Abstract
In this overview, we describe important contributions from the Antibacterial Resistance Leadership Group (ARLG) to patient care, clinical trials design, and mentorship while outlining future priorities. The ARLG research agenda is focused on 3 key areas: gram-positive infections, gram-negative infections, and diagnostics. The ARLG has developed an innovative approach to clinical trials design, the desirability of outcome ranking (DOOR), which uses an ordinal measure of global outcome to assess both benefits and harms. DOOR was initially applied to observational studies to determine optimal dosing of vancomycin for methicillin-resistant Staphylcococcus aureus bacteremia and the efficacy of ceftazidime-avibactam versus colistin for the treatment of carbapenem-resistant Enterobacterales infection. DOOR is being successfully applied to the analysis of interventional trials and, in collaboration with the US Food and Drug Administration (FDA), for use in registrational trials. In the area of diagnostics, the ARLG developed Master Protocol for Evaluating Multiple Infection Diagnostics (MASTERMIND), an innovative design that allows simultaneous testing of multiple diagnostic platforms in a single study. This approach will be used to compare molecular assays for the identification of fluoroquinolone-resistant Neisseria gonorrhoeae (MASTER GC) and to compare rapid diagnostic tests for bloodstream infections. The ARLG has initiated a first-in-kind randomized, double-blind, placebo-controlled trial in participants with cystic fibrosis who are chronically colonized with Pseudomonas aeruginosa to assess the pharmacokinetics and antimicrobial activity of bacteriophage therapy. Finally, an engaged and highly trained workforce is critical for continued and future success against antimicrobial drug resistance. Thus, the ARLG has developed a robust mentoring program targeted to each stage of research training to attract and retain investigators in the field of antimicrobial resistance research.
Keywords: antibacterial resistance, antibacterial agents, bacterial infections, diagnostics, clinical trials
The Antibacterial Resistance Leadership Group conducts innovative clinical research in 3 key areas, gram-positive infections, gram-negative infections, and diagnostics, to inform clinical practice to improve the management and lessen the impact of drug-resistant infections.
Over the past decade, antibacterial resistance (AR) has posed an increasing threat to human health. New varieties of drug-resistant bacteria have emerged and expanded in both hospital and community settings. The coronavirus disease 2019 (COVID-19) pandemic has accelerated these trends. Despite the mounting need, prospects for both new antibiotics and the investigators needed to test them have dwindled as the pharmaceutical industry pursues a mass exodus from antibiotic development.
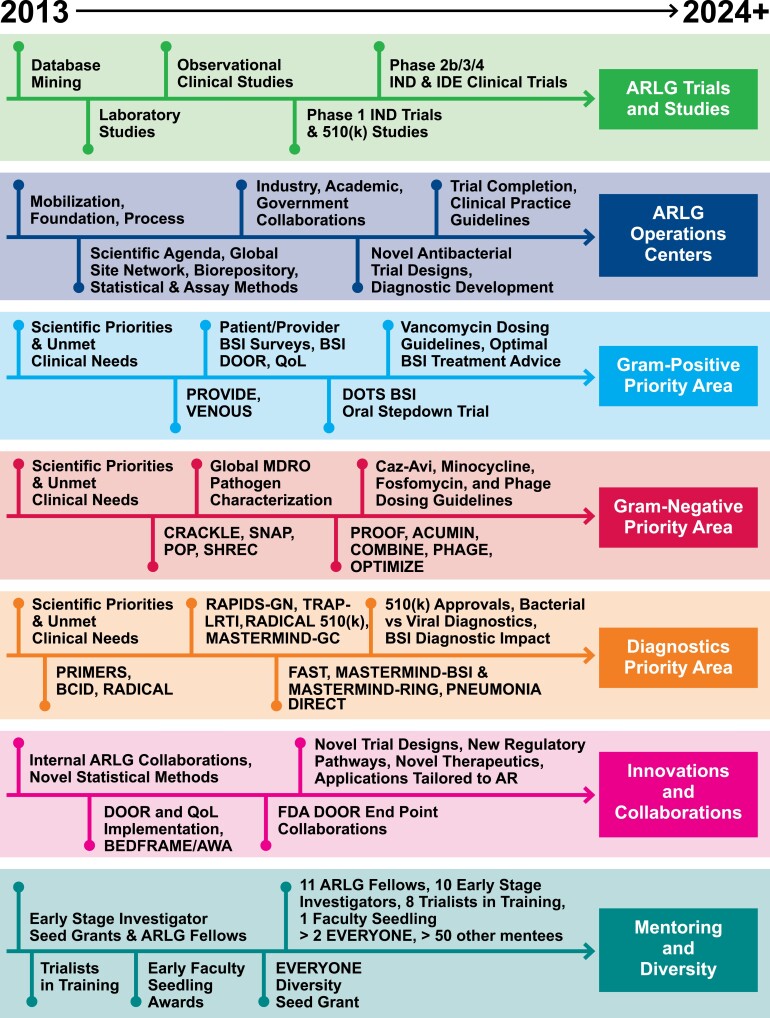
These developments underscore the vital, ongoing need for the Antibacterial Resistance Leadership Group (ARLG). Since its inception in 2013, the ARLG has been tasked by the National Institute of Allergy and Infectious Diseases to support a responsive, adaptable clinical research network on AR and to conduct clinical research that would not otherwise be done. A summary of ARLG priorities and key advancements is provided in Table 1, and a representation of the evolution of ARLG since inception is provided in Figure 1. The purpose of this overview is to make clinicians and investigators aware of some of the ARLG’s important contributions to patient care, clinical trials design, and mentorship during its first decade and to outline programmatic priorities for the future.
Table 1.
