Abstract
The echinocandins are a family of cyclic lipopeptides with potent antifungal activity. These compounds inhibit the synthesis of β-1,3-glucan in fungi. The new semisynthetic echinocandin LY303366 was derivatized to produce a photoactivatable cross-linking echinocandin analog with antifungal activity. This analog was radioiodinated and used as a probe in microsomal membrane preparations of Candida albicans which contain glucan synthase activity. The photoaffinity probe identified two major proteins of 40 and 18 kDa in both membrane preparations. Labeling of these proteins was specific in that it required irradiation with UV light and was effectively competed against with unlabeled echinocandin analogs. In addition, the abilities of echinocandin analogs to compete with the photoaffinity probe correlated to their relative antifungal potencies and glucan synthase inhibition. The 40-kDa protein was isolated, and partial sequences were obtained from internal peptide fragments of the protein. Analysis of the sequences of these internal peptides of the 40-kDa protein revealed that it was a new protein not previously described as being involved in glucan synthesis or the mode of action of echinocandins.
In the past 15 years, there has been a dramatic increase in the number of immunocompromised individuals, which, in part, accounts for the dramatic increase in the incidence of systemic mycoses (36). Candida albicans is the fungal pathogen in a majority of fungal infections, accounting for 72% of nosocomial fungal infections (35). Recently, Candida spp. have surpassed Escherichia coli as the third most frequently isolated organism from nosocomial bloodstream infections (38). Antifungal therapy to treat fungal infections is limited at present. The treatment of choice, with amphotericin B, suffers from the drawbacks of being an intravenous treatment and being limited in total dosage due to its toxicity (19). The oral azole fluconazole is well tolerated, but its antifungal spectrum is limited in that it is not active against some of the other Candida spp. (6). More importantly, the frequency of reports describing resistance to fluconazole in C. albicans is increasing (41). A recent study demonstrated the presence of fluconazole-resistant C. albicans in a patient population which had not been previously treated with the drug (21). In addition, the emergence of new fungal pathogens, such as Candida krusei and Fusarium spp., which are intrinsically less susceptible to fluconazole is of recent clinical concern (36).
The echinocandins are a family of cyclic lipopeptides with intrinsic antifungal activity (3). Recent advances in semisynthetic echinocandins have led to the development of highly potent, orally bioavailable agents, such as LY303366 (Fig. 1) (8). The MICs of this compound for most common fungal pathogens are very low, and this compound has shown oral efficacy in animal models of systemic candidiasis and pneumocystis pneumonia (7). It has also shown efficacy in animal models with Aspergillus fumigatus, one of the more difficult pathogens to treat, when administered by the intravenous route (39). LY303366 is presently in initial clinical trials in humans.
FIG. 1.
Structures of compounds used in this study.
The echinocandins inhibit the synthesis of the major structural glucan in the cell walls of fungi, (1,3)-β-d-glucan. This polymer of glucose is synthesized by the enzyme glucan synthase (EC 2.4.1.34). Crude membrane preparations of C. albicans contain glucan synthase activity, which is inhibited by echinocandin analogs in vitro (42, 46). Substantial effort has gone into the purification of glucan synthase from many fungal organisms, including the model yeast Saccharomyces cerevisiae, Neurospora crassa, and C. albicans (2, 23, 47). In addition to classical biochemical purification, genetic approaches using S. cerevisiae have recently identified genes involved in resistance to the echinocandins. One gene, ETG1, is believed to encode the catalytic subunit of glucan synthase and has been isolated by a number of groups using several different approaches (4, 11, 12, 20, 40). A second gene encoding resistance to echinocandins, GNS1, has also been characterized (15). Direct binding of echinocandin analogs to the products of these genes has not yet been demonstrated.
In this study, a photoactivated cross-linking echinocandin analog was used as a probe for direct binding of an echinocandin analog to proteins in crude microsomal membrane preparations prepared from a synchronously budding yeast culture of C. albicans. The echinocandin analog was prepared by modification of the aminal hydroxyl of LY303366 with ethanolamine and subsequent coupling of an aryl azide photoactivatable cross-linker to the resulting amine. This photoactivated cross-linking analog both has antifungal activity and is competed for in the photocross-linking studies by LY303366. The covalent cross-linking of echinocandin to proteins in membrane preparations of C. albicans, which contain glucan synthase activity, demonstrated interaction of echinocandin with two proteins with apparent molecular masses of 40 and 18 kDa.
MATERIALS AND METHODS
Preparation of LY317854.
Aminoethylation of the aminal hydroxyl of LY303366 was accomplished as previously described for tetrahydroechinocandin B (24).
Preparation of 4-azidobenzamide derivative of LY317854 (LY317854-AB).
One milligram of LY317854 was dissolved in 100 μl of neat dimethylformamide. N-Hydroxysuccinimide ester of 4-azidobenzoic acid (HSAB; Pierce) was dissolved in neat dimethylformamide at a concentration of 10 mg/ml. The HSAB solution (100 μl; 5 molar excess over LY317854) was added to LY317854 in an amber vial. One microliter of triethylamine was added, and the reaction mixture was allowed to stir overnight at room temperature in the dark. The reaction mixture was transferred to a microcentrifuge tube, and the product was precipitated by addition of a 10× volume of cold 10% NaCl. The suspension was kept on ice for 10 min and then centrifuged at 14,000 × g for 10 min at 4°C. The resulting pellet was washed once more with cold 10% NaCl, followed by a wash with cold water. The pellet was then dried in vacuo, yielding 300 μg of product, which was then redissolved in 100% dimethyl sulfoxide (DMSO) at 2 mg/ml. The product was analyzed by high-performance liquid chromatography, and the free amine (LY317854) was not detected.
Radioiodination of LY317854-AB.
In a screw-cap microcentrifuge tube, 5 μl of 317854-AB (2 mg/ml in DMSO) was diluted with 100 μl of acetonitrile, and 20 μl of 0.5 M sodium phosphate, pH 6.8, was added. The solution was mixed vigorously by vortexing. Carrier-free Na125I (5 μl; 500 μCi) was added, and the reaction was initiated by addition of a single IodoBead (Pierce). The reaction was allowed to proceed for 5 min and then the product was purified on a Sep-Pak C18 cartridge equilibrated with 20% acetonitrile-water. The product was eluted with 3 ml of 90% acetonitrile. The solution was transferred to an amber vial, and the solvent was removed in vacuo.
Purification of 125I-labeled LY317854-AB.
