Summary:
Apical extracellular matrix (aECM) constitutes the interface between every tissue and the outside world. It is patterned into diverse tissue-specific structures through unknown mechanisms. Here, we show that a male-specific genetic switch in a single C. elegans glial cell patterns the overlying aECM from a solid sheet to a ~200 nm pore, thus allowing a male sensory neuron to access the environment. Using cell-specific genetic sex-reversal, we find that this switch reflects an inherent sex difference in the glial cell that is independent of the sex identity of the surrounding neurons. Through candidate and unbiased genetic screens, we find that this glial sex difference is controlled by factors shared with neurons (mab-3, lep-2, lep-5) as well as previously unidentified regulators whose effects may be glia-specific (nfya-1, bed-3, jmjd-3.1). The switch results in male-specific glial expression of a secreted Hedgehog-related protein, GRL-18, that we discover localizes to transient nanoscale rings at sites where aECM pores will form. Using electron microscopy, we find that blocking male-specific gene expression in glia prevents pore formation, whereas forcing male-specific glial gene expression induces an ectopic pore. Thus, a switch in gene expression in a single cell is necessary and sufficient to pattern aECM into a specific structure. Our results highlight that aECM is not a simple homogenous meshwork, but instead is composed of discrete local features that reflect the identity of the underlying cells.
Keywords: extracellular matrix, glia, sex differences, C. elegans, cuticle, aECM
eTOC Blurb:
Apical extracellular matrix (aECM) is patterned into tissue-specific nanoscale structures. In C. elegans males, the cuticle aECM forms pores to accommodate sensory neurons used in mating. Fung et al. identify a male-specific switch in gene expression in a glial cell that is necessary and sufficient to induce the formation of these aECM pores.
Graphical Abstract
INTRODUCTION
The apical extracellular matrix (aECM) is a conserved, intricate network of secreted macromolecules that lines the outward or luminal-facing surfaces of epithelial tissues, including the lungs, vasculature, and gut, as well as sense organs like the olfactory epithelium and inner ear1–5. The aECM is often viewed as a static protective barrier, but growing evidence indicates that it is dynamic during development, diverse in composition and form, and plays crucial roles in tissue morphogenesis6. In addition to providing a defense against pathogens and desiccation, distinct aECM structures establish and maintain epithelial tubes, control the function of sense organs by forming nanopores for odor reception or resonating membranes for hearing, and shape organs through the distribution of tensile forces1–5,7,8. The structure of aECM is thus tailored to the needs of a particular tissue or organ. However, the mechanisms that pattern aECM remain largely mysterious.
The C. elegans cuticle is an aECM layer that coats the surface of the animal and provides a model of aECM patterning. As animals transition through four juvenile (larval) stages to the adult stage, they undergo molts in which they shed the old cuticle and form a new cuticle that is specialized for each developmental stage9. We have focused on intricate aECM specializations that are associated with sense organs in the head, midbody, and tail10. Each sense organ contains one or more sensory neurons with long unbranched dendrites that terminate in sensory cilia, as well as two glial cells called the sheath and socket. The sheath glial cell wraps the dendrite endings, while the socket glial cell forms a cellular pore in the skin through which neuronal sensory cilia protrude. This cellular pore is overlaid by cuticle aECM that takes the form of either a closed sheet or an open pore, depending on the function of the sense organ10 (Figure 1A–1B). A closed sheet of aECM is found overlying sense organs that contain a single mechanosensory neuron, whose cilium is embedded directly in the cuticle to sense external forces10 (Figure 1B, left). By comparison, open pores in the cuticle are found at bifunctional sense organs that contain both a mechanosensory and a chemosensory neuron10 (Figure 1B, right). In these sense organs, the chemosensory cilium protrudes through the open pore in the cuticle in order to directly access chemical cues in the external environment10.
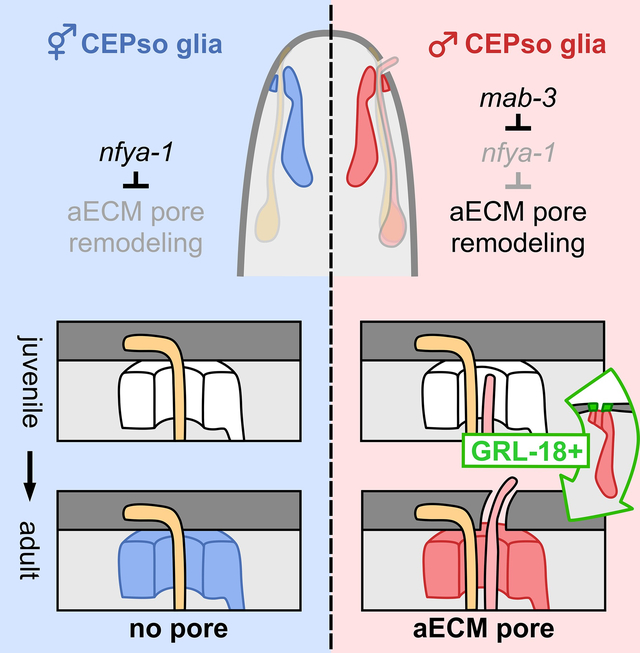
Figure 1. Glia initiate male-specific gene expression at sexual maturity.
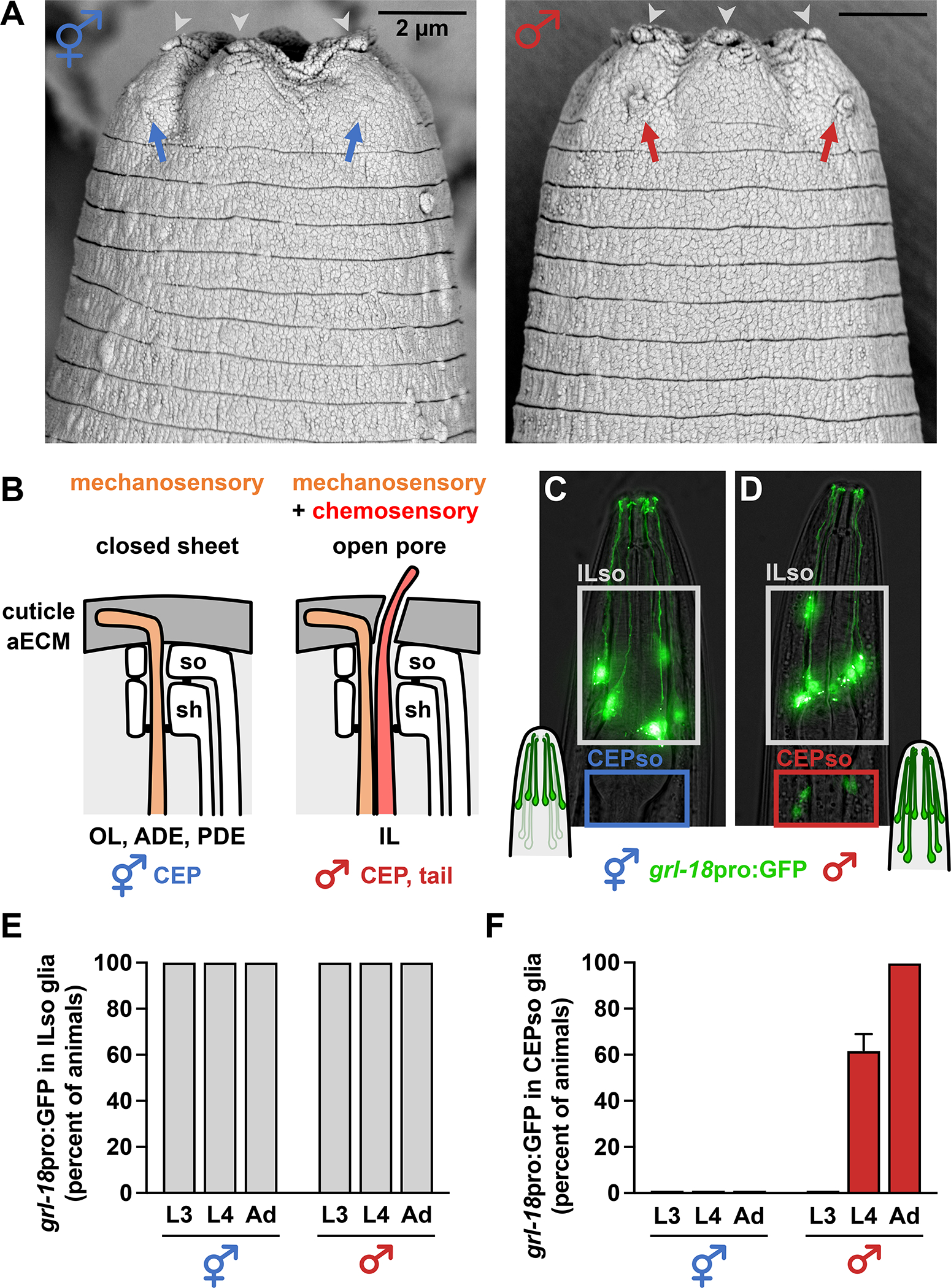
(A) Scanning electron micrographs of the heads of an adult hermaphrodite (left) and male (right). Nose tips are at top. Chemosensory cilia protrude through open cuticle pores associated with the six inner labial (IL) sense organs in both sexes (gray arrowheads; three on each side of the head) and four cephalic (CEP) sense organs in the male (right; red arrows; two on each side of the head). Mechanosensory cilia of the hermaphrodite CEP sense organs are embedded beneath a closed sheet of cuticle and appear as a bump under the surface, with no pore (left; blue arrows). (B) Schematic of C. elegans sense organs showing socket (so) and sheath (sh) glial cells wrapping neuronal endings. Cuticle aECM forms a closed sheet (left) over sense organs with a single mechanosensory neuron (outer labial, OL; deirids, ADE/PDE) or a ~200 nm open pore (right) over sense organs with a mechanosensory and chemosensory neuron pair (inner labial, IL; rays and hook of male tail). Importantly, cuticle aECM of the CEP sense organ undergoes sex-specific remodeling from a closed sheet in hermaphrodites to an open pore in adult males. (C, D) Expression of grl-18pro:GFP in (C) hermaphrodite and (D) male head glia. Nose is up. (E, F) Fraction of hermaphrodites (n=48) and males (n=50) expressing grl-18pro:GFP at third and fourth larval stages (L3, L4) and the 1-day adult stage (Ad) in (E) ILso glia and (F) CEPso glia. The same animals were followed and scored at each stage. Error bars, standard error of the mean (SEM). See also Figure S1.
To better understand how specialized aECM is patterned, we focused on a discrete sex-specific aECM remodeling event associated with the four cephalic (CEP) sense organs in the head. In hermaphrodites, each CEP sense organ contains only a single mechanosensory neuron called CEP and the overlying cuticle forms a closed sheet (Figure 1A, left, blue arrows; Figure 1B, orange, CEP). By contrast, in adult males, each CEP sense organ contains an additional chemosensory neuron called CEM and the overlying cuticle forms a ~200 nm open pore through which the CEM cilium protrudes to detect pheromones from mating partners10–13 and to release extracellular vesicles thought to mediate social communication14 (Figure 1A; right, red arrows; Figure 1B, red, CEM). The CEM neurons undergo embryonic apoptosis in hermaphrodites15 but, in males, they differentiate and form a mature sensory cilium at the onset of sexual maturation in the fourth larval (L4) stage16. Thus, the cuticle remodels from a closed sheet to an open pore during the L4/adult molt in males. It was previously inferred that “the openings in males must be created by the CEM dendrites,”17 because they appeared to be the only cells that differ between the sexes in this sense organ. Surprisingly, we find that one of the glial cells also differs between the sexes, switching on a gene expression program at the L4/adult molt only in males. Remarkably, this glial switch is necessary and sufficient to create the cuticle pore. Overall, we identify a novel sexual dimorphism in a single glial cell, define its upstream regulators, and show that a switch in gene expression in a single cell is enough to induce dramatic remodeling of the aECM.
RESULTS
Glia undergo a sex-specific switch in gene expression
We serendipitously found that CEP socket (CEPso) glia undergo a sex-specific switch in gene expression. Previously, we showed that a 2968-bp transcriptional reporter for the putative secreted protein GRL-18 (grl-18pro:GFP, Figure S1A) is a cell-type specific marker for IL socket (ILso) glia in hermaphrodites (Figure 1C)18,19. ILso glia are associated with a ‘mechanosensory + chemosensory’ organ, with chemosensory cilia protruding through an open pore in the cuticle (Fig. 1A, gray arrowheads). Using this transgene or a reporter engineered at the endogenous locus (grl-18-SL2-YFP:H2B, Figure S1F), we observed expression in ILso glia in both sexes throughout life, as well as in sex-specific reproductive structures – the hermaphrodite vulval epithelial cells and male tail glia – shortly after these cells are born at the L4 stage (Figures 1C–1E, Figures S1B–S1J). Strikingly, we also observed expression in CEPso glia in L4 and adult males, but never in hermaphrodites, indicating that these glia are sexually dimorphic (Figures 1C–1D and 1F, Figures S1G and S1I). Thus, CEPso glia undergo a sex-specific switch in gene expression that coincides with remodeling of the cuticle aECM from a sheet to a pore.
While sexual dimorphism is well-established in C. elegans neurons, only two examples have been reported in glia, both involving the production of male-specific neurons rather than altered function of the glial cell itself20,21. CEPso glia develop embryonically, but our reporter suggests that a sexually dimorphic switch in gene expression occurs specifically at the L4 stage. Due to possible perdurance of the fluorescent reporter, we cannot infer whether this switch is transient or persists throughout adulthood. Notably, there is a nearly perfect correspondence between the glia that express this reporter and those that are associated with narrow cuticle pores (ILso, CEPso, male tail glia)10,17,22, suggesting a shared gene expression program in these glial types.