Antibacterial Resistance Leadership Group Priority Studies and Key Advancements
| Study | Description | Status | Scientific Area |
|---|---|---|---|
| Optimize administration and repurpose existing antibacterial agents | |||
| ACUMIN | Phase 4 open-label PK study of tetracycline antibiotic for injection following a single infusion in critically ill adults | Published [1] | GN, PK |
| COMBINE HFIM | Hollow fiber infection model studies to determine the optimal dosing of the combination of ceftazidime-avibactam with aztreonam against NDM-1–producing Enterobacterales | Published [2] | GN, PK |
| COMBINE | Phase 1 open-label study in healthy adults to evaluate the safety and PK of ceftazidime-avibactam in combination with aztreonam | Published [3, 4] | GN, PK |
| PROOF | Phase 1 study to evaluate PK, safety, and tolerability of 2 dosing regimens of oral fosfomycin tromethamine in healthy adults | Published [5] | GN, PK |
| FOCUS | Phase 4 randomized, double-blinded trial to evaluate the efficacy of oral fosfomycin versus levofloxacin in complicated urinary tract infections | Manuscript submitted | GN |
| PROVIDE | Prospective observational evaluation of the association between the day 2 vancomycin exposure and failure rates among adult hospitalized patients with methicillin-resistant Staphylococcus aureus bloodstream infections | Published [6] | GP, PK |
| OPTIMIZE-GNI | Phase 4 prospective, multicenter, interventional PK study to develop individualized meropenem and cefepime dosing algorithms for critically ill patients with suspected or documented antimicrobial-resistant gram-negative infections and varying degrees of renal function based on observed cystatin C serum concentrations | Study design | GN, PK |
| Evaluate the effectiveness of antibacterial agents alone or in combination | |||
| DOTS | Multicenter, randomized, open-label, assessor-blinded, superiority trial to compare dalbavancin to the standard-of-care treatment for the completion of therapy in participants who have complicated S. aureus bacteremia or right-sided native valve infective endocarditis and who have cleared their baseline bacteremia | Enrollment complete [7] | GP |
| Colistin versus Ceftazidime-Avibactam | Prospective, multicenter, observational study to compare the efficacy and safety of colistin to ceftazidime-avibactam for the treatment of infections due to carbapenem-resistant Enterobacterales | Published [8] | GN |
| Identify and evaluate novel antiinfective agents and treatment strategies for bacterial infections | |||
| PHAGE | Phase 1b/2, multicenter, randomized, double-blind, placebo-controlled trial of the safety and microbiological activity of a single dose of bacteriophage therapy in cystic fibrosis patients colonized with Pseudomonas aeruginosa | Enrolling [9] | GN |
| PHAT | Isolation and characterization of lytic phages that target multidrug-resistant bacteria to generate species-specific panels of phages | Published [10, 11] | GN |
| SCOUT-CAP | Phase 4 double-blind, placebo-controlled, randomized trial to evaluate short-course versus standard-course outpatient therapy of community-acquired pneumonia and microbiome analysis | Published [12, 13] | GP |
| MeChaTeBla | Mechanistic and structural characterization of the interaction of a novel antibiotic with clinically relevant beta-lactamases | Data analysis | GN |
| ARGONAUT | Series of studies for the characterization of the in vitro activity of novel agents against genetically defined clinical isolates of carbapenem-resistant gram-negative bacteria | Data analysis | GN |
| STEP FMT | Study to determine strain temporal engraftment and persistence after FMT by identifying the specific taxa that may confer long-term multidrug-resistant colonization resistance among FMT-treated individuals in a clinical trial of renal transplant recipients undergoing FMT | Data analysis | GN, GP |
| TRAP-LRTI | Multicenter, randomized, placebo-controlled, noninferiority study of azithromycin treatment versus placebo in adults with suspect lower respiratory tract infection and a procalcitonin level of <0.1 ng/mL | Published [14] | GP, Dx |
| Global clinical and molecular epidemiology of MDROs | |||
| CRACKLE | Multicenter, prospective cohort study to define the clinical and molecular epidemiology of carbapenem-resistant Enterobacterales in the United States | Published [15] | GN |
| CRACKLE-2 | International prospective cohort study to define the global clinical and molecular epidemiology of carbapenem-resistant Enterobacterales | Published [16, 17] | GN |
| POP | International prospective cohort study to define the global clinical and molecular epidemiology of carbapenem-resistant P. aeruginosa | Published [18] | GN |
| SNAP | International prospective cohort study to define the global clinical and molecular epidemiology of carbapenem-resistant Acinetobacter baumannii | Manuscript submitted | GN |
| SHREC | Multicenter prospective cohort study that uses the MDRO network to study the clinical and molecular epidemiology of ceftriaxone-resistant Escherichia coli in the United States | Published [19] | GN |
| Identify and evaluate novel diagnostic strategies | |||
| RADICAL | Series of studies to develop and evaluate a host-based assay to distinguish among bacterial, viral, and non-infectious etiologies of acute respiratory tract infections, which could help reduce inappropriate antimicrobial use | Published [20–22] | Dx |
| RADICAL-3 | Prospective, randomized, interventional, pilot study to estimate the clinical utility of the host response-bacterial/viral test run on a novel platform in emergency department patients with acute, febrile respiratory infection | Study design | Dx |
| MASTER RADICAL | MASTER protocol to advance the validation of platforms measuring the RADICAL test, including an evaluation of the impact of specimen type and specimen storage conditions | Data analysis | Dx |
| RADICAL 510(k) | Prospective, multicenter study to assess the performance of host gene expression signature in blood specimens to discriminate between bacterial and viral infections | Study design | Dx |
| MASTERMIND-GC | Cross-sectional study to evaluate the diagnostic accuracy of multiple commercially available NAATs for detection of Neisseria gonorrhoeae and Chlamydia trachomatis from oropharyngeal and rectal sites | Published [23] | Dx |
| MASTERMIND-BSI | MASTER protocol for evaluating multiple rapid diagnostics for identification of bacteria directly from blood samples | Start-up | Dx |
