Abstract
Machine learning potentials have emerged as a means to enhance the accuracy of biomolecular simulations. However, their application is constrained by the significant computational cost arising from the vast number of parameters compared to traditional molecular mechanics. To tackle this issue, we introduce an optimized implementation of the hybrid method (NNP/MM), which combines neural network potentials (NNP) and molecular mechanics (MM). This approach models a portion of the system, such as a small molecule, using NNP while employing MM for the remaining system to boost efficiency. By conducting molecular dynamics (MD) simulations on various protein-ligand complexes and metadynamics (MTD) simulations on a ligand, we showcase the capabilities of our implementation of NNP/MM. It has enabled us to increase the simulation speed by ~5 times and achieve a combined sampling of 1 μs for each complex, marking the longest simulations ever reported for this class of simulation.
Graphical Abstract
Introduction
In the past decade, molecular dynamics (MD) has transitioned from CPU execution to accelerators such as graphical processing units (GPUs). Starting in 2006, CELLMD1 and subsequently ACEMD2 began leveraging GPUs to enhance biomolecular simulations. Numerous other MD codes have either adapted (e.g., AMBER,3 GROMACS,4 NAMD5) or been initially designed to utilize GPUs (e.g., OpenMM,6 HOOMD,7 TorchMD8). This innovation has improved the cost efficiency of MD simulations by two orders of magnitude.9
During the same timeframe, improvements in the accuracy of molecular mechanics (MM) and its force fields (FFs) have not advanced at a comparable pace as the simulation speed. Widely adopted biomolecular FFs, such as AMBER,10,11 CHARMM,12,13 and others, offer parameters for proteins and common biomolecules. However, obtaining accurate parameters for novel drug-like molecules remains a challenging task.14 The recent development of neural network potentials (NNPs) holds promise to address this issue.15 NNPs leverage the characteristic of neural networks (NNs) as universal approximators, which means they can approximate any function with arbitrary precision relative to the training data. In the context of molecular simulations, NNPs are designed to predict the energy and forces of quantum mechanics (QM) calculations.16
Recently, numerous NNPs have been proposed (SchNet,17 TensorMol,18 AIMNet,19 PhysNet,20 DimeNet++,21 OrbNet,22 PaiNN,23 SpookyNet,24 NequIP,25 OrbNet Denali,26 TorchMD-NET,27 MACE,28 etc). One of the most used for organic molecules are ANI29 and its derivatives 30-33 based on a modified Behler-Parrinello (BP) symmetry function.34 For example, the benchmarks of ANI-2x on a set of biaryl fragment, typically found in drug molecules, shows better accuracy than the established general small molecule FFs (CGenFF,35,36 GAFF,37 OPLS,38 and OpenFF39). The mean absolute error for the entire potential energy profile and rotational barrier heights are 0.5 kcal/mol and 1.0 kcal/mol, respectively,40 but it is orders of magnitude faster than its reference QM calculations at the DFT level (ωB97X/6-31G*).33 However, the BP-type NNPs have several limitations. First, the long-range interactions are not properly accounted for. The NNPs only consider the chemical environment around each atom within a given cut-off distance (5.1 Å for ANI-2x). Second, a limited set of elements is supported (H, C, N, O, F, S, and Cl for ANI-2x). Finally, only neutral molecules can be computed.33
Despite the current limitations, NNPs are already improving biomolecular simulations. It has demonstrated that the accuracy for drug-like molecules is improved14 by reparameterizing dihedral angles with ANI-1x.30 Alternatively, the hybrid method of NNP and MM (NNP/MM)41 allows embedding NNP into a simulation. The main idea of NNP/MM is similar to QM/MM:42-44 an important region of a system is modeled with a more accurate method, while a less accurate and computationally cheaper one is used for the rest of the system.
Recently, Lahey and Rowley41 have demonstrated the first application of NNP/MM to protein-ligand complexes. NNP/MM is used to refine binding poses and to compute the conformational free energies. Rufa et al.45 have computed the binding free energies of the Tyk2 congeneric ligand benchmark series46 using alchemical free energy calculations. Instead of using NNP/MM directly, a non-equilibrium switching scheme has been devised to correct the standard MM calculations to NNP/MM accuracy. It reduces the errors from 1.0 kcal/mol to 0.5 kcal/mol. Vant et al.47 have used NNP/MM for the refinement of a protein-ligand complex from cryo-electron microscopy data. The refinement with NNP/MM produces higher-quality models than QM/MM with the semi-empirical PM6 method at a lower computational cost. Xu et al.48 have trained a specialized NNP for zinc and used NNP/MM to simulate zinc-containing proteins. The obtained results are in agreement with QM/MM calculations.
A critical limitation for the wider adoption of NNP/MM is the simulation speed. Despite NNP being much faster than QM, it is still slower than MM. For example, Lahey and Rowley41 and Vant et al.47 have reported the simulations speed of 3.4 ns/day and 0.5 ns/day, respectively, on an NVIDIA TITAN Xp GPU. Also, the longest reported simulation is just 20 ns.47
In this work, we present an optimized implementation of NNP/MM in ACEMD2 based on OpenMM6 and PyTorch.49 First, the method and relevant optimization strategies are introduced. Second, the capability of software is demonstrated by performing metadynamics (MTD)50 simulations of a fragment of erlotinib and molecular dynamics (MD) simulations of four protein-ligand complexes. Finally, the installation and setup of simulations are shown.
