Abstract
INTRODUCTION:
Mutations in INPP5D, which encodes for the SH2-domain-containing inositol phosphatase SHIP-1, have recently been linked to an increased risk of developing late-onset Alzheimer’s disease. While INPP5D expression is almost exclusively restricted to microglia in the brain, little is known regarding how SHIP-1 affects neurobiology or neurodegenerative disease pathogenesis.
METHODS:
We generated and investigated 5xFAD Inpp5dfl/flCx3cr1Ert2Cre mice to ascertain the function of microglial SHIP-1 signaling in response to amyloid beta (Aβ)-mediated pathology.
RESULTS:
SHIP-1 deletion in microglia led to substantially enhanced recruitment of microglia to Aβ plaques, altered microglial gene expression, and marked improvements in neuronal health. Further, SHIP-1 loss enhanced microglial plaque containment and Aβ engulfment when compared to microglia from Cre-negative 5xFAD Inpp5dfl/fl littermate controls.
DISCUSSION:
These results define SHIP-1 as a pivotal regulator of microglial responses during Aβ-driven neurological disease and suggest that targeting SHIP-1 may offer a promising strategy to treat Alzheimer’s disease.
Keywords: Alzheimer’s disease, microglia, SHIP-1, INPP5D, amyloid beta, amyloidosis, neuroimmunology, disease-associated microglia, neurodegenerative disease
1. BACKGROUND
Alzheimer’s disease (AD) is a neurodegenerative disease and the most common form of dementia, affecting millions of people and costing billions of dollars in care [1]. With the prevalence of AD increasing with the aging population and the current lack of disease-modifying treatment options, new avenues of exploration are needed to uncover novel therapeutic targets. AD is classically characterized by the buildup of amyloid beta (Aβ) plaques, neurofibrillary tangles, and neuronal damage that contribute to subsequent cognitive decline and memory loss [2]. Despite this, the biological roots of AD are still in question.
Microglia, the resident macrophages of the central nervous system (CNS), have emerged as major players in AD pathogenesis where they participate in cytokine production and the compaction, containment, phagocytosis, and clearance of Aβ and other forms of neurotoxic material [3,4]. Most notably, recent studies characterizing microglia in the AD brain have shown that microglia adopt a distinct transcriptional profile, termed the “disease-associated microglia” (DAM) profile, that is critical for the microglial response in AD and other neurodegenerative diseases [5,6]. The critical roles of microglia in AD have been further solidified by findings from recent human genome-wide association studies which have identified that a large percentage of late-onset AD (LOAD) risk loci affect genes involved in microglial biology and function [7,8]. While there is an ever-growing appreciation for the critical roles played by microglia in AD, we still lack basic knowledge regarding how many of these recently identified genetic risk factors affect microglial biology in AD.
One recently identified gene locus that has been strongly linked to LOAD risk is INPP5D [7,9]. INPP5D encodes for the SH2-domain-containing inositol phosphatase SHIP-1 and its expression in the brain is almost exclusively restricted to microglia [10,11]. Outside of the brain, SHIP-1 is abundantly expressed by most hematopoietically-derived innate and adaptive immune cells where it functions to dampen immune responses, such as proinflammatory cytokine production, phagocytosis, and reactive oxygen species generation [12–16]. Moreover, SHIP-1 has also been reported to affect the proliferation and survival of circulating myeloid cells [17,18]. The critical function of SHIP-1 as a brake on immune responses in the periphery is best exemplified by the devastating multi-organ inflammatory disease that ensues in mice following SHIP-1 deletion from hematopoietically-derived immune cells [19]. Of particular relevance to AD, SHIP-1 has been reported to associate with and modulate signaling downstream of multiple receptors that have been linked to AD and aging-related dementia, including TREM2, DAP12, CD22, CLEC7A (also known as Dectin-1), and FcγRs [14,15,20–22], although future studies are needed to substantiate if this also occurs in primary microglia during disease.
In contrast to the periphery, very little is currently known about the role of SHIP-1 in the healthy or AD brain. Recent studies have linked the INPP5D single nucleotide polymorphism, rs35349669, to an increased risk of developing LOAD [7,9] and found that SHIP-1 is highly upregulated in plaque-associated microglia in both AD patients and mouse models [23,24]. In addition, recent in vitro studies have also begun to explore how pharmaceutical agents that target SHIP-1 impact in vitro macrophage responses to neuronal damage and exogenous Aβ peptide treatment [11,25]. Despite these early findings linking SHIP-1 to AD, surprisingly little is currently known regarding the effects of SHIP-1 on in vivo microglial responses or Alzheimer’s-related disease progression. This motivated us to investigate the effects of SHIP-1 signaling on in vivo microglial responses in Aβ-mediated pathology, as a deeper understanding of the functions of SHIP-1 may lead to the discovery of novel microglia-specific regulators that can be targeted to treat AD and other neurodegenerative diseases.
2. METHODS
2.1. Mice
All experiments were conducted in accordance with the guidelines and regulations of the University of Virginia and approved by the University of Virginia Animal Care and Use Committee. 5xFAD mice (Stock #: 34848-Jax), Inpp5dfl/fl mice [26] (Stock #: 028255), and Cx3cr1ERT2cre mice [27] (Stock #: 020940) were obtained from The Jackson Laboratory and crossed to generate 5xFAD Inpp5dfl/flCx3cr1ERT2Cre (referred to as 5xFAD Inpp5dΔMG mice) and 5xFAD Inpp5dfl/fl (referred to as 5xFAD mice). At weaning (~3 weeks of age), female 5xFAD Inpp5dΔMG mice and 5xFAD littermate controls were fed tamoxifen diet (Envigo Teklad #TD.130858) ad libitum for two weeks and then returned to normal chow for the remainder of the experiment. Mice were housed under pathogen-free conditions with 12-hour light/dark cycle conditions in rooms equipped with control for temperature (21 ± 1.5°C) and humidity (50 ± 10%).
2.2. Brain sample preparation
Brain samples were collected and processed as previously described in Ennerfelt et al. [28]. Mice were euthanized using CO2 asphyxiation and transcardially perfused with ~25 mL of ice cold 1xPhosphate Buffered Saline (PBS). Brains were dissected out and the full left hemisphere was drop-fixed in 4% paraformaldehyde overnight then transferred to 30% sucrose for 48 hours at 4°C. The cortex from the right hemisphere was microdissected and flash-frozen on dry ice and stored at −80°C. Drop-fixed samples were then frozen in Tissue-Plus OCT (Fisher Scientific) on dry ice and sagittally sectioned at 50 μm in thickness using a cryostat (Leica). Tissue sections were stored in 1xPBS + 0.05% sodium azide (Ricca Chemical) at 4°C for downstream staining and imaging. Flash-frozen cortices were thawed for protein extraction and mechanically homogenized in 250 μL of Tissue Protein Extraction Reagent T-PER (Thermo Fisher, 78–510) containing phosphatase inhibitor cocktail PhosSTOP (Roche, 04–906-845–001) and protease inhibitor cocktail cOmplete (Roche, 11–873-580– 001). Following homogenization, the cortex lysate was spun down at 15,000 RPM for 10 minutes at 4°C and the supernatant (soluble fraction) and pellet were isolated and stored at −80°C for downstream soluble and insoluble protein analyses, respectively. Insoluble protein was further extracted from the pelleted sample using guanidine-extraction in which the samples were incubated 1:6 in 5 M guanidine HCL/50 mM tris (pH = 8.0) at room temperature for 3 hours followed by dilution 1:5 in 1xPBS containing protease inhibitor cocktail cOmplete (Roche, 11–873-580– 001) and centrifugation at 15,000 RPM for 20 minutes at 4°C. The supernatant (insoluble fraction) was collected and stored at −80°C for downstream protein analyses. Protein concentrations for the soluble and insoluble fractions were determined using the Pierce 660 nm Protein Assay Reagent (Thermo Scientific, 22–660). For single-nucleus RNA sequencing (snRNAseq) experiments, the cortex was microdissected from the right hemisphere, flash-frozen on dry ice, and stored at −80°C.
2.3. ELISA
Amyloid beta 40 or 42 Mouse ELISA kits (Thermo Fisher, KMB3481, KMB3441) were used to measure amyloid beta species in soluble and insoluble fraction samples following the manufacturer’s instructions. Thawed soluble and insoluble fractions were standardized to total protein by loading 1,200 μg/mL and 30 μg/mL per sample, respectively.
