Abstract
Background:
Severe bronchiolitis (i.e., bronchiolitis requiring hospitalization) during infancy is a major risk factor for childhood asthma. However, the exact mechanism linking these common conditions remains unclear. We examined the longitudinal relationship between nasal airway miRNAs during severe bronchiolitis and the risk of developing asthma.
Methods:
In a 17-center prospective cohort study of infants with severe bronchiolitis, we sequenced their nasal miRNA at hospitalization. First, we identified differentially expressed miRNAs (DEmiRNAs) associated with the risk of developing asthma by age 6 years. Second, we characterized the DEmiRNAs based on their association with asthma-related clinical features, and expression level by tissue and cell types. Third, we conducted pathway and network analyses by integrating DEmiRNAs and their mRNA targets. Finally, we investigated the association of DEmiRNAs and nasal cytokines.
Results:
In 575 infants (median age=3 months), we identified 23 DEmiRNAs associated with asthma development (e.g., hsa-miR-29a-3p, FDR<0.10), particularly in infants with respiratory syncytial virus infection (FDRinteraction<0.05). These DEmiRNAs were associated with 16 asthma-related clinical features (FDR<0.05)—e.g., infant eczema and corticosteroid use during hospitalization. These DEmiRNAs were also highly expressed in lung tissue and immune cells (e.g., TH cells, neutrophils). Third, DEmiRNAs were negatively correlated with their mRNA targets (e.g., hsa-miR-324-3p/IL13), which were enriched in asthma-related pathways (FDR<0.05)—e.g., toll-like receptor, PI3K-Akt, and FcɛR signaling pathways, and validated by cytokine data.
Conclusion:
In a multicenter cohort of infants with severe bronchiolitis, we identified nasal miRNAs during illness that were associated with major asthma-related clinical features, immune response, and risk of asthma development.
Keywords: bronchiolitis, asthma, viral infection, microRNA, RNA sequencing, epigenetics, immunology
INTRODUCTION
Bronchiolitis is the leading cause of infant hospitalizations in the U.S., accounting for ~110,000 hospitalizations (i.e., severe bronchiolitis) annually [1, 2]. Its chronic morbidity burden is also substantial. Of these infants with severe bronchiolitis, approximately 30% develop asthma in childhood [3–8]. Yet, the underlying mechanisms linking these two common conditions remain unclear, and thereby hinder efforts to prevent asthma in this high-risk population.
MicroRNAs (miRNAs) are small non-coding RNAs consisting of 15 to 22 nucleotides. MiRNAs post-transcriptionally regulate gene expression by directly binding to their mRNA targets. Dysregulated expression of miRNAs leads to aberrant immune function [9, 10] and respiratory outcomes [11, 12], such as prevalent asthma [13–22], asthma severity [23, 24], asthma treatment (e.g., corticosteroids) response [25], and asthma remission [26]. Yet, these earlier reports [13, 15–25]—mostly based on a case-control design testing non-airway specimens from a small sample size of prevalent adult asthma—have precluded researchers from determining the role of airway miRNAs in incident asthma in childhood. Despite the clinical and research significance, no study has investigated miRNA signatures in infants, let alone high-risk infants (i.e., those with severe bronchiolitis), their post-transcriptional regulation of gene expression during the critical period of airway development, and their contribution to the development of asthma.
To address this knowledge gap in the literature, by applying a small RNA sequencing (RNA-seq) approach to a large prospective cohort of infants with severe bronchiolitis , we aimed to identify nasal airway miRNAs that are associated with the development of childhood asthma and to examine their potential mechanisms linking bronchiolitis and asthma.
METHODS
Study Design, Setting, and Participants
The study design and analytic workflow are summarized in Figure 1. We analyzed data from a multicenter prospective cohort study of infants hospitalized for bronchiolitis—the 35th Multicenter Airway Research Collaboration (MARC-35) study [27, 28]. Details of the study design, setting, participants, data collection, testing, and statistical analysis may be found in the Supplementary Methods. At 17 medical centers across 14 U.S. states (Table E1), MARC-35 enrolled infants (age <1 year) who were hospitalized with an attending physician diagnosis of bronchiolitis during three bronchiolitis seasons in 2011-2014. The diagnosis of bronchiolitis was made according to the American Academy of Pediatrics bronchiolitis guidelines, defined as an acute respiratory illness with a combination of rhinitis, cough, tachypnea, wheezing, crackles, or retraction [29]. We excluded infants with preexisting heart or lung disease, immunodeficiency, immunosuppression, or gestational age of <32 weeks. All infants were treated at the discretion of the treating physicians. Of 921 infants enrolled in the MARC-35 longitudinal cohort, the current study investigated 575 infants who have high-quality nasal small RNA-seq data (Figure E1). The institutional review board at each participating hospital approved the study with written informed consent obtained from the parent or guardian.
