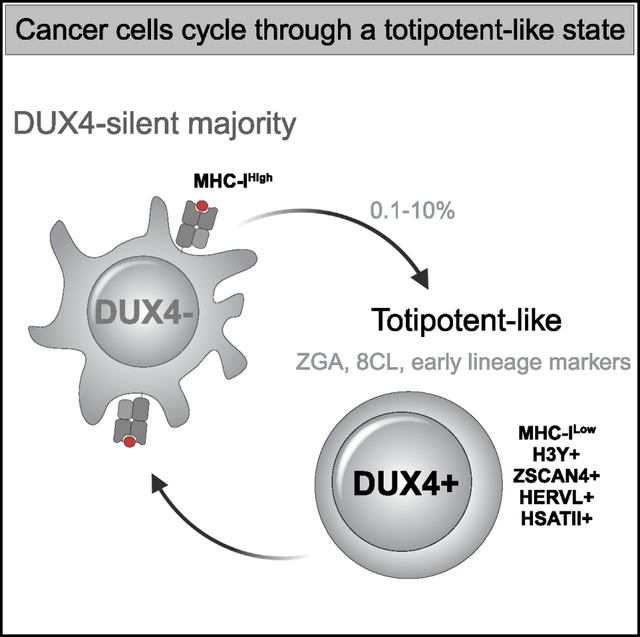
SUMMARY
The transcription factor DUX4 regulates a portion of the zygotic gene activation (ZGA) program in the early embryo. Many cancers express DUX4 but it is unknown whether this generates cells similar to early embryonic stem cells. Here we identified cancer cell lines that express DUX4 and showed that DUX4 is transiently expressed in a small subset of the cells. DUX4 expression activates the DUX4-regulated ZGA transcriptional program, the subsequent 8C-like program, and markers of early embryonic lineages, while suppressing steady-state and interferon-induced MHC class I expression. Although DUX4 was expressed in a small number of cells under standard culture conditions, DNA damage or changes in growth conditions increased the fraction of cells expressing DUX4 and its downstream programs. Our demonstration that transient expression of endogenous DUX4 in cancer cells induces a metastable early embryonic stem cell program and suppresses antigen presentation has implications for cancer growth, progression, and immune evasion.
In brief
Smith et al. identify cancer cell lines that express the early embryonic transcription factor DUX4 and show that it is transiently expressed in a subset of cells. DUX4 expression induces the ZGA and 8C-like transcription programs as well as early lineage markers and suppresses MHC-I expression.
Graphical Abstract
INTRODUCTION
Many studies have identified intra-tumor cell heterogeneity,1–4 including stem cell-like phenotypes,5 as drivers of cancer progression. Our recent identification of DUX4 expression in many different cancers6 raises the possibility that it might generate a cancer cell population similar to early embryonic stem cells in a diverse set of cancers. The transcription factor DUX4, and its mouse ortholog Dux, are normally expressed in the early embryo, where brief expression of DUX4/Dux regulates the initial wave of zygotic gene activation (ZGA) at the 2-cell (2C) stage in mice and 4-cell (4C) stage in humans,7–9 including the activation of classes of retrotransposons and HSATII and GSAT pericentromeric repeats in humans and mice, respectively. In ESC/iPSC cultures, transient expression of the ZGA program in rare cells establishes a naive, or totipotent, subpopulation,10–13 also referred to as 2C-like or 8C-like in mouse and human cells, respectively. Subsequent studies showed that the ZGA program is driven by the transient expression of DUX4/Dux in these cells.10,14
DUX4 was initially characterized because its aberrant expression in skeletal muscle causes facioscapulohumeral dystrophy (FSHD).15–17 The DUX4 retrogene is present in each unit of the D4Z4 macrosatellite array located in the subtelomeric regions of both chromosome 4 and chromosome 10.18 FSHD1 is caused by deletion of a subset of D4Z4 repeats, creating an array of 1–10 D4Z4 units that decreases the epigenetic silencing of the DUX4 locus in somatic cells, whereas FSHD2 is caused by mutations in epigenetic modifiers necessary for repression of the DUX4 locus.17,19,20 Decreased epigenetic repression of the DUX4 locus results in the occasional burst of DUX4 expression in skeletal muscle occurring in ~0.1%–1% of FSHD myoblasts in culture.16,21
In this study, we identified cancer cell lines that express the full-length DUX4 mRNA and show that DUX4 protein and its target gene H3Y are expressed in ~0.1% of cells under standard tissue culture conditions. Single-cell RNA sequencing showed that transient expression of DUX4 activates DUX4-regulated ZGA genes and a portion of the 8C-like transcriptional program, as well as markers of early embryonic lineages, while suppressing MHC Class I (MHC-I) mRNAs. DNA damage resulted in a p53-dependent increase in the percentage of DUX4-expressing cells and subsequent ZGA gene expression. Together, our study shows that the transient expression of endogenous DUX4 in cell lines from different types of cancers induces a metastable early embryonic stem cell program and suppresses MHC-I expression in a subpopulation of cells.
RESULTS
Two classes of DUX4-positive cancer cell lines: DUX4-like and DUX4-full-length
We determined read counts for DUX4 and a panel of 39 genes regulated by DUX4 in the cancer cell lines entered in the Cancer Cell Line Encyclopedia (CCLE).22 Comparing DUX4 mRNA levels to a DUX4-regulated transcriptional signature revealed two different categories of DUX4-expressing cancer cell lines. One category, termed DUX4-like, had relatively high levels of reads mapping to DUX4 but did not have reads mapping to the set of genes regulated by DUX4. The second category, DUX4-full-length (DUX4-FL), had a low but detectable number of reads mapping to DUX4 together with reads mapping to multiple DUX4-target genes (Figure 1A).
Figure 1. DUX4-FL cancer cell lines contain rare DUX4-expressing cells.
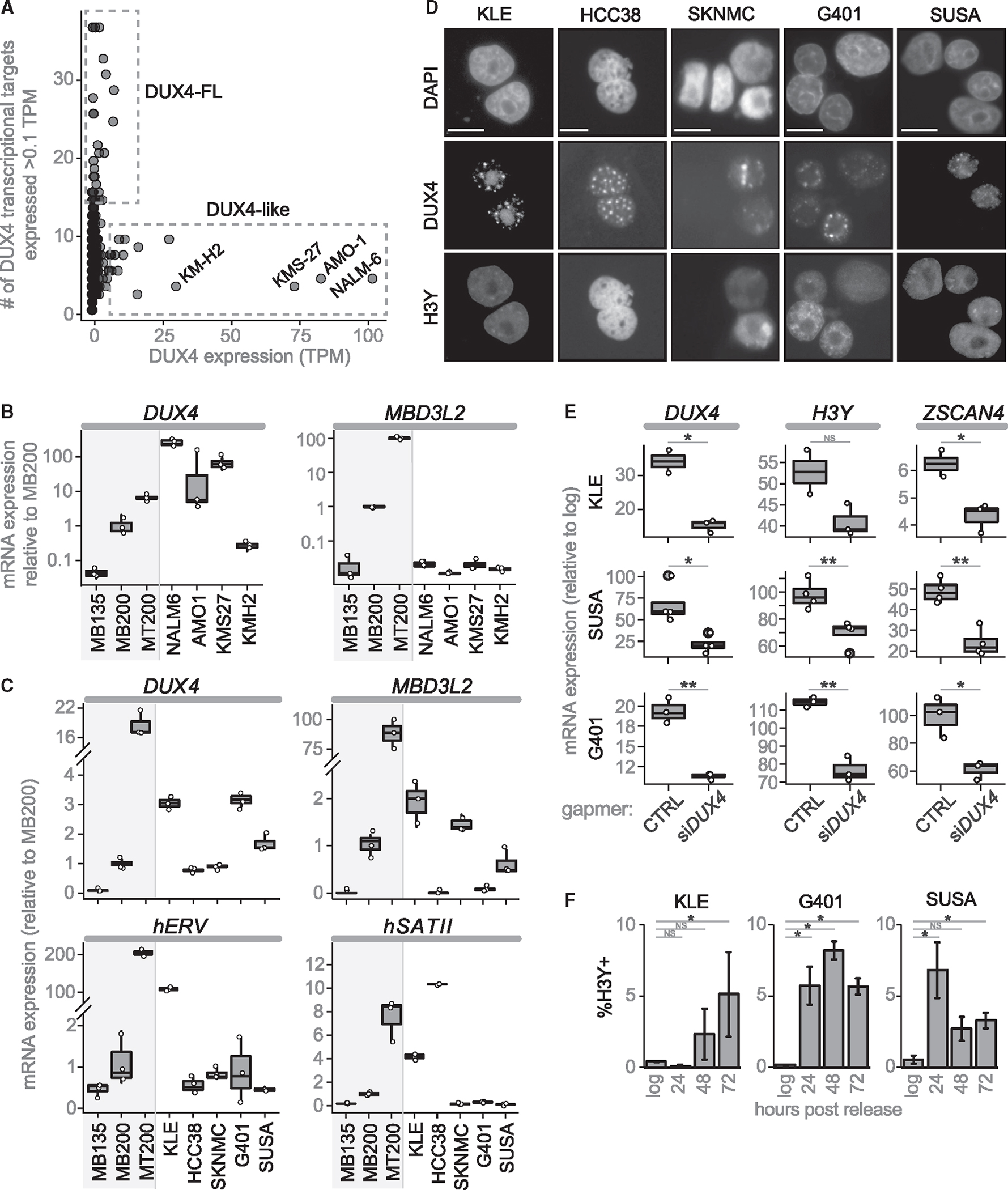
(A) CCLE cancer cell lines plotted by DUX4 expression (TPM) versus number of DUX4 targets expressed (threshold = 0.1TPM).
(B) RT-qPCR of DUX4-like lines normalized to RPL27. Control human myoblasts (MB135) that do not express DUX4, FSHD2 myoblasts (MB200) that express low levels of DUX4, and FSHD2 myotubes (MT200) that express higher levels of DUX4 included for comparison.
(C) RT-qPCR primed with oligo-dT for DUX4 and MBD3L2, or random hexamers for hERV and hSATII.
(D) Immunofluorescence for H3Y and DUX4 (cropped from Figure S1G; scale bars, 50 μM).
(E) RT-qPCR 48 h post release from confluence treated with control or anti-DUX4 MOE gapers, shown as fold-change compared with log-phase cells. p-values: * <0.05; **<0.005.
(F) Percentage of H3Y-positive nuclei at 24, 48, and 72 h post-release, mean ± SEM. For all RT-qPCR experiments, n = 3 biological replicates, except n = 2 for KLE gamper control. For IF, n = 3 biological replicates per timepoint each with five fields. p values calculated with a one-tailed Welch’s t test: * <0.05; **<0.005, ***<0.0005, NS = no significance.