The radioiodinated product was purified by absorption chromatography on a Sephadex LH-20 column. The radioiodinated compound in DMSO was diluted into 1 ml of water. This was loaded on a Sephadex LH-20 column (total column volume, 6 ml) equilibrated with water. The column was then washed with 3 column volumes of water. The bound radioactive material was eluted with 80% acetonitrile-water. Fractions were collected and aliquots were counted in an LKB gamma counter. Peak fractions, examined by high-performance liquid chromatography, gave a single radioactive peak. The specific activity of the compound was 11 mCi/μmol.
Preparation of membranes from C. albicans.
C. albicans 3153 (obtained from H. Buckley) was routinely cultured on Sabouraud’s solid medium at 30°C. Stocks were maintained at 4°C. A single colony from a stock plate was inoculated into 5 ml of Lee’s medium and allowed to shake overnight at room temperature (28). This culture was then used to inoculate 250 ml of Lee’s medium, grown to saturation, and then diluted to obtain synchronously budding cells (32). After synchronous evagination had occurred, cultures were chilled in ice water for 10 min.
Cells were suspended in 50 mM HEPES-Na (pH 8.0)–1 M sucrose–10 mM NaF–5 mM EDTA–1 mM dithiothreitol (DTT) (GS buffer) containing 100 μM 5′-guanylylimidodiphosphate (GMP-PNP), 1 mM phenylmethylsulfonyl fluoride (PMSF), 2 μM pepstatin A, 2 mM benzamidine, and 0.6 μM leupeptin and were disrupted with glass beads. Crude microsomal membranes were prepared as described previously (18). The membranes were resuspended in ice-cold GS buffer without DTT and were transferred to a small Dounce homogenizer on ice and gently homogenized. The membrane suspensions were flash frozen in dry ice-ethanol at a protein concentration of 2 mg/ml and were stored at −90°C.
Preparation of detergent-washed membranes.
Crude microsomal membranes were thawed and diluted with an equal volume of ice-cold GS buffer containing 50 mM NaCl, 100 μM GTP, and 0.02% W-1 detergent. The mixture was gently vortexed and kept on ice. The membrane preparation was then diluted with an equal volume of 50 mM HEPES–5 mM EDTA–10 mM NaF (pH 8.0) containing 50 mM NaCl, 1 mM DTT, 100 μM GTP, and 0.01% W-1 detergent to drop the sucrose concentration to 0.5 M. The membranes were harvested by ultracentrifugation at 200,000 × g at 4°C for 1 h. The pellet was resuspended in cold GS buffer containing 50 mM NaCl, 100 μM GTP, and 0.03% W-1. The resuspended membrane preparation was diluted by addition of an equal volume of the buffer described above, but without sucrose. The membranes were harvested as described above by ultracentrifugation. The pellet was resuspended in GS buffer containing 50 mM NaCl, 100 μM GTP, and 0.03% W-1 detergent and was gently stirred overnight at 4°C. The homogenate was diluted fivefold with buffer without sucrose, and the membranes were harvested by ultracentrifugation as described above. The detergent-washed membranes were homogenized in a Dounce homogenizer in GS buffer without DTT and were stored in aliquots at −90°C.
Determination of protein concentration.
Protein concentration was determined by the bicinchoninic acid method according to the manufacturer’s instructions (Pierce). Samples were read at 590 nm in a Beckman DU-70 spectrophotometer. Standard curves were prepared by using bovine serum albumin (2 mg/ml; Pierce).
SDS-PAGE.
Proteins labeled from the photoaffinity experiments were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using a Mini-protean II system (Bio-Rad) with 12% acrylamide gels containing 4 M urea (27). The stacking gel was 4% acrylamide and contained 4 M urea. Samples for electrophoresis were dissolved in SDS-urea loading buffer, consisting of 0.05 M Tris-HCl (pH 6.8), 10% SDS, 4 M urea, and 0.16 M DTT, and were warmed at 50°C for 5 min, followed by centrifugation at 14,000 × g. Samples were electrophoresed and stained with Coomassie brilliant blue G-250. After destaining and drying, the gels were subjected to autoradiography, and the autoradiograms were quantitated by laser densitometry. Inhibition of photoaffinity labeling by competitor echinocandins was calculated by comparison to a control lacking competitor compounds on a given gel and subsequent autoradiogram. The degree of inhibition is therefore a relative measure compared to the control in any given experiment.
Susceptibility testing.
Susceptibility testing of echinocandin analogs was performed in a microdilution broth assay. C. albicans 3153 was grown overnight in Sabouraud’s dextrose medium (10). Cells were diluted to 105/ml in test medium, Lee’s medium, or Bacto Antibiotic Assay Medium 3 (10). Compounds were prepared as stock solutions in 100% DMSO. Twofold serial dilutions in test medium at twice the final desired concentration in 2% DMSO-test medium were prepared in the microtiter plate (100 μl/well). Cell suspensions were added to each well (100 μl per well). The final compound concentrations ranged by twofold serial dilution from 20 to 0.02 μg/ml in a final concentration of 1% DMSO. Plates were incubated at their respective temperatures for up to 72 h depending on the medium and were read on a Molecular Devices ThermoMax plate reader at 650 nm. The MIC was the lowest concentration of compound which inhibited growth of the organism by more than 90%, as determined by turbidity.
Glucan synthase assays.
Membrane preparations were assayed for glucan synthase activity as described previously (47). The product was identified as 1,3-β-d-glucan by high-performance anion-exchange chromatography and by enzymatic degradation as described previously (48).
Photolabeling of C. albicans microsomal membranes.
Membrane preparations were thawed on ice and diluted 10-fold in GS buffer without DTT. Diluted membrane preparations (200 μg/ml; 25 μl) were transferred to screw-cap microcentrifuge tubes on ice. To these membrane preparations, 20 μl of 62.5 mM sodium [2-(N-morpholino)ethanesulfonic acid] (MES-Na) (pH 5.0)–1 M sucrose–10 mM NaF–5 mM EDTA was added, giving a final pH of 6.7 to 6.8. Echinocandin analogs were dissolved in DMSO at 20 times the stated concentrations. Compound in DMSO or 100% DMSO (2.5 μl) was then added to the membrane preparation on ice. In reduced light, 2.5 μl of the probe echinocandin analog (0.126 nmol; 105 cpm) in DMSO was added. The membranes were gently mixed and kept on ice. The final concentration of the probe echinocandin was 3.4 μg/ml.
Samples prepared as described above were transferred to a water bath at 30°C for 5 min. The membranes were returned to the ice. Samples were transferred to a separate ice tray and individually photolyzed for 45 s at a distance of 3 cm by using a hand-held short-wave UV lamp with the filter removed. Immediately after photolysis, the sample was precipitated by addition of ice-cold 80% methanol (800 μl) and was transferred to −20°C for a minimum of 1 h.