Methods
In the NNP/MM approach, a system is partitioned into NNP and MM regions similarly to QM/MM.42-44 The total potential energy () consists of three terms:
| (1) |
where and are the potential energies of the NNP and MM regions, respectively. is a coupling term; , , and are the atomic position of the entire system, NNP region, and MM region, respectively.
It is required that the NNP potential () is a function of the atomic position () and atomic numbers () of the NNP region. The total charge () can be included if necessary:
| (2) |
Also, it is required that is differentiable with respect to to compute the atomic forces ():
| (3) |
In this work, we adapt the coupling term () proposed by Lahey and Rowley: 41
| (4) |
where is the Coulomb potential, is the Lennard-Jones potential and and are the number of NNP and MM atoms, respectively; and are the atomic charges; and are the Lennard-Jones parameters; is the distance between the atoms; and is the vacuum permittivity (dielectric constant). In the context of QM/MM, this is known as the mechanical embedding scheme.43,44
NNP/MM is implemented in ACEMD2 using several software components. OpenMM,51 a GPU-accelerated MD library, is used to compute MM terms and propagate the MD trajectory. OpenMM-Torch, 52 an OpenMM plugin, is used to compute the NNP term. It uses PyTorch,49 a machine learning framework for NN training and inference on GPUs, to load and execute the NNP on GPU. TorchANI53 is used to create the PyTorch model of ANI-2x.33 NNPOps,54 a library of optimized CUDA kernels for NNP, is used to accelerate critical parts of the computations. Future versions will integrate other NNPs available in TorchMD-NET.27,55
We have optimized the performance of NNP/MM in three ways. First, all the terms of NNP and MM are computed on a GPU. Neither atomic positions nor atomic forces need to be transferred between the CPU and GPU, as is the case with the original implementation.41 Second, the featurizer of ANI has been implemented as a custom CUDA kernel and is available in the NNPOps library.54 The original featurizer in TorchANI is implemented using only standard PyTorch operations, which are an inefficient way of performing this calculation. Third, the computation is parallelized over the NNs (ANI-2x has an ensemble of 8 NNs) and atoms taking advantage that the same molecule is computed repeatedly. The original implementation in TorchANI computes the NNs sequentially. The original TorchANI version is optimized for batch computing, i.e. many molecules are computed simultaneously, while for MD low-latency computing, i.e. one molecule is computed as fast as possible is necessary. The weights and biases of the atomic NNs are replicated and batched in the same order as the atoms in a molecule, allowing a GPU to efficiently parallelize the calculation for a single molecule. The implementation of the optimized NNs is available in the NNPOps library (https://github.com/openmm/nnpops).
Results and Discussion
Simulations of a fragment
We use metadynamics50(MTD) to simulate a fragment (Figure 1) of erlotinib using two models: (1) the conventional MM with GAFF237 parameters for the fragment; and (2) the NNP/MM where the fragment is modeled with ANI-2x.33 Lahey and Rowley41 reported that the fragment has a notable discrepancy between the potential energy surfaces of CGenFF35 and ANI-1ccx.31 In this work, we expand the benchmark by computing the free energy surfaces.
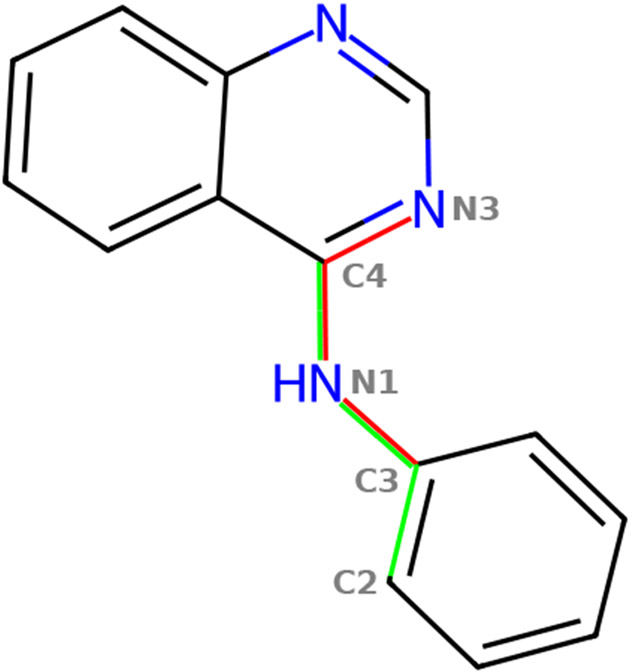
Figure 1:
A fragment of erlotinib. The two dihedral angles used as the collective variable are shown in red and green.
We use the well-tempered MTD56 with two dihedral angles (Figure 1) as collective variables. The MTD simulations use the NVT ensemble (T = 310 K), the time step is set to 4.0 fs for the MM simulations, and to 2.0 fs for the NNP/MM simulations because they are unstable with 4.0 fs. For MTD, PLUMED57 is used. More details are provided in the supplementary information.
The fragment was simulated for 100 ns with each method. This is sufficient to achieve extensive sampling in the collective variable space. The time series of the dihedral angles (Figure 1) are available in the supplementary information (Figure S1-S2).
