Abstract
The prevalence of antibiotic resistance has been increasing globally, and new antimicrobial agents are needed to address this growing problem. We previously reported that a stilbene dimer from Photorhabdus gammaproteobacteria exhibits strong activity relative to its monomer against the multidrug-resistant Gram-positive pathogens methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus faecalis (VRE). Here, we show that related dietary plant stilbene-derived dimers also have activity against these pathogens, and MRSA is unable to develop substantial resistance even after daily non-lethal exposure of the lead compound for a duration of three months. Through a systematic deduction process, we established the mode of action of the lead dimer, which targets the bacterial cell wall. Genome sequencing of modest resistance mutants, mass spectrometry analysis of cell wall precursors, and exogenous lipid II chemical complementation studies support the target as being lipid II itself or lipid II trafficking processes. Given the broad distribution of stilbenes in plants, including dietary plants, we anticipate that our mode of action studies here could be more broadly applicable to multipartite host-bacteria-plant interactions.
Graphical Abstract
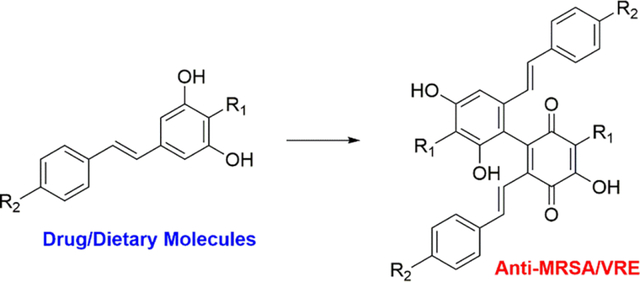
Antibiotics, beginning with the clinical development of penicillin, have had a significant impact on medicine and global health.1, 2 However, bacteria evolve relatively quickly and can develop resistance to most antibiotics.3 The incidence of antibiotic-resistant bacteria is increasing, and multidrug-resistant organisms have become prevalent around the world, representing a growing health concern.4 New antibiotics, particularly new structural classes that do not lead to rapid development of resistant bacteria, are needed to combat this issue. Approximately 69% of antibiotics are natural products or natural product derivatives, and 97% of these have been discovered from bacterial or fungal sources, which likely have evolved as inter-microbial competition strategies.5–7
Stilbenes are a class of phenylpropanoid polyketides that are widespread in plants, including dietary plants such as peanuts, grapes, and blueberries.8 They are also produced by all known species of the gammaproteobacterial genus Photorhabdus.9–11 In Photorhabdus, the most abundant stilbene is an isopropyl-functionalized analog known as tapinarof (1), which has been approved for the treatment of psoriasis and atopic dermatitis.12–15 In addition to immunomodulatory activities, stilbenes (including tapinarof) are known to harbor only moderate antimicrobial activities.14
We previously reported the discovery and structural characterization of two new stilbene dimer scaffolds derived from tapinarof in Photorhabdus.16 One of these dimers, termed duotap-520, exhibits strong antibiotic activity against multidrug resistant Gram-positive pathogens, including methicillin-resistant Staphylococcus aureus (MRSA; minimal inhibitory concentration [MIC] 4 μM or 2 μg/ml) and vancomycin-resistant Enterococcus faecalis (VRE; MIC 6 μM or 3 μg/ml); no activity against selected Gram-negative pathogens was observed.16 We also reported that select plant stilbenes can undergo similar structural transformations.16 Here, we show that the structural transformations of dietary plant stilbenes also lead to functional transformations, endowing these dimeric scaffolds with antibiotic activity. We further establish the mode of action of the lead dimer, duotap-520, which targets the bacterial cell wall and does so without the development of considerable antibiotic resistance in antibiotic challenge studies.
Photorhabdus stilbene 1 can be transformed into dimers 2 (duotap-520) and 7 (carbocyclinone-534) through spontaneous oxidation chemistry; however, we previously showed that a cupin enzyme Plu1886 enhances duotap-520 formation in vitro and in micro-aerobic cell culture studies.16 Indeed, Plu1886 converted the related plant stilbenes pinosylvin (3) and resveratrol (5) into their respective duotap-436 (4) and carbocyclinone-482 (8) products with negligible spontaneous background rates of formation.16 Hence,these types of molecules can be formed spontaneously or biocatalytically. While Plu1886 transformed resveratrol into carbocyclinone-482, we could access its duotap-468 (6) scaffold here through spontaneous chemistry when incubated in the presence of copper, as determined by high-resolution liquid chromatography mass spectrometry (LC/MS; [M+H]+ 469.1297, 1). We isolated this dimerization product and confirmed that it was the resveratrol-derived 6 by 1D and 2D NMR (Figures S2–6). These efforts have established a small panel of stilbene monomers and dimers featuring common stilbene functionalizations for antimicrobial analysis (Figure 1).