2.4. Immunofluorescence staining and microscopy
Brain sections were washed in 1xPBS three times for 5 minutes at room temperature before being blocked with 2% donkey serum, 1% bovine serum albumin, 0.1% Triton-X, and 0.05% Tween-20 in 1xPBS for 2 hours at room temperature. Primary antibody mixes, diluted in the abovementioned block solution, were applied overnight at 4°C. To study microglial number, recruitment, encapsulation, and engulfment, sections were stained with anti-IBA1 (ab5076, Abcam, 1:300) and anti-CD68 (FA-11, MCA1957 BioRad, 1:1000). IBA1 is a widely used marker of microglia that can also stain brain-infiltrating macrophages. Aβ load was probed using anti-Aβ D54D2 (8243, Cell Signaling, 1:300). To investigate proliferation, sections were stained with anti-Ki67-EF660 (SolA15, Invitrogen, 1:100). To explore microglial activation, sections were stained with anti-CLEC7A (R1–8g7, InvivoGen, 1:30). To interrogate neuronal health, sections were stained with anti-APP (Y188, ab326, Abcam, 1:750). Sections were then washed in 1xPBS and 0.05% Tween-20 three times for 5 minutes at room temperature and stained with matching Alexa Fluor 488, 594, 647 donkey anti-rabbit, -goat, and -rat (Thermo Fisher, 1:1000 dilution) secondary antibodies at room temperature for 2 hours. Sections were washed again in 1xPBS and 0.05% Tween-20 three times for 5 minutes at room temperature. Sections were then further stained with DAPI (D9542, Sigma-Aldrich, 1:1000) for 10 min at room temperature or ThioflavinS (Aβ plaques, Sigma-Aldrich, 2 mg/10 mL) for 8 minutes in 50% EtOH followed by three 2-minute washes with 50% ethanol and two 2-minute washes with 1xPBS at room temperature. All sections were then transferred to fresh 1xPBS prior to being mounted on glass slides with ProLongGold antifade reagent (Invitrogen, P36930) and coverslips. Mounted slides were stored at 4°C protected from light and imaged at 10x and 63x magnification on a Leica Stellaris 5 confocal microscope using LAS AF software (Leica Microsystems). Quantification of images were performed using Fiji software or Imaris software (9.5.1).
2.5. Single-nucleus RNA sequencing (snRNAseq)
Flash frozen brain cortices from three individual mice were pooled based on genotype (5xFAD Inpp5dΔMG or 5xFAD, see 2.2) and submitted to GENEWIZ (Azenta Life Sciences) for nuclei extraction and sequencing. Nuclei extraction was performed using the Miltenyi Nuclei Extraction Buffer (Miltenyi Biotec) following the manufacturer’s guidelines with gentle MACS Dissociation and C tubes (Miltenyi Biotec). Following isolation, nuclei were counted using a Countess 3 Automated Cell Counter (Thermo Fisher). Single-nuclei 3’ RNA libraries were generated using the Chromium Single Cell 3’ kit (10X Genomics). For each sample, a target capture of ~3,000 Gel Beads-in-Emulsion per sample was loaded onto the Chromium Controller (10X Genomics) and processed following the standard manufacturer’s specifications. Sequencing libraries were evaluated for quality using the Agilent TapeStation (Agilent Technologies) and quantified using a Qubit 2.0 Fluorometer (Invitrogen). Pooled libraries were quantified using qPCR (Applied Biosystems) and sequenced using a HiSeq 4000 (Illumina). Samples were sequenced at a configuration compatible with the recommended guidelines as outlined by 10X Genomics and raw sequence data were converted into fastq files. Raw fastq files were further processed with the Cell Ranger (7.0.1, 10X Genomics) `cellranger count` pipeline with introns included using the mouse (mm10) genome. Raw feature matrices containing the barcodes, features, and matrix files for each sample were used for further analysis in RStudio (4.2.1).
The Seurat package (4.2.0) in R was used for downstream analyses [29]. For quality control, nuclei containing a low number of reads (<400 UMI) and genes (<350 genes) and >10% mitochondrial reads were removed. Doublet/multiplet nuclei were further filtered out by removing nuclei containing a high number of genes (>5,000 genes). Additionally, nuclei containing a low complexity score (<0.8 log10GenesPerUMI) were removed. After filtering, 20,818 nuclei remained. The samples then underwent normalization, scaling, and integration using SCTransform in Seurat [30]. For SCTransform, the default parameters were used and 3,000 features were utilized in the identification of variable features. Principal component analysis was conducted using the top 3,000 most variable features and uniform manifold approximation and projection (UMAP) reduction was conducted using the top 40 principal components (PCs) based on the ElbowPlot. Clustering was performed using the FindNeighbors and FindClusters functions, which use the K-nearest neighbors graph model and Louvain algorithm. A resolution of 0.1 was chosen for downstream clustering. To determine the identification of each cluster, the FindAllMarkers function was used to extract the differentially expressed signature genes corresponding to each cluster against all other clusters. Following clustering, nuclei populations were arranged in 17 distinct clusters including neuron, microglia, astrocyte, oligodendrocyte, and neurovascular-associated clusters. Nuclei from the microglia cluster were isolated to further analyze microglia subpopulations. Microglia nuclei were re-integrated using SCTrasform as conducted previously. Principal component analysis was performed as previously described and UMAP projection was performed using the top 15 PCs based on the ElbowPlot. Clustering was conducted as previously described and a resolution of 0.1 was chosen for downstream clustering. The resulting four distinct clusters were identified using the FindConservedMarkers, FindMarkers, and FindAllMarkers functions in Seurat. All UMAP, DotPlot, and Heatmap visualizations were performed using Seurat and ggplot2 (3.3.6).
Differential expression analysis between conditions (5xFAD Inpp5dΔMG or 5xFAD) and microglia sub-populations was conducted using the FindMarkers function in Seurat and the DESeq2 algorithm that uses a negative bimodal distribution to identify differentially expressed genes [31]. The average log2(fold-change) represents the average expression between the two conditions tested and the adjusted P value was calculated using Bonferroni correction. Final lists of differentially expressed genes (DEGs) were filtered to remove all genes with adjusted P values >.05 and were used for downstream pathway analysis. For visualization, volcano plots were created using the EnhancedVolcano package (1.14.0) in R and DEGs with adjusted P values <.00001 and average log2(fold-changes) >0.25 or <−0.25 were plotted. Pathway analyses were conducted using the Enrichr web-based application [32] and the WikiPathways Mouse 2019 pathways database. Visualization of enriched pathways was conducted using the Enrichment Analysis Visualization Appyter [33].
Raw sequencing data associated with this study has been deposited in the NCBI Gene Expression Omnibus database under accession code GSE220279. The code used in this study is available at GitHub (https://github.com/LukensLab/5xFAD_INPP5D_Microglia-snRNAseq) and upon reasonable request from the corresponding author.
2.6. Statistical Analysis
Mean values, standard error of the mean (SEM) values, and unpaired Student’s t-tests were calculated using Prism software (GraphPad). For immunohistochemistry and ELISA, P values <.05 were considered significant.
3. RESULTS
3.1. SHIP-1 deletion increases microglial numbers and mobilization to Aβ plaques without appreciably affecting Aβ load
To determine how SHIP-1 signaling in microglia shapes microglial biology and Aβ-mediated neuropathology, we generated Inpp5dfl/flCx3cr1Ert2Cre mice to delete SHIP-1 from microglia and crossed them with 5xFAD mice (further referred to as 5xFAD Inpp5dΔMG mice), which is a widely used AD mouse model of amyloidosis [34]. Cre-negative 5xFAD Inpp5dfl/fl littermate mice (further referred to as 5xFAD mice) were used as controls. All mice received tamoxifen food at weaning for 2 weeks to induce SHIP-1 deletion and then were returned to normal chow to allow for peripheral Cx3cr1-expressing cells to turnover and regain Inpp5d expression, while allowing long-lived microglia to maintain Inpp5d deficiency [27].
Using these newly generated mouse lines, we found that 5xFAD Inpp5dΔMG mice had more microglia in the cortex and dentate gyrus of the hippocampus as measured by the number of IBA1+ microglia and the total IBA1+ volume at 5 months of age (Figure 1A–C, Supplemental Figure 1A–C). Microglia activation is often accompanied by a shift toward a larger and more amoeboid morphology [3,35]. By measuring IBA1+ volume per microglia, we found that SHIP-1 deficient microglia in both the cortex and dentate gyrus were ~2.5-fold larger in size and qualitatively displayed a more amoeboid morphology compared to microglia from 5xFAD littermate control mice (Figure 1D, Supplemental Figure 1D), suggesting enhanced activation. To determine if loss of SHIP-1 influences microglia physiology in the absence of Aβ pathology, we next assessed microglial numbers and morphology in the cortex of Inpp5dΔMG mice that do not express the 5xFAD transgenes (further referred to as wild-type (WT) Inpp5dΔMG mice) and Inpp5dfl/fl littermate controls (further referred to as WT mice). We found that the number of microglia, IBA1+ volume, and microglia size were unchanged when SHIP-1 was deleted from microglia in non-plaque harboring mice (Supplemental Figure 2A–D). Additionally, microglia from both WT Inpp5dΔMG mice and WT littermate controls qualitatively displayed a similar ramified morphology (Supplemental Figure 2A), suggesting that loss of SHIP-1 does not alter microglial state in the absence of Aβ pathology. Taken together, these results suggest that SHIP-1 functions to restrict microglial number and the adoption of a more activated, ameboid morphology in the presence of Aβ pathology.
Figure 1. SHIP-1-deficiency leads to increased mobilization of microglia to Aβ plaques in 5xFAD mice.
