Abstract
Epithelial-to-mesenchymal transition (EMT) is a process by which cells lose their epithelial characteristics and gain mesenchymal phenotypes. In cancer, EMT is thought to drive tumor invasion and metastasis. Recent efforts to understand EMT biology have uncovered that cells undergoing EMT attain a spectrum of intermediate “hybrid E/M” states, which exist along an epithelial-mesenchymal continuum. Here, we summarize recent studies characterizing the epigenetic drivers of hybrid E/M states. We focus on the histone-modification writers, erasers, and readers that assist or oppose the canonical hybrid E/M transcription factors that modulate hybrid E/M state transitions. We also examine the role of chromatin remodelers and DNA methylation in hybrid E/M states. Finally, we highlight the challenges of targeting hybrid E/M pharmacologically, and we propose future directions that might reveal the specific and targetable mechanisms by which hybrid E/M drives metastasis in patients.
INTRODUCTION
Epithelial-to-mesenchymal transition (EMT) has become a hallmark of solid tumor invasion and metastasis (1). Cancer cells transitioning along this epithelial-mesenchymal axis assume a series of hybrid epithelial-mesenchymal (hybrid E/M) states, which are thought to be the primary drivers of tumor metastasis (2, 3). Because hybrid E/M states are dynamic and meta-stable, the regulation of hybrid E/M plasticity is thought to be primarily epigenetic rather than genetic (2). Thus, there is a need to understand the key epigenetic regulators of hybrid E/M states. In this review, we summarize the current body of literature with an emphasis on how histone modification readers, writers, and erasers, as well as chromatin remodelers and DNA methylation enzymes interact with the major hybrid E/M transcription factors (TFs) to promote or inhibit hybrid E/M cell states. We also highlight the potential challenges in targeting hybrid E/M states through epigenetic inhibitors, and propose future directions that might enable new strategies to target these pro-metastatic cell states in patients.
HYBRID EPITHELIAL-MESENCHYMAL CELL STATES
Epithelial-to-mesenchymal transition: discovery and embryologic role
Epithelial-to-mesenchymal transition (EMT) is a dynamic transcriptional process by which epithelial cells lose their cell-cell junctions, and gain mesenchymal characteristics, becoming more migratory and invasive (2). Cells undergo EMT through a series of transcriptional changes that generally include the upregulation of mesenchymal genes, such as N-cadherin (CDH2), fibronectin, and vimentin, and the downregulation of epithelial genes like E-cadherin (CDH1) (3, 4). Morphologically, cells undergoing EMT adopt a spindle shape with front-back polarity (5). Mesenchymal-like cells that have undergone EMT lack the intricate network of tight, gap, and adherens junctions characteristic of epithelial cells, thereby enabling migratory independence. EMT is triggered by a diverse set of signaling molecules, which induce several parallel pathways that culminate in the expression of EMT-associated genes (6). Many EMT-associated genes are transcription factors (TFs), including the six master EMT regulators Twist1/2, Snail1/2, and Zeb1/2, which serve as transcriptional activators and/or repressors of the downstream genes that produce an EMT phenotype (7).
Elizabeth “Betty” Hay first described an EMT-like process in 1968 while observing neural crest migration in chick embryo models (8, 9). Hay also demonstrated that cultured chick epithelial cells suspended in a collagen gel were capable of migrating through the collagen matrix both individually and collectively. Studies in Drosophila revealed that an EMT-like program directed by Twist and Snail mediates embryo dorsoventral polarity (10). Later research showed that EMT also enables other embryologic processes including gastrulation and neural crest migration (11). Additionally, EMT plays a role in wound healing. After an epithelial insult, cells at the wound’s edge undergo EMT and migrate into the damaged extracellular matrix (ECM) (12). The migrating cells then undergo MET and restore normal epithelial architecture within the healing wound. In cancer, EMT may become aberrantly reactivated during tumorigenesis (13). Recent research efforts heavily emphasize EMT as a key driver of solid tumor invasion and metastasis. Thus, EMT and its various intermediate states in this context will be the central focus of this review.
EMT as a spectrum of hybrid epithelial-mesenchymal states
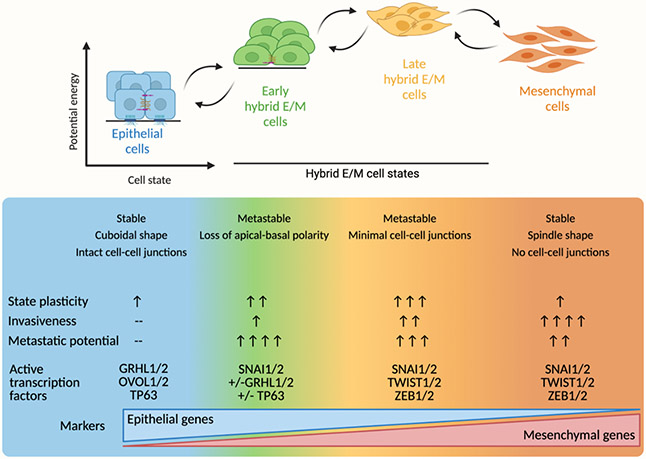
Early investigations characterized EMT as a sharp transition or switch between two binary states. Key epithelial or mesenchymal genes were used as markers of either purely mesenchymal or purely epithelial states, with little attention given to the simultaneous co-expression of both gene types. It is now clear, however, that EMT manifests as a spectrum rather than a switch, and that this spectrum is comprised of several intermediate cell states — collectively termed hybrid epithelial-mesenchymal (hybrid E/M) states (2, 14). Hybrid E/M cells express both epithelial markers (E-cadherin, cytokeratins, claudins, occludins) and mesenchymal markers (N-cadherin, vimentin, fibronectins), and they may display phenotypic characteristics of both cell types (Figure 1). Although there are no standardized biomarkers of hybrid E/M states, analysis of cancer cell lines and patient tumors have demonstrated that cells co-expressing epithelial and mesenchymal genes form the bulk of cells on this spectrum, rather than pure mesenchymal cells (15-17). Importantly, hybrid E/M states are highly plastic and are not fixed in time. Instead, cells transition along the epithelial-mesenchymal axis through a set of intermediate, meta-stable states. “Early” (or more epithelial) hybrid E/M states have a propensity to either revert to an epithelial state or, less commonly, a more mesenchymal-like hybrid E/M state. “Late” hybrid E/M states display the most plasticity and frequently interchange. However, late hybrid E/M cells may also become fully mesenchymal, where they likely remain relatively stable (14). The details of these intermediate states, including their predisposition to metastasize, and their potentially diverse characteristics across different cancers are a major focus of current EMT-related studies (14, 18).
Figure 1. Hybrid E/M states are plastic and phenotypically heterogeneous.
Representative model of hybrid E/M states. Early and late hybrid E/M states display the most plasticity, while mesenchymal and epithelial cells remain relatively stable. Invasiveness is highest in the mesenchymal population. Early hybrid E/M and late hybrid E/M cells likely exhibit the greatest metastatic potential. Hybrid E/M states show simultaneous expression of epithelial and mesenchymal genes. See also: Pastushenko et al. (14), Nieto et al. (2).
Hybrid E/M states in cancer
EMT was first linked to cancer in 1990 when fibroblast growth factor 1 (FGF1) was shown to induce mesenchymal transformation and migration in a rat bladder carcinoma cell line (19). Later studies linked the loss of the adherens junction component E-cadherin to an increased migratory potential, thus prompting investigations into EMT as a driver of tumor invasion and metastasis (20, 21). Additional studies have also implicated various TFs and other hybrid E/M-associated factors in driving the metastatic cascade (22, 23). In patients, tumor mesenchymal gene expression is negatively associated with survival, and correlates with a higher burden of metastatic disease across many cancers (24-29), thus providing additional evidence that EMT drives patient outcomes.
In mouse models, hybrid E/M cells contribute to metastasis while fully mesenchymal cells fail to or rarely establish metastatic colonies (14, 30). Currently, hybrid E/M states are considered the primary drivers of invasion and metastasis over purely mesenchymal cells because hybrid E/M cells demonstrate both epithelial and mesenchymal characteristics, thus enabling them to migrate and travel distantly (mesenchymal phenotypes) but also effectively anchor into and colonize metastatic sites (epithelial phenotypes). For example, hybrid E/M cells invade collectively with some cell-cell junctions still intact, forming tumor projections locally or clusters of circulating tumor cells in blood vessels (31). The collective migration of hybrid E/M cells may give them strength in numbers, thus potentiating their ability to metastasize and subsequently proliferate.
Many genes in the hybrid E/M signature have well-described roles in promoting cell migration. Notably, the matrix metalloproteinases (MMPs) MMP9 and MMP11 are significantly upregulated in cells exhibiting hybrid E/M states (32, 33). MMPs are secreted from malignant cells and cause degradation of the ECM through their collagenase and gelatinase functions, thus promoting cell invasion through dense ECM networks such as basement membranes. Integrins, another group of hybrid E/M effectors, anchor intracellular actomyosin filaments to the ECM and provide the traction necessary for cell migration (34). VIM, which encodes the type III intermediate filament vimentin, is also upregulated in hybrid E/M states and stiffens migrating cells, which produces the structural support necessary for cells to elongate and migrate through tight spaces (35).
During the metastatic cascade, hybrid E/M cells break away from the primary tumor and migrate into local structures, including nearby lymphatic channels, nerves, and blood vessels (36). Cells then travel either independently or in platelet-coated circulating tumor cell (CTC) clusters to distant organs, where they extravasate and migrate into the ECM. A subpopulation of cells may then become more epithelial-like in order to regain their stable cell-cell junctions and proliferative capacity, thus enabling them to anchor at the seeding site, proliferate, and eventually form a stable metastasis (Figure 2).
