Abstract
3D patient tumor avatars (3D-PTAs) hold promise for next-generation precision medicine. Here we describe the benefits and challenges of 3D-PTA technologies and necessary future steps to realize their potential for clinical decision-making. 3D-PTAs require standardization criteria and prospective trials to establish clinical benefits. Innovative trial designs combining omics and 3D-PTA readouts may lead to more accurate clinical predictors, and an integrated platform combining diagnostic and therapeutic development will accelerate new treatments for patients with refractory disease.
Precision medicine, as defined in modern oncology, has focused on the development of therapies that target specific genetic alterations in cancer. Imatinib (Gleevec) for leukemias with BCL-ABL mutations, Trastuzumab (Herceptin) for HER2-overexpressing cancers, and others were promising early demonstrations of this vision. In 2006, the National Institutes of Health (NIH) launched The Cancer Genome Atlas, a landmark cancer genomics program that sequenced over 11,000 primary cancer samples. The precision medicine approach was simple: sequence a patient’s tumor, identify driver mutations, and administer therapies to target those mutations. With tumors dependent on the targeted oncogenes, early successes bolstered collaborations between pharmaceutical companies and academic research for rapid drug development.
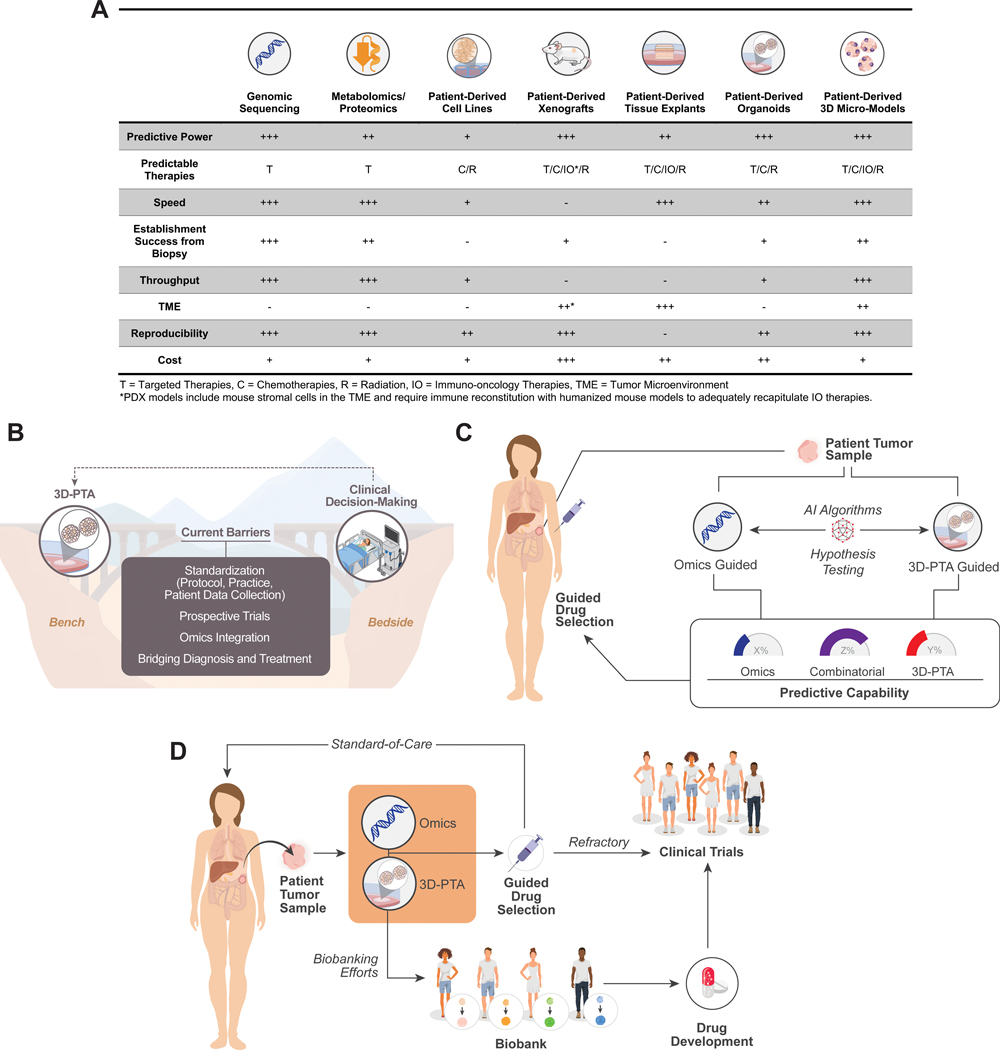
With only a small subset of targetable mutations, a minority of cancer patients are thought to actually benefit from genome-guided therapies1. Given our growing appreciation for other drivers of neoplastic behaviors (metabolic, phenotypic, epigenetic, and microenvironmental) and tumor evolution during treatment, the need for an approach that integrates more biology into therapeutic decision-making is evident. With the support of key national funding entities, including the NIH, biorepositories of patient-derived models have been developed; however, their application in precision medicine has been largely dependent on their ability to adequately recapitulate the clinical response of patient tumors in a laboratory setting. The advantages and limitations of each of these models have been extensively reviewed (Figure 1A).