| MASTERMIND-RING | MASTER protocol for evaluating multiple diagnostics for direct-from-specimen NAATs for detection of ciprofloxacin resistance in N. gonorrhoeae | Protocol development | Dx |
| BCID | Prospective, randomized, controlled trial to evaluate the clinical and economic impact of rapid identification and susceptibility testing of pathogens growing in blood culture bottles | Published [24] | Dx |
| RAPIDS-GN | Multicenter, prospective, randomized, controlled trial to evaluate standard culture and AST versus rapid identification and AST for patients with confirmed gram-negative bacteremia | Published [25] | Dx, GN |
| FAST | International, prospective, randomized trial to evaluate clinical outcomes among patients with gram-negative bacteremia who have blood culture evaluation using standard methods versus rapid antibiotic susceptibility testing | Start-up | Dx, GN |
| DIFFR | Study to optimize a novel molecular diagnostic to detect Clostridioides difficile toxin mRNA expression in stool, determine its analytical performance characteristics, and evaluate the clinical sensitivity and specificity relative to gold-standard methods | Data analysis | Dx, GP |
| PNEUMONIA DIRECT PILOT | Prospective, observational, diagnostic feasibility study to determine the accuracy of pathogen- and host-directed testing for the diagnosis of ventilator-associated pneumonia | Protocol development | Dx, GN, GP |
| GENO-STELLAR | Web-based genomic-epidemiological tool for antimicrobial resistance prediction | Platform launched [26] | Dx, GN |
| REPORT-ABC | Study evaluating laboratory reporting practices of rapid testing performed on positive blood cultures | Published [27] | Dx |
| PST | Comparative evaluation of 2 assays for PST to assess reproducibility of PST methods needed to support clinical trials of phage therapy | Manuscript submitted | Dx, GN |
| Create innovations in clinical trial design [28] | |||
| DOOR | Desirability of outcome ranking: patient-centric benefit-risk evaluation paradigm for the design, analysis, and interpretation of clinical trials of bacterial infection syndromes | … | … |
| QOL | Quality of life: incorporate the patient perspective as an outcome in clinical trials of bacterial infection syndromes | … | … |
| DOOR MAT | Desirability of outcome ranking for the management of antimicrobial therapy: framework for assessing antibiotic selection strategies in the presence of drug resistance | … | … |
| SMART-COMPASS | Sequential, multiple-assignment, randomized trials for comparing personalized antibiotic strategies: a trial design to compare strategies | … | … |
| MASTERMIND | MASTER protocol for simultaneously evaluating multiple infection diagnostics with a single patient’s sample(s), providing efficiencies of specimen collection and characterization | … | … |
| BED-FRAME/AWA | Benefit-risk evaluation of diagnostics: a FRAMEwork and average weighted accuracy: systematic and pragmatic approach to evaluate and compare diagnostic alternatives to aid in clinical decision-making | … | … |
| Clinically adjudicated reference standards | Approach to use clinically adjudicated reference standards for evaluation of diagnostic test accuracy when a predefined gold standard is lacking or the test’s accuracy potentially exceeds that of its predecessors [29] | … | … |
Abbreviations: AST, antimicrobial susceptibility testing; Dx, diagnostics; BSI, bloodstream infection; FMT, fecal microbiota transplantation; GN, gram-negatives; GP, gram-positives; MDRO, multidrug-resistant organism; NAAT, nucleic acid amplification test; NDM, New Delhi metallo-β-lactamase; PK, pharmacokinetic(s); PST, phage susceptibility testing.
Figure 1.
Evolution of the ARLG since inception. See Table 1 and text for names and brief descriptions of trials. Abbreviations: AR, antibacterial resistance; ARLG, Antibacterial Resistance Leadership Group; caz-avi, ceftazidime-avibactam; FDA, US Food and Drug Administration; GC, Gonorrhea-Chlamydia; IND, investigational new drug application; IDE, investigational device exemption; MDRO, multidrug-resistant organism; PRIMERS, platforms for rapid identification of MDR-gram-negative bacteria and evaluation of resistance studies; VENOUS, vancomycin-resistant enterococcal BSI outcomes study.
Innovation and Evolution: Improving Patient Care
The ARLG has systematically conducted a series of studies in its 3 scientific priority areas of gram-positive infections, gram-negative infections, and diagnostics. Each study builds on prior results and informs studies to follow. The ARLG has focused significant efforts on the treatment of methicillin-resistant Staphylococcus aureus (MRSA) infection, the most common type of drug-resistant bacterial infection in the United States [30] and a leading cause of mortality attributable to AR globally [31]. Because vancomycin is the most widely used drug to treat serious MRSA infections, the ARLG undertook the Prospective Observational Evaluation of the Association Between Initial Vancomycin Exposure and Failure Rates Among Adult Hospitalized Patients With Methicillin-Resistant Staphylococcus aureus Bloodstream Infections (PROVIDE) study [6] to examine the relationship between vancomycin dosing and patient outcomes. PROVIDE relied on the desirability of outcome ranking (DOOR), a landmark statistical methodology developed by the ARLG to assess patient outcomes. DOOR uses an ordinal ranking that combines both safety and efficacy to measure the totality of the patient experience for one treatment versus another. DOOR is particularly well suited for determining optimal drug exposure (ie, the exposure that maximizes efficacy and safety). PROVIDE showed that higher vancomycin exposure, measured as area under the drug concentration-time curve (AUC), did not improve efficacy and was associated with a higher rate of acute kidney injury. In contrast, an optimal drug exposure of 24-hour AUC ≤515 mg/L × hour was associated with lower rates of both treatment failure and acute kidney injury. These results directly led to current treatment guidelines for vancomycin dosing for MRSA [32]. The ARLG undertook the Dalbavancin as an Option for Treatment of S. aureus bacteremia (DOTS) trial, a phase 2b superiority randomized, controlled trial (RCT) comparing 2 doses of dalbavancin a week apart with standard intravenous therapy for the treatment of complicated S. aureus bacteremia [7]. DOTS demonstrates the application of DOOR in an interventional study [7]. If successful, DOTS will change treatment for MRSA and methicillin-susceptible S. aureus bacteremia and illustrate the advantages of DOOR in clinical trial design. Enrollment was completed in July 2023.
