Abstract
The prevalence of obesity and overweight is steadily rising, posing a significant global challenge for humanity. The fundamental cause of obesity and overweight lies in the abnormal accumulation of adipose tissue. While numerous regulatory factors related to fat deposition have been identified in previous studies, a considerable number of regulatory mechanisms remain unknown. tRNA-derived small RNAs (tsRNAs), a novel class of non-coding RNAs, have emerged as significant regulators in various biological processes. In this study, we obtained small RNA sequencing data from subcutaneous white adipose tissue and omental white adipose tissue of lean and obese pigs. In addition, we similarly obtained tsRNAs profiles from scapular brown adipose tissue (BAT), inguinal white adipose tissue (iWAT) and epigonadal white adipose tissue (eWAT) of normal mice. Finally, we successfully identified a large number of expressed tsRNAs in each tissue type and identified tsRNAs conserved in different adipose tissues of pigs and mice. These datasets will be a valuable resource for elucidating the epigenetic mechanisms of fat deposition.
Subject terms: Small RNAs, RNA sequencing
Background & Summary
Adipose tissue plays a crucial role in the regulation of the endocrine system and maintaining energy homeostasis in mammals1. The deposition and distribution of adipose tissue are closely linked to obesity-related diseases, including type II diabetes, cardiovascular disease, and metabolic syndrome2,3. Fat deposition is a highly intricate biological process influenced by complex interactions between genetics, epigenetics, and environmental factors, and is accompanied by alterations in cell structure and function4. While researchers have identified numerous key functional genes, such as PPARγ5 and C/EBPα6, involved in the regulation of fat deposition, much remains unknown regarding the epigenetic regulatory mechanisms operating at the pre- and post-transcriptional levels.
In recent years, a growing body of research has highlighted the crucial regulatory role of non-coding RNAs (ncRNAs) in various biological processes7. Among them, tRNA-derived small RNAs (tsRNAs) have emerged as a newly discovered class of ncRNAs derived from tRNAs8. tsRNAs are widely present in a variety of organisms and exhibit functionally regulated ncRNAs with a wide range of biological functions, including protein translation inhibition9, regulation of ribosome biogenesis10, control of mRNA stability11, regulation of cell proliferation12, regulation of apoptosis13, immune regulation14 and epigenetic regulation15. tsRNAs can be categorized into two main types: tRNA halves (tiRNAs) and tRNA-derived fragments (tRFs). Based on their corresponding parental tRNA sequence positions and lengths, tsRNAs can be further classified as follows: (i) tRF-5, originating from the 5′ end of mature tRNA and further divided into tRF-5a, tRF-5b, and tRF-5c based on their length; (ii) tRF-3, derived from the 3′ end of mature tRNA and further subdivided into tRF-3a and tRF-3b; (iii) tRF-1, originating from the 3′ end of precursor tRNA; and (iv) tRF-2, corresponding to an intermediate fragment of mature tRNA. tsRNAs are widely distributed across organisms and play crucial regulatory roles in lipid deposition biology16. For instance, in mice with nonalcoholic steatohepatitis, blueberry monomeric TEC has been shown to inhibit lipid damage and deposition by promoting tRF-4717. Sperm tsRNAs have been implicated as potential mediators for the transmission of maternal high-fat diet-induced addictive behaviors and obesogenic phenotypes to offspring18. In high-fat diet mice, elevated levels of 5′tsRNA-Gly-GCC in mature sperm have been found to promote hepatic gluconeogenesis through the regulation of the Sirt6-FoxO1 pathway, resulting in significant weight gain in F0 mice19. These pieces of evidence underscore the significant regulatory roles of tsRNAs in fat deposition.