Figure 1. Study design and analytic workflow.
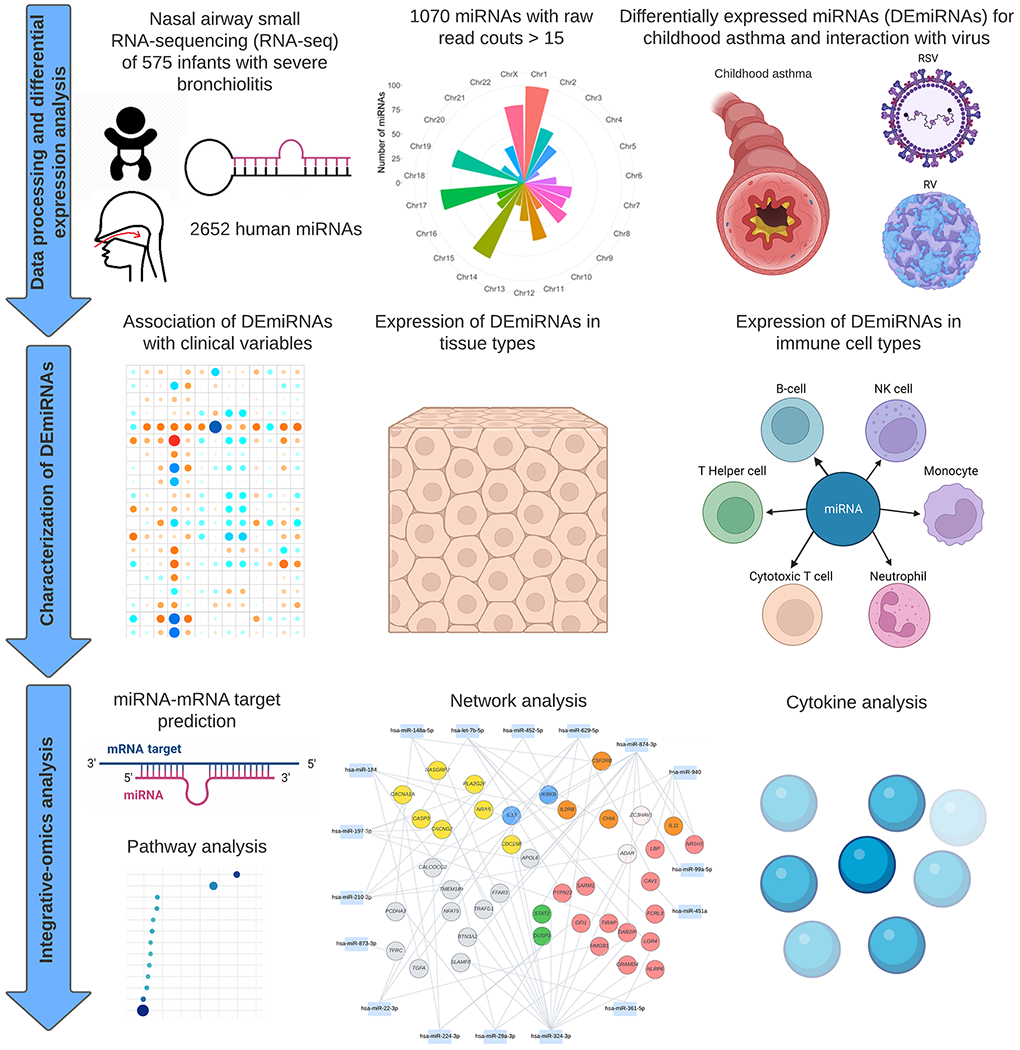
The analytical cohort consists of 575 infants hospitalized for bronchiolitis (severe bronchiolitis) in a multicenter prospective cohort study—the 35th Multicenter Airway Research Collaboration (MARC-35). Fastq files underwent quality control and collapse into unique reads. The trimmed reads were mapped against human miRNA sequences from miRBase V22. A total of 2652 human mature miRNAs were detected. Raw read counts of <15 were filtered out, leading to 1070 high-quality miRNAs for downstream analysis. In Aim 1, the association of 1070 miRNAs with the risk of developing asthma was examined. Effect modification by respiratory syncytial virus (RSV) and rhinovirus (RV) infection on the miRNA-asthma relationship was also investigated. A total of 23 differentially expressed miRNAs (DEmiRNAs) were identified. In Aim 2, the association of DEmiRNAs with asthma-related clinical variables was determined. The between-tissue and -immune cell types expression of the DEmiRNAs were also examined. In Aim 3, the mRNAs targeted by DEmiRNAs were identified using DIANA-microT-CDS and the association of biological pathways with the asthma risk was examined by performing the pathway analyses. The DEmiRNA-mRNA target network was constructed based on negatively correlated pairs.