5xFAD Inpp5dfl/flCx3cr1ERT2Cre (5xFAD Inpp5dΔMG mice) and Cre-negative 5xFAD Inpp5dfl/fl littermate controls (5xFAD mice) received tamoxifen food for 2 weeks beginning at 3 weeks of age and then were returned to regular food for the remainder of the experiment. Mice were harvested at 5 months of age to evaluate microgliosis, microglial association with Aβ plaques, and Aβ plaque burden. (A) Representative immunofluorescence staining of IBA1 (green) and Aβ plaques (ThioS, magenta) in the cortex; zoomed in view of a single plaque (inset 1 and 3) and zoomed in view of a single z-plane from a single plaque (inset 2 and 4) showing the number of IBA1+ microglia interacting with the plaque. (B) Enumeration of the number of IBA1+ cells per field of view (FOV). (C) Quantification of total IBA1 staining volume per FOV. (D) Quantification of microglia size by total IBA1 staining volume per IBA1+ microglia. (E) Quantification of the average number of IBA1+ microglia per ThioS+ Aβ plaque. (F) Quantification of the number of IBA1+ microglia within a 15 μm and 30 μm radius of ThioS+ Aβ plaques. (G) Representative immunofluorescence staining of IBA1 (green), Ki67 (blue), and Aβ plaques (ThioS, magenta) in the cortex. Arrows denote Ki67+ IBA1+ cells. (H) Enumeration of the number of Ki67+ IBA1+ cells per FOV. (I) Representative immunofluorescence staining of IBA1 (green), CLEC7A (grey), and Aβ plaques (ThioS, magenta) in the cortex. (J) Quantification of the percentage of IBA1+ CLEC7A+ staining volume per ThioS+ Aβ plaque. Percent IBA1+ CLEC7A+ staining volume was calculated by dividing the total IBA1+ CLEC7A+ staining volume by the total IBA1 staining volume. (K) Representative immunofluorescence staining of Aβ (anti-Aβ D54D2, red) and DAPI (blue) performed on sagittal brain sections. (L) Quantification of the percent area covered by Aβ across whole sagittal brain sections. For (B-E), (H), and (J), each point represents an individual mouse averaged from 6 images across 3 matching brain sections per mouse. For (F), each point represents an individual mouse with an average of 50 plaques from 3 matching brain sections per mouse. For (L), each point represents an individual mouse averaged from 3 matching brain sections per mouse. Statistical significance between experimental groups was calculated by an unpaired Student’s t-test. Error bars represent mean ± SEM.
In 5xFAD Inpp5dΔMG mice, we also observed a striking microglia “clustering” phenotype where microglia were found to surround Aβ to a greater degree when compared to 5xFAD littermate controls in both the cortex and dentate gyrus (Figure 1A, Supplemental Figure 1A). Indeed, in both the cortex and dentate gyrus, we found that SHIP-1 deletion increased the number of microglia per plaque (Figure 1A,E, Supplemental Figure 1A,E) and plaque-associated microglia within a 15μm and 30μm radius of Aβ plaques (Figure 1A,F, Supplemental Figure 1A,F). We next wanted to know if microglial proliferation contributed to this increase in plaque-associated microglia in SHIP-1-deficient 5xFAD mice. Using Ki67 as a marker of proliferating cells, we found that 5xFAD Inpp5dΔMG mice had more Ki67+ IBA1+ cells in the cortex and dentate gyrus compared to 5xFAD littermate controls (Figure 1G,H, Supplemental Figure 1G,H), suggesting that the increase in SHIP-1-deficient microglia numbers is at least, in part, due to increased proliferation. Microglial cell death may also contribute at some level to the increase in microglia numbers observed in SHIP-1-deficient 5xFAD mice. However, it is important to note that we have not been able to detect appreciable levels of microglia cell death by TUNEL staining in the 5xFAD model in our previous studies [28,36]. Therefore, it is more likely that enhanced microglial proliferation contributes to the increase in microglia numbers seen in SHIP-1-deficeint 5xFAD mice.
Given that SHIP-1-deficient microglia were larger in size (Figure 1D, Supplemental Figure 1D) and associated more closely with Aβ plaques (Figure 1F, Supplemental Figure 1F), we next wanted to further investigate whether SHIP-1 loss enhanced microglial activation. To this end, we stained for CLEC7A, which has been shown to be a canonical marker of microglial activation in multiple models of neurodegenerative disease [5,6,28]. Here we found that microglia from 5xFAD Inpp5dΔMG mice exhibited a ~2.5-fold increase in the amount IBA1+ CLEC7A+ volume per plaque compared to 5xFAD littermate controls (Figure 1I,J), suggesting that loss of SHIP-1 leads to increased microglial activation. Overall, these data point towards SHIP-1 functioning to antagonize microglial numbers, activation, and association with Aβ plaques in the 5xFAD mice model of Aβ amyloidosis.
Given the profound response to Aβ plaques displayed by SHIP-1-deficient microglia, we next wanted to assess the impact of SHIP-1 deletion on Aβ plaque load in these mice. We measured pan-Aβ species by staining for D54D2 (anti-Aβ) and found that Aβ plaque burden did not differ in the brains of 5xFAD Inpp5dΔMG mice when compared to 5xFAD littermate controls (Figure 1K,L). Since microglia are critical for the consolidation of soluble Aβ oligomers that are highly neurotoxic into less toxic, insoluble Aβ fibrils and plaques [37], we extracted Aβ using various solubility-based extraction techniques and evaluated the amount of soluble and insoluble Aβ by ELISA. Similar to our total Aβ load findings (Figure 1K,L), soluble (PBS Fraction) and insoluble (Guanidine Fraction) Aβ1–42 and Aβ1–40 levels were unchanged between 5xFAD Inpp5dΔMG mice and 5xFAD littermate controls (Supplemental Figure 3A,B). Together, these data suggest that SHIP-1 loss in microglia does not affect overall Aβ plaque burden in 5xFAD mice, despite the robust increase in microglia numbers and plaque clustering observed in SHIP-1 deficient microglia.
3.2. Microglia lacking SHIP-1 adopt a unique transcriptional profile in response to Aβ-mediated pathology
Given the striking enhancement in the number of plaque-associated microglia observed in 5xFAD Inpp5dΔMG mice, we were next interested in gaining a comprehensive and unbiased assessment of how deletion of SHIP-1 impacts gene expression in response to Aβ-driven neuropathology. To this end, we conducted single-nucleus RNA sequencing (snRNAseq) on cortical brain samples dissected from 5xFAD Inpp5dΔMG mice and 5xFAD littermate controls at 5 months of age, as this was the area of the brain where we originally saw the greatest differences in plaque-associated microglial responses (Figure 1). A total of 20,818 nuclei across both genotypes were arranged by uniform manifold approximation and projection (UMAP) for visualization, and unsupervised clustering revealed 17 distinct clusters (Figure 2A). Clusters were manually defined based on the expression cell type-specific signature genes as glutamatergic neurons (Clusters 0, 1, 3), GABAergic neurons (Clusters 6, 7, 8), unidentified neurons (Clusters 11, 12, 13, 15), astrocytes (Cluster 2), oligodendrocytes (Cluster 4), microglia (Cluster 5), oligodendrocyte precursor cells (OPCs, Cluster 10), and neurovascular-associated cells including vascular leptomeningeal cells (VLMC, Cluster 9), endothelial cells (Cluster 14), and smooth muscle cells/pericytes (Cluster 16) (Figure 2B, Supplemental Table S1). While neurons made up the majority of the recovered nuclei (~66%, 13,722 nuclei), microglia represented 7.56% (1,573 nuclei) of the total nuclei across both genotypes (Figure 2C). A similar number of nuclei were recovered from both genotypes (5xFAD: 10,424 nuclei, 5xFAD Inpp5dΔMG: 10,394 nuclei) and the relative distribution of nuclei across most clusters was similar (Figure 2D). Notably, however, the microglia cluster exhibited an ~4-fold increase in the number of nuclei in the 5xFAD Inpp5dΔMG cortex (1,264 nuclei) compared to the 5xFAD cortex (309 nuclei, Figure 2D–E), corroborating the increase in microglial number observed histologically in 5xFAD Inpp5dΔMG mice (Figure 1A–C, Supplemental Figure 1A–C). We also observed a slight reduction in the number of GABAergic neuron Cluster 7 nuclei from the 5xFAD Inpp5dΔMG cortex (Figure 2E). Overall, this further suggests that deletion of SHIP-1 increases microglia number and contributes to enhanced microgliosis.
Figure 2. snRNAseq differentiates major brain cell types and reveals enhanced microgliosis following SHIP-1 deletion in 5xFAD mice.

snRNAseq of cortices from 5xFAD Inpp5dΔMG mice and 5xFAD littermate controls harvested at 5 months of age. (A) UMAP rendering of 20,818 nuclei from 5xFAD Inpp5dΔMG and 5xFAD samples showing 17 distinct clusters numbered 0–16. The cell type identity was determined by the expression of cell type-specific markers. (B) Gene expression heatmap depicting signature genes used to identify each cluster in (A). (C) Donut chart depicting the frequency of nuclei recovered per cluster across both genotypes. (D) Bar chart showing the relative nuclei distribution across all clusters in each genotype. (E) Relative frequency of nuclei across all clusters normalized to the number of 5xFAD nuclei per cluster (dashed horizontal line). Cluster 5 (microglia) was highly increased in 5xFAD Inpp5dΔMG mice.
