS2 | p.V717M | VUS | 28.02 | 33.25 | 26.12 |
| Chr15:91295059 | A>C | BLM | p.H281P | VUS | 48.41 | 47.81 | 48.99 |
| Chr9:22008767 | C>G | CDKN2A | p.Q62H | Pathogenic | 48.73 | 44.26 | 42.22 |
| Chr19:10940910 | C>T | DNM2 | p.A800V | VUS | 49.64 | 44.07 | 46.84 |
| Chr12:12022450 | A>G | ETV6 | p.I186V | VUS | 48.20 | 50 | 49.59 |
| Chr15:99451976 | G>A | IGFIR | p.R437H | Nonpathogenic | 47.84 | 48.81 | 51.28 |
| Chr19:36223217 | C>T | KMT2B | p.P1923S | VUS | 48.98 | 49.23 | 50.68 |
| Chr17:5456827 | C>T | NLRP1 | p.V803I | Nonpathogenic | 48.51 | 44.51 | 78.97 |
| Chr14:103342015 | C>7 | TRAF3 | p.R118W | Nonpathogenic | 47.36 | 46.76 | 48.38 |
| Chr19:10478847 | G>A | TYK2 | p.P117S | VUS | 50.26 | 47.78 | 49.09 |
| Chr6:112382222 | C>T | WISP3 | p.P26L | Nonpathogenic | 45.01 | 45.92 | 45.36 |
| Chr6:112382236 | C>A | WISP3 | p.P31T | Nonpathogenic | 45.06 | 45.74 | 44.74 |
AA-change, change that occurred in the peptide sequence; Chr:Pos, chromosome: position; VAF, variant allele frequency.
The patient was treated with neoadjuvant chemotherapy, consisting of 2 cycles of ifosfamide/doxorubicin and 2 cycles of ifosfamide/etoposide according to the regimen of SARC006,9 after which imaging revealed only a minimal decrease in size of the malignant tumor. In preparation for surgery, a right portal vein embolization was performed to allow for compensatory growth of the left liver lobe. He then underwent resection of the tumor with a right hepatectomy, removing the MPSNT entirely encased by plexiform neurofibroma within the right liver lobe. The surgical pathology report revealed a largely viable neoplasm (<5% necrosis) with wide (>3.5 cm) negative margins. No further chemotherapy was recommended based on the apparent lack of response to neoadjuvant chemotherapy. To date, the patient remains in complete remission for 4 years and continues to have clinical and imaging surveillance without evidence of recurrent malignant processes.
Discussion
PNF are benign tumors that occur almost exclusively in patients with NF1. They arise within the nerves and consist of multiple cell types, including Schwann cells, fibroblasts, mast cells, and macrophages.10 While PNF development is thought to require biallelic inactivation of NF1, the NF1+/− microenvironment also contributes to PNF tumorigenesis.11 Previously published studies have shown that LOH, due to a “second hit” somatic mutation, in addition to the germline mutation, is the initiating event in PNF formation.12 However, the pathogenesis and genetic landscape of NF1+/+ plexiform neurofibromas, which is an extremely rare manifestation, is poorly understood and to date, published case reports regarding non-NF1-associated PNF have not reported on molecular analysis of the tumor tissue. Among several published case reports, the oral cavity was the most prevalent site of the non-NF1 PNF and, notably, 5 of these cases describe a single sporadic liver PNF.3–6 To date, none of these cases have reported the malignant transformation of a non-NF1 PNF to MPNST.
PNF in patients with NF1 have an approximate 10–15% lifetime risk of transformation to MPNST.13 The increased lifetime risk of developing MPNST in the NF1 population and the advances in genetic sequencing have aided in understanding the role of specific mutations in MPNST tumorigenesis. To date, loss of CDKN2A/B and TP53 mutations are among the most common alterations recognized in NF1-associated and sporadic MPNST. Further, inactivation of the epigenetic regulatory PRC2 components EED and SUZ12 has been described as a recurring molecular characteristic of NF1-associated MPNST.14 It is well established that the transition of an NF1-associated PNF to atypical neurofibromatous neoplasm of uncertain biological potential (ANNUBP), a premalignant entity that is characterized by specific histological findings,15 is primarily driven by the deletion of CDKN2A/B in addition to NF1 inactivation.16 Further progression to MPNST frequently involves inactivating mutations in the polycomb repressive complex 2 (PRC2) components EED and SUZ12 and these alterations are thought to be more frequent in NF1-MPNST compared to sporadic MPNST.8
The presence of a pre-existing PNF prior to MPNST transformation and the genetic characterization of the MPNST from our patient are consistent with those found in NF1-associated MPNST; however, there is no confirmatory physical or genetic evidence of an underlying NF1 syndrome in our patient. Attempts to prove mosaicism limited to surrounding liver tissue were unsuccessful. Furthermore, the particular genetic alteration in SUZ12 has not previously been definitively classified as pathogenic, but we consider it possible that this variant contributed to tumorigenesis. Further functional studies could validate the role of this particular mutation and this warrants further investigation.
It is worth noting that NF1 variants were not identified in the CALM, blood, normal adjacent liver, nor PNF, excluding the diagnosis of mosaic NF1. Mosaic NF1 is an underdiagnosed condition that has important clinical implications for patients and there are specific diagnostic criteria for the diagnosis. Briefly, one of the following should be present: a pathogenic heterozygous NF1 variant with allele fraction of less than 50% in apparently normal tissue, a pathogenic heterozygous NF1 variant in 2 anatomically independent affected tissues, or a clear segmental distribution of CALM or cutaneous neurofibromas.17 The patient described herein did not meet these criteria.
