Summary
Long non-coding RNAs (lncRNAs) are known to perform important regulatory functions in lipid metabolism. Large-scale whole-genome sequencing (WGS) studies and new statistical methods for variant set tests now provide an opportunity to assess more associations between rare variants in lncRNA genes and complex traits across the genome. In this study, we used high-coverage WGS from 66,329 participants of diverse ancestries with measurement of blood lipids and lipoproteins (LDL-C, HDL-C, TC, and TG) in the National Heart, Lung, and Blood Institute (NHLBI) Trans-Omics for Precision Medicine (TOPMed) program to investigate the role of lncRNAs in lipid variability. We aggregated rare variants for 165,375 lncRNA genes based on their genomic locations and conducted rare-variant aggregate association tests using the STAAR (variant-set test for association using annotation information) framework. We performed STAAR conditional analysis adjusting for common variants in known lipid GWAS loci and rare-coding variants in nearby protein-coding genes. Our analyses revealed 83 rare lncRNA variant sets significantly associated with blood lipid levels, all of which were located in known lipid GWAS loci (in a ±500-kb window of a Global Lipids Genetics Consortium index variant). Notably, 61 out of 83 signals (73%) were conditionally independent of common regulatory variation and rare protein-coding variation at the same loci. We replicated 34 out of 61 (56%) conditionally independent associations using the independent UK Biobank WGS data. Our results expand the genetic architecture of blood lipids to rare variants in lncRNAs.
Keywords: rare variants, lncRNA, whole-genome sequencing, blood lipid, cholesterol, association
Wang and colleagues conducted rare-variant association analyses of long non-coding RNAs (lncRNAs) in 66,000 ancestrally diverse TOPMed participants with whole-genome sequencing and measurements of blood lipids and lipoproteins. The findings suggest an additional genomic element in known lipid gene regions that is distinct from the known lipid-associated genes.
Introduction
Blood lipid levels, including low-density lipoprotein cholesterol (LDL-C), total cholesterol (TC), triglyceride (TG), and high-density lipoprotein cholesterol (HDL-C), are quantitative clinically important traits with well-described monogenic and polygenic bases.1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19 Abnormal blood lipid levels contribute to risk of coronary heart disease (CHD), and in clinical practice, several treatments, including statins and PCSK9 and ANGPTL3 inhibitors,20,21,22 are available to reduce the risk of developing CHD. Each of these therapeutics has supporting evidence of their efficacy from human genetic analysis of blood lipid levels.20,21,22,23
Long non-coding RNAs (lncRNAs) are broadly defined as transcripts greater than 200 nucleotides (nt) in length that biochemically resemble mRNAs but do not code for proteins.24 Compared with protein-coding genes, lncRNAs show lower and more tissue-specific expression.25 lncRNAs are known to perform important regulatory functions in lipid metabolism.26,27,28 For example, lncRNA APOA1-AS can inhibit the transcription of the APO gene cluster that codes for protein components of lipoproteins29; lncRNA LeXis can facilitate interaction between the liver X receptor (LXR) and sterol regulatory element-binding protein transcription factors to regulate hepatic sterol content and serum cholesterol levels.30 Rare variants in lncRNAs have not been systematically explored for their impact on blood lipid levels. In addition, there are difficulties in defining testing units and selecting qualifying variants.31 Rapidly growing knowledge about the regulatory elements of the non-coding genome,32,33,34,35,36,37 large-scale whole-genome sequencing (WGS) studies,38,39,40 and new statistical methods41,42,43 for variant set tests provide the possibility to assess the associations between blood lipid traits and the genome-wide impact of lncRNAs.
We examined the associations of rare variants in lncRNAs from high-coverage WGS of 66,329 participants from diverse ancestry who have blood lipid traits (LDL-C, HDL-C, TC, and TG) in the National Heart, Lung, and Blood Institute (NHLBI) Trans-omics for Precision Medicine (TOPMed) program freeze 8 data.38 We show that the rare noncoding variants in lncRNA genes located near genes associated with Mendelian dyslipidemia disorders contribute to phenotypic variation in lipid levels among unselected individuals from population-based studies independently of common variants associated with blood lipid levels.
Material and methods
Overview
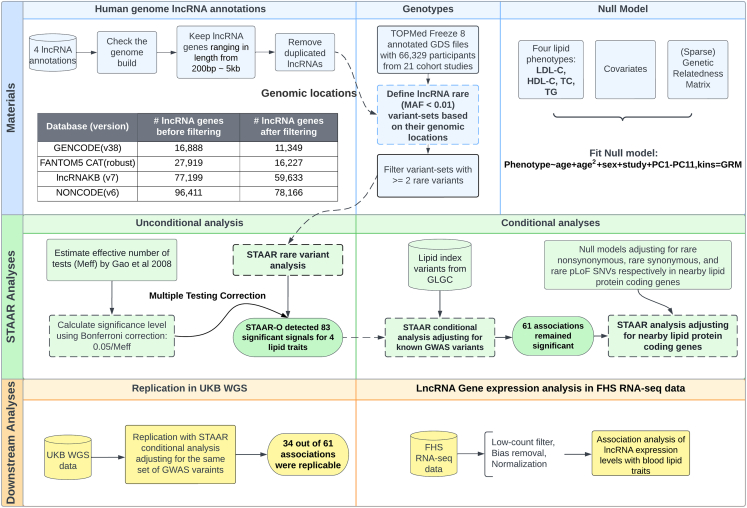
We performed a comprehensive evaluation of the association between quantitative blood lipid traits and rare variants in lncRNA genes across the genome (Figure 1). We systematically curated more than 165,000 lncRNA genes from the union of four human genome lncRNA annotations, including GENCODE,25,32,33 FANTOM5 CAT,34 NONCODE,35 and lncRNAKB.36 We utilized the TOPMed freeze 8 dataset of 66,329 participants from 21 studies with WGS and measured blood lipid levels and performed the rare-variant (minor allele frequency [MAF] <1%) association tests of curated lncRNA genes with four blood lipid phenotypes: LDL-C, HDL-C, TC, and TG. We further conducted conditional analysis adjusting for known genome-wide association study (GWAS) variants from the Global Lipids Genetics Consortium (GLGC).18 Associations between lncRNA genes and lipids that were conditionally independent from the GWAS variants (conditional p value < 6.0 × 10−04) were then tested using the variant-set test for association using annotation information (STAAR) procedure for conditional analysis adjusting for rare nonsynonymous variants (MAF <1%) within the closest protein-coding gene to each lncRNA gene as well as the nearby genes associated with Mendelian lipid disorders. We further performed replication in ∼140,000 genomes from UK Biobank (UKB).44 We intersected our results with the gene expression signatures of lipid traits in 1,505 participants from the Framingham Heart Study (FHS)45 with RNA sequencing (RNA-seq) data and blood lipid levels and observed evidence that the lncRNA rare variants may both influence their gene expression levels and impact lipid traits.
Figure 1.
A schematic illustration of the study
We performed the rare-variant association tests of 165,000 curated lncRNA genes with lipid phenotypes (i.e., LDL-C, HDL-C, TC, and TG) using the TOPMed freeze 8 data. A total of 66,329 participants from 21 studies with WGS and measured blood lipid levels were analyzed using STAAR framework. We further conducted a series of conditional analyses adjusting for known lipid GWAS variants and the nearby protein-coding genes (rare nonsynonymous, rare synonymous, and rare pLoF variants, separately). We replicated the results using an independent UKB WGS cohort. Finally, gene expression levels of the significantly lipid-associated lncRNAs were investigated in FHS RNA-seq data. TOPMed, Trans-Omics for Precision Medicine; UKB, UK Biobank; FHS, Framingham Heart Study; GLGC, Global Lipids Genetics Consortium; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; TC, total cholesterol; TG, triglycerides; lncRNA, long non-coding RNA; GWAS, genome wide association study; STAAR, variant-set test for association using annotation information; pLoF, predicted loss-of-function; MAF, minor allele frequency; SNVs, single-nucleotide variants.
Discovery and replication cohorts
Discovery cohorts
The discovery cohort included 66,329 participants in the NHLBI TOPMed from 21 cohort studies with freeze 8 WGS and blood lipid levels available: Old Order Amish (Amish; n = 1,083), Atherosclerosis Risk in Communities study (ARIC; n = 8,016), Mt. Sinai BioMe Biobank (BioMe; n = 9,848), Coronary Artery Risk Development in Young Adults (CARDIA; n = 3,056), Cleveland Family Study (CFS; n = 579), Cardiovascular Health Study (CHS; n = 3,456), Diabetes Heart Study (DHS; n = 365), FHS (n = 3,992), Genetic Studies of Atherosclerosis Risk (GeneSTAR; n = 1,757), Genetic Epidemiology Network of Arteriopathy (GENOA; n = 1,046), Genetic Epidemiology Network of Salt Sensitivity (GenSalt; n = 1,772), Genetics of Lipid-Lowering Drugs and Diet Network (GOLDN; n = 926), Hispanic Community Health Study - Study of Latinos (HCHS-SOL; n = 7,714), Hypertension Genetic Epidemiology Network and Genetic Epidemiology Network of Arteriopathy (HyperGEN; n = 1,853), Jackson Heart Study (JHS; n = 2,847), Multi-Ethnic Study of Atherosclerosis (MESA; n = 5,290), Massachusetts General Hospital Atrial Fibrillation Study (MGH_AF; n = 683), San Antonio Family Study (SAFS; n = 619), Samoan Adiposity Study (Samoan; n = 1,182), Taiwan Study of Hypertension using Rare Variants (THRV; n = 1,982), and Women’s Health Initiative (WHI; n = 8,263). The discovery cohorts consisted of 29,502 (44.5%) White individuals, 16,983 (25.6%) Black individuals, 13,943 (21.0%) Hispanic individuals, 4,719 (7.1%) Asian individuals, and 1,182 (1.8%) Samoan individuals. More information for study descriptions can be found in the supplemental notes and Table S1.
