Abstract
Developing and implementing the scientific agenda of the Antibacterial Resistance Leadership Group (ARLG) by soliciting input and proposals, transforming concepts into clinical trials, conducting those trials, and translating trial data analyses into actionable information for infectious disease clinical practice is the collective role of the Scientific Leadership Center, Clinical Operations Center, Statistical and Data Management Center, and Laboratory Center of the ARLG. These activities include shepherding concept proposal applications through peer review; identifying, qualifying, training, and overseeing clinical trials sites; recommending, developing, performing, and evaluating laboratory assays in support of clinical trials; and designing and performing data collection and statistical analyses. This article describes key components involved in realizing the ARLG scientific agenda through the activities of the ARLG centers.
Keywords: antibacterial resistance, clinical trial network, infectious disease, laboratory testing, statistics
Developing and implementing the Antibacterial Resistance Leadership Group (ARLG) scientific agenda requires intense creativity, coordination, and collaboration between ARLG operational centers. Their roles in progressing priorities from idea to trial design, conduct, analysis, and recommendations for clinical practice are described.
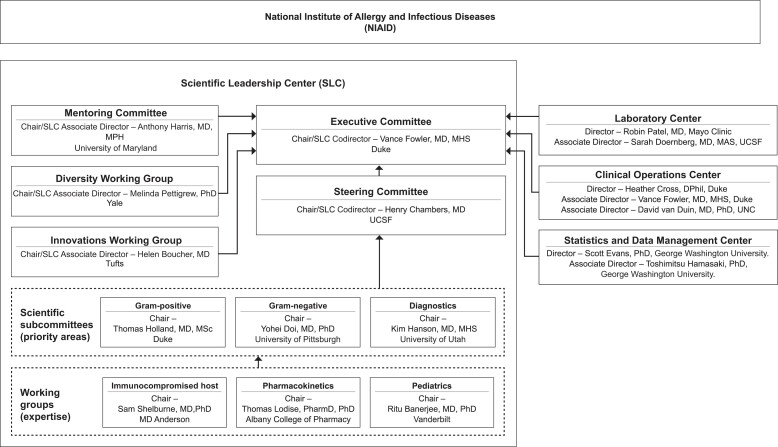
The mission of the Antibacterial Resistance Leadership Group (ARLG) is to prioritize, design, and execute clinical research that will affect the prevention, diagnosis, and treatment of infections caused by antibiotic-resistant bacteria. The Scientific Leadership Center (SLC), along with its various scientific and governance committees (Figure 1), prioritizes and develops the scientific agenda of the ARLG based on input from ARLG leadership and the >120 thought leaders from >50 institutions and 6 countries that comprise the ARLG scientific subcommittees. Prioritization is also informed by the National Institute of Allergy and Infectious Diseases, the scientific community [1], and the ARLG External Advisory Board.
Figure 1.
Organization of committees, subcommittees, and working groups within the Antibacterial Resistance Leadership Group Scientific Leadership Center. Abbreviations: UCSF, University of California, San Francisco; UNC, University of North Carolina.
As a result, the ARLG scientific agenda is dynamic and regularly revisited as new scientific findings emerge. The availability of funding influences the studies undertaken as part of the ARLG scientific agenda. The ARLG Executive Committee makes final decisions on approving studies, incorporating scientific prioritization and guidance from the ARLG Steering Committee, and considering overall budgetary issues as they arise. Operational integration throughout the ARLG is ensured by the inclusion of Clinical Operations Center (COC), Laboratory Center (LC), and Statistical and Data Management Center (SDMC) leadership as voting members in the ARLG Steering and Executive Committees.
Other roles of the SLC include communication; managing and tracking the peer review process for submitted proposals; and supporting the innovations [2], diversity [3], and mentoring [3] components of the ARLG. Communication occurs via the ARLG website (www.arlg.org) [4], newsletters, lay summaries, the X platform (formerly Twitter) (@ARLGnetwork), and ARLG Grand Rounds. This communication is essential to inform the scientific community and the general public about the ARLG's activities and to solicit their input. It is also critical to keep the multiple committee members, center staff, and clinical sites that constitute the ARLG fully informed about all aspects of this large and effective program. More than 50 study proposals have been reviewed by the SLC in the second grant cycle of the ARLG, “ARLG 2.0,” and 24 have been approved or implemented thus far, adding to the 44 studies approved in the initial cycle of the grant, “ARLG 1.0.” The SLC also houses the ARLG Publications Committee, which approves, tracks, and maintains compliance for manuscripts and abstracts generated from ARLG work. To date, 100 manuscripts have been published and 27 abstracts have been presented in ARLG 2.0, adding to the 126 manuscripts and 83 abstracts from ARLG 1.0.