Samples were harvested by centrifugation at 14,000 × g at 4°C for 30 min. The solvent was removed, and the pellet was resuspended in 500 μl of cold acetone. The samples were placed at −20°C for an additional hour and were centrifuged as described above. The pellets were then resuspended in 250 μl of chloroform-methanol (2:1) at room temperature. Pellets were collected by centrifugation and then dried in vacuo. The samples were then dissolved in SDS-urea PAGE loading buffer and subjected to electrophoresis as described above.
Purification of the 40-kDa EBP.
The 40-kDa echinocandin binding protein (EBP) was trace labeled with the 125I-echinocandin photoaffinity probe in crude microsomal membranes of C. albicans 3153. This trace-labeled preparation was diluted with cold 20 mM Tris-HCl (pH 7.9)–150 mM NaCl–300 mM sucrose–1 mM EDTA–1.0% n-octyl-β-d-glucoside (NOG)–0.5% cholamidopropyl-dimethyl-ammonio-1-propane sulfonate (CHAPS) containing 1 mM PMSF, 2 μg of pepstatin A/ml, and 2 μg of aprotinin/ml. The membrane preparation was kept on ice for 20 min and was subsequently centrifuged at 100,000 × g for 1 h at 4°C. The supernatant was collected and stored at −90°C.
C. albicans 3153 cells were grown in 6 liters of Lee’s medium at room temperature overnight. Cells were harvested as previously described, and a crude microsomal membrane preparation was prepared as described above. The membranes were resuspended in 10 ml of cold 20 mM Tris-HCl (pH 7.9)–150 mM NaCl–300 mM sucrose–1 mM EDTA–1.0% NOG–0.5% CHAPS containing 1 mM PMSF, 2 μg of pepstatin A/ml, and 2 μg of aprotinin/ml. The membranes were transferred to a glass Dounce homogenizer on ice and were gently homogenized with 10 strokes of the pestle. Additional buffer was added to adjust the protein concentration to 1 mg/ml. The solubilized membrane preparation was collected after centrifugation at 100,000 × g for 1 h at 4°C. The resulting pellet was resuspended in buffer and homogenized as described above in half the original volume. The supernatant was pooled with the first solubilization fraction after ultracentrifugation as described above.
Trace-labeled membranes (1 ml; 2.4 × 106 cpm) were thawed and added to the solubilized microsomal membranes (25 ml; 7.5 mg of protein). The solubilized membrane preparation was applied to a Sephacryl S-400HR column (2.8 by 45 cm) equilibrated with 20 mM Tris-HCl (pH 7.9)–150 mM NaCl–1 mM EDTA–0.1% NOG–0.05% CHAPS. The column was eluted at 30 ml/h at 4°C. Six fractions per hour were collected, and aliquots were counted in an LKB model 1282 Compugamma counter. The protein concentration in each fraction was determined by the bicinchoninic acid method. Fractions containing a peak of radioactivity and protein were pooled and dialyzed against 20 mM Tris-HCl (pH 7.9)–1 mM EDTA–0.1% NOG–0.05% CHAPS.
The pooled and dialyzed fractions were then loaded onto a DEAE-Sepharose column (0.8 by 7 cm) equilibrated with 20 mM Tris-HCl (pH 7.9)–1 mM EDTA–0.1% NOG–0.05% CHAPS. Bound radioactivity was eluted with a 150-ml linear gradient from 0 to 500 mM NaCl. Fractions were analyzed for radioactivity and protein as described above. Fractions containing a peak of radioactivity were pooled and analyzed by SDS-PAGE as described above.
Sequence analysis of the 40-kDa EBP.
The 40-kDa protein was further purified by SDS-PAGE as described above and transferred to polyvinylidene difluoride (PVDF) membranes (Immobilon-P; Millipore) in 25 mM Tris–192 mM glycine–15% methanol at room temperature (7 V/cm for 1.5 h). Membranes were rinsed with distilled water after transfer to reduce the background and then were stained with 0.025% Coomassie brilliant blue R-250 in 40% methanol for 5 min. The membrane was then destained with 40% methanol and allowed to air dry. Appropriate bands were excised with a razor blade and sent for automated Edman sequencing (30).
Preparation of peptide fragments of the 40-kDa EBP.
Partial proteolysis of the 40-kDa protein was accomplished by the method of Cleveland using endoproteinase-Glu C (endo-Glu C; Boehringer Mannheim) (5). Cleavage with cyanogen bromide was performed within the gel slice according to the method of Sokolov et al. (44). Cleaved fragments of the 40-kDa protein were resolved by standard SDS-PAGE (no urea) and transferred to PVDF membranes as described above.
Sequence analysis.
Peptides were sequenced by gas-phase Edman degradation. Peptide and nucleotide sequence comparisons and analysis were performed with the University of Wisconsin Genetics Computer Group program and by BLAST (1, 9).
RESULTS
Biological activity of LY317854-AB.
The potency of the echinocandin compounds is dependent on the test medium. Generally, the potency of the echinocandins is greater in Bacto Antibiotic Assay Medium 3 than in the standard antifungal susceptibility medium recommended by the National Committee for Clinical Laboratory Standards (37). In the testing of echinocandin derivatives, it has been found that the use of Bacto Antibiotic Assay Medium 3 as a test medium gives a broader range of potency for these compounds, thereby allowing for discrimination of more-potent analogs from less-potent ones. Results of in vitro testing in Bacto Antibiotic Assay Medium 3 also correlated well with the observed potency of these compounds in animal models of experimental Candida infections (data not shown).
The antifungal activity of the aminoethylated compound (LY317854) was lower than that of the parent compound, LY303366, when tested against C. albicans in Lee’s medium and in Bacto Antibiotic Assay Medium 3 (Table 1) (10, 28). Acylation of the amine in LY317854, forming the 4-azidobenzamide derivative, resulted in a decrease in the aqueous solubility of the compound, but the antifungal activity was comparable to that of LY317854 under the given conditions. The introduction of the photoactivatable cross-linking moiety thus had no significant effect on the antifungal activity of LY317854.
TABLE 1.
MICs of echinocandin derivatives against C. albicans 3153 in two media
| Medium (°C) and compound | MIC (μg/ml) |
|---|---|
| Lee’s medium (25) | |
| LY303366 (parent) | 0.02 |
| LY317854 | 0.31 |
| LY317854-AB | 0.31 |
| LY280949a | 0.16 |
| Bacto Antibiotic Assay Medium 3 (30) | |
| LY303366 (parent) | 0.0025 |
| LY280949 | 0.04 |
| LY121019b | 0.31 |
| LY317854 | 0.02 |
| LY317854-AB | 0.04 |
Compound 13b (8).