Data Collection
Clinical data (study participants’ demographic characteristics, and family, environmental, and medical history, and details of the acute illness) were collected via structured interview and chart reviews using a standardized protocol (28, 29). After the index hospitalization for bronchiolitis, trained interviewers began interviewing parents/legal guardians by telephone at 6-month intervals in addition to medical record review by physicians. All data were reviewed at the Emergency Medicine Network Coordinating Center at Massachusetts General Hospital (Boston, MA, USA) [30]. Nasal swab specimens were collected within 24 hours of hospitalization using a standardized protocol [31, 32]. The details of the data collection and measurement methods are described in the Supplementary Methods.
Nasal Small RNA-seq Profiling (for miRNA)
The details of RNA extraction, RNA-seq, and quality control are described in Supplementary Methods. Briefly, after total RNA extraction, we performed small RNA-seq using the PerkinElmer NEXTFLEX® small RNA-seq v3 kit with Unique Dual Indexes (PerkinElmer, Waltham, MA) and sequenced on an Illumina NovaSeq6000 sequencer using an S2 50bp PE Flowcell (Illumina, San Diego, CA). We estimated miRNA detection and abundance using sMETASeq [33]. Fastq files underwent quality control in cutadapt [34] and collapse into unique reads. We mapped trimmed reads against human miRNA sequences from miRBase V22 [35]. We filtered out raw read counts of <15 [25, 36]. Lastly, we normalized the read count by R DESeq2 package [37] using default settings.
Nasopharyngeal RNA-seq Profiling (for mRNA)
The details of RNA-seq, quality control, and transcriptome profiling are described in our previous studies [38–40] and Supplementary Methods. Briefly, after total RNA extraction, DNase treatment, and rRNA reduction, we performed RNA-seq with Illumina NovaSeq6000 (Illumina, San Diego, CA). All RNA-seq samples had high sequence coverage after quality control. The transcript abundances were estimated with Salmon [41] using the human genome (hg38) and the mapping-based mode. A total of 194 infants had both nasal miRNA and nasopharyngeal mRNA data.
Nasal Cytokine Measurement
The details of cytokine measurement and quality control are described in Supplementary Methods. We measured the levels of 10 cytokines (interferon [IFN]-γ, interleukin [IL]-1β, IL-2, IL-4, IL-6, IL-8, IL-10, IL-12p70, IL-13, tumor necrosis factor [TNF]-α) in the nasal swab specimens of infants with severe bronchiolitis, using multiplex the Meso Scale Discovery (MSD) electrochemiluminescent V-Plex multiplex immunoassay (Meso Scale Diagnostics, Rockville, MD), on the MESO QuickPlex SQ 120 system (Meso Scale Diagnostics). The samples with cytokine level less than lower limit of detection were removed. A maximum of 503 infants had both nasal miRNA and nasal cytokine data.
Outcome
The outcome of interest was the development of asthma by age 6 years. The definition of asthma was based on a commonly-used epidemiologic definition of asthma [30, 42]—physician diagnosis of asthma by age 6 years, plus either asthma medication use (e.g., albuterol inhaler, inhaled corticosteroids, montelukast) or asthma-related symptoms in the preceding year.