IMPLEMENTING THE ARLG SCIENTIFIC AGENDA—THE ARLG COC
The COC, hosted at the Duke Clinical Research Institute, is at the core of the ARLG program. COC staff are integrated into the ARLG SLC, LC, and SDMC to maximize communication and continuity. The COC is responsible for the project and financial management of all aspects of the ARLG, including the science, centers, and network. As such, the COC has played a role in all of the >60 ARLG studies and trials to date, including the 24 studies and trials in ARLG 2.0. In addition to operations, the COC contributes to thought leadership in the ARLG, through active participation in all of its committees and working groups [5–7].
The COC becomes involved early in the development of each clinical trial and study to ensure the enrollment, budgetary, and procedural feasibility of the proposal. Immediately following study approval by the Executive Committee, the COC coordinates protocol development and ensures feasibility in a process that involves the SDMC, the LC, the protocol's principal investigator, and the study team. COC staff provide their expertise during the protocol development process to ensure that study procedures, inclusion-exclusion criteria, and other aspects are conducive to timely participant enrollment. For example, in the Pneumonia Direct study currently under development, the COC advised the protocol team to use scavenged endotracheal samples instead of the planned study-specific sampling, which would have led to a more complicated regulatory pathway of an abbreviated investigational device exemption.
In the MASTER-GC study, the COC designed the procedures to be conducive to a waiver of documentation of consent to remove confidentiality concerns as a barrier to participant enrollment [8]. Similarly, for the PROVIDE study, the COC helped design the protocol to require no study-specific participant procedures and therefore enable a waiver of consent, which was important for ensuring both adequate enrollment and inclusivity of a sicker patient population who would not have been able to participate owing to timing if consent from a legally authorized representative had been required [9]. In the MASTERMIND-RING study currently under development, the COC is exploring with sites and the US Food and Drug Administration (FDA) whether the protocol can be designed to ensure that remnant samples can be used to allow testing of only positive samples, which would dramatically reduce the required enrollment.
A key component of the COC is the identification and retention of experienced clinical trials sites. ARLG studies and trials are unique and diverse, involving different drugs, biologics, and diagnostics; multiple body systems and syndromes; multiple participant types, such as healthy volunteer, clinic, emergency department, hospitalized, and intensive care patients; and geographic variation in the prevalence of key resistant pathogens of interest. This complexity presents a major challenge in identifying sites. In the initial cycle of the ARLG grant, an intense effort was dedicated to developing the Multi-Drug Resistant Organism Clinical Trial Network (MDRO-CTN), to address the challenge of identifying sites for the diverse ARLG studies and trials.
The MDRO-CTN Network grew from the Consortium on Resistance Against Carbapenems in Klebsiella pneumoniae and Other Enterobacteriaceae (CRACKLE) [10], which was expanded by the ARLG in a targeted manner to include a number of critical priority resistant pathogens—Pseudomonas aeruginosa, Acinetobacter baumannii, and Escherichia coli [11–14]—and multiple regions globally. MDRO-CTN sites are supplemented by intensive searches of past trials, prior experience databases at the Duke Clinical Research Institute, and communication to potential trial sites that have completed >900 detailed site questionnaires thus far. Site identification is facilitated by innovation; the COC developed a database to allow querying of ARLG feasibility data, which is tailored to each specific ARLG study. A pool of 1128 potential sites have been linked to the ARLG feasibility database, and to date >140 of these clinical trials sites have actively enrolled in ARLG studies, providing real evidence of the ability to navigate regulatory document collection, ethics approvals, contract execution, enrollment, quality maintenance, data cleaning, and sample shipping in the varied landscape of government-funded antibiotic resistance studies.
The ARLG has also collaborated with several networks, such as the Clinical Trials Transformation Initiative, whose PROPHETIC study [15] is providing novel feasibility and study design information, and other qualified sites to inform the design of the ARLG Pneumonia Direct pilot study [16]. Globally, the ARLG collaborates with the European Clinical Research Alliance on Infectious Diseases, formerly the Combatting Bacterial Resistance in Europe group, to provide sites in Europe. Sites in other countries, including Colombia, Chile, China, Argentina, Australia, Nicaragua, Saudi Arabia, Lebanon, Singapore, and New Zealand, are part of the ARLG's own MDRO-CTN Network [12, 17].