Cilofungin.
Photoaffinity labeling of microsomal membranes from C. albicans.
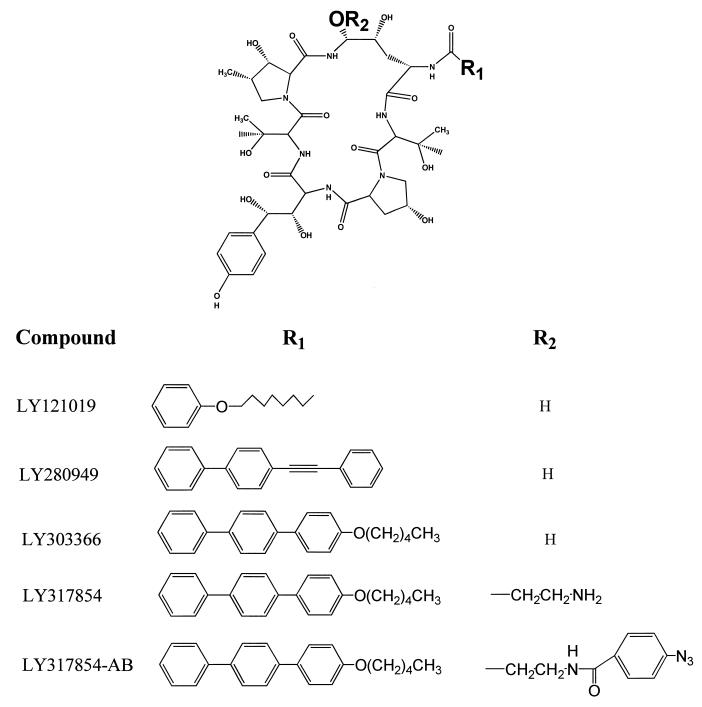
The labeling of membranes from the budding form of C. albicans is shown in Fig. 2A. The first lane represents the photoreaction of the probe molecule in the absence of competitor echinocandin. Two proteins are significantly labeled by the photoaffinity probe: one intensely labeled protein of approximately 40 kDa and a second, less intensely labeled protein with a molecular mass of approximately 18 kDa. The band at the bottom of the gel is at the dye front and is due to the interaction of the probe with lipid components in the crude membrane preparations. The second lane represents the dark control (not photolyzed, but otherwise treated the same as the membranes in lane 1). This control demonstrates that insertion of the photoaffinity probe is due to UV irradiation and subsequent cross-linking and is not the result of insertion through a nonphotolytic event. Lanes 3 to 5 are the patterns resulting from competition by increasing concentrations of LY303366, demonstrating that the photoaffinity analog and the parent compound, LY303366, interact with the same proteins in the membrane preparation. Other proteins, of 32 and 50 kDa, are slightly labeled in this particular experiment, but these bands are not consistently noted when other membrane preparations are used in other experiments. In addition, as mentioned above, significant amounts of the probe echinocandin react with lipid components in the membrane. Thus, some nonspecific background is usually noted in the photoaffinity labeling of membrane preparations. Only the 40- and 18-kDa proteins were reproducibly labeled and inhibited by competitor echinocandins in multiple experiments.
FIG. 2.
Autoradiograms of crude microsomal membranes from C. albicans 3153 reacted by photolysis with 125I-labeled echinocandin photoaffinity probe. Lanes 1, probe alone; lanes 2, nonphotolyzed control. Molecular size markers are in kilodaltons. (A) Lanes 3 to 5, probe in the presence of 1, 10, and 100 μg of LY303366/ml, respectively. (B) Competitors in lanes 3 to 5 are equimolar mixtures of the cyclic-peptide nucleus and the lipophilic side chain of LY303366 at concentrations comparable to echinocandin concentrations in panel A.
The antifungal activity and inhibition of glucan synthase by echinocandins require the cyclic-peptide portion to be covalently linked to the lipophilic side chain. Separation of these two parts of the molecule results in a loss of antifungal activity and glucan synthase inhibition (45). Both the echinocandin cyclic-peptide nucleus and the terphenyl side chain in combination, but not covalently linked, and at comparable concentrations to that of the intact echinocandin, do not significantly inhibit labeling with the photoaffinity reagent (Fig. 2B). Similar lack of competition was found with either the cyclic peptide nucleus or the side chain alone at comparable concentrations (data not shown).
To support the biological significance of the photoaffinity labeling experiments, the competition of the compounds for the 40-kDa protein with the photoaffinity probe was compared to both the antifungal potency of the competitor compounds and their ability to inhibit glucan synthase activity.
The inhibition of glucan synthase activity in crude microsomal membranes by echinocandins of differing potency was also examined in an effort to correlate the photoaffinity labeling results with glucan synthase inhibition. Surprisingly, in crude microsomal membranes, inhibition of glucan synthase by echinocandins did not seem to correlate with antifungal potency. However, when the crude microsomal membranes were washed with detergent, the washed membranes had significantly higher overall glucan synthase activity, similar to previous observations of glucan synthase activity in S. cerevisiae, and the potencies of the echinocandin analogs now correlated with their abilities to inhibit glucan synthase activity (17). It is not clear why this difference in glucan synthase activity between crude and detergent-washed membranes was observed. Dose-response curves of glucan synthase activity in the detergent-washed membranes were obtained with LY303366, LY280949, and LY121019 (cilofungin), and the 50% inhibitory concentration (IC50) was determined for each compound. Detergent-washed membranes were photolabeled with the probe echinocandin as described for the crude microsomal membranes.
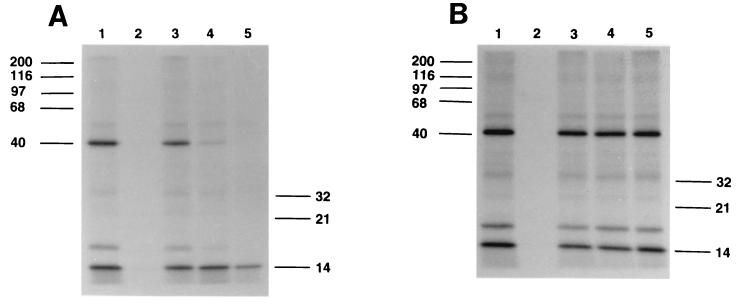
The competition of echinocandins of differing potency with the photoaffinity probe in crude microsomal preparations is shown in Fig. 3. Identical results were obtained with detergent-washed membranes (data not shown). With either type of membrane preparation, the relationship between competition in the photolabeling experiments and antifungal potency was logarithmic (Table 2). The absolute values for inhibition varied slightly from experiment to experiment, but the relative inhibitions of the three competitor compounds in the photolabeling experiments correlated with the antifungal potencies of these compounds in several independent experiments.
