SUMMARY
The 2013 Ebola epidemic in Central and West Africa heralded the emergence of wide-spread, highly pathogenic viruses. The successful recombinant vector vaccine against Ebola (rVSVΔG-ZEBOV-GP) will limit future outbreaks, but identifying mechanisms of protection is essential to protect the most vulnerable. Vaccine-induced antibodies are key determinants of vaccine efficacy, yet the mechanism by which vaccine-induced antibodies prevent Ebola infection remains elusive. Here, we exploit a break in long-term vaccine efficacy in non-human primates to identify predictors of protection. Using unbiased humoral profiling that captures neutralization and Fc-mediated functions, we find that antibodies specific for soluble glycoprotein (sGP) drive neutrophil-mediated phagocytosis and predict vaccine-mediated protection. Similarly, we show that protective sGP-specific monoclonal antibodies have elevated neutrophil-mediated phagocytic activity compared with non-protective antibodies, highlighting the importance of sGP in vaccine protection and monoclonal antibody therapeutics against Ebola virus.
Graphical abstract
In brief
Gunn et al. show that the commercially available Ebola virus vaccine leads to sustained protection in a macaque model of partial protection when antibodies are directed against the soluble glycoprotein of Ebola, a target previously thought to have a minimal role in protection against disease.
INTRODUCTION
The large-scale Ebola virus epidemic in West Africa from 2013 to 2016 and the recurrent outbreaks in the region and across the Democratic Republic of Congo have highlighted that Ebola continues to be a threat to human health. The recombinant vesicular stomatitis virus (rVSV)-vectored vaccine against Ebola virus (rVSVΔG-ZEBOV-GP) has shown high levels of protection in preclinical non-human primate models1 and is highly immunogenic in humans.2–6 rVSVΔG-ZEBOV-GP vaccination demonstrated 100% efficacy in a ring-vaccination trial conducted during the West African epidemic,7 leading to licensure as ERVEBO, and is now administered to at-risk populations in West and Central Africa. While protection has been linked to robust vaccine-induced humoral immune responses with durable neutralizing antibody titers, neutralization alone does not appear to be a strong correlate of protection.8,9 Therefore, the precise mechanism(s) by which vaccine-induced antibodies confer protection remains unclear.
In humans, the vaccine induces neutralizing antibody responses by 21 days post-immunization, and titers have been reported to either remain stable or increase through at least 2 years post-vaccination.2,4,5 Early non-human primate (NHP) studies provide strong evidence for a role for antibodies in rVSVΔG-ZEBOV-GP vaccine-induced protection against lethal Ebola virus challenge.10 Doses as low as 10 plaque-forming units (PFUs) in NHPs elicit detectable levels of neutralizing antibodies by 14 days post-vaccination that peak between 28 and 35 days post-vaccination9,11 and provide complete protection in vaccinated NHPs against Ebola challenge at 42 days post-vaccination,12 with neutralizing antibody levels detectable by 14 days post-vaccination and peaking between 28 and 35 days post-vaccination.9,11 However, mounting data from the monoclonal antibody therapeutics community have highlighted the importance of additional antibody functions in protection against Ebola virus.13–15
In addition to neutralization, antibodies can provide antiviral protection through the induction of innate immune effector functions, including phagocytosis and cellular cytotoxicity mediated by an array of innate immune cells, including monocytes, macrophage, neutrophils, and natural killer (NK) cells. Analysis of a large panel of Ebola virus glycoprotein (GP)-specific monoclonal antibodies demonstrated the critical importance of both neutralization and innate immune effector functions in protection from infection.13,16,17 Specifically, partial or non-neutralizing antibodies able to recruit monocyte phagocytic and NK cell functions were able to provide protection from infection. Moreover, strongly neutralizing antibodies that did not provide protection from infection exhibited compromised innate immune recruiting Fc functions. Collectively, these studies highlighted the critical importance of both the antigen-binding (Fab) and constant (Fc) domain of antibodies in the control of Ebola virus infection. Furthermore, analysis of antibody profiles in a survivor of Ebola virus disease points to the evolution of a unique antibody isotype and immunoglobulin G (IgG) subclass profile among survivors linked to durable and functional humoral immunity.18 Furthermore, these functional antibody profiles of Ebola survivors, mimicked with monoclonal antibodies, demonstrated protection in a post-exposure mouse model.19 However, whether rVSVΔG-ZEBOV-GP-induced antibodies also leverage innate immune effector functions to provide protection from infection is largely unclear. The main antigenic target for Ebola virus-specific vaccines and therapeutics is the viral GP that mediates viral attachment and entry into host cells.20 However, the main protein product of the GP gene is not the full-length GP but rather is a truncated form of GP called soluble GP (sGP). Production of the full-length GP requires the viral polymerase stutters at the 7U editing site, adding another uridine and allowing transcription of the remaining GP gene that includes GP1, the mucin domain, and GP2.21,22 sGP and GP are produced at approximately a 3:1 ratio,21,23 as sGP lacks the transmembrane domain of the GP. It is secreted outside of the infected cell. sGP has profound immunomodulatory functions24 and is thought to act as an antibody sink, thereby acting as a critical factor in viral pathogenesis. Given that GP and sGP share overlapping epitopes,24 antibodies induced by rVSVΔG-ZEBOV-GP can theoretically target both sGP and GP. Interestingly, several sGP-specific monoclonal antibodies have shown some level of protective efficacy in animal models,13,25,26 highlighting the potentially protective role of sGP-specific antibodies, in addition to GP-specific antibodies, against Ebola virus disease. However, whether sGP, GP, or responses to both proteins are associated with vaccine-mediated protection remains unclear.
Here, we have comprehensively and deeply profiled the humoral immune response across three groups of rVSVΔG-ZE-BOV-GP-immunized Mauritian cynomolgus macaques, each challenged with a lethal dose of Ebola virus either at peak immunogenicity at 42 days post-vaccination or at long-term vaccine durability time points of 3 months and 1 year post-vaccination. All 42 days post-vaccination animals survived challenge. Yet in this study, protective vaccine efficacy declined in the 3 month and 1 year post-vaccination animal groups, providing a unique and valuable opportunity to explore the acute and long-term correlates of immunity against Ebola virus infection. Analysis of vaccine-induced humoral features, including neutralizing activity, GP- and sGP-specific antibody isotypes, and subclass titer, and induction of innate immune effector functions demonstrated that surviving animals generate elevated levels of sGP-specific antibodies able to drive antibody-mediated neutrophil phagocytosis. Moreover, multivariate analysis across the immunized animals highlights the critical importance of sGP-specific antibody function in protection from infection over neutralizing activity. Furthermore, comparative analysis between protective and non-protective sGP-specific monoclonal antibodies demonstrated that protective antibodies induced significantly higher levels of sGP-specific neutrophil-phagocytic activity. Together, these data strongly argue for the unexpected but critical role for neutrophil-mediated sGP clearance as a key mechanism for antibody-mediated protection from Ebola infection.
RESULTS
Vaccine efficacy diminished over time in Mauritian cynomolgus macaques and was not associated with antibody titers or neutralization
With increasing Ebola virus breakthroughs among vaccinated populations,27,28 defining the durability profiles of vaccine-induced immunity is a key concern of vaccine-mediated protection. To determine the durability of protection elicited by the rVSVΔG-ZEBOV-GP vaccine, Mauritian cynomolgus macaques were vaccinated and challenged with 1,000 PFU Ebola virus at 42 days, 3 months, and 1 year post-vaccination (Table 1). Consistent with previous studies,1 all animals that were challenged 42 days post-vaccination survived (Figure 1A). Conversely, two of six animals survived challenge at 3 months post-vaccination (33.33% protection), and three of seven animals survived challenge 1 year post-vaccination (42.8% protection). Surviving animals were an average age of 9.97 years old (range 9.01–10.825), whereas non-surviving animals were an average age of 9.58 years old (range 7.5–10.71). The differences in ages between surviving and non-surviving animals were not statistically different by t test. Surviving animals were 33.3% male (3/9) and 66.67% female (6/9), and non-surviving animals were 62.5% male (5/8) and 37.5% female (3/8). The differences in sex between surviving and non-surviving animals were not statistically different by a chi-squared test. Of note, all vaccinated animals were challenged shortly after transport into a BSL-4 facility, potentially introducing additional stress to the challenged animals. Analysis of rVSVΔG-ZEBOV-GP replication at 1, 3, and 7 days post-vaccination showed similar kinetics of vaccine vector replication and clearance across challenge groups (Figure S1) and between survivors and non-survivors. Differences in survival across the groups offered a unique opportunity to mine for correlates of vaccine-mediated immunity induced by rVSVΔG-ZEBOV-GP.
Table 1.
Cohort analyzed in this study including number of animals in each group and vaccine doses in plaque-forming units (PFUs)
| Cohort | No. of animals | Vaccine dose (PFU V920) |
|---|---|---|
|
| ||
| 1 year | 7 | 3 × 106 (n = 3) or 3 × 104 |
| 3 months | 6 | 3 × 106 |
| 42 days | 4 | 3 × 106 |
| Unvaccinated control – 42 days | 2 | 0 |
Figure 1. Durable vaccine efficacy is diminished past 3 months post-vaccination.
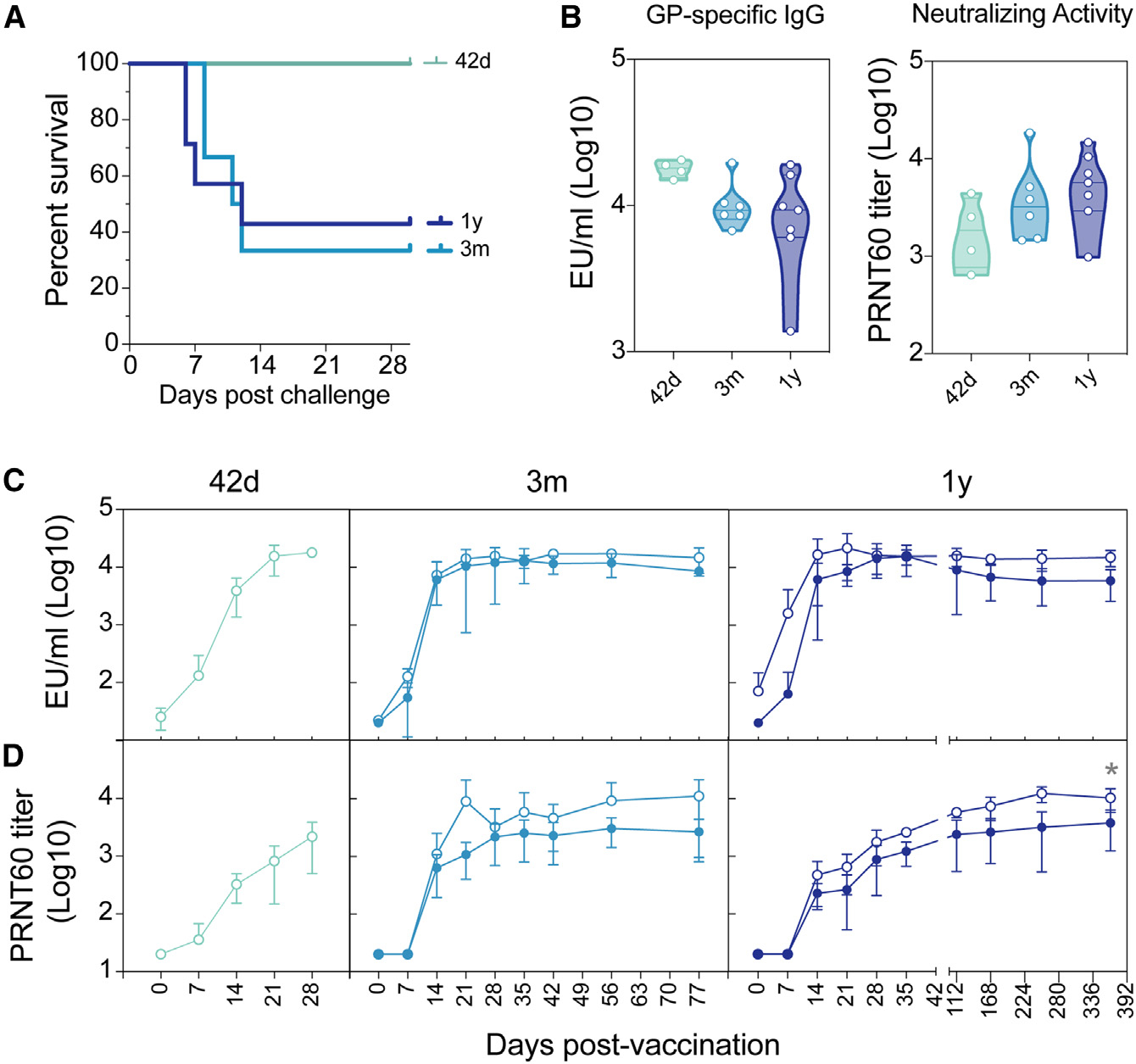
(A) Survival of vaccinated NHP challenged at either 42 days (green line), 3 months (light blue line), or 1 year (dark blue line) post-vaccination.