Once sites are identified, the COC trains sites, assists with site start-up, monitors data and quality, provides site payments, manages vendors, and oversees budgets and timelines. With the frequent “first-in-kind” trials and studies undertaken by the ARLG, the COC often needs to be creative in developing processes that will facilitate data collection at sites to achieve the scientific goals of the ARLG. Examples include facilitating equipment installation for ARLG diagnostic studies, such as MASTER-GC [8] and FAST [16]; navigating multicountry import-export regulations and regulatory requirements for SNAP, CRACKLE, POP [12, 13, 17], and variable multicountry product approval statuses for FAST; developing kits for randomized sample collection order in the MASTERMIND studies [8]; and developing procedures for supervised specimen collection for at-home participant visits for PHAGE [18]. Ingenuity, flexibility, academic collaboration with the SLC, LC, and SDMC, and broad-ranging knowledge of multiple scientific areas are essential for facilitating the ARLG scientific agenda.
THE ARLG LC
The ARLG LC is instrumental in supporting the scientific agenda of the ARLG, in collaboration with the COC and SDMC. ARLG LC-specific expertise includes the provision of leadership and guidance in laboratory aspects of ARLG's research strategies, thought leadership around diagnostic challenges, spearheading of collaborations with diagnostics companies, provision of pathways to bring novel diagnostics to market, and provision of state-of-the-art laboratory testing support to ARLG studies. The ARLG LC also maintains and promotes a specimen and bacterial isolate biorepository.
The ARLG LC has provided leadership to >10 ARLG studies in ARLG 2.0 [1]. The LC reviews all proposals involving laboratory testing, provides protocol development advice for studies involving laboratory testing, ensures diagnostic quality management for ARLG studies (eg, test selection, specimen selection, specimen handling, laboratory standards development, metrics assessment, laboratory processes, and laboratory manual development), and contributes mentorship regarding diagnostic approaches to ARLG investigators.
The ARLG LC also provides thought guidance to the scientific community at large. For example, the ARLG LC, in partnership with the ARLG Diagnostics Subcommittee, collaborated with the Infectious Diseases Society of America to publish a white paper on future urinary tract infection diagnostics [19] and provided diagnostics input to the ARLG Phage Task Force to assist clinicians considering experimental phage therapy for their patients [5]. Members of the ARLG LC also partnered with the ARLG Diagnostics Subcommittee on the REPORT-ABC study, which assessed approaches to performing and reporting results of rapid tests performed on positive blood culture bottles. The analysis of survey responses from REPORT-ABC identified a lack of standardization across US laboratories in the reporting of results from these rapid tests, and best practice recommendations were made [20], informed by ARLG studies [21, 22].
The ARLG LC provides strong connections to industry and academia in the antibacterial resistance (AR) diagnostics space. Through an intake process on the ARLG's website (www.arlg.org), consultative guidance (eg, project design expertise) is provided upon request (Table 1). Through these interactions, the ARLG LC has identified potential companies to participate in ARLG clinical trials, including diagnostics serving as enrollment tools in clinical trials, or to provide assays for evaluation in diagnostic trials (eg, MASTERMIND-BSI, Pneumonia Direct, and MASTERMIND-RING) [16].
Table 1.
Antibacterial Resistance Leadership Group Laboratory Center Collaborations With Industry
| Type of Collaboration | Collaborations, No. |
|---|---|
| Consultations with companies | 44 |
| Companies provided with isolates | 16 |
| Companies collaborating on ARLG studies | 11 |
Abbreviation: ARLG, Antibacterial Resistance Leadership Group.