Figure 2. Hybrid E/M states contribute uniquely to invasion and metastasis.
Both hybrid E/M and mesenchymal cells are able to invade through the extracellular matrix, however the hybrid E/M cells are most likely to seed at a metastatic site. Restoration of epithelial identity in a population of these cells allows for secondary tumor establishment and growth.
The hybrid E/M master transcription factors
The hybrid E/M program is driven primarily by the hybrid E/M master TFs: Snail1/2, Twist1/2, and Zeb1/2. Although these TFs play a crucial role in normal development, once cells have differentiated, hybrid E/M TFs become silenced (37). However, during neoplastic transformation, hybrid E/M TFs can become reactivated and orchestrate malignant cell invasion and migration (22).
Snail1 and Snail2 function primarily as transcriptional repressors that bind the proximal promoter regions of their downstream genes (38). They then associate with repressive complexes, allowing for the transcriptional silencing of their target genes (39). In general, Snail1 and Snail2 block the cell cycle, inhibit apoptosis, and contribute to cell motility through their downstream targets (40). Notably, both Snail1 and Snail2 bind to the promoter of CDH1, as well as other epithelial genes, serving as potent repressors of their expression (39). Conversely, Snail1 and Snail2 can act as transcriptional activators of vimentin and other mesenchymal state-associated genes, thus orchestrating the progression across the hybrid E/M spectrum (41). SNAI1/2 genes are induced primarily by transforming growth factor β (TGF-β) signaling, although endothelial growth factor (EGF) and bone morphogenic protein 4 (BMP4) also contribute to their expression (42). Their post-translational activity is stabilized by TNF-α through the nuclear factor kappa B (NF-kB) pathway, suggesting a role for inflammatory cytokines in promoting hybrid E/M states (43).
The hybrid E/M TFs Twist1 and Twist2 are members of the basic helix-loop-helix (bHLH) family of TFs. Twist1/2 interact with both activating and repressive complexes and thus may upregulate or downregulate their transcriptional targets (44). For example, in hybrid E/M cells, Twist1 silences CDH1 while simultaneously upregulating CDH2, which encodes N-cadherin, a transmembrane protein that plays a crucial role in transendothelial migration and metastasis (45). TWIST1/2 genes become activated by TGF-β, Wnt, and NFkB signaling. Once expressed, Twist1/2 proteins can become diacetylated at coupled lysine residues, which allows them to form complexes with various coactivators, causing upregulation of their downstream genes (46).
Zeb1/2 function as transcriptional activators and repressors depending on the gene and tissue context (47). In their activating mode, Zeb1/2 associate with the p300/CPB-associated factor (PCAF), which promotes the expression of downstream genes through its acetyltransferase activity (48). In contrast, Zeb1/2 also promote transcriptional repression by interacting with C-terminal binding protein 1 (CtBP1) and the Switch/Sucrose Non-Fermentable (SWI/SNF)-associated protein BRG1, which may expose chromatin for Zeb1/2-directed silencing (49). Similar to the TWIST family, Zeb1/2 upregulate mesenchymal genes (VIM, CDH2, FN1), and downregulate epithelial genes (CDH1, occludins, claudins, cytokeratins)(47). ZEB1/2 become upregulated by processes associated with hybrid E/M states, including TGF-β and Wnt/β-catenin signaling, NFkB activity, and hypoxia (50). Interestingly, ZEB1 expression is tightly controlled by both Snail1 and Twist1, suggesting a cooperative feedback loop between the hybrid E/M master TFs (51).
Together, the hybrid E/M master TFs coordinate the expression of downstream mesenchymal and epithelial genes in order to orchestrate cell migration and invasion. Expression levels of the hybrid E/M master TFs is associated with worse survival and adverse clinical features in cancer patients, including local invasion and distant metastasis (52-56). In order to trigger gene expression changes, the hybrid E/M TFs must interact with the cell’s epigenetic machinery to modulate chromatin states. The following section focuses on how both hybrid E/M TFs and pro-epithelial factors recruit general epigenetic effectors in order to promote or inhibit hybrid E/M states.
HISTONE MODIFICATIONS IN HYBRID E/M STATES
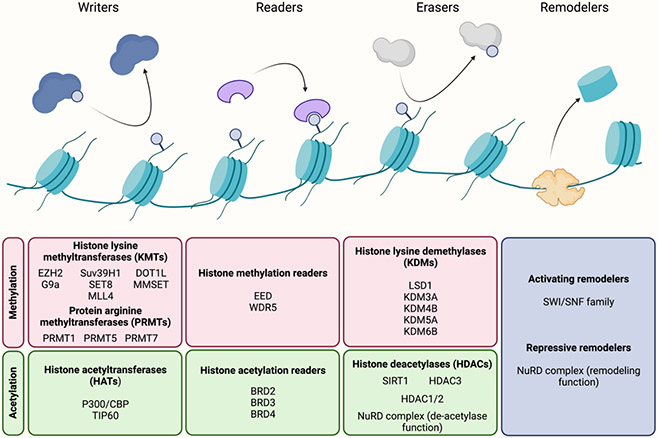
In eukaryotes, histones tightly wrap DNA in order to maintain genomic integrity and to regulate gene expression. The combined DNA-histone structure, the nucleosome, forms the fundamental structural subunit of chromatin, which can generally be found in two states: heterochromatin and euchromatin. Heterochromatin is transcriptionally silent since its condensed structure renders it largely inaccessible to DNA binding proteins. Conversely, euchromatin is less compact and therefore more accessible to transcriptional activators. These chromatin states are, at least in part, regulated by post-translational modifications of the N-terminal tails of the core histone proteins (57). The effectors involved in these processes are divided into three groups: those that modify histones (writers), those that remove modifications (erasers), and those that carry out the function of histone modifications (readers) (58). Additionally, a group of protein complexes known as chromatin remodelers play a crucial role in nucleosome restructuring, movement, and ejection (59). The manipulation of nucleosomes by chromatin remodelers can shield or expose DNA and therefore alter the expression of their target genes. The notable histone-modification effectors in hybrid E/M and their functions are summarized in Figure 3.
Figure 3. Key players in histone-modification biology that contribute to hybrid E/M states.
The histone writers, readers, and erasers modify and carry out functions of covalent histone modifications. The histone remodelers can shift or eject nucleosomes in order to modulate the expression of nearby genes. The listed proteins are those that have been shown to play a role in modulating hybrid E/M states.
Histone modification writers in hybrid E/M
The histone-modification writers can be divided into two broad groups: Methylation and acetylation writers. In general, the histone methylation writers can deposit repressive or permissive methylation marks on their target histones. Thus, histone methylation can be a marker of suppressed or active transcription of nearby genes. In contrast, the histone marks left by the acetylation writers are universally associated with the activation of transcription. The roles of methylation and acetylation writers in hybrid E/M state regulation are listed in Table 1, with some notable examples described below (39, 60-84).
Table 1.
Histone modification writers broadly exhibit pro-hybrid E/M activity.
| Hybrid E/M TF protein interaction |
Hybrid E/M TF gene interaction |
Pro-hybrid E/M state function |
Pro-epithelial state function |
Refs | |
|---|---|---|---|---|---|
| Methylation writers | |||||
| Histone lysine methyltransferases (KMTs) | |||||
| EZH2 (PRC2 complex) | Snai1/2, Twist1 | ZEB2, SNAI2 | Recruited by Snail1/2 and Twist1 to silence expression at epithelial gene promoters. | Involved in silencing key hybrid E/M genes, including ZEB2 and SNAI2. | 39, 60-65 |
| G9a | Snai1/2 | ZEB2 | Recruited by Snail1/2 to silence EpCAM and E-cadherin. | Associates with NuRD complex and downregulates ZEB2. | 66-69, 84 |
| Suv39H1 | Snai1 | -- | Binds to Snail1 and silences epithelial gene expression including E-cadherin. | -- | 70 |
| SET8 | Twist1 | -- | Binds to Twist1; simultaneously silences E-cadherin and upregulates N-cadherin; contributes to oncogenic Wnt signaling. | -- | 71, 72 |
| Histone arginine methyltransferases (PRMTs) | |||||
| PRMT1 | -- | ZEB1 | Catalyzes activating H4R3 methylation at ZEB1 promoter and drives migration and invasion. | -- | 73 |
| PRMT5 | Snai1 | -- | Responds to TGF-β signaling to upregulate mesenchymal genes and downregulate epithelial genes; contributes to Snai1-directed gene silencing. | -- | 74, 61 |
| PRMT7 | -- | -- | Recruits HDAC3 to the E-cadherin promoter. | -- | 75 |
| Permissive histone methyltransferases | |||||
| DOT1L | -- | SNAI1, ZEB1/2 | Catalyzes H3K79 methylation of Snail1 and Zeb1/2 promoters and displaces HDAC1/DNMT1 from loci. | -- | 76 |
| MMSET | -- | TWIST1 | Catalyzes permissive methylation at the Twist1 locus; essential for Twist1-driven invasion. | -- | 77 |
| KMT2B/MLL4 | -- | SNAI1, ZEB1/2 | -- | Binds H3K27 de-methylase UTX and silences SNAI1, ZEB1/2 expression. | 78 |
| Acetylation writers | |||||
| P300/CBP | -- | ZEB1/2, SNAI1 | Mediates TGF-β/Wnt signaling crosstalk; Upregulates several key mesenchymal genes. | -- | 79-81 |
| TIP60 | Twist1, Snai2 | -- | Drives hybrid E/M gene expression through non-histone peptide di-acetylation of Twist1. | Disrupts Snai2-directed gene silencing by destabilizing DNMT1. | 82, 83 |
The histone lysine methyltransferases (KMTs) catalyze the transfer of up to three methyl groups to histone lysine residues, which can repress or permit transcription of nearby genes. (85). Notably, the polycomb repressive complex 2 (PRC2) is an important mediator of repressive histone methylation through its KMT subunits enhancer of zeste 1 polycomb repressive complex 2 (EZH1) and EZH2, which catalyze H3K27 methylation at sites across the genome (86). Arginine residues can also become methylated by protein arginine methyltransferases (PRMTs) (87). Like lysine methylation, arginine methylation can be transcriptionally repressive or permissive.