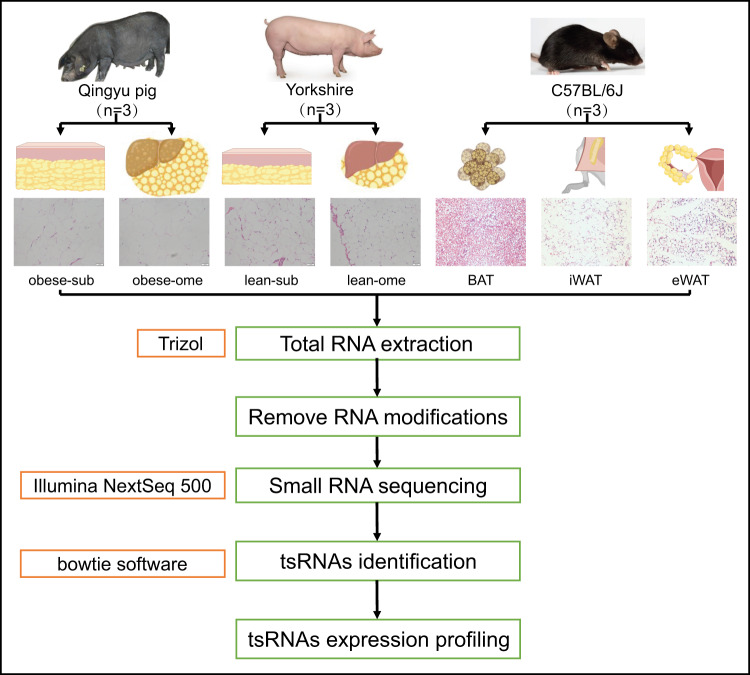
Currently, pigs and mice are the primary animal models used in human medicine. Pigs, being one of the most widely reared livestock species globally20, not only provide a significant source of protein for human consumption but also serve as valuable biological models for studying various human diseases21. This is due to their similarities to humans in terms of cardiovascular system and metabolic characteristics22. Mice, on the other hand, are also extensively utilized as biological models for studying human diseases. They possess a high degree of genetic similarity to humans, exhibit excellent reproductive performance, and are relatively inexpensive to breed and maintain23. However, the biological model of the pig has limitations when it comes to studying the biology of fat deposition. The reason is that brown fat does not exist in pigs. The main function of brown fat is thermogenesis, which is mainly useful in human infancy, but some brown fat remains in the adult body24. And unlike pigs, brown fat is present in mice. So to investigate the role of tsRNAs in fat deposition, we collected subcutaneous white adipose tissue and omental white adipose tissue from both Qingyu pig (obese pig) and Yorkshine (lean pig) and obtained their tsRNAs sequencing data. In addition, we also obtained tsRNAs sequencing data of scapular brown adipose tissue (BAT), inguinal white adipose tissue (iWAT) and epigonadal white adipose tissue (eWAT) from normal C57BL/6j mice (Fig. 1). Detailed information about the samples can be found in Tables 1 and 2. A total of 21 samples were analyzed in this study, including 11 samples that were published25,26, with three biological replicates per group. In this study, we generated 201,097,398 raw reads using the Illumina NextSeq. 500 system. After undergoing Illumina quality control, the sequencing reads were subjected to 5′ and 3′-adaptor trimming, and reads with lengths less than 14 nucleotides or greater than 40 nucleotides were discarded using cutadapt. This resulted in a total of 170,323,089 trimmed reads. Among these, 21,607,372 reads were aligned to tRNA sequences, accounting for 12.7% of the total reads (Table 3). Our study successfully characterized the expression profiles of tsRNAs in subcutaneous and omental adipose tissues of lean and obese pigs, as well as in mouse BAT, iWAT, and eWAT. Furthermore, by comparing the expression profiles of pig and mouse tsRNAs, we identified 117 conserved tsRNAs in both pig and mouse adipose tissues. These findings serve as a valuable resource and foundation for further investigation into the epigenetic mechanisms underlying fat deposition.
Fig. 1.
Illustration of sampled animals, sample collection, sample sectioning and RNA extraction process, small RNA sequencing and data analysis. Scale: 1 bar represents 50 μm.
Table 1.
Basic sample collection information of Pigs.
| Sample | Age (d) | Weight (kg) | Average backfat thickness (mm) | Lean meat percentage (%) |
|---|---|---|---|---|
| Qingyu1 | 300 | 128 | 60.31 | 40.39 |
| Qingyu2 | 300 | 136.5 | 42.98 | 38.89 |
| Qingyu3 | 300 | 135 | 57.36 | 41.35 |
| Yorkshire1 | 180 | 98.5 | 20.69 | 54.88 |
| Yorkshire2 | 180 | 111 | 19.2 | 63.2 |
| Yorkshire3 | 180 | 85.5 | 12.31 | 61.35 |
Table 2.
Basic sample collection information of mice.
| Sample | Age (d) | Weight (g) | BAT (g) | iWAT (g) | eWAT (g) |
|---|---|---|---|---|---|
| C57BL/6J-1 | 56 | 28.1 | 0.101 | 0.145 | 0.525 |
| C57BL/6J-2 | 56 | 26.6 | 0.181 | 0.104 | 0.458 |
| C57BL/6J-3 | 56 | 28.7 | 0.137 | 0.085 | 0.403 |
Table 3.
Mapping summary.
| Item | Number |
|---|---|
| Raw reads | 201097398 |
| Trimmed reads | 170323089 |
| tsRNAs reads | 21607372 |
| tsRNAs reads (%) | 12.69 |
Raw reads: Raw sequencing reads after quality filtering. Trimmed reads: Reads number after 5′, 3′-adaptor trimmed and discarded reads (length < 14nt or > 40nt). tsRNAs reads: Reads number aligned to mature and precursor tRNA. tsRNAs reads(%): The proportion of reads number aligning to mature and precursor tRNA.