Replication cohorts
The UKB is a large, population-based prospective cohort of half a million United Kingdom residents aged 40–69 years that were recruited between 2006 and 2010.46 Consent was previously obtained from each participant regarding storage of biological specimens, genetic sequencing, access to all available electronic health record (EHR) data, and permission to recontact for future studies. All UKB participants gave written informed consent per UKB primary protocol. The UKB WGS data consist of whole genomes of 150,119 UKB participants with an average coverage of 32.5×.44 We used joint-called variant call formats (VCFs) from GraphTyper, which consist of 710,913,648 variants. We sought to replicate the findings using the UKB WGS data for 139,849 genomes with blood lipid traits available, including 116,335 White individuals, 23,335 non-White individuals, and 179 individuals missing reported ancestry (Table S2). We used VCFs provided on the UKB and conducted all the analysis in UKB Research Analysis Platform (UKB RAP).
Ethical regulations
The overall study was approved by the institutional review board (IRB) of the Boston University Medical Center. Individual studies were approved by the appropriate IRBs, and informed consent was obtained from all participants. All UKB participants gave written informed consent per the UKB primary protocol. Secondary use of the UKB data was approved by the Massachusetts General Hospital IRB (protocol 2021P002228) and was facilitated through UKB application 7089.
TOPMed WGS freeze 8 data
Phenotype data
We included four conventionally measured blood lipids in this study: LDL-C, TC, TG, and HDL-C. Detailed phenotype calculation and harmonization were described elsewhere.40 Briefly, LDL-C was either directly measured or calculated by the Friedewald equation when TGs were < 400 mg/dL. We adjusted the TC by dividing by 0.8 and LDL-C by dividing by 0.7 when statins were present.10,39 For TGs, we additionally performed the natural log transformation for analysis because TGs were skewed. We then fitted a linear regression model for each phenotype to obtain the residuals after adjusting for age at lipid measurement, age2, sex, race/ancestry, study, and the first 11 ancestral principal components (PCs) (as recommended by the TOPMed Data Coordinating Center). For Amish participants, we additionally adjusted for APOB c.10580G>A (p.Arg3527Gln; rs5742904) for LDL-C and TC and adjusted for APOC3 c.55C>T (p.Arg19Ter; rs76353203) for HDL-C and TG.47,48,49 The residuals were inverse rank normalized and rescaled by the standard deviation (SD) of the original phenotype within each group.40
Genotype data
WGS data were accessed from the TOPMed freeze 8 release. DNA samples were sequenced at the >30× target coverage at seven centers (Broad Institute of MIT and Harvard, Northwest Genomics Center, New York Genome Center, Illumina Genomic Services, PSOMAGEN [formerly Macrogen], Baylor College of Medicine Human Genome Sequencing Center, and McDonnell Genome Institute [MGI] at Washington University).38 The reads were aligned to human genome build GRCh38 using the BWA-MEM algorithm. The genotype calling was performed using the TOPMed variant-calling pipeline (https://github.com/statgen/topmed_variant_calling). The resulting binary variant call format (BCF) files were converted to SeqArray genomic data storage (GDS) format and were annotated internally by curating data from multiple database sources using functional annotation of variant-online resource (FAVOR [http://favor.genohub.org]).43 The resulting annotated GDS (aGDS) files were used in this study. We computed the genetic relationship matrix (GRM) using R package PC-relate and subtracted GRM from those samples with lipid phenotypes using R package GENESIS.
Human reference genome annotations for lncRNA genes
Multiple lncRNA annotations are available. We obtained four lncRNA annotation resources with different qualities and sizes and merged them to improve comprehensiveness. They include GENCODE,25,32,33 FANTOM5 CAT,34 NONCODE,35 and lncRNAKB.36
GENCODE
GENCODE is the default human reference genome annotation for both Ensembl and UCSC genome browsers. It is also widely adopted by many large-scale genomic consortiums including TOPMed. GENCODE gene sets cover lncRNAs, pseudogenes, and small RNAs in addition to protein-coding genes. The lncRNA annotation in GENCODE is almost entirely manual, which ensures the quality and consistency of the data. We downloaded the GENCODE version 38 (December 2020) human release from https://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_human/release_38/gencode.v38.long_noncoding_RNAs.gtf.gz and kept 17,944 lncRNA genes with a stable identifier and the genomic location information.
FANTOM CAT
The functional annotation of the mammalian genome (FANTOM) CAGE-associated transcriptome (CAT) meta-assembly combines both published sources and in-house short-read assemblies. It utilizes CAGE tags, which mark transcription start sites (TSSs), to identify human lncRNA genes with high-confidence 5′ ends. We acquired the FANTOM CAT (lv3 robust) lncRNAs assembly from https://fantom.gsc.riken.jp/5/suppl/Hon_et_al_2016/data/assembly/lv3_robust/FANTOM_CAT.lv3_robust.only_lncRNA.gtf.gz. Because the FANTOM5 annotations were on genome v.hg19 (GRCh37), we lifted over to genome version hg38 (GRCh38) using the UCSC liftOver tool.50
lncRNAKB
Long non-coding RNA Knowledgebase (lncRNAKB) is an integrated resource for exploring lncRNA biology in the context of tissue specificity and disease association. A systematic integration of annotations using a cumulative stepwise intersection method from six independent databases resulted in 77,199 human lncRNAs. We downloaded the lncRNAKB v.7 from https://osf.io/ru4d2/.
NONCODE
NONCODE database integrates annotations from both literature searches and other public databases. The latest version, NONCODE v.6, is the single largest collection of lncRNAs, describing 96,422 lncRNA genes in humans. Each lncRNA gene in the NONCODE database has been assigned a unique NONCODE ID. We downloaded the whole NONCODE v.6 human data from http://www.noncode.org/datadownload/NONCODEv6_hg38.lncAndGene.bed.gz.
Integration across the lncRNA annotations
We kept only those lncRNA genes ranging in length from 200 nt to 5 kilobases (kb). We limited the maximum length of a lncRNA gene to 5 kb to control for the computational complexity.51 Overlapping lncRNA genes between FANTOM and GENCODE using the Ensembl stable identifier were removed. We split each annotation file into individual files by chromosome with the start and end coordinates of the lncRNA genes. All duplicated lncRNAs between annotation files were removed by checking whether they have the same start and end coordinates. We then used the following intersection order based on experimental validation to merge the four lncRNA annotations: (1) GENCODE, (2) FANTOM5 CAT, (3) NONCODE, and (4) lncRNAKB. Approximately 165,000 lncRNA genes were left for further analysis.
lncRNA rare-variant association test
lncRNA rare-variant sets
We obtained the start and end genomic coordinates (human genome build GRCh38) of the lncRNA genomic regions from our previously curated lncRNA gene list. We then defined aggregation units by using all the rare variants (MAF <1%) based on their genomic locations with respect to the start and end genomic coordinates of the lncRNA genes. We removed lncRNA rare-variant sets that had fewer than two rare variants. For sensitivity analysis, we only aggregated exonic and splicing variants in lncRNA genes provided by GENCODE v.29, which is the default genome annotation employed by TOPMed consortium.38
STAAR unconditional analysis
We applied the STAAR framework to identify rare variants in the lncRNA variant sets that are associated with four quantitative lipid traits (LDL-C, HDL-C, TG, and TC). STAAR is a scalable and powerful variant-set test that uses an omnibus multi-dimensional weighting scheme to incorporate both qualitative functional categories and multiple in silico variant-annotation scores for genetic variants. STAAR accounts for population structure and relatedness, and is scalable for analyzing large WGS studies of continuous and dichotomous traits by fitting linear and logistic mixed models.41 To perform the STAAR unconditional analysis, we first fitted an STAAR null model using fit_null_glmmkin() function to account for sample relatedness with phenotypic data, covariates, and (sparse) GRM as input. For each of the four lipid phenotypes, we adjusted for age, age2, sex, study, and PC1–PC11. We adapted the STAAR gene-centric analysis for lncRNA by grouping all the rare variants (MAF <1%) within each lncRNA region. We calculated the p value for each lncRNA rare-variant set using STAAR-O, an omnibus test in the STAAR framework that combines p values from multiple annotation-weighted burden tests, SKAT, and ACAT-V using the ACAT method. A total of 13 aggregated variant functional annotations were incorporated in STAAR-O, including three integrative scores (CADD,52 LINSIGHT,53 and FATHMM-XF54) and 10 annotation principal components (aPCs)42 (Table S3). All analyses were performed using R packages STAAR (v.0.9.6) and STAARpipeline (v.0.9.6).