The sex-specific switch in gene expression is controlled cell autonomously in glia
Sexually dimorphic CEPso glial gene expression could be induced cell autonomously or non-cell autonomously, for example via male-specific signals from CEM or other neurons. To distinguish these possibilities, we genetically altered the sex identity of neurons or glia through cell-type-specific mis-expression of the masculinizing factor fem-3 or the feminizing factor tra-2(IC) from the sex determination pathway of C. elegans, as previously described12,20,23–25 (Figures 2A and 2D). Neurons and glia were targeted using a pan-neuronal promoter (rab-3pro)12, a pan-glial promoter (mir-228pro)26, and a newly identified CEPso-specific promoter (col-56pro, Figure S2). We found that masculinization of glia induced inappropriate grl-18 expression in hermaphrodite CEPso glia, whereas feminization of glia blocked grl-18 expression in male CEPso glia (Figures 2B and 2E). In contrast, sex-reversal of neurons had no effect (Figures 2B and 2E).
Figure 2. The sex-specific switch in gene expression is controlled cell autonomously in glia and does not depend on male neurons.
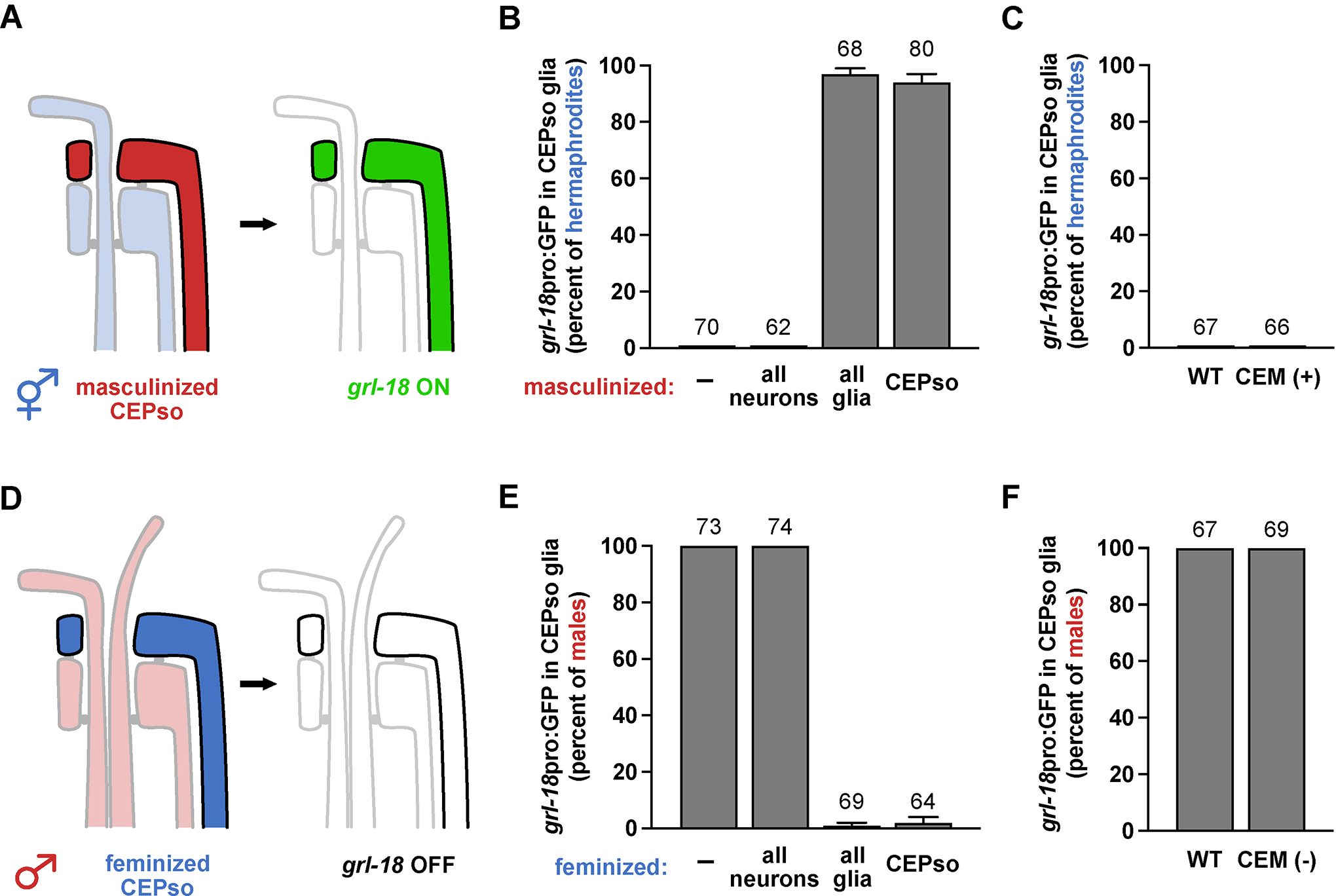
(A-B, D-E) Neurons or glia were (A) masculinized in hermaphrodites or (D) feminized in males via mis-expression of fem-3 or tra-2(IC), respectively, using cell-type-specific promoters (all neurons, rab-3; all glia, mir-228; CEPso glia, col-56, see Figure S2). Fraction of 1-day adult (B) hermaphrodites or (E) males that express grl-18pro:GFP in CEPso glia is shown. (C, F) To test if CEM neurons are sufficient and/or necessary for the switch, grl-18pro:GFP expression was evaluated in (C) ceh-30(n3714gf) hermaphrodites in which CEM neurons inappropriately survive (CEM+), and (F) in ceh-30(n4289lf) males in which CEM neurons inappropriately undergo apoptosis (CEM−)27. Fraction of 1-day adult (C) CEM+ hermaphrodites and (F) CEM− males that express grl-18pro:GFP in CEPso glia. Sample sizes are indicated above the bars of each graph. Error bars, SEM.
We also directly tested whether CEM neurons contribute to the change in glial gene expression by taking advantage of ceh-30 loss-of-function (lf) or gain-of-function (gf) mutants in which CEM neurons inappropriately die in males or survive in hermaphrodites, respectively. ceh-30 encodes a Bar family homeodomain transcription factor that is normally expressed in male CEM neurons and promotes their survival27,28. In the ceh-30(lf) mutant, most of the homeodomain sequence is deleted and the transcription factor is non-functional; whereas in the ceh-30(gf) mutant, a cis-regulatory binding site for the sex determination factor TRA-1 is disrupted, leading to mis-expression of active CEH-30 protein in hermaphrodites27. We observed that grl-18 expression in CEPso glia remains unaffected in ceh-30(lf) males and ceh-30(gf) hermaphrodites (Figures 2C and 2F), showing that CEM neurons are neither necessary nor sufficient for the switch in glial gene expression.
Together, these results suggest that sexually dimorphic gene expression in the glia is controlled cell autonomously by the sex identity of the glial cell itself.
Regulators of sex- and timing-dependent changes in glial gene expression
To identify the regulatory factors that control sexual dimorphism in glia, we used forward and candidate-based screens to isolate mutants that exhibit delayed or absent grl-18 expression in males (OFF mutants; Figure 3A, left), or inappropriate grl-18 expression in hermaphrodites (ON mutants; Figure 3A, right). We identified three classes of genes that regulate the sex-specificity and timing of glial gene expression (Table 1 and Table S1): DM domain transcription factors (class I), heterochronic genes (class II), and novel regulators (class III).
Figure 3. Sex-specific glial gene expression requires the NF-Y repressive complex acting in the glial cell downstream of MAB-3.
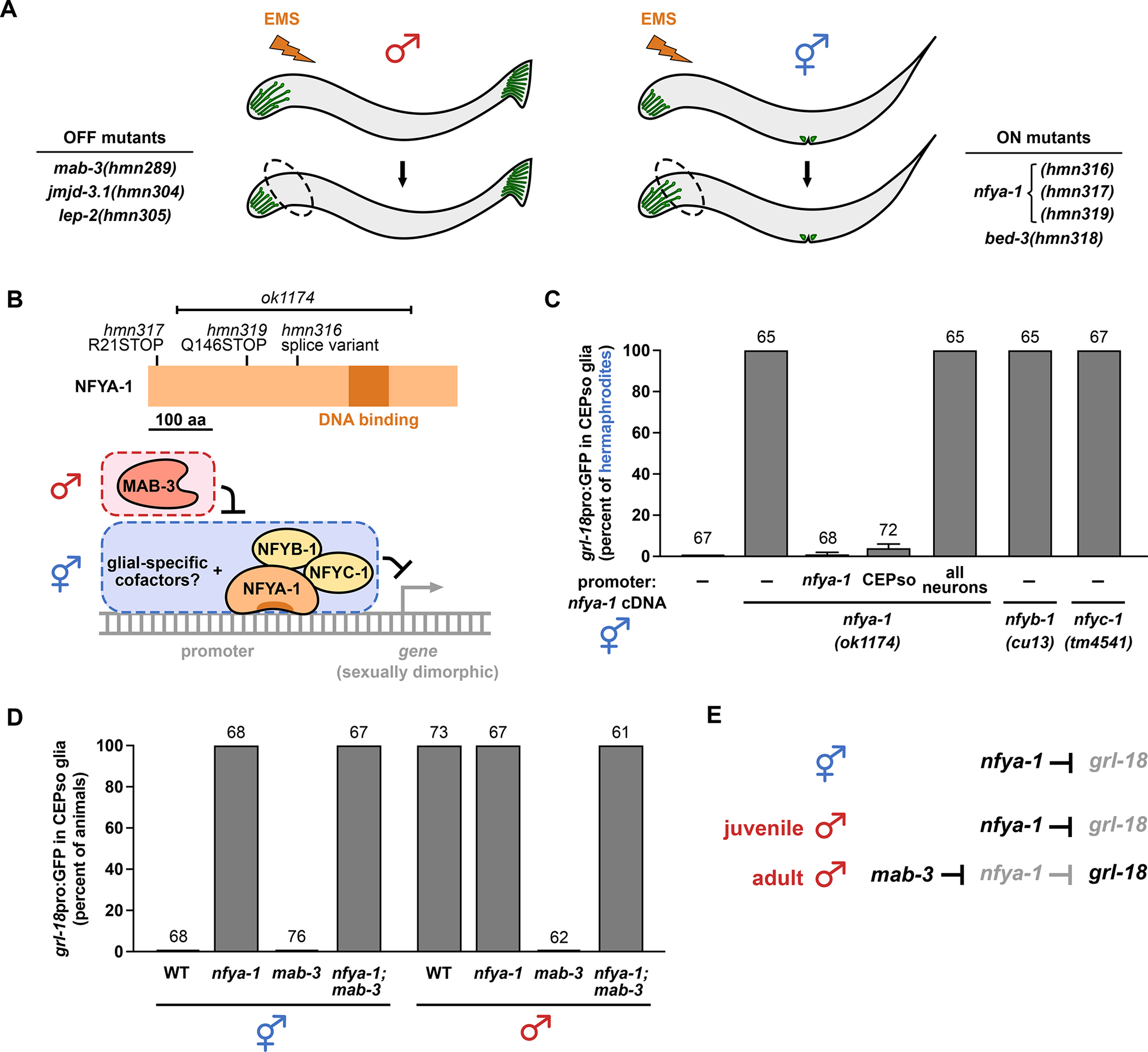
(A) Schematic of forward genetic screens in which animals expressing grl-18 reporters were mutagenized with ethyl methanesulfonate (EMS). Mutant males lacking grl-18 expression in CEPso glia (OFF mutants, left) and mutant hermaphrodites exhibiting inappropriate grl-18 expression in CEPso glia (ON mutants, right) were isolated, and the causal mutations were identified (see also Table 1 and STAR Methods). (B) Schematic of NFYA-1 protein showing effects of novel alleles (hmn316, hmn317, hmn319) and an existing 1837 bp deletion (ok1174). Working model of the NF-Y repressor complex (NFYA-1, NFYB-1, NFYC-1) acting with glial-specific cofactors to inhibit male-specific gene expression, subject to de-repression by MAB-3. (C) Fraction of 1-day adult wild-type or nfya-1, nfyb-1, or nfyc-1 mutant hermaphrodites that express grl-18pro:GFP in CEPso glia. nfya-1 mutant is shown alone (−) or with rescuing nfya-1 cDNA under control of nfya-1, CEPso-specific (col-56), or pan-neuronal (rab-3) promoters. See Figure S3 for quantification of other known sex differences in neurons and glia in nfya-1 mutants. (D) Epistasis analysis showing fraction of 1-day adult mab-3(mu15), nfya-1(ok1174), or mab-3(mu15); nfya-1(ok1174) mutants of each sex that express grl-18pro:GFP in CEPso glia. (E) Genetic model of grl-18 regulation in CEPso glia. Sample sizes are above the bars of each graph. Error bars, SEM.
Table 1. Regulators of sex-specific glial gene expression.