The ARLG LC provides thought and operational leadership to MASTERMIND studies, which deliver diagnostics needed to address AR [23]. The MASTERMIND concept arose from a desire to increase efficiency in diagnostic clinical trials and to expand the availability of approved new diagnostics for AR by providing a unique and affordable avenue to diagnostic companies to facilitate data generation for their FDA submissions. The crux of the MASTERMIND design is to evaluate multiple diagnostics using a sample, or samples, obtained from a single participant or site encounter. For example, in the MASTER-GC study [8], 3 separate swab samples were taken from participants at a single clinic visit and were tested on 3 different diagnostic platforms. The study had a single database, single analysis plan, and a single start-up, contracting, and approval step for enrolling sites, and it resulted in 2 FDA clearances [8]. The efficiency and affordability of the MASTERMIND design promotes AR research that may not otherwise be conducted. In ARLG 2.0, the ARLG LC is assisting with several MASTERMIND-type studies.
Other ARLG LC activities include diagnostic assay development and verification; centralized laboratory services, such as antibacterial phenotypic/genotypic susceptibility testing and phage susceptibility testing; and novel bioinformatic tool development. For the ARLG PHAGE study, for example, the ARLG LC developed and evaluated phage susceptibility testing assays and led a multisite comparative study, alongside performing an assessment of phage stability in sputum to support specimen shipping. The ARLG LC provided centralized reference standard broth microdilution antibacterial susceptibility testing for isolates collected through the MDRO-CTN Network, including the CRACKLE II, POP, SNAP, and SHREC studies [11–14]. ARLG LC–generated antibacterial susceptibility testing data are being used in an ARLG web-based genomic-epidemiological tool for antibiotic resistance prediction, GENO-STELLAR (genostellar.net) [24].
The ARLG LC maintains a biorepository (www.arlgcatalogue.org) [25] to allow members of the research and diagnostics development community to request bacterial isolates or human samples (Table 2). Many isolates are well characterized with associated phenotypic and genotypic information.
Table 2.
Antibacterial Resistance Leadership Group Laboratory Center Biorepository Activities
| Bacterial Isolate/Specimen Biobank Activity | Isolates/Samples, No. |
|---|---|
| Isolates/samples in the ARLG biorepository | 8221 |
| Isolates/samples shared from the ARLG biorepository | 7103 |
Abbreviation: ARLG, Antibacterial Resistance Leadership Group.
The ARLG SDMC
Advancement in the complex landscape of AR clinical research requires the contribution of expert biostatistical and data management leadership with relevant AR experience. In collaboration with clinical and laboratory colleagues, the ARLG SDMC innovates tailoring to AR research challenges. The mission of the ARLG SDMC is to (1) ensure that ARLG studies are designed, conducted, analyzed, and reported with optimal scientific integrity by providing leadership throughout the study lifecycle; (2) advance the ARLG mission by enhancing the scientific value and efficiency of ARLG studies through the development and implementation of innovative practical research methods and tools; and (3) educate and mentor the scientific community regarding clinical trial and research fundamentals [26].
The SDMC achieves this mission by contributing at all study stages from conceptualization to publication, and with comprehensive integration into the ARLG, assimilating into all ARLG scientific and operational project activities. The SDMC has key roles in the ARLG's Steering, Executive, Innovations, and Mentoring Committees, and scientific subcommittees. This allows the SDMC to proactively contribute ideas for innovations and improvements and apply sound fundamentals that enhance the integrity of ARLG studies early in study development. The SDMC develops intricate knowledge of relevant diseases, pathogens, interventions, and diagnostics, promoting effective communication with ARLG investigators and other collaborators.
The SDMC develops and implements transformational strategies for the design, monitoring, analyses, and reporting of AR studies to enhance their utility, validity, and efficiency. Examples include (1) the desirability of outcome ranking (DOOR) [27, 28]; sequential, multiple-assignment, randomized trials for comparing personalized antibiotic strategies (SMART COMPASS) [29]; coprimary clinical and stewardship end-point designs; MASTERMIND; Benefit-Risk Evaluation for Diagnostics: A Framework (BED-FRAME); and DOOR for the management of antimicrobial therapy.
DOOR is a paradigm for the design, analysis, and interpretation of clinical trials and studies based on patient-centric benefit-risk [27, 28]. DOOR has been used in several studies [9, 11–13] and is currently used in the PHAGE [18] and DOTS [30] clinical trials. DOOR has been a focus of the ARLG Innovations Working Group and an FDA and Oak Ridge Institute for Science and Education Fellowship, with collaborations resulting in the development and application of DOOR outcomes for complicated intra-abdominal infection [31] and complicated urinary tract infection [6].