In hybrid E/M, studies have focused mostly on repressive methylation writers—namely through their interaction with the major hybrid E/M TFs (88). EZH2 of the PRC2 complex has been implicated as an essential driver of hybrid E/M states through its ability to bind Snai1/2 and Twist1, which guide PRC2 to epithelial gene promoter regions (39, 61-63). Here, EZH2 catalyzes repressive histone methylation of several epithelial genes, including CDH1 and the pro-epithelial TF SOX2 (18, 60, 89). However, EZH2 and PRC2 have also been described as key guardians of the epithelial state by silencing mesenchymal genes (64). EZH2 binds to and downregulates ZEB2, SNAI2, MMP2, ITGB3, and other key effectors of hybrid E/M state regulation (65). Therefore, inhibition of EZH2 can also induce hybrid E/M states in some cancers (18, 63). Together, the seemingly contradictory stimulation and abrogation of hybrid E/M by EZH2 and PRC2 highlight the complex, context-dependent nature of epigenetic cell-state regulation.
Another group of histone writers involved in regulating hybrid E/M states is the histone acetyltransferases (HATs). P300/CBP are two notable and closely related HATs that upregulate thousands of genes in the human genome. Studies have generally characterized P300/CBP as drivers of hybrid E/M states, with P300 expression levels generally correlating with mesenchymal gene expression (79). CBP may induce hybrid E/M states through its modulation of TGF-β/Wnt signaling crosstalk, which is a critical induction pathway for hybrid E/M (80). Additionally, the inhibition of P300 is associated with a shift toward epithelial-like hybrid E/M states. Under non-hybrid E/M conditions, GRHL2, an important pro-epithelial TF, prevents transactivation of P300, which in turn prevents P300-mediated upregulation of several hybrid E/M markers, including MMPs, fibronectin, and ZEB1 (81).
Histone modification erasers in hybrid E/M
The histone modification erasers are responsible for removing the marks left by histone modification writers. Both histone demethylases (HDMs) and histone deacetylases (HDACs) play important roles in hybrid E/M, and their functions are summarized in Table 2.
Table 2.
Histone modification erasers contribute to both pro-hybrid E/M and pro-epithelial processes.
| Hybrid E/M TF protein interaction |
Hybrid E/M TF gene interaction |
Pro-hybrid E/M state function |
Pro-epithelial state function |
Refs | |
|---|---|---|---|---|---|
| Histone lysine de-methylases (KDMs) | |||||
| LSD1 | Snai1 | -- | Essential for Snai1-mediated transcriptional repression; becomes activated by EGFR signaling and induces migration. | Binds to NuRD complex and silences targets of TGF-β signaling. | 90-93 |
| KDM3A/JMJD1 | -- | SNAI2 | Upregulates SNAI2 in response to hypoxia signaling; de-represses lncRNA MALAT1. | -- | 100, 101 |
| KDM4B/JMJD2B | -- | -- | De-represses mesenchymal genes in response to Wnt/β-catenin signaling. | -- | 102 |
| KDM5A/RBP2 | -- | -- | Responds to TGF-β signaling to silence E-cadherin and upregulate N-cadherin; de-represses TNC. | -- | 103, 104 |
| KDM6B/JMJD3 | Snai1/2, Twist1 | -- | De-represses mesenchymal genes in response to TGF-β signaling. | -- | 105 |
| Histone de-acetylases (HDACs) | |||||
| SIRT1 | Zeb1 | -- | Binds Zeb1 and silences E-cadherin expression; SIRT1 silencing abrogates in vitro migration and in vivo metastasis. | Directly de-acetylates SMAD4 protein to dampen TGF-β-induced hybrid E/M. | 106, 107 |
| HDAC1/2 | Snai1, Zeb1 | SNAI2 | (HDAC1/2) Silence many epithelial genes under hybrid E/M conditions. | (HDAC1) Binds SMAR1 remodeling complex and silences SNAI2 expression. | 95-98 |
| HDAC3 | Twist1, Snai1 | -- | Silences several epithelial genes in response to HIF1α-driven hypoxia signaling; binds to Twist1 and Snai1 | -- | 108 |
| NuRD complex (de-acetylase function) | Twist1 | -- | Promotes Twist1-directed E-cadherin silencing. | Silences targets of TGF-β signaling via LDS1. | 93, 99 |
The histone demethylase LSD1 exhibits opposing functions in hybrid E/M. When bound to Snail1, LSD1 removes activating H3K4 methylation marks from epithelial genes, including CDH1, CLDN7, and KRT8 (90, 91). LSD1 also becomes activated by EGFR signaling, a known stimulator of hybrid E/M, and facilitates EGFR-induced migration (92). However, when bound to the Mi-2/Nucleosome Remodeling Deacetylase (NuRD) repression complex, LSD1 associates with histone de-acetylases and silences targets of TGF-β signaling, which inhibits hybrid E/M states and reduces tumor metastasis in vivo (93). Thus, similar to other histone modifiers that associate with multiple complexes, LSD1 activity and its effect on hybrid E/M are highly context-specific (94).
Histone deacetylation is carried out by the histone deacetylases. HDACs play a role in repressing the expression of genes in acetylated genomic regions. Removal of acetyl groups increases the positive charge of histones and causes DNA to tighten its loops around nucleosomes. The HDAC1/2 de-acetylase complex, which modulates the expression of thousands of genes, is recruited to the CDH1 promoter by both Snail1 and Zeb1 (95, 96). Together, HDAC1 and HDAC2 serve as important silencers of epithelial genes during hybrid E/M transformation. HDAC1, but not HDAC2, may also be required for TGF-β-induced hybrid E/M gene expression and invasion (97). When bound to the SMAR1 chromatin remodeling complex, HDAC1 is recruited to the SNAI2 promoter, where it de-acetylates histones and represses SNAI2 expression, thus dampening hybrid E/M states (98).
The combined nucleosome remodeling and de-acetylase NuRD complex similarly exhibits opposing roles in hybrid E/M. The NuRD complex can be bound by Twist1 and recruited to the CDH1 promoter, thus silencing E-cadherin and promoting hybrid E/M states (99). However, NuRD also interacts with the histone de-methylase LSD1 and silences targets of TGF-β-signaling, which inhibits hybrid E/M and decreases metastasis formation in vivo (93). The effects of two additional HDACs, SIRT1 and HDAC2, on hybrid E/M states are summarized in Table 2 (90-93, 95-108). Thus, similar to the histone de-methylases, HDACs exhibit both pro- and anti-hybrid E/M activity.
Histone modification readers in hybrid E/M
While writers and erasers are the authors of histone marks, the histone-modification readers are the key effectors of those marks. These proteins recognize and bind to specific histone modifications to promote gene upregulation, gene silencing, or chromatin remodeling (109). Both histone methylation and histone acetylation readers contribute to hybrid E/M gene expression, and their roles are outlined in Table 3 (49, 108, 110-123).
Table 3.
Histone modification readers largely exhibit pro-hybrid E/M activity while the chromatin remodelers display opposing functions.
| Hybrid E/M TF protein interaction |
Hybrid E/M TF gene interaction |
Pro-hybrid E/M state function |
Pro-epithelial state function |
Refs | |
|---|---|---|---|---|---|
| Histone methylation readers | |||||
| EED | -- | -- | Recruits PRC2 to catalyze histone methylation at epithelial gene promoters in response to TGF-β signaling. | -- | 110 |
| WDR5 | Twist1, Snai1 | -- | Associates with HDAC3 and SET1/COMPASS complex to silence epithelial genes. | -- | 108 |
| Histone lysine readers | |||||
| BRD4 | Twist1 | SNAI1/2, TWIST1, ZEB1/2 | Maintains activation of SNAI1/2 gene expression; facilitates TGF-β-induced hybrid E/M gene expression; binds di-acetylated Twist1. | Knockdown is associated with downregulation of TWIST1 and ZEB1/2. | 111-116 |
| BRD2 | -- | -- | Positively regulates expression of hybrid E/M genes TFs. | -- | 116 |
| BRD3 | -- | -- | Facilitates expression of hybrid E/M genes. | Represses hybrid E/M gene expression. | 111, 116 |
| Chromatin remodelers | |||||
| SWI/SNF | Zeb1 | -- | BRG1 subunit binds Zeb1 and facilitates Zeb1-directed gene silencing; involved in WNT5A upregulation. | Subunit ARID1A may suppress hybrid E/M gene expression under non-hybrid E/M conditions. | 49, 117-119 |
| NuRD complex (chromatin remodeling function) | Snai1, Zeb1/2 | TWIST1/2, ZEB2 | Recruited by Snail1 and Zeb1/2 for epithelial gene silencing. | Evicts SWI/SNF from hybrid E/M master TF promoters. | 120-123 |
The direct contribution of histone methylation readers to hybrid E/M states has not been extensively studied, and therefore represents a potential direction for future studies. However, one family of histone acetylation readers—the bromodomain and extraterminal domain (BET) family–has become a focus of many studies. The BET family proteins serve as docking sites for transcriptional co-activators as well as histone acetylation complexes in order to sustain gene expression and maintain open chromatin (124).