(B) GP-specific IgG ELISA titers (left graph) and neutralizing antibody titers (right graph) were measured at the final time point before viral challenge. The ELISA lower limit of quantitation (LLOQ) value of 13.62 was used to graph samples that were below the limit of detection (LOD), and the neutralization LLOQ value of 35 was used to graph samples that were below the LOD.
(C) GP-specific IgG ELISA titers were measured indicated time points post-vaccination for vaccinated NHPs that went on to survive challenge (open circles) and NHPs that did not survive challenge (closed circle). LLOQ value of 13.62 was used to graph samples that were below the LOD. Error bars represent the SD of animals within each group.
(D) PRNT60 neutralizing antibody levels were measured using rVSVΔG-ZEBOV-GP vaccination for vaccinated NHPs that went on to survive challenge (open circles) and NHPs that did not survive challenge (closed circles).Statistical analysis was performed using two-way repeated measures (RM) ANOVA with Sidak’s multiple comparisons test. Error bars represent the standard deviation of animals within each group. LLOQ value of 35 was used to graph samples that were below the LOD.
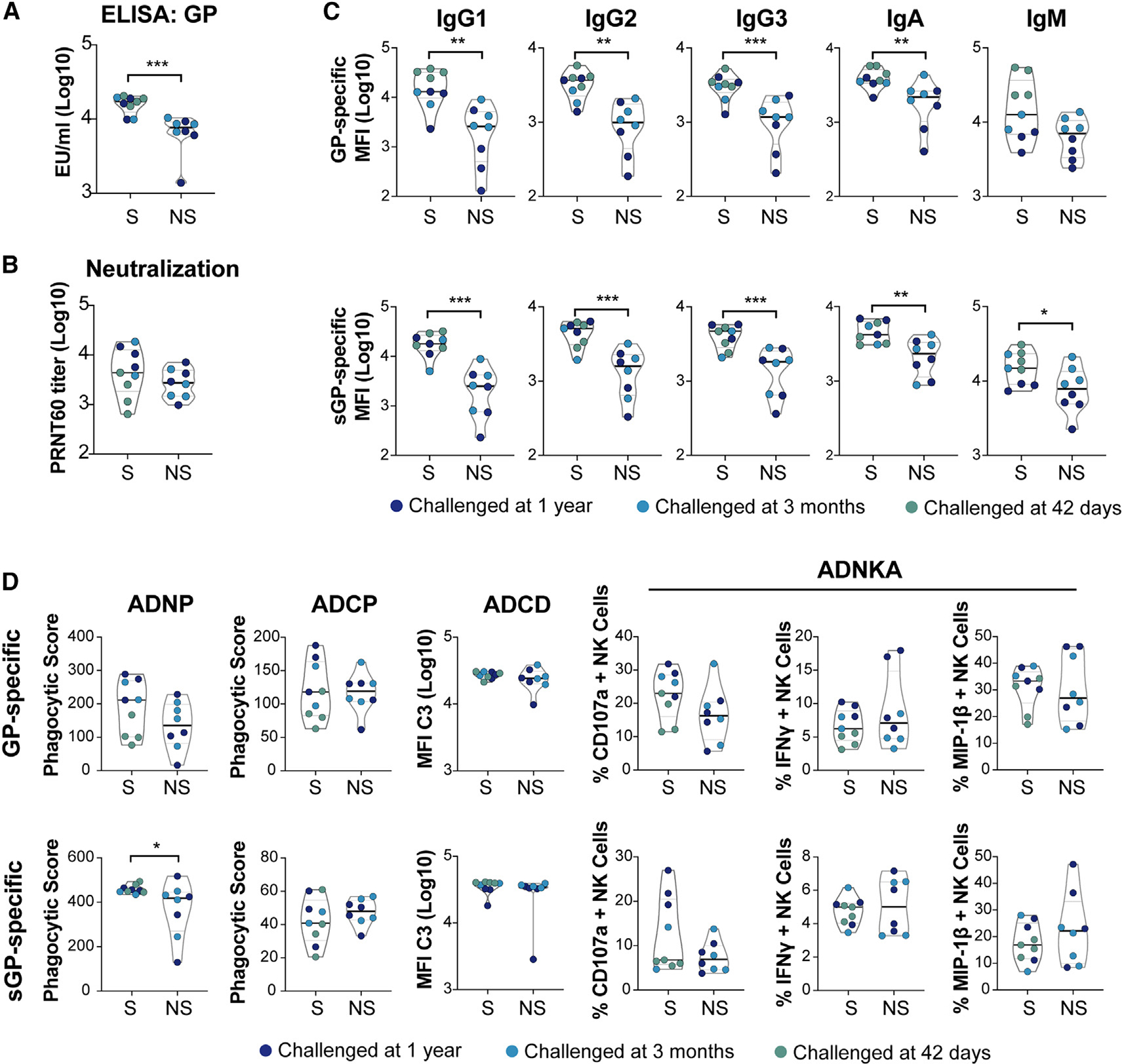
Surprisingly, GP-specific IgG titers and neutralizing titers were nearly equivalent (Figure 1B) across all vaccine groups prior to challenge, highlighting the durability of the humoral response out to 1 year post-vaccination. Survivors showed a trend toward higher antibody responses than non-survivors, but the kinetics and durability of vaccine-specific antibody titers were not statistically different across survivors and non-survivors (Figure 1C). Similarly, neutralizing antibody titers showed a tendency to be higher in survivors among the animals that were challenged at 3 months and were significantly higher among animals that were challenged 1 year following vaccination (Figure 1D), highlighting the important, albeit not perfect, resolving power of neutralization as a correlate of rVSVΔG-ZEBOV-GP-specific immunity against Ebola virus.
sGP-specific, but not GP-specific, antibody subclass variation tracks with protection
Given the functional importance of each antibody isotype/subclass in pathogen-specific immunity and the ability of vaccines to elicit unique antibody humoral profiles,29 we next profiled the subclass-/isotype-specific response across all vaccinated animals. Both GP- and sGP-specific IgG subclasses (IgG1, IgG2, IgG3) and isotypes (IgA, IgM) were quantified at multiple time points following vaccination. Robust GP-specific IgG responses were observed across all immunized animals soon after vaccination (Figure 2A), yet GP-specific titers tended to wane more rapidly in non-survivors compared with survivors in both the 3 month and 1 year challenge groups. Similar profiles were observed in GP-specific IgG2, IgG3, IgA, and IgM responses. In contrast, sGP-specific immunity was strikingly different across survivors and non-survivors, particularly in the 1 year challenge group (Figure 2B). sGP-specific subclass/isotype selection was similar 42 days after challenge in all 3 challenge groups and between survivors and non-survivors. This is consistent with universal protection of animals challenged at this time point. However, at later time points, when only some animals survived the challenge, survivors tended to have higher levels of sGP-specific IgG1, IgG2, IgG3, and IgM before challenge. This was true for animals in both the 3 month and 1 year challenge groups. These differences were most pronounced among animals challenged 1 year following immunization. Lower sGP-specific IgG1, IgG2, IgG3, and IgA responses prior to challenge in non-survivors were all statistically significant (Figure 2B). Surprisingly, non-survivors in the 1 year challenge group diverged in sGP-specific immune profiles from the start of the study, suggesting that, rather than progressive waning of immunity, animals that would not be protected at 1 year post-vaccination had a unique antibody profile that was induced shortly after vaccine administration. This suggests that early sGP responses to vaccination could serve as a predictor of later failure of vaccine-mediated immunity.
Figure 2. Antibody levels against sGP, but not GP, are associated with in durable protection of vaccinated NHP.
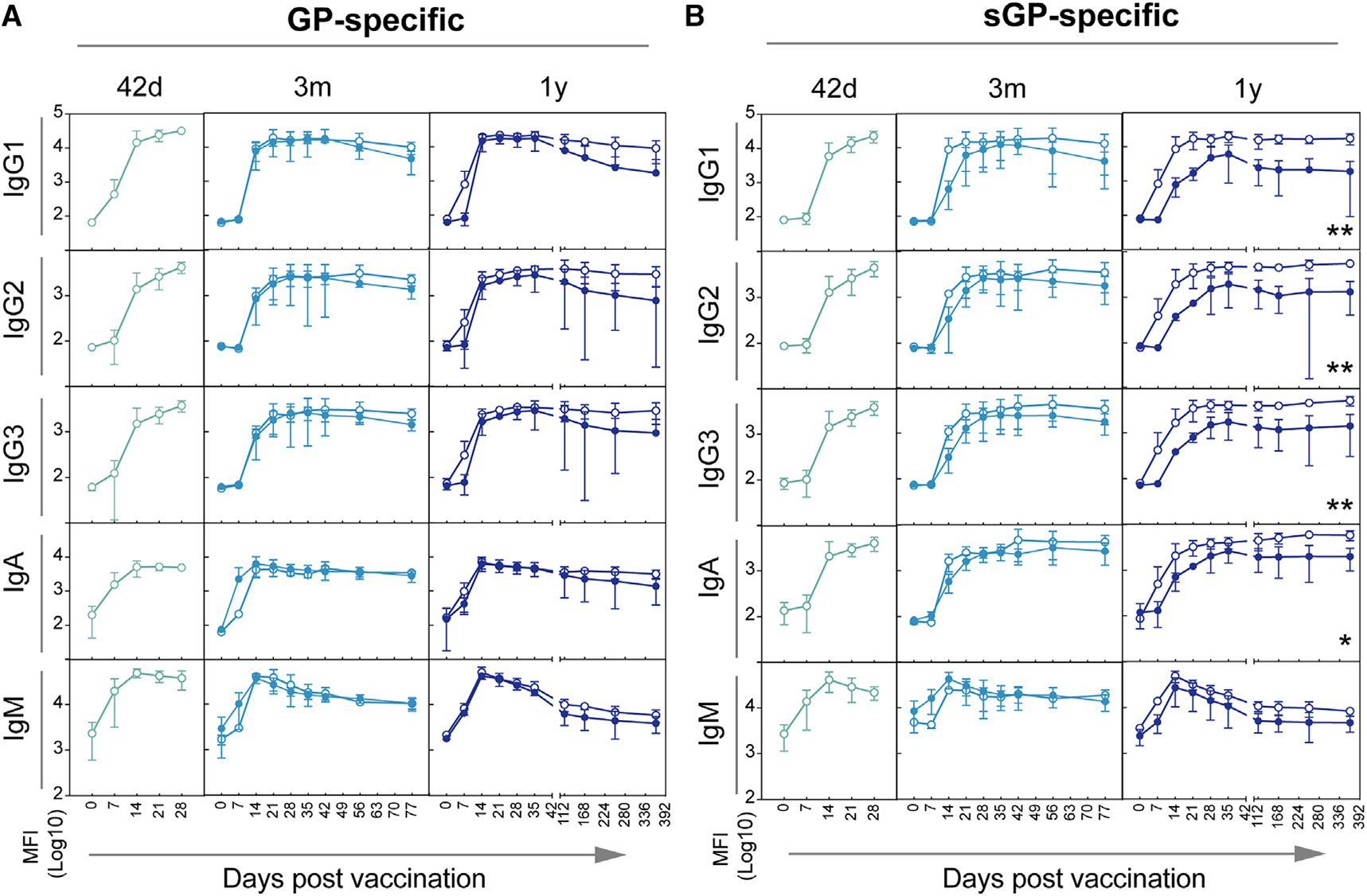
GP-specific (A) and sGP-specific (B) IgG1, IgG2, IgG3, IgA, and IgM levels were measured at indicated time points post-vaccination for vaccinated NHPs that went on to survive (open circles) and NHPs that did not survive challenge (closed circles). Animals challenged at 42 days post vaccination are shown in the left graph (green lines), animals challenged at 3 months post vaccination are shown in the middle graph (light blue lines), and animals challenged at 1 year post-vaccination are shown in the left graph (dark blue lines). Statistical analysis was performed using two-way RM ANOVA with Sidak’s multiple comparisons test. Error bars represent the SD of animals within each group.