Methods
Sample collection and RNA extraction
To ensure the animals’ welfare, three adult female Qingyu pigs (obese type), and three adult female Yorkshire pigs (lean type), were humanely euthanized. Following the slaughtering process, the section of subcutaneous fat from the back of the final rib cage, as well as the omental adipose tissue, were promptly excised and immediately submerged in liquid nitrogen for rapid freezing and subsequent storage. Similarly, normal 8-week-old female C57BL/6j mice were humanely sacrificed, and BAT, iWAT and eWAT were collected and frozen in liquid nitrogen. RNA extraction from the subcutaneous adipose tissue and omental adipose tissue, as well as BAT, eWAT and iWAT, was performed using TRIzol reagent (TaKaRa, Japan) according to the manufacturer’s instructions.
Libraries preparation, sequencing, and tsRNAs mapping
The total RNA samples were assessed for quality using agarose gel electrophoresis and quantified using a Nanodrop spectrophotometer. tsRNAs are heavily decorated by RNA modifications that interfere with small RNA-seq library construction. We do the following treatments before library preparation for total RNA samples: 3′-aminoacyl (charged) deacylation to 3′-OH for 3′adaptor ligation, 3′-cP (2′,3′-cyclic phosphate) removal to 3′-OH for 3′adaptor ligation, 5′-OH (hydroxyl group) phosphorylation to 5′-P for 5′-adaptor ligation, m1A and m3C demethylation for efficient reverse transcription. cDNA was then synthesized and amplified using Illumina’s proprietary RT primers and amplification primers. Subsequently, ~134–160 bp PCR amplified fragments were extracted and purified from the PAGE gel. And finally, the completed libraries were quantified by Agilent 2100 Bioanalyzer. The libraries were denatured and diluted to a loading volume of 1.3 ml and loading concentration of 1.8pM. Raw sequencing data obtained from the Illumina NextSeq. 500 platform underwent a stringent quality control process, which included filtering for Illumina chastity. The trimmed reads, with their 5′ and 3′-adaptor bases removed, were aligned to mature tRNA sequences, allowing for a maximum of 1 mismatch. If any reads failed to align, they were subsequently aligned to precursor tRNA sequences, again permitting only 1 mismatch. Bowtie software was utilized for these alignment processes27. Based on a statistical analysis of the alignments, which considered factors such as mapping ratio, read length, and fragment sequence bias, a determination was made regarding the usability of the results. If deemed suitable, expression profiling and the identification of differentially expressed tsRNAs were performed.
Expression profiling of tsRNAs
The abundance of tsRNAs is assessed by calculating their sequencing counts and normalizing them as counts per million of total aligned reads (CPM). To determine the expression level of each tsRNA, the mapped reads number is utilized and assigned an identification (ID). Subsequently, a filtering step is performed to exclude tsRNAs with a CPM value less than 20 in all samples.The formula for calculating the abundance of tsRNAs is as follows:
i: The i-th read aligned to the tsRNAs region.
n: The number of the reads aligned to the tsRNAs region
ci: The count of the i-th read
mi: The number of tsRNAs generated from the i-th read (mi possibly occur great than one, only when allowing for more than 1 mismatch).
The CPM value of tsRNAs can be calculated with the formula.
N: The total number of reads mapped onto all of the mature or precursor tRNA.
Sequence characteristics of tsRNA profiling of subcutaneous white adipose tissue and omental white adipose tissue of pig
Based on the calculated CPM values, we evaluated the tsRNAs expression profile characteristics of subcutaneous white adipose tissue and visceral adipose tissue in lean and obese pigs. We observed the presence of tsRNAs in the subcutaneous adipose tissue of lean pigs (lean-sub), omental adipose tissue of lean pigs (lean-ome), subcutaneous adipose tissue of obese pigs (obese-sub), and omental adipose tissue of obese pigs (obese-ome) within the length range of 14–40 nucleotides. Among these, tsRNAs with a length of approximately 30–33 nucleotides were found to be the most abundant (Fig. 2a). In total, we identified 485 unique tsRNAs across the subcutaneous and omental adipose tissues of lean and obese pigs. A total of 287 tsRNAs were identified in the adipose tissues of lean pigs, with overlapping tsRNAs representing 56.34% of the total. Similarly, 242 tsRNAs were identified in the adipose tissues of obese pigs, with overlapping tsRNAs accounting for 60.74%. In both omental adipose tissue of lean and obese pigs, a total of 246 tsRNAs were found to overlap, constituting 53.66% of the total. Furthermore, in the subcutaneous adipose tissue of lean and obese pigs, 282 tsRNAs were identified, with overlapping tsRNAs accounting for 62.41% (Fig. 2b). Regarding the classification of tsRNAs, we observed that tRF-5c was the most abundant type in all four groups, followed by tRF-3a (Fig. 2b). In the lean-ome group, Glu-TTC, Gly-GCC, and Gly-CCC were the parental tRNAs of tsRNAs with the highest percentage, accounting for 11.38%, 10.16%, and 5.59%, respectively. In the lean-sub group, Glu-TTC, Gly-GCC, and Ala-TGC were the parental tRNAs of tsRNAs with the highest proportion, accounting for 11.75%, 7.83%, and 5.12%, respectively. In the obese-ome and obese-sub groups, Glu-TTC, Gly-GCC, and Gly-GCC were the parental tRNAs of tsRNAs with the highest proportion, accounting for 8.23%, 8.23%, and 7.82%, and 8.85%, 8.85%, and 5%, respectively (Fig. 2c). In addition, we conducted an analysis of nucleotide deviations in the seed sequence positions of the top 20 expressed tsRNAs within each of the four groups. The findings revealed slight variations in the seed sequence bases among the top 20 tsRNAs in the four groups (Fig. 2d).