BRD4, the most extensively characterized BET protein, has been implicated in facilitating hybrid E/M gene expression. In prostate cancer, BRD4 was shown to bind to the enhancer and promoter regulatory units of SNAI1 and SNAI2, thus driving their expression and promoting hybrid E/M-associated invasive and migratory phenotypes (111). BRD4 inhibition abrogates TGF-β-induced hybrid E/M, suggesting that BRD4 is required for TGF-β signaling and its subsequent gene regulation changes (111, 112). BRD4 also contributes to hybrid E/M states through its protein-protein interaction with di-acetylated Twist1. Once bound to Twist1, BRD4 potently upregulates Twist1 targets, including key mesenchymal genes and WNT5A, an important inducer of hybrid E/M cell states (113-115).
Chromatin remodelers in hybrid E/M
The chromatin remodeling complexes are responsible for moving, ejecting, or restructuring tightly wound nucleosomes (125). Chromatin remodeling activity is indispensable for normal physiologic gene expression as well as changes in cells state in response to external stimuli (126). In eukaryotes, the SWI/SNF and NuRD complexes are the most well-studied complexes to date. In general, the SWI/SNF complex disrupts the equal spacing between nucleosomes, which allows for upregulation of newly exposed genes. Conversely, the NuRD complex simultaneously restructures chromatin and de-acetylates histones in order to downregulate its target genes. The SWI/SNF and NuRD complexes can promote or inhibit hybrid E/M depending on context-specific factors and the co-activators they associate with. Their effects are summarized In Table 3.
The SWI/SNF complex plays a central role in mediating hybrid E/M gene expression changes. Zeb1 binds the SWI/SNF subunit BRG1 and recruits it to Zeb1 target sites. BRG1 is then required for Zeb1-mediated gene silencing, particularly at the CDH1 promoter. BRG1 (SMARCA4) expression levels are associated with pro-hybrid E/M gene expression changes and poor clinical outcomes in several cancers, including colorectal cancer and prostate cancer (49, 117, 118). Interestingly, however, loss of the SWI/SNF subunit ARID1A (which is commonly mutated in numerous cancers), is associated with increased hybrid E/M gene expression, suggesting that the SWI/SNF complex may also play a role in blocking hybrid E/M states (119).
The chromatin remodeling function of the NuRD complex also exhibits pro- and anti-hybrid E/M activity. The NuRD complex binds Snail1, Zeb1, and Twist1 and is recruited to several epithelial-related gene promoters, including CDH1 (99, 120, 121, 127). Emerging evidence also suggests that NuRD can assume an anti-hybrid E/M role by evicting SWI/SNF from the promoters of hybrid E/M master TF genes, thus silencing their expression and abrogating hybrid E/M states (123). Therefore, like the SWI/SNF complex, the NuRD complex can promote or inhibit hybrid E/M depending on context-specific factors and the co-activators it associates with.
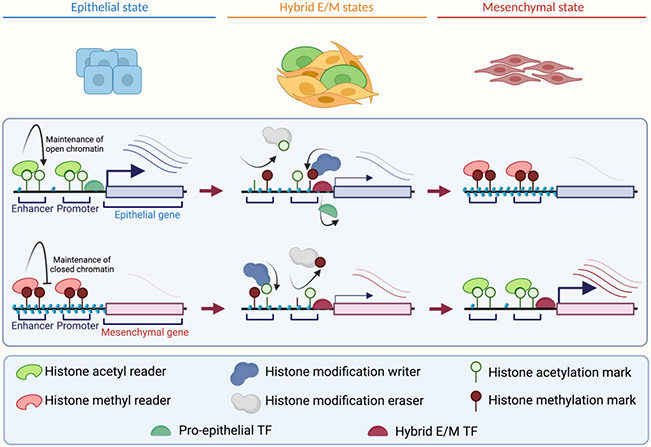
In summary, the effectors of histone modifications contribute to both pro-hybrid E/M and pro-epithelial processes. The transition from epithelial to hybrid E/M states likely relies on the transcriptional changes induced by epigenetic writers and erasers. Once in hybrid E/M states, cells may modulate their expression of both epithelial and mesenchymal genes to progress along the epithelial-mesenchymal axis. Additionally, the sustained activity of histone modification readers likely stabilizes transcription in order to maintain the intermediate hybrid E/M states that drive tumor invasion and metastasis. Ultimately, the cells that reach a full mesenchymal state must exhibit a near-complete silencing of epithelial genes (Figure 4).
Figure 4. Model of the contribution of histone modifications to hybrid E/M state plasticity.
In epithelial cell states, epithelial genes are active and mesenchymal genes are generally silent. Histone modification readers remain bound and maintain open or closed chromatin to stabilize the epithelial state. Additionally, pro-epithelial TFs remain active at epithelial gene promoters. During hybrid E/M induction and in hybrid E/M states, histone writers and erasers silence epithelial genes and de-repress mesenchymal genes in conjunction with pro-mesenchymal TFs. The hybrid E/M states are heterogeneous and highly plastic. In the fully mesenchymal state, histone modification readers maintain open chromatin at mesenchymal genes and closed chromatin at epithelial genes, thus contributing to the relative stability of this state.
DNA METHYLATION AS AN ADDITIONAL REGULATORY MECHANISM IN HYBRID E/M
DNA methylation is an additional epigenetic process that influences gene expression and cell differentiation. The DNA methyltransferase (DNMT) family of enzymes primarily methylates cytosines within CpG dinucleotides (128). DNMT1 maintains CpG methylation of newly synthesized DNA whereas the de novo methyltransferases, DNMT3A and DNMT3B, are primarily responsible for modifying unmethylated cytosines at CpG sites (129, 130). Generally, DNA methylation of promoters silences gene expression (128). Conversely, DNA de-methylation is facilitated by the ten-eleven translocation (TET) family of enzymes. In combination with DNA base excision repair mechanisms, TET enzymes restore an unmethylated cytosine residue to the sequence. Both DNMTs and TETs have been implicated in many human disease processes (131-133). In cancer, global DNA hypomethylation of tumors was observed decades ago (134). Today, it is well-established that aberrant DNA methylation is a hallmark of many tumor types, whereby certain genes are abnormally silenced or expressed (1, 135). The roles of DNMTs and TET enzymes in hybrid E/M are summarized in Table 4 (83, 121, 136-139).
Table 4.
DNA methylation contributes to both hybrid E/M and epithelial state maintenance.
| Hybrid E/M TF protein interaction |
Hybrid E/M TF gene interaction |
Pro-hybrid E/M state function |
Pro-epithelial state function |
Refs | |
|---|---|---|---|---|---|
| DNA methyltransferases (DNMTs) | |||||
| DNMT1 | Snai2 | SNAI1 | Associates with Snail2 and silences target genes. | Associates with ARID2 to silence SNAI1 expression. | 136-138, 83 |
| DNA de-methylases | |||||
| TET1-3 | -- | -- | -- | Maintain active transcription of epithelial genes under non-hybrid E/M conditions | 121, 139 |
Under hybrid E/M conditions, DNMTs generally silence epithelial genes. DNMT1 in particular has demonstrated an important role in driving hybrid E/M states thorough its interaction with Snail1/2 (136, 137). In parallel, TET hydroxylases, which remove repressive DNA methylation marks, become transcriptionally silenced by hybrid E/M signaling pathways (139). Interestingly, DNMT inhibition shows mixed results with regard to hybrid E/M. Pan-DNMT inhibition prevents TGF-β-induced hybrid E/M gene expression and metastatic phenotypes in ovarian and prostate cancer cells (137, 140). However, in lung adenocarcinoma cells, the DNMT inhibitor decitabine blocks hybrid E/M in one cell line but not in the other (138).
Generally, the TET2 and TET3 hydroxylases counteract hybrid E/M states by removing methylation marks on epithelial genes in order to maintain their active transcription. However, hybrid E/M-associated signaling pathways, including TGF-β-signaling, recruit DNMT3A and the NuRD complex to the TET2 and TET3 promoters, silencing their expression and thus exposing epithelial genes for epigenetic silencing (121, 139).
FUTURE CHALLENGES AND STRATEGIES FOR TARGETING EPIGENETIC REGULATORS OF HYBRID E/M
Clinical targeting of hybrid E/M cell states has garnered interest as a way to reduce the burden of metastasis in cancer patients (3). However, inhibiting hybrid E/M states with epigenetic drugs has proven challenging for two key reasons. Firstly, the redundant activities of epigenetic complexes makes targeting specific epigenetic events difficult. For example, the silencing of CDH1 in hybrid E/M cell states is accomplished by the coordination of DNMTs, histone methyltransferases, and HDACs. Therefore, upon inhibition of one of these epigenetic proteins, other silencers may increase their activity and overcome the effects of the drug. Similarly, closely related epigenetic proteins can compensate for each other when one is inhibited. For example, the effect of EZH2 inhibitors is partially reduced by the compensation of EZH1, which can assume the place of EZH2 in the PRC2 complex and fulfill many of its oncogenic roles (141).