We next conducted a deeper analysis of the microglia population. Nuclei from the microglia cluster (Cluster 5) were identified based on the enhanced expression of microglia-specific genes, including Cx3cr1, P2ry12, Hexb, Tmem119, Csf1r, C1qa, Lrmda, and Dock8 (Figure 3A). Differential expression testing revealed a total of 124 differentially expressed genes (DEGs) between 5xFAD Inpp5dΔMG and 5xFAD microglia (Figure 3B, Supplemental Table S2). Notably in 5xFAD Inpp5dΔMG microglia, upregulated genes included Lrmda, Arsb, Cstd, Tgfbr2, Trem2, Ppard, Vav1, Hif1a, and C1qb while Inpp5d, Nfkb1, Tgfbr1, Pi3kr1, and Map4k4 were found to be downregulated. Pathway analysis based on upregulated DEGs in 5xFAD Inpp5dΔMG microglia returned significantly enriched pathways associated with TREM2/DAP12 (TYROBP) signaling and phagocytosis, while pathways including PI3K-AKT-mTOR signaling and lipid metabolism were also enriched but did not reach global significance (Figure 3C, Supplemental Table S3). Additionally, pathway analysis based on downregulated DEGs in 5xFAD Inpp5dΔMG microglia returned significantly enriched pathways associated with IL-3/5/6, TGF-β, MAPK, and EGRF1 signaling (Figure 3D, Supplemental Table S4).
Figure 3. SHIP-1 deletion in 5xFAD mice induces a unique transcriptional shift in microglia.

Microglia cluster analysis following snRNAseq of cortices from 5xFAD Inpp5dΔMG mice and 5xFAD littermate controls harvested at 5 months of age. (A) UMAP rendering of 20,818 nuclei highlighting the microglia cluster (cyan) from Figure 2A expressing microglia signature genes, including Cx3cr1, P2ry12, Hexb, Tmem119, Csf1r, C1qa, Lrmda, and Dock8. (B) Volcano plot depicting DEGs (log2(fold-change) >0.25 or <−0.25 and Bonferroni adjusted P value <.00001) identified between 5xFAD Inpp5dΔMG and 5xFAD microglia. (C) Pathway analysis of DEGs (log2(fold-change) >0 and Bonferroni adjusted P value <.05) that were upregulated in 5xFAD Inpp5dΔMG microglia when compared to 5xFAD microglia. (D) Pathway analysis of DEGs (log2(fold-change) <0 and Bonferroni adjusted P value <.05) that were downregulated in 5xFAD Inpp5dΔMG mice when compared to 5xFAD microglia. (E) UMAP rendering of 1,573 microglia nuclei (from Cluster 5) following re-integration and re-clustering samples showing 4 distinct subclusters numbered 0–3. (F) UMAP rendering of microglia nuclei in (E) split by genotype (5xFAD Inpp5dΔMG: 1,264 nuclei, 5xFAD: 309 nuclei). Subcluster 0 shows an enrichment in 5xFAD Inpp5dΔMG microglia. (G) Bar chart showing the raw frequency of nuclei recovered across all subclusters in each genotype. Subcluster 0 is increased in 5xFAD Inpp5dΔMG microglia. (H) DotPlot colored to show the relative average expression of homeostatic and disease-associated microglia genes. The size of each dot corresponds to the percent of nuclei in each subcluster expressing the gene. (I) Pathway analyses of downregulated DEGs (log2(fold-change) <0 and Bonferroni adjusted P value <.05) enriched in microglia Subcluster 0 versus all other subclusters. For (C), (D), and (I), P values are shown and an asterisk denotes global significance (adjusted P value <.05).
To further explore the composition of the microglia cluster, we re-clustered microglia (1,573 nuclei) and resolved 4 distinct CNS macrophage subclusters (Figure 3E). Strikingly, we found that Subcluster 0 was dominated by microglia from the 5xFAD Inpp5dΔMG cortex (5xFAD Inpp5dΔMG: 1,061 nuclei, 5xFAD: 179 nuclei), a trend similarly seen in Subcluster 1 but to a lesser extent (Figure 3F–G). We compared the average expression of various microglia genes, including homeostatic and DAM genes [5,6,38], across each of the microglia subclusters. Interestingly, Subcluster 0 appeared to closely resemble Subcluster 1 microglia, despite Subcluster 0 displaying enhanced expression of Lrmda. Subcluster 2 appeared to show increased expression of DAM genes, including Ctsb, Ctsd, Apoe, and Trem2, and homeostatic microglia genes, including Cx3cr1, P2ry12, Hexb, Csf1r, and Dock8. Subcluster 3 expressed border-associated macrophage genes, including Mrc1 and F13a1 (Figure 3H). Pathway analysis of DEGs enriched in microglia Subcluster 0 versus all other microglia subclusters (Subclusters 1 and 2, Supplemental Table S5) revealed downregulated pathways pertaining to EGFR1, TNF-α/NF-ĸB, and IL-6 signaling (Figure 3I, Supplemental Table S6). Together, these results suggest that SHIP-1 deletion induces a transcriptional shift in microglia responding to Aβ-mediated pathology that is characterized by increased plaque-response pathways and reduced proinflammatory signaling.
3.3. Loss of SHIP-1 enhances microglial engulfment of Aβ
Since our pathway analysis revealed enhancements in pathways associated with phagocytosis in SHIP-1-deficient microglia compared to microglia from 5xFAD mice (Figure 3C), we predicted that microglia from 5xFAD Inpp5dΔMG mice would exhibit increased engulfment of Aβ. We began by evaluating microglial expression of CD68, which is a canonical marker of microglial phagolysosomes and a commonly used marker of both microglial activation and engulfment [39]. We found that microglial expression of CD68 in the cortex of 5xFAD Inpp5dΔMG mice was significantly increased following SHIP-1 deletion compared to Cre-negative 5xFAD littermate controls (Figure 4A–B). In addition, we observed an ~2.5-fold increase in the amount of CD68 per Aβ plaque (Figure 4A,C). As a secondary approach to evaluate Aβ engulfment in our model, we analyzed the volume of Aβ internalized within IBA1-expressing microglia that colocalized with CD68 using Imaris three-dimensional rendering (Figure 4A) in the cortex. Here, we found that SHIP-1 deletion in microglia resulted in an appreciable increase in the amount of Aβ engulfed within CD68+ phagolysosomes in comparison to microglia from 5xFAD littermate counterparts (Figure 4A,D). In summary, these results further suggest that SHIP-1 antagonizes microglial signaling pathways associated with the internalization of Aβ in 5xFAD mice.
Figure 4. Loss of SHIP-1 in microglia increases CD68 expression and engulfment of Aβ in 5xFAD mice.
5xFAD Inpp5dΔMG mice and 5xFAD littermate controls were harvested at 5 months of age to evaluate IBA1+ microglial CD68 expression and engulfment of Aβ. (A) Representative immunofluorescence staining and Imaris three-dimensional rendering of IBA1 (green), Aβ plaques (ThioS, magenta), and CD68 (blue) in the cortex. The right panel displays the amount of Aβ that localizes with CD68 (orange) in three-dimensional space. (B) Quantification of the total CD68 staining volume per IBA1+ microglia. (C) Quantification of the total CD68 staining volume per ThioS+ Aβ plaque. (D) Quantification of the percentage of Aβ plaque volume that is engulfed within CD68 staining. Each point represents an individual mouse with an average of 50 plaques from 3 matching brain sections per mouse. Statistical significance between experimental groups was calculated by an unpaired Student’s t-test. Error bars represent mean ± SEM.