Percent of males (I, II, III(i)) or hermaphrodites (III(ii)) at the indicated adult age that express grl-18pro:mApple or grl-18pro:GFP in CEPso glia. Expression in ILso glia and male tail glia were unaffected in all cases. Based on the sequence changes involved, the hmn304 and gk384 alleles of jmjd-3.1, the tm4264 allele of nfyc-1, and the hmn318, sy705, and gk996 alleles of bed-3 are unlikely to be nulls. See also Table S1.
| Genotype | Age | grl-18+ CEPso glia (percent of animals) | n |
|---|---|---|---|
|
| |||
| ♂ I. DM Domain Transcription Factors | |||
| wild type males | 1d | 100 | 67 |
| mab-3(hmn289) | 1d | 0 | 71 |
| mab-3(mu15) | 1d | 0 | 66 |
|
| |||
| ♂ II. Heterochronic Genes | |||
| wild type males | 1d | 100 | 42 |
| 2d | 100 | ||
| 3d | 100 | ||
| 4d | 100 | ||
| lep-2(hmn305) | 1d | 0 | 45 |
| 2d | 0 | ||
| 3d | 58 | ||
| 4d | 82 | ||
| lep-2(ok900) | 1d | 0 | 47 |
| 2d | 13 | ||
| 3d | 26 | ||
| 4d | 32 | ||
| lep-5(ny28) | 1d | 0 | 44 |
| 2d | 0 | ||
| 3d | 64 | ||
| 4d | 77 | ||
|
| |||
| III. Novel Regulators | |||
| ♂ i. Positive Regulators - Promote Expression | |||
| wild type males | 1d | 100 | 64 |
| jmjd-3.1(hmn304) | 1d | 3a | 61 |
| jmjd-3.1(gk384) | 1d | 15a | 60 |
| ⚥ ii. Negative Regulators - Inhibit Expression | |||
| wild type hermaphrodites | 1d | 0 | 66 |
| nfya-1(hmn316) | 1d | 100 | 66 |
| nfya-1(hmn317) | 1d | 100 | 60 |
| nfya-1(hmn319) | 1d | 100 | 60 |
| nfya-1(ok1174) | 1d | 100 | 68 |
| nfyb-1(cu13) | 1d | 100 | 65 |
| nfyb-1(tm4257) | 1d | 100 | 68 |
| nfyc-1(tm4541) | 1d | 100 | 67 |
| nfyc-1(tm4264) | 1d | 100b | 67 |
| bed-3(hmn318) | 1d | 100b | 65 |
| bed-3(sy705) | 1d | 76b | 68 |
| bed-3(gk996) | 1d | 87b | 61 |
phenotype is partially penetrant; at least one CEPso fails to express marker
phenotype is partially penetrant; at least one CEPso expresses marker
Class I – We determined that sex-specificity is controlled by the DM domain transcription factor mab-3/DMRT, a conserved protein known to establish sexual dimorphism in vertebrates and invertebrates29–31. In C. elegans, mab-3 is required for promoting male sexual differentiation in neurons and hypodermal cells of the tail32–36. From a screen for mutant males lacking grl-18 expression in CEPso glia (Figure 3A, left), we isolated a R90C mutation in the second DNA-binding domain of MAB-3 (hmn289) and confirmed this phenotype using a previously characterized mab-3 null allele (mu15)30. This phenotype is fully penetrant (Table 1). Mutations in all other DM domain transcription factors of C. elegans were also assessed (except dmd-4, which is lethal) and found to have no effect (Table S1). This result shows that sex identity in glia is controlled by one of the same upstream regulators that is used in neurons33,34,36.
Class II – Heterochronic genes are conserved regulatory factors that control developmental timing and sexual development37–40. We found that male-specific grl-18 expression is absent or delayed in mutants lacking the timing factors lep-2/Makorin41 and lep-5, a long non-coding RNA42 (Table 1). In the screen described above (Figure 3A, left), we isolated a new allele of lep-2(hmn305) that exhibits the same delayed expression phenotype (Table 1). This result shows that the onset of sexual maturation in glia is triggered by the same timing factors that are used in neurons39.
Class III – Finally, we identified novel regulators of sex-specific gene expression in glia that had not been identified in studies of neuronal sex differences. Mutations that disrupt the histone-modifier jmjd-3.1 and the transcription factor bed-3 result in incompletely penetrant defects, where jmjd-3.1 mutants fail to initiate grl-18 expression in male CEPso glia and bed-3 mutants inappropriately initiate grl-18 expression in hermaphrodite CEPso glia (Table 1). Neither jmjd-3.1 or bed-3 has been previously implicated in sexual differentiation of the nervous system, although bed-3 is required for sex-specific development of vulval cells43,44. Most notably, we isolated three alleles of nfya-1 (hmn316, hmn317, hmn319) that result in inappropriate male-like expression of grl-18 in hermaphrodite CEPso glia (Figure 3A, right and Table 1). The phenotype is completely penetrant in all three alleles, as well as in an existing allele (ok1174) (Table 1). Together, these results suggest that sex identity in glia is controlled by genes not previously implicated in sex identity in neurons.
NFYA-1 is a repressor that acts downstream of MAB-3
We chose to focus on nfya-1 because it showed the strongest phenotype. In C. elegans, loss of nfya-1 has previously been shown to affect vulva and male tail morphogenesis, sensory neuron specification, and expression of specific cell fate transcription factors45–48. NFYA-1/NFY-A is a subunit of the conserved, ubiquitously expressed trimeric repressor complex NF-Y/CBF. It associates with the NFYB-1/NFYC-1 dimer and contains the DNA-binding domain of the regulatory complex45,49. All four fully penetrant alleles of nfya-1 carry mutations that are predicted to generate a truncated protein lacking the DNA-binding domain (Figure 3B). The nfya-1 phenotype is rescued when nfya-1 cDNA is expressed using the CEPso-specific promoter, demonstrating that nfya-1 acts cell autonomously to prevent male gene expression in CEPso glia (Figure 3C). As a control, we confirmed that pan-neuronal expression of nfya-1 cDNA does not rescue inappropriate grl-18 expression in nfya-1 mutants (Figure 3C). nfyb-1 and nfyc-1 mutants exhibit the same completely penetrant phenotype as nfya-1 mutants, suggesting the entire complex is required for regulating sex-specific glial gene expression (Figure 3C and Table 1).
We considered the possibility that inappropriate expression of grl-18 in these mutants simply reflects a general defect in transcriptional repression. However, two lines of evidence argue against this. First, we found that loss of nfya-1 does not affect known sex differences in neurons50,51 or other glia20 (Figure S3). Second, while nfya-1, nfyb-1, or nfyc-1 mutant hermaphrodites inappropriately express grl-18 in CEPso glia, they do so only at sexual maturity in the L4/adult stages, suggesting that repression of this gene in juveniles remains intact. Together, these results suggest that the phenotype we observe is specific to loss of sex identity of the CEPso glia.
Interestingly, although both MAB-3 and NFYA-1 are transcriptional repressors, they act in opposite directions to regulate glial gene expression: mab-3 promotes the switch to male-specific expression, whereas nfya-1 prevents this switch. To place them in a genetic pathway, we performed epistasis analysis. We observed that mab-3; nfya-1 males switch on grl-18 with complete penetrance, showing that loss of nfya-1 fully bypasses the requirement for mab-3 (Figure 3D). This leads to the model that, in hermaphrodites and juvenile males, NFYA-1 prevents grl-18 expression in CEPso glia, whereas in sexually mature males, MAB-3 switches off NFYA-1-dependent repression, thus allowing grl-18 to be expressed (Figure 3B and 3E). This model therefore places NFYA-1 genetically downstream of MAB-3.
It is important to note that MAB-3 may not directly act on the NFYA-1 repressive complex (which is expressed broadly in many cell types45) but rather on glial-specific cofactors that may recruit NFYA-1 to cell-type-specific promoters (diagrammed in Figure 3B). Similarly, the NFYA-1 complex may not act directly on grl-18 but instead may control expression of an unidentified activator that then switches on a battery of male-specific glial genes, including grl-18. While it will be important to determine these mechanistic details, MAB-3 and NFYA-1 provide entry points to defining the genetic control of sexual dimorphism in glia.
GRL-18 is a novel aECM protein that localizes to transient nanoscale rings during cuticle patterning
To assess possible functions of the male-specific switch in CEPso glial gene expression, we examined localization of the GRL-18 protein. Using CRISPR/Cas9 genome editing, a sfGFP tag was inserted after the GRL-18 signal sequence at its endogenous locus (see STAR Methods and Table S3). We observed that sfGFP-GRL-18 protein localizes to distinctive rings at the nose tip, corresponding to the positions of the IL and male CEP sense organs (Figure 4A). Similar rings were seen at sense organ endings in the developing male tail (Figure 4B). Notably, each of these head and tail sense organs contains a single chemosensory neuron that protrudes through a narrow cuticle pore in the adult animal, suggesting that GRL-18 marks the sites of cuticle pores.
Figure 4. GRL-18 is a novel aECM protein that transiently localizes to nanoscale rings.
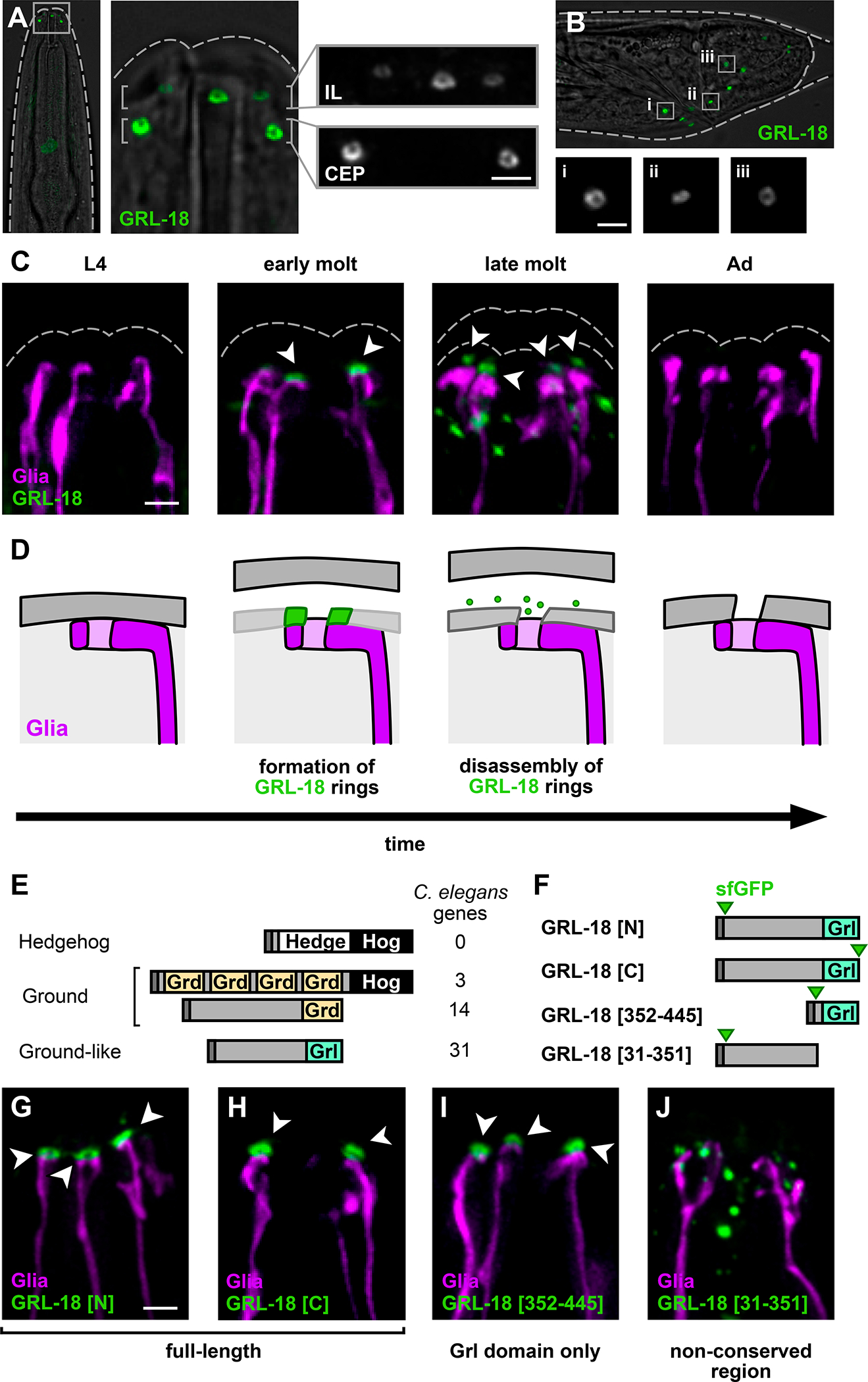
Endogenously-tagged full-length sfGFP-GRL-18 forms rings at the endings of the IL and CEP sense organs in the male head (refer to Figure 1A, right). Views are maximum intensity projections through half of the head nearest the objective, showing three of the six IL sense organs and two of the four CEP sense organs; dashed lines, outline of cuticle. (B) Endogenously-tagged full-length sfGFP-GRL-18 rings are also found at the ray sense organs of the developing male tail during the L4/adult mid-molt stage; dashed lines, outline of cuticle. Insets (i-iii), magnification of rings from the boxed regions. (C) Localization of endogenously-tagged full-length sfGFP-GRL-18 in males through the L4 to adult (Ad) transition, showing early and late molt stages (see Figure S4 for quantification of each localization pattern). Green, sfGFP-GRL-18; magenta, glial endings; dashed lines, outline of cuticle. Arrowheads, sfGFP-GRL-18 rings. (D) Schematic of localization and subsequent disassembly of GRL-18 during patterning of the new cuticle. Cartoons are meant as a guide to the fluorescence images, but the structure and relative positions of the glial ending and cuticle during molting remain unknown. (E) C. elegans gene families distantly related to Hedgehog include ground (grd) and ground-like (grl) genes. (F) Diagram of GRL-18 full-length proteins and fragments containing conserved C-terminal Grl domain [352–445] or non-conserved N-terminal region [31–351] expressed as low-copy transgenes under control of the grl-18 promoter. sfGFP was inserted at the GRL-18 N-terminus after amino acids 1–30 containing the predicted signal sequence (GRL-18[N], GRL-18[31–351]), as an N-terminal fusion preceded by amino acids 1–30 (GRL-18[352–445]), or at the C-terminus. Dark bars, signal sequence; green arrows, sfGFP. (G-J) Localization of sfGFP-GRL-18 at nose tips of L4/adult mid-molt males expressing (G) sfGFP-GRL-18, (H) GRL-18-sfGFP, (I) sfGFP-Grl domain [352–445], and (J) sfGFP-N-terminal region [31–351] transgenes. Green, sfGFP-GRL-18; magenta, glial endings. Arrowheads, sfGFP-GRL-18 rings. All scale bars, 2 μm.