Patient management is dynamic, a sequence of decisions, with tailored therapeutic adjustments made over time as new information becomes available. Empiric and definitive therapies are 2 major treatment selection decision points during the treatment of serious bacterial infections. Clinical decision making will benefit from understanding which strategy—that is, a sequence of decision rules that guide empiric and definitive therapy decisions—optimizes the ultimate patient outcome. The SDMC developed SMART COMPASS for this purpose [29]. The COMPASS concept was used in a multicenter, pragmatic clinical trial evaluating 2 strategies, oral fosfomycin versus oral levofloxacin, for initial or step-down treatment of complicated urinary tract infection without bacteremia, allowing for therapeutic adjustment if resistance or intolerability is observed. SMART COMPASS is valuable in the setting of AR, when therapeutic adjustments are necessary.
Clinical and stewardship outcomes must be considered when evaluating rapid diagnostic tests. RADICAL III is designed to evaluate the effects of a rapid diagnostic test in patients with suspected acute respiratory infection on (1) a DOOR clinical outcome and (2) antibacterial exposure using a coprimary end-point design [16].
The SDMC collaborated with the LC to develop MASTERMIND [23]. MASTERMIND was used to simultaneously evaluate the performance of multiple nucleic acid amplification tests for the detection of Neisseria gonorrhoeae and Chlamydia trachomatis in extragenital sites, resulting in FDA clearance of the first diagnostic tests for these indications [8].
The SDMC developed BED-FRAME [32, 33] and average weighted accuracy [34, 35], providing a systematic and pragmatic approach to evaluate and compare diagnostics to aid in clinical decision making. The results consider the prevalence and relative importance of errors, thereby allowing tailored decision making depending on the local, temporal, and treatment availability context. The methods were used in the design of the RADICAL study [34] and in the evaluation of rapid diagnostics to detect carbapenem resistance in “critical” priority pathogens identified by the World Health Organization [36–38].
During clinical practice, it is desirable to select an antibiotic from the narrowest spectrum for which the target infection is susceptible, to minimize toxicity, cost, and the development of resistance due to selective pressures. The SDMC developed DOOR for the management of antimicrobial therapy, a framework for assessing antibiotic selection strategies (eg, those guided by rapid diagnostic tests) in the presence of drug resistance [39].
The SDMC has developed resources for the scientific community to use in future generations of research studies once methods are conceptually advanced. For example, online tools for DOOR analyses are freely available online (https://methods.bsc.gwu.edu/) [40]. The SDMC improves the conduct and utility of antibacterial studies through the education of researchers regarding application of clinical trial and research fundamentals. For example, the SDMC has developed educational articles on methods and issues in studies of carbapenem-resistant Enterobacteriaceae [41]; adaptive and platform trials [42]; and fundamental issues in antibiotic trials [43].
CONCLUSIONS
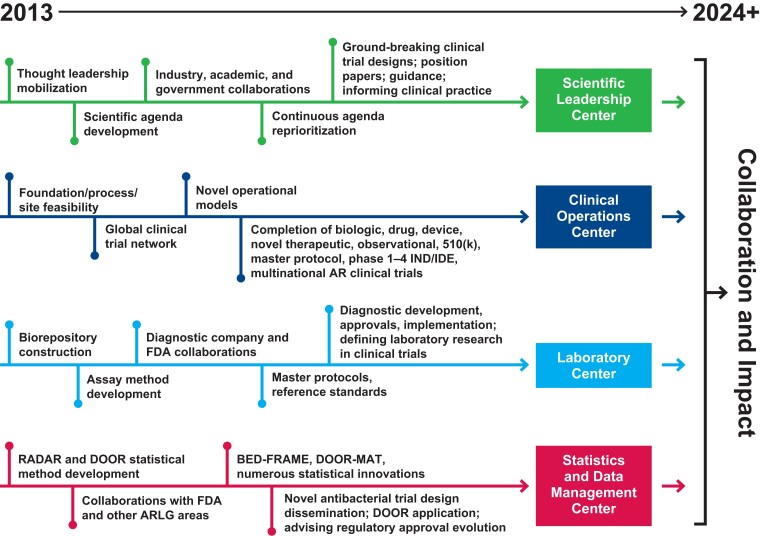
The first iteration of the ARLG grant involved the SLC, COC, SDMC, and LC working together to develop new processes, methods, collaborations, and site networks to create and sustain a research network that was initially focused on laboratory and observational studies and phase I clinical trials. Building from that base, and with an increase in funding, the second cycle of the ARLG grant has been directed to larger pivotal interventional and strategy trials. This evolution is shown in Figure 2. As described in this article, each ARLG center has its own specific expertise, but the collaboration, leadership, and innovation of the ARLG centers collectively were fundamental to this growth. This unique collaboration will be essential as ARLG looks to the future and undertakes the complex and challenging trial designs that are necessary to inform the diagnosis and treatment of elusive syndromes, such as hospital-acquired and ventilator-acquired pneumonia and antibiotic-resistant infections.