DUX4-like cancer cell lines do not encode a full-length DUX4 protein
To determine the origin and structure of the DUX4-like RNAs, we focused on the four cell lines with the highest expression: NALM6 (adult B cell acute lymphoblastic lymphoma), AMO1 (plasma cell myeloma), KMS27 (plasma cell myeloma), and KMH2 (Hodgkin lymphoma). Prior studies showed that NALM6 cells have a translocation that inserts DUX4-coding sequences from the D4Z4 repeat into the immunoglobulin (Ig)H locus and produce a DUX4-IgH fusion transcript that lacks the last 16 amino acids of DUX4, a region necessary for DUX4 transcriptional activity.23,24
For each of these four cell lines, reverse transcription (RT)-qPCR confirmed both expression of an mRNA amplified by primers to DUX4 as well as the absence of DUX4-target gene expression (Figures 1B and S1A). 3-prime rapid amplification on cDNA ends (3′RACE) and sequencing of RT-PCR products confirmed the previously described DUX4-IgH fusion transcript in the NALM6 cells,25 whereas the three other cell lines expressed novel DUX4-like transcripts that mapped to D4Z4-like arrays in the pericentromeric region of chromosome 22 and unplaced genomic regions (Table S1). Based on the sequenced region of these DUX4-like mRNAs and their corresponding genomic sequences, none encoded the full-length DUX4 protein because of frame-shifts and/or stop codons (Table S1). Western blotting with antibodies to the amino-terminus of DUX4 revealed robust expression in NALM6 but not in the other lines (Figure S1B). Together with the absence of DUX4-target gene expression, these data indicate that the DUX4-like group does not express a full-length DUX4 protein that is transcriptionally active.
DUX4-FL cancer cell lines express full-length DUX4 and DUX4-target genes in a subset of cells
We ranked the cancer cell lines in the DUX4-FL group based on the number of reads mapping to the panel of DUX4-target genes (Figure S1C) and focused on characterizing five of the top cell lines: KLE, HCC38, SKNMC, G401, and SUSA (representing endometrial, mammary epithelial, neural epithelial, rhabdoid, and testicular teratoma cancer cell lines, respectively). As negative controls, we used two cell lines in the CCLE that were negative for DUX4 and its targets, Rh30 and MCF7, as well as the commonly used HeLa cell line that was not included in the CCLE.
Oligo-dT primed RT-qPCR demonstrated DUX4-FL lines expressed DUX4 mRNA at levels comparable to FSHD myoblasts and each expressed subsets of DUX4-target genes at similarly low levels (Figures 1C and S1D), whereas cancer cell lines that were not identified as DUX4-FL did not express DUX4 or DUX4-targets (Figure S1E). Furthermore, treating DUX4-FL cell lines with anti-DUX4 MOE gapmers reduced expression of DUX4 targets ZSCAN4 and LEUTX (Figure S1F), confirming that DUX4 is driving expression of these genes. Immunofluorescence identified DUX4 and DUX4-target proteins in a small fraction of the cells (~0.1%), often in pairs or small clusters (Figures 1D and S1G); consistent with a short burst of DUX4 expression, similar to the brief expression of DUX4 or mouse Dux in the early embryo7 and in ES/iPS cells.10 These results indicate that the low levels of DUX4 and DUX4-target mRNAs in the log-phase cultures represents relatively high expression in a small population of cells, similar to DUX4 expression in FSHD muscle cells.16
Release from confluence increases transient DUX4 expression
We tried multiple culture conditions and determined that there was a substantial and transient increase in the expression of DUX4 and DUX4-target RNAs following release from confluence (Figure S1H). Furthermore, DUX4 knockdown confirmed that DUX4-target gene expression was driven by DUX4 (Figure 1E). Immunodetection of the DUX4-target H3Y showed that the increased mRNA levels correlated with a higher percentage of DUX4-target-expressing cells, reaching ~5% of the population, and that the increase in H3Y cells persisted for several days following release from confluence (Figures 1F and S1I).
Transient DUX4 expression activates ZGA, 8CLC, and early embryonic lineage genes
To determine whether transient DUX4 expression in the cancer celllines induced the broader transcriptional program of the cleavage-stage embryo, we performed single-cell RNA sequencing on SUSA and KLE cells in log-phase and at 24, 48, and 72 h following release from confluence. At each time point, 3,000–10,000 cells were sequenced with an average of 3,000–5,000 genes detected per cell (Figure S2A). Data analysis included use of the ScanPy pipeline26 and regression for cell cycle genes (see STAR methods), resulting in UMAPs with 18 clusters in SUSA cells and 21 clusters for KLE cells (Figure 2A; Table S2).
Figure 2. DUX4 activates the ZGA/8CLC programs and suppresses MHC class I expression.
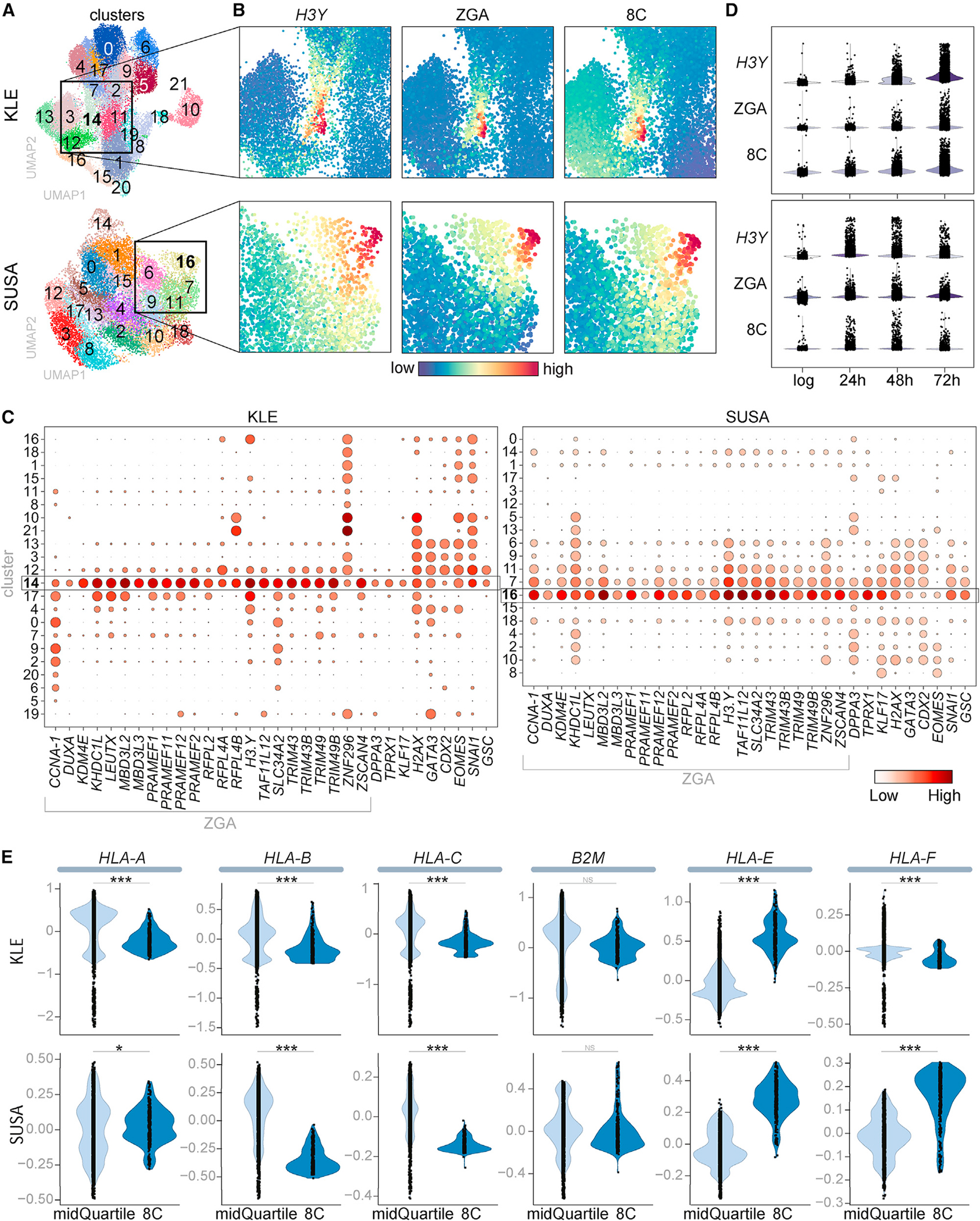
(A–E) Single-cell RNA sequencing of KLE and SUSA lines in log-phase and 24, 48, and 72 h after release from confluence. (A) Phenograph Leiden cell clusters. (B) Heatmap showing relative expression of H3Y and composite scores for ZGA and 8C genes (see Figure S2B for full UMAP images). (C) Dot plot showing the fraction of cells (size) and mean expression (color) of each indicated gene. (D) Relative expression of H3Y, ZGA, and 8C genes. (E) HLA gene expression in the mid quartile or top 350 cells based on imputed 8C gene composite scores. p values calculated with a permutation test (see STAR methods): *<0.05, ***<0.0005.
H3Y mRNA was enriched in KLE cluster 14 and SUSA cluster 16 (Figures 2B and S2B), whereas DUX4 expression was not reliably detected with single-cell sequencing, possibly due to its low abundance and extremely high GC content. EnrichR indicated that these clusters expressed gene sets similar to the eight-cell embryo and primordial germ cells (Figure S2C). Consistent with this, composite scores based on 23 DUX4-regulated ZGA genes10 (Table S3A) identified high-expressing cells in the same cluster as H3Y-expressing cells (Figures 2B and S2B). Expression analysis showed that all 23 genes in this ZGA program were elevated in this cluster (KLE Cluster 14 and SUSA Cluster 16) and, to a lesser extent, in adjacent clusters (KLE Clusters 17 and 12, SUSA Clusters 7 and 11) (Figure 2C). In addition to the DUX4-regulated portion of the ZGA program, genes that characterize 8C-like cells (8CLCs) that are not known to be directly regulated by DUX4 (DPPA3, TPRX1, and KLF17) were expressed in these clusters (Figures 2C and S2D), and a composite score based on 717 8CLC genes10 (Table S3A) identified the expression of the broader 8CLC program in these clusters (Figure S2B). Dividing the 8CLC genes into likely direct targets of DUX4 (64) or not direct (653) (see STAR methods and Table S3B) showed similar although slightly expanded distribution of the putative indirect targets of DUX4 (Figure S2E).
Assessing the different time points showed that cells expressing the ZGA and 8CLC genes were enriched after release from confluence compared with the log-phase cells (Figure 2D). The persistence of the ZGA and 8C populations over 72 h indicated that the transient expression of DUX4 in these cancer cells did not result in cell death, and this was supported by the absence of enrichment for gene expression associated with apoptosis, autophagy, or necrosis pathways in the ZGA/8C enriched cluster (Figure S2F). In addition, immunostaining for the proliferation marker Ki-67 and BrdU labeling for 6 h showed that the H3Y-positive cells had the same proliferative rate as the H3Y-negative population at both 48 and 72 h following release (Figures S3A and S3B).
These data indicate that transient DUX4 expression was associated with activation of the DUX4-regulated ZGA transcriptional program and the subsequent broader 8CLC program in these two cancer cell lines. In addition to this early totipotent program, EnrichR also indicated genes associated with trophectoderm and early line-age specification were enriched in the DUX4/H3Y cluster in KLE cells (Figure S2C). This was supported by expression of genes regulating trophectoderm formation (GATA3, CDX2, EOMES [both KLE and SUSA] and TFAP2C [KLE] or TFAP2A [SUSA]) and genes involved in mesenchymal specification and tumor metastasis (SNAI1, GSC in both and FOXC2 in SUSA) in the DUX4-target and adjacent clusters (Figures 2C, S3C, and S3D). Expression of placental alkaline phosphatase (ALPP), as well as TWIST1 and ZEB1 in SUSA or POU5F1 and DPPA2 in KLE, further support the expression of trophectoderm, mesenchymal, and early embryonic markers that occur following ZGA (Figure S3E), and is consistent with a report that knockout of mouse Dux results in diminished expression of genes involved in trophectoderm development.27 Together, these results indicate at least a partial induction of early embryonic lineage programs that might have relevance to cancer progression.
