Abstract
Introduction
Genetic etiologies are estimated to account for a large portion of chronic kidney diseases (CKD) in children. However, data are lacking regarding the true prevalence of monogenic etiologies stemming from an unselected population screen of children with advanced CKD.
Methods
We conducted a national multicenter prospective study of all Israeli pediatric dialysis units to provide comprehensive “real-world” evidence for the genetic basis of childhood kidney failure in Israel. We performed exome sequencing and assessed the genetic diagnostic yield.
Results
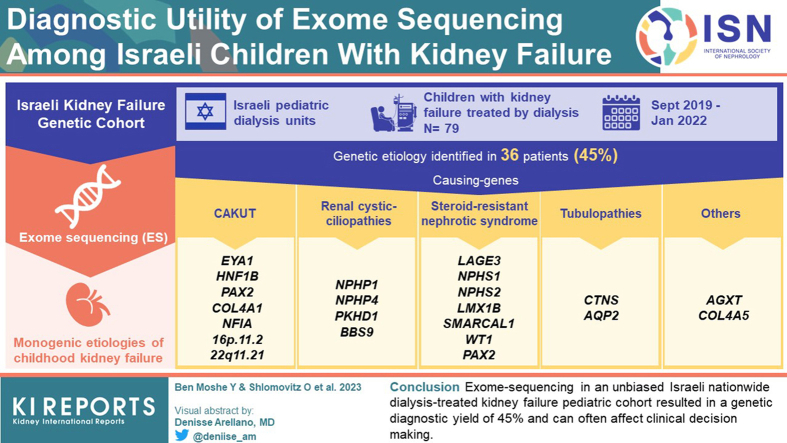
Between 2019 and 2022, we recruited approximately 88% (n = 79) of the children on dialysis from all 6 Israeli pediatric dialysis units. We identified genetic etiologies in 36 of 79 (45%) participants. The most common subgroup of diagnostic variants was in congenital anomalies of the kidney and urinary tract causing genes (e.g., EYA1, HNF1B, PAX2, COL4A1, and NFIA) which together explain 28% of all monogenic etiologies. This was followed by mutations in genes causing renal cystic ciliopathies (e.g., NPHP1, NPHP4, PKHD1, and BBS9), steroid-resistant nephrotic syndrome (e.g., LAGE3, NPHS1, NPHS2, LMX1B, and SMARCAL1) and tubulopathies (e.g., CTNS and AQP2). The genetic diagnostic yield was higher among Arabs compared to Jewish individuals (55% vs. 29%) and in children from consanguineous compared to nonconsanguineous families (63% vs. 29%). In 5 participants (14%) with genetic diagnoses, the molecular diagnosis did not correspond with the pre-exome diagnosis. Genetic diagnosis has a potential influence on clinical management in 27 of 36 participants (75%).
Conclusion
Exome sequencing in an unbiased Israeli nationwide dialysis-treated kidney failure pediatric cohort resulted in a genetic diagnostic yield of 45% and can often affect clinical decision making.
Keywords: children, dialysis, ESKD, exome sequencing, kidney failure, monogenic
Graphical abstract
Childhood CKD covers a wide range of structural and functional disorders that often result from different genetic etiologies. Knowledge about the underlying genetic basis of childhood CKD has dramatically progressed over the last 2 decades with the identification of numerous disease-causing gene alterations and novel pathomechanisms.1,2 Understanding the genetic underpinnings of childhood CKD, which often enables personalized surveillance and therapies, has increased significantly due to the growing availability of genetic testing as well as clinician’s awareness. So far, hundreds of CKD-causing genes have been reported, estimated to explain up to 30% of cases among children1,2 and approximately 10% in adults.3 Nevertheless, the genetic diagnostic yield varies markedly across different study cohorts, depending on preselection criteria, clinical presentation, geographic region, and ethnicity. Furthermore, in clinical practice, genetic kidney diseases might be overlooked due to its variable expressivity, incomplete penetrance, low index of suspicion, lack of overt symptoms at early disease stages, and insufficient availability of next generation sequencing methods. Consequently, data are lacking regarding the true prevalence of monogenic CKD etiologies stemming from a non-selected large scale population screen of children with advanced CKD.
We therefore conducted a national multicenter study of all Israeli pediatric dialysis units to provide comprehensive evidence for monogenic etiologies of childhood kidney failure. We report the results from exome sequencing in 79 children with kidney failure treated by dialysis, comprising 88% of the total of 90 children treated with dialysis in Israel during the study period.
Methods
Study Participants
We conducted a national multicenter collaboration study of all Israeli dialysis units and established the Israeli Kidney Failure Genetic Cohort. The overarching aim of this collaboration is to collect clinical data and DNA samples for genetic studies to provide comprehensive real-world evidence for kidney failure’s genetic basis at a national level. By utilizing the Israeli end-stage kidney disease registry, we identified and located all pediatric units in which Israeli children under the age of 18 years are treated with maintenance dialysis. During the study period (September 2019–January 2022), clinical and pedigree data from all patients were collected by administering a standardized questionnaire, after obtaining informed consent. Blood samples of individuals on dialysis and their parents, if available, were collected. Approval for human subject research was obtained from the Institutional Review Boards of Sheba Medical Center and the Israeli Ministry of Health, as well as from ethics review boards of the participating centers.
Clinical Assessment and Diagnosis
Single or multiple primary clinical diagnoses were recorded for each participant based on their medical records and their primary pediatric nephrologist’s assessment. In addition, all participants underwent a clinical interview at the time of recruitment to the study, which included a comprehensive review of their medical files, imaging studies, and kidney biopsy reports, when available. The primary kidney clinical diagnoses were classified as either congenital anomalies of the kidneys and urinary tract (CAKUT), nephrotic syndrome, renal cystic ciliopathy, tubulointerstitial disease, nephrolithiasis-related kidney disease, and CKD of unknown etiology. Furthermore, information regarding the medical history, family history, ethnicity, age at dialysis initiation, consanguinity, and presence of extrarenal manifestations were obtained (Table 1).