Figure 1:
3D-PTAs as Precision Oncology Tools. (A) Comparison of advantages and disadvantages of current patient tumor models. (B) 3D-PTAs face key barriers to for widespread adoption in clinical decision-making. (C) Paradigm to understand benefits of combinatorial or omics- vs. 3D-PTA-guided drug selection. (D) 3D-PTAs can be evaluated as functional complements to molecular tumor characterization in clinical trials and as a powerful tool for understanding tumor biology for further drug development.
While a mainstay of cancer research over several decades, 2D cell cultures are not ideal models of tumors, often representing only the most rapid-growing cells in a plate rather than the diversity of the neoplastic growth2. Patient-derived xenograft (PDX) models, or murine models of cancer using patient tumor tissue engrafted into immune-compromised mice, have been a critical component of translational modeling. While valuable in their ability to capture genetic diversity and other features of patient tumor physiology, serial PDX modeling through patient treatment and tumor evolution is often not feasible due to substantial cost and time requirements of PDXs. Additionally, the murine stromal tumor microenvironment (TME) and need for immune reconstitution in PDXs makes immuno-oncology studies challenging, thus obligating the further development of other preclinical models. Ex vivo explants, or cultures of whole tissue, capture the 3D architecture and cellular organization of tumor samples, although the amount of required tissue, low throughput, and difficulty in reproducibility limit their scalability for clinical implementation.
In recent years, 3D-PTAs have rapidly emerged as a new model system to explore tumor behaviors. These models, ranging from patient-derived organoids (PDOs) to microscale models like organotypic tumor spheroids (PDOTs), 3D bioprinting, organoids-on-a-chip, and micro-organospheres (MOS), can model cellular behaviors while capturing characteristics true to the source tissue3–7. Several landmark studies demonstrated that PDOs could predict patient tumor response to chemotherapy and radiation8–10. While PDX models often require 6–8 months for development and expansion, PDOs can reduce this time to weeks with higher throughput11. More recently, microscale 3D-PTA technologies leveraging microfabrication or microfluidics have achieved substantially faster establishment and higher throughput.
Although promising, these efforts on 3D-PTAs will require standardization and concerted buy-in from regulatory bodies, clinicians, researchers, and patients to bridge the gap between bench and bedside, and key recommendations to achieve this goal are discussed here (Figure 1B).
Standardizing Practices and Protocols
While the expertise of select groups has demonstrated the viability of 3D-PTAs as tools for modeling cancer, the variability in their creation remains a hurdle for reproducibility and clinical adoption. The skill of the operator remains an important driver of success in establishing both PDOs and ex vivo explants, with the most successful biobanking efforts reporting establishment rates of 70–95% which can decrease in other settings8, 12. Defining and standardizing precise methods to validate whether 3D-PTAs adequately capture the significant inter- and intratumoral heterogeneity of source tissue will also be crucial to deriving insights from this data.