Our prior study showed that DUX4-expressing cancers showed reduced MHC-I Class I gene expression.6 In the current single-cell sequencing data, the cells expressing the 8CLC genes showed lower expression of the canonical MHC-I genes (HLA-A, B, and C) and elevated expression of some of the non-canonical MHC genes (HLA-E and F) (Figure 2E). Similar results were obtained in the cells expressing the ZGA signature (Figure S3F). Therefore, the most profound suppression of MHC-I genes in cancers expressing DUX4 is associated with the DUX4-expressing subpopulation and the subsequent ZGA and 8CLC states.
DUX4 expression is sufficient to induce early embryonic lineage genes in cancer cell lines
To determine whether the transient expression of DUX4 is sufficient to induce early embryonic lineage and extraembryonic programs, the cancer cell lines KLE, SUSA, and G401 were transduced with a doxycycline-inducible DUX4 vector (iDUX4CA).28 Treatment with doxycycline continuously for 24 h in KLE cells (Figures 3A and S4A) or for a 4-h pulse in SUSA cells (Figures 3B and S4B) and G401 cells (Figures 3C and S4C) resulted intransient expression of DUX4 and induction of ZSCAN4 (Figures S4A–S4C), as well as induction of early embryonic lineage genes (GATA3, KLF4, EOMES, CDX2, SNA1, GSC) that persisted for several days (Figures 3A–3C).
Figure 3. DUX4 induces early embryonic lineage markers and is induced by DNA-damaging drugs.
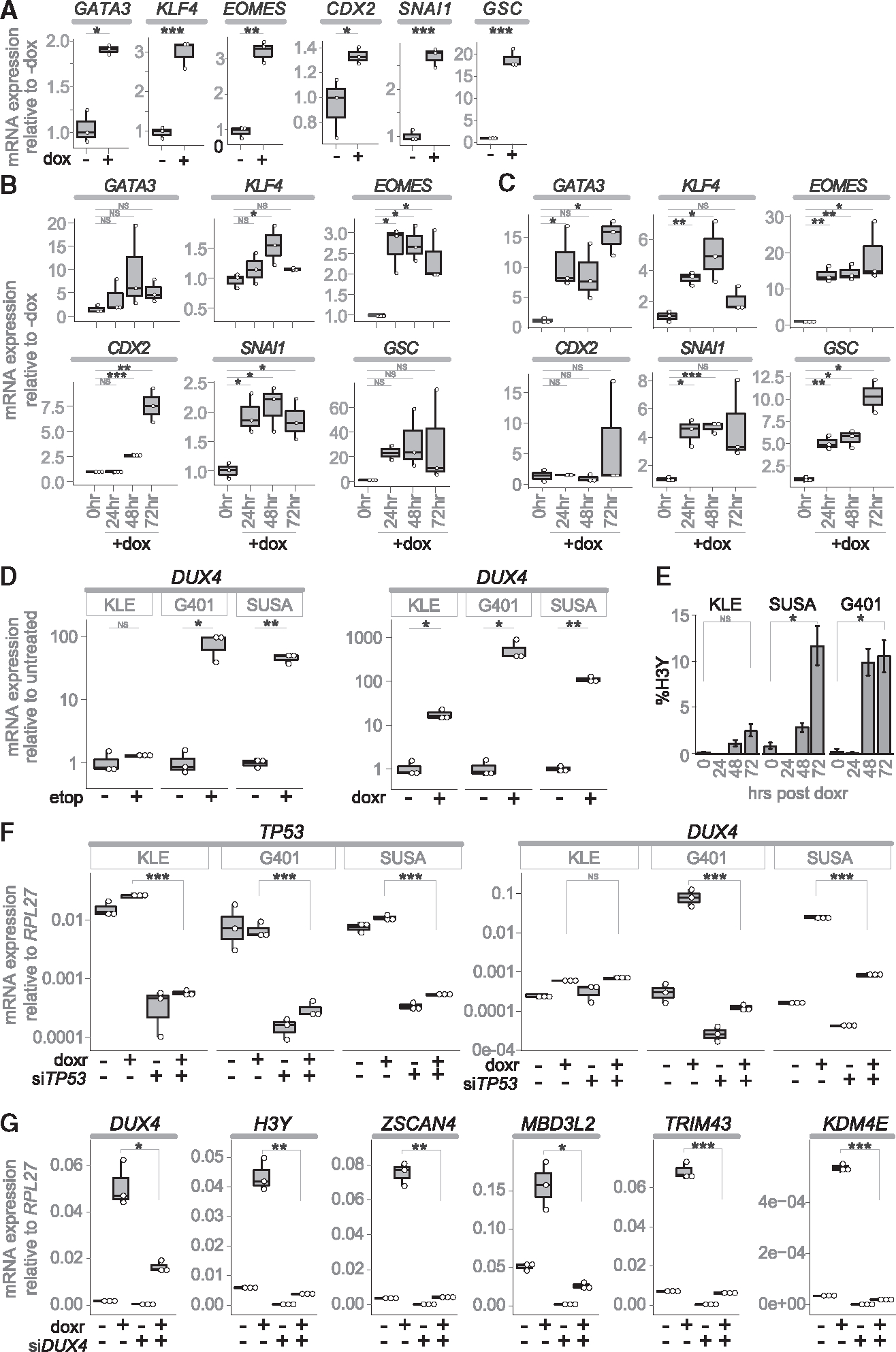
(A–C) RT-qPCR for the indicated trophectoderm and mesenchymal specification genes, after treatment with doxycycline (dox), in: (A) KLE-iDUX4CA, (B) SUSA-iDUX4CA, and (C) G401-iDUX4CA.
(D) RT-qPCR for DUX4 48 h post treatment with etoposide (etop) or doxorubicin (doxr) graphed as the fold-increase relative to untreated.
(E) Percentage of H3Y-positive nuclei expressed as mean ± SEM.
(F) RT-qPCR 24 h post treatment with doxorubicin in cells pretreated with siTP53 or siControl.
(G) RT-qPCR on G401 RNA 24 h post doxorubicin after pretreatment with anti-DUX4 (+) or control (GFP) gapmer (−). For all experiments, n = 3 biological replicates (individual biological samples displayed as white dots). p values calculated with a one-tailed Welch’s t test: * <0.05; **<0.005, ***<0.0005, NS = no significance.
DNA damage enhances DUX4 expression in cancer cells with wild-type p53
A recent study of DUX4 in early development identified p53 as an inducer of DUX4 expression.36 Consistent with those findings, treatment of the two cancer cell lines that have wild-type p53, SUSA, and G401, with etoposide or doxorubicin, topoisomerase II inhibitors that induce p53, showed strong induction of DUX4 (Figure 3D) and a substantial increase in the percentage of H3Y-positive cells (Figure 3E). Knockdown of TP53 with siRNA reduced DUX4 induction in these cells following doxorubicin-induced DNA damage (Figure 3F). In contrast, the KLE cell line, which has a dominant negative TP53 mutation (p.R175H29) that prevents p53 target induction30–32 showed more modest induction of DUX4 following drug-induced DNA damage that was not changed with knockdown of TP53 (Figures 3D–3F). (The DepMap portal reports G401 as having a p.C277F mutation in TP53; however, the source publication29 for this reports wild-type (WT) TP53 in this line and our sequencing did not detect this C277F mutation.) Finally, knockingdown DUX4 withMOE gapmerslargely prevented the induction of ZGA genes (H3Y, ZSCAN4, MBD3L2, TRIM43, KDM4E) following doxorubicin treatment (Figure 3G).
Although etoposide and doxorubicin induced DUX4 expression, as measured by H3Y, in a high fraction of cells (~10%–15%), not all cells showed H3Y expression despite the finding that nearly all cells showed nuclear accumulation of phosphorylated TP53-pS15 (Figure S4D). To determine whether the repressive chromatin state of the D4Z4 conferred a variable degree of permissiveness for DUX4 induction following DNA damage, we doxorubicin treated a set of human myoblasts that have different degrees of D4Z4 chromatin repression based on the number of repeat units on a permissive 4A161 allele: MB135 (74U), MB2401 (11U), and MB073 (8U),33 and the isogenic cell lines derived from a mosaic FSHD individual 54–1 (13U), 54–6 (13U), and 54–2 (3U).34 DUX4 was induced by doxorubicin treatment to some degree in all cell lines; however, the shorter repeat arrays that have decreased chromatin repression35 showed substantially higher induction of DUX4 mRNA and percentages of H3Y positive cells (Figures S4E and S4F), indicating that TP53 induction of DUX4 is constrained by the degree of chromatin repression at the D4Z4 and less chromatin repression correlates with a higher probability of DUX4 expression, as also shown previously.36
Cancer cell populations expressing DUX4 suppress MHC class I presentation
Our single-cell sequencing analysis indicated that expression of DUX4 targets correlated with suppressed steady-state levels of MHC-I mRNAs (see Figure 2E). To determine whether DUX4 induction in these cancer cells would suppress interferon-gamma (IFNg) stimulated MHC-I RNAs and cell surface presentation, we treated SUSA cells released from confluence and G401 cells pretreated with doxorubicin or etoposide with 50 ng/mL IFNg for 16 h (Figures 4A and 4B) and used flow cytometry to measure relative HLA-A/B/C surface expression (Figures 4C–4E). We sorted the treated populations into HLAHigh versus HLALow and analyzed mRNA expression of DUX4-regulated genes that also characterize the ZGA and 8CL states (Figures 4F–4H). RT-qPCR demonstrated that low HLA-A/B/C surface expression correlated with high expression of these DUX4-regulated genes. It is interesting to note that the MHC expression in SUSA appears bimodal suggesting an all-or-none expression of DUX4 whereas the general shift of MHC distribution in G401 following DNA damage is not bimodal, possibly reflecting more of a gradient of DUX4 levels. These data further support that DUX4 expression in cancer cells is associated with suppressed MHC-I presentation. Future studies will be necessary to determine whether this subpopulation preferentially survives immune surveillance.
Figure 4. Low MHC-I surface protein correlates with DUX4 expression.
