Abstract
Antimicrobial susceptibility testing revealed among 150 clinical isolates of Streptococcus pneumoniae 4 pneumococcal isolates with resistance to fluoroquinolones (MIC of ciprofloxacin, ≥32 μg/ml; MIC of sparfloxacin, ≥16 μg/ml). Gene amplification and sequencing analysis of gyrA and parC revealed nucleotide changes leading to amino acid substitutions in both GyrA and ParC of all four fluoroquinolone-resistant isolates. In the case of strains 182 and 674 for which sparfloxacin MICs were 16 and 64 μg/ml, respectively, nucleotide changes were detected at codon 81 in gyrA and codon 79 in parC; these changes led to an Ser→Phe substitution in GyrA and an Ser→Phe substitution in ParC. Strains 354 and 252, for which sparfloxacin MICs were 128 μg/ml, revealed multiple mutations in both gyrA and parC. These strains exhibited nucleotide changes at codon 85 leading to a Glu→Lys substitution in GyrA, in addition to Ser-79→Tyr and Lys-137→Asn substitutions in ParC. Moreover, strain 252 showed additional nucleotide changes at codon 93, which led to a Trp→Arg substitution in GyrA. These results suggest that sparfloxacin resistance could be due to the multiple mutations in GyrA and ParC. However, it is possible that other yet unidentified mutations may also be involved in the high-level resistance to fluoroquinolones in S. pneumoniae.
Streptococcus pneumoniae is the major cause of respiratory tract infections and bacterial meningitis (9). For a long time penicillin was the most effective drug against such infections. However, penicillin-resistant S. pneumoniae was first reported in 1967 (4), followed by the reporting of multiple-drug-resistant pneumococci in the late 1970s in South Africa (5). The incidence of multiple-drug-resistant S. pneumoniae is currently increasing throughout the world (1, 18). These trends have made the selection of optimal antimicrobial therapy for the treatment of infections caused by this organism very difficult.
Sparfloxacin, which has become commercially available in recent years, exhibits improved antimicrobial activity against streptococci including S. pneumoniae (3). The therapeutic use of fluoroquinolones in clinical settings, however, has resulted in the emergence of fluoroquinolone resistance in S. pneumoniae. In fact, we have recently experienced the emergence of clinical isolates of S. pneumoniae with decreased susceptibilities to fluoroquinolones including sparfloxacin (16).
Previous in vitro studies showed that pneumococcal resistance to fluoroquinolones was due to alterations in DNA gyrase and topoisomerase IV (topo IV) (6, 8, 12, 14, 17), and these alterations are similar to the alterations in the DNA gyrase of Escherichia coli that have been shown to reduce the level of binding of fluoroquinolones to the enzyme-DNA complex (19). Mutations in DNA gyrase and topo IV are mainly due to amino acid substitutions in defined regions, that is, quinolone resistance-determining regions (QRDRs) (20), in GyrA subunits of DNA gyrase and ParC subunits of topo IV. However, the mutations in the gyrA and parC genes that result in fluoroquinolone resistance in S. pneumoniae in clinical settings are unknown. Therefore, it remains to be elucidated whether similar mutations are present in clinical isolates of S. pneumoniae with fluoroquinolone resistance.
In the present study, we examined the in vitro activities of fluoroquinolones against clinical isolates of S. pneumoniae, and we describe the mutations identified in the gyrA and parC genes of clinical isolates resistant to fluoroquinolones including sparfloxacin.
MATERIALS AND METHODS
Bacterial strains.
A total of 150 isolates of S. pneumoniae were investigated in the present study. The strains were isolated from various specimens submitted to the clinical laboratory of Ryukyu University Hospital between 1994 and 1996. The isolates were confirmed to be S. pneumoniae by colony morphology, optochin susceptibility, and bile solubility. Bacteria were grown on 5% sheep blood agar (Kyokuto Co., Tokyo, Japan) at 37°C in an atmosphere enriched with 5% CO2. A wild-type fluoroquinolone-susceptible clinical strain, S. pneumoniae 245, was used for sequencing analysis to compare its amino acid sequence with those of the other strains. E. coli DH5α (Clontech Laboratories, Inc., Palo Alto, Calif.) was used to subclone DNA inserts.
Antimicrobial agents.