Defining the extracellular matrix (ECM) scaffolds, media and cocktails of growth factors best suited for different types of 3D-PTAs has been a key effort of many groups, including the NCI-sponsored Patient-Derived Models of Cancer (PDMC) Consortium; however, standard precise culture methods remain elusive and different cancer types will likely require more specific tailoring. Derived from mouse sarcoma, Matrigel tends to have batch-to-batch variability, while the effects of alternative scaffolds such as synthetic gel on 3D-PTA establishment and drug response have not been well characterized. Variations in adding common factors, whether fetal bovine serum (FBS) or antibiotics (penicillin, streptomycin, primocin, etc.) into culture media with supraphysiologic glucose is relatively commonplace. However, the consequent metabolic effects of these combinations are poorly understood but likely of importance. In 3D-PTAs, the need to harvest tissue from donors can also lead to variations in the time and amount of tissue without perfusion, leading to warm vs. cold-ischemic changes that are difficult to characterize. Finally, the post-processing of these tissues – whether using clean-up procedures to remove necrotic tissue, applying specialized media to isolate different immune, stromal, or tumoral components, or mycoplasma surveillance methodologies – adds another layer of variability between studies. As individual labs often optimize growth factor concentrations based on their own experience, results are sometimes hard to compare across publications because of the variabilities discussed above12–14. As standardized protocols are developed for 3D-PTAs, these practices must be codified and shared across research groups.
Following the establishment and experimentation of 3D-PTAs, quantifying results with validated software pipelines is essential to establishing their reproducibility and functional readouts as well15. Further standardization and automation of these processes—handling of source material, culture conditions, validated reproduction of source characteristics, and measurements of endpoints like proliferation and survival—will enhance reproducibility. Together with independent replication studies, these factors will form an important foundation for using 3D-PTAs as companion support tools in research and clinical decision-making.
Standardizing Patient Data Collection
As the number and complexity of tumoral samples collected for large scale 3D-PTA biobanks expand, the clinical factors that are captured should be well-documented to ensure that analytical approaches can draw meaningful conclusions. Specifically, clinical parameters of race and ethnicity, body-mass index, and socio-economic factors have come to be recognized as crucial to overall outcomes for patients but are not routinely available for large-scale biobanks. Other metadata to include in studies should include sex, age, sample collection dates, sample processing, cancer types and full history (initial presentation, metastases, stage/grade, treatment lines), family history, and tobacco exposure. While these parameters have been reported in studies when relevant to the outcomes of interest, capturing that heterogeneity as a dimension of 3D-PTA libraries will enable a more holistic understanding of the predictive capabilities of these functional models.
These requirements for patient data collection need to be established and supported by overseeing bodies–including the NIH, institutions, and journals which publish these studies. Key to achieving this will be to adopt common regulatory frameworks for data exchange and access at a global scale. Building on the example of the PDX Minimal Information Standard (PDX-MI), standards around the clinical information, patient metadata collection, and patient informed consent for 3D-PTAs studies should be developed in the coming years, particularly as we find ourselves at the beginning of this confluence of big data and biorepository development.
Innovating Clinical Trial Designs
While is a well-established framework for genomic testing in accredited laboratories exists, the path for validation of 3D-PTAs for clinical decision-making is yet to be established. As with genome-guided therapy, 3D-PTA-guided therapies must undergo rigorous prospective clinical trials to demonstrate clinical benefits for clinical adoption, regulatory approval, and eventually, payer reimbursements. Currently, many clinical trials are seeking to assess the potential of 3D-PTAs to advance the management of patients with various tumor types in different settings. These clinical studies include observational (non-interventional) “co-clinical trials” aiming to evaluate the feasibility of deriving 3D-PTA-based assays from tissue biopsies in a turnaround time compatible with clinical workflows, as demonstrated by a recent clinical study using the MOS technology6. As a next step, the potential of this platforms for predicting response rates and progression-free survival must be shown with either a prospective validation trial, where clinicians are informed of the 3D-PTA assay prediction when choosing between equipoised approved or investigational drugs, or in the setting of a randomized pilot study with an uninformed arm where 3D-PTA is not performed, and patients are managed as per available best practice. Quantitative measurements of immune, stromal, and tumor cell types over time could help define the temporal fidelity of 3D-PTAs and their utility in modeling tumor heterogeneity and evolution. Ultimately, results from these validation studies will buoy the efforts of early adopters, while providing the basis for further clinical utility trials. These studies, with larger cohort sizes, randomized arms, and clinically-meaningful endpoints of progression-free survival or overall survival, will be the foundation for adoption by the broader community and National Comprehensive Cancer Network (NCCN) guidelines.