Secondly, many epigenetic complexes play seemingly simultaneous pro-epithelial and pro-hybrid E/M roles in the same cells. Given that epigenetic complexes like PRC2 and SWI/SNF carry out their functions across the entire genome, they should be viewed as mediators rather than direct drivers of hybrid E/M gene expression. For example, because gene silencing is crucial for both pro-epithelial and pro-hybrid E/M cell state changes, it follows that PRC2—which is indispensable for genome-wide epigenetic silencing—would play an important role in silencing both epithelial and hybrid E/M genes. Thus, inhibiting these master regulators of transcription is likely to produce broad, global effects rather than hybrid E/M-specific effects.
To overcome the redundant activities of epigenetic complexes, further attention should be directed to combinations of epigenetic therapies that produce larger and more durable transcriptional responses. For example, if targeting epithelial gene silencing in hybrid E/M cells, one might consider combining a DNMT inhibitor with HDAC or histone methyltransferase inhibitors. This approach might help to reduce the likelihood of the development of drug resistance, as it targets two independent but related epigenetic processes. Thus, further studies are needed to determine the most effective combination therapies for targeting hybrid E/M and cancer more broadly.
In order to surmount the challenges presented by the simultaneous pro-hybrid E/M and pro-epithelial functions of many epigenetic complexes, future research efforts should aim to understand the cellular contexts that influence the activity of epigenetic complexes. The rapid advancement of single cell genomics has enabled the study of cell-type specific transcriptional activity, and will likely continue to grow as an indispensable tool to understand how epigenetic complexes function in hybrid E/M versus epithelial-like cells within the same tumor. The majority of existing literature exploring epigenetic regulation of hybrid E/M has been uncovered from bulk cell and tissue RNA samples, which may mask the heterogeneity of hybrid E/M states within cellular subpopulations (142). Thus, what may seem like simultaneous pro-hybrid E/M and pro-epithelial activity from the same epigenetic complex might actually be a collection of unique epigenetic activities stratified by different cell states (i.e. various meta-stable hybrid E/M states). A better understanding of the way cell-state context influences epigenetic complex activity is needed (143). Additionally, future studies should aim to elucidate targets that specifically inhibit pro-hybrid E/M epigenetic activity while permitting pro-epithelial activity. To this end, one approach might be the development of small molecule inhibitors that specifically target the interaction of hybrid E/M TFs with epigenetic complexes. Similarly, a better understanding of how the hybrid E/M TFs interact with transcriptional machinery and epigenetic complexes through deep mutational scanning or small molecule screening might uncover novel methods to target the master hybrid E/M TFs, which to-date have proven difficult to drug.
Finally, in order to successfully target hybrid E/M and metastasis in patients using epigenetic therapies, a better understanding of the intersection between hybrid E/M and its epigenetic regulators with other relevant hallmarks of cancer—including metabolic reprogramming and immune evasion—is needed. For example, hybrid E/M cells have a unique susceptibility to fatty acid receptor inhibitors because of their dependence on lipid metabolism for energy (144). Because epigenetic complexes likely play a role in generating this fatty acid-dependent cell state, they might serve as adjunctive therapy targets to either maximize the effect of metabolic inhibitors or perhaps to reduce the likelihood of adaptive resistance development. Similarly, hybrid E/M cells exhibit a unique ability to both evade and suppress anti-tumor immunity, and they contribute significantly to immune checkpoint blockade (ICB) resistance in tumor mouse models (145, 146). Future investigations might explore the ways that epigenetic complexes are recruited by hybrid E/M TFs to induce ICB resistance, which may occur through several mechanisms including the repression of genes related to antigen presentation and the silencing of potentially immunogenic somatic mutations scattered across the genomes of malignant cells. Many additional examples of the intersectionality between hybrid E/M cell states and other hallmarks of cancer remain to be uncovered. Therefore, future hybrid E/M research should focus on multidisciplinary topics that allow for a more comprehensive understanding of how hybrid E/M cell states affect tumor biology, therapy response and resistance, and ultimately patient outcomes.
CONCLUSION
Invasion and metastasis of solid tumors remains a leading cause of cancer-related death. While countless studies have revealed the epigenetic regulators of hybrid E/M states, to date there are no known therapies that abrogate hybrid E/M and its associated phenotypes in patients. Specific epigenetic regulators and complexes often play opposing roles in promoting and inhibiting hybrid E/M states. Additionally, the gene expression changes associated with hybrid E/M states, such as the silencing of E-cadherin, are the result of redundant and overlapping epigenetic mechanisms. Therefore, future efforts to leverage epigenetic inhibitors as suppressors of hybrid E/M states must focus on potential combination therapies or inhibitors of specific, pro-hybrid E/M protein-protein interactions. Additionally, a better appreciation of the intersection of hybrid E/M and its epigenetic regulators with other hallmarks of cancer might uncover novel therapeutic avenues that lead to more effective and less toxic treatments for cancer patients in the future.
ACKNOWLEDGEMENTS
This work was supported by V Foundation V Scholars Award, Cancer Research Institute Technology Impact Award, Cancer Research Foundation Young Investigator Award, Siteman Cancer Center and Barnes Jewish Foundation, American Cancer Society Institutional Research Grant, K08CA237732 (NIH/NCI), R21DE031072 (NIH/NIDCR) (S.V.P.), R01GM123203 (NIH/NIGMS) (R.D.M.), RF1MH117070, RF1MH126723 (NIH/NIMH) (R.D.M., J.D.D.), R21DE31366 (NIH/NIDCR)(R.D.M.,S.V.P.), and T32HG000045 (NIH/NHGRI) (R.A.S/M.F.N.). Figures created with Biorender.com.
Footnotes
Competing Interests: These authors have no competing interests to declare.
DATA AVAILABILITY STATEMENT
There is no relevant primary data for this publication.
REFERENCES
- 1.Hanahan D. Hallmarks of Cancer: New Dimensions. Cancer Discovery. 2022;12(1):31–46. [DOI] [PubMed] [Google Scholar]
- 2.Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166(1):21–45. [DOI] [PubMed] [Google Scholar]
- 3.Dongre A, Weinberg RA. New insights into the mechanisms of epithelial–mesenchymal transition and implications for cancer. Nature Reviews Molecular Cell Biology. 2019;20(2):69–84. [DOI] [PubMed] [Google Scholar]
- 4.Serrano-Gomez SJ, Maziveyi M, Alahari SK. Regulation of epithelial-mesenchymal transition through epigenetic and post-translational modifications. Molecular Cancer. 2016;15(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moustakas A, Heldin CH. Signaling networks guiding epithelial-mesenchymal transitions during embryogenesis and cancer progression. Cancer Sci. 2007;98(10):1512–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stemmler MP, Eccles RL, Brabletz S, Brabletz T. Non-redundant functions of EMT transcription factors. Nature Cell Biology. 2019;21(1):102–12. [DOI] [PubMed] [Google Scholar]
- 8.Acloque H, Thiery JP, Nieto MA. The physiology and pathology of the EMT. Meeting on the epithelial-mesenchymal transition. EMBO Rep. 2008;9(4):322–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hay B. Organization and fine structure of epithelium and mesenchyme in the developing chick embryos. Proceedings of the 18th Hahnemann Symposium Epithelial-MesenchymalInteractions1968. [Google Scholar]
- 10.Leptin M, Grunewald B. Cell shape changes during gastrulation in Drosophila. Development. 1990;110(1):73–84. [DOI] [PubMed] [Google Scholar]
- 11.Oda H, Tsukita S, Takeichi M. Dynamic behavior of the cadherin-based cell-cell adhesion system during Drosophila gastrulation. Dev Biol. 1998;203(2):435–50. [DOI] [PubMed] [Google Scholar]
- 12.Shaw TJ, Martin P. Wound repair: a showcase for cell plasticity and migration. Curr Opin Cell Biol. 2016;42:29–37. [DOI] [PubMed] [Google Scholar]
- 13.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. [DOI] [PubMed] [Google Scholar]
- 14.Pastushenko I, Brisebarre A, Sifrim A, Fioramonti M, Revenco T, Boumahdi S, et al. Identification of the tumour transition states occurring during EMT. Nature. 2018;556(7702):463–8. [DOI] [PubMed] [Google Scholar]