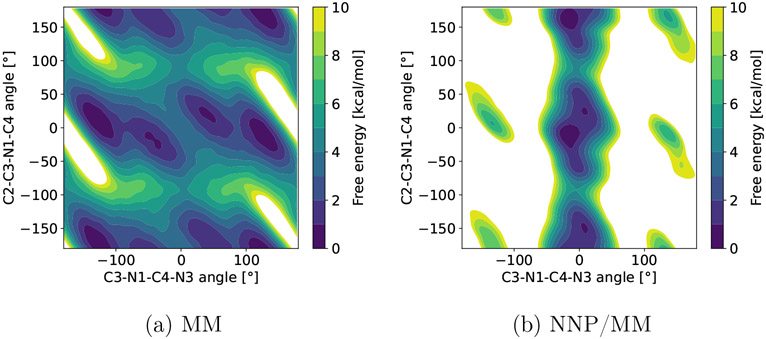
The obtained free energy surfaces (Figure 2) show a significant difference between the models. The dominant conformer of the dihedral angle C3-N1-C4-N3 is predicted by GAFF2 and ANI-2x at ~120° and ~0°, respectively. The fragment has two aromatic rings connected by a conjugated linker, so a planar conformation is expected to be energetically favorable. This is consistent with the potential energy surfaces reported by Lahey and Rowley (see Ref. 41, Figure 3b). Note, the fragment has been chosen for demonstration only and further analysis is beyond the scope of this work.
Figure 2:
Free energy surface of a fragment (Figure 1) of erlotinib computed with MTD using two models MM (a) and NNP/MM (b).
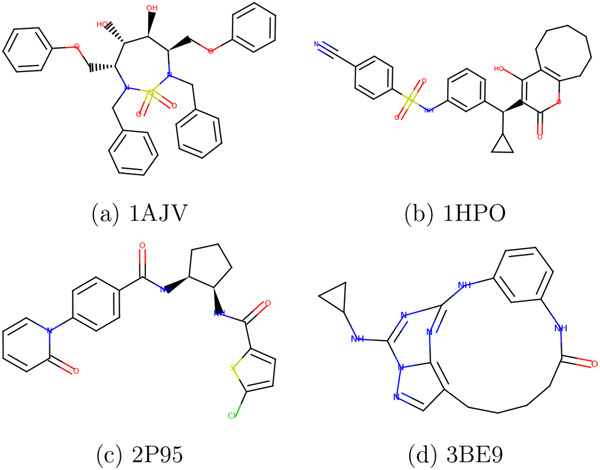
Figure 3:
Ligand structures (a-d) of the selected protein-ligand complexes.
Simulations of protein-ligand complexes
Protein-ligand complexes
We have selected four protein-ligand complexes from PDBbind-201958,59 following these criteria. First, the ligand contains only elements supported by ANI-2x (H, C, N, O, F, S, and Cl)33 and no charged functional groups (amine, carboxylate, etc). Second, the ligand has less than one hundred atoms. Third, the ligand has at least one rotatable bond, and the rotamers energies differ by >3 kcal/mol between GAFF237 and ANI-2x.33 We use the Parameterize tool14 to detect the rotatable bond, scan the dihedral angles of rotatable bonds, and compute the relative rotamer energies. The summary of the protein-ligand complexes is given in Table 1 and the ligand structures are shown in Figure 3. Additionally, the energy profiles of the dihedral angle scan of the ligands are available in the supplementary information (Figure S2-S22).
Table 1:
Summary of protein-ligand complexes.
The protein-ligand complex preparation and equilibration have been carried out with HTMD.64 Each complex has been simulated with two different methods: MM, where the ligand is parameterized with GAFF237 and NNP/MM, where the ligand is modeled with ANI-2x.33 The protein, in both cases, uses AMBER ff14SB11 FF. The MD simulations use the NVT ensemble (T = 310 K), the time step is set to 4.0 fs for the MM simulations, and to 2.0 fs for the NNP/MM simulations because they are unstable with 4.0 fs. For each combination of a complex and method, 10 independent simulations of 100 ns are performed resulting in the combined sampling of 1 μs. More details are provided in the supplementary information.
Analysis of protein-ligand complexes
All the proteins and ligands maintain their structures in the simulations with both methods (MM and NNP/MM). The protein RMSD fluctuates in the range of 1.8-2.8 Å and the residue RMSF have similar magnitudes when comparing the same protein with both methods. The ligand RMSD fluctuates in the range of 0.2-1.7 Å. In the case of 1AJV and 2P95, there is no significant difference between MM and NNP/MM, but, in the case of 1HPO and 3BE9, the fluctuations are larger by ~0.3 Å for NNP/MM. The time series of protein RMSD, residue RMSF, and ligand RMSD are available in the supplementary information (Figure S23-S34). The difference of the ligand RMSD is expected because, as previous works14,40,45 indicates, ANI-2x models the dihedral angles more accurately than GAFF. Also, it is important to note that our simulations are 50 times longer than previously reported41 and have not resulted in any non-physical conformation.
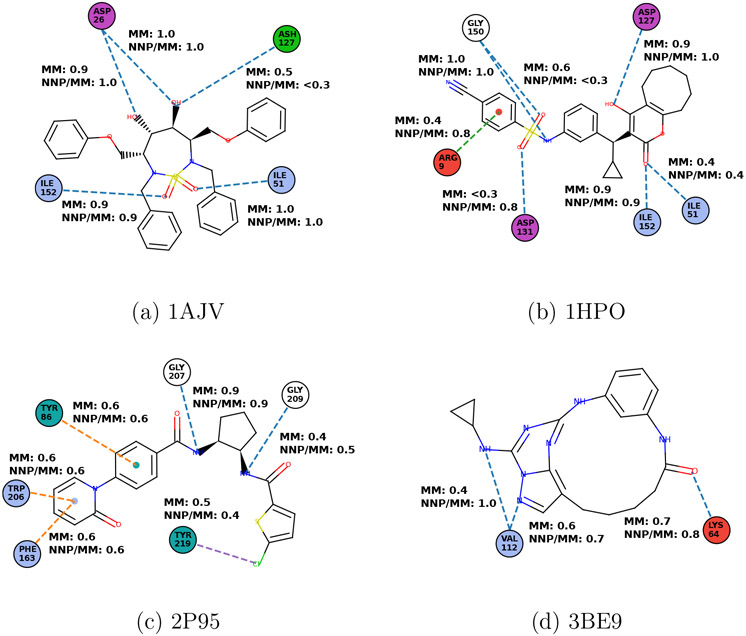
The dominant protein-ligand interactions (Figure 4) qualitatively agree between MM and NNP/MM for all the complexes. The full list of ligand-protein interactions and technical details are available in the supplementary information (Table S1-S8). Note, the protein-ligand systems have been chosen for demonstration only and further analysis is beyond the scope of this work.