STAAR conditional analysis adjusting for known GLGC GWAS variants
We performed conditional analysis to identify lncRNA rare-variant association independent of known lipid-associated variants. We obtained a list of 1,750 significant index variants (Table S4) associated with one or more lipid levels from GLGC’s latest lipid GWAS results.18 Those significant index variants were identified iteratively starting with the most significant variant and grouping the surrounding region into a locus based on the larger of either ±500 kb or ±0.25 cM, followed by a conditional analysis using rareGWAMA, as previously described.18,19,55 The GLGC results were in genome build 37, and thus we lifted over the positions of GLGC index variants to genome build 38 to match the TOPMed data. For each lncRNA gene, we adjusted for the GLGC index variants falling in a ±500-kb window beyond that lncRNA gene.
STAAR rare-variant association test adjusting for nearby protein-coding genes
The unconditional analysis showed that most lncRNA genes associated with lipids are near known lipid genes that cause Mendelian lipid disorders (Table S5). We sought to perform conditional analyses adjusting lncRNA rare-variant sets for nearby protein-coding genes. The adjusted nearby protein-coding genes can be divided into two categories: the closest protein-coding genes to each lncRNA gene and genes associated with Mendelian lipid disorders, including ANGPTL8, APOA1, APOA5, APOB, APOC1, APOC3, APOE, CETP, LDLR, LPA, LPL, PCSK7, PCSK9, PLA2G15, and TM6SF2.19 Our primary analysis was to adjust for only rare nonsynonymous variants (MAF <1%) within nearby protein-coding genes. We did two sensitivity analyses: one adjusted for rare synonymous variants (MAF <1%) within nearby protein-coding genes and another adjusted for rare predicted loss-of-function (pLoF) variants (MAF <1%) within nearby protein-coding genes. For each participant, we created three burden scores separately by combining the minor allele counts of nonsynonymous, synonymous, and pLoF variants with an MAF <1% carried within the closest gene and the nearby lipid monogenic genes in a 250-kb window. We re-fitted null models similar to the unconditional analysis and added all the burden scores of the closest gene and the nearby genes associated with monogenic lipid disorders (if any) as additional covariates for each lipid phenotype. We then repeated the STAAR procedures to calculate the STAAR-O p values after adjusting for rare nonsynonymous, rare synonymous, and rare pLoF variants.
Effective number of independent tests
Although we removed redundant lncRNAs, the remaining lncRNAs can still have overlapping regions across different genome annotations. Therefore, we adopted a principal component analysis (PCA)-based approach, the simpleM method, to calculate the effective number of independent tests.56 For each chromosome, suppose we had tested K lncRNA rare-variant sets (lncRNA1, lncRNA2, …, lncRNAK) for n individuals (1, 2, …, n); we first found the minor allele counts of rare variants (MAF <1%) carried by each individual within each lncRNA rare-variant set that were tested by STAAR and constructed a matrix. We then derived the pairwise lncRNA correlation matrix RKxK that reflected the correlation structure among the tests from the constructed matrix. We calculated the eigenvalues, , from the pairwise lncRNA correlation matrix RKxK. The effective number of tests (Meff) for each chromosome was estimated as , where was a pre-defined parameter that was set to 0.95. We added up the effective number of tests (Meff) by each chromosome, assuming independence between chromosomes. The Bonferroni correction formula was then used to calculate the adjusted significance level as 0.05/Meff as used for unconditional analysis.
lncRNA gene expression analysis
Study participants
This study included 1,505 participants from the FHS Third Generation cohorts.45 Blood samples for RNA-seq were collected from Third Generation participants who attended the second examination cycle (2008–2011). Protocols for participant examinations and collection of genetic materials were approved by the IRB at Boston Medical Center. All participants provided written informed consent for genetic studies. All research was performed in accordance with relevant guidelines/regulations.
RNA-seq data collection, quality control, and data adjustment
The process of collection and isolation of RNA from whole blood was described previously.57 All RNA samples were sequenced by an NHLBI TOPMed program reference laboratory (Northwest Genomics Center) following the TOPMed RNA-seq protocol.38 All RNA-seq data were processed by the University of Washington. The raw reads (in FASTQ files) were aligned using the GRCh38 reference build to generate BAM files. The RNA-SeQC58 software was used for processing of RNA-seq data by the TOPMed RNA-seq pipeline to derive standard quality control metrics from aligned reads. Gene-level expression quantification was provided as read counts and transcripts per million (TPMs). GENCODE v.30 annotation was used for annotating gene-level expression. We performed the trimmed mean of M values (TMM) normalization on the gene read counts of RNA-seq data using the edgeR R/Bioconductor package.59,60 We removed the lowly expressed transcripts that have an SD equal to 0. To minimize confounding, expression residuals were generated by regressing log2(TMM+1) values on technical covariates including year of blood collection, batch (sequencing machine and time, plate, and well), and RNA concentration.
Predicted complete blood count
Because 80% of the participants in this study had directly measured cell count variables and only 20% received imputed variables, partial least squares (PLS) method61 was used to create predicted complete blood count (CBC) data based on the RNA-seq data. To improve the prediction, we set the Basophil percentage (BA_PER) that is greater than 3 as missing. We performed a PLS prediction method with 3-fold cross-validation (2/3 samples for training and 1/3 for validation) to impute these blood-cell components using gene expression from RNA-seq.62 We then tested the accuracy in the testing dataset. Prediction accuracy (R-squared) varied across blood component: white blood cell (WBC), 58%; platelet, 27%; neutrophil percentage, 82%; lymphocyte percentage, 85%; monocyte percentage, 77%; eosinophil percentage, 87%; and BA_PER, 32%.
Statistical analysis
We fitted a linear mixed-effects model for the residuals of the TMM-normalized log2-transformed counts data and the lipid phenotypes adjusting for predicted CBC, constructed surrogate variables (SVs), sex, age, and family structure as variance-covariance matrix using R/Bioconductor package GENESIS.63 SVs are covariates constructed directly from gene expression data to adjust for unknown, unmodeled, or latent sources of noise.64 We estimated the SVs from expression residuals and each lipid phenotype using the R/Bioconductor sva package.65 For each association, we collected the effect estimate (β), T statistics, and p values.
Genome build
All genome coordinates in this manuscript are given in the NCBI GRCh38/UCSC hg38 version of the human genome.
Results
Characteristics of TOPMed participants
We included 66,329 diverse participants from 21 cohort studies in the NHLBI TOPMed consortium with blood lipid levels. The discovery cohorts consisted of 29,502 (44.5%) reported White, 16,983 (25.6%) reported Black, 13,943 (21.0%) reported Hispanic, 4,719 (7.1%) reported Asian, and 1,182 (1.8%) reported Samoan participants (Table S1 and supplemental notes). Among the 66,329 participants, 41,182 (62%) were female. The mean age of the 66,329 participants was 53 years (SD = 15). The mean ages at lipid measurement varied across 21 cohorts from 25 years (SD = 3.56) for the CARDIA to 73 years (SD = 5.38) for the CHS. We observed that the Amish cohort had a higher concentration of LDL-C (140 [SD = 43] mg/dL) and HDL-C (56 [SD = 16] mg/dL) as well as lower TG (median 63 [IQR = 50] mg/dL), consistent with the known founder mutations in APOB and APOC3.39
Identification of rare lncRNA variants associated with blood lipid traits
We defined lncRNA testing units using the available genomic positions in four genome annotation projects described in the material and methods. There were 11,349 lncRNA genes obtained from GENCODE, 16,227 from FANTOM5 CAT, 78,166 from NONCODE, and 59,633 from lncRNAKB. In total, we tested 165,375 lncRNA genes, among which the average number of rare variants in each lncRNA was 483 (SD = 572) and the median number of rare variants in each lncRNA was 241. The minimum and the maximum number of rare variants among the lncRNAs being tested are 2 and 2,947 (Figure S1).
Our aggregation of lncRNAs across four lncRNA resources led to an overlap in the lncRNA units, leading to non-independent tests of association of the lncRNAs with blood lipid levels. We estimated the effective number of tests (Meff) using a PCA-based approach56 because the traditional Bonferroni correction would be too conservative and reduce power to detect association with blood lipid levels.31 Meff was estimated as 111,550, providing a Bonferroni correction significance threshold of 4.5 10−7.
