Abstract
With medical software platforms moving to cloud environments with scalable storage and computing, the translation of predictive artificial intelligence (AI) models to aid in clinical decision-making and facilitate personalized medicine for cancer patients is becoming a reality. Medical imaging, namely radiologic and histologic images, has immense analytical potential in neuro-oncology, and models utilizing integrated radiomic and pathomic data may yield a synergistic effect and provide a new modality for precision medicine. At the same time, the ability to harness multi-modal data is met with challenges in aggregating data across medical departments and institutions, as well as significant complexity in modeling the phenotypic and genotypic heterogeneity of pediatric brain tumors. In this paper, we review recent pathomic and integrated pathomic, radiomic, and genomic studies with clinical applications. We discuss current challenges limiting translational research on pediatric brain tumors and outline technical and analytical solutions. Overall, we propose that to empower the potential residing in radio-pathomics, systemic changes in cross-discipline data management and end-to-end software platforms to handle multi-modal data sets are needed, in addition to embracing modern AI-powered approaches. These changes can improve the performance of predictive models, and ultimately the ability to advance brain cancer treatments and patient outcomes through the development of such models.
Keywords: neuro-oncology, pathomics, pediatrics, radiomics, radio-pathomics
Key Points.
Integrated radio-pathomic analyses can provide novel characterizations of brain tumors
Translational radio-pathomics should be empowered with integrated technology infrastructures and modern analytical solutions
Pediatric central nervous system (CNS) tumors remain the leading cause of cancer mortality in children.1 In the era of precision medicine, the World Health Organization (WHO) Classifications of CNS tumors, the International Society Of Neuropathology, and the International Collaboration on Cancer Reporting all promote an integrated diagnosis in a layered manner, incorporating histopathologic diagnosis, CNS grading, and molecular findings.2–4 Given the emphasis on a tailored approach to diagnosis and treatment, there is an increasing role for advanced data analytics in pediatric neuro-oncology, such as those combining multiple data types (multi-modal; multi-omics) to further enable predictive forecasting and aid in clinical decision-making.
Multi-omic approaches utilizing integrated data sets have promise for providing rich characterizations of different cancer types and outcomes at population and individual levels. There are several types of data collected through standard clinical care for pediatric patients with CNS tumors. This includes clinical (eg, demographic) and qualitative behavioral measures (eg, functional neurological symptoms) as well as radiology imaging scans, which non-invasively capture disease states typically over multiple time points. If there is a surgical collection of tumor tissue as part of a biopsy or resection, there is frequently molecular testing as well as pathological review, and often scanning of the histology slides into digital images, which together provides derivative genomic, transcriptomic (and other -omic; eg, proteomic, metabolomic) and digital pathology data. Only recently has there been a consolidation of large data sets to begin to integrate such data types with predictive analytics like artificial intelligence (AI) methods that require ample training data, though these efforts have largely been restricted to neuro-oncology research on adult populations. In this paper, we focus specifically on the integration of medical imaging data acquired through clinical practice, namely radiology and pathology images, and related applications in pediatric neuro-oncology research.
Utilizing standard radiological imaging with AI and machine learning (AI/ML) methods, radiomics is a rapidly evolving and expanding field. Radiomics entails using advanced computing to extract lesional characteristics from radiology scans for a variety of clinical applications, including predictive modeling of treatment outcomes and classification of tumor type or genetic status.5,6 The steps performed in radiomic analysis may vary on a contextual basis, but overall, radiomics involves image acquisition, pre-processing, tumor region-of-interest segmentation, feature extraction, and finally data analysis (eg, model building and testing). Corresponding quantitative radiomic features encompass both first- (eg, simple signal intensity distribution statistics), second-order (eg, texture, shape), and higher-order (eg, statistical methods after applying image filters or mathematical transforms) image properties, and can span from hundreds to thousands of features for a given patient and time point. The quantitative features extracted and resulting radiologic phenotypes would ideally be reproducible, particularly if formulated using the standard definitions7; however, there are practical limitations to reproducibility caused by differences in image acquisition across scanners and sites.8 In adult brain cancer research, radiomics has shown successful prediction of clinically relevant outcomes and genomic characterizations such as mutational status.9–11 The use of such approaches in pediatrics has been largely limited by a lack of available data sets.8
With the digitization of tissue sectioning and the development of high-resolution whole-slide images (WSIs) afforded by recent advancements in scanning hardware and storage capacities, histopathologic analysis is ripe for advanced computational methods. Much like radiomic analysis of radiologic images, the field of pathomics involves data preparation, cellular segmentation, and extraction of quantitative phenotypic features embedded within WSIs.12 Although the analysis may be done on a nuclear, cellular, or gross sample level, the extracted features can be used to develop a computational model for a variety of purposes including tissue diagnosis, tumor invasion, or tumor classification.12 Substantial work in pathomics focusing on cancers outside of the CNS has advanced rapidly; however, the use of pathomics in neuro-oncology faces particular challenges that have slowed its progress (see Potential Applications of Radio-pathomic Methods in Pediatric Neuro-oncology: Hurdles and Solutions).
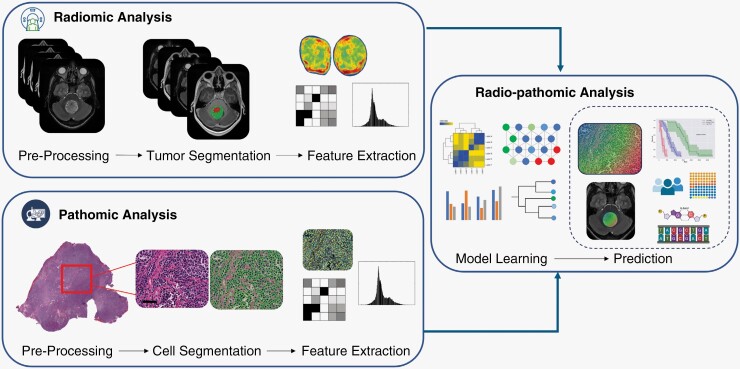
The integration of radiology and pathology data has the unique advantage of capturing tumor properties at complementary macro- and micro-spatial scales. Although WSIs provide a lens into the microscopic cellular makeup and architectural patterns of a tumor and its surrounding regions (microenvironment), MRIs enable characterizing global tumor morphology and appearance, quantitative statistical measurements, and anatomical relationships while also incorporating larger-scale spatial heterogeneity. Together, the combination of radiomic and pathomic features with AI/ML methods (radio-pathomics) can apture complex diagnostic information across multiple spatial scales, providing detailed modeling of both biological and structural elements (Figure 1). Additionally, the marriage of these data types could expand our understanding of the biological underpinnings of imaging features which have been elusive to date.13 Radio-pathomic algorithms could have novel implications not only for personalized treatment pathways but also may aid in pre-operative counseling, development of non-surgical diagnostic tools, and survival prognostication. Some neuro-oncology research studies have employed radio-pathomic methods; however, recent analyses have been limited to adult populations and are nearly non-existent in pediatric research. With a dearth of pathomics and radio-pathomics literature in pediatric neuro-oncology, there is significant potential for advancements in the field and corresponding translational impact in clinical patient care.
Figure 1.
Utilization of radiomic and pathomic analysis into an integrative approach. Images depict MRI (upper panel) and histopathology slide (lower; 20x magnification with hematoxylin and eosin stain, scale bar = 50 μm) of a representative patient with medulloblastoma.
Independently, the fields of radiomics and pathomics involve feature extraction from clinically acquired images and input of those image-based features in a machine or deep learning model for performing a variety of clinical applications. The additive value of integrating across these clinically distinct disciplines, each with immense quantities of extractable data, affords significant potential in improving advanced tools for integrated diagnostics and ultimately facilitating the translational impact of AI/ML models. In this paper, we first briefly review current literature in pediatric brain tumor (PBT) radiomics, pathomics, and radio-pathomics, and then discuss the potential, including the implications and challenges, involved in using these rigorous analyses in practice. Our goal is to highlight and encourage the scientific community to pursue the merging of these rich data sets and create a foundational vision for the potential and challenges of radio-pathomic research. We also highlight the importance of addressing current difficulties in acquiring, integrating, and preparing radiology and pathology data for AI/ML uses in research and clinical domains while outlining potential methodological and analytical solutions for facilitating research in these areas.
Review of the Literature
To examine the literature, we queried Pubmed and Embase for pertinent articles using the search terms “radiomic*” AND “pathomic*” OR “radiopathomic*” published since 2017. With the emergence of radiomic and pathomic analyses, the application and utilization of these modalities are expanding. However, as it pertains to pediatric neuro-oncology, the literature on integrated radio-pathomic analyses is lacking, particularly when compared to its adult counterparts. Our search generated a total of 33 articles, which were selected and filtered based on title and abstract for relevance to yield only a single article pertaining to pediatric neuro-oncology in a post-mortem analysis,14 which further highlights the immense potential for advancement novel exploits of these rigorous data sets.