Figure 4:
Probabilities of protein-ligand interactions for the different systems and methods (a-d). Protein residues are represented as disks (blue: non-polar, green: polar, red:positively charged, magenta: negatively charged, and dark cyan: aromatic). Interactions are depicted as dashed lines (blue: hydrogen bond, green: cation-π, orange: π-π interaction, and violet: σ-hole). The interactions with probabilities lower than 0.3 are excluded.
Simulation speed
On average, NNPOps54 accelerates ANI calculations (energy and forces) 6.5 times (Table 2). The is no strict dependency between the ligand size and the calculation time, which suggests significant overhead is coming from auxiliary operations rather than the computation of NNPs. The overhead mainly comes from PyTorch, which is optimized for batch computing rather than low latency.49
Table 2:
Comparison of ANI-2x inference (energy and forces) time (ms) using the original TorchANI and the TorchANI accelerated with NNPOps (TorchANI/NNPOps). The results were obtained with an NVIDIA RTX 4090 GPU.
| System | TorchANI | TorchANI/NNPOps | Speed-up |
|---|---|---|---|
| 1AJV | 11.5 | 2.17 | 5.3 |
| 1HPO | 11.3 | 1.91 | 5.9 |
| 2P95 | 13.0 | 1.54 | 8.4 |
| 3BE9 | 9.4 | 1.52 | 6.2 |
| Average | 6.5 |
Overall NNP/MM is sped up 5.3 times (Table 3) on average when NNPOps54 is used. Despite this improvement, NNP/MM is still about an order of magnitude slower than the conventional MM (Table 3), but further optimizations are possible. First, ANI-2x33 uses an ensemble of 8 NNs. If only one NN could be used, the simulations would be 2.2 times faster on average (Table 3). Second, the time step for the NNP/MM simulations has to be reduced from 4 fs to 2 fs. If the constraint scheme could be adapted to allow 4 fs timestep, the simulations would be 2 times faster. Finally, not all the software components are already fully optimized. For example, the current implementation of OpenMM-Torch (https://github.com/openmm/openmm-torch) performs the NNP and MM calculations on a GPU sequentially, but it would be more efficient to do that concurrently.
Table 3:
Comparison of MD simulation speed (ns/day) of NNP/MM using the original TorchANI, the TorchANI accelerated with NNPOps (TorchANI/NNPOps), and the TorchANI accelerated with NNPOps and just one model of ANI-2x (1 NN). For reference, MM speed is included. The results were obtained with an NVIDIA RTX 4090 GPU.
| System | NNP/MM (TorchANI)* |
NNP/MM (TorchANI/NNPOps)* |
NNP/MM (1 NN)* |
MM† |
|---|---|---|---|---|
| 1AJV | 12.6 | 60.1 | 155 | 1382 |
| 1HPO | 13.4 | 65.9 | 152 | 1227 |
| 2P95 | 12.2 | 73.5 | 147 | 1006 |
| 3BE9 | 14.0 | 74.2 | 151 | 995 |
2 fs time step
4 fs time step
Extensibility with other NNPs
Our implementation of NNP/MM is agnostic to the NNP model, i.e. it can use any model implemented with PyTorch. As a demonstration, we performed simulations with ANI-1x30 and TorchMD-NET27 trained with the ANI-1 data set.65 The simulation speed benchmarks (Table 4) are available just for 3BE9 because both NNPs are limited to 4 elements (H, C, N, and O).
Table 4:
Comparison of MD simulation speed of ANI-1x and TorchMD-NET and their accuracy in mean absolute error (MAE). The results were obtained with an NVIDIA RTX 4090 GPU.
2 fs time step
Software installation and usage
ACEMD can be installed with the Conda package management system.66 For dependencies, Conda-forge67 is used to ensure compatibility with all major Linux distributions (refer to the ACEMD documentation for details at https://software.acellera.com. The installation command:
$ conda install -c conda-forge \
-c acellera \
-c acellera/label/rc \
acemd=4
For the best performance, it is recommended to have an NVIDIA GPU and its latest drivers installed, but it is possible to run on a CPU only.
The setup of an NNP/MM simulation consists of the following steps. First, a system needs to be prepared for a conventional MM simulation (i.e. initial structure, topology, and force field parameters). Note that the NNP atoms need to be assigned partial charges and Lennard-Jones parameters to compute the coupling term correctly. The system preparation can be easily accomplished with HTMD.64,68 Second, NNP model files need to be generated with prepare-nnp tool included with ACEMD. It needs the initial structure (e.g. structure.pdb), a selection of the NNP atoms (e.g. "resname MOL"), and a name of NNP
$ prepare-nnp structure.pdb --selection "resname MOL" \
--model ANI-2x
The tool generates several files including model.json. Currently, we plan to support the NNP models from TorchANI53 and TorchMD-NET27 but other models will also be supported in the future. Finally, an ACEMD input file needs to be prepared as for a conventional MD simulation (refer to the ACEMD documentation69 for details) and needs just one additional line (nnpfile model.json) to enable NNP/MM.