We applied STAAR framework41 to identify the lncRNA rare-variant sets that associated with quantitative lipid traits (LDL-C, HDL-C, TC, and TG) using TOPMed WGS data. STAAR-O identified 83 genome-wide significant associations (28 with LDL-C, 20 with TC, 19 with HDL-C, and 16 with TG) (Tables 1 and S5). Among the 83 genome-wide significant associations, there are 54 unique lncRNAs. Among 54 unique lncRNAs, 28 are associated with specific lipid levels, 16 are associated with both LDL-C and TC, 7 are associated with both HDL-C and TG, and the remaining 3 lncRNAs (ENSG00000267282.1, NONHSAG026007.2, NONHSAG026009.2) are associated with three lipid traits: LDL-C, TC, and TG. The 3 lncRNAs are all on chromosome 19 neighboring the NECTIN2-TOMM40-APOE-APOC1 region. We observed that all the significant associations in the unconditional analysis were in the known lipid GWAS loci (defined as a ±500-kb window beyond a GLGC index variant) (Table S5). We performed a sensitivity analysis aggregating only exonic and splicing variants in lncRNA genes and observed consistent results to our primary analysis results (Figure S2).
Table 1.
Summary of unconditional analysis, conditional analyses, and replication
| Method | LDL-C | TC | HDL-C | TG | Total number |
|---|---|---|---|---|---|
| STAAR unconditional analysisa | 28 | 20 | 19 | 16 | 83 |
| Conditioning on known lipid GWAS variantsb | 20 | 14 | 15 | 12 | 61 |
| Conditioning on rare nonsynonymous variants within the closest gene and nearby lipid monogenic genesc | 18 | 13 | 15 | 12 | 58 |
| Conditioning on rare synonymous variants within the closest gene and nearby lipid monogenic genesc | 20 | 14 | 15 | 12 | 61 |
| Conditioning on rare pLoF variants within the closest gene and nearby lipid monogenic genesc | 20 | 14 | 15 | 12 | 61 |
| Replication in UKB WGSc | 13 | 7 | 8 | 6 | 34 |
Numbers are count of significant lipid-associated lncRNAs. Results are available in Table S5. STAAR, variant-set test for association using annotation information; GWAS, genome-wide association study; UKB, UK Biobank; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; TC, total cholesterol; TG, triglycerides; lncRNA, long non-coding RNA.
Bonferroni correction level of 0.05/111,550 = 4.5 × 10−07.
Bonferroni correction level of 0.05/83 = 6.0 × 10−04.
Bonferroni correction level of 0.05/61 = 8.2 × 10−04.
Conditional analyses of trait-associated lncRNAs adjusting for known GWAS variants and nonsynonymous variants within the nearby genes associated with monogenic lipid disorders
After conditioning on known lipid-associated variants in a ±500-kb window beyond a variant set, 61 out of 83 associations (73%) remained significant (20 with LDL-C, 14 with TC, 15 with HDL-C, and 12 with TG) at the Bonferroni-corrected level of 0.05/83 = 6.0 10−4, indicating that the associations between the lncRNA genes and lipid levels are distinct from the known GWAS variants (Table S5). The known lipid GWAS variants adjusted for each lncRNA association are shown in Table S5. The most significant association for LDL-C and TC was the lncRNA NONHSAG026007.2 (chr19:44,892,420–44,903,056) near the NECTIN2-TOMM40-APOE-APOC1 region. NONHSAG026007.2 remained significantly associated with LDL-C (p value = 2.44 10−15) and TC (p value = 2.17 10−27) after adjusting for nearby known lipid-associated variants (Figure 2). The most significant associations for HDL-C and TG were NONHSAG063125.1 (chr11:116,790,241–116,805,983) and NONHSAG09700.3 (chr11:116,773,068–116,779,841), respectively, both near APOA5-APOC3-APOA1 region. NONHSAG063125.1 remained similarly associated after conditioning on known lipid GWAS variants, while NONHSAG09700.3 became even more significant (Figure 2). We then conditioned the GWAS-distinct associations on the rare nonsynonymous variants within the closest protein-coding gene and nearby genes associated with monogenic lipid disorders and observed that most (94.9%) of the lncRNA associations with lipid levels remained significant (Table 1; Figure S3). Additionally, when conditioned on the rare synonymous variants or rare pLoF variants within the closest protein-coding gene and nearby genes associated with monogenic lipid disorders, the number of associations remained the same as the number of GWAS-distinct associations (Table 1; Figure S4).
Figure 2.
Significantly associated lncRNAs with four blood lipid traits
The significantly associated lncRNA genes (STAAR-O p value < 4.5 × 10−07) are ordered by chromosome, followed by genomic positions. Dots in red and blue represent the −log10(STAAR-O p value) of the STAAR unconditional and conditional analysis adjusting for known lipid-associated GWAS variants, respectively. The black dashed line is the Bonferroni correction level of 0.05/83 = 6.0 × 10−04. Arrows indicate at least 104-fold change of STAAR-O p values comparing the unconditional analysis and conditional analysis adjusting for known lipid-associated GWAS variants.
Replication of significant lncRNA-blood lipid trait associations
Replication of 61 lncRNAs associated with blood lipid levels was evaluated in 139,849 UKB individuals with WGS and blood lipid levels (Table S2). We replicated 34 out of 61 (56%) lncRNA associations with blood lipid levels at a Bonferroni-corrected threshold of 0.05/61 = 8.2 × 10−04 (Table S5). The most significant associations in the UK Biobank replication were NONHSAG025996.2 (chr19:44,694,720–44,696,054) near APOE-APOC1 region for LDL-C, NONHSAG109604.1 near APOE-APOC1 region for TC, and NONHSAG009700.3 near APOA5-APOC3-APOA1 region for both HDL-C and TG (Table S5), which were consistent with the results from TOPMed.
lncRNA gene expression analysis in FHS RNA-seq data
We overlapped the significant lipid-associated lncRNA genes with the lncRNA genes available in the FHS RNA-seq data generated by TOPMed.57 Because the gene-level expression data in FHS is annotated by GENCODE v.30, we limited the lncRNA genes to those presented in GENCODE. Among the 54 unique lncRNA genes that are significantly associated with either one of the lipid traits using TOPMed WGS data, 10 lncRNA genes are annotated by GENCODE, and 8 out of 10 can be found in the FHS data. We performed association analyses of expression levels of those 8 significant lipid-associated lncRNA genes with blood lipid levels (LDL-C, TC, HDL-C, TG) (Table S6). In total, we tested 12 associations of lncRNA gene expression with blood lipid levels (Table S6). The small proportion of overlapping was partially due to lncRNA genes’ generally lower expression. The lowly expressed genes were filtered out when processing the gene expression data.
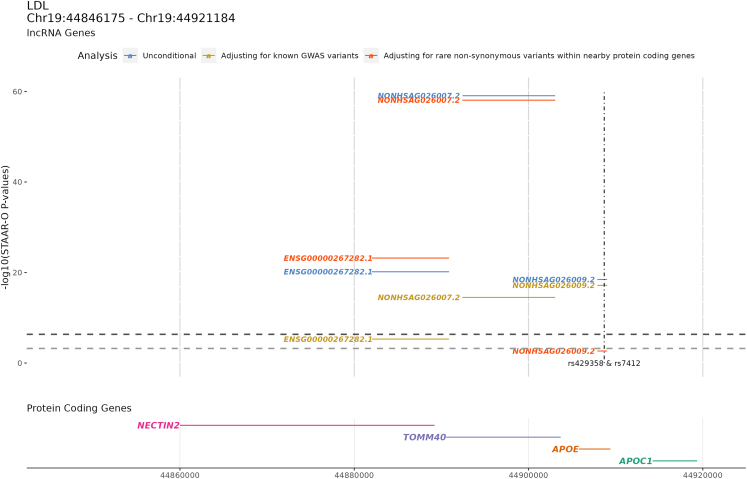
Four associations achieved Bonferroni-adjusted significance, including the gene expression level of ENSG00000267282.1 (chr19:44,881,088–44,890,922) associated with LDL-C, TC, and TG, and the gene expression level of ENSG00000266936.1 (chr19:11,010,917–11,016,011) associated with TC. ENSG00000267282.1 is an antisense of NECTIN2 (also known as PVRL2) (Figure 3). NECTIN2 encodes a single-pass type I membrane glycoprotein and operates as a cholesterol-responsive gene. It was identified in the atherosclerotic arterial wall as one of the genes that was notably downregulated in response to plasma cholesterol lowering (PCL) in atherosclerosis-prone mice with a human-like plasma cholesterol profile.66 Additionally, ENSG00000267282.1 was one of the lncRNA associations that we replicated in the independent UKB (Table S5). We also queried whether the rare variants in this lipid-associated lncRNA led to an alteration of the corresponding lncRNA levels in the blood. However, due to the small number of overlapping individuals between FHS RNA-seq data and TOPMed WGS data (n = 512), the number of rare variants tested in ENSG00000267282.1 for the association of its gene expression level was 59. Compared with the original analysis using all 66,329 individuals for the association with lipid levels, the number of rare variants tested in ENSG00000267282.1 is 1,417. As a result, the association of the rare variants in the ENSG00000267282.1 with ENSG00000267282.1 gene expression levels in blood was not significant (STAAR-O p value = 0.68).