We further determined that these sfGFP-GRL-18 rings are transient, forming adjacent to glial endings at the onset of the L4/adult molt and disassembling within ~1–4 hours as the newly synthesized cuticle is complete and shedding of the old cuticle begins (Figure 4C and Figure S4). The transient localization of GRL-18 is reminiscent of the provisional matrix formed by other C. elegans aECM proteins during molts to pattern the newly synthesized cuticle52. Provisional matrix proteins are thought to provide a temporary structural template or scaffold that is critical for patterning newly synthesized aECM, but they are not stably incorporated into the mature structure8,52–55. Together, our results suggest that GRL-18 is a novel aECM protein that localizes to transient rings at sites where cuticle pores will form (Figure 4D).
Next, we considered what sequences in GRL-18 are required for this localization. grl-18 belongs to a family of ground-like (grl) genes with distant homology to Hedgehog56–58 (Figure 4E). Specifically, Hedgehog has an N-terminal ‘Hedge’ signaling domain and a C-terminal ‘Hog’ catalytic domain that is removed prior to secretion. C. elegans lacks Hedgehog proteins but contains several gene families with a Hog domain, for example Groundhog (Grd) (Figure 4E; also Warthog, Quahog, and Hog, not shown). C. elegans also contains genes that encode a Grd or Grd-like (Grl) domain but lack a Hog domain (Figure 4E). grl-18 is one of 31 Grl domain-containing proteins in the genome, none of which have been assigned a function.
In contrast to the expectation that Grl proteins are diffusible Hedgehog-like signaling molecules, our finding that GRL-18 appears to be a structural aECM protein led us to consider the role of the Grl domain in its localization. To investigate this, we expressed low-copy transgenes consisting of full-length GRL-18 or fragments with or without the Grl domain (Figure 4F). We found that either N- or C-terminally tagged full-length GRL-18 localizes to rings consistent with the endogenously-tagged protein (Figure 4G and 4H; compare Figure 4C), suggesting GRL-18 is unlikely to undergo internal cleavage, consistent with its lack of a Hog domain. The 93-amino acid C-terminal Grl domain localized identically to the full-length protein (Figure 4F and 4I), whereas the non-conserved N-terminal region localized in diffuse puncta and was not observed in rings (Figure 4F and 4J). Localization of the Grl domain transgene is independent of the endogenous full-length GRL-18 protein (Figure S4C). This demonstrates that the conserved Grl domain mediates the localization of GRL-18 to nanoscale rings. It is intriguing to consider whether other Grl domain proteins are also aECM proteins, possibly reflecting an ancestral role for Hedgehog-related proteins in patterning aECM structures58.
Sex-specific changes in glial gene expression remodel the aECM
The male-specific expression of GRL-18 and its localization to cuticle rings led us to wonder if male-specific glial gene expression is required to pattern a cuticle pore for the CEM cilium. To test this possibility, first we assessed the trajectory of CEM chemosensory cilia in animals with genetically feminized CEPso glia. In wild-type males, CEM chemosensory cilia bend outwards and their tips poke past the cuticle, whereas CEP mechanosensory cilia bend inwards and track along the cuticle (Figures 5A and 5B). However, in males with feminized glia, the CEM endings fail to project outwards normally and instead appear to bend inwards, in the same direction as the CEP endings (Figures 5A and 5C), suggesting that cuticle pores may not have properly formed (Figure 5D).
Figure 5. Male-specific glial gene expression is necessary and sufficient to form an aECM pore.
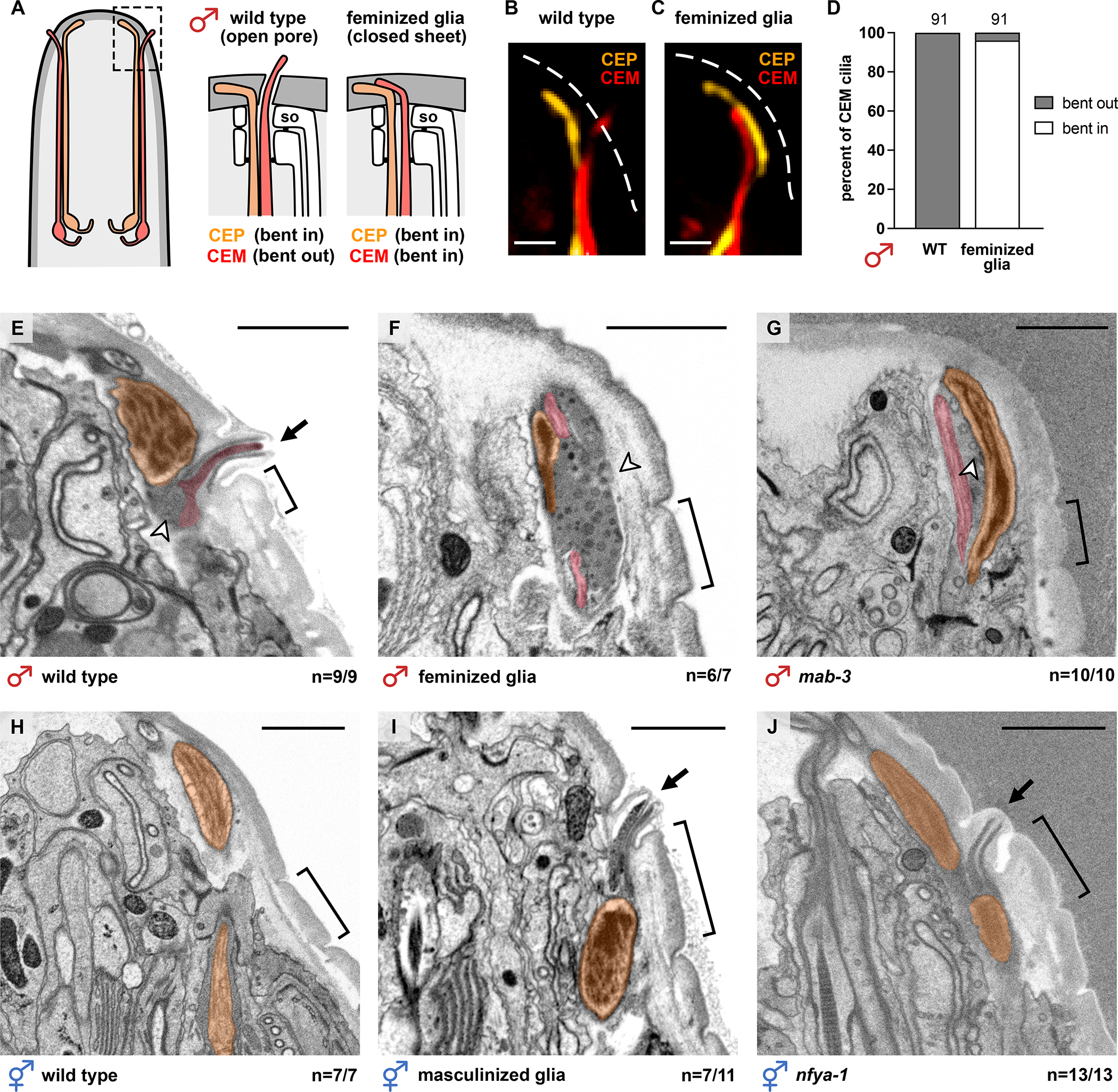
(A-C) Schematic and images of CEP (orange) and CEM (red) cilia in 1-day adult males, either (B) wild-type or (C) with feminized glia (col-56pro:tra-2(IC)). Dashed white lines, outline of cuticle; scale bars, 1 μm. (D) Binary scoring of CEM cilia trajectories. (E-J) Electron micrographs of longitudinal sections through single CEP sense organs in adult (E) wild-type male, (F) male with feminized glia, and (G) mab-3(mu15) mutant male, (H) wild-type hermaphrodite, (I) hermaphrodite with masculinized glia, and (J) nfya-1(ok1174) mutant hermaphrodite. First annulus (bracket) provides a landmark to locate the normal position of the pore. Arrows, cuticle pore; arrowheads, extracellular vesicles. All scale bars, 1 μm. The fraction of CEP sense organs exhibiting the represented phenotype out of the total number scored is shown. See also Figure S5.
Next, to directly test whether male-specific CEPso glial gene expression is required for cuticle pore formation, we used electron microscopy to visualize the aECM overlying the CEP sense organs of wild-type males and males that fail to express male-specific genes in their glia. We obtained longitudinal serial sections through the heads of wild-type males, males with feminized glia, and mab-3 mutant males at the adult stage (~40–100 serial sections per animal, 3–4 animals per genotype; see Figure S5E). CEM neurons were identified based on their position relative to the mechanosensory CEP neuron, their morphology, and the presence of extracellular vesicles in the space surrounding the CEM endings. In wild-type males, we observe that each CEM cilium protrudes through a pore that is consistently located anterior to the first annulus (circumferential ridge9) of the cuticle (Figures 5E and S5E, n=9/9). In males with feminized glia, the CEM cilia are misdirected inward, consistent with the fluorescent images, and pores are absent in most of the sense organs analyzed (Figures 5F and S5E, n=6/7). Similarly, in mab-3 mutant males, CEM cilia are misdirected inward and pores are absent (Figures 5G and S5E, n=10/10). We also observe that there is an overaccumulation of extracellular vesicles (EVs) in males with feminized glia and mab-3 mutant males, suggesting that the EVs are trapped inside the animal and cannot be released into the external environment14. Thus, male-specific gene expression in CEPso glia is necessary for cuticle pore formation.
To determine whether male-specific glial gene expression in CEPso glia is sufficient to remodel the aECM into an open pore, we obtained longitudinal serial sections of adult wild-type hermaphrodites and hermaphrodites with male glial gene expression (~30–100 serial sections per animal, 3–4 animals per genotype; see Figure S5F). Since hermaphrodites do not have CEM neurons, the mechanosensory CEP neuron was used as a landmark to locate the relevant serial sections to analyze (Figures 5H and S5F). Strikingly, hermaphrodites with masculinized glia often have ectopic cuticle pores positioned anterior to the first annulus (Figures 5I and S5F, n=7/11). Ectopic pores are also present in nfya-1 mutant hermaphrodites (Figures 5J and S5F, n=13/13). The pores are properly patterned despite the absence of the CEM neuron. This shows that forced expression of male-specific genes in CEPso glia is sufficient for driving cuticle pore formation.
To test if grl-18 itself is necessary or sufficient for cuticle pore formation, we examined grl-18 mutant males as well as hermaphrodites with forced expression of grl-18 in CEPso. In grl-18 mutant males, we find that the CEM cilia bend outwards normally, suggesting that pores are still formed (Figures S5A and S5B). Using EM, we also find that the CEM cilium protrudes through a cuticle pore, although a subtle alteration in the morphology of the pore is possible (Figures S5C and S5E). We also examined hermaphrodites with forced expression of grl-18 in CEPso and other socket glia, and did not observe ectopic pores (Figures S5D and S5F).
Together, these findings reveal that a sex-specific switch in glial gene expression – likely involving unidentified aECM proteins in addition to GRL-18 – remodels the aECM from a closed sheet to an open pore.
DISCUSSION
A long-standing framework for understanding the formation of biological structures distinguishes between “self-assembly,” where components come together without energy input to attain a stable, thermodynamically-favored conformation, and “self-organization,” where energy input is continuously required to actively maintain a non-equilibrium state59,60. However, a different approach may be needed to understand patterning of aECM structures. Although most aECM structures are highly stable – persisting even after death of the organism – previous studies show that the initial formation of these structures often requires extensive energy input and cellular activity. For example, in Drosophila, epidermal cells and olfactory hair cells form tooth-like denticles and peg-like extensions, respectively, that act as molds to sculpt newly secreted matrix proteins, and then retract as the aECM solidifies4,61. Actin networks also shape aECM pattern through contraction of the cell surface and localized secretion, as in alae of the C. elegans cuticle and taenidia of the Drosophila trachea, respectively52,62. Localized endocytosis has been proposed to promote the formation of cuticle nanopores in Drosophila olfactory sense organs4. Finally, cellular movements apply mechanical forces that can stretch or fold the aECM during tube morphogenesis7,8. In these examples, aECM structures are thought to be sculpted mechanically by active movements of the underlying cells. Similarly, it is possible that the male CEPso glial cell undergoes transient cellular rearrangements during cuticle synthesis that help to shape the aECM pore, analogous to what has been described in Drosophila4,61. Given our results that pore formation is controlled by a transcriptional switch, an intriguing alternative hypothesis is that secreted matrix molecules can “self-assemble” to form a closed sheet or open pore without major cellular rearrangements. In this case, understanding how changes in the biochemical composition of aECM can drive specific structural features might lead to strategies for manipulating matrix structure in vitro and in vivo, for example in tissue engineering, improved drug delivery, or treatment of aECM disorders.