Figure 1.
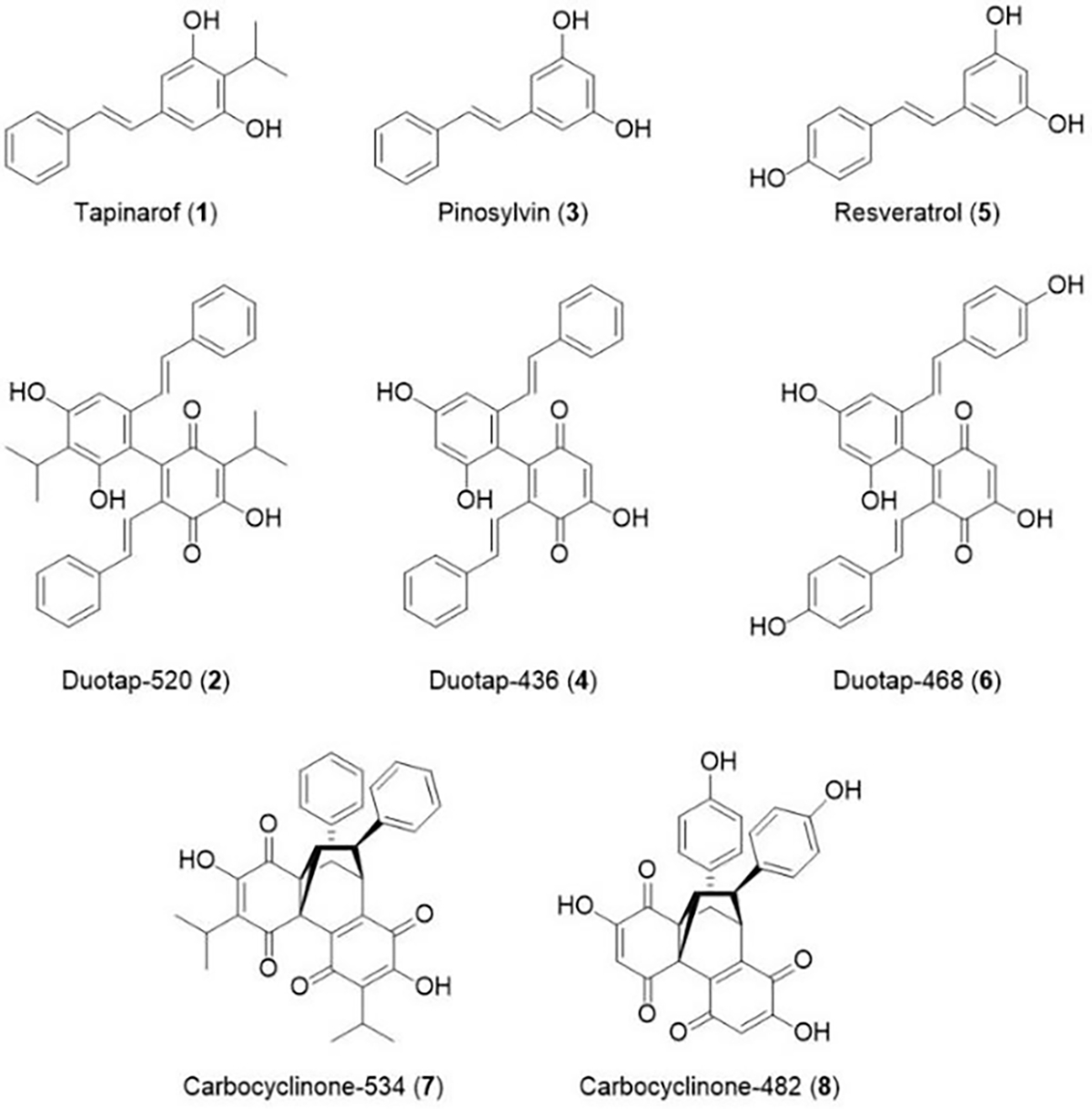
Structures of stilbene monomers (1, 3, and 5), duotap derivatives (2, 4, and 6), and carbocyclinone derivatives (7 and 8). Compound 6 is newly reported in this study.