Table 1.
Clinical characteristics and pre-exome sequencing clinical diagnoses among 79 children with kidney failure
| Characteristics | Value | Percentage |
|---|---|---|
| Gender | ||
| Male | 35 | 44% |
| Female | 44 | 56% |
| Phenotype | ||
| CAKUT | 30 | 38% |
| SRNS | 20 | 25% |
| Renal cystic ciliopathy | 9 | 11% |
| Unknown | 7 | 9% |
| Glomerulopathy | 6 | 8% |
| Tubulopathy | 4 | 5% |
| Nephrolithiasis-related kidney disease | 2 | 3% |
| Other | 1 | 1% |
| Biopsied | ||
| Yes | 22 | 28% |
| No | 57 | 72% |
| Age at dialysis initiation (yrs) | ||
| 0–5 | 28 | 35% |
| 5–10 | 17 | 22% |
| 11–15 | 23 | 29% |
| 15–18 | 11 | 14% |
| Parental consanguinity | ||
| First cousins | 38 | 48% |
| Family history of ESKD | ||
| Yes | 22 | 28% |
| No | 57 | 72% |
CAKUT, congenital anomalies of the kidney and urinary tract; ESKD, end-stage kidney disease; SRNS, steroid-resistant nephrotic syndrome.
Exome Sequencing
DNA was isolated from peripheral blood lymphocytes using the DNeasy Blood and Tissue Kit (Qiagen). Proband exome sequencing was performed on genomic DNA for all affected individuals using an Agilent v5 SureSelect Capture Kit and Illumina 2500 sequencing technology. For each sample, paired end-reads (2 × 150 bp) were obtained, processed, and mapped to the genome. We used the BWA-MEM algorithm (Illumina, San Diego, CA) version 0.7.1216 to align the sequence reads to the human reference genome (hg38). The HaplotypeCaller algorithm of GATK V.3.8 was applied for variant calling, as recommended in the best practice pipeline.4 KGG-seq version 1.1 was used for annotating identified variants.5 In addition, in-house scripts were applied for filtering, based on family pedigree and local datasets of variants detected in previous sequencing projects (in-house cohort of ∼5000 samples) and the Genome Aggregation Database data set (gnomAD),6 as well as from 30,000 exome sequencing data samples of ethnically matched controls (16,000 Jews and 14,000 Arabs) available via the Franklin Platform (Genoox, Tel-Aviv, Israel, available at https://franklin.genoox.com/clinical-db/home).
Diagnostic Analysis
We evaluated exome sequencing data for all rare nonsynonymous and splice variants (minor allele frequencies of 1% or less) in genes present in the OMIM database. In addition, we generated a list of previously reported CKD-causing genes and performed a second targeted analysis for this predefined CKD gene panel (Supplementary Table S1). Mutation calling was performed independently by 3 teams: bioinformaticians, geneticists, and a team of clinician-scientists with expertise in nephrogenetics. Exome sequencing data was filtered using the Franklin Platform (Genoox, Tel-Aviv, Israel, available at https://franklin.genoox.com/clinical-db/home) Exome data were interpreted according to the American College of Genetic and Genomic Medicine guidelines.7 We classified variants as pathogenic, likely pathogenic, variants of unknown significance, likely benign, or benign. American College of Genetic and Genomic Medicine standards and guidelines recommend 28 criteria for variant pathogenicity classification. The Genoox Classification Engine ran 17 of them automatically. The artificial intelligence-based variant classification engine automates rules PVS1, PS1, PM1, PM2, PM4, PM5, PP2, PP3, PP5, BA1, BS1, BS2, BP1, BP3, BP4, BP6, and BP7.8 The remaining rules cannot be automated because they require clinical information specific to the patient genotype, e.g., familial data, de novo evidence (PS2, PM6), segregation data (PP1, BS4), and/or allelic data (PM3, BP2). Therefore, the remaining criteria were manually classified. We evaluated all variants reported pathogenic or likely pathogenic by Clinvar and by the Genoox Classification Engine, all rare (minor allele frequency < 1%) homozygous variants and all rare (minor allele frequency < 1%) heterozygous variants found in our in-house CKD panel. When a trio exome was available, we also performed analysis for de novo variants.
Sanger sequencing was performed to confirm likely pathogenic and pathogenic variants in original DNA samples and when available, to test for familial segregation of the phenotype with the genotype. In addition, copy number variants were called using the DRAGEN software, which identifies copy number variants by comparing the average coverage and improperly paired reads per area of a sample to data from a cohort of normal samples. Areas with significant differences relative to the cohort were considered copy number variants. For further validation, we additionally compared the average coverage per exon of each sample to a cohort of similar samples using in-house scripts. Sequence variants that remained after the exome sequencing evaluation process were examined for segregation in available samples of affected and unaffected family members.
Results
Patient Characteristics
During the study period, 90 children were treated with maintenance dialysis in Israel. The parents of 11 children chose not to participate in the study. The characteristics of the study population, which includes 79 children (45% male) from 78 different families, who underwent exome sequencing are shown in Table 1. Single, duo, and trio exome analysis were performed in 55%, 19%, and 26% of cases, respectively. Fifty-one (65%) children are of Arab descent and 28 (35%) are Jewish. Overall, 22 participants (28%) had a relative with kidney failure. Consanguinity was evident in 38 participants (48%) of the cohort, predominantly among patients of Arab descent. Mean age at dialysis initiation was 8.5 years (range 1 month–18 years; median = 9 years). The most prevalent mode of dialysis was hemodialysis (75% of the cohort). The most common primary clinical diagnosis was CAKUT followed by nephrotic syndrome and renal cystic ciliopathies (Table 1).