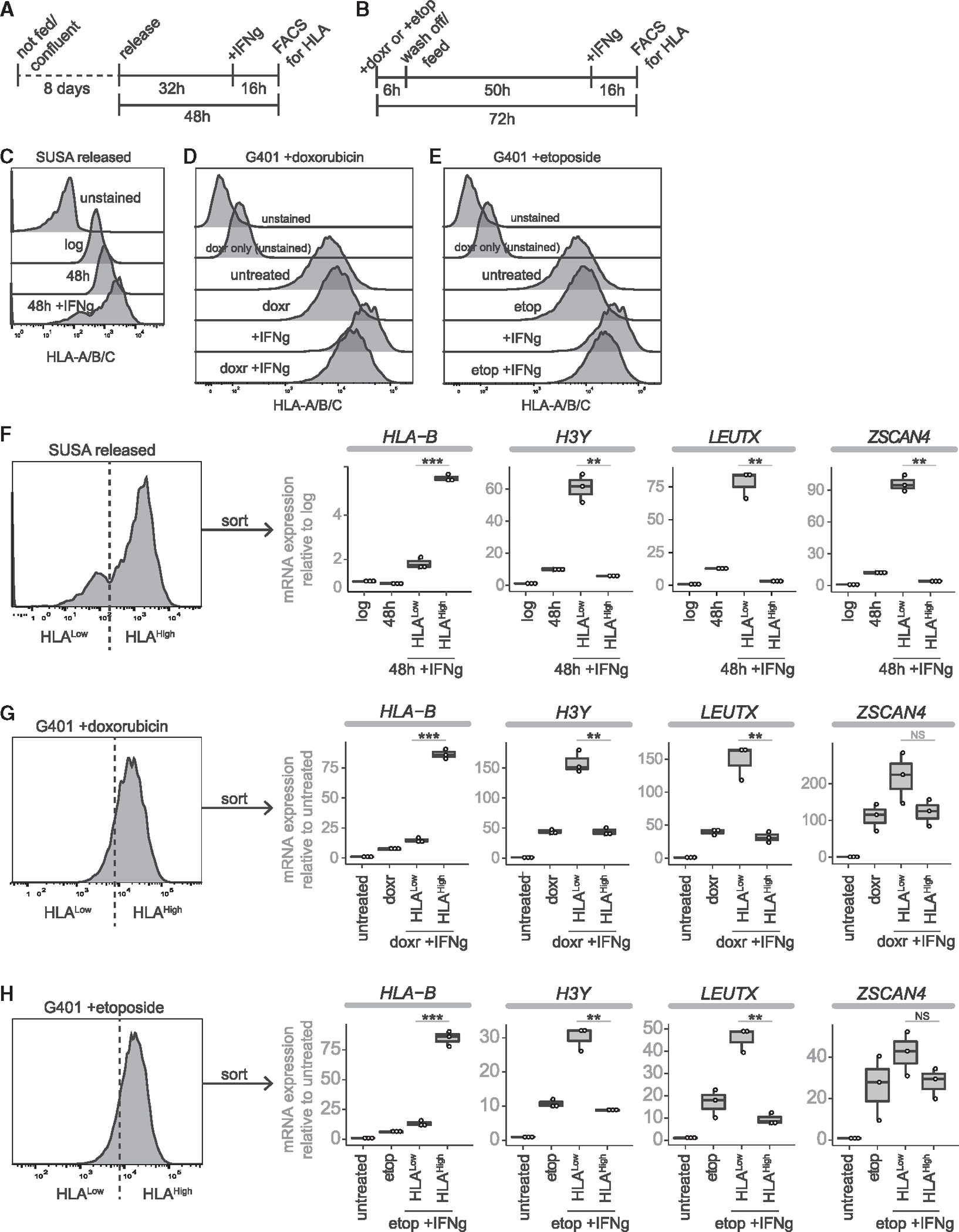
(A and B) Timeline of cell treatments for (A) SUSA experiments and (B) G401 experiments.
(C–E) Histograms of HLA-A/B/C. (C) SUSA 48 h post release stained with BV605 anti HLA-A/B/C. (D and E) G401 72 h post treatment with (D) 1 μM doxorubicin (doxr) or (E) 10 μM etoposide (etop). (D) and (E) were analyzed at the same time and use the same control samples. </p/>(F–H) FACS analysis for HLAHigh versus HLALow cells post-IFNg treatment followed by RT-qPCR for HLA-B and three DUX4-target genes on unsorted and sorted populations of (F) released SUSA, (G) G401 +doxorubicin, or (H) G401 +etoposide. For all RT-qPCR experiments, n = 3 technical triplicates. p values calculated with a one-tailed Welch’s t test: * <0.05; **<0.005, ***<0.0005, NS = no significance.
Cancer cell lines express DUX4 from multiple D4Z4 alleles with decreased DNA methylation
In contrast to DUX4 expression in FSHD muscle cells, which is almost exclusively from the FSHD-permissive 4qA161 allele and polyadenylated in exon 3, 3′RACE and RT-PCR in the DUX4-FL cancer cell lines indicated DUX4 mRNA expression from multiple 4qA, 4qB, and 10q D4Z4 locations that used alternative polyadenylation (pA) sites (Table S1). All of the cell lines expressed DUX4 transcripts with chromosome 10-specific polymorphisms that spliced exon 2 with exon 6 and used a canonical pA signal in exon 6 (Table S1), consistent with DUX4 isoforms previously identified in testis that arise from D4Z4 repeats of both chromosome 4 and 10.16
In addition to this previously described germline DUX4 isoform, 3′RACE identified several DUX4 RNA isoforms with pA sites shared by multiple cancer cell lines, albeit lacking a canonical pA signal motif. Three of the cell lines (G401, HCC38, and SKNMC) had transcripts originating from 4qA and 4qB haplotypes with an exon 2/3a or 2/3b splice junction and polyadenylation at a site in exon 3a. Many of the cancer cell lines (G401, SUSA, SKNMC, and HCC38) also had DUX4 transcripts polyadenylated at identical or nearby sites in intron 1, again without a nearby canonical pA motif. The appearance of the same or similarly located pA sites from multiple cells lines suggests that these might represent alternative pA sites; however, it remains possible that the high GC content in this region creates amplification artifacts and the true pA site might be distal to those mapped in this study, or the RNAs might not be polyadenylated despite priming with oligo-dT.
Decreased DNA methylation in the D4Z4 region is associated with DUX4 expression in FSHD35 and the five DUX4-FL-expressing cancer cell lines showed decreased DNA methylation of the D4Z4 region as determined by the percent cleavage of a methylation sensitive enzyme (FseI), an assay that measures methylation on all haplotypes of both chromosome 4 and 1015,37 (Table S4).
DISCUSSION
Our prior study showed that RNAs encoding DUX4 and DUX4-target genes were detected in a wide variety of cancers.6 However, it remained unknown whether this indicated the presence of a distinct subset of cells that re-activated a broader early embryonic program. In the current study, we identified several cancer cell lines that express DUX4 and DUX4-target genes, albeit at relatively low levels. We showed that the relatively low levels represent the transient expression of DUX4 in a subset of the cancer cells. Single-cell RNA sequencing showed that these cells activated the early DUX4-regulated ZGA program, as well as the broader 8C-like signature. In addition, the associated expression of genes that characterize trophectoderm and epithelial/mesenchymal lineages indicates the progression to components of early lineage specification pathways, and the suppression of steady-state and IFNg-induced MHC-I expression suggests a contribution to immune evasion. Together, our data suggest that DUX4 expression in cancers can transiently induce a metastable early embryonic program that includes genes characteristic of trophectoderm development and epithelial-to-mesenchymal transition that might contribute to aspects of cancer progression.
The transcription program induced by DUX4 was initially described in muscle cells in the context of FSHD where it was shown to regulate an early stem cell program that included induction of ZSCAN4 and classes of retrotransposons.23,38 Subsequent studies showed that transient expression of DUX4 in the 4-cell human embryo, or Dux in the 2-cell mouse embryo, activates an early wave of ZGA gene expression.7–9 Although Dux knockout mice can produce viable embryos, several studies have shown some degree of reduced or delayed ZGA gene expression39,40 and reduction of genes involved in trophectoderm development.27 Earlier studies identified a sub-population of mouse ES cells transiently cycling through a 2C-like naive state that includes ZGA gene expression and metabolic and epigenetic reprogramming.12,13 More recent studies implicate the transient expression of Dux/DUX4 as a regulator of the transient 2C or 8C-like state in mouse or human ES cells, respectively,10,14 and that transient expression of DUX4 in human ES cells can induce the ZGA program and an 8C-like state.41 Our current study builds on and extends this prior work by using the transcriptional programs identified as characterizing the ZGA and 8CLC human ES cell states10 to show that the cells in DUX4-expressing cancers initiate the DUX4-regulated portion of the ZGA program and components of the broader 8CLC gene expression program.
It is interesting to consider why some cancer cell lines transiently express DUX4 in a subset of cells. In ES/iPS cells and the early embryo, Dppa2 and Dppa4 have been shown to bind the Dux locus and regulate its initial expression42 in addition to activation by p53,36 quickly followed by epigenetic suppression by SMCHD1,43,44 resulting in methylation of the locus by antagonizing TET proteins.45 In FSHD, mutations in chromatin modifiers (most commonly SMCHD1 or DNMT3B) or D4Z4 array sizes of 10 or less result in hypomethylation of the D4Z4 region, inefficient epigenetic repression, and episodic expression of DUX4 in FSHD muscle cells,16,17 indicating that D4Z4 hypomethylation and decreased epigenetic repression is sufficient for episodic DUX4 expression in some somatic cells. In addition, multiple chromatin modifying factors (e.g., components of the NuRD, CAF, bromodomain, and polycomb complexes) (reviewed in Campbell et al.46 and Tihaya et al.47) and signaling pathways48,49 can modulate the efficiency of DUX4 suppression. Indeed, our previous study of DUX4-expressing cancers identified loss-of-function mutations in 12 of 23 genes encoding validated or likely repressors of DUX4,6 including SMCHD1 and DNMT3B, as well as high DPPA2 and DPPA4 expression in testicular germ cell tumors. In our current study, the relative hypomethylation at the D4Z4 locus in DUX4-expressing cancer cell lines provides an initial explanation for why these lines are permissive for DUX4 expression. We did not detect higher expression of other macrosatellite repeats, suggesting that the de-repression of D4Z4 does not reflect a genome-wide loss of repeat repression.
The SUSA cell line was isolated from a testicular teratocarcinoma50 and expresses high levels of DPPA2 and DPPA4,51 factors implicated in the developmental regulation of Dux/DUX4 expression.42 As a cancer cell line derived from a teratocarcinoma, it might retain an ability to differentiate into early embryonic lineages and our current study implicates DUX4 expression as regulating the expression of some of these early embryonic genes. KLE cells have mutations in multiple chromatin regulators (DNMT1, BAZ3B, MSX1, CBX, PRDM9, ARID1B, and DPF1), which might individually or in concert alter D4Z4 epigenetic repression; and G401 has an inframe deletion in SMARCA5 (E816del), a gene previously shown to regulate DUX4 expression52; however, the consequences of this particular mutation remain unknown. Together, these data support a multifactorial model of regulating D4Z4 repression in different cancer cell lines, similar to our prior analysis of DUX4-expressing cancers,6 with the common feature of D4Z4 hypomethylation, a feature causative for episodic DUX4 expression in FSHD muscle cells.
In this model, D4Z4 hypomethylation permits induction of DUX4 expression by factors that would be epigenetically blocked in normal somatic tissues, such as p53 or other factors and signaling pathways. For example, increased DUX4 expression following release from confluence occurs independent of p53 status and might reflect activation of stress pathways, such as p38, or as a consequence of other signaling pathways or the chromatin relaxation associated with S-phase. Future studies are needed to determine whether D4Z4 methylation status is a reliable predictor of DUX4 expression in a broad set of cancers.