The following antimicrobial agents, obtained as laboratory-grade powders from their respective manufacturers, were tested: ciprofloxacin (Bayer Yakuhin, Osaka, Japan) and sparfloxacin (Dainippon Pharmaceutical Co., Osaka, Japan).
Antimicrobial susceptibility testing.
Antimicrobial susceptibility was determined by the twofold broth microdilution method according to the guidelines of the National Committee for Clinical Laboratory Standards (10). Cation-adjusted Mueller-Hinton broth (Difco Laboratories, Detroit, Mich.) was supplemented with 3% lysed horse blood and 0.5% yeast extract (Difco). Microdilution trays (final volume, 100 μl per well) were inoculated with an automatic MIC-2000 inoculator (Dynatech Laboratories, Inc., Alexandria, Va.). Final inocula contained approximately 5 × 105 CFU/ml. The MIC of each drug was defined as the lowest concentration resulting in the complete inhibition of visible growth after 18 h of incubation.
Capsular serotyping.
Pneumococcal serotyping was performed by the capsular reaction test as described previously (2) by using the diagnostic pneumococcal antisera (Statens Seruminstitut, Copenhagen, Denmark).
Amplification of the QRDRs of gyrA and parC genes.
Mutations in the QRDRs of the gyrA and parC genes of fluoroquinolone-resistant strains were investigated by the PCR method. Chromosomal DNA was prepared as described previously (7) and was resuspended in distilled water for PCR experiments. The primer sequences used to amplify the gyrA QRDR were as follows: VGA3, 5′-CCGTCGCATTCTTTACG-3′ (gyrA positions 129 to 145), and VGA4, 5′-AGTTGCTCCATTAACCA-3′ (gyrA positions 494 to 510) (12). For the amplification of the parC QRDR, the following oligonucleotide primers were used: Pr-SPGRL3, 5′-ACAACCATGAACCCAGAAAACA-3′ (parE positions 1780 to 1800 of S. pneumoniae), and Pr-SPGRL10, 5′-ATCAAACGGTCATCATCACG-3′ (parC positions 1591 to 1610) (11). The resulting 2.3-kb PCR product encoded a region from residue 434 of ParE to residue 536 of ParC. All amplifications were performed in a 50-μl volume containing 20 pmol of each primer, 100 ng of template DNA, each deoxynucleoside triphosphate at a concentration of 200 μM, 2.5 mM MgCl2, and 2.5 U of Taq polymerase. PCR was performed in a GeneAmp 9600 thermal cycler (Perkin-Elmer Cetus, Norwalk, Conn.) for 30 cycles. The PCR conditions were 30 s at 94°C for denaturation, 1 min at 60°C for annealing, and 5 min at 72°C for extension.
DNA sequencing and analysis.
The 382-bp gyrA PCR products were subcloned into the vector pGEM-T (Promega Co., Madison, Wis.). In the case of the parC gene, the 2.3-kb PCR products, including the QRDR of parC, were digested with the restriction enzymes EcoRI and PstI and were subcloned into the vector pUC19, which had previously been digested with the same endonucleases. DNA fragments for sequencing were generated by digesting insert DNA, resulting in 732-bp EcoRI-PstI fragments encoding a region equivalent to residues 1 to 190 of ParC. Plasmid vectors were introduced into E. coli DH5α. DNA sequences were determined by using the Cy5 Autoread Sequencing Kit and an ALF DNA Sequencer (Pharmacia Biotech, Piscataway, N.J.) according to the instructions provided by the manufacturer. A combination of the M13 universal primer and internal primers that anneal to vector DNA flanking the multicloning site was used to obtain the complete sequence information for both strands. DNA sequences were analyzed by using Genetyx system (Software Development Co., Tokyo, Japan).
RESULTS
Susceptibility test.
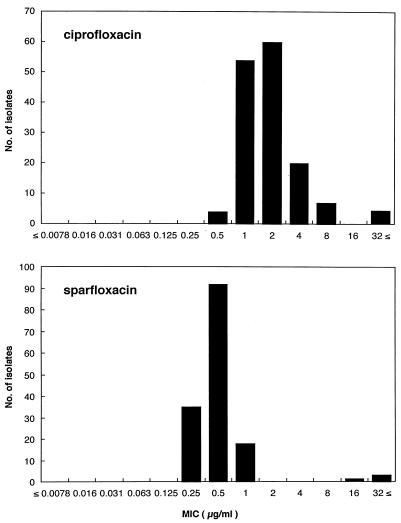
The MICs for all 150 clinical isolates of S. pneumoniae ranged from 0.5 to ≥32 μg/ml for ciprofloxacin and from 0.25 to ≥32 μg/ml for sparfloxacin (Fig. 1). The MIC at which 90% of strains are inhibited was 4.0 μg/ml for ciprofloxacin and 1.0 μg/ml for sparfloxacin. For four isolates ciprofloxacin MICs were 64 μg/ml and sparfloxacin MICs were ≥16 μg/ml; for two of the four isolates sparfloxacin MICs were 128 μg/ml.