Overall, the radiomics literature in PBTs provides initial findings on tasks such as the ability to differentiate between tumor histologies, identify disseminated disease, predict molecular subgroups, and even provide prognostic information.8 For example, ML models using MRI data have been developed to differentiate pilocytic astrocytomas, medulloblastomas and ependymomas, and to predict CSF dissemination in medulloblastoma with reasonable accuracy and internal data set predictive ability.15–18 Furthermore, given the increasing significance and integration of molecular and genomic profiles, not only for WHO diagnosis but also treatment pathways, ML methods have also been developed to predict molecular subtypes and genetic mutations. Predicting BRAF mutations, or medulloblastoma subtypes, this integration of radiomics and molecular or genomics data (radiogenomics) exhibits significant potential.15,19 That said, many of the available radiomic and radiogenomic models have failed to generalize beyond the data sets employed in initial studies and have yet to be translated more broadly in a clinical context.8
Pathomic analyses have been applied in several oncologic disciplines, including adult solid tumors, hematologic malignancies, and CNS lesions.12,20–22 Although similar applications in pediatric neuro-oncology have been limited, several pathomic models have been developed that examine the histologic features of medulloblastoma. In these analyses, WSI were used to extract key features to develop and train machine and deep learning models for the prediction of histologic subtypes of medulloblastoma.23–27 The goal of these studies was automated pathologic evaluation of medulloblastoma for classification purposes to accelerate diagnosis and reduce between and within-observer variability. The data sets were composed of either textural features extracted from WSI, histologic features, or a fusion of textural and spatial histologic features.23,25–27 Utilizing a combination of broad textural features, including gray-level covariance matrix and gray-level run length matrix, Das et al., showed an accuracy of 91.3% in differentiating the four histologic subtypes of medulloblastoma (classic, desmoplastic, large cell, nodular) and an accuracy of 96.7% when applying feature reduction.27 The authors in Bengs et al. showed that the pre-trained EfficientNets deep learning (DL) model could yield an AUC of 0.85 in differentiating between classic and desmoplastic/nodular subtypes with data collected across 12 contributing sites (also see transfer learning approach in Das et al.).24,25 Meanwhile, the DL pipeline presented by Atallah et al., using a combination of neural networks, showed an AUC of 1 in classifying the four medulloblastoma histologic subtypes.23 Beyond predicting medulloblastoma subtypes, a preliminary analysis by Whitney et al. was also able to create a model that predicts clinical outcomes.26 With a study sample of 46 patients, this group was able to use ML classifiers to predict long-term survival using nuclear histomorphometric features. Using a neural network, they were able to predict survival of Group 3 tumors with an AUC of 0.92, showing the clinical potential of pathomic analysis in examining histologic features. Although there is variability in AI/ML techniques and study design, these studies highlight the potential of applying pathomic analyses to other PBTs. Furthermore, pathomic analyses integrated with other -omic data types, including genomics data, could be utilized to augment survival prediction.28
Although radiomic and pathomic analyses have independently shown initial progress, the application of integrated radio-pathomics in pediatric neuro-oncology is lacking. To the best of our knowledge, only a single scientific article exists that employs a radio-pathomic approach for studying PBTs. A recent paper by Ye et al. was able to highlight the synergistic potential of bridging MRI and histologic data.14 In pursuit of a novel image processing technique using MRI for detection and differentiation of tumor histopathology, Ye et al. were able to histologically characterize and classify high-grade PBTs using post-mortem brain tumor specimens. The authors performed an ex-vivo analysis on 45 specimens from 9 patients. By co-registering histologic specimens and MRI images, they obtained input classifiers for a deep neural network algorithm to aid in the development of their novel Diffusion Histology Imaging (DHI) with diffusion basis spectrum imaging (DBSI). The DHI technique achieved an overall accuracy of 85% in classifying tumor histology, and with further validation may be an alternative, non-invasive MRI sequence to provide important histopathologic information to diagnose and monitor high-grade tumors as well as guide surgical interventions.
The studies reviewed varied in their application of cross-validation techniques, and some applied data augmentation to enhance their models’ generalization ability and robustness.23–25 Although most studies implemented methods to support generalizability and reduce over-fitting, none explicitly discussed the use of external test data sets or showcased the generalizability of their models across data sets from different sites or populations with appropriate validation methods (with the exception of Bengs et al24). A summary of the findings is provided in Table 1.
Table 1.
Summary of Recent Pathomic and Radio-pathomic Literature in Pediatric Neuro-Oncology Based on Literature Review
| Authors | Year | Tumor Type | No. of Subjects | No. of Collection Sites | Type of Analysis | Methodology | Validation | Findings/Performance |
|---|---|---|---|---|---|---|---|---|
| Ye et al.14 | 2021 | High Grade Glioma | 9 | 1 | Combined radiomic and pathomic analysis performed post-mortem by employing diffusion basis spectrum imaging on co-registered H&E images | Deep Neural Network | 99,388 imaging voxels split into training,/validation/test (8:1:1 ratio) |
Able to classify tumor histology features with overall accuracy of 85.8% |
| Attallah et al.23 | 2022 | Medulloblastoma | 11 available in source data set,66 no mention of exclusion | 2 | Textural and spatial features from H&E images via pre-trained models are applied for classification of 4 MB histologic subtypes | CNN used for feature extraction. Features then input to 6 classification algorithms (LDA, QDA, SVM, KNN, RF, NB) | No information provided. Pre-trained CNN models trained with original images (59/42//30/23 classic/desnmoplastic/large cell/nodular). |
Reports classification accuracy of 100% when using combined textural and spatial features |
| Bengs et al.24 | 2021 | Medulloblastoma | 161 | 12 | Classification to differentiate between 2 MB histologic subtypes classic and desmoplastic/nodular with pre-trained models | Various pre-trained CNNs (AlexNet, VGG-16, ResNet50, Densenet121, EfficientNets) | 10-fold cross-validation with equal split of patients into test/validation (103 classic, 58 desmoplastic/nodular; 2,769 total patches) | EfficientNet-B5 showed greatest performance with F1-score of 0.80 and AUC 0.85 |
| Das et al. 27 | 2019 | Medulloblastoma | Not stated | 1 | Textural feature analysis to create classification model for binary classification (normal vs. abnormal regions on MB slides) | 6 classifiers (LDA, QDA, SVM, KNN, logistic regression, Decision-Tree) | 5-fold cross-validation with 160 images (80 “normal” patches, 80 MB patches) | 100% accuracy with all 172 texture features (Tree, SVM, KNN algorithms) |
| Das et al. 25 | 2020 | Medulloblastoma | 11 | 2 | Transfer learning for binary (normal vs MB) and multi-class (4 MB histologic subtype) classification | 2 pre-trained models (Alexnet, VGG-16); 2 classification algorithms (SVM, softmax) | Internal data set split training/validation (70:30 ratio) for transfer learning No information provided for classification (1677/888 abnormal/normal patches; 924/798/588/504 classic/desmoplastic/large cell/nodular patches) |
SVM classifier achieved 0.9962 binary &.9338 multi-class accuracy for classifying 4,096 model-extracted features |
| Whitney et al. 26 | 2022 | Medulloblastoma | 46 | 1 | Extracted nuclear histomorphometry features to predict MB molecular subtype (SHH/WNT vs. Group 3/4), and correlative clinical outcomes | 4 classifiers (LDA, SVM, RF, neural network) and 4 feature ranking methods | Patient-separated 3-fold cross-validation | Able to distinguish SHH/WNT from Group 3/4 MB with AUC 0.70 (LDA), and able to predict survival in Group 3/4 with AUC 0.83 (SVM) |
AUC, area under the curve; CNN, convolutional neural network; H&E, hematoxylin and eosin; KNN, k-nearest neighbor; LDA, linear discriminant analysis; MB, medulloblastoma; NB, naïve-bayes analysis; QDA, quadratic discriminant analysis; RF, random forest analysis; SHH, sonic hedgehog; SVM, support vector machine; WNT, wingless-related integration site.
Recent work in adult gliomas can provide a framework for such integrative radio-pathomic methods in pediatrics.29,30 For example, Braman et al. developed a predictive model for glioma survival by integrating radiologic, pathologic, genomic, and clinical data.30 The authors were able to combine such multi-modal data for 176 patients from The Cancer Genome Atlas (TCGA) into an integrative deep learning model, called Deep Orthogonal Fusion. Their novel integrative analyses showed a median concordance index of 0.788, which had superior performance to an unimodal strategy. Braman et al. highlight the practical potential of combining a variety of data modalities into a deep learning model for a specific clinical application. Additionally, several other oncologic disciplines, including rectal, prostate, bladder, and breast cancer, have established success in utilizing radio-pathomic modeling across clinical applications, including diagnosis, prognostication, treatment response assessment, and molecular classification.22,31,32
Potential Applications of Radio-pathomic Methods in Pediatric Neuro-oncology: Hurdles and Solutions
Research on radio-pathomic AI methods in pediatric neuro-oncology has been significantly slower than its adult counterpart. Given the immense synergistic potential of combining these rich data sets, we wish to highlight to the pediatric neuro-oncology community the steps and considerations in pursuing such integrative approaches. With increasing technical and processing capabilities combined with broad aggregation of data via consortiums and multi-institutional cooperation, the opportunity for radio-pathomics research should be pursued and capitalized on. In this section, we review methods in the context of pediatric neuro-oncology research utilizing radio-pathomic AI/ML methods and point to challenges in the ability to acquire, synthesize, and prepare large-scale, uniform data sets suitable for AI/ML modeling, as well as in translating research-developed models into clinical practice. We focus on the use of digital pathology images with AI, as well as those faced in integrating radio-pathomic data. Challenges specific to the use of pediatric radiomic methods are outside of the scope here and are discussed in prior reviews.8,33
Clinical, Histological, Genomic, and Radiological Diversity in Pediatric Brain Tumors
Compared to adults, primary brain tumors in pediatrics consist of a greater number of histopathological and molecular classifications,3 and less is known about the corresponding biological underpinnings of pediatric brain tumor groups. For instance, in adult low-grade gliomas, the prognosis is well-captured by a known gene mutation (IDH1/2) and chromosomal deletion (1p/19q codeletion) status in addition to age (lower survival rates in adults older than 40 years compared to 20-3934), and molecular classification based on these factors has taken precedence to histopathological classification in guiding clinical decision-making.35 In pediatric low-grade gliomas, a greater diversity of mutations, largely based on the Ras/MAPK pathway (eg, germline NF1, BRAF V600E, H3.1/H3.3 K27M, TP53, EGFR), has been established.36 High-grade gliomas (HGG) in pediatrics may histologically resemble adult gliomas, but they are divided into subgroups based on genetic alterations (eg, PDGFRA, TP53, wild type, H3G34, H3K27M, and H3G34R/V mutations)37,38 and tumor location properties (eg, midline vs. pontine). These groups vary in age distribution, mutation types, and survival outcomes.