Figure 3.
lncRNAs in the APOE region associated with LDL-C
Upper shows the −log10(STAAR-O p value) of the STAAR unconditional analysis, STAAR conditional analysis adjusting on known lipid GWAS variants, and STAAR conditional analysis adjusting for rare non-synonymous variants within the closest protein-coding gene and nearby genes associated with monogenic lipid disorders. The bottom is the nearby protein-coding genes with the genomic coordinates. The vertical dashed line is the position of the known GWAS variants that were conditioned on. The black horizontal dashed line is the Bonferroni correction level of 0.05/111,550 = 4.5 × 10−07, and the gray horizontal dashed line is the Bonferroni correction level of 0.05/83 = 6.0 × 10−04.
Lookup for previously reported lncRNA therapeutic target
We further investigated one lncRNA, liver-expressed LXR-induced sequence (LeXis), which is a mediator of the complex effects of LXR signaling on hepatic lipid metabolism to maintain hepatic sterol content and serum cholesterol levels.30,67 A potential ortholog of LeXis in humans, TCONS_00016452 (chr9:104,990,086–104,991,780), is found in a region adjacent to ABCA1. It was not a significant signal for any lipid trait in our study, which might suggest that it was not a functional ortholog of LeXis that substantially influences the blood lipid traits we measured. However, the rapid evolutionary turnover of lncRNAs still hinders the functional identification between species.68
Discussion
In this study, we conducted genome-wide rare-variant associations of 165,000 lncRNAs in ancestrally diverse TOPMed participants (n = 66,329) with measured blood lipid levels. Using rare-variant association tests, we observed 83 lncRNAs significantly associated with blood lipid levels, and of these, 61 (73%) were conditionally distinct from common regulatory variation and rare protein-coding variation at the same loci. Notably, most of these association signals were replicated in an independent WGS dataset, UKB. We also highlighted one trait-associated lncRNA that is close to NECTIN2 and TOMM40, ENSG00000267282.1 (chr19:44,881,088–44,890,922), whose gene expression level was also shown to be associated with lipid levels using RNA-seq data from the FHS data. Together, this systematic assessment of rare lncRNA variants suggests an additional genomic element in known lipid loci that is distinct from the known lipid-associated genes.
Genetic variation for blood lipid levels has been observed across the allelic spectrum with common, rare coding, and rare non-coding variants.40 Blood lipids have been associated with non-coding regulatory variants and coding variation in genes and are now also associated with rare variants in lncRNAs. We show that all the trait-associated lncRNAs are in genomic regions previously associated with blood lipid traits (Table S5), leading to the plausibility of these results. About 75% of the associations are conditionally distinct from common regulatory variation and rare protein-coding variation at the same loci previously identified through GWAS and whole-exome sequencing studies. This indicates that the regulatory variants through lncRNAs additionally contribute to the variation of blood lipid levels.
Despite numerous reports indicating the potential regulatory role of lncRNAs, only a small proportion of them have substantial evidence to support such claims.26,27,68 The fraction of lncRNAs that are functional remains unknown. Through a comprehensive study of over 165,000 lncRNAs, we found that the majority of lncRNAs are not associated with a lipid trait. However, there are still some lncRNAs that harbor variants that predispose individuals to phenotypic differences in blood lipid levels. Our results suggest that investigators should first prioritize individual lncRNAs near the known trait-associated loci (e.g., ANGPTL8, APOA1, APOA5, APOB, APOC1, APOC3, APOE, CETP, LDLR, LPA, LPL, PCSK7, PCSK9, PLA2G15, and TM6SF2) for analysis, which is more likely to yield robust experimental observations.
lncRNAs are involved in diverse aspects of lipid metabolism, including mechanisms with effects at the transcriptional level, post-transcriptional level, and directly on proteins.26 Our results highlight the therapeutic potential of lncRNAs that overlap with nearby protein-coding genes in both the anti-sense and sense direction. Some lncRNAs have already been reported to act in cis to regulate the expression of the neighboring protein-coding genes—for example, APOA1-AS and APOA4-AS.69 Novel therapeutics for lipid-associated lncRNAs could be developed by either targeting DNA by adeno-associated virus (AAV) vectors/CRISPR-Cas9 system or targeting RNA by antisense oligonucleotides (ASOs)/small interfering RNA (siRNA).70
Several limitations of our study should be noted. First, we didn’t consider lncRNAs with slightly different start and end coordinates as duplications when we created the curated list of lncRNAs. Second, our RNA-seq analyses were restricted to GENCODE annotation. The small proportion of overlapping RNA-seq data and WGS data limits the ability to test rare lncRNA variants with their gene expression. Third, we did not correct for the number of tested lipid traits. However, there is a moderate-to-high correlation among the blood lipid levels. For example, using the data from the TOPMed participants, we calculated that the correlation between LDL-C and TC is 0.91 and the correlation between HDL-C and TG is 0.44. Therefore, correcting for the number of tested lipid traits would lead to overcorrection. Fourth, to assess a causal role of the rare lncRNA variants, we need to further show that they are correlated with lncRNA expression but not correlated with altered expression or function of other genes nearby.
In summary, we show in a large ancestrally diverse study that lncRNAs are an additional genomic element in known lipid gene regions associated with blood lipoprotein levels that are distinct from the known genes. We comprehensively evaluated 165,000 lncRNAs for their association with lipid traits and replicated signals in an independent UKB WGS cohort.
Data and code availability
The lncRNA annotations being used in this study are publicly available to download: GENCODE (https://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_human/), FANTOM5 CAT (https://fantom.gsc.riken.jp/cat/), lncRNAKB (https://osf.io/ru4d2/), and NONCODE (http://www.noncode.org/datadownload/). The curated list of lncRNAs is available on GitHub: https://github.com/kyleyxw/lncRNA-paper. Individual whole-genome sequence data for TOPMed and harmonized lipids at individual sample level are available through restricted access via the TOPMed dbGaP Exchange area. Summary-level genotype data from TOPMed are available through the BRAVO browser (https://bravo.sph.umich.edu/). The UK Biobank (UKB) whole-genome sequence data can be accessed through UKB Research Analysis Platform (RAP) through the UKB approval system (https://www.ukbiobank.ac.uk). The dbGaP accessions for TOPMed cohorts are as follows: Old Order Amish (Amish), phs000956 and phs00039; Atherosclerosis Risk in Communities study (ARIC), phs001211 and phs000280; Mt. Sinai BioMe Biobank (BioMe), phs001644 and phs000925; Coronary Artery Risk Development in Young Adults (CARDIA), phs001612 and phs000285; Cleveland Family Study (CFS), phs000954 and phs000284; Cardiovascular Health Study (CHS), phs001368 and phs000287; Diabetes Heart Study (DHS), phs001412 and phs001012; Framingham Heart Study (FHS), phs000974 and phs000007; Genetic Studies of Atherosclerosis Risk (GeneSTAR), phs001218 and phs000375; Genetic Epidemiology Network of Arteriopathy (GENOA), phs001345 and phs001238; Genetic Epidemiology Network of Salt Sensitivity (GenSalt), phs001217 and phs000784; Genetics of Lipid-Lowering Drugs and Diet Network (GOLDN), phs001359 and phs000741; Hispanic Community Health Study - Study of Latinos (HCHS_SOL), phs001395 and phs000810; Hypertension Genetic Epidemiology Network and Genetic Epidemiology Network of Arteriopathy (HyperGEN), phs001293 and phs001293; Jackson Heart Study (JHS), phs000964 and phs000286; Multi-Ethnic Study of Atherosclerosis (MESA), phs001416 and phs000209; Massachusetts General Hospital Atrial Fibrillation Study (MGH_AF), phs001062 and phs001001; San Antonio Family Study (SAFS), phs001215 and phs000462; Samoan Adiposity Study (SAS), phs000972 and phs000914; Taiwan Study of Hypertension using Rare Variants (THRV), phs001387 and phs001387; and Women’s Health Initiative (WHI), phs001237 and phs000200.
All analyses were performed using R Statistical Software (v.3.6.2; R Core Team 2019). R code for implementing the analysis is available at the public GitHub Repository https://github.com/kyleyxw/lncRNA-paper. STAAR is implemented as an open-source R package available at https://github.com/xihaoli/STAAR. STAARpipeline is implemented as an open-source R package available at https://github.com/xihaoli/STAARpipeline.