Fig. 2.
Sequence characteristics of tsRNA profiling of subcutaneous white adipose tissue and omental white adipose tissue of pig. (a) Length distribution of tsRNAs. (b) Venn diagram and type of identified tsRNAs (CPM > 20). (c) Proportion of parental tRNAs of identified tsRNAs. (d) Characterization of the seed sequences of four groups of top20 tsRNAs.
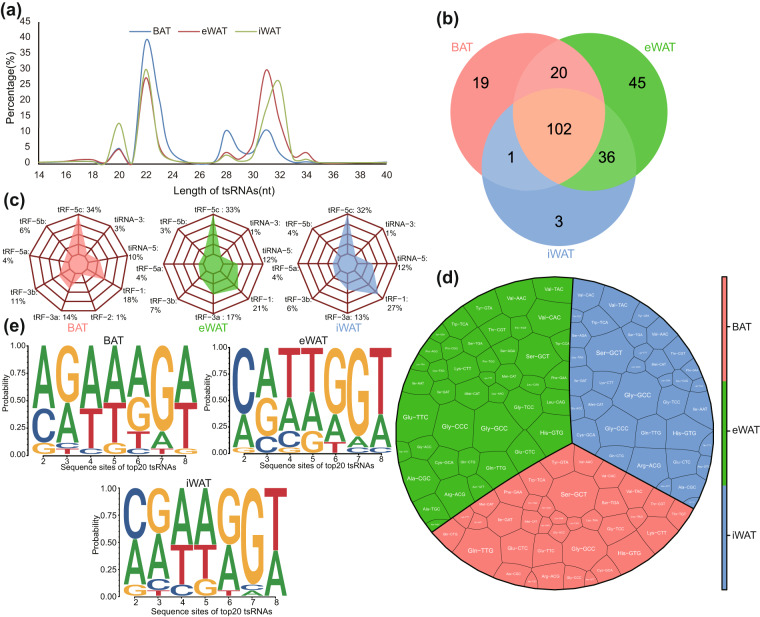
Sequence characteristics of tsRNA profiling of BAT, eWAT and iWAT of mice
Based on the calculated CPM values, we also evaluated the tsRNAs expression profile characteristics of mouse BAT, iWAT, and eWAT. Interestingly, unlike tsRNAs derived from porcine adipose tissue, tsRNAs from mouse adipose tissue showed a predominant length distribution in the ranges of 20–24nt and 28–33nt (Fig. 3a). A total of 278 tsRNAs were identified across the three types of mouse adipose tissues. Among these, we identified 19 BAT-specific tsRNAs, 45 eWAT-specific tsRNAs, and 3 iWAT-specific tsRNAs. The overlapping tsRNAs shared between iWAT and eWAT accounted for 66.67% of their respective expressed tsRNAs. Additionally, the overlapping tsRNAs between BAT and iWAT, as well as between BAT and eWAT, accounted for 56.91% and 52.36% of their expressed tsRNAs, respectively (Fig. 3b). Further analysis of tsRNA types in mouse adipose tissue revealed that, similar to porcine adipose tissue, tRF-5c was the most abundant type of tsRNA. However, unlike porcine adipose tissue, the second most abundant type in mouse adipose tissue was tRF-1 (Fig. 3c). The most common parental tRNAs for tsRNAs were Ser-GCT, Gln-TTG, and Gly-GCC, comprising 8.5%, 7.19%, and 6.54% of the total in the BAT group, respectively. Meanwhile, in the iWAT group, the prevalent parental tRNAs for tsRNAs were Gly-GCC, Ser-GCT, and Ser-GCT, accounting for 8.44%, 7.79%, and 5.84%, respectively. Turning to the eWAT group, the prevailing parental tRNAs for tsRNAs were Gly-GCC, Glu-TTC, and Gly-CCC, accounting for 6.81%, 6.38%, and 5.53% of the total, respectively (Fig. 3d). Additionally, we conducted a similar analysis of the nucleotide deviations in the seed sequence positions of the top 20 expressed tsRNAs in the three mouse adipose tissue groups(Fig. 3e). Interestingly, the nucleotide deviations observed in tsRNAs from mouse adipose tissue and porcine adipose tissue were markedly different.