Other frequent brain tumor histologies found in both pediatric and adult cohorts are ependymoma and medulloblastoma; yet again, these present with distinct tumor location, and biological, genomic, and clinical patterns between age groups. Posterior fossa ependymomas are categorized into groups A (PFA) and B (PFB) with different molecular variants. PFA typically occurs in young children and has lower survival rates compared to PFB found in older children and adolescents with a more favorable prognosis.39 Supratentorial and spinal ependymoma tumors are more frequent in adolescents and adults and have positive outcomes. Spinal ependymoma are typically present with either myxopapillary histology or classic histology with 22q chromosomal deletion, and surgical resection is considered curative.3,40 Supratentorial ependymomas are often grouped by ZFTA and YAP1 fusion types, with the YAP1 group being less frequent but with worse prognosis and appearing in young children compared to ZFTA-fused tumors (more frequent, better prognosis, found in adolescent and young adults).
As an embryonal tumor, medulloblastoma is the most common malignant childhood brain tumor under 14 years, categorized into molecular subtypes with distinct genomic aberrations (Sonic Hedgehog (SHH)-activated TP53 wild type, SHH-activated TP53-mutant, Wingless (WNT), and non-WNT/non-SHH (Group 3/4)) as well as four histology classes (classic, large cell/anaplastic, desmoplastic/nodular, extensive nodularity).41 Adult medulloblastomas are primarily of the SHH subtype (with the remaining split between WNT and Group 4 cases),42 while about 30% of pediatric cases are SHH, 25% Group3, 35% Group 4, and WNT being the least common at about 10%.41,43 Additionally, some work has shown differences between chromosomal aberrations detected with IHC and survival outcomes of WNT and Group 4 subgroups between adult and pediatric medulloblastoma.44 Other embryonal tumors that are less common but occur more frequently in pediatric age groups compared to adults are atypical teratoid rhabdoid tumors (ATRT), craniopharyngioma, pineal tumors, meningioma, germ cell tumors, and others.3
Pediatric brain tumors are not only heterogeneous from histologic, genomic, and clinical perspectives but are also highly diverse in terms of their radiological phenotypes. Unlike adult GBMs that most often appear as solid, contrast-enhancing tumors (higher signal intensity on T1-weighted sequences after contrast injection) located in eloquent brain regions, PBTs are often mixed solid-cystic, have varying patterns of contrast-enhancement, and can frequently present in the posterior fossa and midline.45–50 Pre-trained AI/ML models based on MRIs of adult brain tumors may be trained to include such factors, but are typically evaluated based on the whole tumor (unity of all subcomponents), tumor core (subcomponents excluding edema), and/or enhancing core segmentation performance.8,51 Because of the relatively low frequency of cystic, non-enhancing components in adult data sets used for model development, state-of-the-art deep learning-based tumor segmentation models underperform in these regions.52
In sum, the diverse clinical, radiological, histological, and genomic characteristics of PBTs introduce complexity in uncovering relationships between radiomic and pathomic imaging features and their association with genomic and clinical markers. For some cases in which tumor biology and appearance are similar across adult and pediatric cases, such as in certain high-grade gliomas and SHH medulloblastoma, transfer learning of existing adult radio-pathomic models to pediatric cohorts may be possible. AI/ML approaches for histologies that differ between age groups will require representative data to comprehensively capture different subclasses of PBTs (see Slide Image Acquisition and Data Availability) and their corresponding variations in phenotypic and genotypic properties. This necessitates collaborative data-sharing efforts across institutions, and/or infrastructure development to support large-scale federated learning, in which data files are kept securely within separate, private data centers and only model parameters are passed between participating parties.53 Analytical approaches that show promise to help address cases of limited data for model development are further discussed in Analytical Solutions with Artificial Intelligence and Computer Vision Methods.
Slide Image Acquisition and Data Availability
The use of digital pathology images for pathomic feature extraction and computational analysis is met with limitations in software infrastructure and data availability. Although there have been relatively recent advances in WSI scanning to digitize pathology slides in clinical practice, technological bottlenecks still exist post-scanning. For instance, WSI files are large, can be of various file types (eg, SVS, VMS, SCN, MRXS, BIF, and TIFF), and are typically stored on local departmental servers. This can make it difficult to extract and share these files with researchers more broadly. Variability in scanner vendors utilized across institutions and a lack of established standards for imaging acquisition parameters, such as magnification level, cause variability in generated images that must be accounted for in data preparation and model training. Additionally, there are several types of artifacts that can exist in physical slides which cannot be corrected for in post-processing; for example, frozen sectioning can cause “freezing artifact” altering cellular histoarchitecture and corresponding pathomic features. Other artifacts include pen markings, air bubbles, misalignment and resulting tissue cut-off or folding, and out-of-focus images. To address this, some open-source software tools for (semi-)automated quality control (QC) have been developed, such as HistoQC and PathProfiler, which offer solutions for performing QC and artifact detection at-scale.54–56
Despite the implementation of standardized procedures, histological stains exhibit considerable variability due to slide preparation, which relies heavily on human expertise and the specific devices and reagents employed. Multiple factors influence the ultimate appearance of stained tissue samples in digital pathology images, from the initial surgical excision of the tissue through transportation to the laboratory, fixation, sectioning, staining, scanning, and even the storage conditions of the stained slides and inter-patient differences result in alterations in color and staining intensity.57 This challenge becomes prominent in research studies that involve sharing digital images among different laboratories. Stain normalization algorithms can be used to mitigate such effects on the performance of cell segmentation and AI/ML models, such as color matching and stain separation. However, these conventional algorithms face limitations when stains involve multiple components beyond H&E, and more recently deep learning-based methods have emerged as potent solutions.58,59 For example, utilizing a fusion of color augmentation and stain color normalization techniques can enhance the classification performance of a neural network.60
Another consideration is variable staining techniques in modern practice, and although Hematoxylin and Eosin (H&E) staining is commonplace, ancillary diagnostic methods are needed for the complete characterization of a tumor. For example, some tumor histologies are characterized by small round blue cells, such as medulloblastoma and other types of embryonal tumors, which stain blue/purple with H&E staining but consequently can confound the detection of other cell types such as tumor-infiltrating lymphocytes. Although traditional techniques continue to be regularly utilized, the expanding role of immunohistochemistry, and multiplex antibody-based imaging, is a promising modality to improve phenotypic characterization of the local microenvironment.61 Utilizing novel staining and imaging techniques, including variations of antibody conjugation, direct insights into cellular and sub-cellular tissue domains can be elucidated.62,63 Such methods have the potential to be directly associated with emerging theranostic PET imaging methods, which can capture molecular properties, such as metabolic activity, and are becoming increasingly relevant in imaging of tumor burden and response assessment. For example, comparisons between immunoPET and histopathological staining with multiplex IHC could be used to comprehensively examine the heterogeneous tumor immune microenvironment and its spatial architecture.64
The genotypic and phenotypic diversity of pediatric brain tumors necessitates representative data sets with large numbers of samples per unique class needed to allow machine learning models to make accurate predictions. Combined with a relatively low frequency of brain cancers in the general pediatric population, the application of pathomics with AI algorithms is limited by a lack of large, uniform data sets for model development, particularly in the context of multi-site studies. Even more so, unlike their adult counterparts, there is a dearth of pediatric data sets containing both radiology MRIs and digital pathology images, and large data sets are required for building high-performing AI models to ascertain generalizability. To our knowledge, there is only one available repository providing both clinically acquired radiology MRIs and digital pathology slides, which is the Pediatric Brain Tumor Atlas (PBTA) of the Children’s Brain Tumor Network (CBTN) consortium (cbtn.org).65 The PBTA is a large, multi-site data set with data collected across various cancer types. In addition to MRIs and digital slides, associated clinical annotations are available, and many subjects have paired molecular sequencing data. Aside from the PBTA, we have only found one other available repository of pathology images for PBTs. This is a set of H&E stained slides (202 images at 10x microscopic magnification, 153 images at 100x) acquired from tissue biopsy samples of 11 pediatric patients with medulloblastoma (https://dx.doi.org/10.21227/w0m0-mw21).66
The Digital Imaging and Communications in Medicine (DICOM®) standard is a potential solution to the data infrastructure and organization challenges outlined above. DICOM establishes storage, management, and transfer standards that are implemented in (Picture Archiving and Communications Systems, such as those commonly utilized in clinical radiology departments. Beyond imaging pixel data, these files have built-in placeholders containing metadata relevant to the case. Additionally, the data organization of DICOM provides a way to handle multiple images at various resolutions, which is typical for WSIs, as well as multi-frame images such as for large images that are tiled, which lends to virtual microscopy viewing. Notably, the working group WG-26 aims to develop the DICOM standard to support applications in pathology (https://www.dicomstandard.org/activity/wgs/wg-26). That said, commercially available WSI scanners do not typically output files in DICOM format, necessitating a separate tool for conversion from WSI to DICOM and additional steps to capture and incorporate the image metadata. A related consideration is that while DICOM viewers are readily integrated into clinical radiology workflows, current clinical pathology viewers do not support DICOM file formats. There exist some web-based DICOM WSI viewers (eg, VISILAB, Google Cloud Healthcare API) as well as open-source DICOM servers with integrated viewing tools (eg, Orthanc: https://wsi.orthanc-server.com/demo/) that could be integrated into clinical workflows to address this,67 although this would necessitate medical device certification of the software tools for use in clinical routine.