Exome Sequencing Reveals Monogenic Etiology in Approximately 45% of Children on Dialysis
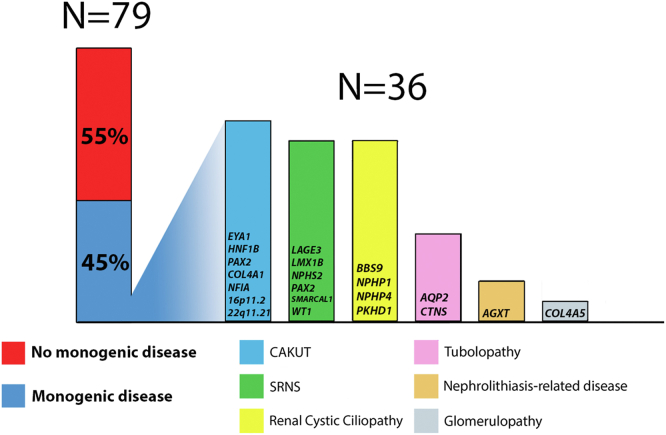
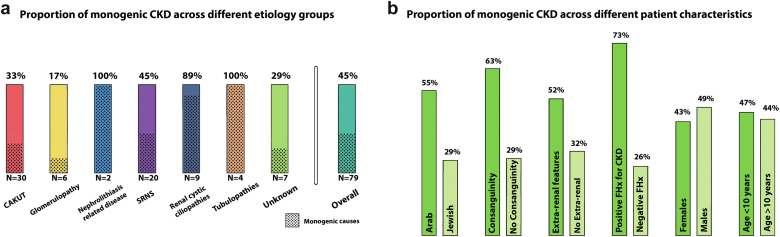
We detected diagnostic single-gene etiologies in 36 of the 79 participants (45%), encompassing 21 distinct monogenic disorders (Table 2, Figure 1). Of these, 13 (36%) are diseases with autosomal dominant inheritance, 22 (61%) with autosomal recessive inheritance, and 1 is an X-linked disease. In addition, 2 participants had compound heterozygous diagnostic pathogenic variants. Among the 36 diagnostic variants that were detected, 24 variants were previously reported as disease-causing and 12 were novel. Overall, 16 of 27 (59%) disease-causing variants had allele frequencies of zero in the Exome Sequencing Project and the Genome Aggregation Database. The mean age of dialysis initiation was comparable between patients with monogenic kidney failure and those with negative exome findings (7.9 vs. 9.05 years, respectively; P = 0.21). The yield of positive molecular diagnosis resulting from exome sequencing for each CKD category is presented in Figure 2a. Whereas the overall genetic diagnostic yield was approximately 45%, clinical diagnoses of ciliopathies, tubulopathies, and nephrolithiasis-related kidney disease exhibited the highest probability for an underlying genetic condition (89%–100% yield). Furthermore, a genetic diagnosis was made in 2 of 7 participants (29%) with kidney failure of unknown cause (Figure 2a). Higher yields of genetic diagnosis were achieved in individuals with syndromic features compared to those without syndromic features (52% vs. 32%, respectively), in individuals with Arabic ethnicity compared to Jewish ethnicity (55% vs. 29%, respectively), and among consanguineous compared to nonconsanguineous families (63% vs. 29%, respectively) (Figure 2b). Nevertheless, patients with none of the above-mentioned risk factors for monogenic kidney disease had an approximately 20% likelihood for an identification of an underlying genetic diagnosis. The distribution of the various monogenic etiologies in relation to age at dialysis initiation is shown in Supplementary Figure S1.
Table 2.
Thirty-six participants with kidney failure for whom exome sequencing yielded a molecular genetic diagnosis
| Patient | Clinical data | Post-ES diagnosis | Gene/region | Annotation | Zygosity | CADD score | Allele frequencya | ACMG class | ACMG criteria |
|---|---|---|---|---|---|---|---|---|---|
| 21–112 | Muslim Arab female with cystic kidney disease; kidney failure at 14 y; no consanguinity | Nephronophthisis 4 | NPHP4 | c.3325C>T p.Arg1109Ter NM_015102.5 |
Homo | 38 | 0.002% | P | PVS1, PM2, PP5 |
| 21–114 | Muslim Arab female with cystic kidney disease and chronic lung disease; kidney failure at 1.8 y; consanguinity | Autosomal recessive polycystic kidney disease | PKHD1 | c.2279G>A p.Arg760His NM_138694.4 |
Homo | 33 | 0.0013% | P | PM2, PP3, PP5 |
| 21–115 | Muslim Arab male with dysplastic kidneys. Kidney failure at 5.5 y; no consanguinity | CAKUT | COL4A1 | c.1807C>T p.Pro603Ser NM_001845.6 |
Het | 25.2 | 0.0019% | LP | PM2, PP3, PM1, PP5 PP2 |
| 21–117 | Muslim Arab female with nephrotic syndrome, dysmorphism, Intellectual disability, short stature, and skeletal deformity; kidney failure at 7 y; consanguinity | Schimke immuno-osseous dysplasia | SMARCAL1 | c.1682G>A p.Arg561His NM_014140.4 |
Homo | 31 | 0% | LP | PP3, PM2, PP4, PP5 |
| 21–139 | Arab Druze female with hypoplastic kidney, polydactyly, obesity, strabismus; kidney failure at 18 y; consanguinity | Bardet-Biedl syndrome type 9 | BBS9 | c.1063C>T p.Gln355Ter NM_198428.3 |
Homo | 40 | 0% | P | PVS1, PM2, PP5 |
| 21–146 | Arab Muslim male with nephronophthisis. Kidney failure at 8 y; consanguinity | Nephronophthisis type 1 | NPHP1 deletion | chr2 deletion: 110,123,780-110,204,978 | Homo | NA | P | ||
| 21–147 | Arab Muslim male with tubulopathy and primary hypothyroidism. Kidney failure at 14 y; consanguinity | Cystinosis | CTNS | c.681G>A p.Glu227Glu NM_004937.3 |
Homo | 23 | 0.0007% | LP | PM2, PP3, PP5, PP4 |
| 21–154 | Arab Muslim male with nephrotic syndrome, Intellectual disability and microcephaly. Kidney failure at 2 y; consanguinity | Galloway-Mowat syndrome | LAGE3 | c.317+4A>G NM_006014.5 |