Subclass/isotype titers are stronger correlates of protection than neutralization at the time of challenge
While only two animals survived following challenge at 3 months post-vaccination, the sGP-binding antibody levels demonstrated a trend toward higher levels in animals that survived compared with the non-surviving animals prior to challenge (Figure 2B). Although this difference did not reach statistical significance when each time point was analyzed separately, we hypothesized that sGP-specific levels of antibodies may be a correlate of protection, regardless of post-vaccination challenge time point, supported by our finding that macaques that were protected at 1 year diverged in their immune response from unprotected macaques shortly after vaccination. Thus, to increase power, we compared the overall levels of GP antibodies by ELISA and neutralization as well as GP- and sGP-specific isotype/subclass levels in surviving and non-surviving animals across all three groups of animals collectively at the final time point prior to challenge.
As observed across groups, GP titers by ELISA were higher in survivors compared with non-survivors (Figure 3A). Conversely, no difference was observed in neutralization across survivors and non-survivors (Figure 3B). Interestingly, while differences were also observed in GP-specific IgG1, IgG2, IgG3, and IgA levels in survivors, these differences were more significant for sGP-specific antibody levels across all the class-switched antibody isotypes/subclasses (Figure 3C). These data point to a broad reduction in the level of humoral immunity in non-protected animals, with an unexpected and more profound reduction in sGP-specific immunity in non-survivors.
Figure 3. Both GP- and sGP-specific antibody titers are associated with survival regardless of challenge time point.
(A) GP-specific IgG ELISA titers were compared between surviving and non-surviving animals at the final time point before challenge. Each point represents an individual animal and is color coded based on challenge time point: 1 year post-vaccination (dark blue circles), 3 months post-vaccination (light blue circles), and 42 days (green circles). ***p < 0.001 indicates the adjusted p values of Mann-Whitney analysis following Benjamini-Hochberg correction for false discovery rate.
(B) PRNT60 titers were compared between surviving and non-surviving animals at the final time point before challenge. Each point represents an individual animal and is color coded based on challenge time point: 1 year post-vaccination (dark blue circles), 3 months post-vaccination (light blue circles), and 42 days (green circles). ***p < 0.001 indicates the adjusted p values of Mann-Whitney analysis following Benjamini-Hochberg correction for false discovery rate.
(C) GP- (top row) and sGP-specific (bottom row) levels of indicated IgG subclasses, IgA or IgM, at the final time point before challenge were compared between survivors and non-survivors. Each point represents an individual animal and is color coded based on challenge time point: 1 year post-vaccination (dark blue circles), 3 months post-vaccination (light blue circles), and 42 days (green circles). **p < 0.005 and ***p < 0.001 indicate the adjusted p values of Mann-Whitney analysis following Benjamini-Hochberg correction for false discovery rate.
(D) The ability of GP- (top row) and sGP-specific (bottom row) antibodies to mediate the indicated Fc-innate immune effector functions at the final time point before challenge were compared between survivors and non-survivors. Each point represents an individual animal and is color coded based on challenge time point: 1 year post-vaccination (dark blue circles), 3 months post-vaccination (light blue circles), and 42 days (green circles). *p < 0.05, **p < 0.005, and ***p < 0.001 indicate the adjusted p values of Mann-Whitney analysis following Benjamini-Hochberg correction for false discovery rate.
Broad Fc-receptor binding and neutrophil phagocytosis are associated with protection. Beyond neutralization, antibodies are able to drive antiviral immunity via the induction of Fc-effector functions, following engagement of Fc receptors present in different combinations on all innate immune cells.30 Thus, we measured the ability of GP- and sGP-specific antibodies to induce NK cell activation, complement deposition, and phagocytosis by neutrophils and monocytes (Figure 3D), as well as the ability to bind to both human (FcγR2A, FcγR2B, FcγR3A, FcγR3B) and NHP Fc (FcγR2A-3, FcγR3A-1, FcγR3A-3) receptors (Figure S2). Although macaque and human Fc receptors (FcRs) have some divergence in their binding capacity to the Fc component of macaque antibodies, their overall binding affinities generally show similar trends regardless of the use of NHP or human FcR,31–33 which was also seen in this study. Across the innate immune effector functions, only sGP-specific neutrophil phagocytic activity was significantly elevated in survivors compared with non-survivors at the final time point before challenge (Figure 3D), suggesting that neutrophils may be able to rapidly clear opsonized sGP.
Unsupervised hierarchical cluster analysis using all antibody features clearly clustered most protected animals (red bars) separately from the non-survivors (blue bars), irrespective of challenge time points (Figure 4). Interestingly, among the features that were selectively enriched in protected animals, broad FcR binding was observed to both GP and sGP as well as elevated sGP-mediated neutrophil phagocytosis and complement deposition (Figure 4). Antibody-dependent cellular phagocytosis (ADCP), a measure of monocyte phagocytosis, NK cell degranulation(NKD), neutralization, and antibody-dependent neutrophil phagocytosis (ADNP) of GP were not associated with protection. When only 3 month and 1 year challenge groups were included in the analysis, NK cell activation and neutralization activity also gained significance (Figure 4).
Figure 4. Neutralizing antibody levels and neutrophil phagocytic activity are elevated in surviving, vaccinated NHPs.
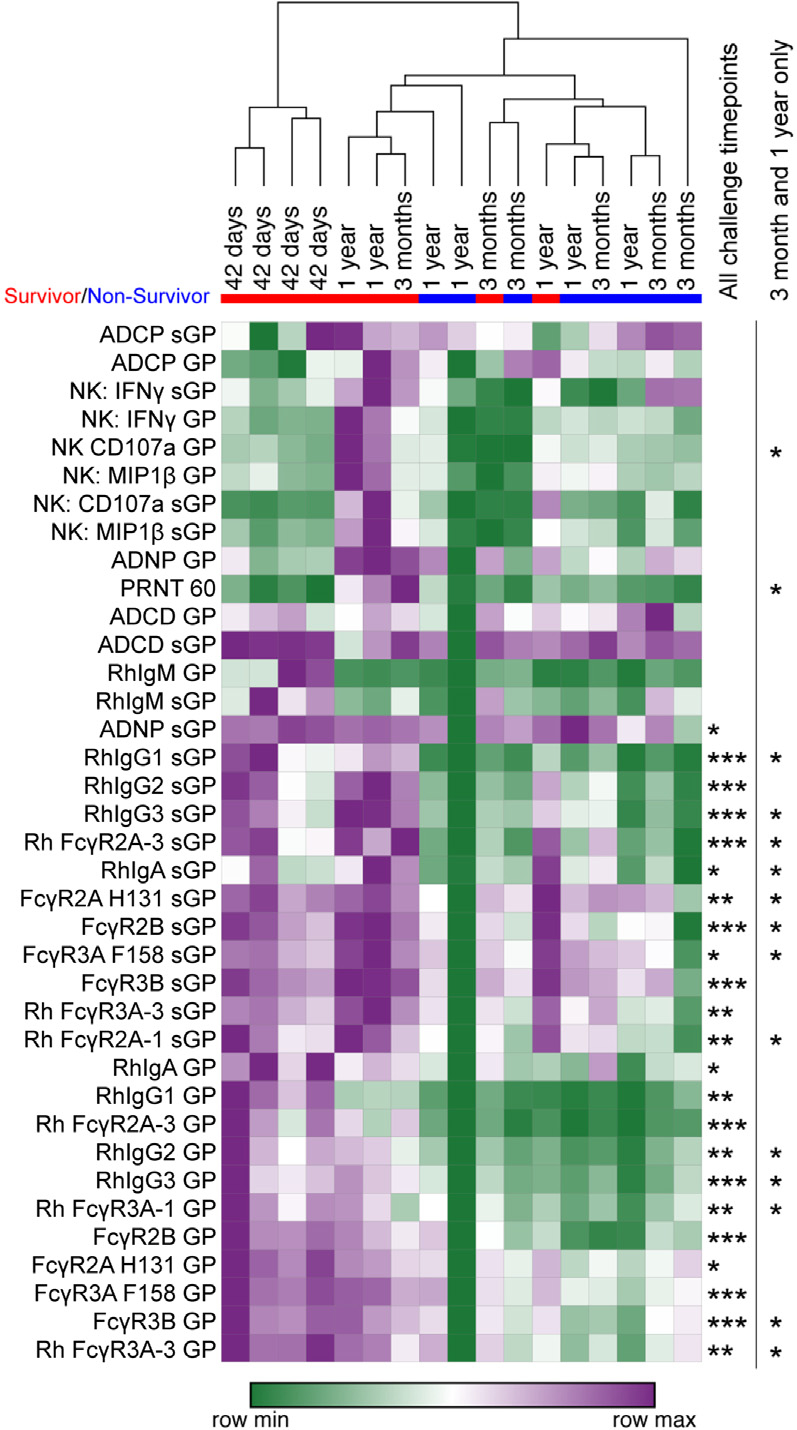
Induction of GP- and sGP-specific antibody-mediated innate immune effector and MIP-1B and sGP-specific antibodies to human Fc receptors (FcγR2a H131, FcγR3A F158, FcγR3B) and rhesus Fc receptors (FcγR2A-3, FcγR3A-1, FcγR3A-3) were measured. Data across all animals and time points were Z scored, and the Z score of each animal at the final time point before challenge was analyzed in an unsupervised hierarchical clustered heatmap (purple is row maximum [max]; green is row minimum [min]). Surviving animals are indicated by a red bar beneath challenge time point, and non-surviving animals are indicated by a blue bar. Statistically significant differences between surviving and non-surviving animals at this final time point are indicated to the right of the heatmap across all challenge time points (left column) or only at the durability time points of 3 months and 1 year post-vaccination (right column). *p < 0.05, **p < 0.005, and ***p < 0.001 by Mann-Whitney analysis with Benjamini-Hochberg correction for false discovery rate.
Fc function, but not antibody neutralization, is a key predictor of protection
To further define biomarkers that were selectively enriched in protected animals, we used an unbiased machine learning strategy to define the minimal correlates of protection against Ebola virus challenge across the vaccinated NHPs at the final time point prior to viral challenge. Specifically, a least absolute shrinkage and selection operator (LASSO) was applied to the 38 antibody features available for all animals to eliminate highly correlated features and define a minimal set of features that explain the overall antibody profile variation across the animals. As few as 5 of the overall 38 antibody features that were collected across all animals were sufficient to separate survivors and non-survivors and predicted survival with a classification accuracy of 73%. The model included both sGP- and GP-specific antibody features that were all selectively enhanced in survivors (Figure 5A). Interestingly, the features did not include neutralization, but instead included sGP- and GP-specific antibody binding to human inhibitory FcγR2B and sGP-specific antibody binding to the activating rhesus FcγR2A-3, as well as sGP-specific IgG1. To validate the LASSO-selected features, we used an orthogonal method and generated an ensemble classification tree using the LASSO-selected features. The receiver operating characteristic (ROC) curve showed an area under the curve of 0.917 (Figure 5B), indicating the robustness of the features in predicting protection across the animals. These data point to the importance of FcR-binding profiles as key correlates of immunity against Ebola virus infection.
Figure 5. sGP-specific antibody features are sufficient to predict durable vaccine-mediated protection.