3D-PTA guided prospective trials can further leverage window-of-opportunity trial designs, which test experimental drugs before standard-of-care (SOC) regimens. Upon biopsy, 3D-PTA assays can be performed to test both experimental drugs and equipoised SOC regimens. During the short period of time prior to SOC treatment, typically ~10–20 days, patients can be treated with experimental therapies and evaluated by a second biopsy and imaging (e.g., PET) or other assays (e.g., circulating tumor DNA) to assess drug on-target activity and early response as well as adaptive resistance programs. These window-of-opportunity trials allow 3D-PTA readouts to be correlated with experimental drug responses while guiding SOC decision-making as well, thus providing valuable insights into the biological effects and mechanisms of action that accelerate development of new drugs and combination regimens. Further, 3D-PTAs may be used to evaluate the sensitivity of non-targeted chemotherapy-based regimens that lack molecular markers to guide patient care and offer additional treatment options to therapy-refractory patients.
Furthermore, 3D-PTAs can serve as valuable tools to guide patient selection and optimize enrollment for specific experimental therapies. While current clinical trials often provide limited benefit for a majority of enrollees, 3D-PTAs can be used to predict patient response and stratify treatment cohorts accordingly, similar to genome-guided umbrella trial designs. Such precision clinical trials could increase the benefits that enrollees derive from experimental therapies—enhancing patient survival, improving quality-of-life, encouraging accruement, reducing trial risk, and utilizing precious clinical resources more efficiently.
Lastly, where current trials often fail to capture racial and socioeconomic diversity adequately, 3D-PTA may provide a more personalized platform to address such disparities and benefit minority and disadvantaged populations by treating them as unique individuals rather than relying on statistics from unrepresentative populations.
Omics- vs. 3D-PTA-Guided Therapeutics
Advances in genome- and function-based assays have occurred largely in parallel; however, the burgeoning confluence of the two has already begun to yield important insights – recapitulating associations with genetic mutations and targeted therapeutic sensitivities and pinpointing mutations which may be of interest for future investigation. A major question remains yet unanswered: can patient response be more accurately predicted by one or both in combination?
As a patient can only receive one treatment at a time, 3D-PTAs offer the opportunity to perform high-throughput screen with library of drugs in parallel and drug combinations, while also guiding lower-throughput in vivo studies. When combined with molecular profiling, these functional models may provide much-needed training datasets to improve the performance of current omics-based predictors. Ultimately, as expanding clinical trials and growing repositories offer increasing statistical power, the combination of the molecular-guided and functional 3D-PTA guided therapies will likely outperform either alone while also providing the most predictive capabilities for future precision medicine efforts (Figure 1C). In this functional precision medicine approach, larger volumes of biopsied tissue can be both molecularly profiled and used to establish PTAs for functional drug response assays. A computational predictor trained using both omics and PTA readouts will be used to predict patient response, which can be gauged against patient endpoints. This integrative approach may be particularly impactful in evaluating clinical approaches to overcome resistance to specific therapies.
In addition to the clinical impact of these predictive computational models, the development of analytical pipelines which can integrate the metabolic, (phospho)proteomic, immune, morphological, and genetic data gathered from libraries of 3D-PTAs may also yield insights into previously unknown associations. Thus, further work on using 3D-PTAs and omics analyses in combination with well-defined computational pipelines to analyze them may better define the precise features of 3D-PTAs that contribute to their predictive value15. To this end, federal funding agencies should leverage relevant consortia such as NCI PDMC, PDXnet, TEC, and CSBC to systematically compare omics-based biomarkers and 3D-PTA drug responses to quantify the predictive power of each alone and in combination. European academic research infrastructures dedicated to the development of preclinical models (i.e., INFRAFRONTIER, EuroPDX) are currently assessing the clinical value of pan-cancer 3D-PTA platforms for advancing precision oncology efforts, both at an academic and translational level. The European initiative, HCA-Organoid, is leveraging single-cell technologies in PDOs to enable therapeutic advances. Ultimately, an integrated approach will create synergy between these global scientific communities, improving omics and 3D-PTA models as the repositories continue to evolve with growing clinical specimens and data.
Integrating Diagnostic and Therapeutic Development
3D-PTAs pose the unique opportunity to perform de novo testing of new, experimental drugs or off-label drugs that lack existing clinical data, an impossible undertaking for clinical -omics predictors that require patient response data to be trained on. The importance of this capability for both drug testing and development is manifold. For one, next-generation precision medicine must be able to identify new treatments for patients who are refractory to existing SOC. As new drugs are increasingly more specific and targeting smaller patient populations, pharmaceutical companies are facing the challenge of finding and accruing the right patients for clinical trials, thus increasing the cost, time, and risk of new drug development. 3D-PTAs offer an opportunity to bring new drugs to market in a safe, expedited manner with preliminary clinical trials carried out using patient surrogates that can increase the likelihood of downstream success. Similarly, 3D-PTAs offer a unique opportunity to expand drug repurposing strategies that may provide solutions to unmet clinical needs.