We examined the antibiotic activities of the dimeric stilbene molecules (2, 4, and 6) against MRSA (Figure S7A–C) and VRE (Figure S7D–F). Similar to the functional transformation observed between 1 and 2, 4 displays significantly enhanced antibiotic activity compared to 3, particularly against VRE (~14 μM). However, 6 showed no enhancement of activity against these pathogens in comparison to 5. Assessment of the relative potencies of these compounds provided some structure activity relationships for the duotap family of antibiotics. Elimination of the aliphatic side chain (analogous to isoprene-functionalization in the arachidins found in peanuts)17 resulted in a slight decrease in antibiotic activity, while substitution of polar hydroxyl groups on the aromatic rings abolished activity against these bacteria at concentrations up to 100 μM (Table S1). Overall, increasing the polarity of the duotap molecules appears to result in diminished activity.
Because of their robust activity against clinically important pathogens, we proceeded to investigate the antibiotic mechanism of action of the lead duotap, 2. We first attempted to develop duotap-resistant MRSA mutants by sub-culturing two independent cultures of bacteria continuously in the presence of sub-lethal concentrations of 2. After 90 days, mutants with only 2-fold (culture 1) and 4-fold (culture 2) changes in MIC relative to wild-type were obtained (Figure 2A, Figure S8). In contrast, attempts to develop resistance to ciprofloxacin under identical conditions led to resistance mutants with an increase in MIC of approximately 37-fold after only 30 days as anticipated (Figure S9). To determine the mutations responsible for the marginal increase in MIC over 90 days, we analyzed MRSA colonies from cultures at 30, 60, and 90 days by whole genome sequencing. Analysis of the sequencing results primarily revealed mutations in genes previously associated with vancomycin-intermediate S. aureus (VISA) that allow bacteria to synthesize a thicker cell wall (Tables S2–S8).20 Relatively few mutations had accumulated through the 90-day subculture process. Of note, frameshifting indels in vraE, vraF, and accessory gene regulator C (agrC, involved in quorum sensing and biofilm formation), as well as a V266F mutation in oligopeptide-binding protein (oppA1, a transmembrane transporter) were observed in populations from both cultures (Figure 2B).20,21 All four genes have previously been implicated in promoting antibiotic resistance.20, 21 The mutants vraF, oppA1, and agrC gradually increased in frequency among the analyzed cells (Figure 2C), whereas vraE mutants were initially enriched at 60 days but fell out of the observed population by 90 days.