Conclusion
We have showcased an optimized implementation of NNP/MM in ACEMD,2 based on OpenMM6 and PyTorch,49 which delivers simulation speeds of approximately 5 times faster than previously reported. While still slower than classical force fields, the enhanced accuracy of NNPs may justify the increased computational expense (see Rufa et al.45). We anticipate this performance gap will continue to shrink in the future. Presently, NNPs have limited applicability due to constraints on charges and elements, but improvements are expected in the near future.
We validated our implementation by conducting metadynamics simulations of an erlotinib fragment and molecular dynamics simulations of four protein-ligand complexes. The fragment simulation results are consistent with prior findings, while the complex simulations exceeded previous durations by over an order of magnitude. These outcomes confirm the effectiveness of our implementation and demonstrate its practical application. Furthermore, NNP/MM can be combined with the enhanced sampling methods (e.g. metadynamics,50 replica exchange,70 steered molecular dynamics,71 etc.) and it holds significant potential for alchemical free energy simulations.45 It is particularly beneficial for drug discovery efforts, where the simulation of novel molecules is routine but accurate force field parameters may be lacking.
Supplementary Material
Acknowledgement
The authors thank the volunteers of GPUGRID.net for donating computing time. This project has received funding from the Torres-Quevedo Programme from the Spanish National Agency for Research (PTQ-17-09078 / AEI / 10.13039/501100011033) (RG); the European Union’s Horizon 2020 research and innovation programme under grant agreement No. 823712 (RG, AV, RF); the Industrial Doctorates Plan of the Secretariat of Universities and Research of the Department of Economy and Knowledge of the Generalitat of Catalonia (AV); the Chan Zuckerberg Initiative DAF (grant number: 2020-219414), an advised fund of Silicon Valley Community Foundation (SD, PE); and the project PID2020-116564GB-I00 has been funded by MCIN / AEI / 10.13039/501100011033. Research reported in this publication was supported by the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health under award number GM140090 (PE, TEM, JDC, GDF). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. JDC is a current member of the Scientific Advisory Board of OpenEye Scientific Software, Redesign Science, Ventus Therapeutics, and Interline Therapeutics, and has equity interests in Redesign Science and Interline Therapeutics. The Chodera laboratory receives or has received funding from multiple sources, including the National Institutes of Health, the National Science Foundation, the Parker Institute for Cancer Immunotherapy, Relay Therapeutics, Entasis Therapeutics, Silicon Therapeutics, EMD Serono (Merck KGaA), AstraZeneca, Vir Biotechnology, Bayer, XtalPi, Interline Therapeutics, the Molecular Sciences Software Institute, the Starr Cancer Consortium, the Open Force Field Consortium, Cycle for Survival, a Louis V. Gerstner Young Investigator Award, and the Sloan Kettering Institute. A complete funding history for the Chodera lab can be found at http://choderalab.org/funding.
Footnotes
Supporting Information Available
- SI.pdf: the time series of the dihedral angles of the fragment; the energy profiles of the dihedral angle scan of the ligands; the protein and ligand RMSD and residue RMSF for all the simulations; and the full list of ligand-protein interactions.
Installation instructions for the software are available at https://software.acellera.com.
References
- (1).De Fabritiis G. Performance of the Cell processor for biomolecular simulations. Comput. Phys. Commun 2007, 176, 660–664. [Google Scholar]
- (2).Harvey MJ; Giupponi G; Fabritiis GD ACEMD: accelerating biomolecular dynamics in the microsecond time scale. J. Chem. Theory Comput 2009, 5, 1632–1639. [DOI] [PubMed] [Google Scholar]
- (3).Salomon-Ferrer R; Case DA; Walker RC An overview of the Amber biomolecular simulation package. Wiley Interdiscip. Rev. Comput. Mol. Sci 2013, 3, 198–210. [Google Scholar]
- (4).Abraham MJ; Murtola T; Schulz R; Páll S; Smith JC; Hess B; Lindahl E GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar]
- (5).Phillips JC; Hardy DJ; Maia JD; Stone JE; Ribeiro JV; Bernardi RC; Buch R; Fiorin G; Hénin J; Jiang W; McGreevy R; Melo MCR; Radak BK; Skeel RD; Singharoy A; Wang Y; Roux B; Aksimentiev A; Luthey-Schulten Z; Kalé LV; Schulten K; Chipot C; Tajkhorshid E Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys 2020, 153, 044130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Eastman P; Pande V OpenMM: A hardware-independent framework for molecular simulations. Comput. Sci. Eng 2010, 12, 34–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Anderson J; Keys A; Phillips C; Dac Nguyen T; Glotzer S HOOMD-blue, general-purpose many-body dynamics on the GPU. APS March Meeting Abstracts. 2010; pp Z18–008. [Google Scholar]