Fig. 3.
Sequence characteristics of tsRNA profiling of BAT, eWAT and iWAT of mice. (a) Length distribution of tsRNAs. (b) Venn diagram of identified tsRNAs (CPM > 20). (c) type of identified tsRNAs. (d) Proportion of parental tRNAs of identified tsRNAs. (e) Characterization of the seed sequences of top20 tsRNAs.
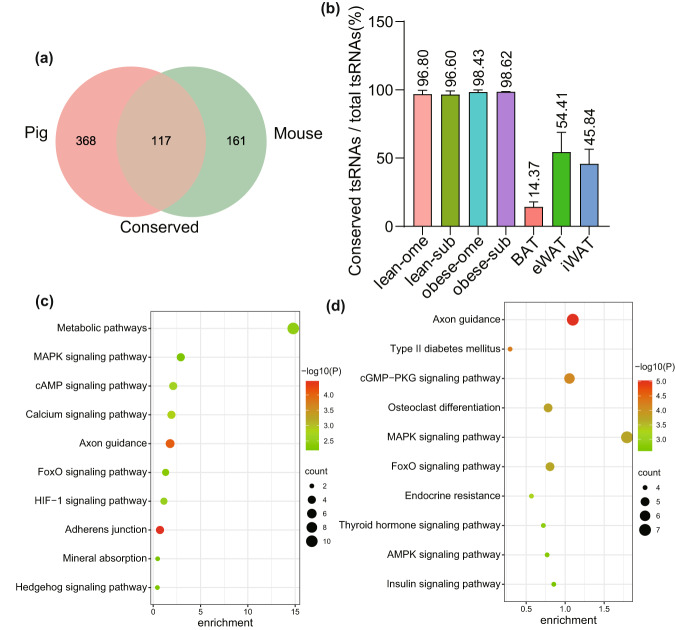
Conserved tsRNAs in adipose tissue of pigs and mice
By comparing the expression profiles of pig and mouse tsRNAs, we identified 117 tsRNAs that were fully conserved between pigs and mice (Supplementary Table S1), accounting for 18.11% of the total tsRNAs identified (Fig. 4a). Moreover, we analyzed the expression percentage of these conserved tsRNAs in each sample. To our surprise, we found that these conserved tsRNAs were highly expressed, accounting for over 96% of the samples in pigs. However, in mice, the expression percentages of these conserved tsRNAs were relatively lower. In iWAT and eWAT tissues, the expression levels accounted for 45.84% and 54.41%, respectively, while in BAT, it was only 14.37% (Fig. 4b). Interestingly, we found that among the conserved tsRNAs, the TOP10 tsRNAs with the highest average expression abundance were mainly derived from two parental tRNAs, Gly-GCC and Glu-CTC. Among them, there were 7 tsRNAs derived from Gly-GCC and 2 tsRNAs derived from Glu-CTC, and the tsRNAs derived from the same parental tRNA had similar seed sequences (Supplementary Table S1). Therefore, we further predicted their potential target genes on the online prediction website Targetscan (https://www.targetscan.org/vert_50/seedmatch.html), and performed Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis on the online tool KOBAS (http://bioinfo.org/kobas). KEGG analysis showed that the potential target genes of tsRNA derived from Gly-GCC were mainly enriched in signal transduction-related pathways, such as Calcium signaling pathway and cAMP signaling pathway, and metabolism-related pathways, such as Metabolic pathways, FoxO signaling pathway and MAPK signaling pathway (Fig. 4c). The potential target genes of tsRNA derived from Glu-CTC were mainly enriched in obesity disease-related pathways, such as Type II diabetes mellitus, and metabolic-related pathways, such as Insulin signaling pathway, MAPK signaling pathway, FoxO signaling pathway, Thyroid hormone signaling pathway and AMPK signaling pathway (Fig. 4d).
Fig. 4. Conserved tsRNAs in adipose tissue of pigs and mice.
(a) Venn diagram illustrating conserved tsRNAs between porcine and mouse adipose tissues. (b) Percentage of conserved tsRNA expression levels in each sample. (c) KEGG analysis of potential target genes of tsRNAs derived from Gly-GCC. (d) KEGG analysis of potential target genes of tsRNAs derived from Glu-CTC.
Data Records
The tsRNA sequencing data for the 12 porcine adipose tissue samples and 9 mouse adipose tissue samples referred to in this article have been uploaded to the NCBI Sequence Read Archive (SRA) under access number SRP40703228 and SRP43999329. The expression profile of tsRNAs in pigs can be obtained in the Gene Expression Omnibus (GEO) database of the NCBI, and the assession number is GSE23508730. The expression profile of mouse tsRNAs can also be obtained in the GEO database, and the assession number is GSE23508831.