Figure 2.
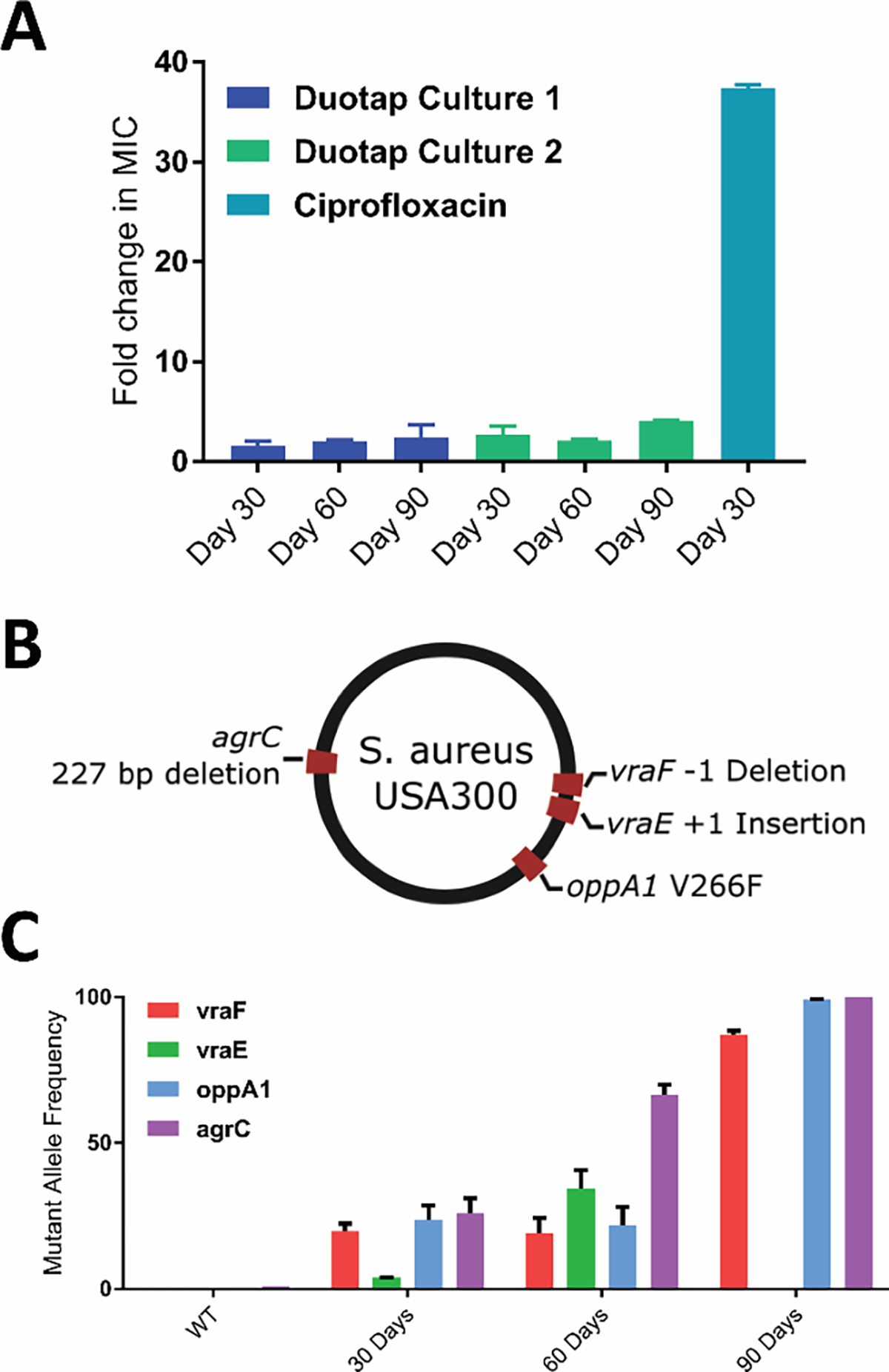
(A) Fold change in MIC for two independent cultures (Duotap-1 and Duotap-2) grown in the presence of sub-lethal levels of duotap-520 after 30, 60, and 90 days. Fold change in MIC for one culture grown in the presence of sub-lethal levels of ciprofloxacin for 30 days is shown as a positive control. Error bars represent the standard deviation of three independent MIC measurements of each strain. (B) Resistance mutations observed by whole genome sequencing for modestly duotap-resistant MRSA. (C) Mutant allele frequencies of candidate resistance genes (quantified as the percent of total whole genome sequencing reads at each position) observed after culturing strains for 30, 60, and 90 days in the presence of duotap.
While we failed to establish robust resistance mutants to support a mode of action for duotap, the genome sequencing data did suggest that the antibiotic may target the bacterial cell wall or cell wall biosynthesis in some way (Figure 3A). This is also consistent with the lack of resistance development observed, as it is more difficult for bacteria to mutate crucial cell wall components compared to other antibiotic targets, such as proteins. For example, teixobactin and malacidin bind to the cell wall precursor lipid II and do not develop resistance under similar experimental conditions.22, 23 A recently reported class of synthetic retinoid derivatives kill Gram-positive bacteria by permeabilizing the cell membrane and also do not select for resistance.24 Finally, the broad-spectrum antibiotic halicin disrupts the electrochemical transmembrane gradient across the bacterial cell wall by dissipating the proton motive force without developing resistance.24 Combined with this precedence and our genome sequencing data, we moved to assess the effects of duotap-520 on the bacterial cell wall. We first looked for an increase in membrane permeability in MRSA treated with SYTOX Green and found that duotap did not appear to alter the integrity of the bacterial membrane (Figure 3B). However, MRSA treated with positive control daptomycin displayed an increase in membrane permeability, indicating that daptomycin and duotap have distinct mechanisms of action. We next assessed the effects of duotap on cell wall biosynthesis by measuring the accumulation of the cell wall precursor molecule UDP-MurNAc-pentapeptide (Figure 3C). Similar to vancomycin, we observed a significant increase in the level of this precursor in cells treated with duotap, suggesting that the target is downstream of this intermediate in cell wall biosynthesis.
Figure 3.