3.4. SHIP-1 deletion in microglia increases microglial encapsulation of Aβ plaques and rescues AD-associated neuronal dystrophy
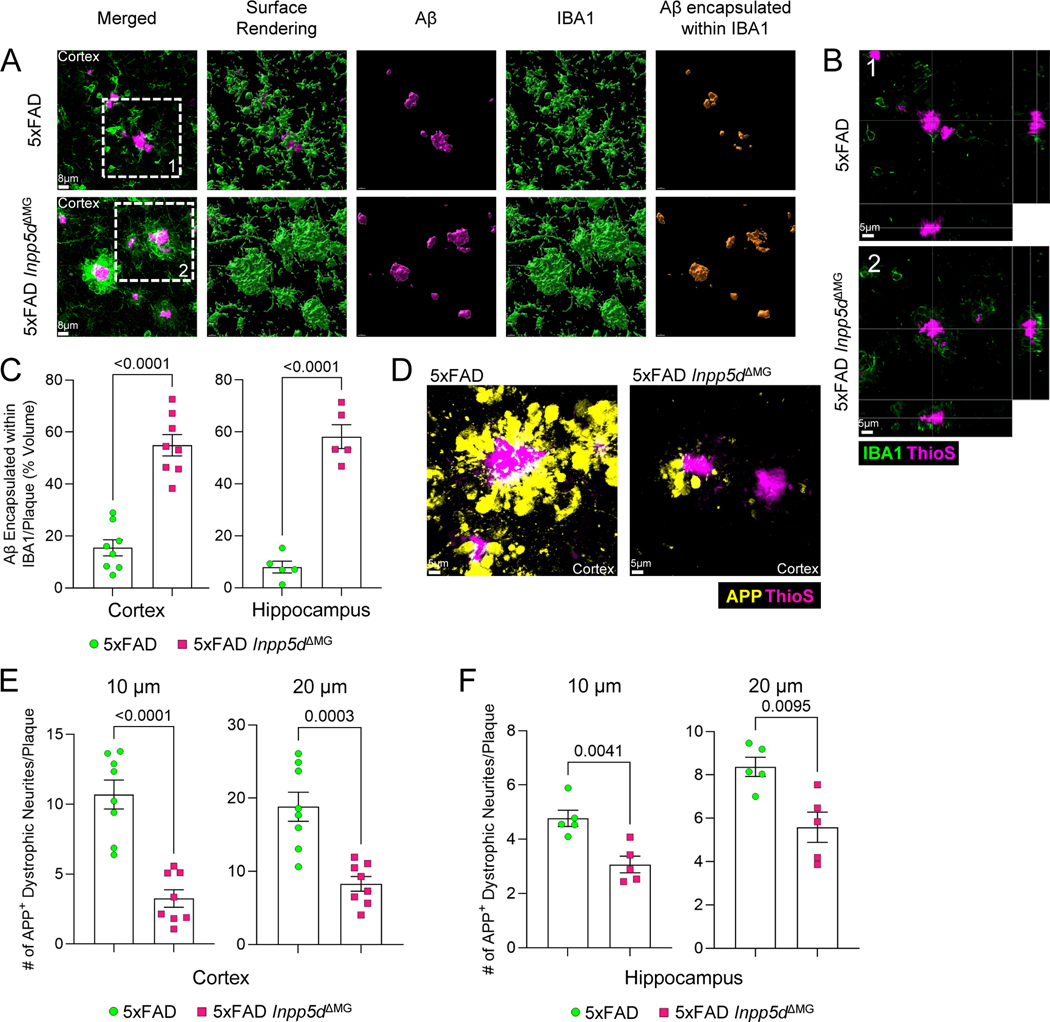
While engulfment has been identified to be a key effector function deployed by microglia to limit Aβ amyloidosis and associated neuropathology [40], the observed increase in Aβ engulfment (Figure 4D) may not fully explain the microglia phenotype observed in 5xFAD Inpp5dΔMG mice. Thus, we were motivated to further explore the striking plaque-associated microglia phenotype observed in microglia from 5xFAD Inpp5dΔMG mice (Figure 1A–F, Supplemental Figure 1A–F) by characterizing the interaction between these microglia and Aβ plaques. Containment of neurotoxic Aβ is a key microglial function that helps to limit neuronal damage caused by neuronal exposure to Aβ aggregate [4,41,42]. Imaris-based three-dimensional rendering of the IBA1+ cells surrounding the plaques indicated that the surface area of Aβ plaques was almost entirely encased by the IBA1+ microglia in 5xFAD Inpp5dΔMG mice (Figure 5A–B) resulting in a ~3-fold increase in the amount of Aβ encapsulated within the microglia projections in the cortex and dentate gyrus of the hippocampus (Figure 5C). In contrast, much of the Aβ staining in 5xFAD littermate controls did not appear to be in direct contact with projections from IBA1-expressing microglia (Figure 5A–B), suggesting that more of the Aβ was exposed to the brain parenchyma.
Figure 5. Deletion of SHIP-1 enhances microglial encapsulation of Aβ plaques and protects against neuronal dystrophy.
5xFAD Inpp5dΔMG and 5xFAD mice were harvested at 5 months of age to evaluate IBA1+ microglia containment and encapsulation of Aβ plaques and neuronal health. (A) Representative immunofluorescence staining and Imaris three-dimensional rendering of IBA1 (green) and ThioS (magenta) in the cortex. The right panel depicts the amount of Aβ that is completely encapsulated by IBA1+ microglia (orange) in three-dimensional space. (B) Representative orthogonal view depicting IBA1+ microglia (green) surrounding Aβ plaques (ThioS, magenta) in the cortex. (C) Quantification of the percentage of the total Aβ plaque volume completely encapsulated by IBA1 staining in the cortex and dentate gyrus of the hippocampus. (D) The formation of dystrophic neurites surrounding plaques in the cortex was determined by staining for APP (yellow) and Aβ (ThioS, magenta). Representative immunofluorescence staining of APP puncta and Aβ plaques in the cortex. (E, F) Quantification of APP+ puncta found within a 10 μm and 20 μm radius of ThioS+ Aβ plaques in the (E) cortex and (F) dentate gyrus of the hippocampus. For cortical analyses, each point represents an individual mouse with an average of 50 plaques from 3 matching brain sections per mouse. For hippocampal analyses, each point represents an individual mouse with an average of 25 plaques from 3 matching brain sections per mouse. Statistical significance between experimental groups was calculated by an unpaired Student’s t-test. Error bars represent mean ± SEM.
It is well known that microglia form barriers around neurotoxic Aβ plaques and that loss of this barrier contributes to increased neuronal damage [2,41]. Since we observed enhanced microglial encapsulation of Aβ plaques in 5xFAD Inpp5dΔMG mice, we hypothesized that SHIP-1 deletion in microglia would lead to improved neuronal health. The buildup of amyloid precursor protein (APP) puncta in neuronal neurites is commonly used as a marker of neuronal dystrophy [28]. Notably, we observed a significant reduction in the number of APP+ dystrophic neurites within a 10 μm and 20 μm radius of Aβ plaques in 5xFAD Inpp5dΔMG mice when compared to 5xFAD littermate controls in both the cortex (Figure 5D–E) and dentate gyrus of the hippocampus (Figure 5F). Therefore, these results suggest that SHIP-1 restricts microglial containment of Aβ plaques and that SHIP-1 deletion in microglia leads to improved neuronal health in 5xFAD mice.
4. DISCUSSION
In this study, we found that SHIP-1 deficiency enhances neuroprotective microglial responses in Aβ-mediated pathology. Conditional deletion of SHIP-1 in microglia led to an enhancement in microglial numbers and association with Aβ plaques in 5xFAD mice. This contributed to a near-complete encapsulation of Aβ plaques by SHIP-1-deficient microglia. Additionally, deletion of SHIP-1 in microglia resulted in an increase in microglial phagocytic capacity, as measured by CD68 immunostaining, and an increase in the amount of Aβ engulfed within microglia, despite overall Aβ load being unchanged. The distinct changes observed in SHIP-1-deficient microglia responding to Aβ plaques led to a dramatic reduction in plaque-associated dystrophic neurites, suggesting that neuronal health was improved following microglial SHIP-1 deletion in 5xFAD mice. Overall, our findings suggest that SHIP-1 functions in CNS-resident microglia to restrict microglial immune and neuroprotective responses to Aβ.
Transcriptional snRNA-seq analysis confirmed that there was an increase in microglia numbers in 5xFAD Inpp5dΔMG mice and further revealed that microglia underwent a unique transcriptional shift following SHIP-1 deletion. Most notably, SHIP-1 deletion in microglia of 5xFAD mice led to the upregulation of pathways associated with TREM2/DAP12 (TYROBP) signaling, phagocytosis, and PI3K-AKT-mTOR signaling. TREM2 and PI3K signaling have been shown to be critical pathways contributing to the activation of microglia and conversion to the DAM phenotype in AD [5,6,38,43]. Further, SHIP-1 deletion in microglia enhanced nuclear lipid metabolism pathways that are thought to be protective in 5xFAD mice [44], suggesting an additional target of microglial SHIP-1 inhibition. Interestingly, SHIP-1 deletion also resulted in the downregulation of IL-6, MAPK, and EGFR1 signaling pathways that are known to be associated with neuroinflammatory processes in microglia [45–47]. TGF-β signaling, a pathway that can antagonize microglial activation and response to Aβ in AD [48], was also reduced in SHIP-1-deficient microglia from 5xFAD mice. Together, these findings suggest that SHIP-1 signaling may regulate proinflammatory processes in microglia and inhibit protective microglial responses to Aβ plaques.
Few studies to date have assessed the roles of SHIP-1 in the in vivo regulation of microglial biology or AD pathogenesis. However, as we were preparing this manuscript, there were two studies published that reported on the effects of Inpp5d deletion on microglia and AD pathology – the first by Castranio et al. [49] using Cx3cr1+ cell-specific Inpp5d knockdown in the APPKM670/671NL/PSEN1Δexon9 mouse model of AD, and the second by Lin et al. [50] using full-body Inpp5d haplodeficiency in 5xFAD mice. All three studies agree that SHIP-1 knockdown in models of Aβ amyloidosis leads to increased microglia numbers, greater microglial association with Aβ plaques, and elevated activation of plaque-associated microglia [49,50]. Together, these studies further highlight that microglial containment and encapsulation of Aβ plaques are likely key effector functions of microglia in response to Aβ-mediated pathology [41,42]. Additionally, spatial transcriptomics and snRNA-seq studies conducted across all three papers found that SHIP-1 deficient microglia were less inflammatory and showed enhanced activation along the DAM phenotype [49,50]. Further, recent in vitro work showed that pharmacological inhibition of SHIP-1 increased microglia-like BV-2 cell phagocytosis of Aβ peptides and dead neurons [11], closely mirroring the enhancement in Aβ engulfment observed in our in vivo studies.