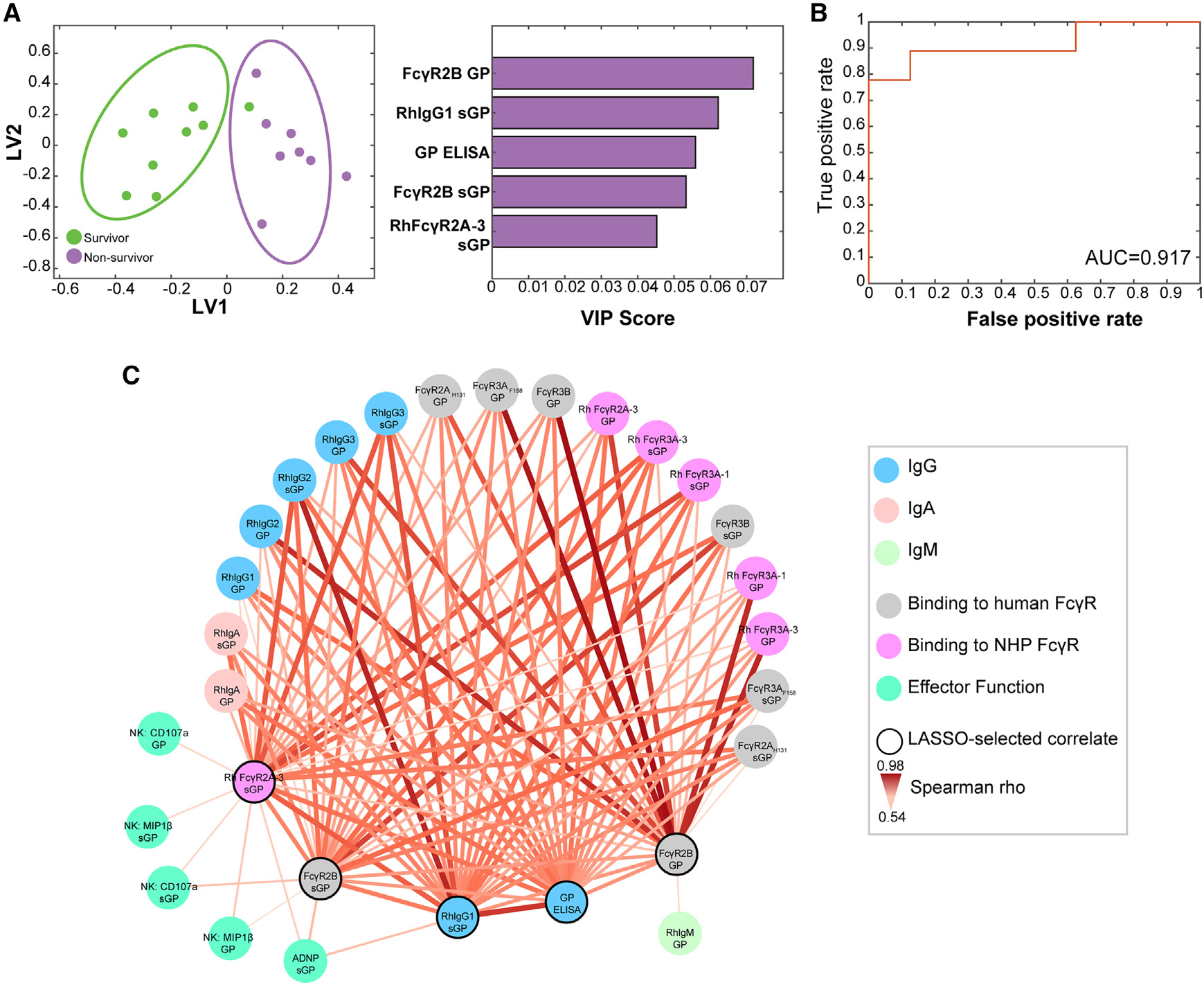
(A) LASSO-PLSDA analysis was used to identify the minimal antibody features measured at the final time point prior to challenge that predict protection for animals challenged at 1 year post-vaccination, 3 months post-vaccination, and 42 days post-vaccination. The variable importance in projection (VIP) rankings are shown on the right.
(B) LASSO-selected features were used to generate an ensemble classification tree, and a receiver operator characteristic (ROC) curve was generated to indicate the robustness of the features in predicting protection.
(C) Co-correlation network analysis of antibody features at the final time point before challenge associated with the LASSO-selected correlates. Each node represents the indicated antibody feature. The connecting lines between features represents statistically significant Spearman rho associations following Benjamini-Hochberg correction for false discovery rate (p < 0.05).
As LASSO selection collapses highly correlated features, we next built a co-correlate network to define additional features that are highly associated with the model-selected features, enriched in protected animals (Figure 5C). Notably, even in this expanded set of features, neutralizing antibodies was not among the network of correlated features. Binding of GP- and sGP-specific antibodies to both FcγRs from humans and NHPs were highly correlated with the LASSO-selected features. sGP-specific ADNP and both GP- and sGP-specific NK cell activation were selectively associated the sGP-specific LASSO-selected features, FcγR2A-3 and FcγR2B, suggesting that these FcRs may have a central role in ADNP and NK cell activation in antibody-mediated protection against Ebola.
sGP-specific monoclonal antibody-mediated neutrophil, but not monocyte, phagocytosis is associated with protection in vivo
Given the unexpected association between sGP-specific functionality with protection following rVSVΔG-ZEBOV-GP vaccination (Figure 5), we next aimed to determine whether sGP-specific monoclonal antibodies able to drive this particular function were associated with post-exposure protection in a stringent Ebola virus challenge model in mice. In this study, a panel of human IgG1 sGP-specific monoclonal antibodies (mAbs), previously analyzed by the Viral Hemorrhagic Fever Immunotherapeutics Consortium (VIC),13 were administered intraperitoneally to 10 mice 2 days after exposure to a 1,000 × LD50 dose of Ebola. Of the 168 human mAbs assembled by VIC, 58 bound to sGP, as previously published and confirmed in our lab. Although all of these mAbs shared a similar epitope, sGP-specific mAbs showed varying levels of post-exposure protection (Figure 6A).
Figure 6. Protective sGP-specific monoclonal antibodies demonstrate elevated binding affinity to sGP and neutrophil phagocytic activity.
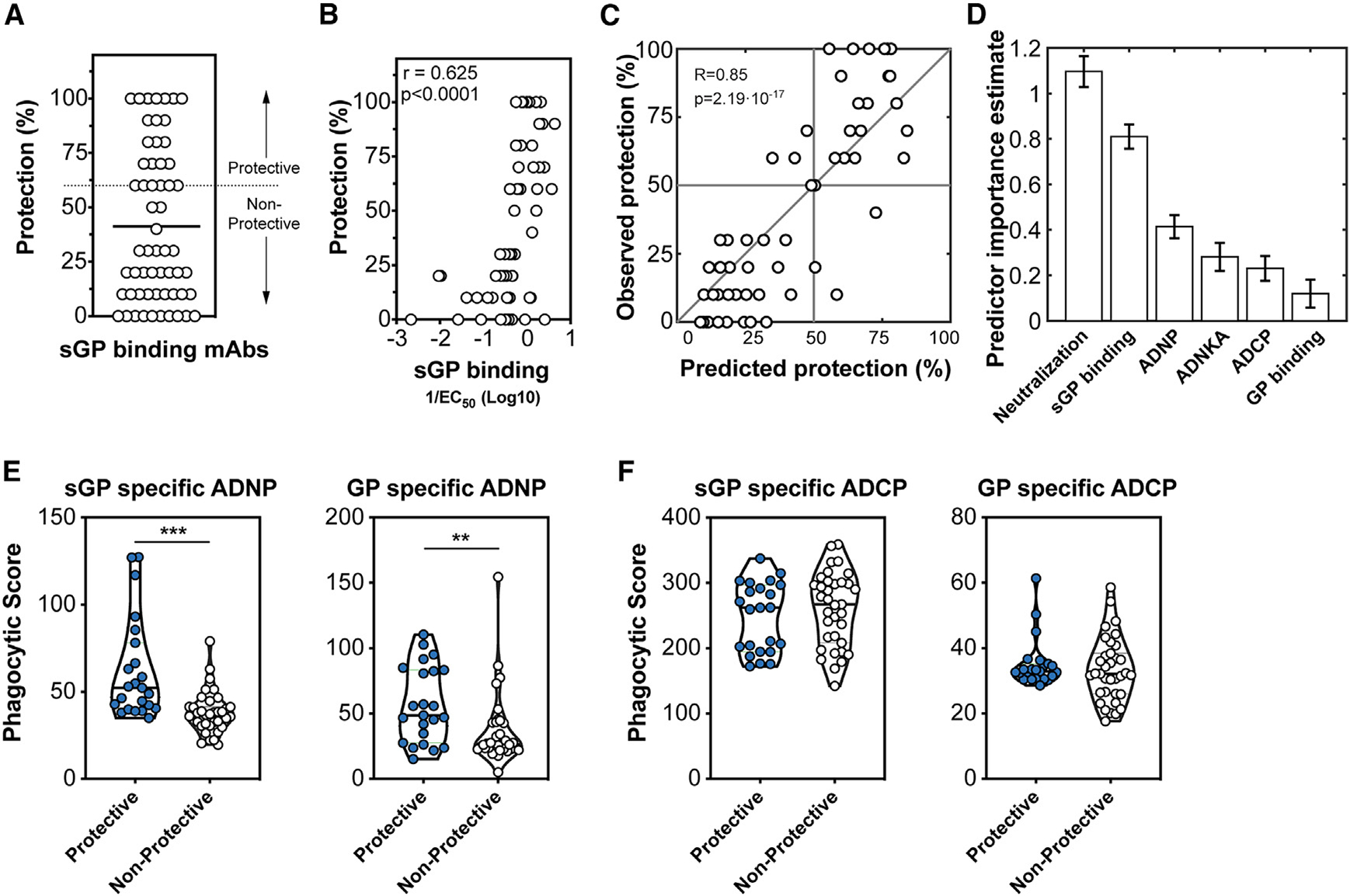
(A) Human IgG1 sGP-specific monoclonal antibodies (n = 58) collected by the Viral Hemorrhagic Fever Immunotherapeutic Consortium were evaluated for the ability to therapeutically protect BALB/c mice 2 days following infection with a mouse-adapted Ebola virus. The protection across all 58 mAbs is shown with the cutoff for protection (60%) is indicated by the dashed line.
(B) Association between the EC50 binding affinity for sGP of each antibody and protection was determined using Spearman rho correlation analysis.
(C) A model using an ensemble of decision trees, applied to antibody features, was predictive of survival as determined by leave-one-out cross validation.
(D) Features were ranked by importance using the permuted predictor delta error from 100 independently constructed models.
(E) Protective and non-protective sGP-specific mAbs were measured for the ability to induce phagocytosis of sGP-coated beads (left) or GP-coated beads (right) by neutrophils. Statistical analysis by Mann-Whitney analysis. ***p < 0.0001.
(F) Protective and non-protective sGP-specific mAbs were measured for the ability to induce phagocytosis of sGP-coated beads (left) or GP-coated beads (right) by monocytes. Statistical analysis by Mann-Whitney analysis. ***p < 0.0001.
To define the features of sGP-specific antibodies that confer protection, affinity, neutralization, and function of each antibody were evaluated. Protection mediated by the sGP-specific mAbs was associated with binding affinity to sGP (Figure 6B), highlighting that sGP is an important target for Ebola immunity. To further define antibody features within the sGP-specific mAbs that predicted protection, we generated a model using an ensemble of decision trees to define the functional correlates of sGP-mediated protection. The predictive power of these features was then assessed with leave-one-out cross validation (Figure 6C). The predictive features were then ranked by permuted predictor delta error (Figure 6D). Interestingly, although neutralization was not predictive in our macaque vaccination model, in this mouse post-exposure model, it ranked as the top feature. This is in agreement with a previously published analysis of the mouse protection model.13 Additional important features of the model include sGP binding with some independent predictive power of GP binding, although GP binding had lower importance to the predictive model. A number of antibody functional responses were also key predictors of protection in the model, including ADNP and NK cell-mediated activation, consistent with the effector function features that were associated with vaccine-mediated protection.
To further differentiate the functional capacity of sGP and GP antibodies that mediate protection, we assessed the capacity of sGP and GP antibodies to drive phagocytic activity. Strikingly, protective sGP- and GP-specific antibodies exhibited significantly higher levels of neutrophil phagocytosis (Figure 6E). but not monocyte phagocytosis (Figure 6F), than their non-protective counterparts. These data, again, support a central role for sGP- and GP-specific antibody-mediated neutrophil phagocytosis as a key mechanism of antibody-mediated protection against Ebola virus.
DISCUSSION
The remarkable, early protection conferred by rVSVΔG-ZEBOV-GP vaccination in NHPs has historically precluded the ability to define a precise correlate of immunity. While neutralizing and non-neutralizing antibody functions have been linked to protection in monoclonal therapeutic transfer studies in mice and NHPs, the precise mechanism by which rVSVΔG-ZEBOV-GP-induced antibodies contribute to protection was unclear. Here, we identify that sGP-targeted antibody responses, especially those that elicit antibody-dependent neutrophil phagocytosis, are key correlates of vaccine-mediated protection in a unique vaccine breakthrough study in NHPs.
Using a comprehensive systems serology profiling approach, unexpected correlates of immunity emerged, largely independent of neutralization. By combining the different NHP vaccination groups, we were able to gain sufficient power to define the functional profiles of antibodies that were selectively augmented in protected animals. Surprisingly, protection was linked to FcR-binding antibodies, able to selectively recruit neutrophil phagocytic activity. Unexpectedly, the antibody response enriched in survivors was largely focused on sGP, the predominant GP transcript of Ebola virus. Corroborated by post-exposure monoclonal antibody studies in mice, these data strongly argue for a critical role for sGP-specific phagocytic antibodies in the clearance of sGP via neutrophils in protection against Ebola virus.