The biological significance of DUX4 and ZGA gene expression in cancers might easily be underestimated because of low levels in bulk RNA. However, our study shows that low levels in bulk RNA represent sub-populations of cells that broadly activate early developmental programs. Our demonstration that growth conditions and DNA damage can substantially increase the percentage of DUX4-expressing cells indicates that environmental factors dynamically modulate DUX4 expression and might provide some advantage during specific bottlenecks in cancer progression. Together with our prior study showing DUX4-mediated suppression of MHC presentation, our current demonstration that DUX4-expressing cancer cells induce components of the 8CLC program and subsequent genes that regulate the specification of embryonic/extra-embryonic tissues and epithelial/mesenchymal specification suggests that DUX4 expression might contribute to many of the major hallmarks of cancer.53,54 Additional studies will be necessary to determine the specific roles of DUX4 expression in cancer progression, metastasis, immune evasion, and response to therapies.
Limitations of the study
The current study shows that expression of DUX4 in cancer cells activates a coherent early embryonic program. The consequences for cancer progression, tissue invasion, immune evasion, and metastasis remain to be addressed. For example, our earlier study indicated that DUX4-expressing cancers had an immune evasion phenotype and MHC suppression. It will be important to determine whether the DUX4-expressing cells and their progeny show selective immune evasion relative to other cells in the population. Similarly, future studies, possibly employing lineage tracing, will be necessary to determine the fate of DUX4-expressing cells, the duration of their transcriptional and possibly epigenetic changes, and their role in cancer progression.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Stephen Tapscott (stapscot@fredhutch.org).
Materials availability
Reagents generated in this study are available from the lead contact with a standard Materials Transfer Agreement.
Data and code availability
Single cell RNA-sequencing data were uploaded to NCBI Gene Expression Omnibus (GEO: GSE223848).
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request
EXPERIMENTAL MODELS
Cell culture
MB135 and MB200 myoblast lines were cultured in F-10 media supplemented with 10% fetal bovine serum and 1x penicillin/streptomycin. For differentiation, myoblasts were cultured in DMEM supplemented with 1x penicillin/streptomycin, insulin (10ug/mL), and transferrin (10ug/mL).
Cancer cell lines were cultured in varying media formulation supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. G401 cells were cultured in McCoy’s 5A (Modified) Medium; KLE and SCC-9 cells were cultured in DMEM/F-12; SKNMC and MCF7 cells were cultured in EMEM; SUSA, HCC38, NALM6, AMO1, KMS27, KMH2, MOLP-2, and OPM-2 cells were cultured in RPMI. HeLa cells were cultured in DMEM.
Release from confluence
Cells were plated at confluence and maintained without media change for 1–2 weeks. Cells were then exposed to fresh media for 2hrs and then passaged 1:6 for post release time course.
Drug treatments
Cells were treated for 6hrs with 1uM doxorubicin, 10uM etoposide, or 5uL DMSO (vehicle) for 6hrs. RNA was harvested 48hrs post treatment using the Machery-Nagel RNA extraction kit.
METHOD DETAILS
Gapmer and siRNA treatments
MOE Gapmers or siRNAs were mixed with OptiMEM and used at a final concentration of 25nM with 3.75uL/mL RNAiMax Lipofectamine (ThermoFisher, 13778075) overnight at 37C in normal media.
Haplotyping and methylation analysis
Number units in D4Z4 macrosatellites on chromosomes 4 and 10 as well as their haplotype was determined with Pulsed Field Gel electrophoresis, Southern blotting and subsequent hybridization of the blots with probes P13E-11, “A” and “B” probes. SSLP length for further haplotyping was determined using an ABI Prism 3100 Genetic analyzer. CpG methylation of D4Z4 macrosatellites on chromosomes 4 and 10 was determined by sensitivity to FseI endonuclease.37
Single-cell RNA sequencing (scRNA seq)
Cells released from confluence were cryobanked at 24, 48, and 72hrs after release in BamBanker (Fisher, NC9582225) at −80°C. Cells were thawed, washed with cold media +10%FBS, then cold PBS with 1% BSA. Cells were filtered through a 35uM filter and resuspended to 1,000cells/ul and kept on ice until processed according to 10X genomics protocol and sequenced by the FHCC genomics core.
Western blot
Cells were harvested in RIPA buffer containing protease and phosphatase inhibitors and sonicated on low for 5 min. The whole cell lysate was centrifuged at 12,0000 RCF for 10 min and the supernatant transferred to a new tube. Protein quantification was performed using the Pierce BCA Protein Assay Kit. Either 5ug or 10ug of whole cell lysate was heated in LDS Sample Buffer with 2.5% β-mercaptoethanol at 70°C for 10 min with shaking. For SDS-PAGE, proteins were loaded onto a 4–12% NuPAGE Bis-Tris gel and run at 100V for about 2 h in NuPAGE MOPS SDS Running Buffer with 300uL NuPAGE antioxidant. Proteins were transferred to a 0.2μm PVDF membrane at 30V for 1 h in NuPAGE Transfer Buffer with 10% methanol. Membranes were blocked in 5% milk in TBST for 1 h, then incubated at 4°C overnight with rocking. Blots were washed three times (15 min each) with TBST and incubated with HRP-conjugated secondary antibody for 1 h. Blots were washed three times (15 min each) with TBST, and bands were detected via chemiluminescence with either SuperSignal West Pico PLUS Substrate, SuperSignal West Femto Substrate, or SuperSignal West Atto Substrate.
Immunofluorescence and immunohistochemistry
Cells were washed with PBS, fixed in 2% paraformaldehyde for 10 min, and washed twice with PBS. Cells were then permeabilized in 0.1% Triton X-100 in PBS for 10 min at room temperature. Cells were then incubated overnight at 4°C with DUX4 [E14–3] primary antibody (1:400 dilution) or H3Y1/2 [8H6–2111] (1:333 dilution) and washed with PBS three times for 10 min each. Cells were incubated in TRITC-conjugated donkey anti-rabbit secondary antibody (1:666 dilution) and TRITC-conjugated donkey anti-rat secondary antibody (1:666 dilution) for 3 h at room temperature. Cells were washed with PBS, stained with DAPI for 10 min at room temperature, and washed with PBS. Cells were imaged with a fluorescence microscope.
Reverse transcription-quantitative PCR (RT-qPCR)
RNA was harvested with the NucleoSpin RNA Kit (Takara) according to the manufacturer’s protocol. RNA quality was verified by NanoDrop 2000 (Thermo Scientific). RNA was treated with DNaseI, Amplification Grade (Invitrogen). Reverse transcription was performed in a 20uL reaction: 200–1000ng whole RNA, 1uL dNTP (10mM), 1uL oligo dT primer (10mM), 4uL 5x SSIV Buffer, 1uL DTT (100mM), 1 μL RNaseOUT, and 1uL SSIV RT enzyme. Thermal cycling conditions for reverse transcription were as follows: 50°C for 40 min, 55°C for 30 min, and 80°C for 10 min. Complementary DNA (cDNA) was treated with 1uL of RNaseH and incubated at 37°C for 20 min, then diluted 1:5 or 1:4 with RNase-free H2O. Quantitative real-time PCR (qPCR) was performed on the QuantStudio 7 Flex Real-Time PCR System in a 10uL reaction: 2uL cDNA, 5uL 2x iTaq Universal SYBR Green Supermix, 0.3uL forward and reverse primer (10uM), and 2.4uL H2O. qPCR primers were synthesized by Integrated DNA Technologies (IDT) and are listed in Star Methods table. Thermal cycling conditions for qPCR were as follows: 50°C for 2 min and 95°C for 10 min; 40 cycles of 95°C for 15 s and 60°C for 60 s.
Generation of cell lines with dox-inducible codon altered DUX4 transgene
A polyclonal KLE-iDUX4CA cell line was generated with lentiviral transduction, selected, and maintained in 3μg/mL puromycin as previously described for MB135-iDUX4CA cells in Jagannathan et al., 2016. SUSA-iDUX4CA and G401-iDUX4CA cell lines were generated using CRISPR/Cas9-mediated genomic integration at the AAVS1 safe harbor locus via homology directed repair. The codon-altered DUX4 transgene was subcloned by restriction enzyme digest into the AfeI and SalI sites of the pMK364 vector backbone (AAVS1-Tet-OsTIR1-AAVS1-Puro; Addgene #158663) containing a dox-inducible promoter and homology arms to the AAVS1 safe harbor locus. SUSA and G401 cells were co-transfected with Cas9/AAVS1-sgRNA expressing plasmid (Addgene #62988) and iDUX4CA plasmid described above at a 1:2 ratio using Lipofectamine 3000 (Fisher #L300015) according to the manufacturer’s protocol. Cells were incubated for two days post-transfection, and subsequently selected and maintained in 3μg/mL puromycin. Monoclonal cell lines were isolated, and iDUX4CA transgene integration at the AAVS1 safe harbor locus was validated with PCR amplification from genomic DNA followed by sanger sequencing.
3′ rapid amplification of cDNA ends (3′ RACE)
RNA was harvested with the NucleoSpin RNA Kit (Takara) according to the manufacturer’s protocol. RNA quality was verified by NanoDrop 2000 (Thermo Scientific). RNA was treated with DNaseI, Amplification Grade (Invitrogen). To enrich for DUX4 transcripts, biotinylated oligonucleotide probes targeting DUX4 exon 1 were denatured for 5 min at 95°C, then hybridized to RNA in hybridization buffer (0.05M Tris-HCl pH7.5, 0.75M NaCl, 1mM EDTA, 10mM DTT, RNaseOUT, and 15% formamide). Thermal cycling conditions for hybridation were 75°C for 2 min, followed by 15 cycles for 10 min each ramping down at −1C per cycle (ranging from 65°C to 50°C). 50uL streptavidin (MyOneC1) beads were washed twice with wash buffer (0.05M Tris-HCl pH7.5, 0.75M NaCl, 1mM EDTA, 10mM DTT, RNaseOUT, and 0.05% Tween 20). Beads were resuspended in 25uL binding buffer (0.01M Tris-HCl pH7.5, 2M NaCl, 1mM EDTA, 10mM DTT, RNaseOUT, and 0.05% Tween 20), and 100uL hybridized RNA sample was added to beads and incubated overnight at room temperature with rotation. Beads were washed twice with 500uL wash buffer. RNA was eluted by resuspending beads in 100uL dissociation buffer (10mM EDTA, 10mM DTT, RNaseOUT, and 95% formamide) and incubating at 95°C for 2 min with shaking. RNA was ethanol precipitated overnight at −80°C, pelleted, air-dried, and resuspended in 20uL of RNase-free H2O.