Our work is consistent with recent evidence that a transient, or provisional, matrix initially patterns the mature aECM and is then removed8,52–55. We propose that GRL-18 is a transient matrix protein that forms temporary rings at sites of future cuticle pores, where it may act as part of a physical plug to keep the cuticle open or as a corral to retain pore-forming components in a restricted region. While GRL-18 by itself is neither necessary nor sufficient to form cuticle pores, it is likely to be part of a module of aECM proteins that are coordinately expressed in male CEPso glia to induce pore formation. The same aECM module may be expressed in other glia to form a cuticle pore around a single chemosensory neuron, including the ILso glia in both sexes and the ray structural cells in males. For example, we previously showed that transcriptional reporters for the collagen-encoding genes col-53 and col-177 are co-expressed in the same glia as grl-1819. Many Grl domain proteins and other Hedgehog-related proteins (Grd, Wrt, Qua domains) of C. elegans are expressed specifically in various socket glia or other cuticle-producing cells58. It is intriguing to consider that this protein family may have a shared role in patterning aECM rather than acting as diffusible signaling molecules – indeed, perhaps Hedgehog proteins themselves evolved from ancient structural components of the aECM.
Finally, we discovered a novel sexual dimorphism in glia and identified some of its regulators. Sex differences have been extensively studied in neurons, but little is known about how these differences are controlled in glia. Glial sex differences are of high interest in the mammalian brain, because glia are a major cell type in the nervous system that sculpts neuronal connections and modulates neuronal activity. Adult male and female microglia have distinct gene expression profiles and can influence sex-specific neuronal structure and function63–65; astrocyte glia exhibit molecular differences early in development and differ in number and morphology in brain regions involved in pheromone-sensing and hormone secretion66–68; and glial functions are disrupted in disease models for sex-biased disorders, including autism and Alzheimer’s disease69,70. In most cases, it remains unclear whether glial sex differences are a response to a sex-specific neuronal environment or reflect cell-intrinsic differences in the glia themselves, and what regulators might control these differences. In C. elegans, two examples have been described in which glia play sex-specific roles as neuronal progenitors20,21. We show that a sex-specific switch can also alter function of the mature glial cell itself. This switch is controlled cell autonomously by some of the same regulators that control sex differences in neurons (sex identity: fem-3, tra-2, mab-3; timing: lep-2, lep-5) but with novel downstream components (nfya-1, bed-3, jmjd-3.1) that may be more glial-specific.
Altogether, our findings underscore that aECM is not a static barrier but can contain highly patterned local structures that are constructed through coordinated regulation of discrete gene expression modules. In this sense, the aECM identity of a cell can be viewed as a defining feature of a cell type, much like neurotransmitter identity is for neurons.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Maxwell G. Heiman (heiman@genetics.med.harvard.edu).
Materials Availability
Plasmids and nematode strains generated in this study will be made available through the lead contact upon request.
Data and Code Availability
All data reported in this study have not been deposited in a public repository but will be shared by the lead contact upon request.
This study does not report original code.
Any additional information required to reanalyze the data reported in this study is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All strains are in an N2 background and, unless otherwise stated, contain him-5(e1490) or him-8(e1489) to generate a higher percentage of male progeny. See Key Resources Table for the genotypes of all strains used in this study. Animals were grown at 20–22°C on nematode growth media (NGM) plates seeded with E. coli OP50 bacteria76. Unless otherwise stated, all animals were picked to sex-segregated plates as L4 hermaphrodites and males based on vulva and tail morphology, respectively, and scored as 1-day adults the next day.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Ethyl Methanesulfonate | Sigma-Aldrich | M0880; CAS: 62–50-0 |
| Sodium Azide | Sigma-Aldrich | S2002; CAS: 26628–22-8 |
| Experimental models: Organisms/strains | ||
| hmnIs82 [grl-18pro:GFP] II | Fung et al., 202019 | CHB3829 |
| hmnIs82 [grl-18pro:GFP] II; him-5(e1490) V | This paper | CHB3843 |
| hmnIs82 [grl-18pro:GFP] II; him-5(e1490) V; hmnEx2172 [prab-3:fem-3-mCherry] | This paper | CHB3925 |
| hmnIs82 [grl-18pro:GFP] II; him-5(e1490) V; hmnEx2193 [mir-228pro:fem-3; unc-122pro:RFP] | This paper | CHB3900 |
| hmnIs82 [grl-18pro:GFP] II; him-5(e1490) V; hmnEx2215 [col-56pro:fem-3; unc-122pro:RFP] | This paper | CHB4032 |
| hmnIs82 [grl-18pro:GFP] II; him-5(e1490) V; hmnEx2208 [prab-3:tra-2-mCherry] | This paper | CHB4030 |
| hmnIs82 [grl-18pro:GFP] II; him-5(e1490) V; hmnEx2204 [mir-228pro:tra-2; unc-122pro:GFP] | This paper | CHB3926 |
| hmnIs82 [grl-18pro:GFP] II; him-5(e1490) V; hmnEx2211 [col-56pro:tra-2; unc-122pro:RFP] | This paper | CHB4031 |
|
nIs131 [pkd-2:GFP] I; hmnIs82 [grl-18pro:GFP] II; him-5(e1490) V |
This paper | CHB4298 |
|
nIs131 [pkd-2:GFP] I; hmnIs82 [grl-18pro:GFP] II; him-5(e1490) V; ceh-30(n3714) X |
This paper | CHB4285 |
|
nIs131 [pkd-2:GFP] I; hmnIs82 [grl-18pro:GFP] II; him-5(e1490) V; ceh-30(n4289) X |
This paper | CHB4338 |
|
hmnIs82 [grl-18pro:GFP] II; him-5(e1490) V; nfya-1(ok1174) X |
This paper | CHB4185 |
|
hmnIs82 [grl-18pro:GFP] II; him-5(e1490) V; nfya-1(ok1174) X; hmnEx4304 [nfya-1pro:nfya-1-cDNA; unc-122pro:RFP] |
This paper | CHB4245 |
|
hmnIs82 [grl-18pro:GFP] II; him-5(e1490) V; nfya-1(ok1174) X; hmnEx2329 [col-56pro:nfya-1-cDNA; unc-122pro:RFP] |
This paper | CHB4303 |
|
hmnIs82 [grl-18pro:GFP] II; him-5(e1490) V; nfya-1(ok1174) X; hmnEx2535 [rab-3pro:nfya-1-cDNA; unc-122pro:RFP] |
This paper | CHB4830 |
|
nfyb-1(cu13) II; him-5(e1490) V; hmnIs78 [grl-18pro:GFP] X |
This paper | CHB4281 |
|
nfyc-1(tm4541) II; him-5(e1490) V; hmnIs78 [grl-18pro:GFP] X |
This paper | CHB4603 |
|
hmnIs82 [grl-18pro:GFP] mab-3(mu15) II; him-5(e1490) V |
This paper | CHB3845 |
| hmnIs82 [grl-18pro:GFP] mab-3(mu15) II; him-5(e1490) V; nfya-1(ok1174) X | This paper | CHB4220 |
|
hmnIs47 [grl-18pro:mApple] I; him-8(e1489) IV; hmnEx2400 [grl-18pro:sfGFP:grl-18-gDNA; rol-6(su1006)] |
This paper | CHB4467 |
|
hmnIs47 [grl-18pro:mApple] I; him-8(e1489) IV; hmnEx2486 [grl-18pro:grl-18-gDNA:sfGFP; rol-6(su1006)] |
This paper | CHB4642 |
| hmnIs47 [grl-18pro:mApple] I; him-8(e1489) IV; hmnEx2398 [grl-18pro:sfGFP:grl-18-gDNA(352–445); rol-6(su1006)] | This paper | CHB4465 |
|
hmnIs47 [grl-18pro:mApple] I; him-8(e1489) IV; hmnEx2394 [grl-18pro:sfGFP:grl-18-gDNA(31–351); rol-6(su1006)] |
This paper | CHB4461 |
|
hmnIs47 [grl-18pro:mApple] I; him-8(e1489) IV; grl-18(syb6299 [sfGFP:grl-18]) V |
This paper | CHB4641 |
|
him-5(e1490) V; hmnEx2422 [pkd-2pro:GFP; dat-1pro:mApple; rol-6(su1006)] |
This paper | CHB4519 |
|
him-5(e1490) V; hmnIs102 [col-56pro:tra-2; unc-122pro:GFP]; hmnEx2422 [pkd-2pro:GFP; dat-1pro:mApple; rol-6(su1006)] |
This paper | CHB4543 |
|
hmnIs82 [grl-18pro:GFP] II; him-5(e1490) V; hmnIs102
[col-56pro:tra-2; unc-122pro:GFP] |
This paper | CHB4369 |
|
hmnIs82 [grl-18pro:GFP] II; him-5(e1490) V; hmnIs105
[col-56pro:fem-3; unc-122pro:GFP] |
This paper | CHB4375 |
| hmnIs47 [grl-18pro:mApple] I; him-5(e1490) V | This paper | CHB3515 |
|
hmnIs47 [grl-18pro:mApple] I; mab-3(hmn289) II; him-5(e1490) V |
This paper | CHB3916 |
|
hmnIs47 [grl-18pro:mApple] I; mab-3(mu15) II; him-5(e1490) V |
This paper | CHB3716 |
|
hmnIs82 [grl-18pro:GFP] II; lep-2(hmn305) IV; him-5(e1490) V |
This paper | CHB4640 |
|
hmnIs82 [grl-18pro:GFP] II; lep-2(ok900) IV; him-5(e1490) V |
This paper | CHB3897 |
|
hmnIs82 [grl-18pro:GFP] II; him-5(e1490) V; lep-5(ny28) X |
This paper | CHB3887 |
|
hmnIs82 [grl-18pro:GFP] II; him-5(e1490) V; jmjd-3.1(hmn304) X |
This paper | CHB4186 |
|
hmnIs82 [grl-18pro:GFP] II; him-5(e1490) V; jmjd-3.1(gk384) X |
This paper | CHB4248 |
| hmnIs82 [grl-18pro:GFP] II; nfya-1(hmn316) X | This paper | CHB4108 |
| hmnIs82 [grl-18pro:GFP] II; nfya-1(hmn317) X | This paper | CHB4109 |
| hmnIs82 [grl-18pro:GFP] II; nfya-1(hmn319) X | This paper | CHB4111 |
|
hmnIs82 [grl-18pro:GFP] II; him-5(e1490) V; nfya-1(ok1174) X; hmnEx2535 [rab-3pro:nfya-1-cDNA; unc-122pro:RFP] |
This paper | CHB4830 |
|
nfyb-1(tm4257) II; him-5(e1490) V; hmnIs78 [grl-18pro:GFP] X |
This paper | CHB4635 |
|
nfyc-1(tm4264) II; him-5(e1490) V; hmnIs78 [grl-18pro:GFP] X |
This paper | CHB4606 |
|
hmnIs82 [grl-18pro:GFP] II; bed-3(hmn318) IV; him-5(e1490) V |
This paper | CHB4289 |
|
hmnIs82 [grl-18pro:GFP] II; bed-3(sy705) IV; him-5(e1490) V |
This paper | CHB4249 |
|
hmnIs82 [grl-18pro:GFP] II; bed-3(gk996) IV; him-5(e1490) V |
This paper | CHB4400 |
|
grl-18(hmn340 [grl-18:SL2:YFP:H2B + loxP]) him-5(e1490) V |
This paper | CHB4470 |
|
hmnEx2223 [col-56pro:GFP; dat-1pro:mApple; rol-6(su1006)] |
This paper | CHB3992 |
|
hmnIs82 [grl-18pro:GFP] II; him-5(e1490) V; myEx696
[pdf-1pro:RFP; unc-122pro:GFP] |
This paper | CHB4284 |
| hmnIs82 [grl-18pro:GFP] II; him-5(e1490) V; nfya-1(ok1174) X; myEx696 [pdf-1pro:RFP; unc-122pro:GFP] | This paper | CHB4246 |
| him-5(e1490) V; kyIs53 [odr-10:GFP] X | This paper | CHB4360 |
| him-5(e1490) V; kyIs53 [odr-10:GFP] nfya-1(ok1174) X | This paper | CHB4352 |
| hmnIs47 [grl-18pro:mApple] I; grl-18(hmn341) him-5(e1490) V; hmnEx2398 [grl-18pro:sfGFP:grl-18-gDNA(352–445); rol-6(su1006)] | This paper | CHB4848 |
| grl-18(hmn341) him-5(e1490) V; hmnEx2422 [pkd-2pro:GFP; dat-1pro:mApple; rol-6(su1006)] | This paper | CHB4567 |
| grl-18(ok2845) him-5(e1490) V; hmnEx2422 [pkd-2pro:GFP; dat-1pro:mApple; rol-6(su1006)] | This paper | CHB4583 |
| unc-119(ox819) III; hmnEx2467 [mam-5pro2:grl-18-gDNA; unc-119(+)] | This paper | CHB4600 |
|
hmnIs47 [grl-18pro:mApple] I; him-8(e1489) IV; mab-23(gk664) V |
This paper | CHB3910 |
|
hmnIs47 [grl-18pro:mApple] I; him-8(e1489) IV; dmd-3(ok1327) V |
This paper | CHB3828 |
|
hmnIs47 [grl-18pro:mApple] I; dmd-5(ok1394) II; him-8(e1489) IV |
This paper | CHB3844 |
|
hmnIs47 [grl-18pro:mApple] I; dmd-6(gk287) IV; him-5(e1490) V |
This paper | CHB3686 |
|
hmnIs47 [grl-18pro:mApple] I; him-8(e1489) IV; dmd-7(ok2276) V |
This paper | CHB3768 |
| hmnIs47 [grl-18pro:mApple] I; dmd-8(ok1294) V | This paper | CHB3769 |
|
hmnIs47 [grl-18pro:mApple] I; dmd-9(ok1438) IV; him-5(e1490) V |
This paper | CHB3717 |
|
hmnIs47 [grl-18pro:mApple] I; him-8(e1489) IV; dmd-10(gk1125) V |
This paper | CHB3901 |
| Oligonucleotides | ||
| See Table S2 for oligonucleotide information. | N/A | |
| Recombinant DNA | ||
| mir-228pro:fem-3 | This paper | pTT03 |
| col-56pro:GFP | This paper | pWF1 |
| mir-228pro:tra-2 | This paper | pWF4 |
| pkd-2pro:GFP | This paper | pWF8 |
| grl-18pro:grl-18-gDNA:sfGFP | This paper | pWF15 |
| grl-18pro:sfGFP:grl-18-gDNA | This paper | pWF16 |
| dat-1pro:mApple | This paper | pWF17 |
| col-56pro:fem-3 | This paper | pWF22 |
| col-56pro:tra-2 | This paper | pWF23 |
| col-56pro:nfya-1-cDNA | This paper | pWF53 |
| nfya-1pro:nfya-1-cDNA | This paper | pWF54 |
| SapTrap_grl-18:SL2:YFP:H2B | This paper | pWF107 |
| grl-18pro:sfGFP:grl-18-gDNA(31–351) | This paper | pWF114 |
| grl-18pro:sfGFP:grl-18-gDNA(352–445) | This paper | pWF115 |
| pU6:grl-18-sgRNA #1 | This paper | pWF125 |
| pU6:grl-18-sgRNA #2 | This paper | pWF126 |
| mam-5pro2:grl-18-gDNA | This paper | pWF136 |
| rab-3pro:nfya-1-cDNA | This paper | pWF147 |
| Software and algorithms | ||
| CRISPOR | Concordet et al., 201871 | http://37.187.154.234/crispor.py |
| MiModD | Wolfgang Maier72 | Version 0.1.9 |
| Galaxy | Afgan et al., 201873 | https://github.com/galaxyproject/galaxy/ |
| Softworx | Applied Precision | Version 5.5 |
| ImageJ (Fiji) | Schindelin et al., 201274 | http://fiji.sc/ |
| Affinity Photo | Serif | Version 1.10.1 |
| Maps | Thermo Fisher Scientific | Versions 3.11 and 3.17 |
| IMOD | Kremer et al., 199675 | https://bio3d.colorado.edu/imod/ |
| GraphPad Prism | GraphPad | Version 9 |
| Other | ||
| DeltaVision | Applied Precision | N/A |
| Helios SEM microscope | Thermo Fisher Scientific | N/A |
METHOD DETAILS
Generation of transgenic strains
Transgenic strains were generated using standard techniques77 with injections of 100 ng/μL DNA (5–80 ng/μL per plasmid). See Table S2 and Key Resources Table for primers and all plasmids generated in this study.