Antibodies targeting sGP have long been considered detrimental, off-target responses that impair development of protective, neutralizing antibody response.24 Prior studies mapping GP epitopes targeted by rVSVΔG-ZEBOV-GP-induced antibodies in vaccinated humans showed broad induction of antibodies to epitopes shared by GP and sGP, including the receptor-binding domain and glycan cap, as well as the more unique epitopes of the mucin-like domain, and the HR2 region of GP2,34 suggesting that humans can elicit differential responses to sGP and GP in vivo. In the study presented here, vaccinated macaques possessed a balanced response to both neutralizing, GP-specific epitopes and non-neutralizing, sGP-specific targets. However, despite the clear importance of neutralization in protection, non-neutralizing, sGP-specific functional immunity was also a consistent, independent predictor of protection from infection. Thus, these data suggest that sGP may not be a distraction but rather a key contributor to durable protection.
sGP is hypothesized to have two critical roles in Ebola virus pathogenesis: (1) as a pathogenic immunomodulator on immune cells35 and 2) as a decoy and antibody sink,36 drawing GP-specific antibodies away from infected cells or virus. Thus, an sGP-specific humoral immune response, able to drive rapid clearance of sGP via neutrophils, the most abundant innate immune cell in the blood and first responder at sites of infection, may allow additional GP-specific antibodies to neutralize additional virus in the system or mark infected cells for targeted destruction by innate immune cells. Linked to the enhanced protection observed with sGP-phagocytic monoclonal antibodies in mice, these data strongly argue that sGP clearance itself may be beneficial in the ultimate control of Ebola virus. This is supported by our post-exposure mouse model, where sGP-targeted antibodies with similar functionality to those elicited in macaques with sustained protection after vaccination were, indeed, able to protect against Ebola-mediated death. However, it remains to be determined whether additional antibody functions (neutralization/killing of cells) are required to ultimately eliminate the virus or whether elimination of the immunomodulatory activity of sGP is sufficient. The larger contribution of NK effector functions in our mouse model, but not the macaque model, suggests that there may also be differences in protective effector cell functions of antibodies targeted against the same antigen at different stages of Ebola infection. Indeed, future passive transfer studies with monoclonal antibodies that are specific to sGP and lack GP cross-reactivity are likely to uncover the ultimate mechanistic basis of this correlate of immunity.
Binding to the FcRs FcγR2A and -2B was selected by LASSO as a strong predictor of protection. FcγR2A is known to facilitate neutrophil-mediated antibody complex clearance.37 However, FcγR2B is the sole inhibitory FcγR in humans,38 implicated in regulating phagocytosis and dampening inflammation.39,40 In Ebola infection, cytokine storm and immune-mediated pathology are associated with severe Ebola virus disease. In particular, innate immune cells, including neutrophils, have been implicated as key mediators of tissue damage, resulting in hemorrhaging and ultimate organ failure.41 Thus, antibody-mediated pathogen clearance, in the context of Ebola virus infection, may require a balanced antipathogen response to minimize host tissue damage. While neutrophils express low constitutive levels of FcγR2B,42 after activation, they upregulate this specific FcγR, limiting the inflammatory potential of sGP- and GP-targeted immune responses. However, the ligation of FcγR2B may also result in an immunomodulatory activation of dendritic cells and macrophages, FcR functions that were not directly tested in this study, limiting pathology and facilitating resolution of the disease. Thus, antibody binding to FcγR2B may represent a self-regulating mechanism that prevents prolonged immune activation and tissue damage while still allowing for rapid clearance of sGP early in infection.
Limitations of the study
One potential caveat in this study is that a follow-up study using the same vaccine platform in Cambodian cynomolgus macaques challenged at 4 months post-vaccination did not replicate the variability in vaccine efficacy observed here in the Mauritian macaques (A.S., United States Army Medical Research Institute of Infectious Diseases). Mauritian cynomolgus macaques have more rapid disease progression after Ebola virus infection compared with cynomolgus macaques of Cambodian origin.43 This is believed to be mediated by differences in immune cell populations and microbiota,43–45 with a likely contribution of their divergent genetics, which may include divergence in FcR sequence and functionality.46–48 However, the reduced efficacy observed in this study does replicate observations in humans of reduced vaccine efficacy at later time points and presents a unique opportunity to investigate antibody-mediated correlates of protection after rVSVΔG-ZEBOV-GP vaccination.
It is plausible that profiles more proximal to the time of challenge could provide additional insights. However, in this study, due to the experimental planning at the time of the intense rush to develop the VSV vaccine, samples were only collected at peak immunogenicity. Thus, in this analysis, we were only able to look at peak immunogenicity predictors of protection. Interestingly, we did find common signatures of protection at peak immunogenicity across animals challenged early and later, likely reflecting a proportional decay in particular functional antibody responses. Therefore, if an animal starts with a higher response, it will likely have higher levels after several months, as has been noted across many functional durability vaccine studies performed over the pandemic.49–52 Most importantly, it is critical to note that neutralization at peak immunogenicity, even though it also decays proportionally, did not discriminate the groups.
Our assays described here focus on antibody functional interactions with free virus or antigens and not Ebola virus-infected cells. At this time, the relative importance of antibody activity against free virus/floating antigens and infected cells is uncertain in Ebola virus infection. Further comparison of soluble antigen vs. GP-expressing cell-based assays would be of great interest to further define the mechanics of antibody-mediated protection against Ebola virus.
Additionally, T cells are not within the scope of this study, partly due to the unavailability of peripheral blood mononuclear cells (PBMCs) from vaccinated macaques. While previous studies aimed at defining immunogenicity of Ebola virus vaccines have noted the presence of T cells following vaccination, these responses have not tracked consistently with protection or may be influenced by co-infecting agents or dosing.53–58 We did not evaluate T cell responses within this study as PBMCs from these animals were not available. A full analysis of the adaptive responses to Ebola virus vaccination very much be of interest in future vaccination studies.
Conclusions
As increasing numbers of breakthrough Ebola virus infections have been observed in previously vaccinated individuals,27,28 there is an urgent need to define biomarkers and correlates of protective vaccine-mediated immunity against Ebola virus. Collectively, our studies point to the importance of both quantitative and qualitative features of the GP- and sGP-specific humoral response in rVSVΔG-ZEBOV-GP-mediated protection. Future validation and mechanistic studies will provide greater insights into vaccine mechanism of action. In addition, similar analyses of human breakthrough samples will provide enhanced resolution of the applicability of the NHP model in advancing correlate/biomarker discovery for human vaccines against Ebola virus.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the data point of contact, Ryan McNamara (ryan.mcnamara@mgh.harvard.edu).
Materials availability
All reagents used in this study are commercially available. This study did not generate new, unique reagents.
Data and code availability
Any data reported in this paper is available from the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Macaques
Nineteen cynomolgus macaques of Mauritian origin, 9 males and 10 females, were included in the study. The animals were maintained in a colony at New Iberia Research Center (New Iberia, Louisiana USA) where the vaccination phase of the study was conducted. At time of vaccination the age of the animals was 6–10 years old and body weight ranged from 4–7 kg. The animals were transferred to the United States Army Medical Research Institute of Infectious Diseases (USAMRIID) in Ft. Detrick (Maryland, USA) 6 days prior to EBOV challenge and were placed directly into BSL-4 containment for the remainder of the study. Procedures involving the care and use of animals in the study were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) at Merck & Co., Inc. and at USAMRIID. During the study, the care and use of animals were conducted in compliance with the Animal Welfare Act, Public Health Service (PHS) Policy and other Federal statutes and regulations relating to animals and experiments involving animals. The facilities where this research was conducted are accredited by the Association for Assessment and Accreditation of Laboratory Animal Care, International and adhere to principles stated in the Guide for the Care and Use of Laboratory Animals, National Research Council, 2011.
Mice
Mouse studies included in this study were previously described in detail.13 Briefly, monoclonal antibodies were donated in sets and purified from either cell lines or splenocytes. Female BALB/c mice, aged 6 to 8 weeks-old, were housed in microisolator cages. All animal work involving infectious live virus was performed in the BSL-4 laboratories at USAMRIID or PHAC. At USAMRIID, all work with animals was conducted in compliance with the Animal Welfare Act and other Federal statutes and regulations relating to animals and experiments involving animals and adhered to the principles stated in the Guide for the Care and Use of Laboratory Animals, NRC Publication, 1996 edition. All procedures were reviewed and approved by the appropriate Institutional Animal Care and Use Committee at USAMRIID. Work performed at PHAC was approved by the Canadian Science Centre for Human and Animal Health Animal Care Committee following the guidelines of the Canadian Council on Animal Care.
METHOD DETAILS
Vaccination of cynomolgus macaques
The animals were vaccinated with a single intramuscular (IM) inoculation of clinical trial grade V920 at nominal doses of 3×104 or 3×106 plaque-forming units (pfu) approximately one year, three months or six weeks (42 days) prior to challenge, as summarized in Table 1.
Macaque challenge
Animals were challenged intramuscularly with a target dose of 1000 PFU of Ebola Zaire Kikwit (EBOV R4415, a 7U variant) in an approved BSL-4 facility at USAMRIIID. Technical personnel evaluating challenged animals for vaccine efficacy and signs of EBOV disease were blinded to vaccine group designation. Health status checks were performed at regular intervals throughout the challenge phase of this study and animals were assigned a responsiveness score of 0–5 at each observation. Physical examination under anesthesia was performed on Days 0, 3, 5, 7, 10, 14, 21, and 28. On all scheduled days animals were weighed and rectal temperatures were acquired. Blood was collected during the scheduled physical examinations for immune response analysis, clinical pathology, and virology. When moribund, or at the end of the in-life phase, animals were euthanized and necropsied. Gross pathology was noted at the time of necropsy and tissues were processed for histology and immunohistochemistry. Moribund animals were euthanized under deep anesthesia in accordance with USAMRIID approved procedures by a qualified member of the study team.
Mice post-exposure protection model
The mouse studies used in this paper are previously published and described in detail.13 Briefly, mouse-adapted EBOV mouse model, developed at the U.S. Army Medical Research Institute of Infectious Diseases (USAMRIID) was used.60,61 Assays were performed either at USAMRIID or Public Health Agency of Canada (PHAC). Mouse-adapted EBOV/Mayinga (EBOV/M.mus-tc/COD/76/Yambuku-Mayinga) was for in vivo protection studies.
On day 0 of the assay, the mice were transferred to a biosafety level (BSL) level 4 containment area and challenged, intraperitoneally, with mouse-adapted EBOV (Mayinga isolate, Yambuku variant, 100 p.f.u. (USAMRIID) or 1,000 × LD50 (PHAC)). On day 2 post-infection, groups of 10 mice were treated intraperitoneally with 100 μg (~5 mg/kg) mAb per mouse. Mice were observed daily and group weights were recorded daily through day 14 and again on days 21 and 28. Protection is expressed as the percentage of mice surviving at the end of a 28 day period.
Determination of GP-specific antibody titers by ELISA
The levels of GP-specific IgG antibodies in serum samples collected prior to challenge were determined by Q2 Solutions (San Juan Capistrano, CA) using a validated ELISA assay originally developed by the Filovirus Animal Non-Clinical Group (FANG) and subsequently validated for both human and NHP samples.4,62,63 Transmembrane deleted GP and sGP were generated by the Ollmann-Saphire lab. Quality control was performed to ensure correct protein conformation and multimerization for sGP. Antigens were used within 2 weeks of delivery to ensure high quality analytics. Briefly, sera are serially diluted 1:50–1:1,600 and incubated in recombinant ZEBOV GP coated wells of a microtiter plate. A reference standard curve, generated from a pool of vaccinated donors, is included on each plate. Following sample incubation, wells are washed and incubated with goat anti-human IgG horseradish peroxidase, and developed using tetramethylbenzidine substrate, and stopped by addition of sulfuric acid. Sample concentrations are determined from the reference standard curve using a 4-Parameter Logistic equation (4-PL) curve fit, and reported at ELISA Units (EU)/ml. The lower limit of quantitation (LLOQ) in the assay is 13.62 EU/ml.