The next generation of precision medicine must incorporate the heterogeneity of the patient population into every step of the diagnosis-treatment cascade. While current diagnostic and therapeutic developments are largely siloed, 3D-PTA functional precision medicine can serve as an integrated workflow to enhance cancer care and expedite clinical development of new drugs (Figure 1D). The clinical biomarkers captured by the omics profiling combined with the functional readout from the 3D-PTA can guide the patient to either SOC or new therapeutics in clinical trials. Compared to conventional clinical trial accrual, functional precision medicine may represent a more effective approach in selecting appropriate therapies, thereby improving quality and length of life while reducing clinical resource wastage and unnecessary toxicity from ineffective therapies.
Furthermore, the 3D-PTAs from the diagnostic assays can be further passaged and preserved to form 3D-PTA biobanks with diverse clinical response data that capture patient heterogeneity. These are already becoming invaluable resources for pre-clinical drug and biomarker discovery as well as AI-based learning algorithms, which can then aid development of both new diagnostics and therapies. Finally, aggregating the data derived from these studies and making it accessible to the broader scientific community via resources like PDCM Finder (www.cancermodels.org) will maximize the utility of 3D-PTAs for precision oncology.
Conclusion
3D-PTAs hold tremendous promise for next-generation precision medicine, with support from initiatives around the globe. However, for these technologies to fulfill their envisioned goals as clinical decision-making tools, further work is necessary to build on what the scientific communities have accomplished so far. The remaining challenges to incorporation of 3D-PTAs include standardization of techniques and patient metadata collection, analytical tools, and the development of new clinical trial designs, all of which require a concerted community-wide effort guided by the best practices and standards proposed here. Physician scientists, or clinical key opinion leaders, from around the world will set the standards to develop and ensure 3D-PTAs can be incorporated into clinical practice and drive patient care that is truly personalized.
Acknowledgements
We thank the NCI 3D Workshop, Patient-Derived Model of Cancer (PDMC) Program, and PDXNet for constructive discussions, recommendations, and purposeful efforts to advance the practice of precision oncology.
S.B., M.B., H.M., Z.P., C.A.M., K.M.N., M.F.P., R.C.S. and J.H.C. have no competing interests to declare. S.E.K. is an advisor to Xilis, Inc. F.M.B. declares consulting/advisory fees from AbbVie, Aduro BioTech Inc., Alkermes, AstraZeneca, Daiichi Sankyo Co. Ltd., DebioPharm, eFFECTOR Therapeutics, F. Hoffman-La Roche Ltd., Genentech Inc., Harbinger Health, IBM Watson, Infinity Pharmaceuticals, Jackson Laboratory, Kolon Life Science, Lengo Therapeutics, OrigiMed, PACT Pharma, Parexel International, Pfizer Inc., Protai Bio Ltd, Samsung Bioepis, Seattle Genetics Inc., Tallac Therapeutics, Tyra Biosciences, Xencor, Zymeworks, Black Diamond, Biovica, Eisai, FogPharma, Immunomedics, Inflection Biosciences, Karyopharm Therapeutics, Loxo Oncology, Mersana Therapeutics, OnCusp Therapeutics, Puma Biotechnology Inc., Seattle Genetics, Sanofi, Silverback Therapeutics, Spectrum Pharmaceuticals, Zentalis; sponsored research to her institution from Aileron Therapeutics, Inc. AstraZeneca, Bayer Healthcare Pharmaceutical, Calithera Biosciences Inc., Curis Inc., CytomX Therapeutics Inc., Daiichi Sankyo Co. Ltd., Debiopharm International, eFFECTOR Therapeutics, Genentech Inc., Guardant Health Inc., Klus Pharma, Takeda Pharmaceutical, Novartis, Puma Biotechnology Inc., Taiho Pharmaceutical Co. and honoraria for a speaking engagement from Chugai Biopharmaceuticals. S.K. is a consultant for FGH BioTech and has received research funding from FGH BioTech and Systems Oncology. K.A.P. is co-inventor on a patent for EGFRT790M mutation testing issued, licensed, and with royalties paid from Molecular Diagnostics/Memorial Sloan Kettering Cancer Center. She reports research funding to the institution from AstraZeneca; Roche/Genentech, Boehringer Ingelheim, and D2G Oncology; and consulting for AstraZeneca and Jannssen. M.G.C. reports grants from NeoImmuneTech, as well as other support from Orbus Therapeutics, Incyte, Merck, and UpToDate outside the submitted work; in