- 15.Lambert AW, Pattabiraman DR, Weinberg RA. Emerging Biological Principles of Metastasis. Cell. 2017;168(4):670–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell. 2017;171(7):1611–24.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andriani F, Bertolini G, Facchinetti F, Baldoli E, Moro M, Casalini P, et al. Conversion to stem-cell state in response to microenvironmental cues is regulated by balance between epithelial and mesenchymal features in lung cancer cells. Mol Oncol. 2016;10(2):253–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pastushenko I, Mauri F, Song Y, de Cock F, Meeusen B, Swedlund B, et al. Fat1 deletion promotes hybrid EMT state, tumour stemness and metastasis. Nature. 2021;589(7842):448–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vallés AM, Boyer B, Badet J, Tucker GC, Barritault D, Thiery JP. Acidic fibroblast growth factor is a modulator of epithelial plasticity in a rat bladder carcinoma cell line. Proc Natl Acad Sci U S A. 1990;87(3):1124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsuyoshi N, Hamaguchi M, Taniguchi S, Nagafuchi A, Tsukita S, Takeichi M. Cadherin-mediated cell-cell adhesion is perturbed by v-src tyrosine phosphorylation in metastatic fibroblasts. Journal of Cell Biology. 1992;118(3):703–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vincent-Salomon A, Thiery JP. Host microenvironment in breast cancer development: epithelial-mesenchymal transition in breast cancer development. Breast Cancer Res. 2003;5(2):101–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fazilaty H, Rago L, Kass Youssef K, Ocaña OH, Garcia-Asencio F, Arcas A, et al. A gene regulatory network to control EMT programs in development and disease. Nature Communications. 2019;10(1):5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tran HD, Luitel K, Kim M, Zhang K, Longmore GD, Tran DD. Transient SNAIL1 expression is necessary for metastatic competence in breast cancer. Cancer research. 2014;74(21):6330–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao R, Yuan L, Ma B, Wang G, Qiu W, Tian Y. An EMT-related gene signature for the prognosis of human bladder cancer. J Cell Mol Med. 2020;24(1):605–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung CH, Parker JS, Ely K, Carter J, Yi Y, Murphy BA, et al. Gene expression profiles identify epithelial-to-mesenchymal transition and activation of nuclear factor-kappaB signaling as characteristics of a high-risk head and neck squamous cell carcinoma. Cancer Res. 2006;66(16):8210–8. [DOI] [PubMed] [Google Scholar]
- 26.Foroutan M, Cursons J, Hediyeh-Zadeh S, Thompson EW, Davis MJ. A Transcriptional Program for Detecting TGFβ-Induced EMT in Cancer. Mol Cancer Res. 2017;15(5):619–31. [DOI] [PubMed] [Google Scholar]
- 27.Gröger CJ, Grubinger M, Waldhör T, Vierlinger K, Mikulits W. Meta-analysis of gene expression signatures defining the epithelial to mesenchymal transition during cancer progression. PLoS One. 2012;7(12):e51136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang RY, Wong MK, Tan TZ, Kuay KT, Ng AH, Chung VY, et al. An EMT spectrum defines an anoikis-resistant and spheroidogenic intermediate mesenchymal state that is sensitive to e-cadherin restoration by a src-kinase inhibitor, saracatinib (AZD0530). Cell Death Dis. 2013;4(11):e915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiong T, Wang M, Zhao J, Liu Q, Yang C, Luo W, et al. An esophageal squamous cell carcinoma classification system that reveals potential targets for therapy. Oncotarget. 2017;8(30):49851–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lüönd F, Sugiyama N, Bill R, Bornes L, Hager C, Tang F, et al. Distinct contributions of partial and full EMT to breast cancer malignancy. Dev Cell. 2021;56(23):3203–21.e11. [DOI] [PubMed] [Google Scholar]
- 31.Saxena K, Jolly MK, Balamurugan K. Hypoxia, partial EMT and collective migration: Emerging culprits in metastasis. Transl Oncol. 2020;13(11):100845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roy R, Yang J, Moses MA. Matrix metalloproteinases as novel biomarkers and potential therapeutic targets in human cancer. J Clin Oncol. 2009;27(31):5287–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141(1):52–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hood JD, Cheresh DA. Role of integrins in cell invasion and migration. Nature Reviews Cancer. 2002;2(2):91–100. [DOI] [PubMed] [Google Scholar]
- 35.Xuan B, Ghosh D, Jiang J, Shao R, Dawson MR. Vimentin filaments drive migratory persistence in polyploidal cancer cells. Proceedings of the National Academy of Sciences. 2020;117(43):26756–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu W, Kang Y. Epithelial-Mesenchymal Plasticity in Cancer Progression and Metastasis. Dev Cell. 2019;49(3):361–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Batlle E, Sancho E, Francí C, Domínguez D, Monfar M, Baulida J, et al. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2(2):84–9. [DOI] [PubMed] [Google Scholar]
- 39.Herranz N, Pasini D, Díaz VM, Francí C, Gutierrez A, Dave N, et al. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol Cell Biol. 2008;28(15):4772–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Virtakoivu R, Mai A, Mattila E, De Franceschi N, Imanishi SY, Corthals G, et al. Vimentin-ERK Signaling Uncouples Slug Gene Regulatory Function. Cancer Res. 2015;75(11):2349–62. [DOI] [PubMed] [Google Scholar]
- 42.Peinado H, Quintanilla M, Cano A. Transforming growth factor beta-1 induces snail transcription factor in epithelial cell lines: mechanisms for epithelial mesenchymal transitions. J Biol Chem. 2003;278(23):21113–23. [DOI] [PubMed] [Google Scholar]
- 43.Wu Y, Deng J, Rychahou PG, Qiu S, Evers BM, Zhou BP. Stabilization of snail by NF-kappaB is required for inflammation-induced cell migration and invasion. Cancer Cell. 2009;15(5):416–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wallerand H, Robert G, Pasticier G, Ravaud A, Ballanger P, Reiter RE, et al. The epithelial-mesenchymal transition-inducing factor TWIST is an attractive target in advanced and/or metastatic bladder and prostate cancers. Urol Oncol. 2010;28(5):473–9. [DOI] [PubMed] [Google Scholar]
- 45.Ramis-Conde I, Chaplain MA, Anderson AR, Drasdo D. Multi-scale modelling of cancer cell intravasation: the role of cadherins in metastasis. Phys Biol. 2009;6(1):016008. [DOI] [PubMed] [Google Scholar]
- 46.Wang L-T, Wang S-N, Chiou S-S, Liu K-Y, Chai C-Y, Chiang C-M, et al. TIP60-dependent acetylation of the SPZ1-TWIST complex promotes epithelial–mesenchymal transition and metastasis in liver cancer. Oncogene. 2019;38(4):518–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang P, Sun Y, Ma L. ZEB1: at the crossroads of epithelial-mesenchymal transition, metastasis and therapy resistance. Cell Cycle. 2015;14(4):481–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mizuguchi Y, Specht S, Lunz JG 3rd, Isse K, Corbitt N, Takizawa T, et al. Cooperation of p300 and PCAF in the control of microRNA 200c/141 transcription and epithelial characteristics. PLoS One. 2012;7(2):e32449–e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sánchez-Tilló E, Lázaro A, Torrent R, Cuatrecasas M, Vaquero EC, Castells A, et al. ZEB1 represses E-cadherin and induces an EMT by recruiting the SWI/SNF chromatin-remodeling protein BRG1. Oncogene. 2010;29(24):3490–500. [DOI] [PubMed] [Google Scholar]
- 50.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7(6):415–28. [DOI] [PubMed] [Google Scholar]
- 51.Dave N, Guaita-Esteruelas S, Gutarra S, Frias À, Beltran M, Peiró S, et al. Functional cooperation between Snail1 and twist in the regulation of ZEB1 expression during epithelial to mesenchymal transition. J Biol Chem. 2011;286(14):12024–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blechschmidt K, Sassen S, Schmalfeldt B, Schuster T, Höfler H, Becker KF. The E-cadherin repressor Snail is associated with lower overall survival of ovarian cancer patients. British Journal of Cancer. 2008;98(2):489–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shioiri M, Shida T, Koda K, Oda K, Seike K, Nishimura M, et al. Slug expression is an independent prognostic parameter for poor survival in colorectal carcinoma patients. British Journal of Cancer. 2006;94(12):1816–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang J, Mani SA, Weinberg RA. Exploring a new twist on tumor metastasis. Cancer Res. 2006;66(9):4549–52. [DOI] [PubMed] [Google Scholar]
- 55.Aigner K, Dampier B, Descovich L, Mikula M, Sultan A, Schreiber M, et al. The transcription factor ZEB1 (deltaEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene. 2007;26(49):6979–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spaderna S, Schmalhofer O, Wahlbuhl M, Dimmler A, Bauer K, Sultan A, et al. The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer Res. 2008;68(2):537–44. [DOI] [PubMed] [Google Scholar]
- 57.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Research. 2011;21(3):381–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tarakhovsky A. Tools and landscapes of epigenetics. Nature Immunology. 2010;11(7):565–8. [DOI] [PubMed] [Google Scholar]
- 59.Saha A, Wittmeyer J, Cairns BR. Chromatin remodelling: the industrial revolution of DNA around histones. Nat Rev Mol Cell Biol. 2006;7(6):437–47. [DOI] [PubMed] [Google Scholar]
- 60.Battistelli C, Cicchini C, Santangelo L, Tramontano A, Grassi L, Gonzalez FJ, et al. The Snail repressor recruits EZH2 to specific genomic sites through the enrollment of the lncRNA HOTAIR in epithelial-to-mesenchymal transition. Oncogene. 2017;36(7):942–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hou Z, Peng H, Ayyanathan K, Yan KP, Langer EM, Longmore GD, et al. The LIM protein AJUBA recruits protein arginine methyltransferase 5 to mediate SNAIL-dependent transcriptional repression. Mol Cell Biol. 2008;28(10):3198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Langer EM, Feng Y, Zhaoyuan H, Rauscher FJ 3rd, Kroll KL, Longmore GD. Ajuba LIM proteins are snail/slug corepressors required for neural crest development in Xenopus. Dev Cell. 2008;14(3):424–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cardenas H, Zhao J, Vieth E, Nephew KP, Matei D. EZH2 inhibition promotes epithelial-to-mesenchymal transition in ovarian cancer cells. Oncotarget. 2016;7(51):84453–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Y, Donaher JL, Das S, Li X, Reinhardt F, Krall JA, et al. Genome-wide CRISPR screen identifies PRC2 and KMT2D-COMPASS as regulators of distinct EMT trajectories that contribute differentially to metastasis. Nat Cell Biol. 2022;24(4):554–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gallardo A, Molina A, Asenjo HG, Lopez-Onieva L, Martorell-Marugán J, Espinosa-Martinez M, et al. EZH2 endorses cell plasticity to non-small cell lung cancer cells facilitating mesenchymal to epithelial transition and tumour colonization. Oncogene. 2022. [DOI] [PubMed] [Google Scholar]