FIG. 3.
Autoradiogram of crude microsomal membranes from C. albicans 3153. Experimental conditions and lanes 1 and 2 were the same as for Fig. 2. Lanes 3 to 5, probe in the presence of 10 μg of LY121019 (cilofungin), LY280949, and LY303366/ml, respectively.
TABLE 2.
Correlation of inhibition of photoaffinity labeling with biological activities of echinocandins
In addition, competition in the photoaffinity labeling experiments correlated with glucan synthase inhibition in detergent-washed membranes (Table 2). Although the IC50s for glucan synthase inhibition with the echinocandin analogs fell within a relatively narrow range, there was a linear relationship between glucan synthase inhibition and ability to compete in the photoaffinity labeling experiments.
Purification of the 40-kDa EBP.
Two-dimensional gel analysis of the labeled 40-kDa EBP indicated that the pI of the protein was approximately 5.6 (data not shown). Initial attempts at sequencing the protein from two-dimensional gels failed, suggesting that the amino-terminal end may be blocked. To obtain sequence from internal fragments, the protein was purified by conventional methods by using the 125I-labeled EBP from photolabeling experiments as a tracer.
Photoaffinity labeling of the purified 40-kDa EBP.
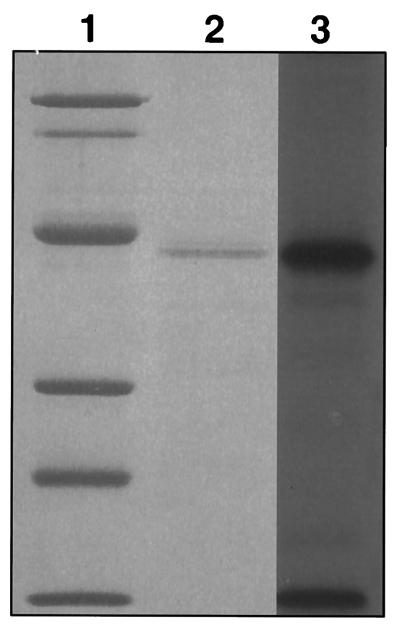
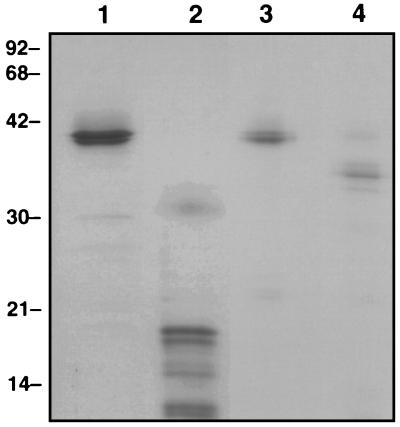
To confirm that the isolated protein was the desired 40-kDa EBP, the partially purified protein (10 μg) was photoaffinity labeled. The isolated protein reacts strongly with the echinocandin probe. Competition with increasing concentrations of LY303366 demonstrated that the isolated protein is the 40-kDa EBP (Fig. 4).
FIG. 4.
Photoaffinity labeling of isolated 40-kDa protein. Lane 1, molecular weight markers stained with Coomassie brilliant blue G-250: phosphorylase B (92 kDa), bovine serum albumin (68 kDa), ovalbumin (42 kDa), carbonic anhydrase (30 kDa), soybean trypsin inhibitor (21 kDa), and lysozyme (14 kDa). Lane 2, partially purified 40-kDa protein from C. albicans stained with Coomassie brilliant blue G-250; lane 3, corresponding autoradiogram of photoaffinity-labeled partially purified 40-kDa protein from C. albicans.
Partial proteolysis of the 40-kDa EBP by endo-Glu C.
The partially purified 40-kDa protein was further purified by SDS-PAGE, and gel slices were subjected to proteolytic digestion with endo-Glu C as described previously (5). By using increasing amounts of protease, progressive digestion of the 40-kDa EBP was demonstrated (Fig. 5, lane 2). The resulting doublet bands at 18 and ∼10 kDa were subjected to amino acid sequence analysis. The doublet bands at 18 kDa resulted in no sequence information. The doublet bands at ∼10 kDa gave a single sequence: IGELEDQYIDKYDQYRIXLK. The sequence was compared to the database sequences by using the BLAST algorithm. A single high-scoring segment was found to match pir:S52527, a hypothetical protein from the S. cerevisiae genome (13 of 17 amino acids; 76% identity). This hypothetical yeast protein is the putative translation product of the yeast open reading frame YPL004c, located on chromosome XVI.
FIG. 5.
SDS-PAGE of fragmented 40 kDa protein. Lanes 1 and 3, isolated 40-kDa protein stained with Coomassie brilliant blue G-250; lane 2, endo-Glu C partial digest of the 40-kDa protein; lane 4, cyanogen bromide digest of the 40-kDa protein. Molecular size markers (in kilodaltons) are shown on the left.
Cyanogen bromide digestion of the 40-kDa EBP.
Digestion of the 40-kDa EBP with CNBr was accomplished in the gel slice. Separation of the resulting polypeptides by SDS-PAGE demonstrated a single major band with an approximate molecular mass of 33 kDa (Fig. 5, lane 4). The digestions were repeated, and the resulting polypeptide was transferred to PVDF membranes and sequenced. The 33-kDa band gave a single amino acid sequence: RAVEVTSRER(K/T)DV. The sequence was compared to database sequences by using the BLAST algorithm. The highest-scoring match for this peptide (when residue 11 was T) was pir:B552872, human neurofibromatosis type 1 protein (NF-1) (9 of 12 amino acids; 75% identity). The second-highest-scoring segment was the hypothetical yeast protein pir:S52527, described above (8 of 16 amino acids; 50% identity).
The high level of homology between the C. albicans polypeptides and the translation product of YPL004c prompted a search of the databases by BLAST using the entire putative translation product of YPL004c. A second highly homologous protein was detected in the S. cerevisiae genome, corresponding to a second hypothetical yeast protein of unknown function translated by the open reading frame YGR086c, located on chromosome VII. Both proteins are highly homologous (69% identical, 82% similar), but there are no recognizable functional motifs and their function is unknown. There were also several lower-scoring segments indicative of low-level homology to tropomyosins from several sources. The significance of these homologies from a functional perspective is not clear. Figure 6 shows the amino acid sequences of the translation products of YPL004c and YGR086c, with a comparison of the homologous C. albicans peptides sequenced from the 40-kDa EBP.