The ARLG has also focused on critical unmet needs posed by multidrug-resistant (MDR) gram-negative bacteria by establishing the Multidrug Resistant Organism (MDRO) Network. Through the MDRO Network, the ARLG characterized the clinical and molecular epidemiology of the following critical priority pathogens identified by the World Health Organization (WHO): carbapenem-resistant Enterobacterales (CRE), carbapenem-resistant Pseudomonas aeruginosa, and carbapenem-resistant Acinetobacter baumannii complex.
The ARLG established the Consortium on Resistance Against Carbapenems in Klebsiella and Other Enterobacteriaceae (CRACKLE) to characterize the clinical and molecular epidemiology of CRE in the United States and globally (CRACKLE-). In these studies, outcome data were collected for patients treated with colistin, considered at the time to be the first-line agent, or ceftazidime-avibactam, of promising but unproven efficacy for CRE. DOOR was used to perform efficacy, safety, and risk–benefit analyses for colistin compared with ceftazidime-avibactam. The differences were striking, with a more than 3-fold reduction in 30-day mortality (32% for colistin, 9% for ceftazidime-avibactam) and a 64% probability of a better overall outcome with ceftazidime-avibactam [8]. Since its publication in 2018, this article has been cited more than 500 times and has served as the basis for treatment guidelines to use ceftazidime-avibactam instead of colistin for CRE [33].
Unfortunately, ceftazidime-avibactam is not an option for New Delhi metallo-β-lactamase (NDM)-producing Enterobacterales, high rates of which were identified in the CRACKLE studies. This reinforced the need for effective therapies for infections caused by these pathogens. Building on initial reports from ARLG investigators suggesting that coadministration of aztreonam and ceftazidime-avibactam could represent a treatment option for NDM-producing Enterobacterales infections [34], the ARLG initially undertook hollow fiber infection model (HFIM) studies to identify optimal doses of these drug combinations [2]. Building on these results, the ARLG conducted a phase 1 trial of healthy volunteers to confirm dosing schedules predicted in the HFIM work [3, 4], providing clinicians with guidance for dosing and safety considerations. Future studies are planned to test the clinical effectiveness of aztreonam + ceftazidime-avibactam as well as other novel antibiotics in patients with serious infections caused by NDM-producing pathogens.
The ARLG’s Carbapenem-resistant Pseudomonas aeruginosa (CRPA) and associated carbapenemases (POP) cohort study sought to define the clinical characteristics and frequency of CRPA infections globally [18]. Identification of dramatic regional differences in the mechanisms and clinical impact of carbapenem resistance in P. aeruginosa globally confirmed the need to pursue multiple treatment options, including nontraditional ones, for this pathogen. This discovery motivated the ARLG to evaluate bacteriophages as a possible treatment for P. aeruginosa.
A central concept guiding the ARLG approach to phages is that if they are intended to be used as a drug, they must be tested and developed as such to advance the field beyond anecdotal reports. To that end, the ARLG published guidance outlining clinical, laboratory, and pharmacokinetic considerations of phage therapy [35]. Additionally, with industry and government collaborators, the ARLG helped to advance methods for susceptibility testing. This document, which has been downloaded more than 11 500 times in the 2 years since its publication, provides the basis for the ARLG’s phase 1b/2, multicentered, randomized, double-blind, placebo-controlled trial of the Safety and Microbiological Activity of a Single Dose of Bacteriophage Therapy in Cystic Fibrosis Subjects Colonized with Pseudomonas aeruginosa (PHAGE) trial (ClinicalTrials.gov identifier: NCT05453578) [9]. Currently enrolling, PHAGE will allow a comprehensive evaluation of phage safety, antibacterial activity, pharmacokinetics, susceptibility testing, and emergence of resistance to therapy.
Innovation and Evolution: Diagnostics
As 1 of its 3 scientific priority areas, the ARLG is addressing the need for better diagnostics in the AR sphere. The ARLG pioneered the use of MASTERMIND, an innovative trial design that allows simultaneous testing of multiple diagnostic platforms in a single study [36]. In brief, MASTERMIND studies involve a study protocol that provides equitable sample access for each diagnostic platform involved in the study. Care is taken to ensure that optimal assay conditions are met for each tested platform. At the completion of a MASTERMIND study, each company receives the necessary performance data for its diagnostic platform to support possible regulatory filing. Using this approach, MASTERMIND-GC supported FDA approval for 2 molecular assays for the detection of extragenital N. gonorrhoeae or Chlamydia trachomatis [23] in a single clinical trial. Additional MASTERMIND studies are being designed for other indications. MASTERMIND-RING will evaluate molecular tests to predict ciprofloxacin resistance or susceptibility in N. gonorrhoeae, and MASTERMIND-BSI will evaluate rapid diagnostics for bloodstream infections.
Viral upper respiratory infections are one of the leading reasons for unnecessary antibiotic prescriptions. To address this, the ARLG has focused on methods to differentiate bacterial from viral etiologies of respiratory infection. The ARLG has partnered with Biomeme and is pursuing 510k approval from the FDA for the Rapid Diagnostics in Categorizing Acute Lung Infections (RADICAL) platform, which assays host gene expression patterns to differentiate bacterial and viral infection. Ultimately, the ARLG plans to test the clinical utility of the RADICAL assay by randomizing patients who present to the emergency department with acute respiratory infection to either standard care or RADICAL-guided treatment. The Targeted Reduction of Antibiotics using Procalcitonin in Lower Respiratory Tract Infection (TRAP-LRTI) study, a double-blind, noninferiority RCT, compared azithromycin with placebo in outpatients with clinically suspected nonpneumonia LRTIs and procalcitonin levels ≤0.25 ng/mL [14]. The primary analysis, which used the standard frequentist approach, found placebo not to be noninferior to azithromycin for clinical improvement at day 5. Interestingly, the secondary DOOR analysis of a global patient outcome based on clinical benefits and harms showed no significant difference between treatment groups, but azithromycin was associated with more solicited adverse events [14].