Cell Annotations, Nuclei Detection and Segmentation
Once digital slide data is collected and curated, it must be prepared prior to input to AI/ML model training to reduce the impact of unrelated or undesired noise on learning performance. This often involves pixel intensity histogram normalization, color normalization, or gray scaling (for texture features), and resampling to a common resolution. Note that downsampling (resampling to a lower resolution) can result in loss of cellular-level spatial detail but can required, for example, if there are images of various resolutions in a data set. Next, tumor-involved areas of an image are selected for further analysis. This involves viewing and determining regions of high tumor cell density in each slide.
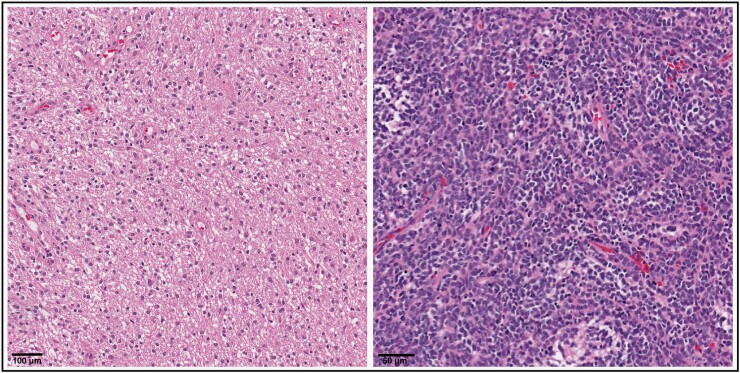
To utilize quantitative pathomic features in AI/ML approaches, individual cellular segmentations must be generated. This is often accomplished using semi-automated methods. Using standalone software such as QuPath,68 a user will examine a region of tumor on a slide and adjust auto-detection parameters until all tumor cells are detected and segmented within the region (based on visual assessment). As there is no standard protocol for cell segmentation, there can be ample inter- and intra-observer variance in manual segmentations due to differing or non-uniform assessment techniques. Inter-observer variability presents a well-known challenge in achieving diagnostic consensus within the field of pathology,69,70 and pediatric cases are particularly susceptible due to their diverse histological appearance.71 Strategies such as molecular or immunohistochemical staining, central reviews, establishing widely agreed-upon and adopted standards, and seeking second opinions in complex or rare cases can elevate diagnostic accuracy, foster consensus among experts, and mitigate variability (also see analytical solutions in Future Directions for Translational Radio-pathomics in Pediatrics). Furthermore, the advent of WSI for converting entire glass slides into digital images has enabled pathologists worldwide to collaborate on the analysis of intricate cases.72 Given the variation in tumor characteristics, such as cellularity, invasiveness, and cellular morphology, there also exists variation in segmentation patterns between tissue and tumor types. For example, the dense, high nuclear/cytoplasm ratio, and less invasive pattern in medulloblastoma pose unique segmentation patterns when compared to the diffuse, and less compact histologic pattern of low-grade glioma (Figure 2). This variation in histologic patterns becomes of greater importance when attempting to create and adopt fully automatic segmentation methods. Some automatic segmentation methods have been proposed using adult data73; future work should focus on generalizing these methods to pediatric settings and integrating them into user-friendly software. Following segmentation, features such as shape morphology, spatial texture patterns, cell count, density, etc., are extracted and can be used as input to AI/ML models to perform tasks such as classification.
Figure 2.
Histopathology slides from two representative patients highlighting the structural and architectural differences between low-grade glioma (left; 20x magnification with hematoxylin and eosin stain, scale bar =100 μm) and medulloblastoma (right; 20x magnification with hematoxylin and eosin stain, scale bar =50 μm).
To date, the process to prepare digital pathology images for use in AI remains manual and time-consuming. The development of standardized, automated pre-processing pipelines that generalize across data sets should help to reduce the technical expertise required to perform analyses with pathomic features. This underscores the development of requisite representative data sets for model building and generalization that would allow for the integration of auto-segmentation models into fully automated pipelines. For instance, some deep learning patch-based approaches have been shown to automatically select local regions of a WSI that are representative tiles for learning tasks such as adult cancer classification,74,75 without the need for manual intervention; however, these nonetheless require an initial set of ample input training data with manual annotations to perform well (but see analytical solutions in Future Directions for Translational Radio-pathomics in Pediatrics).
Spatial Alignment of MRI and Pathology Slides via Co-registration
Broadly speaking, applications of radio-pathomics in oncology research have been particularly successful when MRI scans and digital slides can be spatially aligned. One pertinent example of this is in studies on prostate cancer. Surgical resection of the entire prostate (radical prostatectomy) can be part of the standard of care for patients with prostate cancer, and subsequent co-registration of the histology slides with the pre-operative anatomical MRI can be accomplished with high accuracy. This is because 3D-printed molds can be created based on the MRI scans such that the resected tissue can be sectioned to match the orientation and thickness of the MRI slices.76 Subsequently, co-registration of the MRI with the digitized slide images can be performed. This precise spatial alignment allows for direct comparisons between the two image types, and thus the ability to harness the complementary macro- and micro-scale features that each affords.
Oncologic surgery in the CNS is predicated on maximal tumor resection with preservation of non-tumoral elements. In highly eloquent, and challenging anatomical locations, such as midline structures involved by diffuse midline gliomas (DMG), only a small sample of the tumor is collected via biopsy for diagnostic histological assessment. In either case, spatial alignment of resulting digital slides and pre-surgical MRIs is often difficult due to a lack of spatial landmarks and post-surgical changes. With the advent and increasing adoption of intraoperative MRI, real-time spatial variations caused by surgical manipulation, fluid shifts, and CSF egress may be accounted for.77 This may also potentially be alleviated with image-guided biopsy methods, such that a pre-operative MRI is used to guide the sampling of tissue during biopsy and thus aid in subsequent alignment. Furthermore, supramaximal en bloc resections may also facilitate radiologic and histologic co-registrations, when technically and clinically applicable, via more direct spatial alignment of histology and radiology data (such as described by Roodakker et al.78). Additionally, it is possible for an MRI-based 3D-printed mold to be utilized to slice a resected tumor; however, the implementation of this method in clinical practice is not standard. Notably, 3D model-based registration of histology to MRI has been performed in autopsy cases in adult brain cancer research. With this type of approach, one study found that a radio-pathomic model could detect disease infiltration (based on hypercellularity) beyond spatial boundaries defined by traditional radiological features alone.79
Alternative analytical approaches can be employed to avoid the need to co-register MRI and pathology slides but still utilize features from both modalities in AI/ML applications. One method involves extracting radiomic and pathomic image features separately and then inputting the combined feature set into the AI/ML model.29 Another method involves inputting each feature type into separate models, and then combining their predictions into a single output. In both cases, the ability to acquire information from the spatial overlap of histopathological images and MRI images is lost, and in this way, we cannot make direct inferences about their relationship. Relatedly, this prevents the use of AI methods to perform spatial image tasks, such as predicting the extent of disease infiltration or locations of recurrence, modeling tumor appearance, and corresponding spatial and texture characteristics such as in the analysis of tumor microenvironment. The importance of spatial heterogeneity characteristics of a tumor’s cellular makeup has been acknowledged but has yet to be fully elucidated. For example, tumor microenvironment features obtained from topographical spatial analyses in adult gliomas can highlight immunotherapy target responses and cell signaling pathways.73,80,81 Ideally, radiomic and pathomic co-registration methods should be further pursued to address the limitations of spatial alignment and consequently enable spatial comparisons between the imaging modalities. On the other hand, utilizing informative radiomic and pathomic features for clinically relevant tasks does not necessarily require spatial alignment to perform well. This can include classification tasks such as diagnosis and molecular subtyping, as well as predictions of clinical outcome variables such as in risk assessments of survival or assessment of treatment response. In sum, the co-registration of MRI and pathology images is difficult in practice, particularly in the context of clinically acquired data, and is often impossible with retrospectively collected data due to a lack of corresponding spatial landmarks across the image types. Thus, if researchers aim to study spatial image tasks and metrics across radiology and pathology images, they can prospectively design studies such that alignment across modalities is possible.