Homo | 22 | 0% | LP | PM2, PP3, PP5, PM6, PP4 |
| 21–174 | Jewish male with CAKUT; kidney failure at 2 y; no consanguinity | Branchiootic syndrome 1 | EYA1 | c.1597+2T>C NM_000503.6 |
Het | 33 | 0% | LP | PVS1, PM2 |
| 21–182 | Arab Muslim male with nephrogenic DI, vesicoureteral reflux, uropathy, recurrent UTIs; kidney failure at 16 y; consanguinity | Nephrogenic DI | AQP2 | c.83T>C p.Leu28Pro NM_000486.6 |
Homo | 27 | 0% | LP | PM2, PM1 PP3, PP2, PP5 |
| 21–183 | Arab Muslim male with nephrotic syndrome; kidney failure at 6 y; no consanguinity | Nephrotic syndrome type 2 | NPHS2 | c.714G>T p.Arg238Ser NM_014625.4 c.412C>T p.Arg138Ter NM_014625.4 |
Comp het | 25.2 44 |
0.0032% 0.0006% |
P P |
PS1, PM2, PP3, PP2, PP5 PSV1, PM2, PP5 |
| 21–213 | Arab Muslim female with kidney and liver cystic disease; kidney failure at 3 y; consanguinity | Autosomal recessive polycystic kidney disease | PKHD1 | c.2279G>A p.Arg760His NM_138694.4 |
Homo | 33 | 0.0013% | P | PM2, PP3, PP5 |
| 21–218 | Arab Muslim male with CKDu, Intellectual disability, short stature, skeletal deformity, convulsions; kidney failure at 15 y; consanguinity | CAKUT | TBX6 + 29 genes deletion | chr16 deletion: 29,802,071-30,199,161 (16p11.2) | Het | NA | P | ||
| 21–219 | Arab Muslim male with polycystic kidney, developmental delay, elevated liver enzymes; kidney failure at 1.5 y; no consanguinity | HNF1B- related disease | HNF1B + 28 genes deletion | chr17 deletion: 36,459,737-38,222,841 (17q12-q21.2) |
Het | NA | P | ||
| 21–225 | Jewish female with renal aplasia and imperforated anus; kidney failure at 2 months; no consanguinity | Branchiootic syndrome 1 | EYA1 | c.1586C>A p.Ala529Glu NM_000503.6 |
Het | 29 | 0% | LP | PP3, PM2 |
| 21–226 | Arab Muslim female with cystinosis; kidney failure at 5.5 y; consanguinity | Cystinosis | CTNS | c.587dup p.Asn196fs NM_004937.3 |
Homo | NA | 0% | P | PVS1, PM2, PP5, PP4 |
| 21–250 | Arab Muslim female with cystinosis; kidney failure at 8 y; consanguinity | Cystinosis | CTNS | c.587dup p.Asn196fs NM_004937.3 |
Homo | NA | 0% | P | PVS1,PM2, PP5, PP4 |
| 21–253 | Arab Muslim female with suspected nephronophthisis; kidney failure at 3 y; no consanguinity | CAKUT | NFIA | c.39C>G p.Tyr13Ter NM_001145512.2 |
Het | 31 | 0% | LP | PVS1, PM2 |
| 21–262 | Arab Muslim male with hyperoxaluria, skeletal deformity, developmental delay; kidney failure at 3.5 months; consanguinity | Primary hyperoxaluria type 1 | AGXT | c.997A>T p.Arg333Ter NM_000030.3 |
Homo | 43 | 0% | P | PVS1, PM2, PP5, PP4 |
| 21–263 | Arab Muslim male with glomerulopathy and deafness; kidney failure at 14 y; no consanguinity | Alport syndrome | COL4A5 | c.3319_3335del p.Gly1107fs NM_033380.3 |
Hemi | NA | 0% | LP | PVS1, PM2 |
| 21–264 | Arab Muslim male with hyperoxaluria, skeletal deformity, severe growth delay, splenomegaly; kidney failure at 2.5 mo; consanguinity | Primary hyperoxaluria type 1 | AGXT | c.358+1G>A NM_000030.3 |
Homo | 33 | 0% | P | PVS1, PM2, PP4 |
| 21–314 | Arab Muslim female with cystic kidney disease; kidney failure at 17 y; consanguinity | Nephronophthisis 1 | NPHP1 deletion | chr2 deletion: 110,881,357-110,962,555 | Homo | NA | P | ||
| 21–315 | Jewish male with CAKUT and Intellectual disability; kidney failure at 14 y; no consanguinity | CAKUT | COL4A1 deletion | chr13 deletion: 110,152,323-110,214,025 ring chromosome 13 |
Het | NA | LP | ||
| 21–354 | Jewish male with CAKUT; kidney failure at 2.5 y; no consanguinity | CAKUT | AIFM3 + 76 genes duplication | chr22 duplication: 18516490-21046135 (22q11.21 dup) |
Het | NA | P | ||
| 21–481 | Arab Muslim male nephrotic syndrome; kidney failure at 18 months; consanguinity | Nephrotic syndrome, type 1 | NPHS1 | c.2104G>A p.Gly702Arg NM_004646.4 |
Homo | 23.6 | 0% | LP | PM2, PP1 PS1 |
| 21–482 | Arab Muslim male with cystic kidney disease, liver cysts and bronchiectasis; kidney failure at 14.5 y; consanguinity | Autosomal recessive polycystic kidney disease | PKHD1 | c.2279G>A p.Arg760His NM_138694.4 c.4870C>T p.Arg1624Trp NM_138694.4 |
Comp het | 33 26 |
0.0013% 0.0059% |
P LP |
PM2, PP3, PP5 PM2, PP5 |
| 21–483 | Jewish male with nephrotic syndrome; kidney failure at 11.5 y; no consanguinity | Nail-patella syndrome | LMX1B | c.737G>A p.Arg246Gln NM_001174147.2 |