Delays in the clinical availability of antimicrobial susceptibility results for patients with bloodstream infections (BSI) potentially contribute to antibiotic overuse. Additionally, patients may not receive antibiotics active against the bacterium with which they are infected. The ARLG conducted 2 RCTs to evaluate the clinical impact of rapid diagnostics for positive blood culture bottles. In the Blood Culture Identification (BCID) study, use of a rapid multiplex polymerase chain reaction panel reported with templated comments reduced treatment of contaminants and use of broad-spectrum antibiotics when compared with standard blood culture bottle processing [24]. Benefits were most evident in patients with gram-positive bacteremia, which was hypothesized to be due to the dearth of clinically actionable information on the susceptibility of gram-negative bacteria. Accordingly, the Rapid Identification and Susceptibility Testing for Gram-negative Bacteremia (RAPIDS-GN) trial evaluated organism identification and phenotypic antimicrobial susceptibility testing using the Accelerate PhenoTest BC Kit (Accelerate Diagnostics, Tucson, AZ; RAPID) directly on positive blood culture bottles. In patients with gram-negative bacteremia, the median time to antibiotic escalation (for resistant bacteria) and deescalation (for nonresistant bacteria) was significantly shorter in the RAPID diagnostic arm [25]. Based on the results from the BCID and RAPIDS-GN studies, the ARLG recently published best practice guidelines supporting use of rapid tests on positive blood culture bottles with guidance on rapid reporting [27]. However, neither the BCID nor RAPIDS-GN platform was shown to improve the outcomes of patients with bloodstream infections. This finding is potentially due to the low rates of MDR gram-negative bacteria, for which a shorter time to effective antibiotic therapy would presumably afford the greatest potential benefit in either of the 2 US-based studies. As a result, the Fast Antibiotic Susceptibility Testing for Gram-negative Bacteremia (FAST) trial is designed to test whether rapid phenotypic susceptibility testing of positive blood culture bottles leads to improved outcomes in patients with gram-negative bacterial bloodstream infections in clinical settings with high rates of AR. FAST is leveraging a metabolomic-based rapid phenotypic susceptibility platform.
Innovation and Evolution: Clinical Trials Design
Advances in AR research and study design have the potential to improve not just one study but a generation of studies. In addition to improving patient care and clinical practice, the ARLG has focused on enhancing the ways clinical trials are conducted and analyzed. For example, the ARLG began work with the novel clinical trials end point DOOR [28] through analyses of observational studies but now has applied this methodology to RCTs. The longer-term goal is to bring these innovations into the mainstream. To this end, the ARLG has established the Innovations Working Group, a unique National Institutes of Health (NIH)–FDA collaboration tasked with generating publicly available deliverables that could be used to support FDA approval and package insert language for novel antibacterials. Projects fall into 2 workstreams: DOOR-type end points and health-related quality-of-life measures for the 4 most common licensing indications for antibacterial agents: acute bacterial skin and skin structure infections, complicated urinary tract infection (cUTI), complicated intraabdominal infection (cIAI), and hospital-acquired/ventilator-associated bacterial pneumonia (HABP/VABP). This collaboration has been further strengthened by the creation of an FDA Antibacterial Drug Resistance (DOOR) Fellowship made possible by the FDA through the Oak Ridge Institute for Science and Education (ORISE). To date, DOOR end points have been published for cUTI [37] and cIAI [38]. A manuscript describing DOOR for HABP/VABP is being finalized. The ARLG has also designed a DOOR app, freely available online to the scientific community for use in performing DOOR-type analyses (https://methods.bsc.gwu.edu/) [39].
The ARLG has also developed several innovative advances to execute and analyze studies that involve diagnostic platforms. In addition to the MASTERMIND approach, the Benefit-Risk Evaluation of Diagnostics: A Framework (BED-FRAME) provides a means of communicating the expected impact of diagnostic application and trade-offs of diagnostic alternatives to guide decision-making [40]. The ARLG has pioneered application of RCTs to diagnostics to inform when and how they should be used, and this guidance is being adopted by the scientific community [25]. Additionally, the ARLG has assembled a web-searchable biorepository of well-characterized MDR bacteria that are available without cost to the scientific community (https://arlg.org/laboratory-center-strain-access/) [41].
Collaborations
Collaborations with industry partners, governmental agencies, and academic study sites around the world have been vital to ARLG’s mission over the past decade. The ARLG has provided companies with epidemiologic data on MDR pathogens, well-characterized bacterial isolates from the ARLG Biorepository, consultative advice for a sponsor’s clinical trial, clinical sample matrices for diagnostic development, and clinical trial infrastructure to execute MASTERMIND-based collaborative studies. In other projects, the ARLG has received study drug or diagnostic tests for interventional trials, industry expertise on antibiotics or diagnostic platforms involved in ARLG studies, and access to industry-owned datasets from phase 2 and 3 clinical trials to advance DOOR development. The ARLG has also fostered intergovernmental agency collaborations. One example is the FDA-NIH collaboration to develop DOOR end points. The FDA has also interacted with the ARLG Laboratory Center (eg, MASTERMIND-RING, Phage Task Force) and with the ARLG Innovations Working Group through regular meetings with patient representatives and (with appropriate firewall protection given FDA involvement) industry members. This collaboration has led to the publication of 2 DOOR end points for antibacterial indications [37, 38] and the creation of an ORISE fellowship by the FDA to support a trainee to work at the FDA on DOOR end point development. Importantly, this partnership is fully consistent with the CARB National Action Plan for a concerted response to AR [42].