FIG. 1.
Distribution of ciprofloxacin and sparfloxacin MICs for 150 clinical isolates of S. pneumoniae. MICs were determined by the twofold broth microdilution method according to the guidelines of the National Committee for Clinical Laboratory Standards (10).
Fluoroquinolone-resistant clinical isolates.
On the basis of the distributions of the MICs of ciprofloxacin and sparfloxacin, four isolates for which the ciprofloxacin MICs were ≥32 μg/ml and the sparfloxacin MICs were ≥16 μg/ml were used to classify the fluoroquinolone resistance. The drug susceptibility profiles of these strains are presented in Table 1.
TABLE 1.
Profiles of clinical isolates of sparfloxacinresistant S. pneumoniaea
| Strain | Relevant characteristics | MIC (μg/ml)b
|
||
|---|---|---|---|---|
| PCG | CPFX | SPFX | ||
| 182 | Isolate from sputum of a 76-yr-old male patient treated with sparfloxacin and levofloxacinc | 1.0 | 64 | 16 |
| 674 | Isolate from sputum of a 74-yr-old male patient during CS-940 treatmentd | 0.25 | 64 | 64 |
| 354 | Isolate from sputum of a 74-yr-old male patient after CS-940 treatmentd | 0.25 | 64 | 128 |
| 252 | Isolate from blood of a 33-yr-old male patient treated with ampicillin, imipenem-cilastatin, clindamycin, and vancomycin | 0.25 | 64 | 128 |
All strains were of serotype 23.
PCG, penicillin G; CPFX, ciprofloxacin; SPFX, sparfloxacin.
MIC of levofloxacin, 16 μg/ml.
CS-940 is a newly developed fluoroquinolone agent (MIC, 8 μg/ml).
Strain 182 was isolated from the sputum of a male patient during levofloxacin (the first 3 days) and sparfloxacin (the following 3 weeks) treatment for pneumococcal pneumonia. Strains 674 and 354 were sequential isolates obtained from the same patient treated with CS-940, a newly developed fluoroquinolone agent in Japan, for an acute exacerbation of chronic bronchitis; strain 674 was isolated during CS-940 treatment, and strain 354 was isolated 7 days after the end of chemotherapy. Strain 252 was cultured from a blood sample from a patient during treatment with vancomycin for infective endocarditis caused by Staphylococcus aureus. Prior to vancomycin treatment, this patient had been treated with ampicillin, imipenem-cilastatin, and clindamycin. No fluoroquinolone agent was used for this patient after he had visited a clinic. Clinical cure was obtained by vancomycin treatment.
Nucleotide sequence of PCR products encompassing the QRDRs of gyrA and parC genes from fluoroquinolone-susceptible S. pneumoniae 245.
A 382-bp fragment of the gyrA gene spanning amino acids 46 to 172 of GyrA was obtained by PCR and was partially sequenced (Fig. 2). PCR was also used to amplify a segment of the topo IV gene encompassing the QRDR of ParC. A 732-bp EcoRI-PstI fragment of the parC gene was obtained by digesting the PCR product. The partial sequence of the parC gene and the deduced amino acid sequence of ParC (residues 44 to 163) are shown in Fig. 3. These amino acid sequences of GyrA and ParC including the QRDRs showed identity with those of previously reported GyrA and ParC proteins of S. pneumoniae 7785 (12, 13).
FIG. 2.
DNA sequence of the PCR product encompassing the QRDR in the gyrA gene from wild-type fluoroquinolone-susceptible clinical isolate S. pneumoniae 245. Letters under the nucleotide sequence indicate the deduced protein sequence. The numbering of the GyrA sequence for S. pneumoniae was taken from Pan et al. (12).
FIG. 3.