RACE reverse transcription was performed with RNA in a 20uL reaction: 7.5uL RNA, 2uL dNTP (10mM), 1uL long dT anchor primer (37.5uM), 4uL 5x SSIV Buffer, 0.5uL DTT (100mM), 0.5uL RNaseOUT, 1uL SSIV RT enzyme. Thermal cycling conditions for reverse transcription were as follows: 50°C for 40 min, 55°C for 30 min, and 80°C for 10 min. Complementary DNA (cDNA) was treated with 1uL of RNaseH and incubated at 37°C for 20 min, then purified with the Macherey-Nagel NucleoSpin Gel and PCR Clean-up Kit according to the manufacturer’s protocol. A nested RACE-PCR was then performed in a 20uL reaction: 1uL template DNA, 1uL forward and reverse primer (10uM), 10uL 2x Phusion Plus PCR Master Mix, 4uL 5x Phusion GC Enhancer, and 3uL ddH2O. Primary PCR was performed with the 1st set of nested primers the following thermal cycling conditions: initial denaturation at 98°C for 2 min; 25 cycles of 98°C for 30 s, 60°C for 30 s, and 72°C for 30 s; and final extension at 72°C for 7 min. PCR products were purified and a 2nd PCR was performed with the 2nd set of nested primers with the following thermal cycling conditions: initial denaturation at 98°C for 2 min; 30 cycles of 98°C for 30 s, 60°C for 30 s, and 72°C for 30 s; and final extension at 72°C for 7 min. PCR products were run on a 1% agarose TBE gel, and products of interest were gel excised and purified with the Macherey-Nagel NucleoSpin Gel and PCR Clean-up Kit according to the manufacturer’s protocol. Products were A-tailed in a 20uL reaction: 10uL purified PCR product, 2uL 10X PCR Buffer (−MgCl2), 0.25uL MgCl2 (50mM), 0.25uL dATP (100mM), and 0.25uL Taq polymerase. Reaction was incubated at 72°C for 20 min. A-tailed PCR products were then TA TOPO cloned in a 4uL reaction: 2uL A-tailed PCR product, 0.5uL salt solution, and 0.5uL pCR4-TOPO TA vector. Reaction was incubated at room temperature for 30 min, then transformed in chemically competent DH5α E. coli cells. Colonies were grown up in 3mL LB-carbenicillin and Minipreps performed with the PureLink Quick Plasmid Miniprep Kit (Invitrogen) according to the manufacturer’s protocol. Minipreps were test digested with EcoRI at 37°C for 1 h and products visualized on a 1% agarose TBE gel. Minipreps were submitted for Sanger sequencing with the M13 Forward primer.
Staining for MHC-I and FACS
Cells were treated with 50 ng/mL IFNg (R&D Systems, 285-IF/CF) for 16hrs before lifting with 0.25% Trypsin-EDTA. Cells were pelleted at 300RCF and resuspended in 100uL PBS 1%FBS +1:50 APC/FIRE or BV605 mouse anti-human HLA-A/B/C (Biolegend) and incubated 20–30 min at 4°C in dark. Cells were washed with PBS and filtered through 35uM strainer caps before analyzing and sorting with the BD Symphony 6. Cells were sorted into 4°C media and pelleted at 300RCF before being processed for RT-qPCR as above. Flow analyses were repeated for publication using FlowJo10.
Determining direct DUX4 targets from the 8CLC gene list
The 8CLC gene list was adopted from Taubenschmid-Stowers et al.10 Direct targets of DUX4 were determined by the presence of a DUX4 ChIP-seq peak adjacent to the DUX4-induced TSS based on prior data from DUX4 expression in MB135 myoblasts8,23,28,38 and prior observation that DUX4 induces transcription at its binding site.8,23,38 Genes induced by DUX4 with a ChIP-seq peak adjacent to the TSS were categorized as direct DUX4 targets, whereas all other genes were categorized as not direct DUX4 targets.
QUANTIFICATION AND STATISTICAL ANALYSIS
For RTqPCR analyses
Each Biological replicate was run in technical triplicates, with -RT controls. Median CT values of the technical triplicates were used for analysis. Gene expression was normalized to the housekeeping gene RPL27 (ribosomal protein L27). p-values were calculated with an unpaired, one-tailed Welch’s t test (assuming unequal variance).
Immunofluorescent quantification
At least 5 random fields were acquired for each sample. DAPI+ cells were counted along with other markers indicated. p-values were calculated with an unpaired, one-tailed Welch’s t test (assuming unequal variance).
Single-cell RNA data analysis
Sequencing data were aligned and quantified using 10X Genomic’s Cell Ranger v 6.1.255 using the reference human GRCh38 genome and GRCh38–2020-A transcriptome supplied with Cell Ranger. Raw feature matrices were initially analyzed independently using ScanPy v. 1.9.1,26 where genes not expressed in at least 1 cell and cell barcodes without at least 200 unique genes expressed were removed. Cell barcodes were then filtered with the following thresholds: the top 2.5% of cells by UMI counts were removed as potential doublets; cells with log10(UMI) less than 2.8 were removed as low quality; and cells with more than 25% UMIs aligning to mitochondrial reads were removed, also considered poor quality. After filtering, ScanPy was used to normalize and log transform read counts. The top 2500 highly variable genes were calculated and used to perform dimensional reduction via principal component analysis (PCA) with subsequent visualization by PC variance plot to determine optimal number of principal components (PCs) to include in downstream analyses. Next, we calculated the neighborhood graph and performed non-linear dimensional reduction using UMAP and the first 30 PCs. Finally, clusters were calculated using scanpy.tl.leiden() and cluster. Sources of technical bias were plotted on UMAPs to ensure that clustering was not being driven by artifact. After individual samples were processed, they were concatenated using anndata’s concatenate function and analyzed in general method as above, including per-cell normalization, log transformation, PCA, neighborhood and UMAP dimensionality reduction using the first 30 PCs. Deviations from the above workflow for the merged dataset include calculating an S and G2M cell cycle gene module score using scanpy.tl.score_genes_cell_cycle, a simple linear regression using scanpy.pp.regress_out() to remove technical noise associated with UMI and mitochondrial counts as well as gene expression associated with S and G2M gene module score, gene expression imputation using scanpy.external.pp.magic(), and clustering using Phenograph ver. 1.5.7.56 Sources of technical bias were again plotted on the UMAP to ensure that the clustering was driven by biological processes. The count matrix was exported and Seurat (ver. 4.1.1)57,58 was used to determine cluster marker genes via FindAllMarkers(). Marker genes in EnrichR59–61 were used to query the CellMarker_Augmented_2021 database.62 Imputed counts were used for all single gene visualizations, and Z score normalized imputed counts were used for gene module scores calculated via scanpy.tl.score_genes(). Dot plot clustering dendrogram was calculated using ZGA genes alone. Cells were ranked by 8C-like score or ZGA score and MAGIC imputed expression for MHC and B2M genes for the cells in the top 350 and the middle quartile (37.5%–62.5%) of ranked expression. These expression levels were plotted as violin plots using ggplot263 and statistical significance was assessed using permutationTest2() from the resample64 package with 500,000 permutations of the difference between the mean of the top 350 and middle quartile groups.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Anti-GAPDH (6C5) | GeneTex | Cat#GTX28245; RRID: AB_370675 |
| Anti-DUX4 (E14-3) | Geng et al.23 | N/A |
| Anti-Histone H3Y1/2 (8H6-2111) | Active Motif | Cat#61161, RRID: AB_2793533 |
| Anti-beta-Actin | Cell Signaling | Cat#4967, RRID:AB_330288 |
| BV605 mouse anti-human HLA-A/B/C | BioLegend | Cat#311431, RRID: AB_2566150 |
| APC/FIRE mouse anti-human HLA-A/B/C | BioLegend | Cat#311444, RRID: AB_2629629 |
| anti-Ki67 (D2H10) | Cell Signaling | RRID: AB_2636984 |
| anti-BrdU (ZBU30) | Fischer Scientific | RRID: AB_2532917 |
| Fluorescein (FITC) AffiniPure Donkey Anti-Rabbit IgG | Jackson ImmunoResearch | Cat#711-095-152, RRID:AB_2315776 |
| Fluorescein (FITC) AffiniPure Donkey Anti-Rat IgG | Jackson ImmunoResearch | Cat#712-095-150, RRID:AB_2340651 |
| Fluorescein (FITC) AffiniPure Donkey Anti-Mouse IgG | Jackson ImmunoResearch | Cat#715-095-151, RRID:AB_2335588 |
| Goat Anti-Rabbit IgG (H + L) Superclonal Recombinant Secondary Antibody, HPR | Invitrogen/Thermo Fisher | Cat#A27036, RRID:AB_2536099 |
| Goat Anti-Mouse IgG (H + L) Superclonal Recombinant Secondary Antibody, HPR | Invitrogen/Thermo Fisher | Cat#A28177, RRID:AB_2536163 |
| Rhodamine (TRITC) Donkey Anti-Rabbit IgG | Jackson ImmunoResearch | Cat#711-025-152, RRID:AB_2340588 |
| Rhodamine (TRITC) Donkey Anti-Rat IgG | Jackson ImmunoResearch | Cat#712-025-150, RRID:AB_2340635 |
| Rhodamine (TRITC) Donkey Anti-Mouse IgG | Jackson ImmunoResearch | Cat#715-025-151, RRID:AB_2340767 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Recombinant human basic fibroblast growth factor | PeproTech | Cat#100018B |
| Dexamethasone | Sigma-Aldrich | Cat#D4902 |
| Puromycin dihydrochloride | Sigma-Aldrich | Cat#P8833 |
| Penicillin/streptomycin | Gibco/Thermo Fisher | Cat#15140122 |
| Ham’s F-10 | Gibco/Thermo Fisher | Cat#11550043 |
| RPMI 1640 | Gibco/Thermo Fisher | Cat#11875093 |
| McCoy’s 5A | Gibco/Thermo Fisher | Cat#16600-082 |
| DMEM | Gibco/Thermo Fisher | Cat#11965092 |
| Opti-MEM I Reduced Serum Media | Gibco/Thermo Fisher | Cat#31985070 |
| PBS, pH 7.4 | Gibco/Thermo Fisher | Cat#10010023 |
| Fetal Bovine Serum | Hyclone | Cat#SH30071.03 |
| Insulin | Sigma-Aldrich | Cat#I1882 |
| Transferrin | Sigma-Aldrich | Cat#T0665 |
| Doxorubicin hydrochloride | Sigma-Aldrich | Cat#44583-1MG |
| Etoposide | Sigma-Aldrich | Cat#E1383-25MG |
| 0.25% Trypsin-EDTA | Gibco/Thermo Fisher | Cat#25200056 |
| RNase OUT | Invitrogen/Thermo Fisher | Cat#100000840 |
| Isopropanol, DNAse, RNAse and Protease free | Thermo Fisher | Cat#AC327272500 |
| EDTA (0.5 M), pH 8.0, RNase-free | Invitrogen/Thermo Fisher | Cat#AM9260G |
| 1 M MgCl2, RNase-free | Invitrogen/Thermo Fisher | Cat#AM9530G |
| 5 M NaCI, RNase-free | Invitrogen/Thermo Fisher | Cat#AM9760G |
| UltraPure Ethidium Bromide | Invitrogen/Thermo Fisher | Cat#15585011 |
| GeneRuIer 100 bp DNA Ladder | Thermo Fisher | Cat#SM0242 |
| GeneRuIer 1 kb Plus DNA Ladder | Thermo Fisher | Cat#SM1331 |
| PageRuIer Prestained Protein Ladder | Thermo Fisher | Cat#26616 |
| Triton X-100 Detergent, Molecular Biology Grade | Sigma-Aldrich | Cat#648466 |
| Tween 20 | Fisher Chemical | Cat#BP337500 |
| 16% Paraformaldehyde | Electron Microscopy Sciences | Cat#100503-917 |