Central to ARLG’s mission has been collaboration with thought leaders, both domestic and international, to successfully define the clinical and molecular epidemiology of WHO critical pathogens. As a result, the ARLG currently involves more than 120 investigators from 6 countries. Having actively worked with Combatting Bacterial Resistance in Europe (COMBACTE), the ARLG is currently reestablishing a memorandum of understanding for COMBACTE’s replacement organization, the European Clinical Research Alliance on Infectious Diseases. In summary, the ARLG believes that the global threat of AR will continue to require an ongoing collaborative approach with international investigators, companies, and government entities.
Mentoring
Not to be overlooked is the need to address the declining workforce of investigators pursuing careers in AR and antiinfective drug development. To this end, the ARLG has established a strong mentoring program. This investment is beginning to deliver returns as 6 of 12 (50%) trainees from the first cycle of ARLG have secured K23s or other career development awards. Even more encouraging, several current ARLG studies, including MASTERMIND-BSI, DOTS, and FAST, are currently being led by trainees from the initial grant cycle.
ARLG: The Future
Progress in the design and conduct of clinical trials and diagnostics, recently accentuated by the necessities of the COVID-19 pandemic, offers new approaches for evaluating antibacterial compounds, treatments, and tests. As more antibacterials active against MDR gram-negative bacteria become available, platform trials comparing multiple interventions, including unconventional approaches such as phage therapy, become possible. Such trials would leverage local resources and international collaborations developed within the MDRO Network. Optimal management of S. aureus bloodstream infections, particularly those caused by MRSA, remains elusive. New diagnostic technologies, such as the detection of microbial cell-free DNA in plasma, have the potential to reduce antibiotic use by individualizing antibiotic therapy duration for patients with bloodstream infections [43]. Advances in artificial intelligence and access to ever-larger clinical databases make real-world data ever more powerful, particularly when results of RCTs are unavailable.
Diagnostics for detecting bacterial infections and identifying antibacterial resistance remain a top priority for the ARLG. The ARLG’s MASTERMIND studies are well suited for evaluating diagnostic platforms in terms of performance, informed by advanced reference standards [29]; RCTs are valuable clinical utility studies particularly as diagnostics provide progressively faster genotypic and phenotypic data for bacterial infections. Marrying the host response with pathogen detection has the potential to improve the diagnosis and, ultimately, outcomes of ventilator-associated pneumonia and other infections.
The ARLG will continue to adapt and evolve to respond to the threat of AR by building on its greatest assets: its team of investigators, its collaborative relationships, and those in the greater scientific community [44].
Contributor Information
Henry F Chambers, Division of Infectious Diseases, Department of Medicine, University of California –San Francisco, San Francisco, California, USA.
Heather R Cross, Duke Clinical Research Institute, Duke University School of Medicine, Durham, North Carolina, USA.
Maria Souli, Duke Clinical Research Institute, Duke University School of Medicine, Durham, North Carolina, USA.
Scott R Evans, Department of Biostatistics, George Washington University, Washington, DC, USA.
Robin Patel, Division of Infectious Diseases, Mayo Clinic, Rochester, Minnesota, USA.
Vance G Fowler, Jr., Duke Clinical Research Institute, Duke University School of Medicine, Durham, North Carolina, USA Division of Infectious Diseases, Department of Medicine, Duke University School of Medicine, Durham, North Carolina, USA.
for the Antibacterial Resistance Leadership Group:
Thomas Lodise, Nancie Deckard, Carl Schuler, Ivra Bunn, Thomas Holland, Nicholas Turner, Smitha Zaharoff, Shrabani Sharma, Cathy Wickward, Jason Waller, Holly Wilson, David van Duin, Keri Baum, Lauren Komarow, Minggui Wang, Beth Evans, Deborah Hopkins, Lizhao Ge, Abhigya Giri, Weixiao Dai, Guoqing Diao, Tamara Fidler, Wanying Shao, Nyssa Schwager, Robert Bonomo, Donald Mau, Michael Satlin, Yixuan Li, Pranita Tamma, Robert Schooley, Toshimitsu Hamasaki, Zoe Sund, Grant Booth, Leslie Estes, Kerryl Greenwood-Quaintance, Krupa Mukesh Parmar, Scott Cunningham, Sarah Doernberg, Andrew Dodd, Ephraim Tsalik, Gayani Tillekeratne, Praneeta Raza, Lijuan Zeng, Ritu Banerjee, Erin Abbenante, Elizabeth Mocka, Heather King, Tori Kinamon, Jessica Howard-Anderson, Helen Boucher, Holly Geres, Yijie He, Maureen Mehigan, Varduhi Ghazaryan, Seema Nayak, Erica Raterman, Tamika Samuel, and Marina Lee
Notes
Author Contributions . H. F. C. and V. G. F. wrote the initial manuscript and the final version. H. R. C. and M. S. collected data, designed the figure, and provided edits. S. R. E. and R. P. contributed and edited text on clinical trials design and diagnostics.
Acknowledgments. The authors thank the members of all Antibacterial Resistance Leadership Group (ARLG) committees, subcommittees, working groups, and task forces for their invaluable contributions to the ARLG program. The authors also thank the following study team members for their contributions, which made the studies and results possible: Thomas Lodise, Nancie Deckard, Carl Schuler, Ivra Bunn, Thomas Holland, Nicholas Turner, Smitha Zaharoff, Shrabani Sharma, Cathy Wickward, Jason Waller, Holly Wilson, David van Duin, Keri Baum, Lauren Komarow, Minggui Wang, Beth Evans, Deborah Hopkins, Lizhao Ge, Abhigya Giri, Weixiao Dai, Guoqing Diao, Tamara Fidler, Wanying Shao, Nyssa Schwager, Robert Bonomo, Donald Mau, Michael Satlin, Yixuan Li, Pranita Tamma, Robert Schooley, Toshimitsu Hamasaki, Zoe Sund, Grant Booth, Leslie Estes, Kerryl Greenwood-Quaintance, Krupa Mukesh Parmar, Scott Cunningham, Sarah Doernberg, Andrew Dodd, Ephraim Tsalik, Gayani Tillekeratne, Praneeta Raza, Lijuan Zeng, Ritu Banerjee, Erin Abbenante, Elizabeth Mocka, Heather King, Tori Kinamon, Jessica Howard-Anderson, Helen Boucher, Holly Geres, Yijie He, Maureen Mehigan, Varduhi Ghazaryan, Seema Nayak, Erica Raterman, Tamika Samuel, Marina Lee, other Division of Microbiology and Infectious Diseases team members, and the members of the Emmes and Clinical Research Operations and Management System teams. The authors also thank all study site staff and study participants, without whom this work would not be possible.