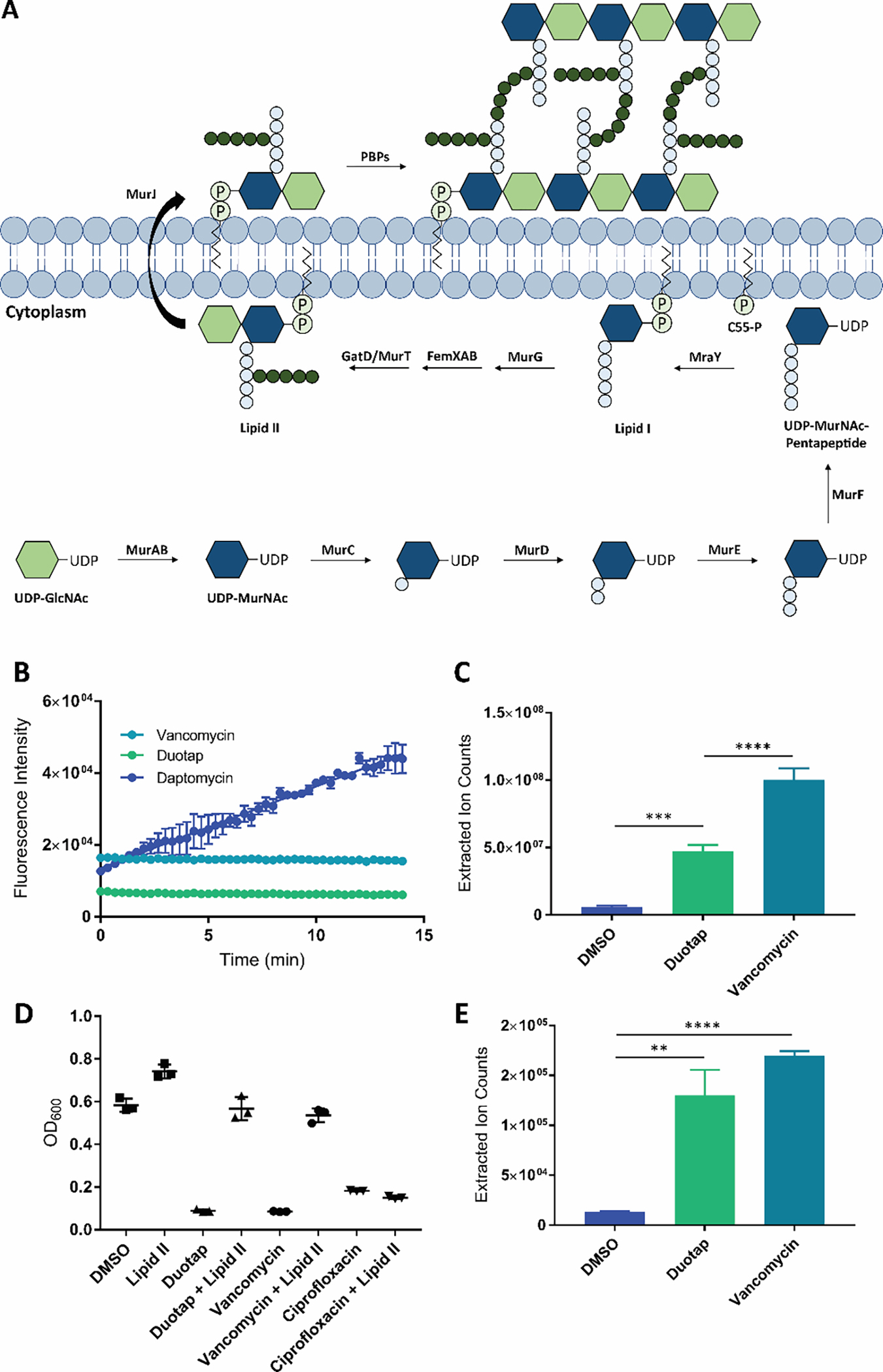
(A) MRSA cell wall biosynthesis. The intermediate lipid II is synthesized, flipped outside of the cell membrane, and polymerized into peptidoglycan by the penicillin binding proteins (PBPs).18, 19 (B) SYTOX Green fluorescence assay for antibiotic-induced bacterial membrane permeability. Vancomycin was used as a negative control, and daptomycin was used as a positive control. Error bars represent the standard error of the mean across three biological replicates. (C) Accumulation of the cell wall precursor UDP-MurNAc-pentapeptide in MRSA treated with duotap-520 compared to cells treated with vehicle DMSO. Vancomycin was used as a positive control. The UDP-MurNAc-pentapeptide was identified by mass spectrometry, with an [M+H]+ of 1150.3590. Error bars represent the standard deviation of three biological replicates. (D) Supplementation of lipid II in the growth medium can restore growth of MRSA exposed to duotap-520 and vancomycin (known to bind lipid II) to antibiotic-free levels. Lipid II cannot rescue bacteria grown in the presence of negative control ciprofloxacin (a DNA gyrase inhibitor). Data from three biological replicates are shown. (E) Accumulation of lipid II in MRSA treated with duotap-520 compared to cells treated with vehicle DMSO. Vancomycin was used as a positive control. Lipid II was boiled to remove the lipid tail, and de-lipidated lipid II was quantified by mass spectrometry, with an [M+H]+ of 1331.5362. Error bars represent the standard deviation of three biological replicates.