FIG. 6.
Alignment of homologous S. cerevisiae translation products of YPL004c and YGR086c and comparison to the peptides sequenced from the C. albicans 40-kDa EBP.
DISCUSSION
The echinocandins are noncompetitive inhibitors of glucan synthase (46). Although a number of genes involved in glucan synthase activity have been identified, there has been no evidence of a direct interaction of the echinocandins or pneumocandins with any of these molecules. In the photoaffinity labeling studies presented herein, it has been demonstrated that the echinocandins interact directly with at least two polypeptides in crude and detergent-washed microsomal membrane preparations of C. albicans, the 40- and 18-kDa proteins.
In competition studies using LY303366, there is a clear dose-response relationship in the inhibition of labeling of the 40-kDa protein with the photoaffinity probe at a fixed concentration. In addition, compounds of differing antifungal potency demonstrated a correlation between competition for labeling of the 40-kDa protein and both potency and glucan synthase inhibition. Both experimental results indirectly support the role of the 40-kDa protein in the mechanism of action of the echinocandins and as part of the glucan synthase complex. Specific enrichment of a 39-kDa protein along with other higher-molecular-mass proteins has been reported in the partial purification of the glucan synthase complex from C. albicans by product entrapment (16). This enriched 39-kDa protein could be the 40-kDa protein identified in this work.
GNS1 is a gene identified through resistance to pneumocandin in S. cerevisiae (15). Gns1p is believed to be involved in glucan synthesis, as disruption of the gene resulted in a 90% loss of glucan synthase specific activity, although the β-glucan content of whole cells appeared normal. The predicted product of the GNS1 gene is a 40-kDa protein with five to six putative transmembrane domains. The authors suggested that Gns1p is a subunit of the glucan synthase complex, but GNS1 has recently been shown to be allelic with ELO2, a gene involved in very-long-chain fatty acid elongation (34). Isolation of the 40-kDa EBP from Candida and subsequent sequencing of internal peptide fragments demonstrated that the 40-kDa EBP is not the gene product of a GNS1 homolog in C. albicans. By using peptide sequences, two homologous hypothetical proteins were identified in the S. cerevisiae genome, but no functional role has been assigned to these genes. We have cloned the two homologous genes from S. cerevisiae, and their characterization will be reported elsewhere. We are currently isolating the gene coding for the 40-kDa EBP from C. albicans in order to construct a C. albicans strain with a deletion of this gene.
The nucleotide GTP is known to activate glucan synthase in vitro in S. cerevisiae and other fungi (43). In S. cerevisiae, the GTP-activating component of the glucan synthase complex may be separated from the catalytic component of the enzyme by extraction with salt and detergent (33). The GTP-binding protein associated with glucan synthase activity in S. cerevisiae has been characterized as a 20-kDa protein and has been identified as the translation product of the RHO1 gene (14). The role of Rho1p in glucan synthase activity has been supported by inactivation of glucan synthase activity by ADP ribosylation with exoenzyme C3 from Clostridium botulinum, which specifically targets Rho1p (31).
One clue to the possible function of the 40-kDa EBP is the homology of the CNBr fragment to the human protein NF-1. Protein NF-1 is a GTPase-activating protein (GAP) involved in small GTPase cycling (49). One of the targets of protein NF-1 is the small GTPase ras p21 (29). This suggests a potential role of the 40-kDa EBP as a regulator of glucan synthase activity, possibly through a GAP-like function, although the region of homology of the CNBr fragment of the 40-kDa EBP to NF-1 is outside the ras-GAP-related domain of NF-1 (22). Given the importance of GTP as a regulator of glucan synthesis as described above, it is intriguing to speculate that the 40-kDa EBP is involved in GTPase cycling.
A similar relationship between labeling and antifungal activity is also qualitatively observable for the 18-kDa protein, but due to the significantly less intense labeling of the protein, the relationship is not readily quantified. In addition, the physical amount of the 18-kDa protein is very small compared to that of the 40-kDa protein. We were therefore unable to directly sequence this protein from gels. Due to its low labeling efficiency and low physical abundance, we have been unable to identify this protein by classical biochemical methods.
The role of GTP in glucan synthase activity, the homology of the 40-kDa EBP to a ras-GAP protein, and the molecular size of the 18-kDa protein suggested a possible role for the 18-kDa protein as a small GTPase, possibly the Candida homolog of Rho1p. The identity of the 18-kDa protein and its possible functional role remain to be determined, but preliminary evidence obtained from ADP ribosylation with exoenzyme C3 in Candida suggests that the 18-kDa protein is not a Rho1p homolog in C. albicans (unpublished data). This does not exclude the possibility that the 18-kDa protein is another type of small GTPase.
Genetic studies with S. cerevisiae, using a phenotype of resistance to pneumocandins or echinocandins, suggest that the target for these cyclic lipopeptides is encoded by the ETG1 gene (11, 13). This gene encodes a large 215-kDa membrane protein with 16 putative transmembrane segments. This gene has also been isolated through other phenotypes, such as resistance to papulacandin B, another glucan synthase inhibitor (PBR1), hypersensitivity to calcofluor white (CWH53), hypersensitivity to the immune suppressant FK506 in a calcineurin-deficient background (FKS1), and synthetic lethality with calcineurin mutants (CND1) (4, 12, 20, 40). In addition, the protein encoded by the ETG1 locus was identified biochemically through selective enrichment of glucan synthase by glucan product entrapment (23). Homologs of the ETG1 locus have been identified in C. albicans and Aspergillus nidulans (25, 26). Surprisingly, there was no evidence of direct interaction of the probe with a large polypeptide corresponding to the putative gene product of the C. albicans ETG1 homolog. This result could be an artifact of the positioning of the cross-linker group on the cyclic-peptide nucleus or could possibly be due to a low abundance of the C. albicans ETG1 gene product in crude membranes. Alternatively, it may be that noncompetitive inhibition of glucan synthase is mediated by proteins other than the ETG1 gene product.
In summary, using an analog of LY303366, we have constructed a photoaffinity probe for examining the direct interaction of echinocandins with polypeptides in membrane preparations of C. albicans. The photoaffinity experiments described in this work detected two polypeptides but did not detect the gene product of the C. albicans homolog of ETG1, previously identified through genetic and biochemical studies to be the putative catalytic domain of glucan synthase. The major labeled polypeptide of 40 kDa possesses limited homology to the human NF-1 protein, a protein involved in regulation of small GTPase cycling, and therefore may be involved in GTP regulation of glucan synthase activity.