Het | 31 | 0% | P | PM2, PM1, PP2, PM5, PP3, PP5, PM6 |
| 21–484 | Arab Muslim female with nephrotic syndrome, failure to thrive, osseous dysplasia; kidney failure at 11 y; consanguinity | Schimke immuno-osseous dysplasia | SMARCAL1 | c.2290C>T p.Arg764Trp NM_014140.4 |
Homo | 25.2 | 0.0006% | P | PM2, PM5, PP3, PP5, PP4 |
| 21–487 | Arab Muslim female with glomerulopathy, coloboma, cardiomyopathy; kidney failure at 6 y; consanguinity | Papillorenal syndrome | PAX2 | c.76dup p.Val26fs NM_000278.5 |
Het | NA | 0% | P | PVS1, PM2, PP5 |
| 21–509 | Jewish female with cystinosis; kidney failure at 13.5 y; no consanguinity | Cystinosis | CTNS | c.1015G>A p.Gly339Arg NM_004937.3 |
Homo | 26.6 | 0.0032% | P | PP3, PM2, PP5, PP4 |
| 21–680 | Arab Muslim male with hydroureteronephrosis and dysplastic kidneys, mild Intellectual disability; kidney failure at 17.5 y; consanguinity | Bardet-Biedl syndrome 9 | BBS9 | c.385C>T p.His129Tyr NM_198428.3 |
Homo | 23 | 0% | VUSb | PM2, PP3 |
| 21–763 | Jewish female (per phenotype) with XY DSD, proteinuria, CNS cystic lesions; kidney failure at birth; consanguinity | Denys-Drash syndrome | WT1 | c.1316G>T p.Arg439Leu NM_024426.6 |
Het | 32 | 0% | LP | PM2, PM5, PM1, PP2, PP5 |
| 21–848 | Arab Muslim female with nephrotic syndrome; kidney failure at 15 y; consanguinity | Nephrotic syndrome, type 2 | NPHS2 | c.851C>T p.Ala284Val NM_014625.4 |
Homo | 26 | 0.0039% | P | PM2, PM1, PP2, PP3, PP5 |
| 21–878 | Jewish female with large hyperechoic kidneys and severe oligohydramnios; kidney failure at birth; no consanguinity | Autosomal recessive polycystic kidney disease | PKHD1 | c.3766del p.Gln1256fs NM_138694.4 |
Homo | NA | 0.02% | P | PVS1, PM2, PP5, |
| 21–511 | Arab Muslim male with renal dysplasia, elevated CK, hyperlipidemia, fatty liver, short stature, GH deficiency, normal eye examination. Kidney failure at 13.5 y; consanguinity | Papillorenal syndrome | PAX2 | c.76dup p.Val26fs NM_000278.5 |
Het | NA | 0% | P | PVS1, PM2, PP5, |
| 22–511 | Arab Muslim male with renal hypoplasia, elevated CK, normal eye examination; kidney failure at 2.5 y; consanguinity | Papillorenal syndrome | PAX2 | c.76dup p.Val26fs NM_000278.5 |
Het | NA | 0% | P | PVS1, PM2, PP5, |
ACMG, American College of Medical Genetics and Genomics; CADD, combined annotation dependent depletion; CAKUT, congenital anomalies of the kidney and urinary tract; CK, creatine kinase; CKDu, chronic kidney disease of unknown origin; Comp, compound; DI, diabetes insipidus; DSD, disorder of sex development; ES, exome sequencing; GH, growth hormone; Hemi, hemizygous; Het, heterozygous; Homo, homozygous; LP, likely pathogenic; P, pathogenic; UTI, urinary tract infection; y, years.
Allele frequency was determined using the genome AD website (https://gnomad.broadinstitute.org).
Although this variant is designated as VUS per ACMG criteria, in the context of the specific clinical findings and absence of controls, it was considered as disease-causing.
Figure 1.
Diagnostic yield of exome sequencing in Israeli children on dialysis. In 36 (45%) participants (blue), we identified monogenic CKD-related diagnostic variants. For the 36 solved cases, the different genes of the diagnostic variants are grouped into subcategories and displayed on the right side. CAKUT, congenital anomalies of the kidney and the urinary tract; CKD, chronic kidney disease; SRNS, steroid-resistant nephrotic syndrome.
Figure 2.
Proportion of monogenic CKD etiologies across different clinical categories. (a) Proportion of monogenic CKD across different etiology groups. Among patients with tubulopathies, nephrolithiasis-related kidney disease and renal cystic ciliopathies, the yield was very high ∼90% to 100% compared with lower yields of ∼17% to 30% among patients with glomerulopathy, CAKUT, and CKDu. One patient with kidney failure secondary to acute kidney injury (AKI) is not displayed in the left panel. (b) Proportion of monogenic CKD across different patient’s characteristics. CAKUT, congenital anomalies of the kidney and the urinary tract; CKD, chronic kidney disease; CKDu, CKD of unknown etiology; FHx, family history; SRNS, steroid-resistant nephrotic syndrome.