Determination of neutralizing antibody titers by plaque reduction neutralization titer (PRNT)
Sera were tested for neutralizing activity using a validated PRNT60 assay.4 Briefly, heat-inactivated sera is serially diluted (1:5 to 1: 10,240) and incubated with an equal volume of diluted rVSVΔG-ZEBOV-GP for 20 hours at 4°C before incubation with Vero cells for 1 hour at 37°C. Cells are then incubated with a methylcellulose overlay for 2 days, and plaques are visualized by staining with crystal violet. The PRNT60 (60% neutralizing titer) is calculated by linear regression based on the percent reduction in viral plaques in the presence of sera compared to no serum controls. The limit of detection (LOD) in the assay is 20 and the lower limit of quantitation (LLOQ) is 35.
Evaluation of induction of GP-specific innate immune effector function
Immune effector function was studied, using human effector cells, as per prior studies.64–68
Antibody-mediated cellular phagocytosis by human monocytes (ADCP)
This assay utilizes THP-1 cells, human-derived monocytes, that reproduce phagocytic activity of primary monocytes but have divergent cytokine profiles on stimulation. Recombinant EBOV GPΔTM (Mayinga strain; IBT Bioservices) was biotinylated using LC-LC-Sulfo-NHS Biotin (ThermoScientific). Excess biotin was removed using a Zeba desalting column (ThermoScientific). Biotinylated GP antigen was then coupled to 1μm yellow-green Neutravidin beads (ThermoScientific) by incubating beads and antigen overnight at 4°C. Beads were washed twice with PBS containing 0.1% bovine serum albumin (BSA). Samples were diluted 1:100 in culture medium and incubated with GP-coated beads for 2h at 37°C. Unbound antibodies were removed by centrifugation prior to the addition of THP-1 cells at 2.5×104cells/well. Cells were fixed with 4% paraformaldehyde and analyzed by flow cytometry. A phagocytic score was determined using the following formula: (percentage of FITC+ cells)*(geometric mean fluorescent intensity (gMFI) of the FITC+ cells)/10,000.
Antibody-mediated phagocytosis by human neutrophils (ADNP)
Recombinant EBOV GPΔTM was biotinylated and coupled to yellow-green Neutravidin beads (ThermoScientific) as described above for ADCP. Samples were diluted 1:100 in culture medium and incubated with GP-coated beads for 2h at 37°C. Freshly isolated white blood cells from human donor peripheral blood (5×104cells/well) were incubated for 1h at 37°C. Cells were stained for CD66b (Clone G10F5; Biolegend), CD3 (Clone UCHT1; BD Biosciences), and CD14 (Clone MφP9; BD Biosciences), fixed with 4% paraformaldehyde, and analyzed by flow cytometry. Neutrophils were defined as SSC-Ahigh/CD66b+/CD3neg/CD14neg. A phagocytic score was determined using the following formula: (percentage of FITC+ cells)*(geometric mean fluorescent intensity (gMFI) of the FITC+ cells)/10,000.
Antibody-mediated complement deposition (ADCD)
This assay is designed to replicate in vivo interaction of complement/antibody complexes with free antigens and/or virus. Recombinant EBOV GPΔTM was biotinylated and coupled to red Neutravidin beads (ThermoScientific) as described above for ADCP. Heat-inactivated sera was diluted 1:10 in culture medium and incubated with GP-coated beads for 2h at 37°C. Unbound antibodies were removed by centrifugation prior to the addition of reconstituted guinea pig complement (Cedarlane Labs) diluted in veronal buffer supplemented with calcium and magnesium (Boston Bioproducts) for 20 min at 37°C. Beads were washed with PBS containing 15mM EDTA, and stained with an FITC-conjugated anti-guinea pig C3 antibody (MP Biomedicals, Santa Ana, CA). C3 deposition onto beads was analyzed by flow cytometry. The gMFI of FITC of all beads was measured.
Antibody-dependent NK cell degranulation and activation
Recombinant EBOV GPΔTM antigen was coated onto a MaxiSorp 96-well plates (Nunc) at 300ng/well at 4°C overnight. Wells were washed with PBS and blocked with 5% BSA prior to addition of samples diluted 1:50 for 2h at 37°C. Unbound antibodies were removed by PBS wash, and NK cells enriched from the peripheral blood of human donors were added at 5×104 cells/well in the presence of 4μg/ml brefeldin A (Sigma Aldrich), 5μg/ml GolgiStop (Life Technologies) and anti-CD107a antibody (Clone H4A3, BD Biosciences) for 5h. Cells were fixed and permeabilized with Fix/Perm (Life Technologies) according to manufacturer’s instructions to stain for intracellular IFNγ (Clone B27, BD Biosciences) and MIP-1β (Clone D21–1351, BD Biosciences). Cells were analyzed on a BD LSRII flow cytometer.
Determination of antigen-specific antibody reactivity
Recombinant EBOV GP and EBOV soluble GP (sGP; IBT Bioservices) were coupled to MagPlex beads (Luminex) via sulfo-NHS coupling chemistry. Heat-inactivated samples were diluted 1:50 in 1X PBS + 0.1% bovine serum albumin (BSA) + 0.05% Tween20 and incubated with antigen-coupled beads for 2h. Beads were washed, and secondary antibodies specific for non-human primate antibody subclasses IgG1 (clone 7H11; NHP Resource Center), IgG2 (clone 3C10; NHP Resource Center), IgG3 (clone 2G11; NHP Resource Center) and isotypes (IgM, IgA) were detected by incubating with 0.65μg/ml of secondary antibodies (Southern Biotech) for 1h at room temperature. Beads were washed 6X with assay buffer and incubated with a PE-labeled anti-mouse IgG1 (1μg/ml) for 1 hour at room temperature. Beads were washed analyzed on a Flexmap 3D instrument (Luminex). The median fluorescent intensity of 30 beads/region was recorded. Sera from naive cynomolgus macaques were used to establish baseline reactivity and thresholds for positivity.
Determination of antigen-specific antibody binding to Fc-receptors
Recombinant EBOV GP or EBOV soluble GP (sGP; IBT Bioservices) were coupled to MagPlex beads (Luminex) via sulfo-NHS coupling chemistry. Three activating FcγR receptors (FcγR2A, FcγR3A, FcγR3B) and one inhibitory receptor (FcγR2B). NHP have polymorphic receptors and among the four rhesus FcγR2A receptors, only FcγR2A-2 and FcγR2A-3 show similar IgG binding, and within the three known FcγR3A receptors (-1, -2, -3) (69), FcγR3A-1 and FcγR3A-3 have distinct IgG binding profiles, and thus binding antibodies to rhesus FcγR2A-3, FcγR3A-1 and FcγR3A-3 was measured. Heat-inactivated samples were diluted 1:50 (rhesus FcRs) or 1:500 (human FcRs) in 1X PBS + 0.1% bovine serum albumin (BSA) + 0.05% Tween20 and incubated with antigen-coupled beads for 2h. Beads were washed and incubated with recombinant biotinylated Fc-receptors that were tetramerized via Streptavidin-PE for 1 hour at room temperature. Beads were washed analyzed on a Flexmap 3D instrument (Luminex). The median fluorescent intensity of 30 beads/region was recorded. Sera from naive cynomolgus macaques were used to establish baseline reactivity and thresholds for positivity.
QUANTIFICATION AND STATISTICAL ANALYSIS
Co-correlation networks
Associations between antibody features were determined using non-parametric spearman correlation coefficient. Statistically significant associations after Bonferroni correction for multiple comparisons (adjusted p-value<0.05) were used to generate networks in Cytoscape (v3.4.0).
Statistical analysis
Univariate analyses were performed using Prism9 software. A two-way RM ANOVA with Sidak’s multiple comparisons test was used for longitudinal analysis between survivors and non-survivors, and Mann-Whitney analysis with Benjamini-Hochberg correction for false discovery rate was used for comparison between survivors and non-survivors.
Machine learning methods
Least absolute shrinkage and selection operator (LASSO) feature selection. LASSO is a supervised regularized linear regression model. The model enforces a regularization penalty, the L1 norm, during optimization. This allows for predictions to made using only a sparse set of features. LASSO regression was performed in a leave-one-out cross validation framework with 100 repetitions. Feature selection within folds was performed by including only those features having non-zero beta coefficient values in the fitted LASSO regression model. The most informative features were selected by ranking features by the frequency with which they were selected across folds and repetitions.
LASSO-PLSDA model
A LASSO-PLSDA model is constructed to select a minimal set of antibody features that together are most predictive of protection. Specifically, first a LASSO model is fit to the data using the lasso MATLAB function and antibody features that are assigned non-zero regression coefficients are selected. Next, a PLSDA model is fit to the subset of LASSO-selected data. We visualize the first two latent variables in a bi-plot and show the set of LASSO-selected antibody features ranked by their respective.
VIP scores
Bagged decision trees ROC. To validate the ability of the set of selected antibody features to accurately discriminate NHPs based on protection, we employ an orthogonal Bagged Decision Tree model. Specifically, we fit a Bagged Decision Tree regression model to the subset of LASSO-selected data using the TreeBagger MATLAB function with the NumPredictorsToSample argument set to “all”. Subsequently we contract a receiver operating characteristic (ROC) curve using the perfcurve MATLAB function to evaluate the performance of this model.
Ensemble-model of survival
A model was constructed as an ensemble of decision trees using the TreeBagger function (MATLAB ver. R2019a). Neutralization was computed as an average of 4 different neutralization metrics and NK-score as an average of 3 different z-scored metrics of NK activity. Feature importance was assessed by the permutated predictor delta error, which measures the increase in out of bag error when permuting the values of the feature and reported as the average and standard deviation of 100 independently constructed models. Leave one out cross validation was performed by leaving out one sample at a time, the outcome for each sample was predicted by a model constructed from data from the remaining samples.
Hierarchical clustering analysis
Unsupervised hierarchical clustering analysis was performed in JMP Pro 15, using the Ward method.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Mouse anti-human CD66β (clone G10F5), Pacific Blue | BioLegend | RRID:AB_2563294 |
| Mouse anti-human CD3 (clone UCHT1), Alexa Fluor 700 | BD Biosciences | RRID:AB_396952 |
| Mouse anti-human CD107α (clone H4A3), PE-Cy5 | BD Biosciences | RRID:AB_396136 |
| Mouse anti-human IFNγ (clone B27), FITC | BD Biosciences | RRID: AB_398580 |
| Mouse anti-human MIP-1β (clone D21–1351), PE | BD Biosciences | RRID: AB_393549 |
| Mouse anti-human CD56 (clone B159), PE-Cy7 | BD Biosciences | RRID:AB_396853 |
| Mouse anti-human CD14 (clone MφP9), APC-Cy7 | BD Biosciences | RRID:AB_396889 |
| Mouse anti-human CD16 (clone 3G8), APC-Cy7 | BD Biosciences | RRID:AB_396864 |
| Anti-guinea pig complement C3 goat IgG fraction, FITC | MP Biomedicals | RRID:AB_2334913 |
| Anti-human IgG Fc antibody | Jackson ImmunoResearch | RRID: AB_2888996 |
| Mouse anti-rhesus IgG1 (clone 7H11) | NHP Resource Center | RRID:AB_2819310 |
| Mouse anti-rhesus IgG2 (clone 2C10) | NHP Resource Center | PR-3100 |
| Mouse anti-rhesus IgG3 (clone 2G11) | NHP Resource Center | PR-1130 |
| VIC panel of mAbs | Saphire et al.13,17 | N/A |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| Ebola Zaire Kikwit (EBOV R4415, a 7U variant) | Kugelman et al.59 | N/A |
|
| ||
| Biological samples | ||
|
| ||
| Recombinant Vesicular Stomatitis Virus △G- Zaire Ebola glycoprotein vaccine | Merck & Co | N/A |
| LowTox Guinea Pig Complement | CedarLane Labs | Cat# CL4051 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| LC-LC-Sulfo-NHS Biotin | ThermoFisher | Cat# A35358 |
| Brefeldin A | Sigma Aldrich | Cat# B7651 |
| GolgiStop | BD Biosciences | Cat# 554724 |
| EBOV GP△TM | Laboratory of E.O. Saphire | N/A |
| Recombinant EBOV Soluble GP (sGP) | Laboratory of E.O. Saphire | N/A |
| Recombinant FcγRs (human and rhesus) | Duke University Human Vaccine Institute | N/A |
| Streptavidin-R-Phycoerythrin | Prozyme | Cat# PJ31S |
|
| ||
| Critical commercial assays | ||
|
| ||
| RosetteSep NK cell enrichment kit | Stem Cell Technologies | Cat# 15025 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| THP-1 monocytes | ATCC | RRID: CVCL_0006 |
| VeroE6 | ATCC | RRID: CVCL_0574 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Cynomolgus macaques (Mauritius) | New Iberia Research Center | N/A |
| BALB/c mice | Charles River Laboratory | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| GraphPad Prism 8 | GraphPad Software, Inc. | RRID:SCR_002798 |
| R Studio | R Project for Statistical Computing | RRID:SCR_000432 |
| Flow Jo | BD Bioscience | RRID:SCR_008520 |
| JMP Pro 15 | JMP | RRID:SCR_014242 |
| MATLAB ver. R2019a | MathWorks | RRID:SCR_001622 |
|
| ||
| Other | ||
|
| ||
| FluoSpheres® NeutrAvidin®-Labeled Microspheres, 1.0 μm, yellow-green fluorescent (505/515), 1% solids | Life Technologies | Cat# F-8776 |
| FluoSpheres® NeutrAvidin®-Labeled Microspheres, 1.0 μm, red fluorescent (580/605), 1% solids | Life Technologies | Cat# F8775 |
| MagPlex microspheres | Luminex corporation | Cat# MC12001 –01, MCI12040–01, MCI10077–01 |
Highlights.