Contrary to the results presented in our paper and the findings reported by Lin et al. [50], SHIP-1 deletion in Cx3cr1-expressing cells from APPKM670/671NL/PSEN1Δexon9 mice appeared to increase fibrillar Aβ plaque burden while neuronal health remained unchanged [49]. This is in contrast to our findings which showed no changes in Aβ burden and reduced neuronal dystrophy, as well as the work by Lin et al., which reported decreased Aβ load and improved neuronal health [50]. Taken together, these findings highlight the complexity surrounding SHIP-1-mediated signaling in AD pathology. There are multiple potential explanations that could help to reconcile these seemingly divergent results. For one, our deletion strategy targeted Inpp5d in microglia prior to disease onset (at 3–4 weeks of age) whereas the Castranio et al. [49] study induced knockdown of Inpp5d expression after disease onset (at 3 months of age), which suggests that differences in the effect of SHIP-1 deletion on neuropathology may be partially influenced by disease state. As a result, inhibiting SHIP-1 signaling at different points during the course of disease may lead to distinct clinical outcomes and this will be important to carefully consider if SHIP-1-targeted approaches are pursued in future AD patient studies. Secondly, both our study and the Castranio et al. [49] work utilized Cx3cr1ERT2Cre Inpp5dfl/fl deletion strategies to target Inpp5d deletion specifically to Cx3cr1-expressing cells, although it is important to note that different Cx3cr1ERT2Cre deleter mouse strains were used between our studies. In contrast, the Lin et al. [50] study used a full-body Inpp5d haplodeficiency model to knockdown Inpp5d expression globally. Inpp5d is known to be expressed by multiple peripheral immune cell lineages including T cells, B cells, and various peripherally-derived myeloid cell populations [12–16]. Therefore, the deletion of SHIP-1 outside of the brain in full-body Inpp5d haplodeficient 5xFAD mice may lead to systemic inflammatory responses or infiltration of the brain by peripheral-derived immune cells which could both influence various aspects of disease progression in 5xFAD mice. Lastly, inherent differences in genetic AD mouse models may contribute at some level to the divergent results observed in Aβ plaque load and neuronal health between studies from Castranio et al. which used APPKM670/671NL/PSEN1Δexon9 mice [49] and the combined work from our group and Lin et al. [50] which employed 5xFAD mice. Therefore, exploring the effects of SHIP-1 modulation in additional AD mouse models will be critical to understanding how SHIP-1 signaling impacts AD pathology.
5. CONCLUSION
In summary, we show that microglia-specific deletion of SHIP-1 in the 5xFAD mouse model of Aβ amyloidosis leads to enhanced numbers of microglia as well as increased microglial mobilization to plaques. Interestingly, SHIP-1 deletion in microglia was not found to significantly affect overall Aβ plaque burden in 5xFAD mice. Further characterization of microglia by single-nucleus RNA sequencing revealed that SHIP-1-deficient microglia in 5xFAD mice adopt a unique transcriptional profile that is characterized by an increase in pathways associated with DAM signaling and phagocytosis. While a modest enhancement in the engulfment of Aβ was observed in SHIP-1-deficient microglia, we found that SHIP-1 deletion in microglia led to almost complete encapsulation of Aβ plaques by microglial processes and a reduction of AD-associated neuropathology. Overall, these findings show that SHIP-1 restricts plaque-associated microglial responses to Aβ plaques and that its deletion reduces neuronal dystrophy in the 5xFAD mouse model of AD, thus identifying SHIP-1 as a critical inhibitor of neuroprotective microglial responses in Aβ-mediated pathology.
Supplementary Material
Supplemental Figure 1. SHIP-1-deficiency leads to increased mobilization of microglia to Aβ plaques in the hippocampus of 5xFAD mice. 5xFAD Inpp5dfl/flCx3cr1ERT2Cre (5xFAD Inpp5dΔMG mice) and Cre-negative 5xFAD Inpp5dfl/fl littermate controls (5xFAD mice) received tamoxifen food for 2 weeks beginning at 3 weeks of age and then were returned to regular food for the remainder of the experiment. Mice were harvested at 5 months of age to evaluate microgliosis and microglial association with Aβ plaques. (A) Representative immunofluorescence staining of IBA1 (green) and Aβ plaques (ThioS, magenta) in the dentate gyrus of the hippocampus; zoomed in view of a single plaque (inset 1 and 3) and zoomed in view of a single z-plane from a single plaque (inset 2 and 4) showing the number of IBA1+ microglia interacting with the plaque. (B) Enumeration of the number of IBA1+ cells per field of view (FOV). (C) Quantification of total IBA1 staining volume per FOV. (D) Quantification of microglia size by total IBA1 staining volume per IBA1+ microglia. (E) Quantification of the average number of IBA1+ microglia per ThioS+ Aβ plaque. (F) Quantification of the number of IBA1+ microglia within a 15 μm and 30 μm radius of ThioS+ Aβ plaques. (G) Representative immunofluorescence staining of IBA1 (green), Ki67 (blue), and Aβ plaques (ThioS, magenta) in the dentate gyrus of the hippocampus. Arrows denote Ki67+ IBA1+ cells. (H) Enumeration of the number of Ki67+ IBA1+ cells per field of view. For (B-E) and (H), each point represents an individual mouse averaged from 3 images across 3 matching brain sections per mouse. For (F), each point represents an individual mouse with an average of 25 plaques from 3 matching brain sections per mouse. Statistical significance between experimental groups was calculated by an unpaired Student’s t-test. Error bars represent mean ± SEM.
Supplemental Figure 3. Deletion of SHIP-1 in microglia does not alter soluble or insoluble Aβ plaque levels in 5xFAD mice. 5xFAD Inpp5dfl/flCx3cr1ERT2Cre (5xFAD Inpp5dΔMG mice) and Cre-negative 5xFAD Inpp5dfl/fl littermate controls (5xFAD mice) received tamoxifen food for 2 weeks beginning at 3 weeks of age and then were returned to regular food for the remainder of the experiment. Mice were harvested at 5 months of age to evaluate soluble and insoluble Aβ plaque load by ELISA in the cortex. (A) Quantification of the amount of soluble (PBS Fraction) and insoluble (Guanidine Fraction) Aβ1–42 measured by ELISA and normalized to total protein. (B) Quantification of the amount of soluble (PBS Fraction) and insoluble (Guanidine Fraction) Aβ1–40 measured by ELISA and normalized to total protein. Each point represents an individual mouse. Statistical significance between experimental groups was calculated by an unpaired Student’s t-test. Error bars represent mean ± SEM.
Supplemental Figure 2. Loss of SHIP-1 in the absence of Aβ pathology does not alter microglia number or state. Inpp5dfl/flCx3cr1ERT2Cre (WT Inpp5dΔMG mice) and Cre-negative Inpp5dfl/fl littermate controls (WT mice) received tamoxifen food for 2 weeks beginning at 3 weeks of age and then were returned to regular food for the remainder of the experiment. Mice were harvested at 5 months of age to evaluate microgliosis and microglial association with Aβ plaques. (A) Representative immunofluorescence staining of IBA1 (green) and Aβ plaques (ThioS, magenta) in the cortex. (B) Enumeration of the number of IBA1+ cells per field of view (FOV). (C) Quantification of total IBA1 staining volume per FOV. (D) Quantification of microglia size by total IBA1 staining volume per IBA1+ microglia. Each point represents an individual mouse averaged from 6 images across 3 matching brain sections per mouse. Statistical significance between experimental groups was calculated by an unpaired Student’s t-test. Error bars represent mean ± SEM.
ACKNOWLEDGEMENTS
We thank members of the Lukens Lab, Price Lab, and Center for Brain Immunology and Glia (BIG) at the University of Virginia for valuable discussions.
FUNDING SOURCES
This work was supported by The National Institutes of Health/National Institute of Aging (1RF1AG071996-01; awarded to J.R.L.), The National Institutes of Health/National Institute of Neurological Disorders and Stroke (R01NS106383; awarded to J.R.L. and R01NS111102; awarded to R.J.P.), The National Institutes of Health/National Institute of Biomedical Imaging and Bioengineering (R01EB030744; awarded to R.J.P.), The Alzheimer’s Association (ADSF-21-816651; awarded to J.R.L.), the Cure Alzheimer’s Fund (awarded to J.R.L.), and The Owens Family Foundation (Awarded to J.R.L.). A.M.J. was supported by the University of Virginia Brain Immunology and Glia NIH T32 (5T32NS115657). A.E.W was supported by the University of Virginia Biotechnology NIH T32 (5T32GM008715). J.D.S. was supported by a University of Virginia Brain Institute Presidential Fellowship. We thank the University of Virginia Brain Institute for funding to support the Leica Stellaris 5 confocal microscope.
Abbreviations:
- Aβ
amyloid beta
- AD
Alzheimer’s disease
- APP
amyloid precursor protein
- CNS
central nervous system
- DAM
disease-associated microglia
- DEG
differentially expressed gene
- FOV
field of view
- LOAD
late-onset AD
- MG
microglia
- OPC
oligodendrocyte precursor cell
- PBS
phosphate buffered saline
- PC
principal component
- snRNAseq
single-nucleus RNA sequencing
- SHIP-1
SH2-domain-containing inositol phosphatase 1
- SEM
standard error of the mean
- ThioS
Thioflavin S
- UMAP
uniform manifold approximation and projection
- UMI
unique molecular identifier
- VLMC
vascular leptomeningeal cell
- WT
wild type
Footnotes
CONSENT STATEMENT
This work did not involve the use of material from human subjects.