addition, M.G.C. has a patent for Zika virus strains for the treatment of glioblastoma pending. E.E.’s full disclosures are given here: www.bit.ly/3xuWMer. J.T. reports personal financial interest in form of scientific consultancy role for Array Biopharma, AstraZeneca, Bayer, Boehringer Ingelheim, Chugai, Daiichi Sankyo, F. Hoffmann-La Roche Ltd, Genentech Inc, HalioDX SAS, Hutchison MediPharma International, Ikena Oncology, Inspirna Inc, IQVIA, Lilly, Menarini, Merck Serono, Merus, MSD, Mirati, Neophore, Novartis, Ona Therapeutics, Orion Biotechnology, Peptomyc, Pfizer, Pierre Fabre, Samsung Bioepis, Sanofi, Scandion Oncology, Scorpion Therapeutics, Seattle Genetics, Servier, Sotio Biotech, Taiho, Tessa Therapeutics and TheraMyc. Stocks: Oniria Therapeutics and also educational collaboration with Imedex/HMP, Medscape Education, MJH Life Sciences, PeerView Institute for Medical Education and Physicians Education Resource (PER). D.A.T. is a member of the Scientific Advisory Board and receives stock options from Leap Therapeutics, Surface Oncology, Cygnal Therapeutics, Mestag Therapeutics and Xilis, Inc. outside the submitted work. D.A.T. is scientific co-founder of Mestag Therapeutics. D.A.T. has received research grant support from Fibrogen, Mestag, and ONO Therapeutics. D.A.T. receives grant funding from the Lustgarten Foundation, the NIH, and the Thompson Foundation. None of this work is related to the publication. No other disclosures were reported. C.D.W. is a part-time consultant for LifeNet Health and receives grant funding from AACR-Novocure and Varian Medical Systems. X.S. and H.C. are cofounders of Xilis, Inc. and inventors on patents related to organoid research and micro-organospheres. X.S. is CEO of Xilis, Inc. H.C.’s full disclosure is given here: www.uu.nl/staff/JCClevers/Additionalfunctions.
Footnotes
Declarations of Interest
M.S., B.J.R., P.R., X.L., B.E.W., A.L.W. do not report any conflicts.
References
- 1.Marquart J, Chen EY & Prasad V. Estimation of the percentage of US patients with cancer who benefit from genome-driven oncology. JAMA oncology 4, 1093–1098 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Letai A Functional precision cancer medicine—moving beyond pure genomics. Nature medicine 23, 1028 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Jenkins RW et al. Ex Vivo Profiling of PD-1 Blockade Using Organotypic Tumor SpheroidsEx Vivo Profiling of Immune Checkpoint Blockade. Cancer discovery 8, 196–215 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sato T et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141, 1762–1772 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Park SE, Georgescu A & Huh D Organoids-on-a-chip. Science 364, 960–965 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding S et al. Patient-derived micro-organospheres enable clinical precision oncology. Cell Stem Cell 29, 905–917. e906 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Langer EM et al. Modeling tumor phenotypes in vitro with three-dimensional bioprinting. Cell reports 26, 608–623. e606 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bose S, Clevers H & Shen X Promises and challenges of organoid-guided precision medicine. Med 2, 1011–1026 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vlachogiannis G et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 359, 920–926 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ganesh K et al. A rectal cancer organoid platform to study individual responses to chemoradiation. Nature medicine 25, 1607–1614 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hidalgo M et al. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer discovery 4, 998–1013 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Tienderen GS et al. Hepatobiliary tumor organoids for personalized medicine: a multicenter view on establishment, limitations, and future directions. Cancer cell 40, 226–230 (2022). [DOI] [PubMed] [Google Scholar]
- 13.Xie AW & Murphy WL Engineered biomaterials to mitigate growth factor cost in cell biomanufacturing. Current Opinion in Biomedical Engineering 10, 1–10 (2019). [Google Scholar]
- 14.Veninga V. & Voest EE Tumor organoids: opportunities and challenges to guide precision medicine. Cancer Cell 39, 1190–1201 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Kong J et al. Network-based machine learning in colorectal and bladder organoid models predicts anti-cancer drug efficacy in patients. Nature communications 11, 1–13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]