ACKNOWLEDGMENTS
We thank Paul Skatrud, Michele Smith, and William Current for suggestions regarding the preparation of the manuscript. The technical assistance of Jenny Vessels was also greatly appreciated. In addition, we thank Mel Johnson and Gerry Becker for the peptide sequence information.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Awald P, Zugel M, Monks C, Frost D, Selitrennikoff C P. Purification of 1,3-β-d-glucan synthase from Neurospora crassa by product entrapment. Exp Mycol. 1993;17:130–141. [Google Scholar]
- 3.Balkovec J M. Lipopeptide antifungal agents. Expert Opin Invest Drugs. 1994;3:65–82. [Google Scholar]
- 4.Castro C, Ribas J C, Valdivieso M H, Varona R, del Rey F, Duran A. Papulacandin B resistance in budding and fission yeasts: isolation and characterization of PBR1, a gene involved in (1,3)-β-d-glucan synthesis in Saccharomyces cerevisiae. J Bacteriol. 1995;177:5732–5739. doi: 10.1128/jb.177.20.5732-5739.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cleveland D W. Peptide mapping in one dimension by limited proteolysis of sodium dodecyl sulfate-solubilized proteins. Methods Enzymol. 1983;96:222–229. doi: 10.1016/s0076-6879(83)96020-2. [DOI] [PubMed] [Google Scholar]
- 6.Como J A, Dismukes W E. Oral azole drugs as systemic antifungal therapy. N Engl J Med. 1994;330:263–272. doi: 10.1056/NEJM199401273300407. [DOI] [PubMed] [Google Scholar]
- 7.Current W L, Boylan C, Raab P. Presented at the 33rd Interscience Conference on Antimicrobial Agents and Chemotherapy, New Orleans, La., 17 to 20 Oct. 1993. 1993. [Google Scholar]
- 8.Debono M, Turner W W, LaGrandeur L, Burkhardt F J, Nissen J S, Nichols K K, Rodriguez M J, Zweifel M J, Zeckner D J, Gordee R S, Tang J, Parr T R J. Semisynthetic chemical modification of the antifungal lipopeptide echinocandin B (ECB): structure-activity studies of the lipophilic and geometric parameters of polyarylated analogs of ECB. J Med Chem. 1995;38:3271–3281. doi: 10.1021/jm00017a012. [DOI] [PubMed] [Google Scholar]
- 9.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Difco Laboratories. Difco manual. 10th ed. Detroit, Mich: Difco Laboratories; 1984. [Google Scholar]
- 11.Dixon C K, Ma D. Characterization of ECB LY280949 resistant mutants in Saccharomyces cerevisiae. Yeast. 1995;11:S538. [Google Scholar]
- 12.Douglas C M, Foor F, Marrinan J A, Morin N, Nielsen J B, Dahl A M, Mazur P, Baginsky W, Li W, El-Sherbeini M, Clemas J A, Mandala S M, Frommer B R, Kurtz M B. The Saccharomyces cerevisiae FKS1 (ETG1) gene encodes an integral membrane protein which is a subunit of 1,3-β-d-glucan synthase. Proc Natl Acad Sci USA. 1994;91:12907–12911. doi: 10.1073/pnas.91.26.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Douglas C M, Marrinan J A, Li W, Kurtz M B. A Saccharomyces cerevisiae mutant with echinocandin-resistant 1,3-β-d-glucan synthase. J Bacteriol. 1994;176:5686–5696. doi: 10.1128/jb.176.18.5686-5696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drgnova J, Drgon T, Tanaka K, Kollar R, Chen G-C, Ford R A, Chan C S M, Takai Y, Cabib E. Rho1p, a yeast protein at the interface between cell polarization and morphogenesis. Science. 1996;272:277–281. doi: 10.1126/science.272.5259.277. [DOI] [PubMed] [Google Scholar]
- 15.El-Sherbeini M, Clemas J A. Cloning and characterization of GNS1: a Saccharomyces cerevisiae gene involved in synthesis of 1,3-β-glucan in vitro. J Bacteriol. 1995;177:3227–3234. doi: 10.1128/jb.177.11.3227-3234.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frost D, Brandt K, Estill C, Goldman R. Partial purification of (1,3)-β-glucan synthase from Candida albicans. FEMS Microbiol Lett. 1997;146:255–261. doi: 10.1111/j.1574-6968.1997.tb10202.x. [DOI] [PubMed] [Google Scholar]
- 17.Frost D, Drake R R, Wasserman B P. (1,3)-β-Glucan synthase from Saccharomyces cerevisiae: in vitro activation by β-lactoglobulin or Brij-35, and photoaffinity labeling of enriched microsomal fractions with 5-azido-UDP-Glc and 8-azido-GTP. Curr Microbiol. 1992;24:295–300. [Google Scholar]
- 18.Frost D J, Brandt K, Capobianco J, Goldman R. Characterization of (1,3)-β-glucan synthase in Candida albicans: microsomal assay from the yeast or mycelial morphological forms and a permeabilized whole-cell assay. Microbiology. 1994;140:2239–2246. doi: 10.1099/13500872-140-9-2239. [DOI] [PubMed] [Google Scholar]
- 19.Gallis H A, Drew R H, Pickard W W. Amphotericin B: 30 years of clinical experience. Rev Infect Dis. 1990;12:308–329. doi: 10.1093/clinids/12.2.308. [DOI] [PubMed] [Google Scholar]
- 20.Garrett-Engele P, Moilanen B, Cyert M S. Calcineurin, the Ca2+/calmodulin-dependent protein phosphatase, is essential in yeast mutants with cell integrity defects and in mutants that lack a functional vacuolar H+-ATPase. Mol Cell Biol. 1995;15:4103–4114. doi: 10.1128/mcb.15.8.4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goff D A, Koletar S L, Buesching W J, Barnisham J, Fass R J. Isolation of fluconazole-resistant Candida albicans from human immunodeficiency virus-negative patients never treated with azoles. Clin Infect Dis. 1995;20:77–83. doi: 10.1093/clinids/20.1.77. [DOI] [PubMed] [Google Scholar]
- 22.Gutmann D H, Boguski M, Marchuk D, Wigler M, Collins F S, Ballester R. Analysis of the neurofibromatosis type 1 (NF1) GAP-related domain by site directed mutagenesis. Oncogene. 1993;8:761–769. [PubMed] [Google Scholar]
- 23.Inoue S B, Takewaki N, Takasuka T, Mio T, Adachi M, Fujii Y, Miyamoto C, Arisawa M, Furuichi Y, Watanabe T. Characterization and gene cloning of 1,3-β-d-glucan synthase from Saccharomyces cerevisiae. Eur J Biochem. 1995;231:845–854. doi: 10.1111/j.1432-1033.1995.tb20770.x. [DOI] [PubMed] [Google Scholar]
- 24.Keller-Juslen, C., and M. Kuhn. August 1977. German patent 2,704,030.