Computing Environments and Standardized Pipelines for Image Analysis
The development of AI/ML models, together with the high storage and processing demands of radiology and pathology data, requires high-performance computing environments. Cloud-based infrastructures offer such storage and compute services while offering HIPAA-compliance and eliminating the need for maintenance of physical on-premise servers, such as locally deployed high-performance clusters that are common in academic research settings. One recently released platform that was developed for integrated imaging analysis is the Platform for Imaging in Precision Medicine (PRISM), which offers services for the management of integrated radiology and pathology data sets and feature sets, and tools for cohort creation and clinical data exploration while being deployed in either on-premise or cloud environments.82
In clinical practice, some radiology software platforms have migrated to cloud-based deployments, but clinical pathology software is still most often deployed on local departmental systems. Additionally, while some platforms have begun to support predictive analytics and AI model deployment through integrated tooling, these are still in nascent phases of development. Thus, there is a need for high-computing, analytical platforms that can offer multi-modal data integration across departments in clinical practice and direct application of derived models.
Aside from the need for computational resources, there remains the burden of acquiring and preparing radiology and pathology data to make it AI-ready via large-scale collection, curation, and pre-processing. Currently, the onus of this lies on each researching individual and team that desires to perform studies with radio-pathomic methods. There are no automated processing pipelines developed specifically for pediatric radiomic, pathomic, or radio-pathomic data. Moreover, although there have been automated pipelines developed in the context of adult cancer research, none have been validated using pediatric data and few have deployed in cloud-based clinical platforms. One open-source WSI pre-processing pipeline that is deployed on a research cloud platform is the “SBG Histology Whole Slide Image Preprocessing” toolkit developed by SevenBridges Genomics and available on CAVATICA (cavatica.org). This pipeline performs tissue versus background segmentation and tile generation. Another CAVATICA pipeline is the “HoVer-Net Inference” which implements the HoVer-Net deep learning model for nuclei segmentation and classification of H&E slide images.83 With these types of workflow implementations, researchers have the ability to rapidly and efficiently prepare their digital slide images at-scale with standardized, uniform processes. Furthermore, the adoption of such workflows can support generalizability in downstream analytics. Future work should aim to validate such pipelines with pediatric data and strive towards developing integrated user-friendly software and platforms that empower researchers to utilize existing data and models and flexibly develop their own.
Analytical Solutions with Artificial Intelligence and Computer Vision Methods
The genotypic and phenotypic diversity of pediatric brain tumors leads to complexity of building clinically relevant AI/ML models. Although data aggregation efforts are made difficult due to regulatory, operational, and technical barriers, analytical solutions may help accelerate the use of predictive modeling. Recent advancements in deep learning and computer vision with transfer learning methods show significant promise for alleviating some of the aforementioned challenges in supervised deep learning approaches such as limited data sets and annotations. Self-supervised pre-training in particular provides a technique for initializing a model based on large, unlabeled data sets before translating the model to a different but related task or domain. During pre-training, a model is trained on an initial task using data augmentation, such as rotation or context prediction, to optimize internal parameters and learn useful representations in the images, such as visual patterns of textures, shapes, and structures, in a data-driven manner. The pre-trained model can then be fine-tuned on a different task with labeled data (supervised learning) and theoretically, since model parameters have been initialized with relevant data, learning performance on this secondary task can be boosted.
Self-supervised pre-training has been shown effective in a variety of medical imaging tasks. In the context of histopathological image tasks, it can reduce the amount of labeled training data. In one related study, a deep learning model was pre-trained using a contrastive learning task (discriminating positive and negative pairs of augmented images) on images from 32,529 publicly available WSIs of various cancer types and locations.84 The model was then evaluated on three supervised tasks: (1) classification of colorectal cancer tissue types and normal tissue from 86 WSIs; (2) breast cancer detection from 400 WSIs; and (3) classification of benign and precancerous colorectal polyps from 328 WSIs. The pre-trained model (TransPath; https://github.com/Xiyue-Wang/TransPath) outperformed a model with randomized initial parameters and a model pre-trained on ImageNet85 database. This study indicates great potential for self-supervised methods in pediatric histopathology, particularly to overcome the challenges of limited labeled data, data imbalance, and class fragmentation (significant within-class variation) in pediatric neuro-oncology tasks (eg, tumor detection, cell classification, cancer grading).
Weakly supervised learning methods have also shown a potential to alleviate the challenges of limited annotations and training data with imaging data sets. This method uses partially labeled data, and assuming these labels might be noisy, incomplete, or inaccurate, reduces the influence of observer variability.86 For histopathology images with limited annotations, weakly supervised deep learning has shown efficacy in tasks like cancer classification, localization of regions of interest, tumor subtyping or grading, genetic mutation prediction, and detection of metastasis.87–93 One recent radio-pathomic study, evaluated in the CPM-RadPath 2020 challenge,94 employed weak supervision to predict glioma subtypes (adult oligodendroglioma 1p/19q codeletion, adult astrocytoma IDH-mutant, adult glioblastoma IDH-wildtype) from WSIs without cellular annotations.95 These subtype predictions were then compared with MRI-based class predictions to obtain a confidence measure based on the agreement between data types.
Many of these studies additionally divide WSIs into local spatial regions, that is, patches, and with this approach, spatial probability maps can be generated from trained deep learning models in order to visualize the regions and characteristics that contribute to model predictions. This offers both the efficient use of information across the global WSI for model development, as opposed to only local regions that are commonly selected in manual approaches, and crucially lends to the interpretability of relevant, data-driven information extracted from the images. Notably, these approaches may address the challenges of high false positive and/or negative rates in several weakly supervised deep learning methods, particularly in localization tasks.87 Lastly, combining self-supervised pre-training and weakly supervised methods can significantly reduce the amount of labeled data and boost model performance with histopathology images.96
In radio-pathomic analytics with high-dimensional imaging feature spaces, joint representation learning techniques for multi-modal learning can be used.97 These methods learn a shared representation by integrating features from multiple modalities, eg, histology and radiology image features, as well as genomic and/or clinical factors. Different methods can be used to learn a new representation of the input feature sets, such as directly combining into a single representation (multi-modal fusion), association in a network graph (graph-based fusion), or training on each separately followed by alignment of the shared latent space (variational autoencoders). Across approaches, each modality type can contribute to the shared representation space, and in turn, the result can be a “joint” representation that captures mutual information between disparate data sources. Joint representation learning is a relatively new analytical approach and initial work shows promise with multi-modal biomedical data, such as using chest radiographs and corresponding radiology text reports for image classification and segmentation,98 as well as WSIs, genome, and transcriptome data for determining patient prognosis.28
Future Directions for Translational Radio-pathomics in Pediatrics
The translation of radio-pathomic models to clinical practice settings is an avenue with immense potential for facilitating personalized treatment decision-making and improved patient outcomes. Direct clinical applications include distinguishing between various types of brain tumors, measuring tumor proliferation, infiltration and margins, identification of molecular characteristics, and integrating such factors to aid in understanding the mechanisms of tumor progression, metastasis, and response to treatments. In many types of pediatric brain tumors, the standard of care involves combination therapies of surgery, radiation, and chemotherapy; however, for many pediatric histologies clinical treatment protocols have not changed in decades and high rates of recurrence and low survival accompanied by a lack of effective therapies remain.99,100 For instance, atypical teratoid rhabdoid tumors have an overall 5-year survival rate of around 25-50%.101 New advancements in targeted therapies and immunotherapy approaches can potentially expand treatment options for specific genetic mutations or critical pathway alterations.100,102 Capturing nuanced information in the global-local spatial heterogeneity and quantifying the tumor microenvironment and mutational burden with combined radio-pathomics could be used to tailor such treatments to the individual patient. One study demonstrated that spatial maps of tumor-infiltrating lymphocytes (TIL) could be generated with H&E WSIs across multiple cancer types, which captured lymphocytic infiltration within the tumor and surrounding regions that were related to genomic TIL estimates and survival outcomes.103 Immune cell infiltration in pediatric brain tumors is variable within and across cancer types underscoring the need for individualized approaches,102,104 particularly with combined imaging and genomic analysis. Additionally, some preliminary work has shown the ability to characterize immune microenvironment subgroups in pediatric central nervous system tumors that are related to characterizations from IHC and methylation measures.104
Radio-pathomic models could also lead to more precise and complex modeling of tumor growth and change over time. Pediatric tumors are particularly heterogeneous and different parts of a tumor may grow at different rates and behave differently to treatment. Some tumors, such as ependymoma, can be characterized by intratumoral heterogeneity as various genetic alterations and histological patterns can be comprised within the same tumor.105,106 Anatomical information about surrounding brain structures, tumor location, and global morphology extracted from radiology imaging in combination with microscopic information about cellular subpopulations from histopathology imaging could lead to sophisticated 3D models of the non-uniform evolution of a given tumor over time. In this way, radio-pathomics may better capture the spatial heterogeneity of pediatric brain tumors and be used to construct individualized predictive modeling of tumor growth and infiltration. This could significantly advance the ability to target specific regions more effectively with treatment and lead to more precise assessments of treatment response compared to current standards that rely on visual and 2D volumetric measurements based on MRI.
Discussion
Current literature employing radio-pathomic features with predictive AI models in the field of pediatric neuro-oncology is limited compared to adult brain and other cancer types. We reviewed related challenges that arise in radio-pathomic research across data collection, data management, data preparation, multi-modal analysis, and translation back to clinical practice. These factors should be considered when designing radio-pathomic studies, as limitations that arise in data generation may dictate the types of analytical approaches that can be used, and subsequently their clinical relevance and interpretation of findings. For example, unsupervised ML requires a significant amount of data to perform properly, but supervised methods require image annotations (eg, cell segmentations) that can be time-consuming to generate. Self-supervised pre-training and weakly supervised methods on the other hand have been shown to alleviate the need for large, labelled data sets using data-driven approaches. Additionally, the ability to co-register radiology and pathology images will inform the types of AI/ML analyses that can be performed (ie, spatial vs. nonspatial tasks).