CONFLICTS OF INTERESTS: The authors declare no competing financial interests.
REFERENCES
- [1].Alzheimer’s Association. More Than Normal Aging: Understanding Mild Cognitive Impairment. Facts and Figures 2022. [Google Scholar]
- [2].Long JM, Holtzman DM. Alzheimer Disease: An Update on Pathobiology and Treatment Strategies. Cell 2019;179:312–39. 10.1016/J.CELL.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hickman S, Izzy S, Sen P, Morsett L, el Khoury J. Microglia in neurodegeneration. Nature Neuroscience 2018 21:10 2018;21:1359–69. 10.1038/s41593-018-0242-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Song WM, Colonna M. The identity and function of microglia in neurodegeneration. Nature Immunology 2018 19:10 2018;19:1048–58. 10.1038/s41590-018-0212-1. [DOI] [PubMed] [Google Scholar]
- [5].Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK, et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 2017;169:1276–1290.e17. 10.1016/J.CELL.2017.05.018/ATTACHMENT/63D76284-8C53-4D00-93AA-E1E70B1AE6DE/MMC7.XLSX. [DOI] [PubMed] [Google Scholar]
- [6].Deczkowska A, Keren-Shaul H, Weiner A, Colonna M, Schwartz M, Amit I. Disease-Associated Microglia: A Universal Immune Sensor of Neurodegeneration. Cell 2018;173:1073–81. 10.1016/J.CELL.2018.05.003. [DOI] [PubMed] [Google Scholar]
- [7].Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 2013;45:1452. 10.1038/NG.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bellenguez C, Küçükali F, Jansen IE, Kleineidam L, Moreno-Grau S, Amin N, et al. New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nature Genetics 2022 54:4 2022;54:412–36. 10.1038/s41588-022-01024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Jing H, Zhu J-X, Wang H-F, Zhang W, Zheng Z-J, Kong L-L, et al. INPP5D rs35349669 polymorphism with late-onset Alzheimer’s disease: A replication study and meta-analysis. Oncotarget 2016;7:69225–30. 10.18632/ONCOTARGET.12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ando K, Erneux C, Homa M, Houben S, de Fisenne MA, Brion JP, et al. Dysregulation of Phosphoinositide 5-Phosphatases and Phosphoinositides in Alzheimer’s Disease. Front Neurosci 2021;15:126. 10.3389/FNINS.2021.614855/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Pedicone C, Fernandes S, Dungan OM, Dormann SM, Viernes DR, Adhikari AA, et al. Pan-SHIP1/2 inhibitors promote microglia effector functions essential for CNS homeostasis. J Cell Sci 2020;133. 10.1242/JCS.238030/266158/AM/PAN-SHIP1-2-INHIBITORS-PROMOTE-MICROGLIA-EFFECTOR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Pauls SD, Marshall AJ. Regulation of immune cell signaling by SHIP1: A phosphatase, scaffold protein, and potential therapeutic target. Eur J Immunol 2017;47:932–45. 10.1002/EJI.201646795. [DOI] [PubMed] [Google Scholar]
- [13].Mondal S, Subramanian KK, Sakai J, Bajrami B, Luo HR. Phosphoinositide lipid phosphatase SHIP1 and PTEN coordinate to regulate cell migration and adhesion. Mol Biol Cell 2012;23:1219. 10.1091/MBC.E11-10-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Blanco-Menéndez N, del Fresno C, Fernandes S, Calvo E, Conde-Garrosa R, Kerr WG, et al. SHIP-1 Couples to the Dectin-1 hemITAM and Selectively Modulates Reactive Oxygen Species Production in Dendritic Cells in Response to Candida albicans. The Journal of Immunology 2015;195:4466–78. 10.4049/JIMMUNOL.1402874/-/DCSUPPLEMENTAL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Saz-Leal P, del Fresno C, Brandi P, Martínez-Cano S, Dungan OM, Chisholm JD, et al. Targeting SHIP-1 in Myeloid Cells Enhances Trained Immunity and Boosts Response to Infection. Cell Rep 2018;25:1118–26. 10.1016/J.CELREP.2018.09.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kamen LA, Levinsohn J, Cadwallader A, Tridandapani S, Swanson JA. SHIP-1 Increases Early Oxidative Burst and Regulates Phagosome Maturation in Macrophages. The Journal of Immunology 2008;180:7497–505. 10.4049/JIMMUNOL.180.11.7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Liu Q, Sasaki T, Kozieradzki I, Wakeham A, Itie A, Dumont DJ, et al. SHIP is a negative regulator of growth factor receptor-mediated PKB/Akt activation and myeloid cell survival. Genes Dev 1999;13:786. 10.1101/GAD.13.7.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Qin S, Li J, Zhou C, Privratsky B, Schettler J, Deng X, et al. SHIP-1 Regulates Phagocytosis and M2 Polarization Through the PI3K/Akt–STAT5–Trib1 Circuit in Pseudomonas aeruginosa Infection. Front Immunol 2020;11:307. 10.3389/FIMMU.2020.00307/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ye X, Zhang F, Zhou L, Wei Y, Zhang L, Wang L, et al. Selective deletion of SHIP-1 in hematopoietic cells in mice leads to severe lung inflammation involving ILC2 cells. Scientific Reports 2021 11:1 2021;11:1–14. 10.1038/s41598-021-88677-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Poe JC, Fujimoto M, Jansen PJ, Miller AS, Tedder TF. CD22 Forms a Quaternary Complex with SHIP, Grb2, and Shc. Journal of Biological Chemistry 2000;275:17420–7. 10.1074/jbc.m001892200. [DOI] [PubMed] [Google Scholar]
- [21].Mkaddem SB, Benhamou M, Monteiro RC. Understanding Fc receptor involvement in inflammatory diseases: From mechanisms to new therapeutic tools. Front Immunol 2019;10:811. 10.3389/FIMMU.2019.00811/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Peng Q, Malhotra S, Torchia JA, Kerr WG, Coggeshall KM, Humphrey MB. TREM2- and DAP12-dependent activation of PI3K requires DAP10 and is inhibited by SHIP1. Sci Signal 2010;3. 10.1126/SCISIGNAL.2000500/SUPPL_FILE/3_RA38_SM.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Tsai AP, Lin PBC, Dong C, Moutinho M, Casali BT, Liu Y, et al. INPP5D expression is associated with risk for Alzheimer’s disease and induced by plaque-associated microglia. Neurobiol Dis 2021;153:105303. 10.1016/J.NBD.2021.105303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Castillo E, Leon J, Mazzei G, Abolhassani N, Haruyama N, Saito T, et al. Comparative profiling of cortical gene expression in Alzheimer’s disease patients and mouse models demonstrates a link between amyloidosis and neuroinflammation. Scientific Reports 2017 7:1 2017;7:1–16. 10.1038/s41598-017-17999-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Pedicone C, Fernandes S, Matera A, Meyer ST, Loh S, Ha JH, et al. Discovery of a novel SHIP1 agonist that promotes degradation of lipid-laden phagocytic cargo by microglia. IScience 2022;25:104170. 10.1016/J.ISCI.2022.104170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wang JW, Howson JM, Ghansah T, Desponts C, Ninos JM, May SL, et al. Influence of SHIP on the NK repertoire and allogeneic bone marrow transplantation. Science (1979) 2002;295:2094–7. 10.1126/SCIENCE.1068438/SUPPL_FILE/1068440S3_THUMB.GIF. [DOI] [PubMed] [Google Scholar]
- [27].Yona S, Kim KW, Wolf Y, Mildner A, Varol D, Breker M, et al. Fate Mapping Reveals Origins and Dynamics of Monocytes and Tissue Macrophages under Homeostasis. Immunity 2013;38:79–91. 10.1016/J.IMMUNI.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ennerfelt H, Frost EL, Shapiro DA, Holliday C, Zengeler KE, Voithofer G, et al. SYK coordinates neuroprotective microglial responses in neurodegenerative disease. Cell 2022;185:4135–52. 10.1016/J.CELL.2022.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, et al. Integrated analysis of multimodal single-cell data. Cell 2021;184:3573–3587.e29. 10.1016/J.CELL.2021.04.048/ATTACHMENT/1E5EB5C1-59EE-4B2B-8BFA-14B48A54FF8F/MMC3.XLSX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol 2019;20:1–15. 10.1186/S13059-019-1874-1/FIGURES/6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:1–21. 10.1186/S13059-014-0550-8/FIGURES/9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Xie Z, Bailey A, Kuleshov M v., Clarke DJB, Evangelista JE, Jenkins SL, et al. Gene Set Knowledge Discovery with Enrichr. Curr Protoc 2021;1:e90. 10.1002/CPZ1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Clarke DJB, Jeon M, Stein DJ, Moiseyev N, Kropiwnicki E, Dai C, et al. Appyters: Turning Jupyter Notebooks into data-driven web apps. Patterns 2021;2:100213. 10.1016/j.patter.2021.100213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Richard BC, Kurdakova A, Baches S, Bayer TA, Weggen S, Wirths O. Gene Dosage Dependent Aggravation of the Neurological Phenotype in the 5XFAD Mouse Model of Alzheimer’s Disease. Journal of Alzheimer’s Disease 2015;45:1223–36. 10.3233/JAD-143120. [DOI] [PubMed] [Google Scholar]