GP and sGP antibodies are induced by rVSVΔ G-ZEBOV-GP vaccine
Neutrophil phagocytosis is leveraged by sGP-targeted antibodies
Non-neutralizing antibody functions to sGP are correlated with EBOV protection
ACKNOWLEDGMENTS
We would like to thank Tom Monath, Rick Nichols, Brian Martin, Gray Heppner, and Tracy Kemp from NewLink Genetics and George Lewis at the University of Maryland for initial support of analysis of NHP studies using rVSVΔG-ZEBOV-GP. We also thank Jay Wells, Kelly Stuthman, Ginger Lynn, Nicole Lackemeyer, Sean Van Tongeren, Jesse Steffens, and the Pathology Division at USAMRIID for their assistance with the NHP study and collection of the samples for the assays described herein. We would also like to thank the members of the Alter Laboratory for critical discussion. This work was supported by funding from the Defense Threat Reduction Agency (HDTRA1–15-C-0058) and the MGH Research Institute Scholars Fund (awarded to G.A.), U19-AI135995 (D.L.), and U19-AI142790 (E.O.S.). Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the US Department of the Army, the US Department of Defense, or the US Department of Health and Human Services or of the institutions and companies affiliated with the authors.
Footnotes
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
DECLARATION OF INTERESTS
S.D., M.E., B.-A.C., and J.K.S. are employees of Merck & Co., T.J.S. is an employee of SeromYx, Inc., and G.A. is the founder of SeromYx, Inc.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112402.
REFERENCES
- 1.Marzi A, Robertson SJ, Haddock E, Feldmann F, Hanley PW, Scott DP, Strong JE, Kobinger G, Best SM, and Feldmann H (2015). EBOLA VACCINE. VSV-EBOV rapidly protects macaques against infection with the 2014/15 Ebola virus outbreak strain. Science 349, 739–742. 10.1126/science.aab3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Regules JA, Beigel JH, Paolino KM, Voell J, Castellano AR, Hu Z, Munoz P, Moon JE, Ruck RC, Bennett JW, et al. (2017). A recombinant vesicular stomatitis virus Ebola vaccine. N. Engl. J. Med. 376, 330–341. 10.1056/NEJMoa1414216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agnandji ST, Huttner A, Zinser ME, Njuguna P, Dahlke C, Fernandes JF, Yerly S, Dayer JA, Kraehling V, Kasonta R, et al. (2016). Phase 1 trials of rVSV Ebola vaccine in Africa and Europe. N. Engl. J. Med. 374, 1647–1660. 10.1056/NEJMoa1502924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heppner DG Jr., Kemp TL, Martin BK, Ramsey WJ, Nichols R, Dasen EJ, Link CJ, Das R, Xu ZJ, Sheldon EA, et al. (2017). Safety and immunogenicity of the rVSVG-ZEBOV-GP Ebola virus vaccine candidate in healthy adults: a phase 1b randomised, multicentre, double-blind, placebo-controlled, dose-response study. Lancet Infect. Dis. 17, 854–866. 10.1016/S1473-3099(17)30313-4. [DOI] [PubMed] [Google Scholar]
- 5.Halperin SA, Das R, Onorato MT, Liu K, Martin J, Grant-Klein RJ, Nichols R, Coller BA, Helmond FA, and Simon JK; V920–012 Study Team (2019). Immunogenicity, lot consistency, and extended safety of rVSVDeltaG-ZEBOV-GP vaccine: a phase 3 randomized, double-blind, placebo-controlled study in healthy adults. J. Infect. Dis. 220, 1127–1135. 10.1093/infdis/jiz241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kennedy SB, Bolay F, Kieh M, Grandits G, Badio M, Ballou R, Eckes R, Feinberg M, Follmann D, Grund B, et al. (2017). Phase 2 placebo-controlled trial of two vaccines to prevent Ebola in Liberia. N. Engl. J. Med. 377, 1438–1447. 10.1056/NEJMoa1614067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henao-Restrepo AM, Camacho A, Longini IM, Watson CH, Edmunds WJ, Egger M, Carroll MW, Dean NE, Diatta I, Doumbia M, et al. (2017). Efficacy and effectiveness of an rVSV-vectored vaccine in preventing Ebola virus disease: final results from the Guinea ring vaccination, open-label, cluster-randomised trial (Ebola Ca Suffit!). Lancet 389, 505–518. 10.1016/S0140-6736(16)32621-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grais R, Kennedy SB, Mahon BE, Dubey SA, Grant-Klein RJ, Liu K, Hartzel J, Coller BA, Welebob C, Hanson ME, and Simon JK (2021). Estimation of the correlates of protection of the rVSVΔG-ZEBOV-GP Zaire ebolavirus vaccine: a post-hoc analysis of data from phase 2/3 clinical trials. Lancet Microbe 2, e70–e78. 10.1016/S2666-5247(20)30198-1. [DOI] [PubMed] [Google Scholar]
- 9.Medaglini D, Santoro F, and Siegrist CA (2018). Correlates of vaccine-induced protective immunity against Ebola virus disease. Semin. Immunol. 39, 65–72. 10.1016/j.smim.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Marzi A, Engelmann F, Feldmann F, Haberthur K, Shupert WL, Brining D, Scott DP, Geisbert TW, Kawaoka Y, Katze MG, et al. (2013). Antibodies are necessary for rVSV/ZEBOV-GP-mediated protection against lethal Ebola virus challenge in nonhuman primates. Proc. Natl. Acad. Sci. USA 110, 1893–1898. 10.1073/pnas.1209591110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones SM, Feldmann H, Stroher U, Geisbert JB, Fernando L, Grolla A, Klenk HD, Sullivan NJ, Volchkov VE, Fritz EA, et al. (2005). Live attenuated recombinant vaccine protects nonhuman primates against Ebola and Marburg viruses. Nat. Med. 11, 786–790. 10.1038/nm1258. [DOI] [PubMed] [Google Scholar]
- 12.Marzi A, Reynolds P, Mercado-Hernandez R, Callison J, Feldmann F, Rosenke R, Thomas T, Scott DP, Hanley PW, Haddock E, and Feldmann H (2019). Single low-dose VSV-EBOV vaccination protects cynomolgus macaques from lethal Ebola challenge. EBioMedicine 49, 223–231. 10.1016/j.ebiom.2019.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saphire EO, Schendel SL, Fusco ML, Gangavarapu K, Gunn BM, Wec AZ, Halfmann PJ, Brannan JM, Herbert AS, Qiu X, et al. (2018). Systematic analysis of monoclonal antibodies against Ebola virus GP defines features that contribute to protection. Cell 174, 938–952.e13. 10.1016/j.cell.2018.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wec AZ, Bornholdt ZA, He S, Herbert AS, Goodwin E, Wirchnianski AS, Gunn BM, Zhang Z, Zhu W, Liu G, et al. (2019). Development of a human antibody cocktail that deploys multiple functions to confer pan-ebolavirus protection. Cell Host Microbe 25, 39–48.e5. 10.1016/j.chom.2018.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Misasi J, Gilman MS, Kanekiyo M, Gui M, Cagigi A, Mulangu S, Corti D, Ledgerwood JE, Lanzavecchia A, Cunningham J, et al. (2016). Structural and molecular basis for Ebola virus neutralization by protective human antibodies. Science 351, 1343–1346. 10.1126/science.aad6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gunn BM, Yu WH, Karim MM, Brannan JM, Herbert AS, Wec AZ, Halfmann PJ, Fusco ML, Schendel SL, Gangavarapuet K, et al. (2018). A role for Fc function in therapeutic monoclonal antibody-mediated protection against Ebola virus. Cell Host Microbe 24, 221–233.e5. 10.1016/j.chom.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saphire EO, Schendel SL, Gunn BM, Milligan JC, and Alter G (2018). Antibody-mediated protection against Ebola virus. Nat. Immunol. 19, 1169–1178. 10.1038/s41590-018-0233-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gunn BM, Roy V, Karim MM, Hartnett JN, Suscovich TJ, Goba A, Momoh M, Sandi JD, Kanneh L, Andersen KG, et al. (2019). Survivors of Ebola virus disease develop polyfunctional antibody responses. J. Infect. Dis. 221, 156–161. 10.1093/infdis/jiz364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gunn BM, Lu R, Slein MD, Ilinykh PA, Huang K, Atyeo C, Schendel SL, Kim J, Cain C, Roy V, et al. (2021). A Fc engineering approach to define functional humoral correlates of immunity against Ebola virus. Immunity 54, 815–828.e5. 10.1016/j.immuni.2021.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JE, and Saphire EO (2009). Ebolavirus glycoprotein structure and mechanism of entry. Future Virol. 4, 621–635. 10.2217/fvl.09.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez A, Trappier SG, Mahy BW, Peters CJ, and Nichol ST (1996). The virion glycoproteins of Ebola viruses are encoded in two reading frames and are expressed through transcriptional editing. Proc. Natl. Acad. Sci. USA 93, 3602–3607. 10.1073/pnas.93.8.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanchez A, Yang ZY, Xu L, Nabel GJ, Crews T, and Peters CJ (1998). Biochemical analysis of the secreted and virion glycoproteins of Ebola virus. J. Virol. 72, 6442–6447. 10.1128/JVI.72.8.6442-6447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mehedi M, Falzarano D, Seebach J, Hu X, Carpenter MS, Schnittler HJ, and Feldmann H (2011). A new Ebola virus nonstructural glycoprotein expressed through RNA editing. J. Virol. 85, 5406–5414. 10.1128/JVI.02190-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de La Vega MA, Wong G, Kobinger GP, and Qiu X (2015). The multiple roles of sGP in Ebola pathogenesis. Viral Immunol. 28, 3–9. 10.1089/vim.2014.0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson JA, Hevey M, Bakken R, Guest S, Bray M, Schmaljohn AL, and Hart MK (2000). Epitopes involved in antibody-mediated protection from Ebola virus. Science 287, 1664–1666. 10.1126/science.287.5458.1664. [DOI] [PubMed] [Google Scholar]
- 26.Qiu X, Wong G, Audet J, Bello A, Fernando L, Alimonti JB, Fausther-Bovendo H, Wei H, Aviles J, Hiatt E, et al. (2014). Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature 514, 47–53. 10.1038/nature13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kasereka MC, Ericson AD, Conroy AL, Tumba L, Mwesha OD, and Hawkes MT (2020). Prior vaccination with recombinant Vesicular Stomatitis Virus - Zaire Ebolavirus vaccine is associated with improved survival among patients with Ebolavirus infection. Vaccine 38, 3003–3007. 10.1016/j.vaccine.2020.02.044. [DOI] [PubMed] [Google Scholar]
- 28.Ilunga Kalenga O, Moeti M, Sparrow A, Nguyen VK, Lucey D, and Ghebreyesus TA (2019). The ongoing Ebola epidemic in the Democratic Republic of Congo, 2018–2019. N. Engl. J. Med. 381, 373–383. 10.1056/NEJMsr1904253. [DOI] [PubMed] [Google Scholar]
- 29.Chung AW, Kumar MP, Arnold KB, Yu WH, Schoen MK, Dunphy LJ, Suscovich TJ, Frahm N, Linde C, Mahan AE, et al. (2015). Dissecting polyclonal vaccine-induced humoral immunity against HIV using systems Serology. Cell 163, 988–998. 10.1016/j.cell.2015.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gunn BM, and Alter G (2016). Modulating antibody functionality in infectious disease and vaccination. Trends Mol. Med. 22, 969–982. 10.1016/j.molmed.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan YN, Boesch AW, Osei-Owusu NY, Emileh A, Crowley AR, Cocklin SL, Finstad SL, Linde CH, Howell RA, Zentner I, et al. (2016). IgG binding characteristics of rhesus macaque FcγR. J. Immunol. 197, 2936–2947. 10.4049/jimmunol.1502252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown EP, Dowell KG, Boesch AW, Normandin E, Mahan AE, Chu T, Barouch DH, Bailey-Kellogg C, Alter G, and Ackerman ME (2017). Multiplexed Fc array for evaluation of antigen-specific antibody effector profiles. J. Immunol. section 443, 33–44. 10.1016/j.jim.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boesch AW, Miles AR, Chan YN, Osei-Owusu NY, and Ackerman ME (2017). IgG Fc variant cross-reactivity between human and rhesus macaque FcγRs. mAbs 9, 455–465. 10.1080/19420862.2016.1274845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khurana S, Fuentes S, Coyle EM, Ravichandran S, Davey RT Jr., and Beigel JH (2016). Human antibody repertoire after VSV-Ebola vaccination identifies novel targets and virus-neutralizing IgM antibodies. Nat. Med. 22, 1439–1447. 10.1038/nm.4201. [DOI] [PubMed] [Google Scholar]