Our results demonstrate significant phenotypic and genetic heterogeneity in childhood-onset CKD. For example, in 3 unrelated Arab Muslim participants, we detected the PKHD1 p.Arg760His alteration, potentially suggesting a founder mutation effect. This mutation exhibits phenotypic heterogeneity in terms of kidney failure timing and the resulting dialysis initiation age, which was different for each individual (1.5 and 3 years). Similarly, 3 participants from 2 different Arab Muslim families, were found to harbor the heterozygous PAX2 p.Val26fs alteration, known to reside in a genetic hotspot.9,10 Whereas 2 participants presented clinically with CAKUT, the third participant presented with steroid-resistant nephrotic syndrome and focal segmental glomerulosclerosis.11
Clinical Implications of Molecular Diagnosis
Overall, the molecular genetic diagnosis corresponded with the primary pediatric nephrologists’ pre-exome clinical diagnoses in 31 of 36 (∼86%) cases, often confirming the diagnosis of a specific CKD etiology. Furthermore, for all genetically solved cases, the molecular diagnosis enabled validation of a final diagnosis and the performance of targeted formal genetic counseling to the participants and their family members. This also allowed for the provision of specific therapies, if available, which require confirmed molecular diagnoses. For example, establishing a genetic diagnosis of hyperoxaluria type 1 enables the initiation of the RNA interference–based therapy that is approved for this subtype of hyperoxaluria.12 Although the diagnosis can also be suggested upon biochemical parameters, genetic diagnosis should always be established for hyperoxaluria.13,14
Altogether, in 27 of 36 (75%) genetically confirmed cases, the molecular diagnosis has a potential influence on the clinical management (Table 3). The molecular genetic diagnosis guided clinical decision making in terms of disease surveillance, kidney transplant related regimen, introduction of disease-specific treatments, and performance of preimplantation genetic diagnosis for disease prevention in the context of family planning (Table 3).
Table 3.
Twenty-seven patients (75%) for whom the molecular genetic diagnosis influenced subsequent clinical management
| Patient | Gene | Clinical management | |
|---|---|---|---|
| 21–112 | NPHP4 | Changed the initial pre-ES clinical diagnosis of CAKUT. Subsequently, the patient was referred for ophthalmologic evaluation to monitor for the potential development of retinitis pigmentosa. | Surveillance |
| 21–139 | BBS9 | The genetic diagnosis prompted specialized ophthalmologic surveillance at a tertiary hospital due to a lack of local expertise. | Surveillance |
| 21–174 | EYA1 | Surveillance and ENT evaluation to identify and address any potential hearing problems. | Surveillance |
| 21–253 | NFIA | Prompted the referral for brain MRI. | Surveillance |
| 21–263 | COL4A5 | Surveillance and ENT evaluation to identify and address any potential hearing problems. Necessitates regular ophthalmologic evaluations to monitor for the potential development of ophthalmic diseases such as anterior lenticonus. | Surveillance |
| 21–354 | 22q11.21 dup | Led to completion of a comprehensive multidisciplinary evaluation, comprising of immunology and cardiology investigations, which ultimately led to a better comprehension of the entire clinical profile of chronic lung disease and bronchiectasis, which the patient exhibited. | Surveillance |
| 21–763 | WT1 | The genetic diagnosis, in this orthodox Jewish family, revealed the actual gender of the offspring, impacting matters related to Jewish legal practices and customs related to gender within their community and led to comprehensive multidisciplinary targeted evaluation and monitoring. | Surveillance |
| 21–117 | SMARCAL1 | The diagnosis led to a change in the patient’s pretransplant regimen. Instead of using Tacrolimus, Cyclosporine was used, as patients with this condition are at an elevated risk for developing infections due to their weakened immunity. Additionally, there is ongoing research and discussion about the potential benefits of combining a bone marrow transplant with a kidney transplant in this population to further improve results. | Pharmacological treatment Transplant |
| 21–262 | AGXT | Led to the implementation of a specific pharmacological treatment (Lumasiran), and prompted the need for ongoing surveillance by a multidisciplinary team. | Pharmacological treatment Surveillance |
| 21–264 | AGXT | Led to the implementation of specific pharmacological treatment (Lumasiran), and prompted the need for ongoing surveillance by a multidisciplinary team. | Pharmacological treatment Surveillance |
| 21–147 | CTNS | Led to the implementation of a specific pharmacological treatment (Cysteamine), and prompted the need for ongoing surveillance by a multidisciplinary team. | Pharmacological treatment Surveillance |
| 21–226 | CTNS | Led to the implementation of a specific pharmacological treatment (Cysteamine), and prompted the need for ongoing surveillance by a multidisciplinary team. | Pharmacological treatment Surveillance |
| 21–250 | CTNS | Led to the implementation of a specific pharmacological treatment (Cysteamine), and prompted the need for ongoing surveillance by a multidisciplinary team. | Pharmacological treatment Surveillance |
| 21–509 | CTNS | Led to the implementation of a specific pharmacological treatment (Cysteamine), and prompted the need for ongoing surveillance by a multidisciplinary team. | Pharmacological treatment Surveillance |
| 21–182 | AQP2 | Led to the implementation of a specific pharmacological treatment. | Pharmacological treatment |
| 21–183 | NPHS2 | The diagnosis obviated the need for immunosuppressive drugs. | Pharmacological treatment |
| 21–848 | NPHS2 | The diagnosis obviated the need for immunosuppressive drugs. | Pharmacological treatment |
| 21–481 | NPHS1 | The diagnosis obviated the need for immunosuppressive drugs. | Pharmacological treatment |
| 21–483 | LMX1B | The diagnosis obviated the need for immunosuppressive drugs. | Pharmacological treatment |
| 21–213 | PKHD1 | Impacted the evaluation process prior to transplantation by requiring a thorough examination of the liver. Led to the determination to undertake a combined transplantation of both the liver and kidney. | Transplant |
| 21–219 | HNF1B | Following the genetic diagnosis, genetic testing of the parents was conducted, and the father was identified as an asymptomatic carrier of the mutation. As a result, the mother was chosen as the living donor for the transplant. | Transplant |