Acknowledgments
Whole-genome sequencing (WGS) for the Trans-Omics in Precision Medicine (TOPMed) program was supported by the National Heart, Lung, and Blood Institute (NHLBI). G.M.P. is supported by NIH grants R01HL142711 and R01HL127564. P.N. is supported by grants from the National Heart, Lung, and Blood Institute (R01HL142711, R01HL148050, R01HL151283, R01HL148565, R01HL135242, and R01HL151152), Fondation Leducq (TNE-18CVD04), and Massachusetts General Hospital (Paul and Phyllis Fireman Endowed Chair in Vascular Medicine). X. Lin is supported by grants R35-CA197449, U19-CA203654, R01-HL113338, and U01-HG009088. We would like to acknowledge all the grants that supported this study: R01 HL121007, U01 HL072515, R01 AG18728, X01HL134588, HL 046389, HL113338, and 1R35HL135818, K01 HL135405, R03 HL154284, U01HL072507, R01HL087263, R01HL090682, P01HL045522, R01MH078143, R01MH078111, R01MH083824, U01DK085524, R01HL113323, R01HL093093, R01HL133040, R01HL140570, R01HL142711, R01HL127564, R01HL148050, R01HL148565, HL105756, and Leducq TNE-18CVD04. The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute; the National Institutes of Health; or the U.S. Department of Health and Human Services. We gratefully acknowledge the studies and participants who provided biological samples and data for TOPMed and UK Biobank. The full study-specific acknowledgments and NHLBI TOPMed Fellowship acknowledgment are detailed in Supplementary Notes.
Author contributions
Y.W., P.N., and G.M.P. designed the study. Y.W. carried out all the primary analysis with critical input from P.N. and G.M.P. M.S.S. carried out the replication analysis. Y.W. and J.A.H. carried out the secondary analysis. Y.W., M.S.S., X. Li, Z.L., A.K.D., J.C.B., J.B., E.B., D.W.B., B.E.C., J.C.C., A.P.C., Y.C., J.E.C., P.S.D., S.K.D., P.T.E., J.S.F., M.F., B.I.F., S. Gabriel, S. Germer, R.A.G., X.G., J.H., N.H., B.H., L.H., M.R.I., R.J., R.C.K., S.L.R.K., T.N.K., R.K., C.K., B.G.K., D.L., C. Li, C. Liu, D.L.-J., R.J.F.L., M.C.M., L.W.M., R.A.M., R.L.M., B.D.M., M.E.M., A.C.M., J.M.M., T.N., J.R.O., N.D.P., M.H.P., B.M.P., L.M.E., D.C.R., S.R., A.P.R., S.S.R., M.R., W.H.-H.S., J.A.S., A.S., H.K.T., M.Y.T., K.A.V., Z.W., L.R.Y., W.Z., J.I.R., X. Lin., P.N., and G.M.P. acquired, analyzed, or interpreted data. G.M.P. and P.N. and NHLBI TOPMed Lipids Working Group provided administrative, technical, or material support. Y.W. and G.M.P. wrote the first draft of the manuscript and revised it according to suggestions by the coauthors. All authors critically reviewed the manuscript, suggested revisions as needed, and approved the final version.
Declaration of interests
P.N. reports research grants from Allelica, Apple, Amgen, Boston Scientific, Genentech/Roche, and Novartis; personal fees from Allelica, Apple, AstraZeneca, Blackstone Life Sciences, Eli Lilly & Co, Foresite Labs, Genentech/Roche, GV, HeartFlow, Magnet Biomedicine, and Novartis; scientific advisory board membership of Esperion Therapeutics, Preciseli, and TenSixteen Bio; scientific co-founder of TenSixteen Bio; equity in MyOme, Preciseli, and TenSixteen Bio; and spousal employment at Vertex Pharmaceuticals, all unrelated to the present work. B.M.P. serves on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. L.M.R., S.S.R., and R.M. are consultants for the TOPMed Administrative Coordinating Center (through Westat). M.E.M. receives funding from Regeneron Pharmaceutical Inc. unrelated to this work. X. Lin is a consultant of AbbVie Pharmaceuticals and Verily Life Sciences. P.T.E. receives sponsored research support from Bayer AG, IBM Research, Bristol Myers Squibb, Pfizer, and Novo Nordisk; he has also served on advisory boards or consulted for Bayer AG, MyoKardia, and Novartis. A.P.C. previously received investigator-initiated grant support from Amgen, Inc. unrelated to the present work.
Published: October 5, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2023.09.003.
Supplemental information
References
- 1.Diabetes Genetics Initiative of Broad Institute of Harvard and MIT Lund University and Novartis Institutes of BioMedical Research. Saxena R., Voight B.F., Lyssenko V., Burtt N.P., de Bakker P.I.W., Chen H., Roix J.J., Kathiresan S., Hirschhorn J.N., et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 2.Kathiresan S., Manning A.K., Demissie S., D’Agostino R.B., Surti A., Guiducci C., Gianniny L., Burtt N.P., Melander O., Orho-Melander M., et al. A genome-wide association study for blood lipid phenotypes in the Framingham Heart Study. BMC Med. Genet. 2007;8 doi: 10.1186/1471-2350-8-S1-S17. S17–S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kathiresan S., Melander O., Anevski D., Guiducci C., Burtt N.P., Roos C., Hirschhorn J.N., Berglund G., Hedblad B., Groop L., et al. Polymorphisms Associated with Cholesterol and Risk of Cardiovascular Events. N. Engl. J. Med. 2008;358:1240–1249. doi: 10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
- 4.Teslovich T.M., Musunuru K., Smith A.V., Edmondson A.C., Stylianou I.M., Koseki M., Pirruccello J.P., Ripatti S., Chasman D.I., Willer C.J., et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asselbergs F.W., Guo Y., Van Iperen E.P.A., Sivapalaratnam S., Tragante V., Lanktree M.B., Lange L.A., Almoguera B., Appelman Y.E., Barnard J., et al. Large-scale gene-centric meta-analysis across 32 studies identifies multiple lipid loci. Am. J. Hum. Genet. 2012;91:823–838. doi: 10.1016/j.ajhg.2012.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Albrechtsen A., Grarup N., Li Y., Sparsø T., Tian G., Cao H., Jiang T., Kim S.Y., Korneliussen T., Li Q., et al. Exome sequencing-driven discovery of coding polymorphisms associated with common metabolic phenotypes. Diabetologia. 2013;56:298–310. doi: 10.1007/s00125-012-2756-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tachmazidou I., Dedoussis G., Southam L., Farmaki A.E., Ritchie G.R.S., Xifara D.K., Matchan A., Hatzikotoulas K., Rayner N.W., Chen Y., et al. A rare functional cardioprotective APOC3 variant has risen in frequency in distinct population isolates. Nat. Commun. 2013;4:1–6. doi: 10.1038/ncomms3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willer C.J., Schmidt E.M., Sengupta S., Peloso G.M., Gustafsson S., Kanoni S., Ganna A., Chen J., Buchkovich M.L., Mora S., et al. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holmen O.L., Zhang H., Fan Y., Hovelson D.H., Schmidt E.M., Zhou W., Guo Y., Zhang J., Langhammer A., Løchen M.L., et al. Systematic evaluation of coding variation identifies a candidate causal variant in TM6SF2 influencing total cholesterol and myocardial infarction risk. Nat. Genet. 2014;46:345–351. doi: 10.1038/ng.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peloso G.M., Auer P.L., Bis J.C., Voorman A., Morrison A.C., Stitziel N.O., Brody J.A., Khetarpal S.A., Crosby J.R., Fornage M., et al. Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56,000 whites and blacks. Am. J. Hum. Genet. 2014;94:223–232. doi: 10.1016/j.ajhg.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Surakka I., Horikoshi M., Mägi R., Sarin A.P., Mahajan A., Lagou V., Marullo L., Ferreira T., Miraglio B., Timonen S., et al. The impact of low-frequency and rare variants on lipid levels. Nat. Genet. 2015;47:589–597. doi: 10.1038/ng.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang C.S., Zhang H., Cheung C.Y.Y., Xu M., Ho J.C.Y., Zhou W., Cherny S.S., Zhang Y., Holmen O., Au K.W., et al. Exome-wide association analysis reveals novel coding sequence variants associated with lipid traits in Chinese. Nat. Commun. 2015;6:1–9. doi: 10.1038/ncomms10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu D.J., Peloso G.M., Yu H., Butterworth A.S., Wang X., Mahajan A., Saleheen D., Emdin C., Alam D., Alves A.C., et al. Exome-wide association study of plasma lipids in >300,000 individuals. Nat. Genet. 2017;49:1758–1766. doi: 10.1038/ng.3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu X., Peloso G.M., Liu D.J., Wu Y., Zhang H., Zhou W., Li J., Tang C.S.M., Dorajoo R., Li H., et al. Exome chip meta-analysis identifies novel loci and East Asian–specific coding variants that contribute to lipid levels and coronary artery disease. Nat. Genet. 2017;49:1722–1730. doi: 10.1038/ng.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffmann T.J., Theusch E., Haldar T., Ranatunga D.K., Jorgenson E., Medina M.W., Kvale M.N., Kwok P.Y., Schaefer C., Krauss R.M., et al. A large electronic-health-record-based genome-wide study of serum lipids. Nat. Genet. 2018;50:401–413. doi: 10.1038/s41588-018-0064-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klarin D., Damrauer S.M., Cho K., Sun Y.V., Teslovich T.M., Honerlaw J., Gagnon D.R., DuVall S.L., Li J., Peloso G.M., et al. Genetics of blood lipids among ∼300,000 multi-ethnic participants of the Million Veteran Program. Nat. Genet. 