Figure 2.
Evolution of the Antibacterial Resistance Leadership Group (ARLG) centers. Abbreviations: AR, antibacterial resistance; BED-FRAME, Benefit-Risk Evaluation for Diagnostics: A Framework; DOOR, desirability of outcome ranking; DOOR-MAT, DOOR for the management of antimicrobial therapy; FDA, US Food and Drug Administration; IND/IDE, investigational new drug/investigational device exemption; RADAR, response adjusted for duration of antibiotic risk.
Contributor Information
Heather R Cross, Duke Clinical Research Institute, Duke University School of Medicine, Durham, North Carolina, USA.
Kerryl E Greenwood-Quaintance, Division of Clinical Microbiology, Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, Minnesota, USA.
Maria Souli, Duke Clinical Research Institute, Duke University School of Medicine, Durham, North Carolina, USA.
Lauren Komarow, Biostatistics Center, Department of Biostatistics and Bioinformatics, George Washington University, Rockville, Maryland, USA.
Holly S Geres, Duke Clinical Research Institute, Duke University School of Medicine, Durham, North Carolina, USA.
Toshimitsu Hamasaki, Biostatistics Center, Department of Biostatistics and Bioinformatics, George Washington University, Rockville, Maryland, USA.
Henry F Chambers, Division of Infectious Diseases, Department of Medicine, University of California, San Francisco, California, USA.
Vance G Fowler, Jr, Duke Clinical Research Institute, Duke University School of Medicine, Durham, North Carolina, USA; Division of Infectious Diseases, Department of Medicine, Duke University School of Medicine, Durham, North Carolina, USA.
Scott R Evans, Biostatistics Center, Department of Biostatistics and Bioinformatics, George Washington University, Rockville, Maryland, USA.
Robin Patel, Division of Clinical Microbiology, Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, Minnesota, USA; Division of Public Health, Infectious Diseases, and Occupational Medicine, Department of Medicine, Mayo Clinic, Rochester, Minnesota, USA.
Notes
Author contributions. Concept and design: H. R. C. Acquisition, analysis, or interpretation of data: H. R. C. and M. S. Drafting of the manuscript: H. R. C., K. E. G. Q., and S. R. E. Critical revision of the manuscript for important intellectual content: H. F. C. and V. G. F. Statistical analysis: L. K., T. H., and S. R. E. Obtained funding: H. R. C., H. F. C., and V. G. F. Administrative, technical, or material support: K. E. G. Q., M. S., and H. S. G. Supervision: H. R. C., H. F. C., V. G. F., S. R. E., and R. P.
Acknowledgments. The authors thank all the faculty and staff of the Scientific Leadership, Clinical Operations, Laboratory, and Statistics and Data Management Centers for their tireless efforts making the Antibacterial Resistance Leadership Group program, and its constituent studies and trials, possible.
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (grant UM1AI104681).
Supplement sponsorship. This article appears as part of the supplement “The Antibacterial Resistance Leadership Group (ARLG): Innovation and Evolution,” sponsored by the Antibacterial Resistance Leadership Group.
References
- 1. Chambers HF, Evans SR, Patel R, et al. Antibacterial Resistance Leadership Group 2.0: back to business. Clin Infect Dis 2021; 73:730–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Evans SR, Patel R, Hamasaki T, et al. The future ain’t what it used to be…out with the old…in with the better: Antibacterial Resistance Leadership Group (ARLG) innovations. Clin Infect Dis 2023; 77:321–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Harris AD, Souli M, Pettigrew MM; for the Antibacterial Resistance Leadership Group. The next generation: mentoring and diversity in the Antibacterial Resistance Leadership Group.Clin Infect Dis 2023; 77:331–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Antibacterial Resistance Leadership Group. Available at: https://arlg.org. Accessed 14 June 2023.