- 25.Kelly R, Register E, Hsu M, Kurtz M, Nielsen J. Isolation of a gene involved in a 1,3-β-glucan synthesis in Aspergillus nidulans and purification of the corresponding protein. J Bacteriol. 1996;178:4381–4391. doi: 10.1128/jb.178.15.4381-4391.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurtz M B, Abruzzo G, Flattery A, Bartizal K, Marrinan J A, Li W, Milligan J, Nollstadt K, Douglas C M. Characterization of echinocandin-resistant mutants of Candida albicans: genetic, biochemical, and virulence studies. Infect Immun. 1996;64:3244–3251. doi: 10.1128/iai.64.8.3244-3251.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Lee K L, Buckley H R, Campbell C C. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia. 1975;13:148–153. doi: 10.1080/00362177585190271. [DOI] [PubMed] [Google Scholar]
- 29.Martin G, Viskochil D, Bollag G, McCabe P C, Crosler W J, Haubruck H, Conroy L, Clark R, O’Connell P, Cawthon R M, Innis M A, McCormick F. The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell. 1990;63:843–849. doi: 10.1016/0092-8674(90)90150-d. [DOI] [PubMed] [Google Scholar]
- 30.Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 31.Mazur P, Baginsky W. In vitro activity of 1,3-β-glucan synthase requires the GTP-binding protein Rho1. J Biol Chem. 1996;271:14604–14609. doi: 10.1074/jbc.271.24.14604. [DOI] [PubMed] [Google Scholar]
- 32.Mitchell L H, Soll D R. Commitment to germ tube or bud formation during release from stationary phase in Candida albicans. Exp Cell Res. 1979;120:167–179. doi: 10.1016/0014-4827(79)90547-0. [DOI] [PubMed] [Google Scholar]
- 33.Mol P C, Park H, Mullins J T, Cabib E. A GTP-binding protein regulates the activity of (1,3)-β-glucan synthase, an enzyme directly involved in yeast cell wall morphogenesis. J Biol Chem. 1994;269:31267–31274. [PubMed] [Google Scholar]
- 34.Oh C, Toke D A, Mandala S, Martin C E. ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J Biol Chem. 1997;272:17376–17384. doi: 10.1074/jbc.272.28.17376. [DOI] [PubMed] [Google Scholar]
- 35.Pfaller M, Wenzel R. Impact of the changing epidemiology of fungal infections in the 1990s. Eur J Clin Microbiol Infect Dis. 1992;11:287–291. doi: 10.1007/BF01962067. [DOI] [PubMed] [Google Scholar]
- 36.Pfaller, M. A. 1994. Epidemiology and control of fungal infections. Clin. Infect. Dis. 19(Suppl. 1):S8–S13. [DOI] [PubMed]
- 37.Pfaller M A, Messer S A, Coffman S. In vitro susceptibilities of clinical yeast isolates to a new echinocandin derivative, LY3033566, and other antifungal agents. Antimicrob Agents Chemother. 1997;41:763–766. doi: 10.1128/aac.41.4.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pittet D, Wenzel R P. Nosocomial bloodstream infections. Arch Intern Med. 1995;155:1177–1184. doi: 10.1001/archinte.155.11.1177. [DOI] [PubMed] [Google Scholar]
- 39.Raab P, Zeckner D J, Boyll B, Boylan C, Current W. Presented at the 34th Interscience Conference on Antimicrobial Agents and Chemotherapy, Orlando, Fla., 4 to 7 Oct. 1994. 1994. [Google Scholar]
- 40.Ram A F J, Brekelmans S S C, Oehlen L J W M, Klis F M. Identification of two cell cycle regulated genes affecting the β1,3-glucan content of cell walls in Saccharomyces cerevisiae. FEBS Lett. 1995;358:165–170. doi: 10.1016/0014-5793(94)01418-z. [DOI] [PubMed] [Google Scholar]
- 41.Rex J H, Rinaldi M G, Pfaller M A. Resistance of Candida species to fluconazole. Antimicrob Agents Chemother. 1995;39:1–8. doi: 10.1128/aac.39.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sawistowska-Schroder E T, Kerridge D, Perry H. Echinocandin inhibition of 1,3-beta-d-glucan synthase from Candida albicans. FEBS Lett. 1984;173:134–138. doi: 10.1016/0014-5793(84)81032-7. [DOI] [PubMed] [Google Scholar]
- 43.Shematek E M, Cabib E. Biosynthesis of the yeast cell wall. II. Regulation of β-(1,3)glucan synthetase by ATP and GTP. J Biol Chem. 1980;255:895–902. [PubMed] [Google Scholar]
- 44.Sokolov B P, Sher B M, Kalinin V N. Modified method for peptide mapping of collagen chains using cyanogen bromide-cleavage of protein within polyacrylamide gels. Anal Biochem. 1989;176:365–367. doi: 10.1016/0003-2697(89)90324-2. [DOI] [PubMed] [Google Scholar]
- 45.Taft C S, Selitrennikoff C P. Cilofungin inhibition of (1,3)-β-glucan synthase: the lipophilic side chain is essential for inhibition of enzyme activity. J Antibiot. 1990;43:433–437. doi: 10.7164/antibiotics.43.433. [DOI] [PubMed] [Google Scholar]
- 46.Taft C S, Stark T, Selitrennikoff C P. Cilofungin ( LY121019) inhibits Candida albicans (1-3)-β-d-glucan synthase activity. Antimicrob Agents Chemother. 1988;32:1901–1903. doi: 10.1128/aac.32.12.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang J, Parr T R., Jr W-1 solubilization and kinetics of inhibition by cilofungin of Candida albicans (1,3)-β-d-glucan synthase. Antimicrob Agents Chemother. 1991;35:99–103. doi: 10.1128/aac.35.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vessels J M, Radding J A. Oligosaccharide mapping of fungal glucan synthase product by high-performance anion-exchange chromatography. Anal Biochem. 1993;215:150–155. doi: 10.1006/abio.1993.1567. [DOI] [PubMed] [Google Scholar]
- 49.Xu G, O’Connell P, Viskochil D, Cawthon R, Robertson M, Culver M, Dunn D, Stevens J, Gesteland R, White R, Weiss R. The neurofibromatosis type 1 gene encodes a protein related to GAP. Cell. 1990;62:599–608. doi: 10.1016/0092-8674(90)90024-9. [DOI] [PubMed] [Google Scholar]