Disclaimer. The content is solely the authors’ responsibility and does not necessarily represent the official views of the National Institutes of Health (NIH).
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award UM1AI104681.
Supplement sponsorship. This article appears as part of the supplement “The Antibacterial Resistance Leadership Group (ARLG): Innovation and Evolution,” sponsored by the Antibacterial Resistance Leadership Group.
References
- 1. Lodise TP, Van Wart S, Sund ZM, et al. . Pharmacokinetic and pharmacodynamic profiling of minocycline for injection following a single infusion in critically ill adults in a phase IV open-label multicenter study (ACUMIN). Antimicrob Agents Chemother 2021; 65:e01809-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lodise TP, Smith NM, O’Donnell N, et al. . Determining the optimal dosing of a novel combination regimen of ceftazidime/avibactam with aztreonam against NDM-1-producing Enterobacteriaceae using a hollow-fibre infection model. J Antimicrob Chemother 2020; 75:2622–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lodise TP, O’Donnell JN, Raja S, et al. . Safety of ceftazidime-avibactam in combination with aztreonam (COMBINE) in a phase I, open-label study in healthy adult volunteers. Antimicrob Agents Chemother 2022; 66:e0093522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lodise TP, O’Donnell JN, Balevic S, et al. . Pharmacokinetics of ceftazidime-avibactam in combination with aztreonam (COMBINE) in a phase 1, open-label study of healthy adults. Antimicrob Agents Chemother 2022; 66:e0093622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wenzler E, Bleasdale SC, Sikka M, et al. . Phase I study to evaluate the pharmacokinetics, safety, and tolerability of two dosing regimens of oral fosfomycin tromethamine in healthy adult participants. Antimicrob Agents Chemother 2018; 62:e00464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lodise TP, Rosenkranz SL, Finnemeyer M, et al. . The emperor’s new clothes: prospective observational evaluation of the association between initial vancomycIn exposure and failure rates among adult hospitalized patients with methicillin-resistant Staphylococcus aureus bloodstream infections (PROVIDE). Clin Infect Dis 2020; 70:1536–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Turner NA, Zaharoff S, King H, et al. . Dalbavancin as an option for treatment of S. aureus bacteremia (DOTS): study protocol for a phase 2b, multicenter, randomized, open-label clinical trial. Trials 2022; 23:407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. van Duin D, Lok JJ, Earley M, et al. . Colistin versus ceftazidime-avibactam in the treatment of infections due to carbapenem-resistant Enterobacteriaceae. Clin Infect Dis 2018; 66:163–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tamma PD, Souli M, Billard M, et al. . Safety and microbiological activity of phage therapy in persons with cystic fibrosis colonized with Pseudomonas aeruginosa: study protocol for a phase 1b/2, multicenter, randomized, double-blind, placebo-controlled trial. Trials 2022; 23:1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Finney AG, Perry JM, Evans DR, et al. . Isolation and characterization of lytic bacteriophages targeting diverse Enterobacter spp. clinical isolates. Phage (New Rochelle) 2022; 3:50–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nordstrom HR, Evans DR, Finney AG, et al. . Genomic characterization of lytic bacteriophages targeting genetically diverse Pseudomonas aeruginosa clinical isolates. iScience 2022; 25:104372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Williams DJ, Creech CB, Walter EB, et al. . Short- vs standard-course outpatient antibiotic therapy for community-acquired pneumonia in children: the SCOUT-CAP randomized clinical trial. JAMA Pediatr 2022; 176:253–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pettigrew MM, Kwon J, Gent JF, et al. . Comparison of the respiratory resistomes and microbiota in children receiving short versus standard course treatment for community-acquired pneumonia. mBio 2022; 13:e0019522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tsalik EL, Rouphael NG, Sadikot RT, et al. . Efficacy and safety of azithromycin versus placebo to treat lower respiratory tract infections associated with low procalcitonin: a randomised, placebo-controlled, double-blind, non-inferiority trial. Lancet Infect Dis 2023; 23:484–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van Duin D, Perez F, Rudin SD, et al. . Surveillance of carbapenem-resistant Klebsiella pneumoniae: tracking molecular epidemiology and outcomes through a regional network. Antimicrob Agents Chemother 2014; 58:4035–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van Duin D, Arias CA, Komarow L, et al. . Molecular and clinical epidemiology of carbapenem-resistant Enterobacterales in the USA (CRACKLE-2): a prospective cohort study [published correction appears in Lancet Infect Dis. 2020 Apr 23]. Lancet Infect Dis 2020; 20:731–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang M, Earley M, Chen L, et al. . Clinical outcomes and bacterial characteristics of carbapenem-resistant Klebsiella pneumoniae complex among patients from different global regions (CRACKLE-2): a prospective, multicentre, cohort study. Lancet Infect Dis 2022; 22:401–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Reyes J, Komarow L, Chen L, et al. . Global epidemiology and clinical outcomes of carbapenem-resistant Pseudomonas aeruginosa and associated carbapenemases (POP): a prospective cohort study. Lancet Microbe 2023; 4:e159–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tamma PD, Komarow L, Ge L, et al. . Clinical impact of ceftriaxone resistance in Escherichia coli bloodstream infections: a multicenter prospective cohort study. Open Forum Infect Dis 2022; 9:ofac572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu Y, Tsalik EL, Jiang Y, et al. . Average weighted accuracy: pragmatic analysis for a rapid diagnostics in categorizing acute lung infections (RADICAL) study. Clin Infect Dis 2020; 70:2736–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tsalik EL, Henao R, Montgomery JL, et al. . Discriminating bacterial and viral infection using a rapid host gene expression test. Crit Care Med 2021; 49:1651–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tsalik EL, Henao R, Nichols M, et al. . Host gene expression classifiers diagnose acute respiratory illness etiology. Sci Transl Med 2016; 8:322ra11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Doernberg SB, Komarow L, Tran TTT, et al. . Simultaneous evaluation of diagnostic assays for pharyngeal and rectal Neisseria gonorrhoeae and Chlamydia trachomatis using a master protocol. Clin Infect Dis 2020; 71:2314–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Banerjee R, Teng CB, Cunningham SA, et al. . Randomized trial of rapid multiplex polymerase chain reaction-based blood culture identification and susceptibility testing. Clin Infect Dis 2015; 61:1071–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Banerjee R, Komarow L, Virk A, et al. . Randomized trial evaluating clinical impact of RAPid IDentification and susceptibility testing for gram-negative bacteremia: RAPIDS-GN. Clin Infect Dis 2021; 73:e39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. GENO-STELLAR. GENOmics, Sequencing-based Typing, EpidemioLogy, Linkage, and Antimicrobial Resistance Tool. Available at: https://genostellar.net. Accessed 26 June 2023.