| 21–482 | PHKD1 | Impacted the evaluation process prior to transplantation requiring a thorough examination of the liver. Led to a combined liver and kidney transplant. | Transplant |
| 21–680 | BBS9 | An inadequate evaluation of the bladder during the initial transplant procedure led to implant failure due to urological complications. It was not until 5 years after the initial transplant that the underlying urological issues were identified and addressed. In retrospect, the molecular diagnosis of a BBS variant known to cause urological problems could have prompted a more thorough urological evaluation and surveillance, potentially preventing the failure of the initial transplant and the need for a second transplant. | Transplant Surveillance |
| 21–154 | LAGE3 | The genetic diagnosis facilitated the successful use of PGD in the following pregnancy. | Pre-implantation Genetic diagnosis |
| 21–511 | PAX2 | The genetic diagnosis facilitated the successful use of PGD in the third child, preventing the transmission of the disorder. Additionally, the patient was sent for an ophthalmological evaluation to identify and address any potential visual problems. | Pre-implantation Genetic diagnosis Surveillance |
| 22–511 | PAX2 | The genetic diagnosis facilitated the successful use of PGD in the third child, preventing the transmission of the disorder. Additionally, the patient was sent for an ophthalmological evaluation to identify and address any potential visual problems. | Pre-implantation Genetic diagnosis Surveillance |
| 21–878 | PKHD1 | The genetic diagnosis facilitated the successful use of PGD in a following pregnancy. | Pre-implantation Genetic diagnosis |
BBS, Bardet-Biedl Syndrome; CAKUT, congenital anomalies of the kidney and urinary tract; Dup, duplication; ENT, ear, nose, and throat; MRI, magnetic resonance imaging; PGD, preimplantation genetic diagnosis.
In 5 cases, the genetic diagnosis did not correspond with the pre-exome clinical diagnosis of the primary nephrologist (participants no. 21–218, 21–182, 21–680, 21–253, and 21–112; Table 2). These included cases with an unknown diagnosis and cases where the distinction between CAKUT and renal cystic ciliopathies was challenging solely based on the phenotype and imaging. Consequently, ciliopathies were incorrectly classified as CAKUT and vice-versa.
Discussion
In the present nationwide exome sequencing study involving nearly all dialysis-treated Israeli children with kidney failure, we detected diagnostic variants in 36 of 79 participants (45%). The study cohort included children from different ethnicities who were all exclusively treated in 1 of 6 Israeli pediatric dialysis units across the country. These findings show a high prevalence of monogenic etiologies in childhood-onset kidney failure and emphasize the extent to which genetic testing can help resolve underlying diagnostic etiologies. This allows for gene-specific management and monitoring of the affected children with advanced CKD and their families.
Our study has several limitations that should be considered. First, we did not perform a chromosomal microarray-based copy-number variation analysis. This, to some extent, may affect our ability to detect hemizygous conditions or microdeletion syndromes and potentially, might have resulted in an underestimation of the overall genetic diagnostic yield. Nonetheless, using the exome data, we were still able to detect medium and large size gene deletions in 6 patients. Second, trio exome analysis was not available for all probands. This can affect, to some degree, the detection of novel de novo mutations. Specifically, it can result in a more conservative exome sequencing molecular diagnostic yield, because identifying de novo variants can increase the strength of evidence to classify variants as pathogenic. Ideally, a trio genome sequencing study would be most effective because it could uncover additional variants such as mitochondrial genome variants, copy number variants, structural variants, and single nucleotide variants in noncoding regions. Finally, our study was limited to Israeli children with different ethnic backgrounds and with relatively high rates of consanguinity; therefore, its generalizability may be limited.
The strength of our study includes its nationwide and comprehensive nature, which included a non-selected cohort reflecting “real world” clinical utility of exome sequencing of all dialysis-treated Israeli children.
The observed genetic detection of 45% is a relative high yield compared with that previously reported for single-center or selected pediatric CKD cohorts.1,15,16 However, this relatively high genetic diagnostic yield is comparable to other Israeli nationwide exome sequencing studies performed in children with neurodevelopmental disorders.17 Moreover, we found high percentages of monogenic cases in children even when there was no positive family history, consanguinity, or syndromic features (26%, 29%, and 32% respectively). This implies that a significant proportion of all childhood-onset CKD can be attributed to genetic causes, which are often overlooked given the lack of overt risk factors for an underlying genetic condition. Similarly, in a single-center experience of exome sequencing in 104 pediatric recipients of kidney transplantation, Mann et al. reported a genetic diagnostic yield of 32.7%.16
International comparisons demonstrate that Israel is one of the leading countries in childhood kidney failure prevalence mainly among non-Jewish minorities.18,19 We have previously hypothesized that this, in part, is secondary to widespread and underappreciated genetic CKD etiologies in Israeli minorities.18 The current study results support this hypothesis and highlight the need for targeted screening and prevention strategies for high-risk populations.
Genetic diagnoses enabled identification of potential prospective consequences for many patients and could have enabled earlier diagnosis in the course of their disease, which might have mitigated negative outcomes. Detection of monogenic causes of CKD enables personalized surveillance and treatments, spares unnecessary diagnostic procedures such as kidney biopsies, prevents inappropriate therapy use, and might prevent incompatible transplantation from affected relatives. In addition, it allows family genetic consulting and early diagnosis of asymptomatic family members. As a result, we and others have recently launched with the Israeli Ministry of Health, a national government supported pilot for publicly funded trio exome sequencing testing for all Israeli pediatric patients with kidney failure.