2018;50:1514–1523. doi: 10.1038/s41588-018-0222-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spracklen C.N., Chen P., Kim Y.J., Wang X., Cai H., Li S., Long J., Wu Y., Wang Y.X., Takeuchi F. Association analyses of East Asian individuals and trans-ancestry analyses with European individuals reveal new loci associated with cholesterol and triglyceride levels. Hum. Mol. Genet. 2018;27:1122. doi: 10.1093/hmg/ddx439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graham S.E., Clarke S.L., Wu K.-H.H., Kanoni S., Zajac G.J.M., Ramdas S., Surakka I., Ntalla I., Vedantam S., Winkler T.W., et al. The power of genetic diversity in genome-wide association studies of lipids. Nature. 2021;600:675–679. doi: 10.1038/s41586-021-04064-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanoni S., Graham S.E., Wang Y., Surakka I., Ramdas S., Zhu X., Clarke S.L., Bhatti K.F., Vedantam S., Winkler T.W., et al. Implicating genes, pleiotropy, and sexual dimorphism at blood lipid loci through multi-ancestry meta-analysis. Genome Biol. 2022;23:268. doi: 10.1186/s13059-022-02837-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grundy S.M., Stone N.J., Bailey A.L., Beam C., Birtcher K.K., Blumenthal R.S., Braun L.T., de Ferranti S., Faiella-Tommasino J., Forman D.E., et al. 2018 AHA/ACC/AACVPR/AAPA/ABC/ACPM/ADA/AGS/APhA/ASPC/NLA/PCNA Guideline on the Management of Blood Cholesterol: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Circulation. 2019;139 doi: 10.1161/CIR.0000000000000625. E1082–e1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Musunuru K., Pirruccello J.P., Do R., Peloso G.M., Guiducci C., Sougnez C., Garimella K.V., Fisher S., Abreu J., Barry A.J., et al. Exome Sequencing, ANGPTL3 Mutations, and Familial Combined Hypolipidemia. N. Engl. J. Med. 2010;363:2220–2227. doi: 10.1056/NEJMoa1002926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen J.C., Boerwinkle E., Mosley T.H., Hobbs H.H. Sequence Variations in PCSK9, Low LDL, and Protection against Coronary Heart Disease. N. Engl. J. Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 23.Kathiresan S., Myocardial Infarction Genetics Consortium A PCSK9 Missense Variant Associated with a Reduced Risk of Early-Onset Myocardial Infarction. N. Engl. J. Med. 2008;358:2299–2300. doi: 10.1056/NEJMc0707445. [DOI] [PubMed] [Google Scholar]
- 24.Uszczynska-Ratajczak B., Lagarde J., Frankish A., Guigó R., Johnson R. Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet. 2018;19:535–548. doi: 10.1038/s41576-018-0017-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G., et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Solingen C., Scacalossi K.R., Moore K.J. Long noncoding RNAs in lipid metabolism. Curr. Opin. Lipidol. 2018;29:224–232. doi: 10.1097/MOL.0000000000000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muret K., Désert C., Lagoutte L., Boutin M., Gondret F., Zerjal T., Lagarrigue S. Long noncoding RNAs in lipid metabolism: literature review and conservation analysis across species. BMC Genom. 2019;20:882. doi: 10.1186/s12864-019-6093-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Statello L., Guo C.J., Chen L.L., Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2020;22:96–118. doi: 10.1038/s41580-020-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Halley P., Kadakkuzha B.M., Faghihi M.A., Magistri M., Zeier Z., Khorkova O., Coito C., Hsiao J., Lawrence M., Wahlestedt C. Regulation of the apolipoprotein gene cluster by a long noncoding RNA. Cell Rep. 2014;6:222–230. doi: 10.1016/j.celrep.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sallam T., Jones M.C., Gilliland T., Zhang L., Wu X., Eskin A., Sandhu J., Casero D., Vallim T.Q.D.A., Hong C., et al. Feedback modulation of cholesterol metabolism by the lipid-responsive non-coding RNA LeXis. Nature. 2016;534:124–128. doi: 10.1038/nature17674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bocher O., Génin E. Rare variant association testing in the non-coding genome. Hum. Genet. 2020;139:1345–1362. doi: 10.1007/s00439-020-02190-y. [DOI] [PubMed] [Google Scholar]
- 32.Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S., et al. GENCODE: The reference human genome annotation for the ENCODE project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frankish A., Diekhans M., Jungreis I., Lagarde J., Loveland J.E., Mudge J.M., Sisu C., Wright J.C., Armstrong J., Barnes I., et al. GENCODE 2021. Nucleic Acids Res. 2021;49:D916–D923. doi: 10.1093/nar/gkaa1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hon C.C., Ramilowski J.A., Harshbarger J., Bertin N., Rackham O.J.L., Gough J., Denisenko E., Schmeier S., Poulsen T.M., Severin J., et al. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature. 2017;543:199–204. doi: 10.1038/nature21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao L., Wang J., Li Y., Song T., Wu Y., Fang S., Bu D., Li H., Sun L., Pei D., et al. NONCODEV6: An updated database dedicated to long non-coding RNA annotation in both animals and plants. Nucleic Acids Res. 2021;49:D165–D171. doi: 10.1093/nar/gkaa1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seifuddin F., Singh K., Suresh A., Judy J.T., Chen Y.C., Chaitankar V., Tunc I., Ruan X., Li P., Chen Y., et al. lncRNAKB, a knowledgebase of tissue-specific functional annotation and trait association of long noncoding RNA. Sci. Data. 2020;7:326. doi: 10.1038/s41597-020-00659-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ellingford J.M., Ahn J.W., Bagnall R.D., Baralle D., Barton S., Campbell C., Downes K., Ellard S., Duff-Farrier C., FitzPatrick D.R., et al. Recommendations for clinical interpretation of variants found in non-coding regions of the genome. Genome Med. 2022;14:73. doi: 10.1186/s13073-022-01073-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taliun D., Harris D.N., Kessler M.D., Carlson J., Szpiech Z.A., Torres R., Taliun S.A.G., Corvelo A., Gogarten S.M., Kang H.M., et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590:290–299. doi: 10.1038/s41586-021-03205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Natarajan P., Peloso G.M., Zekavat S.M., Montasser M., Ganna A., Chaffin M., Khera A.V., Zhou W., Bloom J.M., Engreitz J.M., et al. Deep-coverage whole genome sequences and blood lipids among 16,324 individuals. Nat. Commun. 2018;9:3391. doi: 10.1038/s41467-018-05747-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Selvaraj M.S., Li X., Li Z., Pampana A., Zhang D.Y., Park J., Aslibekyan S., Bis J.C., Brody J.A., Cade B.E., et al. Whole genome sequence analysis of blood lipid levels in >66,000 individuals. Nat. Commun. 2022;13:1–18. doi: 10.1038/s41467-022-33510-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X., Li Z., Zhou H., Gaynor S.M., Liu Y., Chen H., Sun R., Dey R., Arnett D.K., Aslibekyan S., et al. Dynamic incorporation of multiple in silico functional annotations empowers rare variant association analysis of large whole-genome sequencing studies at scale. Nat. Genet. 2020;52:969–983. doi: 10.1038/s41588-020-0676-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Z., Li X., Zhou H., Gaynor S.M., Selvaraj M.S., Arapoglou T., Quick C., Liu Y., Chen H., Sun R., et al. A framework for detecting noncoding rare-variant associations of large-scale whole-genome sequencing studies. Nat. Methods. 2022;19:1599–1611. doi: 10.1038/s41592-022-01640-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou H., Arapoglou T., Li X., Li Z., Zheng X., Moore J., Asok A., Kumar S., Blue E.E., Buyske S., et al. FAVOR: functional annotation of variants online resource and annotator for variation across the human genome. Nucleic Acids Res. 2023;51:D1300–D1311. doi: 10.1093/nar/gkac966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Halldorsson B.V., Eggertsson H.P., Moore K.H.S., Hauswedell H., Eiriksson O., Ulfarsson M.O., Palsson G., Hardarson M.T., Oddsson A., Jensson B.O., et al. The sequences of 150,119 genomes in the UK Biobank. Nature. 2022;607:732–740. doi: 10.1038/s41586-022-04965-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Splansky G.L., Corey D., Yang Q., Atwood L.D., Cupples L.A., Benjamin E.J., D’Agostino R.B., Fox C.S., Larson M.G., Murabito J.M., et al. The Third Generation Cohort of the National Heart, Lung, and Blood Institute’s Framingham Heart Study: Design, Recruitment, and Initial Examination. Am. J. Epidemiol. 2007;165:1328–1335. doi: 10.1093/aje/kwm021. [DOI] [PubMed] [Google Scholar]