- (8).Doerr S; Majewski M; Pérez A; Krämer A; Clementi C; Noe F; Giorgino T; De Fabritiis G Torchmd: A deep learning framework for molecular simulations. J. Chem. Theory Comput 2021, 17, 2355–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Stone JE; Hardy DJ; Ufimtsev IS; Schulten K GPU-accelerated molecular modeling coming of age. J. Mol. Graph. Model 2010, 29, 116–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Cornell WD; Cieplak P; Bayly CI; Gould IR; Merz KM; Ferguson DM; Spellmeyer DC; Fox T; Caldwell JW; Kollman PA A Second Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules. J. Am. Chem. Soc 1995, 117, 5179–5197. [Google Scholar]
- (11).Maier JA; Martinez C; Kasavajhala K; Wickstrom L; Hauser KE; Simmerling C ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput 2015, 11, 3696–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).MacKerell AD; Bashford D; Bellott M; Dunbrack RL; Evanseck JD; Field MJ; Fischer S; Gao J; Guo H; Ha S; Joseph-McCarthy D; Kuchnir L; Kuczera K; Lau FTK; Mattos C; Michnick S; Ngo T; Nguyen DT; Prodhom B; Reiher WE; Roux B; Schlenkrich M; Smith JC; Stote R; Straub J; Watanabe M; Wiórkiewicz-Kuczera J; Yin D; Karplus M All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J. Phys. Chem. B 1998, 102, 3586–3616. [DOI] [PubMed] [Google Scholar]
- (13).Huang J; MacKerell AD Jr CHARMM36 All-Atom Additive Protein Force Field: Validation Based on Comparison to NMR Data. J. Comput. Chem 2013, 34, 2135–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Galvelis R; Doerr S; Damas JM; Harvey MJ; De Fabritiis G A Scalable Molecular Force Field Parameterization Method Based on Density Functional Theory and Quantum-Level Machine Learning. J. Chem. Inf. Model 2019, 59, 3485–3493. [DOI] [PubMed] [Google Scholar]
- (15).Noé F; Tkatchenko A; Müller K-R; Clementi C Machine learning for molecular simulation. Annu. Rev. Phys. Chem 2020, 71, 361–390. [DOI] [PubMed] [Google Scholar]
- (16).Behler J. Constructing high-dimensional neural network potentials: A tutorial review. Int. J. Quantum Chem 2015, 115, 1032–1050. [Google Scholar]
- (17).Schütt KT; Sauceda HE; Kindermans P-J; Tkatchenko A; Müller K-R SchNet–A deep learning architecture for molecules and materials. J. Chem. Phys 2018, 148, 241722. [DOI] [PubMed] [Google Scholar]
- (18).Yao K; Herr JE; Toth DW; Mckintyre R; Parkhill J The TensorMol-0.1 model chemistry: a neural network augmented with long-range physics. Chem. Sci 2018, 9, 2261–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Zubatyuk R; Smith JS; Leszczynski J; Isayev O Accurate and transferable multitask prediction of chemical properties with an atoms-in-molecules neural network. Sci. Adv 2019, 5, eaav6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Unke OT; Meuwly M PhysNet: A neural network for predicting energies, forces, dipole moments, and partial charges. J. Chem. Theory Comput 2019, 15, 3678–3693. [DOI] [PubMed] [Google Scholar]
- (21).Klicpera J; Giri S; Margraf JT; Günnemann S Fast and Uncertainty-Aware Directional Message Passing for Non-Equilibrium Molecules. arXiv preprint arXiv:2011.14115 2020, [Google Scholar]
- (22).Qiao Z; Welborn M; Anandkumar A; Manby FR; Miller TF III OrbNet: Deep learning for quantum chemistry using symmetry-adapted atomic-orbital features. J. Chem. Phys 2020, 153, 124111. [DOI] [PubMed] [Google Scholar]
- (23).Schütt K; Unke O; Gastegger M Equivariant message passing for the prediction of tensorial properties and molecular spectra. International Conference on Machine Learning. 2021; pp 9377–9388. [Google Scholar]
- (24).Unke OT; Chmiela S; Gastegger M; Schütt KT; Sauceda HE; Müller K-R SpookyNet: Learning force fields with electronic degrees of freedom and nonlocal effects. Nat. Commun 2021, 12, 7273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Batzner S; Smidt TE; Sun L; Mailoa JP; Kornbluth M; Molinari N; Kozinsky B SE(3)-equivariant graph neural networks for data-efficient and accurate interatomic potentials. arXiv preprint arXiv:2101.03164 2021, [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Christensen AS; Sirumalla SK; Qiao Z; O’Connor MB; Smith DG; Ding F; Bygrave PJ; Anandkumar A; Welborn M; Manby FR; Miller TF III OrbNet Denali: A machine learning potential for biological and organic chemistry with semi-empirical cost and DFT accuracy. J. Chem. Phys 2021, 155. [DOI] [PubMed] [Google Scholar]
- (27).Thölke P; De Fabritiis G Equivariant transformers for neural network based molecular potentials. International Conference on Learning Representations. 2022. [Google Scholar]
- (28).Batatia I; Kovacs DP; Simm G; Ortner C; Csányi G MACE: Higher order equivariant message passing neural networks for fast and accurate force fields. Advances in Neural Information Processing Systems 2022, 35, 11423–11436. [Google Scholar]
- (29).Smith JS; Isayev O; Roitberg AE ANI-1: an extensible neural network potential with DFT accuracy at force field computational cost. Chem. Sci 2017, 8, 3192–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Smith JS; Nebgen B; Lubbers N; Isayev O; Roitberg AE Less is more: Sampling chemical space with active learning. J. Chem. Phys 2018, 148, 241733. [DOI] [PubMed] [Google Scholar]
- (31).Smith JS; Nebgen BT; Zubatyuk R; Lubbers N; Devereux C; Barros K; Tretiak S; Isayev O; Roitberg AE Approaching coupled cluster accuracy with a general-purpose neural network potential through transfer learning. Nat. Commun 2019, 10, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Stevenson JM; Jacobson LD; Zhao Y; Wu C; Maple J; Leswing K; Harder E; Abel R Schr\" odinger-ANI: An Eight-Element Neural Network Interaction Potential with Greatly Expanded Coverage of Druglike Chemical Space. arXiv preprint arXiv:1912.05079 2019, [Google Scholar]