DNA sequence of the PCR product encompassing the QRDR in the parC gene from wild-type fluoroquinolone-susceptible clinical isolate S. pneumoniae 245. Letters under the nucleotide sequence indicate the deduced protein sequence. The numbering of ParC sequence for S. pneumoniae was taken from Pan and Fisher (13).
Detection of mutations in QRDRs of gyrA and parC genes in fluoroquinolone-resistant clinical isolates of S. pneumoniae.
Sequencing of the region encoding the QRDRs of GyrA and ParC was carried out to investigate the involvement of gene mutations in fluoroquinolone-resistant clinical isolates. The mutations in gyrA and parC of four fluoroquinolone-resistant isolates are summarized in Table 2. These results of sequencing analysis were reproducible.
TABLE 2.
Mutations identified in the GyrA and ParC containing QRDRs in sparfloxacin-resistant clinical isolates of S. pneumoniae
| Strain | MIC (μg/ml)
|
Amino acid (nucleotide) at the indicated position:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| CPFXa | SPFXb | GyrAc
|
ParCd
|
||||||
| 75 | 81 | 83 | 85 | 93 | 79 | 137 | |||
| 245 | 0.5 | 0.25 | Tyr (TAT) | Ser (TCC) | Ile (ATT) | Glu (GAA) | Trp (TGG) | Ser (TCT) | Lys (AAG) |
| 182 | 64 | 16 | —e | Phe (TTC) | — | — | — | Phe (TTT) | — |
| 674 | 64 | 64 | — | Phe (TTC) | — | — | — | Phe (TTT) | — |
| 354 | 64 | 128 | Tyr (TAC) | — | — | Lys (AAA) | — | Tyr (TAT) | Asn (AAT) |
| 252 | 64 | 128 | Tyr (TAC) | — | Ile (ATC) | Lys (AAA) | Arg (CGG) | Tyr (TAT) | Asn (AAT) |
Strain 182, for which the sparfloxacin MIC was 16 μg/ml, exhibited nucleotide changes at codon 81 in gyrA and codon 79 in parC leading to a Ser (TCC)→Phe (TTC) substitution in GyrA and a Ser (TCT)→Phe (TTT) substitution in ParC, respectively (the underscored bases are the substitutions). Strain 674, for which the sparfloxacin MIC was 64 μg/ml, showed mutations in gyrA and parC genes identical to those in strain 182.
Strains 354 and 252, for which the sparfloxacin MICs were 128 μg/ml, revealed noticeable mutations in both gyrA and parC genes. These strains exhibited nucleotide changes at codon 75 (TAT→TAC) and codon 85, leading to a Glu→Lys (GAA→AAA) substitution in GyrA, in addition to Ser-79→Tyr (TCT→TAT) and Lys-137→Asn (AAG→AAT) substitutions in ParC. Moreover, strain 252 showed additional nucleotide changes at codon 83 (ATT→ATC) and codon 93, leading to a Trp→Arg (TGG→CGG) substitution in GyrA.
DISCUSSION
In the present study, we have characterized the mutations in the gyrA and parC genes of four sparfloxacin-resistant clinical isolates of S. pneumoniae. In the case of two strains for which the sparfloxacin MICs were 16 and 64 μg/ml, respectively, the gene mutations that were identified are similar to those detected in fluoroquinolone-resistant S. pneumoniae obtained by in vitro stepwise selection (6, 8, 12, 14). Nucleotide changes leading to a Ser-81→Phe substitution in GyrA and a Ser-79→Phe substitution in ParC of S. pneumoniae also appear to be encouraged by changes in GyrA and ParC associated with the acquisition of resistance to fluoroquinolones in clinical isolates.
We were greatly concerned with the implication that mutational alterations in GyrA and ParC may generate further resistance to sparfloxacin (MIC, 128 μg/ml) in strains 354 and 252. The gene mutations detected in these strains were characteristic in some respects. As indicated in Table 2, sequencing analysis demonstrated multiple gene mutations in these strains. To our knowledge, these multiple mutational alterations of GyrA and ParC in S. pneumoniae have never been described previously. Of particular interest is that strain 252 exhibited double amino acid substitutions in GyrA (Glu-85→Lys and Trp-93→Arg) and ParC (Ser-79→Tyr and Lys-137→Asn) and that these multiple mutations are associated with the acquisition of further resistance to sparfloxacin.