Lipid II is a crucial peptidoglycan building block downstream of UDP-MurNAc-pentapeptide and is the target of several antibiotics, including vancomycin (MIC 1.6 μM or 2.3 μg/ml, Figure S10).26 We next grew bacteria in the presence of lethal concentrations of antibiotics with and without supplementation of lipid II in the medium. Chemical complementation of functional lipid II in the presence of duotap and vancomycin restored MRSA growth to antibiotic-free levels (Figure 3D). In contrast, addition of lipid II was not able to rescue bacterial growth in the presence of control antibiotic ciprofloxacin, which is a DNA gyrase inhibitor and does not target the cell wall. Based on these results, the target of duotap-520 lies between UDP-MurNAc-pentapeptide and lipid II in cell wall biosynthesis. We considered the possibility that duotap could bind directly to precursors in the pathway, namely lipid II itself or its precursors undecaprenyl phosphate (C55-P) and undecaprenyl pyrophosphate (C55-PP). These lipids are the targets of numerous antibiotics.26, 27 Compounds that bind to lipid II are known to result in its accumulation in cells, while compounds that bind to C55-P or C55-PP result in a reduction in lipid II levels.28 We therefore looked at the effects of duotap-520 on lipid II pools in bacteria. It has previously been reported that the lipid tail of lipid II can be removed by boiling in acidic conditions, allowing quantification by LC-MS.18 Following this protocol, we observed an accumulation of a mass matching de-lipidated lipid II when bacteria were treated with either duotap or vancomycin (Figure 4E). The identity of this molecule was confirmed by tandem mass spectrometry and comparison to published fragmentation data (Figure S11).18 Because chemical complementation of exogenously supplied lipid II rescues bacterial growth in the presence of duotap and duotap also results in the accumulation of lipid II, the antibiotic target cannot be before or after lipid II in the biosynthesis of the cell wall. When taken together, these results support that duotap likely targets the bacterial cell wall by binding to lipid II itself and preventing its subsequent polymerization and cross-linking into peptidoglycan, or deleteriously affecting the trafficking of lipid II (e.g., flippase MurJ involved in transport of lipid II from the cytoplasm to the periplasm).
The bacterial cell wall is an important target of many antibiotics, and our results here add stilbene dimers to this growing list.26 However, the emergence of drug resistance to cell wall-targeting compounds is a major problem.20, 29 Strikingly, duotap targets the bacterial cell wall without selecting for considerable antibiotic resistance even after 90-days of sub-lethal treatments, although some cytotoxicity in human cells treated with the compound at concentrations above the MIC was observed (Figure S12). While other cell wall-targeting antibiotics like vancomycin bind directly to lipid II, duotap is active against VRE, in addition to MRSA. How duotap engages the structure or trafficking of lipid II remains an open question, but it likely occupies a different binding site than vancomycin, maintaining efficacy against vancomycin resistant strains with mutated D-Ala-D-Ala motifs in lipid II. It is notable that plant stilbenes can be converted into these dimeric scaffolds, suggesting that the structural and functional traits reported here may have been evolutionarily selected in stilbene-producing plants and bacteria. Furthermore, stilbenes are commonly consumed in the human diet or as dietary supplements, so it is conceivable that this class of antimicrobials could influence the composition of the gut microbiome.30, 31 While the specific dimers that we have identified have not yet been reported in plants to our knowledge, the well-established plant precursors can form these dimers through spontaneous and enzymatic chemistries. The dietary impact of stilbenes is an active area of research, and spontaneous and/or biocatalytic conversion of these molecules into antibiotics could have interesting implications at the host-bacteria-plant interface.
Supplementary Material
ACKNOWLEDGMENT
The authors would like to thank Professor Suzanne Walker of Harvard Medical School for providing the lipid II compound used in these studies.
Funding Sources
This work was in part supported by the National Institutes of Health (1DP2-CA186575 and R00-GM097096), the Burroughs Wellcome Fund (1016720), and the Camille & Henry Dreyfus Foundation (TC-17-011). T.N.G. and J.P. were in part supported by the National Institutes of Health (5T32GM06754 3-12).