- 66.Chen MW, Hua KT, Kao HJ, Chi CC, Wei LH, Johansson G, et al. H3K9 histone methyltransferase G9a promotes lung cancer invasion and metastasis by silencing the cell adhesion molecule Ep-CAM. Cancer Res. 2010;70(20):7830–40. [DOI] [PubMed] [Google Scholar]
- 67.Liu S, Ye D, Guo W, Yu W, He Y, Hu J, et al. G9a is essential for EMT-mediated metastasis and maintenance of cancer stem cell-like characters in head and neck squamous cell carcinoma. Oncotarget. 2015;6(9):6887–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dong C, Wu Y, Yao J, Wang Y, Yu Y, Rychahou PG, et al. G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. J Clin Invest. 2012;122(4):1469–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu Y, Zheng Y, Dai M, Wang X, Wu J, Yu B, et al. G9a and histone deacetylases are crucial for Snail2-mediated E-cadherin repression and metastasis in hepatocellular carcinoma. Cancer Sci. 2019;110(11):3442–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dong C, Wu Y, Wang Y, Wang C, Kang T, Rychahou PG, et al. Interaction with Suv39H1 is critical for Snail-mediated E-cadherin repression in breast cancer. Oncogene. 2013;32(11):1351–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang F, Sun L, Li Q, Han X, Lei L, Zhang H, et al. SET8 promotes epithelial-mesenchymal transition and confers TWIST dual transcriptional activities. Embo j. 2012;31(1):110–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li Z, Nie F, Wang S, Li L. Histone H4 Lys 20 monomethylation by histone methylase SET8 mediates Wnt target gene activation. Proc Natl Acad Sci U S A. 2011;108(8):3116–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gao Y, Zhao Y, Zhang J, Lu Y, Liu X, Geng P, et al. The dual function of PRMT1 in modulating epithelial-mesenchymal transition and cellular senescence in breast cancer cells through regulation of ZEB1. Sci Rep. 2016;6:19874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen H, Lorton B, Gupta V, Shechter D. A TGFβ-PRMT5-MEP50 axis regulates cancer cell invasion through histone H3 and H4 arginine methylation coupled transcriptional activation and repression. Oncogene. 2017;36(3):373–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yao R, Jiang H, Ma Y, Wang L, Wang L, Du J, et al. PRMT7 induces epithelial-to-mesenchymal transition and promotes metastasis in breast cancer. Cancer Res. 2014;74(19):5656–67. [DOI] [PubMed] [Google Scholar]
- 76.Cho MH, Park JH, Choi HJ, Park MK, Won HY, Park YJ, et al. DOT1L cooperates with the c-Myc-p300 complex to epigenetically derepress CDH1 transcription factors in breast cancer progression. Nat Commun. 2015;6:7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ezponda T, Popovic R, Shah MY, Martinez-Garcia E, Zheng Y, Min DJ, et al. The histone methyltransferase MMSET/WHSC1 activates TWIST1 to promote an epithelial-mesenchymal transition and invasive properties of prostate cancer. Oncogene. 2013;32(23):2882–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Choi HJ, Park JH, Park M, Won HY, Joo HS, Lee CH, et al. UTX inhibits EMT-induced breast CSC properties by epigenetic repression of EMT genes in cooperation with LSD1 and HDAC1. EMBO Rep. 2015;16(10):1288–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hou X, Gong R, Zhan J, Zhou T, Ma Y, Zhao Y, et al. p300 promotes proliferation, migration, and invasion via inducing epithelial-mesenchymal transition in non-small cell lung cancer cells. BMC Cancer. 2018;18(1):641. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 80.Zhou B, Liu Y, Kahn M, Ann DK, Han A, Wang H, et al. Interactions between β-catenin and transforming growth factor-β signaling pathways mediate epithelial-mesenchymal transition and are dependent on the transcriptional co-activator cAMP-response element-binding protein (CREB)-binding protein (CBP). J Biol Chem. 2012;287(10):7026–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pifer PM, Farris JC, Thomas AL, Stoilov P, Denvir J, Smith DM, et al. Grainyhead-like 2 inhibits the coactivator p300, suppressing tubulogenesis and the epithelial-mesenchymal transition. Mol Biol Cell. 2016;27(15):2479–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang LT, Wang SN, Chiou SS, Liu KY, Chai CY, Chiang CM, et al. TIP60-dependent acetylation of the SPZ1-TWIST complex promotes epithelial-mesenchymal transition and metastasis in liver cancer. Oncogene. 2019;38(4):518–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang Y, Subbaiah VK, Rajagopalan D, Tham CY, Abdullah LN, Toh TB, et al. TIP60 inhibits metastasis by ablating DNMT1-SNAIL2-driven epithelial-mesenchymal transition program. J Mol Cell Biol. 2016;8(5):384–99. [DOI] [PubMed] [Google Scholar]
- 84.Si W, Huang W, Zheng Y, Yang Y, Liu X, Shan L, et al. Dysfunction of the Reciprocal Feedback Loop between GATA3- and ZEB2-Nucleated Repression Programs Contributes to Breast Cancer Metastasis. Cancer Cell. 2015;27(6):822–36. [DOI] [PubMed] [Google Scholar]
- 85.Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142(6):967–80. [DOI] [PubMed] [Google Scholar]
- 86.Hyun K, Jeon J, Park K, Kim J. Writing, erasing and reading histone lysine methylations. Experimental & Molecular Medicine. 2017;49(4):e324–e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zurita-Lopez CI, Sandberg T, Kelly R, Clarke SG. Human protein arginine methyltransferase 7 (PRMT7) is a type III enzyme forming ω-NG-monomethylated arginine residues. The Journal of biological chemistry. 2012;287(11):7859–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lin Y, Dong C, Zhou BP. Epigenetic regulation of EMT: the Snail story. Curr Pharm Des. 2014;20(11):1698–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tiwari N, Tiwari VK, Waldmeier L, Balwierz PJ, Arnold P, Pachkov M, et al. Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer Cell. 2013;23(6):768–83. [DOI] [PubMed] [Google Scholar]
- 90.Lin T, Ponn A, Hu X, Law BK, Lu J. Requirement of the histone demethylase LSD1 in Snai1-mediated transcriptional repression during epithelial-mesenchymal transition. Oncogene. 2010;29(35):4896–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lin Y, Wu Y, Li J, Dong C, Ye X, Chi YI, et al. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. Embo j. 2010;29(11):1803–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shao G, Wang J, Li Y, Liu X, Xie X, Wan X, et al. Lysine-specific demethylase 1 mediates epidermal growth factor signaling to promote cell migration in ovarian cancer cells. Sci Rep. 2015;5:15344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang Y, Zhang H, Chen Y, Sun Y, Yang F, Yu W, et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009;138(4):660–72. [DOI] [PubMed] [Google Scholar]
- 94.Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007;446(7138):882–7. [DOI] [PubMed] [Google Scholar]
- 95.Peinado H, Ballestar E, Esteller M, Cano A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol. 2004;24(1):306–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Aghdassi A, Sendler M, Guenther A, Mayerle J, Behn CO, Heidecke CD, et al. Recruitment of histone deacetylases HDAC1 and HDAC2 by the transcriptional repressor ZEB1 downregulates E-cadherin expression in pancreatic cancer. Gut. 2012;61(3):439–48. [DOI] [PubMed] [Google Scholar]
- 97.Lei W, Zhang K, Pan X, Hu Y, Wang D, Yuan X, et al. Histone deacetylase 1 is required for transforming growth factor-beta1-induced epithelial-mesenchymal transition. Int J Biochem Cell Biol. 2010;42(9):1489–97. [DOI] [PubMed] [Google Scholar]
- 98.Adhikary A, Chakraborty S, Mazumdar M, Ghosh S, Mukherjee S, Manna A, et al. Inhibition of epithelial to mesenchymal transition by E-cadherin up-regulation via repression of slug transcription and inhibition of E-cadherin degradation: dual role of scaffold/matrix attachment region-binding protein 1 (SMAR1) in breast cancer cells. J Biol Chem. 2014;289(37):25431–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fu J, Qin L, He T, Qin J, Hong J, Wong J, et al. The TWIST/Mi2/NuRD protein complex and its essential role in cancer metastasis. Cell Res. 2011;21(2):275–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ahn HJ, Moon B, Park M, Kim JA. KDM3A regulates Slug expression to promote the invasion of MCF7 breast cancer cells in hypoxia. Oncol Lett. 2020;20(6):335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tee AE, Ling D, Nelson C, Atmadibrata B, Dinger ME, Xu N, et al. The histone demethylase JMJD1A induces cell migration and invasion by up-regulating the expression of the long noncoding RNA MALAT1. Oncotarget. 2014;5(7):1793–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhao L, Li W, Zang W, Liu Z, Xu X, Yu H, et al. JMJD2B promotes epithelial-mesenchymal transition by cooperating with β-catenin and enhances gastric cancer metastasis. Clin Cancer Res. 2013;19(23):6419–29. [DOI] [PubMed] [Google Scholar]