- 46.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J., et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soria L.F., Ludwig E.H., Clarke H.R., Vega G.L., Grundy S.M., McCarthy B.J. Association between a specific apolipoprotein B mutation and familial defective apolipoprotein B-100. Proc. Natl. Acad. Sci. USA. 1989;86:587–591. doi: 10.1073/pnas.86.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen H., Damcott C.M., Rampersaud E., Pollin T.I., Horenstein R.B., McArdle P.F., Peyser P.A., Bielak L.F., Post W.S., Chang Y.-P.C., et al. Familial Defective Apolipoprotein B-100 and Increased Low-Density Lipoprotein Cholesterol and Coronary Artery Calcification in the Old Order Amish. Arch. Intern. Med. 2010;170:1850–1855. doi: 10.1001/archinternmed.2010.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pollin T.I., Damcott C.M., Shen H., Ott S.H., Shelton J., Horenstein R.B., Post W., Mclenithan J.C., Bielak L.F., Peyser P.A., et al. A Null Mutation in Human APOC3 Confers a Favorable Plasma Lipid Profile and Apparent Cardioprotection ∗ NIH Public Access. Science. 2008;322:1702–1705. doi: 10.1126/science.1161524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Casper J., Zweig A.S., Villarreal C., Tyner C., Speir M.L., Rosenbloom K.R., Raney B.J., Lee C.M., Lee B.T., Karolchik D., et al. The UCSC Genome Browser database: 2018 update. Nucleic Acids Res. 2018;46:D762–D769. doi: 10.1093/nar/gkx1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lumley T., Brody J., Peloso G., Morrison A., Rice K. FastSKAT: Sequence kernel association tests for very large sets of markers. Genet. Epidemiol. 2018;42:516–527. doi: 10.1002/gepi.22136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang Y.F., Gulko B., Siepel A. Fast, scalable prediction of deleterious noncoding variants from functional and population genomic data. Nat. Genet. 2017;49:618–624. doi: 10.1038/ng.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kircher M., Witten D.M., Jain P., O’roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rogers M.F., Shihab H.A., Mort M., Cooper D.N., Gaunt T.R., Campbell C. FATHMM-XF: Accurate prediction of pathogenic point mutations via extended features. Bioinformatics. 2018;34:511–513. doi: 10.1093/bioinformatics/btx536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramdas S., Judd J., Graham S.E., Kanoni S., Wang Y., Surakka I., Wenz B., Clarke S.L., Chesi A., Wells A., et al. A multi-layer functional genomic analysis to understand noncoding genetic variation in lipids. Am. J. Hum. Genet. 2022;109:1366–1387. doi: 10.1016/j.ajhg.2022.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gao X., Starmer J., Martin E.R. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet. Epidemiol. 2008;32:361–369. doi: 10.1002/gepi.20310. [DOI] [PubMed] [Google Scholar]
- 57.Liu C., Joehanes R., Ma J., Wang Y., Sun X., Keshawarz A., Sooda M., Huan T., Hwang S.-J., Bui H., et al. Whole genome DNA and RNA sequencing of whole blood elucidates the genetic architecture of gene expression underlying a wide range of diseases. Sci. Rep. 2022;12 doi: 10.1038/s41598-022-24611-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deluca D.S., Levin J.Z., Sivachenko A., Fennell T., Nazaire M.D., Williams C., Reich M., Winckler W., Getz G. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012;28:1530–1532. doi: 10.1093/bioinformatics/bts196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robinson M.D., Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR : a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nguyen D.V. Partial least squares dimension reduction for microarray gene expression data with a censored response. Math. Biosci. 2005;193:119–137. doi: 10.1016/j.mbs.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 62.Joehanes R., Zhang X., Huan T., Yao C., Ying S.X., Sturcke A., Nguyen Q.T., Demirkale C.Y., Feolo M.L., Sharopova N.R., et al. Integrated genome-wide analysis of expression quantitative trait loci aids interpretation of genomic association studies. Genome Biol. 2017;18:16. doi: 10.1186/s13059-016-1142-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gogarten S.M., Sofer T., Chen H., Yu C., Brody J.A., Thornton T.A., Rice K.M., Conomos M.P. Genetic association testing using the GENESIS R/Bioconductor package. Bioinformatics. 2019;35:5346–5348. doi: 10.1093/bioinformatics/btz567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leek J.T., Storey J.D. Capturing Heterogeneity in Gene Expression Studies by Surrogate Variable Analysis. PLoS Genet. 2007;3:1724–1735. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leek J.T., Johnson W.E., Parker H.S., Jaffe A.E., Storey J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rossignoli A., Shang M.-M., Gladh H., Moessinger C., Foroughi Asl H., Talukdar H.A., Franzén O., Mueller S., Björkegren J.L.M., Folestad E., Skogsberg J. Poliovirus Receptor–Related 2. Arterioscler. Thromb. Vasc. Biol. 2017;37:534–542. doi: 10.1161/ATVBAHA.116.308715. [DOI] [PubMed] [Google Scholar]
- 67.Tontonoz P., Wu X., Jones M., Zhang Z., Salisbury D., Sallam T. Long Noncoding RNA Facilitated Gene Therapy Reduces Atherosclerosis in a Murine Model of Familial Hypercholesterolemia. Circulation. 2017;136:776–778. doi: 10.1161/CIRCULATIONAHA.117.029002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ponting C.P., Haerty W. Genome-Wide Analysis of Human Long Noncoding RNAs: A Provocative Review. Annu. Rev. Genomics Hum. Genet. 2022;23:153–172. doi: 10.1146/annurev-genom-112921-123710. [DOI] [PubMed] [Google Scholar]
- 69.Huang S.F., Peng X.F., Jiang L., Hu C.Y., Ye W.C. LncRNAs as Therapeutic Targets and Potential Biomarkers for Lipid-Related Diseases. Front. Pharmacol. 2021;12:729745. doi: 10.3389/fphar.2021.729745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen R., Lin S., Chen X. The promising novel therapies for familial hypercholesterolemia. J. Clin. Lab. Anal. 2022;36:e24552. doi: 10.1002/jcla.24552. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The lncRNA annotations being used in this study are publicly available to download: GENCODE (https://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_human/), FANTOM5 CAT (https://fantom.gsc.riken.jp/cat/), lncRNAKB (https://osf.io/ru4d2/), and NONCODE (http://www.noncode.org/datadownload/). The curated list of lncRNAs is available on GitHub: https://github.com/kyleyxw/lncRNA-paper. Individual whole-genome sequence data for TOPMed and harmonized lipids at individual sample level are available through restricted access via the TOPMed dbGaP Exchange area. Summary-level genotype data from TOPMed are available through the BRAVO browser (https://bravo.sph.umich.edu/). The UK Biobank (UKB) whole-genome sequence data can be accessed through UKB Research Analysis Platform (RAP) through the UKB approval system (https://www.ukbiobank.ac.uk). The dbGaP accessions for TOPMed cohorts are as follows: Old Order Amish (Amish), phs000956 and phs00039; Atherosclerosis Risk in Communities study (ARIC), phs001211 and phs000280; Mt. Sinai BioMe Biobank (BioMe), phs001644 and phs000925; Coronary Artery Risk Development in Young Adults (CARDIA), phs001612 and phs000285; Cleveland Family Study (CFS), phs000954 and phs000284; Cardiovascular Health Study (CHS), phs001368 and phs000287; Diabetes Heart Study (DHS), phs001412 and phs001012; Framingham Heart Study (FHS), phs000974 and phs000007; Genetic Studies of Atherosclerosis Risk (GeneSTAR), phs001218 and phs000375; Genetic Epidemiology Network of Arteriopathy (GENOA), phs001345 and phs001238; Genetic Epidemiology Network of Salt Sensitivity (GenSalt), phs001217 and phs000784; Genetics of Lipid-Lowering Drugs and Diet Network (GOLDN), phs001359 and phs000741; Hispanic Community Health Study - Study of Latinos (HCHS_SOL), phs001395 and phs000810; Hypertension Genetic Epidemiology Network and Genetic Epidemiology Network of Arteriopathy (HyperGEN), phs001293 and phs001293; Jackson Heart Study (JHS), phs000964 and phs000286; Multi-Ethnic Study of Atherosclerosis (MESA), phs001416 and phs000209; Massachusetts General Hospital Atrial Fibrillation Study (MGH_AF), phs001062 and phs001001; San Antonio Family Study (SAFS), phs001215 and phs000462; Samoan Adiposity Study (SAS), phs000972 and phs000914; Taiwan Study of Hypertension using Rare Variants (THRV), phs001387 and phs001387; and Women’s Health Initiative (WHI), phs001237 and phs000200.
All analyses were performed using R Statistical Software (v.3.6.2; R Core Team 2019). R code for implementing the analysis is available at the public GitHub Repository https://github.com/kyleyxw/lncRNA-paper. STAAR is implemented as an open-source R package available at https://github.com/xihaoli/STAAR. STAARpipeline is implemented as an open-source R package available at https://github.com/xihaoli/STAARpipeline.