- (33).Devereux C; Smith JS; Davis KK; Barros K; Zubatyuk R; Isayev O; Roitberg AE Extending the Applicability of the ANI Deep Learning Molecular Potential to Sulfur and Halogens. J. Chem. Theory Comput 2020, 16, 4192–4202. [DOI] [PubMed] [Google Scholar]
- (34).Behler J; Parrinello M Generalized neural-network representation of high-dimensional potential-energy surfaces. Phys. Rev. Lett 2007, 98, 146401. [DOI] [PubMed] [Google Scholar]
- (35).Vanommeslaeghe K; Hatcher E; Acharya C; Kundu S; Zhong S; Shim J; Darian E; Guvench O; Lopes P; Vorobyov I; MacKerell AD Jr CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem 2010, 31, 671–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Vanommeslaeghe K; Raman EP; MacKerell AD Jr Automation of the CHARMM General Force Field (CGenFF) II: assignment of bonded parameters and partial atomic charges. J. Chem. Inf. Model 2012, 52, 3155–3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Wang J; Wolf RM; Caldwell JW; Kollman PA; Case DA Development and testing of a general amber force field. J. Comput. Chem 2004, 25, 1157–1174. [DOI] [PubMed] [Google Scholar]
- (38).Jorgensen WL; Tirado-Rives J Potential energy functions for atomic-level simulations of water and organic and biomolecular systems. Proc. Natl. Acad. Sci 2005, 102, 6665–6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Mobley DL; Bannan CC; Rizzi A; Bayly CI; Chodera JD; Lim VT; Lim NM; Beauchamp KA; Slochower DR; Shirts MR; Gilson MK; Eastman PK Escaping atom types in force fields using direct chemical perception. J. Chem. Theory Comput 2018, 14, 6076–6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Lahey S-LJ; Thien Phuc TN; Rowley CN Benchmarking Force Field and the ANI Neural Network Potentials for the Torsional Potential Energy Surface of Biaryl Drug Fragments. J. Chem. Inf. Model 2020, [DOI] [PubMed] [Google Scholar]
- (41).Lahey S-LJ; Rowley CN Simulating protein–ligand binding with neural network potentials. Chem. Sci 2020, 11, 2362–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Warshel A; Levitt M Theoretical studies of enzymic reactions: dielectric, electrostatic and steric stabilization of the carbonium ion in the reaction of lysozyme. J. Mol. Biol 1976, 103, 227–249. [DOI] [PubMed] [Google Scholar]
- (43).Lin H; Truhlar DG QM/MM: what have we learned, where are we, and where do we go from here? Theor. Chem. Acc 2007, 117, 185–199. [Google Scholar]
- (44).Senn HM; Thiel W QM/MM methods for biomolecular systems. Angew. Chem 2009, 48, 1198–1229. [DOI] [PubMed] [Google Scholar]
- (45).Rufa DA; Macdonald HEB; Fass J; Wieder M; Grinaway PB; Roitberg AE; Isayev O; Chodera JD Towards chemical accuracy for alchemical free energy calculations with hybrid physics-based machine learning/molecular mechanics potentials. BioRxiv 2020, [Google Scholar]
- (46).Wang L; Wu Y; Deng Y; Kim B; Pierce L; Krilov G; Lupyan D; Robinson S; Dahlgren MK; Greenwood J; Romero DL; Masse C; Knight JL; Steinbrecher T; Beuming T; Damm W; Harder E; Sherman W; Brewer M; Wester R; Murcko M; Frye L; Farid R; Lin T; Mobley DL; Jorgensen WL; Berne BJ; Friesner RA; Abel R Accurate and reliable prediction of relative ligand binding potency in prospective drug discovery by way of a modern free-energy calculation protocol and force field. J. Am. Chem. Soc 2015, 137, 2695–2703. [DOI] [PubMed] [Google Scholar]
- (47).Vant JW; Lahey S-LJ; Jana K; Shekhar M; Sarkar D; Munk BH; Kleinekathöfer U; Mittal S; Rowley C; Singharoy A Flexible Fitting of Small Molecules into Electron Microscopy Maps Using Molecular Dynamics Simulations with Neural Network Potentials. J. Chem. Inf. Model 2020, [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Xu M; Zhu T; Zhang JZ Automatically Constructed Neural Network Potentials for Molecular Dynamics Simulation of Zinc Proteins. Front. Chem 2021, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Paszke A; Gross S; Massa F; Lerer A; Bradbury J; Chanan G; Killeen T; Lin Z; Gimelshein N; Antiga L; Desmaison A; Köpf A; Yang E; DeVito Z; Raison M; Tejani A; Chilamkurthy S; Steiner B; Fang L; Bai J; Chintala S Pytorch: An imperative style, high-performance deep learning library. Advances in neural information processing systems 2019, 32. [Google Scholar]
- (50).Barducci A; Bonomi M; Parrinello M Metadynamics. Wiley Interdiscip. Rev.: Comput. Mol. Sci 2011, 1, 826–843. [Google Scholar]
- (51).Eastman P; Swails J; Chodera JD; McGibbon RT; Zhao Y; Beauchamp KA; Wang L-P; Simmonett AC; Harrigan MP; Stern CD; Wiewiora RP; Brooks BR; Pande VS OpenMM 7: Rapid development of high performance algorithms for molecular dynamics. PLoS Comput. Biol 2017, 13, e1005659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Eastman P. OpenMM-Torch. https://github.com/openmm/openmm-torch, 2021; accessed 2023/05/21. [Google Scholar]
- (53).Gao X; Ramezanghorbani F; Isayev O; Smith JS; Roitberg AE TorchANI: A free and open source PyTorch-based deep learning implementation of the ANI neural network potentials. J. Chem. Inf. Model 2020, 60, 3408–3415. [DOI] [PubMed] [Google Scholar]
- (54).Eastman P; Galvelis R NNPOps. https://github.com/openmm/nnpops, 2021; accessed 2023/05/21. [Google Scholar]
- (55).TorchMD-NET. https://github.com/torchmd/torchmd-net, accessed 2023/05/21.