Footnotes
The authors declare no competing financial interests
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Experimental methods, supporting figures, and resistance mutations tables.
REFERENCES
- 1.Fleming A (1929) On the Antibacterial Action of Cultures of a Penicillium, with Special Reference to Their Use in the Isolation of B. Influenzae. Brit. J. Exp. Pathol. 10(3), 226–236. [Google Scholar]
- 2.Gould K (2016) Antibiotics: from prehistory to the present day. J. Antimicrob. Chemoth. 71(3), 572–575. [DOI] [PubMed] [Google Scholar]
- 3.Blair JMA; Webber MA; Baylay AJ; Ogbolu DO; Piddock LJV (2015) Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 13(1), 42–51. [DOI] [PubMed] [Google Scholar]
- 4.Mulani MS; Kamble EE; Kumkar SN; Tawre MS; Pardesi KR (2019) Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review. Front. Microbiol. 10, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patridge E; Gareiss P; Kinch MS; Hoyer D (2016) An analysis of FDA-approved drugs: natural products and their derivatives. Drug Discov. Today. 21(2), 204–207. [DOI] [PubMed] [Google Scholar]
- 6.Newman DJ; Cragg GM (2007) Natural products as sources of new drugs over the last 25 years. J. Nat. Prod. 70(3), 461–477. [DOI] [PubMed] [Google Scholar]
- 7.Newman DJ; Cragg GM (2016) Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 79(3), 629–661. [DOI] [PubMed] [Google Scholar]
- 8.Shen T; Wang XN; Lou HX (2009) Natural stilbenes: an overview. Nat. Prod. Rep. 26(7), 916–935. [DOI] [PubMed] [Google Scholar]
- 9.Fuchs SW; Bozhuyuk KAJ; Kresovic D; Grundmann F; Dill V; Brachmann AO; Waterfield NR; Bode HB (2013) Formation of 1,3-Cyclohexanediones and Resorcinols Catalyzed by a Widely Occuring Ketosynthase. Angew. Chem. Int. Ed. 52(15), 4108–4112. [DOI] [PubMed] [Google Scholar]
- 10.Vizcaino MI; Guo X; Crawford JM (2014) Merging chemical ecology with bacterial genome mining for secondary metabolite discovery. J. Ind. Microbiol. Biot. 41(2), 285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi YM; Bode HB (2018) Chemical language and warfare of bacterial natural products in bacteria-nematode-insect interactions. Nat. Prod. Rep. 35(4), 309–335. [DOI] [PubMed] [Google Scholar]
- 12.Paul VJ; Frautschy S; Fenical W; Nealson KH (1981) Antibiotics in Microbial Ecology - Isolation and Structure Assignment of Several New Anti-Bacterial Compounds from the Insect-Symbiotic Bacteria Xenorhabdus Spp. J. Chem. Ecol. 7(3), 589–597. [DOI] [PubMed] [Google Scholar]
- 13.Richardson WH; Schmidt TM; Nealson KH (1988) Identification of an Anthraquinone Pigment and a Hydroxystilbene Antibiotic from Xenorhabdus-Luminescens. Appl. Environ. Microb. 54(6), 1602–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi DS; An R; Zhang WB; Zhang GL; Yu ZG (2017) Stilbene Derivatives from Photorhabdus temperata SN259 and Their Antifungal Activities against Phytopathogenic Fungi. J. Agr. Food Chem. 65(1), 60–65. [DOI] [PubMed] [Google Scholar]
- 15.Furue M; Hashimoto-Hachiya A; Tsuji G (2019) Aryl Hydrocarbon Receptor in Atopic Dermatitis and Psoriasis. Int. J. Mol. Sci. 20(21), 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park HB; Goddard TN; Oh J; Patel J; Wei Z; Perez CE; Mercado BQ; Wang R; Wyche TP; Piizzi G; Flavell RA; Crawford J (2020) Bacterial Autoimmune Drug Metabolism Transforms an Immunomodulator into Structurally and Functionally Divergent Antibiotics. Angew. Chem. Int. Ed. 59, 2–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brents LK; Medina-Bolivar F; Seely KA; Nair V; Bratton SM; Nopo-Olazabal L; Patel RY; Liu HN; Doerksen RJ; Prather PL; Radominska-Pandya A (2012) Natural prenylated resveratrol analogs arachidin-1 and −3 demonstrate improved glucuronidation profiles and have affinity for cannabinoid receptors. Xenobiotica. 