Generation of alleles by genome editing
For genome editing with the CRISPR/Cas9 system, the CRISPOR web tool71 was used to identify potential guide RNA sequences (gRNAs) within the target gene sequence.
To generate the endogenous grl-18 transcriptional reporter (grl-18-SL2-YFP:H2B), a SapTrap plasmid78,79 was assembled to contain the gRNA (5’GAGTATCAAAGTTTGAATCA-3’) and repair template. The repair template includes ~530 bp 5’ and 3’ homology arms flanking an SL2-YFP:H2B fragment, a loxP-flanked positive selection marker, sqt-1, that causes a roller (Rol) phenotype, and a sequence encoding heat-shock-inducible Cre recombinase. A total of 65 ng/μL SapTrap plasmid was co-injected with a panel of markers used to select against extrachromosomal arrays (2 ng/μL pCFJ90, 4 ng/μL pGH8, 4 ng/μL pCFJ104, and 25 ng/μL pBlueScript)78 into the strain EG9888, which carries an integrated transgene for germline Cas9 expression80. A single genome-edited line was isolated exhibiting the Rol phenotype and confirmed by Sanger sequencing to contain the SL2-YFP:H2B fragment inserted after the stop codon of the endogenous grl-18 locus (Table S3). Finally, the integrated Cas9 transgene was removed by outcrossing to wild-type animals and the positive-selection marker was removed by Cre induction via heat-shock (34°C for 4 h).
To generate a null allele of grl-18, a co-CRISPR81,82 and dual-sgRNA83 approach was used. Two gRNAs were selected to target the 5’ and 3’ ends of the grl-18 gene (5’-AGTTTACCGAATCCAAGTTG-3’ and 5’-GAGTATCAAAGTTTGAATCA-3’, respectively) and cloned into separate plasmids. To screen and select for genome-edited animals, unc-58 was targeted to introduce a gain-of-function allele corresponding to unc-58(e665gf)81. 25 ng/μL of each sgRNA plasmid was co-injected with 25 ng/μL pJA50 (unc-58 sgRNA plasmid), 600 ng/μL unc-58(e665) repair oligo, and 25 ng/μL pBlueScript into the strain EG9888. Animals carrying a deletion in grl-18 were identified via PCR using primers flanking the gRNA target sites. The hmn341 allele was recovered as a 960 bp deletion that removes the conserved Grl domain sequences of the endogenous grl-18 locus (Table S3).
The allele grl-18(syb6299) was generated by SunyBiotech (Fuzhou, China). The first intron of the endogenous grl-18 locus was deleted and sfGFP was inserted between the first and second exon. Synonymous mutations were introduced to prevent re-cutting by Cas9/gRNA (Table S3).
Forward genetic screens for altered CEPso glial gene expression
Novel alleles in Table 1 were isolated through two clonal screens (hmn289, hmn304, and hmn305) and one nonclonal screen (hmn316, hmn317, hmn318, and hmn319). In all cases, animals were mutagenized with 70 mM ethyl methanesulfonate (EMS, Sigma) at 22°C for 4 h. In each of two independent clonal screens, animals of genotype hmnIs47 (grl-18pro:mApple) I; him-5 V were mutagenized, ~600 F1 progeny were picked to individual plates, and plates of F2 siblings were screened using a fluorescence stereomicroscope to identify the presence of adult males lacking grl-18pro:mApple expression in CEPso glia. When such males were found, the mutant allele was recovered by picking ~6–12 L4 hermaphrodite siblings to individual plates. Siblings that produced male progeny lacking grl-18pro:mApple in CEPso glia carried the mutant allele of interest. In a single nonclonal screen, animals of genotype hmnIs82 (grl-18pro:GFP) II were mutagenized and pooled F2 progeny were screened en masse using a fluorescence stereomicroscope to identify and isolate adult hermaphrodites that inappropriately express grl-18pro:GFP in CEPso glia. Mutant alleles generated in this study are in Table S3.
Genetic mapping and identification of causal mutations
Causal mutations in hmn289 and hmn305 were identified by candidate gene analysis based on distinctive tail morphology defects observed in mutant males. hmn289 males exhibit missing tail sensory rays, a phenotype that is characteristic of mab (male abnormal) mutants84, and showed non-complementation with mab-3(mu15) (from the cross mu15/+ x hmn289, 34/60 1-day adult males failed to express grl-18pro:mApple in CEPso glia), which is known to affect male-specific phenotypes32–36. hmn305 males exhibit a lep (leptoderan) male tail phenotype, characterized by retention of the juvenile tail tip in the adult stage85, and showed non-complementation with lep-2(ok900) (from the cross ok900/+ x hmn305, 26/53 1-day adult males failed to express grl-18pro:mApple in CEPso glia), which is known to affect the timing of sexual maturation39,41. Sanger sequencing showed that hmn289 encodes a R90C missense mutation in the second DNA-binding domain of MAB-3 and that hmn305 encodes a C216Y missense mutation in the conserved RING finger domain of LEP-2. The reference alleles mab-3(mu15) and lep-2(ok900) recapitulated the grl-18 reporter defects of hmn289 and hmn305 (Table 1). Together, these data indicate that hmn289 and hmn305 are alleles of mab-3 and lep-2, respectively.
To identify the causal mutation in hmn304, a series of crosses were first used to determine that the mutation is X-linked. Whole-genome sequencing identified a W831STOP nonsense mutation in jmjd-3.1 that is predicted to truncate the protein within the conserved JmjC DNA-binding domain. The jmjd-3.1(gk387) mutation recapitulates the hmn304 phenotype (Table 1) and a transgene bearing the fosmid WRM0610dB04, which consists of the 4.4 kb jmjd-3.1 gene with ~15 kb upstream and ~13 kb downstream sequences, rescues the hmn304 mutant phenotype (41/42 1-day adult males express grl-18pro:GFP in at least one CEPso glia). Together, these data indicate that hmn304 is an allele of jmjd-3.1.
To identify the causal mutation in hmn318, whole-genome sequencing was performed. Among the mutations in this strain, we identified a C144G missense mutation in the DNA-binding BED domain of BED-3, a transcriptional repressor important in vulval development43. We noted that hmn318 hermaphrodites exhibit weak vulval defects, consistent with loss of bed-3 function. Further, we found that two existing alleles of bed-3, sy705 and gk996, recapitulate the hmn318 defects in grl-18pro:GFP expression (Table 1). A transgene bearing the fosmid WRM0624bB06, which consists of the 5.4 kb bed-3 gene with ~19 kb upstream and ~7 kb downstream sequences, rescues the hmn318 mutant phenotype (9/67 1-day adult hermaphrodites inappropriately express grl-18pro:GFP in at least one CEPso glia). Together, these data indicate that hmn318 is an allele of bed-3.
To identify the causal mutations in hmn316, hmn317, and hmn319, one-step mapping was performed by crossing each mutant to the polymorphic strain CB4856 (“Hawaiian”), selecting F2 recombinants that exhibit inappropriate grl-18 expression in CEPso glia, and analyzing their pooled progeny by whole-genome sequencing86. Sequencing results were analyzed using MiModD (v0.1.9)72 through a local Galaxy interface73. All three mutations were mapped to an interval between 6 Mb and 14 Mb on chromosome X. Each mutant was found to have probable loss-of-function sequence changes in nfya-1: hmn316, g>a splice site donor mutation between exons 4 and 5; hmn317, R21STOP nonsense mutation; and hmn319, Q146STOP nonsense mutation. A strain bearing the reference allele nfya-1(ok1174) recapitulated the grl-18 reporter expression defect of all three mutants (Table 1). In complementation tests, hmn316 failed to complement hmn317, hmn319, or ok1174 (41/42, 27/27, and 26/26 1-day adult hermaphrodites inappropriately express grl-18pro:GFP in CEPso glia, respectively). Finally, the hmn316 mutant phenotype was rescued with the WRM0637aE09 fosmid, which consists of the 2.4 kb nfya-1 gene with ~14 kb upstream and ~17 kb downstream sequences (5/76 1-day adult hermaphrodites inappropriately express grl-18pro:GFP in CEPso glia), and the ok1174 mutant phenotype was rescued with nfya-1 cDNA (Figure 3C). Together, these results indicate that hmn316, hmn317, and hmn319 are alleles of nfya-1.
Fluorescence microscopy and image processing
Animals were washed and immobilized in M9 solution containing 50 mM sodium azide and mounted on 2% agarose pads containing 50 mM sodium azide. Image stacks were collected on a DeltaVision Core imaging system (Applied Precision) with a UApo 40x/1.35 NA, PlanApo 60x/1.42 NA, or UPlanSApo 100x/1.40 NA oil immersion objective and a CoolSnap HQ2 camera. Images were deconvolved using Softworx (Applied Precision) and maximum intensity projections were generated in ImageJ (Fiji)74. The brightness and contrast of each projection were linearly adjusted in Affinity Photo 1.10.1. Fluorescent signals were pseudo-colored, and merged images were generated using the Screen layer mode in Affinity Photo 1.10.1.
Developmental staging for time course experiments
For time course experiments, single animals were tracked across development from larval to adult stages. L3 hermaphrodites and males were identified based on vulva and tail morphology, respectively, picked to individual plates, and scored. The same individuals were scored after ~10 h (late L4), again after ~12 h (1-day adult), and then at 24 h intervals (2-, 3-, 4-day adults). Animals that crawled off the plate or died before the experiment was complete were not included in the data. Fluorescent reporter expression was scored visually on a Nikon SMZ1500 stereomicroscope with an HR Plan Apo 1.6x objective.
To image animals during the L4/adult molt, L4 males were selected based on tail morphology and either imaged immediately or monitored up to 4 h until the cuticle was shed and the adult tail was formed. Animals were imaged as described above (“Fluorescence microscopy and image processing”). Staging within the molt was determined based on male tail morphology85,87 as follows: L4 – pointed tail, no retraction of hypodermal cells; early molt – rounded tail, hypodermal cells hyp9 and hyp10 have retracted from the L4 cuticle; late molt – further retraction of hypodermal cells hyp8–11 from the L4 cuticle, rays beginning to form; adult – L4 cuticle has been shed, fans and rays are fully formed.
Binary scoring of CEM cilia trajectories
CEM and CEP cilia were imaged using fluorescent markers for each neuron (CEM, pkd-2pro:GFP; CEP, dat-1pro:mApple). L4 animals were selected based on tail morphology and imaged as 1-day adults as described above (“Fluorescence microscopy and image processing”). A sense organ was only scored if both CEM and CEP were labeled and oriented flat with their cilia in the x-y plane, not the z plane. In rare cases, either one or both the CEM and CEP cilia were short or abnormally shaped; these were not scored. The CEP cilium was used as a reference for a trajectory that bends inward. If the CEM cilium traveled in the same direction as the CEP cilium, then it was scored as “bent in”; if it traveled in the direction orthogonal to the CEP cilium, then it was scored as “bent out”. Because the CEM cilium is extremely thin and the fluorescent signal at the tip is very dim, the brightness and contrast of the images were adjusted until the cilia were visible, which often saturated the signal in the rest of the CEM ending.