Technical Validation
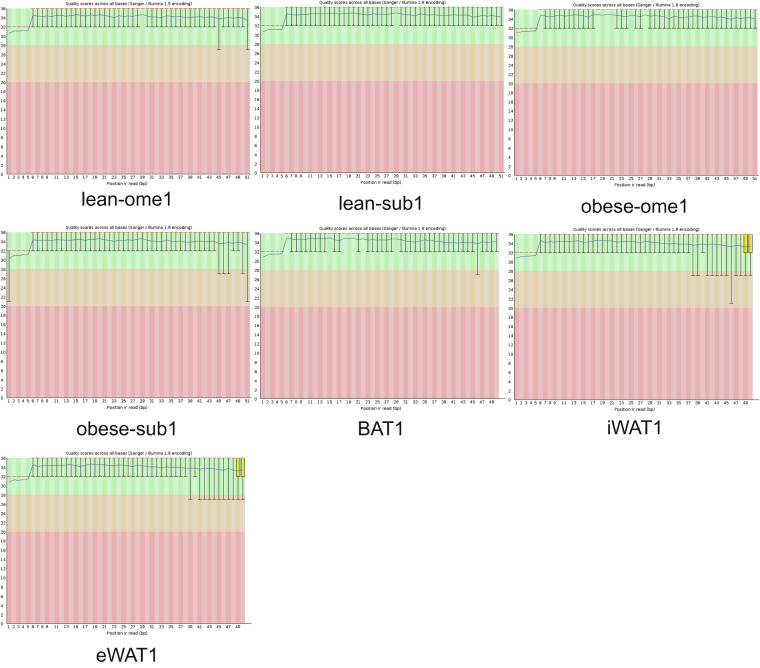
Sequencing quality control
Raw data files in FASTQ format were obtained from the Illumina sequencer. To assess the quality of the sequencing data, we performed quality control using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). We generated plots displaying the quality scores for each sample (Fig. 5; Supplementary Figures S1). The quality score, denoted as Q, is logarithmically associated with the probability of base calling errors (P).
Fig. 5.
tsRNA-seq quality score plot. The X-axis displays the position in the read, while the Y-axis represents the Q value. The median Q score is shown as a red line, and the mean Q score is depicted as a blue line. The inter-quartile range is represented by the boxplot, and the whiskers extend to the 10% and 90% points. Data with a Q score exceeding 30, indicating accuracy above 99.9%, is considered high-quality.
For example, Q30 means the incorrect base calling probability to be 0.001 or 99.9% base calling accuracy. All samples of Q30 are shown in Table 4.
Table 4.
Quality score of the sample used in our study.
| Sample | TotalRead | TotalBase | BaseQ. 30 | BaseQ. 30 (%) |
|---|---|---|---|---|
| obese-sub1 | 11833544 | 603510744 | 567352963 | 94.01 |
| obese-sub2 | 8256383 | 421075533 | 391482779 | 92.97 |
| obese-sub3 | 10399443 | 530371593 | 493002831 | 92.95 |
| obese-ome1 | 8079564 | 412057764 | 379340758 | 92.06 |
| obese-ome2 | 9529406 | 485999706 | 452234929 | 93.05 |
| obese-ome3 | 10168082 | 518572182 | 482763022 | 93.09 |
| lean-sub1 | 11373476 | 580047276 | 537160780 | 92.61 |
| lean-sub2 | 11199763 | 571187913 | 532242409 | 93.18 |
| lean-sub3 | 12980769 | 662019219 | 609990267 | 92.14 |
| lean-ome1 | 13108635 | 668540385 | 622927286 | 93.18 |
| lean-ome2 | 12112953 | 617760603 | 575238627 | 93.12 |
| lean-ome3 | 8544746 | 435782046 | 400493079 | 91.90 |
| BAT1 | 10395534 | 519776700 | 486707550 | 93.64 |
| BAT2 | 8114960 | 405748000 | 371737506 | 91.62 |
| BAT3 | 8898480 | 444924000 | 410684324 | 92.30 |
| eWAT1 | 7084165 | 354208250 | 326496003 | 92.18 |
| eWAT2 | 7755765 | 387788250 | 357414689 | 92.17 |
| eWAT3 | 5759943 | 287997150 | 280278763 | 97.32 |
| iWAT1 | 7835103 | 391755150 | 360597465 | 92.05 |
| iWAT2 | 8814977 | 440748850 | 404587758 | 91.80 |
| iWAT3 | 8851707 | 442585350 | 405879110 | 91.71 |
Sample: Sample ID. TotalRead: Total sequencing reads post quality filtering. TotalBase: Total bases post quality filtering. BaseQ. 30: Bases with Q score greater than 30 post quality filtering. BaseQ. 30 (%): Percentage of bases (Q ≥ 30) post quality filtering.