| 1 M DTT, BioUltra, 10mL | Sigma-Aldrich | Cat#43816 |
| SuperSignal West Pico PLUS Chemiluminescent (ECL) Substrate | Thermo Fisher | Cat#34577 |
| SuperSignal West Femto Chemiluminescent (ECL) Substrate | Thermo Fisher | Cat# 34096 |
| SuperSignal West Atto Chemiluminescent (ECL) Substrate | Thermo Fisher | Cat#38554 |
| Lipofectamine 3000 Transfection Reagent | Invitrogen/Thermo Fisher | Cat#L3000001 |
| Lipofectamine RNAiMAX | Invitrogen/Thermo Fisher | Cat#13778150 |
| DAPI | Sigma-Aldrich | Cat#D9542 |
| Recombinant Human Interferon-gamma Protein, Carrier Free (IFNg) | R&D Systems | Cat#285-IF/CF |
|
| ||
| Critical commercial assays | ||
|
| ||
| Pierce BCA Protein Assay Kit | Thermo Fisher | Cat#23225 |
| DNase I, Amplification Grade | Gibco/Thermo Fisher | Cat#18068015 |
| SuperScript IV First-Strand Synthesis System | Invitrogen/Thermo Fisher | Cat#18091050 |
| iTaq SYBR Green Supermix | Bio-Rad | Cat#1725124 |
| NucleoSpin RNA Kit | Machery-Nagel | Cat#740955 |
| NucleoSpin Gel and PCR Clean-up Kit | Machery-Nagel | Cat#740609 |
| PureLink Quick Plasmid Miniprep Kit | Invitrogen | Cat#K210011 |
| GeneJET Genomic DNA Purification Kit | Thermo Fisher | Cat#K0722 |
| Chromium 10X Gel Beads 3’v3.1 (−80C) | 10X Genomics | Cat#2000164 |
| Chromium 10X Chip G | 10X Genomics | Cat#2000177 |
| Chromium 10X Next GEM kit 3’v3.1 | 10X Genomics | Cat#1000123 |
| Chromium 10X Library Construction kit | 10X Genomics | Cat#1000190 |
|
| ||
| Deposited data | ||
|
| ||
| single-cell RNA-sequencing data | GEO# GSE223848 | N/A |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| MB135 cells (female) | (Snider et al.)16 | N/A |
| MB200 cells (FSHD2, male) | Fields Center for FSHD and Neuromuscular Research, University of Rochester Medical Center | (https://www.urmc.rochester.edu/neurology/fields-center.aspx) |
| KLE cells (female) | ATCC | Cat#CRL-1622, RRID:CVCL_1329 |
| G401 cells (male) | ATCC | Cat#CRL-1441, RRID:CVCL_0270 |
| SUSA cells (male) | DSMZ | ACC 747; RRID: CVCL_L280 |
| SKNMC cells (female) | ATCC | Cat#HTB-10, RRID:CVCL_0530 |
| HCC38 cells (female) | ATCC | Cat#CRL-2314, RRID:CVCL_1267 |
| NALM6 cells (male) | ATCC | Cat#CRL-3273, RRID:CVCL_0092 |
| AMO1 cells (female) | DSMZ | Cat# ACC 538, RRID:CVCL_1806 |
| KMS27 cells (male) | JCRB, NIBIOHN | Cat# JCRB1188, RRID:CVCL_2993 |
| KMH2 cells (male) | DSMZ | Cat# ACC 8, RRID:CVCL_1330 |
| MCF7 cells (female) | ATCC | Cat#HTB-22, RRID:CVCL_3397 |
| HeLa cells (female) | ATCC | Cat#CCL-2 RRID:CVCL_0030 |
|
| ||
| Oligonucleotides | ||
|
| ||
| Biotinylated probe 1: DUX4 exon1, 5’:GGGAGGGTGCTGTCCGAGGGTGTCGGGAGGGCCAT | This paper | N/A |
| Biotinylated probe 2: DUX4 exon1, 3’:TCCTAAAGCTCCTCCAGCAGAGCCCGGTATTCTTC | This paper | N/A |
| 3’RACE _extended 3’ anchor for reverse transcription_FCTAATACGACTCACTATAGGACCACGCGTATCGATGTCGACTTTTTTTTTTTTTTTTV | This paper | N/A |
| control MOE-gapmer (GFP): GAGAAAGTGTGACAAGTG | This paper | N/A |
| DUX4 MOE-gapmer +258: TTCCGCTCAAAGCAGGCT | This paper | N/A |
| M13F for sequencing clones: GTAAAACGACGGCCAG | This paper | N/A |
| ON-TARGETplus Non-targeting Control Pool | Horizon | Cat#D-001810-10-20 |
| ON-TARGETplus Human TP53 siRNA SMARTPool | Horizon | Cat#L-003329-00-0005 |
| See Table S4B for RT-PCR and RTqPCR primers | This paper | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| pCW57.1-iDUX4CA | Addgene | Cat#99281 |
| pMD2.G | Addgene | Cat#12259 |
| psPAX2 | Addgene | Cat#12260 |
| pAAVS1-Tet-OsTIR1-AAVS1-Puro | Addgene | Cat#158663 |
| pCas9/AAVS1-sgRNA | Addgene | Cat#62988 |
|
| ||
| Software and algorithms | ||
|
| ||
| Python, V3.9.6 | Python.org | http://www.python.org |
| R,2022 | R Foundation for Statistical Computing, Vienna, Austria | https://www.R-project.org/ |
| FlowJo10 | BD Biosciences | N/A |
|
| ||
| Other | ||
|
| ||
| QuantStudio 7 Flex | Applied Biosystem | N/A |
| Zeiss Axiophot fluorescent microscope | Zeiss | N/A |
| BD Symphony 6 | Becton Dickinson | N/A |
Highlights.
DUX4 is transiently expressed in subpopulations of cells in some cancer cell lines
DUX4 activates ZGA and 8C-like gene expression as well as early lineage markers
Cells that express DUX4 have decreased MHC-I expression
DNA damage increases the number of DUX4-expressing cells through TP53 activation
ACKNOWLEDGMENTS
We thank Bradley Cairns, Robert Bradley, Eric Holland, and the Tapscott lab for critical reading of the manuscript, as well as the Fred Hutchinson Cancer Center Flow Cytometry Core and Genomics Core for technical assistance. This work was funded by NIH grants R01AR045203 (S.J.T.), F32-CA254805 (D.C.H.), and funding from the Friends of FSH Research and the Chris Carrino Foundation for FSHD (S.J.T.).
Footnotes
DECLARATION OF INTERESTS
S.J.T. is an inventor on the FHCC patent application Methods and Composition for Treating Cancer and a member of the Renogenyx Board of Directors, a company developing FSHD therapeutics.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.113114.
REFERENCES
- 1.Meacham CE, and Morrison SJ (2013). Tumour heterogeneity andcancer cell plasticity. Nature 501, 328–337. 10.1038/nature12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dagogo-Jack I, and Shaw AT (2018). Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 15, 81–94. 10.1038/nrclinonc.2017.166. [DOI] [PubMed] [Google Scholar]
- 3.Batlle E, and Clevers H (2017). Cancer stem cells revisited. Nat. Med. 23, 1124–1134. 10.1038/nm.4409. [DOI] [PubMed] [Google Scholar]
- 4.Prasetyanti PR, and Medema JP (2017). Intra-tumor heterogeneity from a cancer stem cell perspective. Mol. Cancer 16, 41. 10.1186/s12943-017-0600-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, and Weinberg RA (2008). An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 40, 499–507. 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chew GL, Campbell AE, De Neef E, Sutliff NA, Shadle SC, Tapscott SJ, and Bradley RK (2019). DUX4 Suppresses MHC Class I to Promote Cancer Immune Evasion and Resistance to Checkpoint Blockade. Dev. Cell 50, 658–671.e7. 10.1016/j.devcel.2019.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hendrickson PG, Doráis JA, Grow EJ, Whiddon JL, Lim JW, Wike CL, Weaver BD, Pflueger C, Emery BR, Wilcox AL, et al. (2017). Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat. Genet. 49, 925–934. 10.1038/ng.3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whiddon JL, Langford AT, Wong CJ, Zhong JW, and Tapscott SJ (2017). Conservation and innovation in the DUX4-family gene network. Nat. Genet. 49, 935–940. 10.1038/ng.3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Iaco A, Planet E, Coluccio A, Verp S, Duc J, and Trono D (2017). DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat. Genet. 49, 941–945. 10.1038/ng.3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taubenschmid-Stowers J, Rostovskaya M, Santos F, Ljung S, Argelaguet R, Krueger F, Nichols J, and Reik W (2022). 8C-like cells capture the human zygotic genome activation program in vitro. Cell Stem Cell 29, 449–459.e6. 10.1016/j.stem.2022.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ren W, Gao L, Mou Y, Deng W, Hua J, and Yang F (2022). DUX: One Transcription Factor Controls 2-Cell-like Fate. Int. J. Mol. Sci. 23, 2067. 10.3390/ijms23042067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eckersley-Maslin MA, Svensson V, Krueger C, Stubbs TM, Giehr P, Krueger F, Miragaia RJ, Kyriakopoulos C, Berrens RV, Milagre I, et al. (2016). MERVL/Zscan4 Network Activation Results in Transient Genome-wide DNA Demethylation of mESCs. Cell Rep. 17, 179–192. 10.1016/j.celrep.2016.08.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akiyama T, Xin L, Oda M, Sharov AA, Amano M, Piao Y, Cadet JS, Dudekula DB, Qian Y, Wang W, et al. (2015). Transient bursts of Zscan4 expression are accompanied by the rapid derepression of heterochromatin in mouse embryonic stem cells. DNA Res. 22, 307–318. 10.1093/dnares/dsv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu X, Djekidel MN, and Zhang Y (2020). A transcriptional roadmap for 2C-like-to-pluripotent state transition. Sci. Adv. 6, eaay5181. 10.1126/sciadv.aay5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lemmers RJLF, van der Vliet PJ, Klooster R, Sacconi S, Camaño P, Dauwerse JG, Snider L, Straasheijm KR, van Ommen GJ, Padberg GW, et al. (2010). A unifying genetic model for facioscapulohumeral muscular dystrophy. Science 329, 1650–1653. 10.1126/science.1189044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Snider L, Geng LN, Lemmers RJLF, Kyba M, Ware CB, Nelson AM, Tawil R, Filippova GN, van der Maarel SM, Tapscott SJ, and Miller DG (2010). Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet. 6, e1001181. 10.1371/journal.pgen.1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tawil R, van der Maarel SM, and Tapscott SJ (2014). Facioscapulohumeral dystrophy: the path to consensus on pathophysiology. Skelet. Muscle 4, 12. 10.1186/2044-5040-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gabriëls J, Beckers MC, Ding H, De Vriese A, Plaisance S, van der Maarel SM, Padberg GW, Frants RR, Hewitt JE, Collen D, and Belayew A (1999). Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene 236, 25–32. 10.1016/s0378-1119(99)00267-x. [DOI] [PubMed] [Google Scholar]
- 19.Daxinger L, Tapscott SJ, and van der Maarel SM (2015). Genetic and epigenetic contributors to FSHD. Curr. Opin. Genet. Dev. 33, 56–61. 10.1016/j.gde.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Statland JM, and Tawil R (2016). Facioscapulohumeral Muscular Dystrophy. Continuum 22,1916–1931. 10.1212/CON.0000000000000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rickard AM, Petek LM, and Miller DG (2015). Endogenous DUX4 expression in FSHD myotubes is sufficient to cause cell death and disrupts RNA splicing and cell migration pathways. Hum. Mol. Genet. 24, 5901–5914. 10.1093/hmg/ddv315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV, Sonkin D, et al. (2012). The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607. 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geng LN, Yao Z, Snider L, Fong AP, Cech JN, Young JM, van der Maarel SM, Ruzzo WL, Gentleman RC, Tawil R, and Tapscott SJ (2012). DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev. Cell 22, 38–51. 10.1016/j.devcel.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi SH, Gearhart MD, Cui Z, Bosnakovski D, Kim M, Schennum N, and Kyba M (2016). DUX4 recruits p300/CBP through its C-terminus and induces global H3K27 acetylation changes. Nucleic Acids Res. 44, 5161–5173. 10.1093/nar/gkw141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yasuda T, Tsuzuki S, Kawazu M, Hayakawa F, Kojima S, Ueno T, Imoto N, Kohsaka S, Kunita A, Doi K, et al. (2016). Recurrent DUX4 fusions in B cell acute lymphoblastic leukemia of adolescents and young adults. Nat. Genet. 48, 569–574. 10.1038/ng.3535. [DOI] [PubMed] [Google Scholar]