Electron microscopy
For serial thin-section scanning electron microscopy, samples were high-pressure frozen and quick freeze substituted as previously described88. Epon flat-embedded samples were carefully oriented for longitudinal sectioning through the head, except for a wild-type male and hermaphrodite that were each oriented for transverse sectioning88–90. To target the region of interest and to minimize artifacts, excess resin was removed using a 90° trimming tool (Diatome, Switzerland). Samples were sectioned using an ATS knife (Diatome, Switzerland) mounted on a Leica UC7 ultramicrotome (Leica, Austria). Approximately 100 to 300 sections were obtained per animal and transferred to a 2×4 cm silicon wafer89,91. Wafers were dried at ambient temperature by evaporation and subsequently incubated in a 60°C oven for further fixation as previously described89,91,92. Wafers were analyzed with a Helios SEM microscope (Thermo Fisher Scientific) at 2keV landing energy and 0.8 nA beam current at 2 mm distance using a Mirror Detector (MD-BSA)89,91. Images were collected manually or automatically with 4–6 μs dwell time using Maps 3.11 software (Thermo Fisher Scientific)89,91,92. Images were collected at 3 mm/s (1024×886) or 5 mm/s (6084×2044) to generate 20 nm and 5 nm resolution images, respectively. To cover a larger area of the sample, several images were collected at a given resolution and stitched together using Maps 3.17 software (Thermo Fisher Scientific)89,91. Finally, serial sections were aligned using the IMOD program (University of Colorado)75.
For wholemount scanning electron microscopy of wild-type animals (N2 Bristol), samples were fixed with 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 PB buffer. Samples were subsequently post-fixed/contrasted using 2% osmium and dehydrated using increasing concentrations of ethanol. After three changes of 100% ethanol, samples were processed in a critical point dryer apparatus (Leica) for 2 h. Then, samples were mounted on aluminum stubs and sputter coated with 20 nm gold (Baltec, Switzerland). Samples were analyzed with the Helios SEM microscope using the same conditions as above for thin sections.
QUANTIFICATION AND STATISTICAL ANALYSIS
GraphPad Prism 9 software was used to generate all graphs and to perform statistical analysis. Data in all graphs are depicted as percentages with error bars representing the standard error of the mean (SEM). For pairwise comparisons in Figure S3, The Mann-Whitney test was used to determine statistical significance as previously described39,50,51 (see Figure S3 legend for more details). The exact value of n for each experiment is indicated either above the bars of the graphs or in the figure legends.
Supplementary Material
Highlights:
A switch in gene expression occurs in C. elegans glia as males reach sexual maturity
mab-3 promotes male gene expression in glia by relieving nfya-1-mediated repression
Male glia secrete GRL-18, which forms transient rings at sites of aECM pore formation
Male gene expression in glia is necessary and sufficient to create aECM cuticle pores
Acknowledgments:
We thank Douglas Portman, Maureen Barr, and members of the Scott Kennedy laboratory for reagents; Lisa Goodrich, Joshua Kaplan, Constance Cepko, and Norbert Perrimon for advice; members of the Heiman laboratory for comments on the manuscript; and Wormbase. Some strains were provided by the C. elegans Genetics Center [CGC, funded by National Institutes of Health Office of Research Infrastructure Program (P40 OD010440)], the International C. elegans Gene Knockout Consortium, and the National BioResource Project of Japan for C. elegans. This work was supported by NIH grants R01NS124879 and R01NS112343 to M.G.H. and by NIH grant F31NS122139 to W.F.
Footnotes
Declaration of Interests: The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Whitsett JA, Wert SE, and Weaver TE (2015). Diseases of Pulmonary Surfactant Homeostasis. Annu. Rev. Pathol. Mech. Dis. 10, 371–393. 10.1146/annurev-pathol-012513-104644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaudette S, Hughes D, and Boller M (2020). The endothelial glycocalyx: Structure and function in health and critical illness. Journal of Veterinary Emergency and Critical Care 30, 117–134. 10.1111/vec.12925. [DOI] [PubMed] [Google Scholar]
- 3.Johansson MEV, Sjövall H, and Hansson GC (2013). The gastrointestinal mucus system in health and disease. Nat Rev Gastroenterol Hepatol 10, 352–361. 10.1038/nrgastro.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ando T, Sekine S, Inagaki S, Misaki K, Badel L, Moriya H, Sami MM, Itakura Y, Chihara T, Kazama H, et al. (2019). Nanopore Formation in the Cuticle of an Insect Olfactory Sensillum. Current Biology 29, 1512–1520.e6. 10.1016/j.cub.2019.03.043. [DOI] [PubMed] [Google Scholar]
- 5.Goodyear RJ, and Richardson GP (2018). Chapter Six - Structure, Function, and Development of the Tectorial Membrane: An Extracellular Matrix Essential for Hearing. In Current Topics in Developmental Biology Extracellular Matrix and Egg Coats., Litscher ES and Wassarman PM, eds. (Academic Press; ), pp. 217–244. 10.1016/bs.ctdb.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Li Zheng S, Adams JG, and Chisholm AD (2020). Form and function of the apical extracellular matrix: new insights from Caenorhabditis elegans, Drosophila melanogaster, and the vertebrate inner ear. Fac Rev 9, 27. 10.12703/r/9-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong B, and Hayashi S (2015). Shaping of biological tubes by mechanical interaction of cell and extracellular matrix. Current Opinion in Genetics & Development 32, 129–134. 10.1016/j.gde.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 8.Gill HK, Cohen JD, Ayala-Figueroa J, Forman-Rubinsky R, Poggioli C, Bickard K, Parry JM, Pu P, Hall DH, and Sundaram MV (2016). Integrity of Narrow Epithelial Tubes in the C. elegans Excretory System Requires a Transient Luminal Matrix. PLOS Genetics 12, e1006205. 10.1371/journal.pgen.1006205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Page A (2007). The cuticle. WormBook. 10.1895/wormbook.1.138.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ward S, Thomson N, White JG, and Brenner S (1975). Electron microscopical reconstruction of the anterior sensory anatomy of the nematode caenorhabditis elegans. Journal of Comparative Neurology 160, 313–337. 10.1002/cne.901600305. [DOI] [PubMed] [Google Scholar]
- 11.Chasnov JR, So WK, Chan CM, and Chow KL (2007). The species, sex, and stage specificity of a Caenorhabditis sex pheromone. Proceedings of the National Academy of Sciences 104, 6730–6735. 10.1073/pnas.0608050104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.White JQ, Nicholas TJ, Gritton J, Truong L, Davidson ER, and Jorgensen EM (2007). The Sensory Circuitry for Sexual Attraction in C. elegans Males. Current Biology 17, 1847–1857. 10.1016/j.cub.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 13.Srinivasan J, Kaplan F, Ajredini R, Zachariah C, Alborn HT, Teal PEA, Malik RU, Edison AS, Sternberg PW, and Schroeder FC (2008). A blend of small molecules regulates both mating and development in Caenorhabditis elegans. Nature 454, 1115–1118. 10.1038/nature07168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J, Silva M, Haas LA, Morsci NS, Nguyen KCQ, Hall DH, and Barr MM (2014). C. elegans Ciliated Sensory Neurons Release Extracellular Vesicles that Function in Animal Communication. Current Biology 24, 519–525. 10.1016/j.cub.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sulston JE, Schierenberg E, White JG, and Thomson JN (1983). The embryonic cell lineage of the nematode Caenorhabditis elegans. Developmental Biology 100, 64–119. 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- 16.Akella JS, Silva M, Morsci NS, Nguyen KC, Rice WJ, Hall DH, and Barr MM (2019). Cell type-specific structural plasticity of the ciliary transition zone in C. elegans. Biol. Cell 111, 95–107. 10.1111/boc.201800042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perkins LA, Hedgecock EM, Thomson JN, and Culotti JG (1986). Mutant sensory cilia in the nematode Caenorhabditis elegans. Developmental Biology 117, 456–487. 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- 18.Cebul ER, McLachlan IG, and Heiman MG (2020). Dendrites with specialized glial attachments develop by retrograde extension using SAX-7 and GRDN-1. Development, dev.180448. 10.1242/dev.180448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fung W, Wexler L, and Heiman MG (2020). Cell-type-specific promoters for C. elegans glia. Journal of Neurogenetics 34, 335–346. 10.1080/01677063.2020.1781851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sammut M, Cook SJ, Nguyen KCQ, Felton T, Hall DH, Emmons SW, Poole RJ, and Barrios A (2015). Glia-derived neurons are required for sex-specific learning in C. elegans. Nature 526, 385–390. 10.1038/nature15700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Molina-García L, Lloret-Fernández C, Cook SJ, Kim B, Bonnington RC, Sammut M, O’Shea JM, Gilbert SP, Elliott DJ, Hall DH, et al. (2020). Direct glia-to-neuron transdifferentiation gives rise to a pair of male-specific neurons that ensure nimble male mating. eLife 9, e48361. 10.7554/eLife.48361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sulston JE, Albertson DG, and Thomson JN (1980). The Caenorhabditis elegans male: Postembryonic development of nongonadal structures. Developmental Biology 78, 542–576. 10.1016/0012-1606(80)90352-8. [DOI] [PubMed] [Google Scholar]
- 23.Mehra A, Gaudet J, Heck L, Kuwabara PE, and Spence AM (1999). Negative regulation of male development in Caenorhabditis elegans by a protein–protein interaction between TRA-2A and FEM-3. Genes Dev 13, 1453–1463. 10.1101/gad.13.11.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee K, and Portman DS (2007). Neural Sex Modifies the Function of a C. elegans Sensory Circuit. Current Biology 17, 1858–1863. 10.1016/j.cub.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 25.Mowrey WR, Bennett JR, and Portman DS (2014). Distributed Effects of Biological Sex Define Sex-Typical Motor Behavior in Caenorhabditis elegans. J. Neurosci. 34, 1579–1591. 10.1523/JNEUROSCI.4352-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pierce ML, Weston MD, Fritzsch B, Gabel HW, Ruvkun G, and Soukup GA (2008). MicroRNA-183 family conservation and ciliated neurosensory organ expression. Evolution & Development 10, 106–113. 10.1111/j.1525-142X.2007.00217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwartz HT, and Horvitz HR (2007). The C. elegans protein CEH-30 protects male-specific neurons from apoptosis independently of the Bcl-2 homolog CED-9. Genes & Development 21, 3181–3194. 10.1101/gad.1607007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peden E, Kimberly E, Gengyo-Ando K, Mitani S, and Xue D (2007). Control of sex-specific apoptosis in C. elegans by the BarH homeodomain protein CEH-30 and the transcriptional repressor UNC-37/Groucho. Genes Dev. 21, 3195–3207. 10.1101/gad.1607807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burtis KC, and Baker BS (1989). Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell 56, 997–1010. 10.1016/0092-8674(89)90633-8. [DOI] [PubMed] [Google Scholar]
- 30.Raymond CS, Shamu CE, Shen MM, Seifert KJ, Hirsch B, Hodgkin J, and Zarkower D (1998). Evidence for evolutionary conservation of sex-determining genes. Nature 391, 691–695. 10.1038/35618. [DOI] [PubMed] [Google Scholar]
- 31.Raymond CS, Murphy MW, O’Sullivan MG, Bardwell VJ, and Zarkower D (2000). Dmrt1, a gene related to worm and fly sexual regulators, is required for mammalian testis differentiation. Genes Dev. 14, 2587–2595. 10.1101/gad.834100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen MM, and Hodgkin J (1988). mab-3, a gene required for sex-specific yolk protein expression and a male-specific lineage in C. elegans. Cell 54, 1019–1031. 10.1016/0092-8674(88)90117-1. [DOI] [PubMed] [Google Scholar]
- 33.Yi W, Ross JM, and Zarkower D (2000). mab-3 is a direct tra-1 target gene regulating diverse aspects of C. elegans male sexual development and behavior. 13. 10.1242/dev.127.20.4469. [DOI] [PubMed] [Google Scholar]
- 34.Ross JM, Kalis AK, Murphy MW, and Zarkower D (2005). The DM Domain Protein MAB-3 Promotes Sex-Specific Neurogenesis in C. elegans by Regulating bHLH Proteins. Developmental Cell 8, 881–892. 10.1016/j.devcel.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 35.Mason DA, Rabinowitz JS, and Portman DS (2008). dmd-3, a doublesex-related gene regulated by tra-1, governs sex-specific morphogenesis in C. elegans. Development 135, 2373–2382. 10.1242/dev.017046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fagan KA, Luo J, Lagoy RC, Schroeder FC, Albrecht DR, and Portman DS (2018). A Single-Neuron Chemosensory Switch Determines the Valence of a Sexually Dimorphic Sensory Behavior. Current Biology 28, 902–914.e5. 10.1016/j.cub.2018.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moss EG (2007). Heterochronic Genes and the Nature of Developmental Time. Current Biology 17, R425–R434. 10.1016/j.cub.2007.03.043. [DOI] [PubMed] [Google Scholar]