The significant prevalence of monogenic etiologies among children on dialysis seen here implies an even greater number of undiagnosed patients in the considerably more prevalent antecedent CKD stages.
Overall, our study suggests that exome sequencing should be a standard and routine diagnostic tool for all children with advanced CKD.
Disclosure
All the authors declared no competing interests.
Acknowledgments
We thank all the families and referring physicians who submitted samples for testing. This work was carried out in partial fulfillment of the PhD degree requirements for OS. This research was supported by the Israel Science Foundation (grant No. 2773/19). AV is supported by an European Research Council Starting Grant (StG ERC grant #101040267-GeneCKD). The funding organizations had no role in the design and conduct of the study, collection, management, analysis, and interpretation of the data, preparation, review, or approval of the manuscript, and decision to submit the manuscript for publication.
Footnotes
Figure S1. Age of kidney failure for each genetic etiology.
Table S1. Monogenic CKD gene panel.
STROBE Statement.
Supplementary Materials
Figure S1. Age of kidney failure for each genetic etiology.
Table S1. Monogenic CKD gene panel.
STROBE Statement.
References
- 1.Vivante A., Hildebrandt F. Exploring the genetic basis of early-onset chronic kidney disease. Nat Rev Nephrol. 2016;12:133–146. doi: 10.1038/nrneph.2015.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Connaughton D.M., Hildebrandt F. Personalized medicine in chronic kidney disease by detection of monogenic mutations. Nephrol Dial Transplant. 2020;35:390–397. doi: 10.1093/ndt/gfz028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Groopman E.E., Marasa M., Cameron-Christie S., et al. Diagnostic utility of exome sequencing for kidney disease. N Engl J Med. 2019;380:142–151. doi: 10.1056/NEJMoa1806891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McKenna A., Hanna M., Banks E., et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li M.X., Gui H.S., Kwan J.S.H., Bao S.Y., Sham P.C. A comprehensive framework for prioritizing variants in exome sequencing studies of Mendelian diseases. Nucleic Acids Res. 2012;40:e53. doi: 10.1093/nar/gkr1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Q., Pierce-Hoffman E., Cummings B.B., et al. Landscape of multi-nucleotide variants in 125,748 human exomes and 15,708 genomes. Nat Commun. 2020;11:2539. doi: 10.1038/s41467-019-12438-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nykamp K., Anderson M., Powers M., et al. Sherloc: a comprehensive refinement of the ACMG-AMP variant classification criteria. Genet Med. 2017;19:1105–1117. doi: 10.1038/gim.2017.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Einhorn Y., Einhorn M., Kamshov A., et al. Gene-specific artificial intelligence-based variant classification engine: results of a time-capsule experiment. Europe PMC. 2019 doi: 10.21203/rs.2.11834/v1. [DOI] [Google Scholar]
- 9.Lv N., Wang Y., Zhao M., Dong L., Wei H. The role of PAX2 in neurodevelopment and disease. Neuropsychiatr Dis Treat. 2021;17:3559–3567. doi: 10.2147/NDT.S332747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang X., Li Y., Fang Y., et al. Phenotypic spectrum and genetics of PAX2-related disorder in the Chinese cohort. BMC Med Genomics. 2021;14:250. doi: 10.1186/s12920-021-01102-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vivante A., Chacham O.S., Shril S., et al. Dominant PAX2 mutations may cause steroid-resistant nephrotic syndrome and FSGS in children. Pediatr Nephrol. 2019;34:1607–1613. doi: 10.1007/s00467-019-04256-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garrelfs S.F., Frishberg Y., Hulton S.A., et al. Lumasiran, an RNAi therapeutic for primary hyperoxaluria Type 1. N Engl J Med. 2021;384:1216–1226. doi: 10.1056/NEJMoa2021712. [DOI] [PubMed] [Google Scholar]
- 13.Groothoff J.W., Metry E., Deesker L., et al. Clinical practice recommendations for primary hyperoxaluria: an expert consensus statement from ERKNet and OxalEurope. Nat Rev Nephrol. 2023;19:194–211. doi: 10.1038/s41581-022-00661-1. [DOI] [PubMed] [Google Scholar]
- 14.Mandrile G., Beck B., Acquaviva C., et al. Genetic assessment in primary hyperoxaluria: why it matters. Pediatr Nephrol. 2023;38:625–634. doi: 10.1007/s00467-022-05613-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao M., Yu F., Dong R., et al. Diagnostic application of exome sequencing in Chinese children with suspected inherited kidney diseases. Front Genet. 2022;13 doi: 10.3389/fgene.2022.933636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mann N., Braun D.A., Amann K., et al. Whole-exome sequencing enables a precision medicine approach for kidney transplant recipients. J Am Soc Nephrol. 2019;30:201–215. doi: 10.1681/ASN.2018060575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pode-Shakked B., Barel O., Singer A., et al. A single center experience with publicly funded clinical exome sequencing for neurodevelopmental disorders or multiple congenital anomalies. Sci Rep. 2021;11 doi: 10.1038/s41598-021-98646-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Regev-Epstein L.C., Frishberg Y., Davidovits M., et al. Dialysis in Israeli children between 1990 and 2020: trends and international comparisons. Clin J Am Soc Nephrol. 2023;18:363–373. doi: 10.2215/CJN.0000000000000063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stel V.S., de Jong R.W., Kramer A., et al. Supplemented ERA-EDTA Registry data evaluated the frequency of dialysis, kidney transplantation, and comprehensive conservative management for patients with kidney failure in Europe. Kidney Int. 2021;100:182–195. doi: 10.1016/j.kint.2020.12.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.