- 103.Wang S, Wang Y, Wu H, Hu L. RBP2 induces epithelial-mesenchymal transition in non-small cell lung cancer. PLoS One. 2013;8(12):e84735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liang X, Zeng J, Wang L, Shen L, Ma X, Li S, et al. Histone demethylase RBP2 promotes malignant progression of gastric cancer through TGF-β1-(p-Smad3)-RBP2-E-cadherin-Smad3 feedback circuit. Oncotarget. 2015;6(19):17661–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ramadoss S, Chen X, Wang CY. Histone demethylase KDM6B promotes epithelial-mesenchymal transition. J Biol Chem. 2012;287(53):44508–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Byles V, Zhu L, Lovaas JD, Chmilewski LK, Wang J, Faller DV, et al. SIRT1 induces EMT by cooperating with EMT transcription factors and enhances prostate cancer cell migration and metastasis. Oncogene. 2012;31(43):4619–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen IC, Chiang WF, Huang HH, Chen PF, Shen YY, Chiang HC. Role of SIRT1 in regulation of epithelial-to-mesenchymal transition in oral squamous cell carcinoma metastasis. Mol Cancer. 2014;13:254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wu MZ, Tsai YP, Yang MH, Huang CH, Chang SY, Chang CC, et al. Interplay between HDAC3 and WDR5 is essential for hypoxia-induced epithelial-mesenchymal transition. Mol Cell. 2011;43(5):811–22. [DOI] [PubMed] [Google Scholar]
- 109.Yun M, Wu J, Workman JL, Li B. Readers of histone modifications. Cell Research. 2011;21(4):564–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Oktyabri D, Tange S, Terashima M, Ishimura A, Suzuki T. EED regulates epithelial-mesenchymal transition of cancer cells induced by TGF-β. Biochem Biophys Res Commun. 2014;453(1):124–30. [DOI] [PubMed] [Google Scholar]
- 111.Shafran JS, Jafari N, Casey AN, Győrffy B, Denis GV. BRD4 regulates key transcription factors that drive epithelial-mesenchymal transition in castration-resistant prostate cancer. Prostate Cancer Prostatic Dis. 2021;24(1):268–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tan Y-F, Wang M, Chen Z-Y, Wang L, Liu X-H. Inhibition of BRD4 prevents proliferation and epithelial–mesenchymal transition in renal cell carcinoma via NLRP3 inflammasome-induced pyroptosis. Cell Death & Disease. 2020;11(4):239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shi J, Wang Y, Zeng L, Wu Y, Deng J, Zhang Q, et al. Disrupting the interaction of BRD4 with diacetylated Twist suppresses tumorigenesis in basal-like breast cancer. Cancer Cell. 2014;25(2):210–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Qin ZY, Wang T, Su S, Shen LT, Zhu GX, Liu Q, et al. BRD4 Promotes Gastric Cancer Progression and Metastasis through Acetylation-Dependent Stabilization of Snail. Cancer Res. 2019;79(19):4869–81. [DOI] [PubMed] [Google Scholar]
- 115.Lu L, Chen Z, Lin X, Tian L, Su Q, An P, et al. Inhibition of BRD4 suppresses the malignancy of breast cancer cells via regulation of Snail. Cell Death Differ. 2020;27(1):255–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Andrieu GP, Denis GV. BET Proteins Exhibit Transcriptional and Functional Opposition in the Epithelial-to-Mesenchymal Transition. Mol Cancer Res. 2018;16(4):580–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lin S, Jiang T, Ye L, Han Z, Liu Y, Liu C, et al. The chromatin-remodeling enzyme BRG1 promotes colon cancer progression via positive regulation of WNT3A. Oncotarget. 2016;7(52):86051–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bai J, Mei P, Zhang C, Chen F, Li C, Pan Z, et al. BRG1 is a prognostic marker and potential therapeutic target in human breast cancer. PLoS One. 2013;8(3):e59772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tomihara H, Carbone F, Perelli L, Huang JK, Soeung M, Rose JL, et al. Loss of ARID1A Promotes Epithelial-Mesenchymal Transition and Sensitizes Pancreatic Tumors to Proteotoxic Stress. Cancer Res. 2021;81(2):332–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Manshouri R, Coyaud E, Kundu ST, Peng DH, Stratton SA, Alton K, et al. ZEB1/NuRD complex suppresses TBC1D2b to stimulate E-cadherin internalization and promote metastasis in lung cancer. Nat Commun. 2019;10(1):5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gao J, Liu R, Feng D, Huang W, Huo M, Zhang J, et al. Snail/PRMT5/NuRD complex contributes to DNA hypermethylation in cervical cancer by TET1 inhibition. Cell Death Differ. 2021;28(9):2818–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nihan Kilinc A, Sugiyama N, Reddy Kalathur RK, Antoniadis H, Birogul H, Ishay-Ronen D, et al. Histone deacetylases, Mbd3/NuRD, and Tet2 hydroxylase are crucial regulators of epithelial-mesenchymal plasticity and tumor metastasis. Oncogene. 2020;39(7):1498–513. [DOI] [PubMed] [Google Scholar]
- 123.Mohd-Sarip A, Teeuwssen M, Bot AG, De Herdt MJ, Willems SM, Baatenburg de Jong RJ, et al. DOC1-Dependent Recruitment of NURD Reveals Antagonism with SWI/SNF during Epithelial-Mesenchymal Transition in Oral Cancer Cells. Cell Rep. 2017;20(1):61–75. [DOI] [PubMed] [Google Scholar]
- 124.Sabari BR, Zhang D, Allis CD, Zhao Y. Metabolic regulation of gene expression through histone acylations. Nature Reviews Molecular Cell Biology. 2017;18(2):90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Clapier CR, Iwasa J, Cairns BR, Peterson CL. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat Rev Mol Cell Biol. 2017;18(7):407–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463(7280):474–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Xu Y, Qin L, Sun T, Wu H, He T, Yang Z, et al. Twist1 promotes breast cancer invasion and metastasis by silencing Foxa1 expression. Oncogene. 2017;36(8):1157–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Moore LD, Le T, Fan G. DNA Methylation and Its Basic Function. Neuropsychopharmacology. 2013;38(1):23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet. 2009;10(11):805–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–57. [DOI] [PubMed] [Google Scholar]
- 131.Greenberg MVC, Bourc’his D. The diverse roles of DNA methylation in mammalian development and disease. Nature Reviews Molecular Cell Biology. 2019;20(10):590–607. [DOI] [PubMed] [Google Scholar]
- 132.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nature Reviews Genetics. 2013;14(3):204–20. [DOI] [PubMed] [Google Scholar]
- 133.Nishiyama A, Nakanishi M. Navigating the DNA methylation landscape of cancer. Trends in Genetics. 2021;37(11):1012–27. [DOI] [PubMed] [Google Scholar]
- 134.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301(5895):89–92. [DOI] [PubMed] [Google Scholar]
- 135.Saghafinia S, Mina M, Riggi N, Hanahan D, Ciriello G. Pan-Cancer Landscape of Aberrant DNA Methylation across Human Tumors. Cell Rep. 2018;25(4):1066–80.e8. [DOI] [PubMed] [Google Scholar]
- 136.Jiang H, Cao HJ, Ma N, Bao WD, Wang JJ, Chen TW, et al. Chromatin remodeling factor ARID2 suppresses hepatocellular carcinoma metastasis via DNMT1-Snail axis. Proc Natl Acad Sci U S A. 2020;117(9):4770–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lee E, Wang J, Yumoto K, Jung Y, Cackowski FC, Decker AM, et al. DNMT1 Regulates Epithelial-Mesenchymal Transition and Cancer Stem Cells, Which Promotes Prostate Cancer Metastasis. Neoplasia. 2016;18(9):553–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhang N, Liu Y, Wang Y, Zhao M, Tu L, Luo F. Decitabine reverses TGF-β1-induced epithelial-mesenchymal transition in non-small-cell lung cancer by regulating miR-200/ZEB axis. Drug Des Devel Ther. 2017;11:969–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gong F, Guo Y, Niu Y, Jin J, Zhang X, Shi X, et al. Epigenetic silencing of TET2 and TET3 induces an EMT-like process in melanoma. Oncotarget. 2017;8(1):315–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Cardenas H, Vieth E, Lee J, Segar M, Liu Y, Nephew KP, et al. TGF-β induces global changes in DNA methylation during the epithelial-to-mesenchymal transition in ovarian cancer cells. Epigenetics. 2014;9(11):1461–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nepali K, Liou J-P. Recent developments in epigenetic cancer therapeutics: clinical advancement and emerging trends. Journal of Biomedical Science. 2021;28(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pal A, Barrett TF, Paolini R, Parikh A, Puram SV. Partial EMT in head and neck cancer biology: a spectrum instead of a switch. Oncogene. 2021;40(32):5049–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Puram SV, Mints M, Pal A, Qi Z, Reeb A, Gelev K, et al. Cellular states are coupled to genomic and viral heterogeneity in HPV-related oropharyngeal carcinoma. Nature Genetics. 2023;55(4):640–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pascual G, Avgustinova A, Mejetta S, Martín M, Castellanos A, Attolini CS, et al. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature. 2017;541(7635):41–5. [DOI] [PubMed] [Google Scholar]
- 145.Dongre A, Rashidian M, Eaton EN, Reinhardt F, Thiru P, Zagorulya M, et al. Direct and Indirect Regulators of Epithelial-Mesenchymal Transition-Mediated Immunosuppression in Breast Carcinomas. Cancer Discov. 2021;11(5):1286–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mullins RDZ, Pal A, Barrett TF, Heft Neal ME, Puram SV. Epithelial-Mesenchymal Plasticity in Tumor Immune Evasion. Cancer Res. 2022;82(13):2329–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
There is no relevant primary data for this publication.