42(2), 139–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiao Y; Srisuknimit V; Rubino F; Schaefer K; Ruiz N; Walker S; Kahne D (2017) Lipid II overproduction allows direct assay of transpeptidase inhibition by beta-lactams. Nat. Chem. Biol. 13(7), 793–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarick M; Bertsche U; Stahl M; Schultz D; Methling K; Lalk M; Stigloher C; Steger M; Schlosser A; Ohlsen K (2018) The serine/threonine kinase Stk and the phosphatase Stp regulate cell wall synthesis in Staphylococcus aureus. Sci. Rep. 8(13693), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howden BP; Davies JK; Johnson PDR; Stinear TP; Grayson ML (2010) Reduced Vancomycin Susceptibility in Staphylococcus aureus, Including Vancomycin-Intermediate and Heterogeneous Vancomycin-Intermediate Strains: Resistance Mechanisms, Laboratory Detection, and Clinical Implications. Clin. Microbiol. Rev. 23(1), 99–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Acosta MBR; Ferreira RCC; Padilla G; Ferreira LCS; Costa SOP (2000) Altered expression of oligopeptide-binding protein (OppA) and aminoglycoside resistance in laboratory and clinical Escherichia coli strains. J. Med. Microbiol. 49(5), 409–413. [DOI] [PubMed] [Google Scholar]
- 22.Ling LL; Schneider T; Peoples AJ; Spoering AL; Engels I; Conlon BP; Mueller A; Schaberle TF; Hughes DE; Epstein S; Jones M; Lazarides L; Steadman VA; Cohen DR; Felix CR; Fetterman KA; Millett WP; Nitti AG; Zullo AM; Chen C; Lewis K (2015) A new antibiotic kills pathogens without detectable resistance. Nature. 517(7535), 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hover BM; Kim SH; Katz M; Charlop-Powers Z; Owen JG; Ternei MA; Maniko J; Estrela AB; Molina H; Park S; Perlin DS; Brady SF (2018) Culture-independent discovery of the malacidins as calcium-dependent antibiotics with activity against multidrug-resistant Gram-positive pathogens. Nat. Microbiol. 3(4), 415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim W; Zhu WP; Hendricks GL; Van Tyne D; Steele AD; Keohane CE; Fricke N; Conery AL; Shen S; Pan W; Lee K; Rajamuthiah R; Fuchs BB; Vlahovska PM; Wuest WM; Gilmore MS; Gao HJ; Ausubel FM; Mylonakis E (2018) A new class of synthetic retinoid antibiotics effective against bacterial persisters. Nature. 556(7699), 103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Breukink E; de Kruijff B (2006) Lipid II as a target for antibiotics. Nat. Rev. Drug Discov. 5(4), 321–332. [DOI] [PubMed] [Google Scholar]
- 26.Stokes JM; Yang K; Swanson K; Jin W; Cubillos-Ruiz A; Donghia NM; MacNair CR; French S; Carfrae LA; Bloom- Ackerman Z.; Tran VM.; Chiappino-Pepe A.; Badran A.; Andrews IW.; Chory EJ.; Church GM.; Brown ED.; Jaakkola TS.; Barzilay R.; Collins JJ. (2020) A Deep Learning Approach to Antibiotic Discovery. Cell. 180, 688–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muller A; Klockner A; Schneider T (2017) Targeting a cell wall biosynthesis hot spot. Nat. Prod. Rep. 34(7), 909–932. [DOI] [PubMed] [Google Scholar]
- 28.Santiago M; Lee W; Abou Fayad A; Coe KA; Rajagopal M; Do T; Hennessen F; Srisuknimit V; Muller R; Meredith TC; Walker S (2018) Genome-wide mutant profiling predicts the mechanism of a Lipid II binding antibiotic. Nat. Chem. Biol. 14(6), 601–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernandes R; Amador P; Prudencio C (2013) beta-Lactams: chemical structure, mode of action and mechanisms of resistance. Rev. Med. Microbiol. 24(1), 7–17. [Google Scholar]
- 30.Cardona F; Andres-Lacueva C; Tulipani S; Tinahones FJ; Queipo-Ortuno MI (2013) Benefits of polyphenols on gut microbiota and implications in human health. J. Nutr. Biochem. 24(8), 1415–1422. [DOI] [PubMed] [Google Scholar]
- 31.Ozdal T; Sela DA; Xiao JB; Boyacioglu D; Chen F; Capanoglu E (2016) The Reciprocal Interactions between Polyphenols and Gut Microbiota and Effects on Bioaccessibility. Nutrients. 8(2) 1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