- (56).Barducci A; Bussi G; Parrinello M Well-tempered metadynamics: a smoothly converging and tunable free-energy method. Phys. Rev. Lett 2008, 100, 020603. [DOI] [PubMed] [Google Scholar]
- (57).Tribello GA; Bonomi M; Branduardi D; Camilloni C; Bussi G PLUMED 2: New feathers for an old bird. Comput. Phys. Commun 2014, 185, 604–613. [Google Scholar]
- (58).Wang R; Fang X; Lu Y; Wang S The PDBbind database: Collection of binding affinities for protein- ligand complexes with known three-dimensional structures. J. Med. Chem 2004, 47, 2977–2980. [DOI] [PubMed] [Google Scholar]
- (59).Liu Z; Su M; Han L; Liu J; Yang Q; Li Y; Wang R Forging the basis for developing protein–ligand interaction scoring functions. Acc. Chem. Res 2017, 50, 302–309. [DOI] [PubMed] [Google Scholar]
- (60).Bäckbro K; Löwgren S; Österlund K; Atepo J; Unge T; Hultén J; Bonham NM; Schaal W; Karlén A; Hallberg A Unexpected binding mode of a cyclic sulfamide HIV-1 protease inhibitor. J. Med. Chem 1997, 40, 898–902. [DOI] [PubMed] [Google Scholar]
- (61).Skulnick HI; Johnson PD; Aristoff PA; Morris JK; Lovasz KD; Howe WJ; Watenpaugh KD; Janakiraman MN; Anderson DJ; Reischer RJ; Schwartz TM; Banitt LS; Tomich PK; Lynn JC; Horng M-M; Chong K-T; Hinshaw RR; Dolak LA; Seest EP; Schwende FJ; Rush BD; Howard GM; Toth LN; Wilkinson KR; Kakuk TJ; Johnson CW; Cole SL; Zaya RM; Zipp GL; Possert PL; Dalga RJ; Zhong W-Z; Williams MG; Romines KR Structure-based design of nonpeptidic HIV protease inhibitors: the sulfonamide-substituted cyclooctylpyranones. J. Med. Chem 1997, 40, 1149–1164. [DOI] [PubMed] [Google Scholar]
- (62).Qiao JX; Chang C-H; Cheney DL; Morin PE; Wang GZ; King SR; Wang TC; Rendina AR; Luettgen JM; Knabb RM; Wexler RR; Lam PY SAR and X-ray structures of enantiopure 1, 2-cis-(1R, 2S)-cyclopentyldiamine and cyclohexyldiamine derivatives as inhibitors of coagulation Factor Xa. Bioorg. Med. Chem. Lett 2007, 17, 4419–4427. [DOI] [PubMed] [Google Scholar]
- (63).Nie Z; Perretta C; Erickson P; Margosiak S; Lu J; Averill A; Almassy R; Chu S Structure-based design and synthesis of novel macrocyclic pyrazolo [1, 5-a][1, 3, 5] triazine compounds as potent inhibitors of protein kinase CK2 and their anticancer activities. Bioorg. Med. Chem. Lett 2008, 18, 619–623. [DOI] [PubMed] [Google Scholar]
- (64).Doerr S; Harvey M; Noé F; De Fabritiis G HTMD: high-throughput molecular dynamics for molecular discovery. J. Chem. Theory Comput 2016, 12, 1845–1852. [DOI] [PubMed] [Google Scholar]
- (65).Smith JS; Isayev O; Roitberg AE ANI-1, A data set of 20 million calculated off-equilibrium conformations for organic molecules. Sci. Data 2017, 4, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Conda. https://docs.conda.io/, accessed 2023/05/21.
- (67).conda-forge. https://conda-forge.org/, accessed 2023/05/21.
- (68).HTMD documentation. https://software.acellera.com/htmd, accessed 2023/05/21.
- (69).ACEMD documentation. https://software.acellera.com/acemd, acessed 2023/05/21.
- (70).Sugita Y; Okamoto Y Replica-exchange molecular dynamics method for protein folding. Chem. Phys. Lett 1999, 314, 141–151. [Google Scholar]
- (71).Izrailev S; Stepaniants S; Isralewitz B; Kosztin D; Lu H; Molnar F; Wriggers W; Schulten K Steered molecular dynamics. Computational Molecular Dynamics: Challenges, Methods, Ideas: Proceedings of the 2nd International Symposium on Algorithms for Macromolecular Modelling, Berlin, May 21–24, 1997. 1999; pp 39–65. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.