- 5. Suh GA, Lodise TP, Tamma PD, et al. Considerations for the use of phage therapy in clinical practice. Antimicrob Agents Chemother 2022; 66:e0207121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Howard-Anderson J, Hamasaki T, Dai W, et al. Improving traditional registrational trial end points: development and application of a desirability of outcome ranking end point for complicated urinary tract infection clinical trials. Clin Infect Dis 2023; 76:e1157–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. King HA, Doernberg SB, Miller J, et al. Patients’ experiences with Staphylococcus aureus and gram-negative bacterial bloodstream infections: a qualitative descriptive study and concept elicitation phase to inform measurement of patient-reported quality of life. Clin Infect Dis 2021; 73:237–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Doernberg SB, Komarow L, Tran TTT, et al. Simultaneous evaluation of diagnostic assays for pharyngeal and rectal Neisseria gonorrhoeae and Chlamydia trachomatis using a Master Protocol. Clin Infect Dis 2020; 71:2314–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lodise TP, Rosenkranz SL, Finnemeyer M, et al. The emperor's new clothes: prospective observational evaluation of the association between initial vancomycin exposure and failure rates among adult hospitalized patients with methicillin-resistant Staphylococcus aureus bloodstream infections (PROVIDE). Clin Infect Dis 2020; 70:1536–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. van Duin D, Perez F, Rudin SD, et al. Surveillance of carbapenem-resistant Klebsiella pneumoniae: tracking molecular epidemiology and outcomes through a regional network. Antimicrob Agents Chemother 2014; 58:4035–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van Duin D, Arias CA, Komarow L, et al. Molecular and clinical epidemiology of carbapenem-resistant Enterobacterales in the USA (CRACKLE-2): a prospective cohort study. Lancet Infect Dis [published correction appears in Lancet Infect Dis 2020; 20:e116]2020; 20:731–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang M, Earley M, Chen L, et al. Clinical outcomes and bacterial characteristics of carbapenem-resistant Klebsiella pneumoniae complex among patients from different global regions (CRACKLE-2): a prospective, multicentre, cohort study. Lancet Infect Dis 2022; 22:401–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Reyes J, Komarow L, Chen L, et al. Global epidemiology and clinical outcomes of carbapenem-resistant Pseudomonas aeruginosa and associated carbapenemases (POP): a prospective cohort study. Lancet Microbe 2023; 4:e159–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tamma PD, Komarow L, Ge L, et al. Clinical impact of ceftriaxone resistance in Escherichia coli bloodstream infections: a multicenter prospective cohort study. Open Forum Infect Dis 2022; 9:ofac572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bergin SP, Coles A, Calvert SB, et al. PROPHETIC: Prospective Identification of Pneumonia in Hospitalized Patients in the ICU. Chest 2020; 158:2370–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hanson KE, Banerjee R, Doernberg SB, et al. Priorities and progress in diagnostic research by the Antibacterial Resistance Leadership Group. Clin Infect Dis 2023; 77:314–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van Duin D, Gu P, Dong J, et al. China–United States research collaborations in antimicrobial resistance. Clin Infect Dis [published correction appears in Clin Infect Dis 2019; 68:1254]2018; 67(suppl 2):S142–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tamma PD, Souli M, Billard M, et al. Safety and microbiological activity of phage therapy in persons with cystic fibrosis colonized with Pseudomonas aeruginosa: study protocol for a phase 1b/2, multicenter, randomized, double-blind, placebo-controlled trial. Trials 2022; 23:1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Patel R, Polage CR, Dien Bard J, et al. Envisioning future urinary tract infection diagnostics. Clin Infect Dis 2022; 74:1284–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Simner PJ, Dien Bard J, Doern C, et al. Reporting of antimicrobial resistance from blood cultures, an Antibacterial Resistance Leadership Group survey summary: resistance marker reporting practices from positive blood cultures. Clin Infect Dis 2023; 76:1550–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Banerjee R, Teng CB, Cunningham SA, et al. Randomized trial of rapid multiplex polymerase chain reaction-based blood culture identification and susceptibility testing. Clin Infect Dis 2015; 61:1071–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Banerjee R, Komarow L, Virk A, et al. Randomized trial evaluating clinical impact of rapid identification and susceptibility testing for gram-negative bacteremia: RAPIDS-GN. Clin Infect Dis 2021; 73:e39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Patel R, Tsalik EL, Petzold E, et al. MASTERMIND: bringing microbial diagnostics to the clinic. Clin Infect Dis 2017; 64:355–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. GENOmics Sequencing-based Typing, EpidemioLogy, Linkage, and Antimicrobial Resistance tool. Available at: https://genostellar.net. Accessed 14 June 2023.