Conclusions
The patient presented herein highlights a unique case of a sporadic liver PNF without a known molecular etiology, which subsequently progressed to MPNST with the typical genomic alterations seen in NF1-associated malignancy, in the absence of clinical findings to meet the diagnostic criteria of constitutional or mosaic NF1. With the currently available molecular testing, we were unable to identify discrete genomic evidence of a germline NF1 diagnosis despite the seemingly classic presentation of NF1-associated MPNST.
Additional preclinical and genomic studies are needed to identify less prevalent genetic constitutional variants in human PNF. This effort is vital to define the molecular landscape of these rare tumors, and collaborative efforts among multiple institutions are required to maximize the information gathered from sporadic PNF and their risk for malignant transformation.
Contributor Information
Maria Ioannou, Department of Neurology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Lindy Zhang, Department of Oncology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Krista Schatz, Department of Genetic Medicine, Johns Hopkins Hospital, Baltimore, Maryland, USA.
Fausto J Rodriguez, Department of Pathology, University of California Los Angeles, Los Angeles, California, USA.
Shivani Ahlawat, Russell H. Morgan Department of Radiology & Radiological Science, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Christopher D Gocke, Department of Oncology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA; Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Daniel S Rhee, Department of Pediatric Surgery, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Verena Staedtke, Department of Neurology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA; Department of Oncology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Christine A Pratilas, Department of Oncology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA; Department of Pediatrics, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Conflict of Interest
None declared.
Funding
None.
Ethics Statement
The patient has provided consent to publication of relevant information presented in the manuscript.
References
- 1. Zhu Y, Ghosh P, Charnay P, Burns DK, Parada LF.. Neurofibromas in NF1: Schwann cell origin and role of tumor environment. Science. 2002;296(5569):920–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Korf BR. Plexiform neurofibromas. Am J Med Genet. 1999;89(1):31–37. [DOI] [PubMed] [Google Scholar]
- 3. Ji G, Wang K, Jiao C, Lu Z, Li X.. Solitary plexiform neurofibroma of the hepatic artery. J Gastrointest Surg. 2018;22(4):757–758. [DOI] [PubMed] [Google Scholar]
- 4. Nebiki H, Hiramatsu S, Sakata Y, et al. A rare case of plexiform neurofibroma of the liver in a patient without neurofibromatosis type 1. Clin J Gastroenterol. 2020;13(6):1297–1302. [DOI] [PubMed] [Google Scholar]
- 5. Jungmann J, Heydt C, Bohle R, et al. Genetic basis of a solitary familial plexiform neurofibroma without verified associated neurofibromatosis. J Dtsch Dermatol Ges. 2016;14(5):525–527. [DOI] [PubMed] [Google Scholar]
- 6. Beert E, Brems H, Renard M, et al. Biallelic inactivation of NF1 in a sporadic plexiform neurofibroma. Genes Chromosomes Cancer 2012;51(9):852–857. [DOI] [PubMed] [Google Scholar]
- 7. Scheurkogel M, Koshy J, Cohen K, Huisman T, Bosemani T.. Diagnosis and management of an isolated pediatric plexiform neurofibroma involving the hepatic and celiac plexus using multimodality approach: problem solving with diffusion-weighted magnetic resonance imaging. Eur J Pediatr Surg Rep. 2013;01(01):005–008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang M, Wang Y, Jones S, et al. Somatic mutations of SUZ12 in malignant peripheral nerve sheath tumors. Nat Genet. 2014;46(11):1170–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Higham CS, Steinberg SM, Dombi E, et al. SARC006: Phase II trial of chemotherapy in sporadic and neurofibromatosis type 1 associated chemotherapy-naive malignant peripheral nerve sheath tumors. Sarcoma. 2017;2017:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Management of neurofibromatosis type 1-associated plexiform neurofibromas. Neuro-Oncology. Accessed February 5, 2023. https://academic.oup.com/neuro-oncology/article/24/11/1827/6601427?login=false [DOI] [PMC free article] [PubMed]
- 11. Kluwe L, Friedrich RE, Mautner VF.. Allelic loss of the NF1 gene in NF1-associated plexiform neurofibromas. Cancer Genet Cytogenet. 1999;113(1):65–69. [DOI] [PubMed] [Google Scholar]
- 12. Yang FC, Ingram DA, Chen S, et al. Nf1-dependent tumors require a microenvironment containing Nf1+/− and c-kit-dependent bone marrow. Cell. 2008;135(3):437–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Higham CS, Dombi E, Rogiers A, et al. The characteristics of 76 atypical neurofibromas as precursors to neurofibromatosis 1 associated malignant peripheral nerve sheath tumors. Neuro-Oncology. 2018;20(6):818–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim A, Stewart DR, Reilly KM, et al. Malignant peripheral nerve sheath tumors state of the science: leveraging clinical and biological insights into effective therapies. Sarcoma. 2017;2017:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Miettinen MM, Antonescu CR, Fletcher CDM, et al. Histopathologic evaluation of atypical neurofibromatosis tumors and their transformation into malignant peripheral nerve sheath tumors in neurofibromatosis 1 patients—a consensus review. Hum Pathol. 2017;67:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pemov A, Hansen NF, Sindiri S, et al. ; National Intramural Sequencing Center (NISC) Comparative Sequencing Program. Low mutation burden and frequent loss of CDKN2A/B and SMARCA2, but not PRC2, define premalignant neurofibromatosis type 1–associated atypical neurofibromas. Neuro-Oncology. 2019;21(8):981–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Legius E, Messiaen L, Wolkenstein P, et al. ; International Consensus Group on Neurofibromatosis Diagnostic Criteria (I-NF-DC). Revised diagnostic criteria for neurofibromatosis type 1 and Legius syndrome: an international consensus recommendation. Genet Med. 2021;23(8):1506–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]