Overall, while the development of AI algorithms with radio-pathomics has gained momentum in adult neuro-oncology research, it remains severely limited in pediatrics. This is largely accounted for by limited data availability and a lack of standardized workflows for image acquisition and data processing. Researchers are required to gain domain and technical expertise and expend significant effort on data collection, curation, and preparation, and even so, are limited in their ability to generate generalizable models. In order to accelerate pediatric radio-pathomic research and related AI applications in computer-aided clinical decision-making, significant advancements must be made in data repository generation as well as the software and infrastructure that support clinical data management and AI model deployment. The collection of digital pathology slide images across sites using standard acquisition protocols and release to the research community could greatly facilitate the use of histopathology images in developing predictive models for pediatric brain tumors. Clinical software systems and workflows should generate research-ready data by adopting imaging standards and integrating with centralized data repositories. Researchers must also strive to develop models with direct clinical application and provide them in formats such that they can be implemented in clinical workflows and corresponding software platforms. The most direct way to accomplish this bi-directional approach is through collaborations via multi-site consortia consisting of clinicians, pathologists, radiologists, and researchers.
Conclusion
The rise in computational potential has led to machine and deep learning models utilized in a variety of clinical applications. Although pediatric brain tumor research has shown progress in radiomics, there remains significant work to be done with pathomic and radio-pathomic modeling. Along with the inherent limitations in the size of this population, other challenges in radio-pathomic research include molecular, radiological, and histological heterogeneity in pediatrics, as well as image standardization, data organization, computational resources, and pipeline development. At the same time, integrating these two imaging data types has considerable potential for rich characterizations of disease across spatial scales, by combining localized features of a tumor’s cellular and architectural properties with global statistical and morphological properties. Such multi-modal characterizations provide similar potential for better prediction of molecular and clinical outcome factors compared to any one data type alone. Additionally, such developments are necessary to accomplish tailored precision medicine and an understanding of individual variability compared to larger populations.
Contributor Information
Ariana M Familiar, Center for Data-Driven Discovery in Biomedicine, Children’s Hospital of Philadelphia, Philadelphia, PA, USA.
Aria Mahtabfar, Center for Data-Driven Discovery in Biomedicine, Children’s Hospital of Philadelphia, Philadelphia, PA, USA; Department of Neurosurgery, Thomas Jefferson University Hospital, Philadelphia, PA, USA.
Anahita Fathi Kazerooni, Center for Data-Driven Discovery in Biomedicine, Children’s Hospital of Philadelphia, Philadelphia, PA, USA; Department of Neurosurgery, Children’s Hospital of Philadelphia, Philadelphia, PA, USA; Department of Neurosurgery, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA.
Mahsa Kiani, Center for Data-Driven Discovery in Biomedicine, Children’s Hospital of Philadelphia, Philadelphia, PA, USA; Department of Radiology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA.
Arastoo Vossough, Center for Data-Driven Discovery in Biomedicine, Children’s Hospital of Philadelphia, Philadelphia, PA, USA; Department of Radiology, Children’s Hospital of Philadelphia, Philadelphia, PA, USA; Department of Radiology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA.
Angela Viaene, Department of Pathology and Laboratory Medicine, Children’s Hospital of Philadelphia, Philadelphia, PA, USA; Department of Pathology and Laboratory Medicine, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA.
Phillip B Storm, Center for Data-Driven Discovery in Biomedicine, Children’s Hospital of Philadelphia, Philadelphia, PA, USA; Department of Neurosurgery, Children’s Hospital of Philadelphia, Philadelphia, PA, USA; Department of Neurosurgery, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA.
Adam C Resnick, Center for Data-Driven Discovery in Biomedicine, Children’s Hospital of Philadelphia, Philadelphia, PA, USA; Department of Neurosurgery, Children’s Hospital of Philadelphia, Philadelphia, PA, USA.
Ali Nabavizadeh, Center for Data-Driven Discovery in Biomedicine, Children’s Hospital of Philadelphia, Philadelphia, PA, USA; Department of Radiology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA.
Funding
NIH NCI Childhood Cancer Data Initiative (CCDI) 75N91019D00024.
Conflict of Interest Statement
None declared.
Authorship Statement
Conceptualization: A.M.F., A.M., A.F.K., A.N. Resources: P.B.S., A.C.R., A.N. Writing – original draft preparation: A.M.F., A.M., A.F.K., M.K. Writing – review and editing: A.M.F., A.M., A.F.K., M.K., A.Vo., A.Vi., P.B.S., A.C.R., A.N. All authors have read and agreed to the published version of the manuscript.
References
- 1. Ostrom QT, Patil N, Cioffi G, et al. CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2013-2017. Neuro Oncol. 2020;22(12 Suppl 2):iv1–iv96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bale TA, Rosenblum MK.. The 2021 WHO classification of tumors of the central nervous system: An update on pediatric low-grade gliomas and glioneuronal tumors. Brain Pathol. 2022;32(4):e13060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Louis DN, Perry A, Wesseling P, et al. The 2021 WHO classification of tumors of the central nervous system: A summary. Neuro Oncol. 2021;23(8):1231–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Louis D, Perry A, Burger P, et al. International society of neuropathology--Haarlem consensus guidelines for nervous system tumor classification and grading. Brain Pathol. 2014;24(5):429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gillies RJ, Kinahan PE, Hricak H.. Radiomics: Images are more than pictures, they are data. Radiology. 2016;278(2):563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14(12):749–762. [DOI] [PubMed] [Google Scholar]
- 7. Zwanenburg A, Vallières M, Abdalah M, et al.. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology. 2020;295:328–338. Accessed August 25, 2023. https://pubs.rsna.org/doi/full/10.1148/radiol.2020191145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Madhogarhia R, Haldar D, Bagheri S, et al. Radiomics and radiogenomics in pediatric neuro-oncology: A review. Neuro-Oncol Adv. 2022;4(1):vdac083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li Z, Wang Y, Yu J, Guo Y, Cao W.. Deep Learning based Radiomics (DLR) and its usage in noninvasive IDH1 prediction for low grade glioma. Sci Rep. 2017;7(1):5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kickingereder P, Burth S, Wick A, et al. Radiomic profiling of glioblastoma: Identifying an imaging predictor of patient survival with improved performance over established clinical and radiologic risk models. Radiology. 2016;280(3):880–889. [DOI] [PubMed] [Google Scholar]
- 11. Fathi Kazerooni A, Saxena S, Toorens E, et al. Clinical measures, radiomics, and genomics offer synergistic value in AI-based prediction of overall survival in patients with glioblastoma. Sci Rep. 2022;12(1):8784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gupta R, Kurc T, Sharma A, Almeida JS, Saltz J.. The emergence of pathomics. Curr Pathobiol Rep 2019;7:73–84. [Google Scholar]
- 13. Tomaszewski MR, Gillies RJ.. The biological meaning of radiomic features. Radiology. 2021;298(3):505–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ye Z, Srinivasa K, Meyer A, et al. Diffusion histology imaging differentiates distinct pediatric brain tumor histology. Sci Rep. 2021;11(1):4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Iv M, Zhou M, Shpanskaya K, et al. MR Imaging–based radiomic signatures of distinct molecular subgroups of medulloblastoma. AJNR. 2019;40(1):154–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Valvi S, Hansford JR.. Radiomics-A new age of presurgical assessment to improve outcomes in pediatric neuro-oncology. Neuro Oncol. 2022;24(6):995–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grist JT, Withey S, MacPherson L, et al. Distinguishing between paediatric brain tumour types using multi-parametric magnetic resonance imaging and machine learning: A multi-site study. Neuroimage Clin. 2020;25:102172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fetit AE, Novak J, Rodriguez D, et al. Radiomics in paediatric neuro-oncology: A multicentre study on MRI texture analysis. NMR Biomed. 2018;31(1):e3781. [DOI] [PubMed] [Google Scholar]
- 19. Wagner MW, Hainc N, Khalvati F, et al. Radiomics of pediatric low-grade gliomas: Toward a pretherapeutic differentiation of BRAF-mutated and BRAF-fused tumors. AJNR. 2021;42(4):759–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lu C, Shiradkar R, Liu Z.. Integrating pathomics with radiomics and genomics for cancer prognosis: A brief review. Chin J Cancer Res. 2021;33(5):563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shao L, Liu Z, Feng L, et al. Abstract 2014: Radiopathomics strategy combining multiparametric MRI with whole-slide image for pretreatment prediction of tumor regression grade to neoadjuvant chemoradiotherapy in rectal cancer. Cancer Res. 2020;80(Suppl 16):2014. [Google Scholar]