- 38.Abreu AP, Macedo DB, Brito VN, Kaiser UB, and Latronico AC (2015). A new pathway in the control of the initiation of puberty: the MKRN3 gene. Journal of Molecular Endocrinology 54, R131–R139. 10.1530/JME-14-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lawson H, Vuong E, Miller RM, Kiontke K, Fitch DH, and Portman DS (2019). The Makorin lep-2 and the lncRNA lep-5 regulate lin-28 to schedule sexual maturation of the C. elegans nervous system. eLife 8, e43660. 10.7554/eLife.43660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pereira L, Aeschimann F, Wang C, Lawson H, Serrano-Saiz E, Portman DS, Großhans H, and Hobert O (2019). Timing mechanism of sexually dimorphic nervous system differentiation. 31. 10.7554/eLife.42078.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herrera RA, Kiontke K, and Fitch DHA (2016). Makorin ortholog LEP-2 regulates LIN-28 stability to promote the juvenile-to-adult transition in Caenorhabditis elegans. Development 143, 799–809. 10.1242/dev.132738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kiontke KC, Herrera RA, Vuong E, Luo J, Schwarz EM, Fitch DHA, and Portman DS (2019). The Long Non-Coding RNA lep-5 Promotes the Juvenile-to-Adult Transition by Destabilizing LIN-28. Developmental Cell 49, 542–555.e9. 10.1016/j.devcel.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Inoue T, and Sternberg PW (2010). C. elegans BED domain transcription factor BED-3 controls lineage-specific cell proliferation during organogenesis. Developmental Biology 338, 226–236. 10.1016/j.ydbio.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goh KY, and Inoue T (2018). A large transcribed enhancer region regulates C. elegans bed-3 and the development of egg laying muscles. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms 1861, 519–533. 10.1016/j.bbagrm.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 45.Deng H, Sun Y, Zhang Y, Luo X, Hou W, Yan L, Chen Y, Tian E, Han J, and Zhang H (2007). Transcription factor NFY globally represses the expression of the C. elegans Hox gene Abdominal-B homolog egl-5. Developmental Biology 308, 583–592. 10.1016/j.ydbio.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 46.Milton AC, Packard AV, Clary L, and Okkema PG (2013). The NF-Y complex negatively regulates Caenorhabditis elegans tbx-2 expression. Developmental Biology 382, 38–47. 10.1016/j.ydbio.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aklilu S, Krakowiak M, Frempong A, Wilson K, Powers C, and Fantz D (2022). Nfya-1 functions as a substrate of ERK-MAP kinase during Caenorhabditis elegans vulval development. Cells & Development 169, 203757. 10.1016/j.cdev.2021.203757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heo W, Hwang H, Kim J, Oh SH, Yu Y, and Kim J-HL& K. (2023). The CCAAT-box transcription factor, NF-Y complex, mediates the specification of the IL1 neurons in C. elegans. BMB Reports 56, 153–159. 10.5483/BMBRep.2022-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dolfini D, Gatta R, and Mantovani R (2012). NF-Y and the transcriptional activation of CCAAT promoters. Critical Reviews in Biochemistry and Molecular Biology 47, 29–49. 10.3109/10409238.2011.628970. [DOI] [PubMed] [Google Scholar]
- 50.Ryan DA, Miller RM, Lee K, Neal SJ, Fagan KA, Sengupta P, and Portman DS (2014). Sex, Age, and Hunger Regulate Behavioral Prioritization through Dynamic Modulation of Chemoreceptor Expression. Current Biology 24, 2509–2517. 10.1016/j.cub.2014.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wexler LR, Miller RM, and Portman DS (2020). C. elegans Males Integrate Food Signals and Biological Sex to Modulate State-Dependent Chemosensation and Behavioral Prioritization. Current Biology 30, 2695–2706.e4. 10.1016/j.cub.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Katz SS, Barker TJ, Maul-Newby HM, Sparacio AP, Nguyen KCQ, Maybrun CL, Belfi A, Cohen JD, Hall DH, Sundaram MV, et al. (2022). A transient apical extracellular matrix relays cytoskeletal patterns to shape permanent acellular ridges on the surface of adult C. elegans. PLOS Genetics 18, e1010348. 10.1371/journal.pgen.1010348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Öztürk-Çolak A, Moussian B, and Araújo SJ (2016). Drosophila chitinous aECM and its cellular interactions during tracheal development. Developmental Dynamics 245, 259–267. 10.1002/dvdy.24356. [DOI] [PubMed] [Google Scholar]
- 54.Cohen JD, Sparacio AP, Belfi AC, Forman-Rubinsky R, Hall DH, Maul-Newby H, Frand AR, and Sundaram MV (2020). A multi-layered and dynamic apical extracellular matrix shapes the vulva lumen in Caenorhabditis elegans. eLife 9, e57874. 10.7554/eLife.57874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cohen JD, and Sundaram MV (2020). C. elegans Apical Extracellular Matrices Shape Epithelia. Journal of Developmental Biology 8, 23. 10.3390/jdb8040023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bürglin TR (1996). Warthog and Groundhog, novel families related to Hedgehog. Current Biology 6, 1047–1050. 10.1016/S0960-9822(02)70659-3. [DOI] [PubMed] [Google Scholar]
- 57.Aspöck G, Kagoshima H, Niklaus G, and Bürglin TR (1999). Caenorhabditis elegans Has Scores of hedgehog Related Genes: Sequence and Expression Analysis. Genome Res. 9, 909–923. 10.1101/gr.9.10.909. [DOI] [PubMed] [Google Scholar]
- 58.Hao L, Johnsen R, Lauter G, Baillie D, and Bürglin TR (2006). Comprehensive analysis of gene expression patterns of hedgehog-related genes. BMC Genomics 7, 280. 10.1186/1471-2164-7-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Misteli T (2001). The concept of self-organization in cellular architecture. J Cell Biol 155, 181–186. 10.1083/jcb.200108110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marshall WF (2020). Pattern Formation and Complexity in Single Cells. Current Biology 30, R544–R552. 10.1016/j.cub.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fernandes I, Chanut-Delalande H, Ferrer P, Latapie Y, Waltzer L, Affolter M, Payre F, and Plaza S (2010). Zona Pellucida Domain Proteins Remodel the Apical Compartment for Localized Cell Shape Changes. Developmental Cell 18, 64–76. 10.1016/j.devcel.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 62.Öztürk-Çolak A, Moussian B, Araújo SJ, and Casanova J (2016). A feedback mechanism converts individual cell features into a supracellular ECM structure in Drosophila trachea. Elife 5, e09373. 10.7554/eLife.09373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guneykaya D, Ivanov A, Hernandez DP, Haage V, Wojtas B, Meyer N, Maricos M, Jordan P, Buonfiglioli A, Gielniewski B, et al. (2018). Transcriptional and Translational Differences of Microglia from Male and Female Brains. Cell Reports 24, 2773–2783.e6. 10.1016/j.celrep.2018.08.001. [DOI] [PubMed] [Google Scholar]
- 64.Villa A, Gelosa P, Castiglioni L, Cimino M, Rizzi N, Pepe G, Lolli F, Marcello E, Sironi L, Vegeto E, et al. (2018). Sex-Specific Features of Microglia from Adult Mice. Cell Reports 23, 3501–3511. 10.1016/j.celrep.2018.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lenz KM, Nugent BM, Haliyur R, and McCarthy MM (2013). Microglia Are Essential to Masculinization of Brain and Behavior. Journal of Neuroscience 33, 2761–2772. 10.1523/JNEUROSCI.1268-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rurak GM, Simard S, Freitas-Andrade M, Lacoste B, Charih F, Geel AV, Stead J, Woodside B, Green JR, Coppola G, et al. (2022). Sex differences in developmental patterns of neocortical astroglia: A mouse translatome database. Cell Reports 38. 10.1016/j.celrep.2022.110310. [DOI] [PubMed] [Google Scholar]
- 67.Amateau SK, and McCarthy MM (2002). Sexual Differentiation of Astrocyte Morphology in the Developing Rat Preoptic Area. Journal of Neuroendocrinology 14, 904–910. 10.1046/j.1365-2826.2002.00858.x. [DOI] [PubMed] [Google Scholar]
- 68.Schwarz JM, and Bilbo SD (2012). Sex, glia, and development: Interactions in health and disease. Hormones and Behavior 62, 243–253. 10.1016/j.yhbeh.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Neniskyte U, and Gross CT (2017). Errant gardeners: glial-cell-dependent synaptic pruning and neurodevelopmental disorders. Nat Rev Neurosci 18, 658–670. 10.1038/nrn.2017.110. [DOI] [PubMed] [Google Scholar]
- 70.Vegeto E, Villa A, Della Torre S, Crippa V, Rusmini P, Cristofani R, Galbiati M, Maggi A, and Poletti A (2020). The Role of Sex and Sex Hormones in Neurodegenerative Diseases. Endocr Rev 41, 273–319. 10.1210/endrev/bnz005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Concordet J-P, and Haeussler M (2018). CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Research 46, W242–W245. 10.1093/nar/gky354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maier Wolfgang (Ralf Baumeister lab, University of Freiburg). Mapping-by-sequencing: identification of a phenotype-causing mutation in a nematode genome — MiModD 0.1.9 documentation. [Google Scholar]
- 73.Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Čech M, Chilton J, Clements D, Coraor N, Grüning BA, et al. (2018). The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Research 46, W537–W544. 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kremer JR, Mastronarde DN, and McIntosh JR (1996). Computer Visualization of Three-Dimensional Image Data Using IMOD. Journal of Structural Biology 116, 71–76. 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 76.Brenner S (1974). The Genetics of Caenorhabditis elegans. Genetics 77, 71–94. 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mello C, and Fire A (1995). Chapter 19 DNA Transformation. In Methods in Cell Biology Cuenorhubditis elegans: Modern Biologcal Analysis of an Organism., Epstein HF and Shakes DC, eds. (Academic Press; ), pp. 451–482. 10.1016/S0091-679X(08)61399-0. [DOI] [PubMed] [Google Scholar]
- 78.Dickinson DJ, Pani AM, Heppert JK, Higgins CD, and Goldstein B (2015). Streamlined Genome Engineering with a Self-Excising Drug Selection Cassette. Genetics 200, 1035–1049. 10.1534/genetics.115.178335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schwartz ML, and Jorgensen EM (2016). SapTrap, a Toolkit for High-Throughput CRISPR/Cas9 Gene Modification in Caenorhabditis elegans. Genetics 202, 1277–1288. 10.1534/genetics.115.184275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schwartz ML, Davis MW, Rich MS, and Jorgensen EM (2021). High-efficiency CRISPR gene editing in C. elegans using Cas9 integrated into the genome. PLOS Genetics 17, e1009755. 10.1371/journal.pgen.1009755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Arribere JA, Bell RT, Fu BXH, Artiles KL, Hartman PS, and Fire AZ (2014). Efficient Marker-Free Recovery of Custom Genetic Modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198, 837–846. 10.1534/genetics.114.169730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim H, Ishidate T, Ghanta KS, Seth M, Conte D, Shirayama M, and Mello CC (2014). A co-CRISPR strategy for efficient genome editing in Caenorhabditis elegans. Genetics 197, 1069–1080. 10.1534/genetics.114.166389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen X, Xu F, Zhu C, Ji J, Zhou X, Feng X, and Guang S (2014). Dual sgRNA-directed gene knockout using CRISPR/Cas9 technology in Caenorhabditis elegans. Sci Rep 4, 7581. 10.1038/srep07581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hodgkin J (1983). Male Phenotypes and Mating Efficiency in Caenorhabditis elegans. Genetics 103, 43–64. 10.1093/genetics/103.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nguyen CQ, Hall DH, Yang Y, and Fitch DHA (1999). Morphogenesis of the Caenorhabditis elegans Male Tail Tip. Developmental Biology 207, 86–106. 10.1006/dbio.1998.9173. [DOI] [PubMed] [Google Scholar]
- 86.Doitsidou M, Poole RJ, Sarin S, Bigelow H, and Hobert O (2010). C. elegans Mutant Identification with a One-Step Whole-Genome-Sequencing and SNP Mapping Strategy. PLOS ONE 5, e15435. 10.1371/journal.pone.0015435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Emmons SW (2005). Male development. WormBook. 10.1895/wormbook.1.33.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kolotuev I (2014). Positional Correlative Anatomy of Invertebrate Model Organisms Increases Efficiency of TEM Data Production. Microscopy and Microanalysis 20, 1392–1403. 10.1017/S1431927614012999. [DOI] [PubMed] [Google Scholar]
- 89.Burel A, Lavault M-T, Chevalier C, Gnaegi H, Prigent S, Mucciolo A, Dutertre S, Humbel BM, Guillaudeux T, and Kolotuev I (2018). A targeted 3D EM and correlative microscopy method using SEM array tomography. Development 145. 10.1242/dev.160879. [DOI] [PubMed] [Google Scholar]
- 90.Kato M, Kolotuev I, Cunha A, Gharib S, and Sternberg PW (2021). Extrasynaptic acetylcholine signaling through a muscarinic receptor regulates cell migration. Proc Natl Acad Sci U S A 118, e1904338118. 10.1073/pnas.1904338118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Franke T, and Kolotuev I (2021). Array Tomography Workflow for the Targeted Acquisition of Volume Information using Scanning Electron Microscopy. 173, 28. [DOI] [PubMed] [Google Scholar]
- 92.Santella A, Kolotuev I, Kizilyaprak C, and Bao Z (2022). Cross-modality synthesis of EM time series and live fluorescence imaging. eLife 11, e77918. 10.7554/eLife.77918. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this study have not been deposited in a public repository but will be shared by the lead contact upon request.
This study does not report original code.
Any additional information required to reanalyze the data reported in this study is available from the lead contact upon request.