Reproducibility validation
To assess the reproducibility of the biological replicates in our sample set, we conducted correlation analysis on the 12 samples obtained from pigs and mice. The correlation heatmap revealed high correlation coefficients among the majority of biological replicates (Fig. 6a,b). Additionally, principal component analysis (PCA) demonstrated that most of the biological samples clustered together (Fig. 6c,d). These results provide strong evidence for the high confidence and reliability of our study data.
Fig. 6.
Assessment of reproducibility across biological replicates. (a,b) Heatmap of correlation of pig and mice samples respectively. (c,d) PCA analysis of pig and mice samples respectively.
Supplementary information
Acknowledgements
This work was supported by National Natural Science Foundation of China (31972524, 32372844); Sichuan Science and Technology Program (2021YFYZ0007, 2021ZDZX0008, 2021YFYZ0030, sccxtd-2023-08-09); China Agriculture Research System (CARS-35).
Author contributions
L.S. and L.Z. designed the experiment. Y.L., Y.W. and L.C. collected samples and constructed a sequencing library. T.L. and M.G. analyzed the data and wrote the paper. L.S. and L.Z. revised the paper. All authors reviewed the manuscript.
Code availability
All software used in this study species are in the public domain, except for those explicitly described in the text and methods. No custom scripts or code was used during the curation and validation of the dataset in this study. Also the following software tools were used:
tRNA sequences of cytoplasmic were downloaded from GtRNAdb32,33.
tRNA sequences of mitochondrial were predicted with tRNAscan-SE34,35 software.
Principal Component Analysis (PCA), Correlation Analysis, Pie plots, Venn plots, and radar plots were performed in the online analysis tool Bioinformatics (www.bioinformatics.com).
The seed sequence motif was done using the BioLadder online tool (www.bioladder.cn/).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Tianci Liao, Mailin Gan.
Contributor Information
Linyuan Shen, Email: shenlinyuan@sicau.edu.cn.
Li Zhu, Email: zhuli@sicau.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41597-023-02624-y.
References
- 1.Koenen M, Hill MA, Cohen P, Sowers JR. Obesity, Adipose Tissue and Vascular Dysfunction. Circ Res. 2021;128:951–968. doi: 10.1161/CIRCRESAHA.121.318093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen J, et al. Sirtuins: Key players in obesity-associated adipose tissue remodeling. Frontiers in immunology. 2022;13:1068986. doi: 10.3389/fimmu.2022.1068986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tran TT, Yamamoto Y, Gesta S, Kahn CR. Beneficial effects of subcutaneous fat transplantation on metabolism. Cell metabolism. 2008;7:410–420. doi: 10.1016/j.cmet.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petrov MS, Taylor R. Intra-pancreatic fat deposition: bringing hidden fat to the fore. Nat Rev Gastroenterol Hepatol. 2022;19:153–168. doi: 10.1038/s41575-021-00551-0. [DOI] [PubMed] [Google Scholar]
- 5.Gu H, et al. Targeted overexpression of PPARγ in skeletal muscle by random insertion and CRISPR/Cas9 transgenic pig cloning enhances oxidative fiber formation and intramuscular fat deposition. FASEB J. 2021;35:e21308. doi: 10.1096/fj.202001812RR. [DOI] [PubMed] [Google Scholar]
- 6.Tang QQ, Lane MD. Adipogenesis: from stem cell to adipocyte. Annual review of biochemistry. 2012;81:715–736. doi: 10.1146/annurev-biochem-052110-115718. [DOI] [PubMed] [Google Scholar]
- 7.Xue C, et al. The Mechanism Underlying the ncRNA Dysregulation Pattern in Hepatocellular Carcinoma and Its Tumor Microenvironment. Frontiers in immunology. 2022;13:847728. doi: 10.3389/fimmu.2022.847728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schimmel P. The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis. Nature reviews. Molecular cell biology. 2018;19:45–58. doi: 10.1038/nrm.2017.77. [DOI] [PubMed] [Google Scholar]
- 9.Sobala A, Hutvagner G. Small RNAs derived from the 5′ end of tRNA can inhibit protein translation in human cells. RNA Biol. 2013;10:553–563. doi: 10.4161/rna.24285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim HK, et al. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature. 2017;552:57–62. doi: 10.1038/nature25005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shao Y, et al. tRF-Leu-CAG promotes cell proliferation and cell cycle in non-small cell lung cancer. Chem Biol Drug Des. 2017;90:730–738. doi: 10.1111/cbdd.12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maute RL, et al. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:1404–1409. doi: 10.1073/pnas.1206761110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tao E-W, et al. A specific tRNA half, 5′tiRNA-His-GTG, responds to hypoxia via the HIF1α/ANG axis and promotes colorectal cancer progression by regulating LATS2. Journal of experimental & clinical cancer research: CR. 2021;40:67. doi: 10.1186/s13046-021-01836-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kazimierczyk M, et al. tRNA-derived fragments from the Sus scrofa tissues provide evidence of their conserved role in mammalian development. Biochem Biophys Res Commun. 2019;520:514–519. doi: 10.1016/j.bbrc.2019.10.062. [DOI] [PubMed] [Google Scholar]