Studies by Pan and Fisher (14) indicate that GyrA is the primary target of sparfloxacin and that the amino acid substitution at position Ser-81 or Glu-85 in GyrA is responsible for the resistance to sparfloxacin in vitro. The gyrA nucleotide sequence of strain 252 was identical to that of strain 354 except that it had additional mutations at codon 83 (ATT→ATC; no amino acid change) and codon 93 leading to a Trp→Arg substitution in GyrA. The contribution of the Trp-93→Arg change in GyrA to the in vitro fluoroquinolone resistance has not been ascertained, whereas the Gly-85→Lys change in GyrA has been shown to confer resistance to fluoroquinolones (6, 12, 14). Considering our finding that there were no differences in the sparfloxacin MICs for strains 354 and 252, it is suggested that the mutations at codon 83 and 93 in GyrA would not confer the changes in the MICs of sparfloxacin and that the mutation at codon 85 could be a cause of the increase in the MICs of sparfloxacin.
In regard to the mutations in parC, Tankovic et al. (17) mentioned the important point that S. pneumoniae acquires a clinical level of resistance to sparfloxacin after the occurrence of two mutations in GyrA and ParC. Strains 354 and 252 possessed double amino acid substitutions (Ser-79→Tyr and Lys-137→Asn) in ParC, whereas strains 182 and 674 showed only Ser-79→Phe substitutions. Previous studies by Muñoz and De La Campa (8) showed that the Lys-137→Asn amino acid change in ParC protein is not involved in ciprofloxacin resistance in vitro, although ParC is the primary target of ciprofloxacin in S. pneumoniae. Our observations are consistent with that conception. A more likely explanation is that the Lys-137→Asn substitution in ParC would confer the resistance to sparfloxacin, not to ciprofloxacin, and the difference in the deduced amino acid would be due to nucleotide changes at codon 79.
The results described here suggest that sparfloxacin-resistance could be mediated by the multiple mutations in GyrA and ParC. However, we consider that further investigation of the inhibitory activities of fluoroquinolones against altered DNA gyrase or topo IV with multiple amino acid changes, as indicated in the presented study, in addition to the three-dimensional structural features of DNA gyrase and topo IV with altered subunits, is needed.
Of additional interest is the fact that strain 674 contains the mutations in gyrA and parC identical to those in strain 182, whereas the respective MICs of sparfloxacin are different, which indicates that the presence of additional undetected mutations gives rise to resistance to sparfloxacin in strain 674. Recent studies by Perichon et al. (15) showed that a mutation in ParE, the alternative component of topo IV, is also responsible for fluoroquinolone resistance in S. pneumoniae. In addition, Zeller et al. (21) recently reported that an efflux mechanism may contribute to fluoroquinolone resistance in S. pneumoniae.
Finally, whether the high-level resistance to sparfloxacin is associated with mutational alterations in only gyrA or parC is still unclear. Thus, it is possible that other yet unidentified mutations in other portions of the genes encoding subunits of DNA gyrase, topo IV, etc., may also be involved in the high-level resistance to fluoroquinolones in S. pneumoniae.
ACKNOWLEDGMENTS
We are grateful to Y. Onodera, K. Sato, and I. Hayakawa (Daiichi Pharmaceutical Co., Ltd.) for helpful advice. We also thank the staff of the Division of Bacteriology, Clinical Laboratory, at Ryukyu University Hospital.