- 22. Tran WT, Jerzak K, Lu FI, et al. Personalized breast cancer treatments using artificial intelligence in radiomics and pathomics. J Med Imaging Radiat Sci. 2019;50(4 Suppl 2):S32–S41. [DOI] [PubMed] [Google Scholar]
- 23. Attallah O, Zaghlool S.. AI-based pipeline for classifying pediatric medulloblastoma using histopathological and textural images. Life. 2022;12(2):232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bengs M., Bockmayr M., Schüller U., Schlaefer A.. Medulloblastoma tumor classification using deep transfer learning with multi-scale EfficientNets. In: Medical Imaging 2021: Digital Pathology. Vol. 11603: SPIE; 2021:70–75. [Google Scholar]
- 25. Das D, Mahanta LB, Ahmed S, Baishya BK.. Classification of childhood medulloblastoma into WHO-defined multiple subtypes based on textural analysis. J Microsc. 2020;279(1):26–38. [DOI] [PubMed] [Google Scholar]
- 26. Whitney J, Dollinger L, Tamrazi B, et al. Quantitative nuclear histomorphometry predicts molecular subtype and clinical outcome in medulloblastomas: Preliminary findings. J Pathol Inform. 2022;13:100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Das D, Mahanta LB, Ahmed S, Baishya BK, Haque I.. Automated classification of childhood brain tumours based on texture feature. Songklanakarin J Sci Technol. 2019;41(5):1014–1020. [Google Scholar]
- 28. Chen RJ, Lu MY, Wang J, et al. Pathomic fusion: An integrated framework for fusing histopathology and genomic features for cancer diagnosis and prognosis. IEEE Transac Med Imaging. 41(4), 757–770. doi: 10.48550/arXiv.1912.08937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rathore S, Chaddad A, Iftikhar MA, Bilello M, Abdulkadir A.. Combining MRI and histologic imaging features for predicting overall survival in patients with glioma. Radiol Imaging Cancer. 2021;3(4):e200108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Braman N, Gordon JWH, GoossensET, Willis C, Stumpe MC, Venkataraman J.. Deep orthogonal fusion: Multimodal prognostic biomarker discovery integrating radiology, pathology, genomic, and clinical data. In: de Bruijne M, Cattin PC, Cotin S, et al. , eds. Medical Image Computing and Computer Assisted Intervention – MICCAI 2021. Lecture Notes in Computer Science: Springer International Publishing; 2021:667–677. [Google Scholar]
- 31. Feng L, Liu Z, Li C, et al. Development and validation of a radiopathomics model to predict pathological complete response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer: A multicentre observational study. Lancet Digit Health. 2022;4(1):e8–e17. [DOI] [PubMed] [Google Scholar]
- 32. McGarry SD, Bukowy JD, Iczkowski KA, et al. Radio-pathomic mapping model generated using annotations from five pathologists reliably distinguishes high-grade prostate cancer. J Med Imaging. 2020;7(5):054501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shaari H, Kevrić J, Jukić S, et al. Deep learning-based studies on pediatric brain tumors imaging: Narrative review of techniques and challenges. Brain Sci. 2021;11(6):716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Johnson DR, Brown PD, Galanis E, Hammack JE.. Pilocytic astrocytoma survival in adults: Analysis of the surveillance, epidemiology, and end results program of the National Cancer Institute. J Neurooncol. 2012;108(1):187–193. [DOI] [PubMed] [Google Scholar]
- 35. The Cancer Genome Atlas Research Network. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. NEJM. 2015;372(26):2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ryall S, Tabori U, Hawkins C.. Pediatric low-grade glioma in the era of molecular diagnostics. Acta Neuropathol Commun. 2020;8(1):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rallis KS, George AM, Wozniak AM, et al. Molecular genetics and targeted therapies for paediatric high-grade glioma. Cancer Genomics Proteomics. 2022;19(4):390–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Roux A, Pallud J, Saffroy R, et al. High-grade gliomas in adolescents and young adults highlight histomolecular differences from their adult and pediatric counterparts. Neuro-Oncol. 2020;22(8):1190–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pajtler KW, Witt H, Sill M, et al. Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell. 2015;27(5):728–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Santi M, Viaene AN, Hawkins C.. Ependymal tumors. Pediatr Dev Pathol. 2022;25(1):59–67. [DOI] [PubMed] [Google Scholar]
- 41. Cotter JA, Hawkins C.. Medulloblastoma: WHO 2021 and beyond. Pediatr Dev Pathol. 2022;25(1):23–33. [DOI] [PubMed] [Google Scholar]
- 42. Zhao F, Ohgaki H, Xu L, et al. Molecular subgroups of adult medulloblastoma: A long-term single-institution study. Neuro-Oncol. 2016;18(7):982–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Khatua S, Song A, Sridhar DC, Mack SC.. Childhood medulloblastoma: Current therapies, emerging molecular landscape and newer therapeutic insights. Curr Neuropharmacol. 2018;16(7):1045–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Remke M, Hielscher T, Northcott PA, et al. Adult medulloblastoma comprises three major molecular variants. JCO. 2011;29(19):2717–2723. [DOI] [PubMed] [Google Scholar]
- 45. Fangusaro J, Witt O, Hernáiz Driever P, et al. Response assessment in paediatric low-grade glioma: Recommendations from the Response Assessment in Pediatric Neuro-Oncology (RAPNO) working group. Lancet Oncol. 2020;21(6):e305–e316. [DOI] [PubMed] [Google Scholar]
- 46. Erker C, Tamrazi B, Poussaint TY, et al. Response assessment in paediatric high-grade glioma: Recommendations from the Response Assessment in Pediatric Neuro-Oncology (RAPNO) working group. Lancet Oncol. 2020;21(6):e317–e329. [DOI] [PubMed] [Google Scholar]
- 47. Cooney TM, Cohen KJ, Guimaraes CV, et al. Response assessment in diffuse intrinsic pontine glioma: Recommendations from the Response Assessment in Pediatric Neuro-Oncology (RAPNO) working group. Lancet Oncol. 2020;21(6):e330–e336. [DOI] [PubMed] [Google Scholar]
- 48. Lindsay HB, Massimino M, Avula S, et al. Response assessment in paediatric intracranial ependymoma: Recommendations from the Response Assessment in Pediatric Neuro-Oncology (RAPNO) working group. Lancet Oncol. 2022;23(8):e393–e401. [DOI] [PubMed] [Google Scholar]
- 49. Hoffman LM, Jaimes C, Mankad K, et al. Response assessment in pediatric craniopharyngioma: Recommendations from the Response Assessment in Pediatric Neuro-Oncology (RAPNO) Working Group. Neuro-Oncol. 2023;25(2):224–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Warren KE, Vezina G, Poussaint TY, et al. Response assessment in medulloblastoma and leptomeningeal seeding tumors: Recommendations from the Response Assessment in Pediatric Neuro-Oncology committee. Neuro-Oncol. 2018;20(1):13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ghaffari M, Sowmya A, Oliver R.. Automated brain tumor segmentation using multimodal brain scans: A survey based on models submitted to the BraTS 2012–2018 challenges. IEEE Rev Biomed Eng. 2020;13:156–168. [DOI] [PubMed] [Google Scholar]
- 52. Fathi Kazerooni A, Arif S, Madhogarhia R, et al. Automated tumor segmentation and brain tissue extraction from multiparametric MRI of pediatric brain tumors: A multi-institutional study. Neuro-Oncol Adv. 2023;5(1):vdad027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Camajori Tedeschini B, Savazzi S, Stoklasa R, et al. Decentralized federated learning for healthcare networks: A case study on tumor segmentation. IEEE Access. 2022;10:8693–8708. [Google Scholar]
- 54. Haghighat M, Browning L, Sirinukunwattana K, et al.. Automated quality assessment of retrospective histopathology whole-slide image cohorts by artificial intelligence. Sci Rep. 2022;12:5002. Published online September 27, 2021:2021.09.24.21263762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Janowczyk A, Zuo R, Gilmore H, Feldman M, Madabhushi A.. HistoQC: An open-source quality control tool for digital pathology slides. JCO Clinic Cancer Informatics. 2019;3:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zarella MD, Rivera Alvarez K.. High‐throughput whole‐slide scanning to enable large‐scale data repository building. J Pathol. 2022;257(4):383–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bancroft JD, Gamble M.. Theory and Practice of Histological Techniques. Amsterdam, The Netherlands: Elsevier Health Sciences; 2008. [Google Scholar]
- 58. Azevedo Tosta TA, de Faria PR, Neves LA, do Nascimento MZ.. Computational normalization of H&E-stained histological images: Progress, challenges and future potential. Artif Intell Med. 2019;95:118–132. [DOI] [PubMed] [Google Scholar]
- 59. Zanjani FG, Zinger S, Bejnordi BE, van der Laak JA.. With PHN de. Histopathology Stain-Color Normalization Using Deep Generative Models. In; 2022. https://openreview.net/forum?id=SkjdxkhoG
- 60. Tellez D, Litjens G, Bándi P, et al. Quantifying the effects of data augmentation and stain color normalization in convolutional neural networks for computational pathology. Med Image Anal. 2019;58:101544. [DOI] [PubMed] [Google Scholar]
- 61. Alvi E, Gupta R, Borok RZ, Escobar-Hoyos L, Shroyer KR.. Overview of established and emerging immunohistochemical biomarkers and their role in correlative studies in MRI. J Magn Reson Imaging. 2020;51(2):341–354. [DOI] [PubMed] [Google Scholar]