- 26.Wolf FA, Angerer P, and Theis FJ (2018). SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15. 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bosnakovski D, Gearhart MD, Ho Choi S, and Kyba M (2021). Dux facilitates post-implantation development, but is not essential for zygotic genome activationdagger. Biol. Reprod. 104, 83–93. 10.1093/biolre/ioaa179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jagannathan S, Shadle SC, Resnick R, Snider L, Tawil RN, van der Maarel SM, Bradley RK, and Tapscott SJ (2016). Model systems of DUX4 expression recapitulate the transcriptional profile of FSHD cells. Hum. Mol. Genet. 25, 4419–4431. 10.1093/hmg/ddw271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghandi M, Huang FW, Jané-Valbuena J, Kryukov GV, Lo CC, McDonald ER 3rd, Barretina J, Gelfand ET, Bielski CM, Li H, et al. (2019). Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569, 503–508. 10.1038/s41586-019-1186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freed-Pastor WA, and Prives C (2012). Mutant p53: one name, many proteins. Genes Dev. 26, 1268–1286. 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willis A, Jung EJ, Wakefield T, and Chen X (2004). Mutant p53 exerts a dominant negative effect by preventing wild-type p53 from binding to the promoter of its target genes. Oncogene 23, 2330–2338. 10.1038/sj.onc.1207396. [DOI] [PubMed] [Google Scholar]
- 32.Boettcher S, Miller PG, Sharma R, McConkey M, Leventhal M, Krivtsov AV, Giacomelli AO, Wong W, Kim J, Chao S, et al. (2019). A dominant-negative effect drives selection of TP53 missense mutations in myeloid malignancies. Science 365, 599–604. 10.1126/science.aax3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Campbell AE, Shadle SC, Jagannathan S, Lim JW, Resnick R, Tawil R, van der Maarel SM, and Tapscott SJ (2018). NuRD and CAF-1-mediated silencing of the D4Z4 array is modulated by DUX4-induced MBD3L proteins. Elife 7, e31023. 10.7554/eLife.31023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krom YD, Dumonceaux J, Mamchaoui K, den Hamer B, Mariot V, Negroni E, Geng LN, Martin N, Tawil R, Tapscott SJ, et al. (2012). Generation of isogenic D4Z4 contracted and noncontracted immortal muscle cell clones from a mosaic patient: a cellular model for FSHD. Am. J. Pathol. 181, 1387–1401. 10.1016/j.ajpath.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lemmers RJLF, Goeman JJ, van der Vliet PJ, van Nieuwenhuizen MP, Balog J, Vos-Versteeg M, Camano P, Ramos Arroyo MA, Jerico I, Rogers MT, et al. (2015). Inter-individual differences in CpG methylation at D4Z4 correlate with clinical variability in FSHD1 and FSHD2. Hum. Mol. Genet. 24, 659–669. 10.1093/hmg/ddu486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grow EJ, Weaver BD, Smith CM, Guo J, Stein P, Shadle SC, Hendrickson PG, Johnson NE, Butterfield RJ, Menafra R, et al. (2021). p53 convergently activates Dux/DUX4 in embryonic stem cells and in facioscapulohumeral muscular dystrophy cell models. Nat. Genet. 53, 1207–1220. 10.1038/s41588-021-00893-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Overveld PGM, Lemmers RJFL, Sandkuijl LA, Enthoven L, Winokur ST, Bakels F, Padberg GW, van Ommen GJB, Frants RR, and van der Maarel SM (2003). Hypomethylation of D4Z4 in 4qlinked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat. Genet. 35, 315–317. 10.1038/ng1262. [DOI] [PubMed] [Google Scholar]
- 38.Young JM, Whiddon JL, Yao Z, Kasinathan B, Snider L, Geng LN, Balog J, Tawil R, van der Maarel SM, and Tapscott SJ (2013). DUX4 binding to retroelements creates promoters that are active in FSHD muscle and testis. PLoS Genet. 9, e1003947. 10.1371/journal.pgen.1003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Z, and Zhang Y (2019). Loss of DUX causes minor defects in zygotic genome activation and is compatible with mouse development. Nat. Genet. 51, 947–951. 10.1038/s41588-019-0418-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Iaco A, Verp S, Offner S, Grun D, and Trono D (2020). DUX Is a Non-essential Synchronizer of Zygotic Genome Activation. Development 147. 10.1242/dev.177725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshihara M, Kirjanov I, Nykänen S, Sokka J, Weltner J, Lundin K, Gawriyski L, Jouhilahti EM, Varjosalo M, Tervaniemi MH, et al. (2022). TransientDUX4 expression in human embryonic stem cells induces blastomere-like expression program that is marked by SLC34A2. Stem Cell Rep. 17, 1743–1756. 10.1016/j.stemcr.2022.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eckersley-Maslin M, Alda-Catalinas C, Blotenburg M, Kreibich E, Krueger C, and Reik W (2019). Dppa2 and Dppa4 directly regulate the Dux-driven zygotic transcriptional program. Genes Dev. 33, 194–208. 10.1101/gad.321174.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dion C, Roche S, Laberthonnière C, Broucqsault N, Mariot V, Xue S, Gurzau AD, Nowak A, Gordon CT, Gaillard MC, et al. (2019). SMCHD1 is involved in de novo methylation of the DUX4-encoding D4Z4 macrosatellite. Nucleic Acids Res. 47, 2822–2839. 10.1093/nar/gkz005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruebel ML, Vincent KA, Schall PZ, Wang K, and Latham KE (2019). SMCHD1 terminates the first embryonic genome activation event in mouse two-cell embryos and contributes to a transcriptionally repressive state. Am. J. Physiol. Cell Physiol. 317, C655–C664. 10.1152/ajpcell.00116.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang Z, Yu J, Cui W, Johnson BK, Kim K, and Pfeifer GP (2021). The chromosomal protein SMCHD1 regulates DNA methylation and the 2c-like state of embryonic stem cells by antagonizing TET proteins. Sci. Adv. 7, eabb9149. 10.1126/sciadv.abb9149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campbell AE, Belleville AE, Resnick R, Shadle SC, and Tapscott SJ (2018). Facioscapulohumeral dystrophy: activating an early embryonic transcriptional program in human skeletal muscle. Hum. Mol. Genet. 27, R153–R162. 10.1093/hmg/ddy162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tihaya MS, Mul K, Balog J, de Greef JC, Tapscott SJ, Tawil R, Statland JM, and van der Maarel SM (2023). Facioscapulohumeral muscular dystrophy: the road to targeted therapies. Nat. Rev. Neurol. 19, 91–108. 10.1038/s41582-022-00762-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oliva J, Galasinski S, Richey A, Campbell AE, Meyers MJ, Modi N, Zhong JW, Tawil R, Tapscott SJ, and Sverdrup FM (2019). Clinically Advanced p38 Inhibitors Suppress DUX4 Expression in Cellular and Animal Models of Facioscapulohumeral Muscular Dystrophy. J. Pharmacol. Exp. Ther. 370, 219–230. 10.1124/jpet.119.259663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cruz JM, Hupper N, Wilson LS, Concannon JB, Wang Y, Oberhauser B, Patora-Komisarska K, Zhang Y, Glass DJ, Trendelenburg AU, and Clarke BA (2018). Protein kinase A activation inhibits DUX4 gene expression in myotubes from patients with facioscapulohumeral muscular dystrophy. J. Biol. Chem. 293, 11837–11849. 10.1074/jbc.RA118.002633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hogan B, Fellous M, Avner P, and Jacob F (1977). Isolation of a human teratoma cell line which expresses F9 antigen. Nature 270, 515–518. 10.1038/270515a0. [DOI] [PubMed] [Google Scholar]
- 51.Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, Gill S, Harrington WF, Pantel S, Krill-Burger JM, et al. (2017). Defining a Cancer Dependency Map. Cell 170, 564–576.e16. 10.1016/j.cell.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Himeda CL, Jones TI, Virbasius CM, Zhu LJ, Green MR,and Jones PL (2018). Identification of Epigenetic Regulators of DUX4-fl for Targeted Therapy of Facioscapulohumeral Muscular Dystrophy. Mol. Ther. 26, 1797–1807. 10.1016/j.ymthe.2018.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hanahan D, and Weinberg RA (2011). Hallmarks of cancer: the next generation. Cell 144, 646–674. 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 54.Hanahan D, and Weinberg RA (2000). The hallmarks of cancer. Cell 100, 57–70. 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 55.Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. (2017). Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049. 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Levine JH, Simonds EF, Bendall SC, Davis KL, Amir E.a.D., Tadmor MD, Litvin O, Fienberg HG, Jager A, Zunder ER, et al. (2015). Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell 162, 184–197. 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al. (2021). Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29. 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.P H (2022). SeuratDisk: Interfaces for HDF5-Based Single Cell File Formats. https://mojaveazure.github.io/seurat-disk/.https://github.com/mojaveazure/seurat-disk.
- 59.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, and Ma’ayan A (2013). Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 14, 128. 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97. 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xie Z, Bailey A, Kuleshov MV, Clarke DJB, Evangelista JE, Jenkins SL, Lachmann A, Wojciechowicz ML, Kropiwnicki E, Jagodnik KM, et al. (2021). Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 1, e90. 10.1002/cpz1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu C, Li T, Xu Y, Zhang X, Li F, Bai J, Chen J, Jiang W, Yang K, Ou Q, et al. (2023). Cell Marker 2.0: an updated database of manually curated cell markers in human/mouse and web tools based on scRNA-seq data. Nucleic Acids Res. 51, D870–D876. 10.1093/nar/gkac947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wickham H (2016). Ggplot2: Elegant Graphics for Data Analysis. https://ggplot2.tidyverse.org.
- 64.Hesterberg T (2022). Resample: Resampling Functions. https://CRAN.R-project.org/package=resample.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single cell RNA-sequencing data were uploaded to NCBI Gene Expression Omnibus (GEO: GSE223848).
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request