REFERENCES
- 1.Appelbaum P C. Antimicrobial resistance in Streptococcus pneumoniae: an overview. Clin Infect Dis. 1992;15:77–83. doi: 10.1093/clinids/15.1.77. [DOI] [PubMed] [Google Scholar]
- 2.Austrian R. Quellung reaction, a neglected microbiologic technique. Mount Sinai J Med. 1976;43:699–709. [PubMed] [Google Scholar]
- 3.Baquero, F., and R. Canton. 1996. In-vitro activity of sparfloxacin in comparison with currently available antimicrobials against respiratory tract pathogens. J. Antimicrob. Chemother. 37(Suppl. A):1–18. [DOI] [PubMed]
- 4.Hansman D, Bullen M M. A resistant pneumococcus. Lancet. 1967;ii:264–265. doi: 10.1016/s0140-6736(75)91547-0. [DOI] [PubMed] [Google Scholar]
- 5.Jacobs M R, Koornhof H J, Robins-Browne R M, Stevenson C M, Vermaak Z A, Freiman I, Miller G B, Witcomb M A, Isaäcson M, Ward J I, Austrian R. Emergence of multiply resistant pneumococci. N Engl J Med. 1978;299:735–740. doi: 10.1056/NEJM197810052991402. [DOI] [PubMed] [Google Scholar]
- 6.Janoir C, Zeller V, Kitzis M, Moreau N, Gutmann L. High-level fluoroquinolone resistance in Streptococcus pneumoniae requires mutations in parC and gyrA. Antimicrob Agents Chemother. 1996;40:2760–2764. doi: 10.1128/aac.40.12.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laible G, Hakenbeck R, Sicard M A, Joris B, Ghuysen J M. Nucleotide sequences of the pbpX genes encoding the penicillin-binding proteins 2x from Streptococcus pneumoniae R6 and a cefotaxime-resistant mutant, C506. Mol Microbiol. 1989;3:1337–1348. doi: 10.1111/j.1365-2958.1989.tb00115.x. [DOI] [PubMed] [Google Scholar]
- 8.Muñoz R, De La Campa A G. ParC subunit of DNA topoisomerase IV of Streptococcus pneumoniae is a primary target of fluoroquinolones and cooperates with DNA gyrase A subunit in forming resistance phenotype. Antimicrob Agents Chemother. 1996;40:2252–2257. doi: 10.1128/aac.40.10.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Musher D M. Streptococcus pneumoniae. In: Mandell G L, Bennett J E, Dolin R, editors. Principles and practice of infectious diseases. 4th ed. New York, N.Y: Churchill Livingstone; 1995. pp. 1811–1826. [Google Scholar]
- 10.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A4. 4th ed. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 11.Onodera Y, Tanaka M, Sato K, Hayakawa I. Program and abstracts of the 36th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1996. Molecular cloning of Streptococcus pneumoniae DNA topoisomerase IV, abstr. C4; p. 34. [Google Scholar]
- 12.Pan X S, Ambler J, Mehtar S, Fisher L M. Involvement of topoisomerase IV and DNA gyrase as ciprofloxacin targets in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1996;40:2321–2326. doi: 10.1128/aac.40.10.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan X S, Fisher L M. Cloning and characterization of the parC and parE genes of Streptococcus pneumoniae encoding DNA topoisomerase IV: role in fluoroquinolone resistance. J Bacteriol. 1996;178:4060–4069. doi: 10.1128/jb.178.14.4060-4069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan X S, Fisher L M. Targeting of DNA gyrase in Streptococcus pneumoniae by sparfloxacin: selective targeting of gyrase or topoisomerase IV by quinolones. Antimicrob Agents Chemother. 1997;41:471–474. doi: 10.1128/aac.41.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perichon B, Tankovic J, Courvalin P. Characterization of a mutation in the parE gene that confers fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:1166–1167. doi: 10.1128/aac.41.5.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taba H, Shinzato T, Higa F, Tateyama M, Kusano N, Saito A. Program and abstracts of the 37th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1997. In vitro antimicrobial activities of new fluoroquinolones against clinical isolates of Streptococcus pneumoniae, abstr. E60; p. 124. [Google Scholar]
- 17.Tankovic J, Perichon B, Duval J, Courvalin P. Contribution of mutations in gyrA and parC genes to fluoroquinolone resistance of mutants of Streptococcus pneumoniae obtained in vivo and in vitro. Antimicrob Agents Chemother. 1996;40:2505–2510. doi: 10.1128/aac.40.11.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomasz, A. 1997. Antibiotic resistance in Streptococcus pneumoniae. Clin. Infect. Dis. 24(Suppl. 1):S85–S88. [DOI] [PubMed]
- 19.Willmott C J, Maxwell A. A single point mutation in the DNA gyrase A protein greatly reduces binding of fluoroquinolones to the gyrase-DNA complex. Antimicrob Agents Chemother. 1993;37:126–127. doi: 10.1128/aac.37.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshida H, Bogaki M, Nakamura M, Nakamura S. Quinolone resistance determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob Agents Chemother. 1990;34:1271. doi: 10.1128/aac.34.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeller V, Janoir C, Kitzis M D, Gutmann L, Moreau N J. Active efflux as a mechanism of resistance to ciprofloxacin in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:1973–1978. doi: 10.1128/aac.41.9.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]