- 62. Hickey JW, Neumann EK, Radtke AJ, et al. Spatial mapping of protein composition and tissue organization: A primer for multiplexed antibody-based imaging. Nat Methods. 2022;19(3):284–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mund A, Coscia F, Kriston A, et al. Deep visual proteomics defines single-cell identity and heterogeneity. Nat Biotechnol. 2022;40(8):1231–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Manafi-Farid R, Ataeinia B, Ranjbar S, et al. ImmunoPET: Antibody-based PET imaging in solid tumors. Front Med. 2022;9:916693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lilly JV, Rokita JL, Mason JL, et al. The children’s brain tumor network (CBTN) - Accelerating research in pediatric central nervous system tumors through collaboration and open science. Neoplasia. 2023;35:100846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Das D, Mahanta LB.. Childhood Medulloblastoma Microscopic Images. IEEE Dataport. Published online August 25, 2020. [Google Scholar]
- 67. Lajara N, Espinosa-Aranda JL, Deniz O, Bueno G.. Optimum web viewer application for DICOM whole slide image visualization in anatomical pathology. Comput Methods Programs Biomed. 2019;179:104983. [DOI] [PubMed] [Google Scholar]
- 68. Bankhead P, Loughrey MB, Fernández JA, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep. 2017;7(1):16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Albayrak A, Akhan AU, Calik N, et al. A whole-slide image grading benchmark and tissue classification for cervical cancer precursor lesions with inter-observer variability. Med Biol Eng Comput. 2021;59(7):1545–1561. [DOI] [PubMed] [Google Scholar]
- 70. Kang C, Lee C, Song H, Ma M, Pereira S.. Variability matters: Evaluating inter-rater variability in histopathology for robust cell detection. In: Karlinsky L, Michaeli T, Nishino K, eds. Computer Vision – ECCV 2022 Workshops. Lecture Notes in Computer Science. Springer Nature Switzerland; 2023:552–565. [Google Scholar]
- 71. Pfister S, Hartmann C, Korshunov A.. Histology and molecular pathology of pediatric brain tumors. J Child Neurol. 2009;24(11):1375–1386. [DOI] [PubMed] [Google Scholar]
- 72. van den Bent MJ. Interobserver variation of the histopathological diagnosis in clinical trials on glioma: A clinician’s perspective. Acta Neuropathol. 2010;120(3):297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kurc T, Bakas S, Ren X, et al. Segmentation and classification in digital pathology for glioma research: Challenges and deep learning approaches. Front Neurosci. 2020;14:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Barker J, Hoogi A, Depeursinge A, Rubin DL.. Automated classification of brain tumor type in whole-slide digital pathology images using local representative tiles. Med Image Anal. 2016;30:60–71. [DOI] [PubMed] [Google Scholar]
- 75. Kong J, Zhang P, Liang Y, et al. Robust cell segmentation for histological images of glioblastoma. Proc IEEE Int Symp Biomed Imaging. 2016;2016:1041–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Baldi D, Aiello M, Duggento A, Salvatore M, Cavaliere C.. MR imaging-histology correlation by tailored 3D-printed slicer in oncological assessment. Contrast Media Mol Imaging. 2019;2019:1071453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Matsumae M, Nishiyama J, Kuroda K.. Intraoperative MR imaging during glioma resection. Magn Reson Med Sci. 2022;21(1):148–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Roodakker KR, Alhuseinalkhudhur A, Al-Jaff M, et al. Region-by-region analysis of PET, MRI, and histology in en bloc-resected oligodendrogliomas reveals intra-tumoral heterogeneity. Eur J Nucl Med Mol Imaging. 2019;46(3):569–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bobholz SA, Lowman AK, Brehler M, et al. Radio-pathomic maps of cell density identify brain tumor invasion beyond traditional MRI-defined margins. AJNR. 2022;43(5):682–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Coy S, Wang S, Stopka SA, et al. Single cell spatial analysis reveals the topology of immunomodulatory purinergic signaling in glioblastoma. Nat Commun. 2022;13:4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Shekarian T, Zinner CP, Bartoszek EM, et al. Immunotherapy of glioblastoma explants induces interferon-γ responses and spatial immune cell rearrangements in tumor center, but not periphery. Sci Adv. 2022;8(26):eabn9440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Sharma A, Tarbox L, Kurc T, et al. PRISM: A platform for imaging in precision medicine. JCO Clin Cancer Inform. 2020;4:491–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Graham S, Vu QD, Raza SEA, et al. Hover-Net: Simultaneous segmentation and classification of nuclei in multi-tissue histology images. Med Image Anal. 2019;58:101563. [DOI] [PubMed] [Google Scholar]
- 84. Wang X, Yang S, Zhang J, et al. TransPath: Transformer-based self-supervised learning for histopathological image classification. In: de Bruijne M, Cattin PC, Cotin S, et al. , eds. Medical Image Computing and Computer Assisted Intervention – MICCAI 2021. Springer International Publishing; 2021:186–195. [Google Scholar]
- 85. Deng J, Dong W, Socher R, Li LJ, Li K, Fei-FeiL.. ImageNet: A large-scale hierarchical image database. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition; 2009:248–255. [Google Scholar]
- 86. Tizhoosh HR, Diamandis P, Campbell CJV, et al. Searching images for consensus: Can AI remove observer variability in pathology? Am J Pathol. 2021;191(10):1702–1708. [DOI] [PubMed] [Google Scholar]
- 87. Rony J, Belharbi S, Dolz J, et al. Deep weakly-supervised learning methods for classification and localization in histology images: A survey. Melba. 2023;2(March 2023):96–150. [Google Scholar]
- 88. Lu MY, Williamson DFK, Chen TY, et al. Data-efficient and weakly supervised computational pathology on whole-slide images. Nat Biomed Eng. 2021;5(6):555–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Campanella G, Hanna MG, Geneslaw L, et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med. 2019;25(8):1301–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ghaffari Laleh N, Muti HS, Loeffler CML, et al. Benchmarking weakly-supervised deep learning pipelines for whole slide classification in computational pathology. Med Image Anal. 2022;79:102474. [DOI] [PubMed] [Google Scholar]
- 91. Srinidhi CL, Kim SW, Chen FD, Martel AL.. Self-supervised driven consistency training for annotation efficient histopathology image analysis. Med Image Anal. 2022;75:102256. [DOI] [PubMed] [Google Scholar]
- 92. Anand D, Yashashwi K, Kumar N, et al. Weakly supervised learning on unannotated H&E-stained slides predicts BRAF mutation in thyroid cancer with high accuracy. J Pathol. 2021;255(3):232–242. [DOI] [PubMed] [Google Scholar]
- 93. Silva-Rodríguez J, Colomer A, Naranjo V.. WeGleNet: A weakly-supervised convolutional neural network for the semantic segmentation of Gleason grades in prostate histology images. Comput Med Imaging Graph. 2021;88:101846. [DOI] [PubMed] [Google Scholar]
- 94. Farahani K, Kurc T, Bakas S, et al. Computational Precision Medicine Radiology-Pathology Challenge on Brain Tumor Classification 2020. In: 23rd International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2020). Zenodo; 2020;2020:3718894. Published 2020. 10.5281/zenodo.3718894 [DOI] [Google Scholar]
- 95. Hsu WW, Guo JM, Pei L, et al. A weakly supervised deep learning-based method for glioma subtype classification using WSI and mpMRIs. Sci Rep. 2022;12(1):6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Schirris Y, Gavves E, Nederlof I, Horlings HM, Teuwen J.. DeepSMILE: Contrastive self-supervised pre-training benefits MSI and HRD classification directly from H&E whole-slide images in colorectal and breast cancer. Med Image Anal. 2022;79:102464. [DOI] [PubMed] [Google Scholar]
- 97. Guo W, Wang J, Wang S.. Deep multimodal representation learning: A survey. IEEE Access. 2019;7:63373–63394. [Google Scholar]
- 98. Huang SC, Shen L, Lungren MP, Yeung S.. GLoRIA: A multimodal global-local representation learning framework for label-efficient medical image recognition. In: 2021 IEEE/CVF International Conference on Computer Vision (ICCV). IEEE; 2021:3922–3931. [Google Scholar]
- 99. Cronin KA, Scott S, Firth AU, et al. Annual report to the nation on the status of cancer, part 1: National cancer statistics. Cancer. 2022;128(24):4251–4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Norris RE, Adamson PC.. Challenges and opportunities in childhood cancer drug development. Nat Rev Cancer. 2012;12(11):776–782. [DOI] [PubMed] [Google Scholar]
- 101. Upadhyaya SA, Robinson GW, Onar-Thomas A, et al. Relevance of molecular groups in children with newly diagnosed atypical teratoid rhabdoid tumor: Results from prospective St. Jude multi-institutional trials. Clin Cancer Res. 2021;27(10):2879–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Yao B, Delaidelli A, Vogel H, Sorensen PH.. Pediatric brain tumours: Lessons from the immune microenvironment. Current Oncol. 2023;30(5):5024–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Saltz J, Gupta R, Hou L, et al. ; Cancer Genome Atlas Research Network. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 2018;23(1):181–193.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Nabbi A, Beck P, Delaidelli A, et al. Transcriptional immunogenomic analysis reveals distinct immunological clusters in pediatric nervous system tumours. Genome Med. 2023;15:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Vinci M, Burford A, Molinari V, et al. Functional diversity and cooperativity between subclonal populations of pediatric glioblastoma and diffuse intrinsic pontine glioma cells. Nat Med. 2018;24(8):1204–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Liu SJ, Magill ST, Vasudevan HN, et al. Multiplatform molecular profiling reveals epigenomic intratumor heterogeneity in ependymoma. Cell Rep. 2020;30(5):1300–1309.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]