- [35].Nagayach A, Patro N, Patro I. Microglia in the Physiology and Pathology of Brain. Proceedings of the National Academy of Sciences, India Section B: Biological Sciences 2015 86:4 2015;86:781–94. 10.1007/S40011-015-0585-Y. [DOI] [Google Scholar]
- [36].Kumar S, Budhathoki S, Oliveira CB, Kahle AD, Calhan OY, Lukens JR, et al. Role of the Caspase-8/RIPK3 axis in Alzheimer’s disease pathogenesis and Aβ-induced NLRP3 inflammasome activation. JCI Insight 2023. 10.1172/JCI.INSIGHT.157433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Sengupta U, Nilson AN, Kayed R. The Role of Amyloid-β Oligomers in Toxicity, Propagation, and Immunotherapy. EBioMedicine 2016;6:42. 10.1016/J.EBIOM.2016.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhou Y, Song WM, Andhey PS, Swain A, Levy T, Miller KR, et al. Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nature Medicine 2020 26:1 2020;26:131–42. 10.1038/s41591-019-0695-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Walker DG, Lue LF. Immune phenotypes of microglia in human neurodegenerative disease: Challenges to detecting microglial polarization in human brains. Alzheimers Res Ther 2015;7:1–9. 10.1186/S13195-015-0139-9/FIGURES/2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Huang Y, Happonen KE, Burrola PG, O’Connor C, Hah N, Huang L, et al. Microglia use TAM receptors to detect and engulf amyloid β plaques. Nature Immunology 2021 22:5 2021;22:586–94. 10.1038/s41590-021-00913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Yuan P, Condello C, Keene CD, Wang Y, Bird TD, Paul SM, et al. TREM2 Haplodeficiency in Mice and Humans Impairs the Microglia Barrier Function Leading to Decreased Amyloid Compaction and Severe Axonal Dystrophy. Neuron 2016;90:724–39. 10.1016/J.NEURON.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Condello C, Yuan P, Schain A, Grutzendler J. Microglia constitute a barrier that prevents neurotoxic protofibrillar Aβ42 hotspots around plaques. Nature Communications 2015 6:1 2015;6:1–14. 10.1038/ncomms7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Wang S, Sudan R, Peng V, Zhou Y, Du S, Yuede CM, et al. TREM2 drives microglia response to amyloid-β via SYK-dependent and -independent pathways. Cell 2022;185:4153–4169.e19. 10.1016/J.CELL.2022.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Malm T, Mariani M, Donovan LJ, Neilson L, Landreth GE. Activation of the nuclear receptor PPARδ is neuroprotective in a transgenic mouse model of Alzheimer’s disease through inhibition of inflammation. J Neuroinflammation 2015;12:1–15. 10.1186/S12974-014-0229-9/FIGURES/9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Bachstetter AD, Xing B, de Almeida L, Dimayuga ER, Watterson DM, van Eldik LJ. Microglial p38α MAPK is a key regulator of proinflammatory cytokine up-regulation induced by toll-like receptor (TLR) ligands or beta-amyloid (Aβ). J Neuroinflammation 2011;8:1–12. 10.1186/1742-2094-8-79/FIGURES/6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Lyra e Silva NM, Gonçalves RA, Pascoal TA, Lima-Filho RAS, Resende E de PF, Vieira ELM, et al. Pro-inflammatory interleukin-6 signaling links cognitive impairments and peripheral metabolic alterations in Alzheimer’s disease. Translational Psychiatry 2021 11:1 2021;11:1–15. 10.1038/s41398-021-01349-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Qu W, Tian D, Guo Z, Fang J, Zhang Q, Yu Z, et al. Inhibition of EGFR/MAPK signaling reduces microglial inflammatory response and the associated secondary damage in rats after spinal cord injury. J Neuroinflammation 2012;9:1–14. 10.1186/1742-2094-9-178/FIGURES/7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Huang WC, Yen FC, Shie FS, Pan CM, Shiao YJ, Yang CN, et al. TGF-β1 blockade of microglial chemotaxis toward Aβ aggregates involves SMAD signaling and down-regulation of CCL5. J Neuroinflammation 2010;7:1–11. 10.1186/1742-2094-7-28/FIGURES/5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Castranio EL, Hasel P, Haure-Mirande J-V, Ramirez Jimenez A v, Wade Hamilton B, Kim RD, et al. Microglial INPP5D limits plaque formation and glial reactivity in the PSAPP mouse model of Alzheimer’s disease. Alzheimer’s & Dementia 2022. 10.1002/ALZ.12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Lin PB- C, Tsai AP- Y, Soni D, Lee-Gosselin A, Moutinho M, Puntambekar SS, et al. INPP5D deficiency attenuates amyloid pathology in a mouse model of Alzheimer’s disease. Alzheimer’s & Dementia 2022. 10.1002/ALZ.12849. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. SHIP-1-deficiency leads to increased mobilization of microglia to Aβ plaques in the hippocampus of 5xFAD mice. 5xFAD Inpp5dfl/flCx3cr1ERT2Cre (5xFAD Inpp5dΔMG mice) and Cre-negative 5xFAD Inpp5dfl/fl littermate controls (5xFAD mice) received tamoxifen food for 2 weeks beginning at 3 weeks of age and then were returned to regular food for the remainder of the experiment. Mice were harvested at 5 months of age to evaluate microgliosis and microglial association with Aβ plaques. (A) Representative immunofluorescence staining of IBA1 (green) and Aβ plaques (ThioS, magenta) in the dentate gyrus of the hippocampus; zoomed in view of a single plaque (inset 1 and 3) and zoomed in view of a single z-plane from a single plaque (inset 2 and 4) showing the number of IBA1+ microglia interacting with the plaque. (B) Enumeration of the number of IBA1+ cells per field of view (FOV). (C) Quantification of total IBA1 staining volume per FOV. (D) Quantification of microglia size by total IBA1 staining volume per IBA1+ microglia. (E) Quantification of the average number of IBA1+ microglia per ThioS+ Aβ plaque. (F) Quantification of the number of IBA1+ microglia within a 15 μm and 30 μm radius of ThioS+ Aβ plaques. (G) Representative immunofluorescence staining of IBA1 (green), Ki67 (blue), and Aβ plaques (ThioS, magenta) in the dentate gyrus of the hippocampus. Arrows denote Ki67+ IBA1+ cells. (H) Enumeration of the number of Ki67+ IBA1+ cells per field of view. For (B-E) and (H), each point represents an individual mouse averaged from 3 images across 3 matching brain sections per mouse. For (F), each point represents an individual mouse with an average of 25 plaques from 3 matching brain sections per mouse. Statistical significance between experimental groups was calculated by an unpaired Student’s t-test. Error bars represent mean ± SEM.
Supplemental Figure 3. Deletion of SHIP-1 in microglia does not alter soluble or insoluble Aβ plaque levels in 5xFAD mice. 5xFAD Inpp5dfl/flCx3cr1ERT2Cre (5xFAD Inpp5dΔMG mice) and Cre-negative 5xFAD Inpp5dfl/fl littermate controls (5xFAD mice) received tamoxifen food for 2 weeks beginning at 3 weeks of age and then were returned to regular food for the remainder of the experiment. Mice were harvested at 5 months of age to evaluate soluble and insoluble Aβ plaque load by ELISA in the cortex. (A) Quantification of the amount of soluble (PBS Fraction) and insoluble (Guanidine Fraction) Aβ1–42 measured by ELISA and normalized to total protein. (B) Quantification of the amount of soluble (PBS Fraction) and insoluble (Guanidine Fraction) Aβ1–40 measured by ELISA and normalized to total protein. Each point represents an individual mouse. Statistical significance between experimental groups was calculated by an unpaired Student’s t-test. Error bars represent mean ± SEM.
Supplemental Figure 2. Loss of SHIP-1 in the absence of Aβ pathology does not alter microglia number or state. Inpp5dfl/flCx3cr1ERT2Cre (WT Inpp5dΔMG mice) and Cre-negative Inpp5dfl/fl littermate controls (WT mice) received tamoxifen food for 2 weeks beginning at 3 weeks of age and then were returned to regular food for the remainder of the experiment. Mice were harvested at 5 months of age to evaluate microgliosis and microglial association with Aβ plaques. (A) Representative immunofluorescence staining of IBA1 (green) and Aβ plaques (ThioS, magenta) in the cortex. (B) Enumeration of the number of IBA1+ cells per field of view (FOV). (C) Quantification of total IBA1 staining volume per FOV. (D) Quantification of microglia size by total IBA1 staining volume per IBA1+ microglia. Each point represents an individual mouse averaged from 6 images across 3 matching brain sections per mouse. Statistical significance between experimental groups was calculated by an unpaired Student’s t-test. Error bars represent mean ± SEM.