- 25. ARLG Laboratory Center. Virtual biorepository—strain catalogue. Available at: https://www.arlgcatalogue.org/arlgCatalogue. Accessed 14 June 2023.
- 26. Huvane J, Komarow L, Hill C, et al. Fundamentals and catalytic innovation: the statistical and data management center of the Antibacterial Resistance Leadership Group. Clin Infect Dis 2017; 64(suppl 1):S18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Evans SR, Rubin D, Follmann D, et al. Desirability of outcome ranking (DOOR) and response adjusted for duration of antibiotic risk (RADAR). Clin Infect Dis [published correction appears in Clin Infect Dis 2023 ; 76:182]2015; 61:800–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Evans SR, Follmann D. Using outcomes to analyze patients rather than patients to analyze outcomes: a step toward pragmatism in benefit:risk evaluation. Stat Biopharm Res 2016; 8:386–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Evans SR, Follmann D, Liu Y, et al. Sequential, multiple-assignment, randomized trials for COMparing Personalized Antibiotic StrategieS (SMART-COMPASS). Clin Infect Dis 2019; 68:1961–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Turner NA, Zaharoff S, King H, et al. Dalbavancin as an option for treatment of S. aureus bacteremia (DOTS): study protocol for a phase 2b, multicenter, randomized, open-label clinical trial. Trials 2022; 23:407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kinamon T, Gopinath R, Waack U, et al. Exploration of a potential desirability of outcome ranking endpoint for complicated intra-abdominal infections using 9 registrational trials for antibacterial drugs. Clin Infect Dis 2023; 77;649–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Evans SR, Pennello G, Pantoja-Galicia N, et al. Benefit-Risk Evaluation for Diagnostics: A Framework (BED-FRAME). Clin Infect Dis 2016; 63:812–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pennello G, Pantoja-Galicia N, Evans S. Comparing diagnostic tests on benefit-risk. J Biopharm Stat 2016; 26:1083–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu Y, Tsalik EL, Jiang Y, et al. Average weighted accuracy: pragmatic analysis for a Rapid Diagnostics in Categorizing Acute Lung Infections (RADICAL) study. Clin Infect Dis 2020; 70:2736–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jiang Y, Pan Q, Liu Y, Evans S. A statistical review: why average weighted accuracy, not accuracy or AUC? Biostat Epidemiol 2021; 5:267–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Evans SR, Hujer AM, Jiang H, et al. Rapid molecular diagnostics, antibiotic treatment decisions, and developing approaches to inform empiric therapy: PRIMERS I and II. Clin Infect Dis 2016; 62:181–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Evans SR, Tran TTT, Hujer AM, et al. Rapid molecular diagnostics to inform empiric use of ceftazidime/avibactam and ceftolozane/tazobactam against Pseudomonas aeruginosa: PRIMERS IV. Clin Infect Dis 2019; 68:1823–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Evans SR, Hujer AM, Jiang H, et al. Informing antibiotic treatment decisions: evaluating rapid molecular diagnostics to identify susceptibility and resistance to carbapenems against Acinetobacter spp. n PRIMERS III. J Clin Microbiol [published correction appears in J Clin Microbiol 2017; 55:1970]2016; 55:134–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wilson BM, Jiang Y, Jump RLP, et al. Desirability of outcome ranking for the management of antimicrobial therapy (DOOR MAT): a framework for assessing antibiotic selection strategies in the presence of drug resistance. Clin Infect Dis 2021; 73:344–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. DOOR analyses: standard edition. Available at: https://methods.bsc.gwu.edu. Accessed 14 June 2023.
- 41. Evans SR, Harris AD. Methods and issues in studies of CRE. Virulence 2017; 8:453–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huskins WC, Fowler VG Jr, Evans S. Adaptive designs for clinical trials: application to healthcare epidemiology research. Clin Infect Dis 2018; 66:1140–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Evans SR, Follmann D. Comment: fundamentals and innovation in antibiotic trials. Stat Biopharm Res 2015; 7:331–6. [DOI] [PMC free article] [PubMed] [Google Scholar]