- 35.Escudero-Perez B, Volchkova VA, Dolnik O, Lawrence P, and Volchkov VE (2014). Shed GP of Ebola virus triggers immune activation and increased vascular permeability. PLoS Pathog. 10, e1004509. 10.1371/journal.ppat.1004509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohan GS, Li W, Ye L, Compans RW, and Yang C (2012). Antigenic subversion: a novel mechanism of host immune evasion by Ebola virus. PLoS Pathog. 8, e1003065. 10.1371/journal.ppat.1003065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lood C, Arve S, Ledbetter J, and Elkon KB (2017). TLR7/8 activation in neutrophils impairs immune complex phagocytosis through shedding of FcgRIIA. J. Exp. Med. 214, 2103–2119. 10.1084/jem.20161512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nimmerjahn F, and Ravetch JV (2006). Fcgamma receptors: old friends and new family members. Immunity 24, 19–28. 10.1016/j.immuni.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 39.Anthony RM, Kobayashi T, Wermeling F, and Ravetch JV (2011). Intravenous gammaglobulin suppresses inflammation through a novel T(H)2 pathway. Nature 475, 110–113. 10.1038/nature10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boruchov AM, Heller G, Veri MC, Bonvini E, Ravetch JV, and Young JW (2005). Activating and inhibitory IgG Fc receptors on human DCs mediate opposing functions. J. Clin. Invest. 115, 2914–2923. 10.1172/JCI24772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eisfeld AJ, Halfmann PJ, Wendler JP, Kyle JE, Burnum-Johnson KE, Peralta Z, Maemura T, Walters KB, Watanabe T, Fukuyama S, et al. (2017). Multi-platform ‘Omics analysis of human Ebola virus disease pathogenesis. Cell Host Microbe 22, 817–829.e8. 10.1016/j.chom.2017.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bruhns P (2012). Properties of mouse and human IgG receptors and their contribution to disease models. Blood 119, 5640–5649. 10.1182/blood-2012-01-380121. [DOI] [PubMed] [Google Scholar]
- 43.Niemuth NA, Fallacara D, Triplett CA, Tamrakar SM, Rajbhandari A, Florence C, Ward L, Griffiths A, Carrion R Jr., Goez-Gazi Y, et al. (2021). Natural history of disease in cynomolgus monkeys exposed to Ebola virus Kikwit strain demonstrates the reliability of this non-human primate model for Ebola virus disease. PLoS One 16, e0252874. 10.1371/journal.pone.0252874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sibley L, Daykin-Pont O, Sarfas C, Pascoe J, White AD, and Sharpe S (2021). Differences in host immune populations between rhesus macaques and cynomolgus macaque subspecies in relation to susceptibility to Mycobacterium tuberculosis infection. Sci. Rep. 11, 8810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sibley L, Gooch K, Wareham A, Gray S, Chancellor A, Dowall S, Bate S, Marriott A, Dennis M, White AD, et al. (2019). Differences in monocyte: lymphocyte ratio and Tuberculosis disease progression in genetically distinct populations of macaques. Sci. Rep. 9, 3340. 10.1038/s41598-019-39819-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ng J, Trask JS, Smith DG, and Kanthaswamy S (2015). Heterospecific SNP diversity in humans and rhesus macaque (Macaca mulatta). J. Med. Primatol. 44, 194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kanthaswamy S, Ng J, Ross CT, Trask JS, Smith DG, Buffalo VS, Fass JN, and Lin D (2013). Identifying human-rhesus macaque gene orthologs using heterospecific SNP probes. Genomics 101, 30–37. 10.1016/j.ygeno.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kanthaswamy S, Ng J, Satkoski Trask J, George DA, Kou AJ, Hoffman LN, Doherty TB, Houghton P, and Smith DG (2013). The genetic composition of populations of cynomolgus macaques (Macaca fascicularis) used in biomedical research. J. Med. Primatol. 42, 120–131. 10.1111/jmp.12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beaudoin-Bussières G, Chen Y, Ullah I, Prévost J, Tolbert WD, Symmes K, Ding S, Benlarbi M, Gong SY, Tauzin A, et al. (2022). A Fc-enhanced NTD-binding non-neutralizing antibody delays virus spread and synergizes with a nAb to protect mice from lethal SARS-CoV-2 infection. Cell Rep. 38, 110368. 10.1016/j.celrep.2022.110368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prévost J, Gasser R, Beaudoin-Bussières G, Richard J, Duerr R, Laumaea A, Anand SP, Goyette G, Benlarbi M, Ding S, et al. (2020). Cross-sectional evaluation of humoral responses against SARS-CoV-2 spike. Cell Rep. Med. 1, 100126. 10.1016/j.xcrm.2020.100126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laing ED, Weiss CD, Samuels EC, Coggins SA, Wang W, Wang R, Vassell R, Sterling SL, Tso MS, Conner T, et al. (2022). Durability of antibody response and frequency of SARS-CoV-2 infection 6 Months after COVID-19 vaccination in healthcare workers. Emerg. Infect. Dis. 28, 828–832. 10.3201/eid2804.212037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pegu A, O’Connell SE, Schmidt SD, O’Dell S, Talana CA, Lai L, Albert J, Anderson E, Bennett H, Corbett KS, et al. (2021). Durability of mRNA-1273 vaccine-induced antibodies against SARS-CoV-2 variants. Science 373, 1372–1377. 10.1126/science.abj4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bowyer G, Sharpe H, Venkatraman N, Ndiaye PB, Wade D, Brenner N, Mentzer A, Mair C, Waterboer T, Lambe T, et al. (2020). Reduced Ebola vaccine responses in CMV+ young adults is associated with expansion of CD57+KLRG1+ T cells. J. Exp. Med. 217, e20200004. 10.1084/jem.20200004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ledgerwood JE, DeZure AD, Stanley DA, Coates EE, Novik L, Enama ME, Berkowitz NM, Hu Z, Joshi G, Ploquin A, et al. (2017). Chimpanzee adenovirus vector Ebola vaccine. N. Engl. J. Med. 376, 928–938. 10.1056/NEJMoa1410863. [DOI] [PubMed] [Google Scholar]
- 55.Meyer M, Gunn BM, Malherbe DC, Gangavarapu K, Yoshida A, Pietzsch C, Kuzmina NA, Saphire EO, Collins PL, Crowe JE, et al. (2021). Ebola vaccine-induced protection in nonhuman primates correlates with antibody specificity and Fc-mediated effects. Sci. Transl. Med. 13, eabg6128. 10.1126/scitranslmed.abg6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meyer M, Malherbe DC, and Bukreyev A (2019). Can Ebola virus vaccines have universal immune correlates of protection? Trends Microbiol. 27, 8–16. 10.1016/j.tim.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cooper CL, Martins KA, Stronsky SM, Langan DP, Steffens J, Van Tongeren S, and Bavari S (2017). T-cell-dependent mechanisms promote Ebola VLP-induced antibody responses, but are dispensable for vaccine-mediated protection. Emerg. Microb. Infect. 6, e46. 10.1038/emi.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Menicucci AR, Jankeel A, Feldmann H, Marzi A, and Messaoudi I (2019). Antiviral innate responses induced by VSV-EBOV vaccination contribute to rapid protection. mBio 10, e00597–19. 10.1128/mBio.00597-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kugelman JR, Rossi CA, Wiley MR, Ladner JT, Nagle ER, Pfeffer BP, Garcia K, Prieto K, Wada J, Kuhn JH, and Palacios G (2016). Informing the Historical Record of Experimental Nonhuman Primate Infections with Ebola Virus: Genomic Characterization of USAMRIID Ebola Virus/H.sapiens-tc/COD/1995/Kikwit-9510621 Challenge Stock “R4368” and Its Replacement “R4415. PLoS One 11, e0150919. 10.1371/journal.pone.0150919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bray M, Davis K, Geisbert T, Schmaljohn C, and Huggins J (1998). A mouse model for evaluation of prophylaxis and therapy of Ebola hemorrhagic fever. J. Infect. Dis. 178, 651–661. 10.1086/515386. [DOI] [PubMed] [Google Scholar]
- 61.Gibb TR, Bray M, Geisbert TW, Steele KE, Kell WM, Davis KJ, and Jaax NK (2001). Pathogenesis of experimental Ebola Zaire virus infection in BALB/c mice. J. Comp. Pathol. 125, 233–242. 10.1053/jcpa.2001.0502. [DOI] [PubMed] [Google Scholar]
- 62.Niemuth NA, Rudge TL Jr., Sankovich KA, Anderson MS, Skomrock ND, Badorrek CS, and Sabourin CL (2020). Method feasibility for cross-species testing, qualification, and validation of the Filovirus Animal Nonclinical Group anti-Ebola virus glycoprotein immunoglobulin G enzyme-linked immunosorbent assay for non-human primate serum samples. PLoS One 15, e0241016. 10.1371/journal.pone.0241016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rudge TL Jr., Sankovich KA, Niemuth NA, Anderson MS, Badorrek CS, Skomrock ND, Cirimotich CM, and Sabourin CL (2019). Development, qualification, and validation of the Filovirus Animal Nonclinical Group anti-Ebola virus glycoprotein immunoglobulin G enzyme-linked immunosorbent assay for human serum samples. PLoS One 14, e0215457. 10.1371/journal.pone.0215457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McMahan K, Yu J, Mercado NB, Loos C, Tostanoski LH, Chandrashekar A, Liu J, Peter L, Atyeo C, Zhu A, et al. (2021). Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature 590, 630–634. 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gorman MJ, Patel N, Guebre-Xabier M, Zhu AL, Atyeo C, Pullen KM, Loos C, Goez-Gazi Y, Carrion R Jr., Tian JH, et al. (2021). Fab and Fc contribute to maximal protection against SARS-CoV-2 following NVX-CoV2373 subunit vaccine with Matrix-M vaccination. Cell Rep. Med. 2, 100405. 10.1016/j.xcrm.2021.100405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Siddiqui SM, Bowman KA, Zhu AL, Fischinger S, Beger S, Maron JS, Bartsch YC, Atyeo C, Gorman MJ, Yanis A, et al. (2022). Serological markers of SARS-CoV-2 reinfection. mBio 13, e0214121. 10.1128/mbio.02141-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Irvine EB, O’Neil A, Darrah PA, Shin S, Choudhary A, Li W, Honnen W, Mehra S, Kaushal D, Gideon HP, et al. (2021). Robust IgM responses following intravenous vaccination with Bacille Calmette-Guérin associate with prevention of Mycobacterium tuberculosis infection in macaques. Nat. Immunol. 22, 1515–1523. 10.1038/s41590-021-01066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alter G, Yu WH, Chandrashekar A, Borducchi EN, Ghneim K, Sharma A, Nedellec R, McKenney KR, Linde C, Broge T, et al. (2020). Passive transfer of vaccine-elicited antibodies protects against SIV in rhesus macaques. Cell 83, 185–196.e14. 10.1016/j.cell.2020.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nguyen DC, Scinicariello F, and Attanasio R (2011). Characterization and allelic polymorphisms of rhesus macaque (Macaca mulatta) IgG Fc receptor genes. Immunogenetics 63, 351–362. 10.1007/s00251-011-0514-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Any data reported in this paper is available from the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