- 27. Simner PJ, Dien Bard J, Doern C, et al. . Reporting of antimicrobial resistance from blood cultures, an Antibacterial Resistance Leadership Group survey summary: resistance marker reporting practices from positive blood cultures. Clin Infect Dis 2023; 76:1550–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Evans SR, Patel R, Hamasaki T, et al. . The future ain’t what it used to be…out with the old…in with the better: Antibacterial Resistance Leadership Group (ARLG) innovations. Clin Infect Dis 2023; 77:321–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Patel R, Tsalik EL, Evans S, Fowler VG. Doernberg SB; Antibacterial Resistance Leadership Group. Clinically adjudicated reference standards for evaluation of infectious diseases diagnostics. Clin Infect Dis 2023; 76:938–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jernigan JA, Hatfield KM, Wolford H, et al. . Multidrug-resistant bacterial infections in U.S. hospitalized patients, 2012–2017. N Engl J Med 2020; 382:1309–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Antimicrobial Resistance Collaborators . Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis [published correction appears in Lancet. 2022 Oct 1; 400(10358):1102]. Lancet 2022; 399:629–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rybak MJ, Le J, Lodise TP, et al. . Therapeutic monitoring of vancomycin for serious methicillin-resistant Staphylococcus aureus infections: a revised consensus guideline and review by the American Society of Health-System Pharmacists, the Infectious Diseases Society of America, the Pediatric Infectious Diseases Society, and the Society of Infectious Diseases Pharmacists. Am J Health Syst Pharm 2020; 77:835–64. [DOI] [PubMed] [Google Scholar]
- 33. Tamma PD, Aitken SL, Bonomo RA, Mathers AJ, van Duin D, Clancy CJ. Infectious Diseases Society of America guidance on the treatment of extended-spectrum β-lactamase producing Enterobacterales (ESBL-E), carbapenem-resistant Enterobacterales (CRE), and Pseudomonas aeruginosa with difficult-to-treat resistance (DTR-P. aeruginosa). Clin Infect Dis 2021; 72:1109–16. [DOI] [PubMed] [Google Scholar]
- 34. Marshall S, Hujer AM, Rojas LJ, et al. . Can ceftazidime-avibactam and aztreonam overcome β-lactam resistance conferred by metallo-β-lactamases in Enterobacteriaceae? Antimicrob Agents Chemother 2017; 61:e02243-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Suh GA, Lodise TP, Tamma PD, et al. . Considerations for the use of phage therapy in clinical practice. Antimicrob Agents Chemother 2022; 66:e0207121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Patel R, Tsalik EL, Petzold E, et al. . MASTERMIND: bringing microbial diagnostics to the clinic. Clin Infect Dis 2017; 64:355–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Howard-Anderson J, Hamasaki T, Dai W, et al. . Improving traditional registrational trial end points: development and application of a desirability of outcome ranking end point for complicated urinary tract infection clinical trials. Clin Infect Dis 2023; 76:e1157–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kinamon T, Gopinath R, Waack U, et al. . Exploration of a potential DOOR endpoint for complicated intra-abdominal infections using nine registrational trials for antibacterial drugs. Clin Infect Dis 2023; 22:649–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. DOOR Analyses: Standard edition. Available at: https://methods.bsc.gwu.edu. Accessed 12 June 2023.
- 40. Evans SR, Pennello G, Pantoja-Galicia N, et al. . Benefit-risk evaluation for diagnostics: a framework (BED-FRAME). Clin Infect Dis 2016; 63:812–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Antibacterial Resistance Leadership Group . ARLG biorepository. Available at: https://arlg.org/laboratory-center-strain-access/. Accessed 12 June 2023.
- 42. Federal Task Force on Combating Antibiotic-Resistant Bacteria . National Action Plan for Combating Antibiotic-Resistant Bacteria 2020–2025. Available at: https://www.hhs.gov/sites/default/files/carb-national-action-plan-2020-2025.pdf. Accessed 19 May 2023.
- 43. Mourad A, Fowler VG Jr, Holland TL. Which trial do we need? Next-generation sequencing to individualize therapy in Staphylococcus aureus bacteraemia. Clin Microbiol Infect 2023: 29:955–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wenzler E, Bleasdale SC, Sikka M, et al. . Phase I study to evaluate the pharmacokinetics, safety, and tolerability of two dosing regimens of oral fosfomycin tromethamine in healthy adult participants. Antimicrob Agents Chemother 2018; 62:e00464-18. doi: 10.1128/AAC.00464-18 [DOI] [PMC free article] [PubMed] [Google Scholar]