- 15.Chen Q, et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science (New York, N.Y.) 2016;351:397–400. doi: 10.1126/science.aad7977. [DOI] [PubMed] [Google Scholar]
- 16.Li, S., Xu, Z. & Sheng, J. tRNA-Derived Small RNA: A Novel Regulatory Small Non-Coding RNA. Genes9, 10.3390/genes9050246 (2018). [DOI] [PMC free article] [PubMed]
- 17.Zhu J, et al. The monomer TEC of blueberry improves NASH by augmenting tRF-47-mediated autophagy/pyroptosis signaling pathway. J Transl Med. 2022;20:128. doi: 10.1186/s12967-022-03343-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarker G, et al. Maternal overnutrition programs hedonic and metabolic phenotypes across generations through sperm tsRNAs. Proceedings of the National Academy of Sciences of the United States of America. 2019;116:10547–10556. doi: 10.1073/pnas.1820810116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang B, et al. Paternal High-Fat Diet Altered Sperm 5′tsRNA-Gly-GCC Is Associated With Enhanced Gluconeogenesis in the Offspring. Frontiers in molecular biosciences. 2022;9:857875. doi: 10.3389/fmolb.2022.857875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knox RV. Impact of swine reproductive technologies on pig and global food production. Adv Exp Med Biol. 2014;752:131–160. doi: 10.1007/978-1-4614-8887-3_7. [DOI] [PubMed] [Google Scholar]
- 21.Spurlock ME, Gabler NK. The development of porcine models of obesity and the metabolic syndrome. J Nutr. 2008;138:397–402. doi: 10.1093/jn/138.2.397. [DOI] [PubMed] [Google Scholar]
- 22.Lunney JK, et al. Importance of the pig as a human biomedical model. Sci Transl Med. 2021;13:eabd5758. doi: 10.1126/scitranslmed.abd5758. [DOI] [PubMed] [Google Scholar]
- 23.Walsh NC, et al. Humanized Mouse Models of Clinical Disease. Annu Rev Pathol. 2017;12:187–215. doi: 10.1146/annurev-pathol-052016-100332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Søberg S, et al. Altered brown fat thermoregulation and enhanced cold-induced thermogenesis in young, healthy, winter-swimming men. Cell Rep Med. 2021;2:100408. doi: 10.1016/j.xcrm.2021.100408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang, L. et al. tsRNA Landscape and Potential Function Network in Subcutaneous and Visceral Pig Adipose Tissue. Genes14, 10.3390/genes14040782 (2023). [DOI] [PMC free article] [PubMed]
- 26.Gu, H. et al. Differential Expression Analysis of tRNA-Derived Small RNAs from Subcutaneous Adipose Tissue of Obese and Lean Pigs. Animals: an open access journal from MDPI12, 10.3390/ani12243561 (2022). [DOI] [PMC free article] [PubMed]
- 27.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.2022. Sequence Read Archive. SRP407032
- 29.2023. Sequence Read Archive. SRP439993
- 30.2023. Gene Expression Omnibus. GSE235087
- 31.2023. Gene Expression Omnibus. GSE235088
- 32.Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic acids research. 2009;37:D93–D97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan PP, Lowe TM. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic acids research. 2016;44:D184–D189. doi: 10.1093/nar/gkv1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lowe TM, Chan PP. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic acids research. 2016;44:W54–W57. doi: 10.1093/nar/gkw413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic acids research. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- 2022. Sequence Read Archive. SRP407032
- 2023. Sequence Read Archive. SRP439993
- 2023. Gene Expression Omnibus. GSE235087
- 2023. Gene Expression Omnibus. GSE235088
Supplementary Materials
Data Availability Statement
All software used in this study species are in the public domain, except for those explicitly described in the text and methods. No custom scripts or code was used during the curation and validation of the dataset in this study. Also the following software tools were used:
tRNA sequences of cytoplasmic were downloaded from GtRNAdb32,33.
tRNA sequences of mitochondrial were predicted with tRNAscan-SE34,35 software.
Principal Component Analysis (PCA), Correlation Analysis, Pie plots, Venn plots, and radar plots were performed in the online analysis tool Bioinformatics (www.bioinformatics.com).
The seed sequence motif was done using the BioLadder online tool (www.bioladder.cn/).