Statistical Analysis
The analytic workflow is summarized in Figure 1. First, to investigate the relationship of the miRNAs with the risk of developing asthma, we performed miRNA differential expression analysis using the negative binomial generalized linear model from DESeq2 R package [37]. Based on a priori-defined hypothesis, we also examined the effect modification by the respiratory syncytial virus (RSV), rhinovirus (RV), RV-A, and RV-C infection on the risk of developing asthma. In the differential expression analysis, we adjusted for potential confounders (i.e., age, sex, number of previous breathing problems, and IgE sensitization) based on a priori knowledge and clinical plausibility [30, 39, 43, 44]. We corrected multiple testing using the Benjamini-Hochberg false discovery rate (FDR) method [45]. We defined differentially expressed miRNA (DEmiRNA) as those miRNA significantly associated with asthma development at an FDR<0.10.
Second, we examined the relationship of DEmiRNAs with asthma-related clinical variables [30, 39, 43, 44], including RSV, RSV-A, RSV-B, RV, RV-A, RV-B, RV-C, parental history of asthma and eczema, number of previous breathing problems, infant history of corticosteroids use, eczema, IgE sensitization, blood eosinophil count, need for positive pressure ventilation (PPV), and intensive care use (PPV use and/or intensive care unit adimission). To provide biological insights into the identified nasal DEmiRNAs, we investigated DEmiRNAs across 15 asthma-related tissue types (e.g., adipocyte, lung, lymph nodes, muscle, pancreas, skin) [46] using the publicly available human miRNA TissueAtlas data [47]. We also examined DEmiRNAs across six immune cell types [B cell, cytotoxic T (TC) cell, helper T (TH) cell, monocyte, natural killer (NK) cell, and neutrophil cell] using publicly available single-cell small RNA-seq data from 162 healthy subjects [48]. We normalized the miRNA data using the variance stabilizing transformation approach implemented in DESeq2 [37].
Third, we identified the mRNAs targeted by DEmiRNAs using DIANA-microT-CDS [49] and conducted a gene-set enrichment analysis to investigate the enrichment of these targeted mRNA in the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways using DIANA miRPath v.3.0 [50]. We also performed the correlation analysis based on upper airway miRNA and mRNA data (Supplementary Methods) given that the main biological function of miRNAs is to degrade the target mRNAs. We constructed network based on negatively correlated (Spearman correlation coefficient<−0.10) miRNA-mRNA pairs and prior knowledge of identified immune-related pathways [13, 20, 24, 51–54]. Finally, we investigated the association of DEmiRNA with ten cytokines using the negative binomial generalized linear model from DESeq2 R package [37].
RESULTS
Of the 921 infants with severe bronchiolitis enrolled into the MARC-35 longitudinal cohort, the current study focused on 575 infants who had high quality nasal miRNA data. The analytic and non-analytic cohorts did not differ in patient characteristics (P≥0.05; Table E2), except for RSV and RV-B infection. Among the analytic cohort, the median age was 3 (IQR, 2-6) months, 41% were female, 47% were non-Hispanic White, 28% were Hispanic, and 22% were non-Hispanic Black. Subsequently, 27% developed asthma by age 6 years (Tables 1 and E3). In addition, 76% of infants with RSV infection (vs. 87% with no RSV infection) and 28% with RV infection (vs. 16% with no RV infection) developed asthma. For miRNA profiling, a total of 2,652 human mature miRNAs were identified. Among these, 1,070 miRNAs had raw read counts of more than 15 reads and were included in the subsequent analysis (Figure 2A).
Table 1.
Baseline characteristics and clinical course of 575 infants hospitalized for bronchiolitis
| Characteristics | Overall (n=575) |
|---|---|
| Demographics | |
| Age (month), median (IQR) | 3 (2–6) |
| Female sex | 234 (41) |
| Race/ethnicity | |
| Non-Hispanic White | 268 (47) |
| Non-Hispanic Black | 126 (22) |
| Hispanic | 162 (28) |
| Other or unknown | 19 (3) |
| Prematurity (32–36.9 weeks) | 96 (17) |
| Birth weight (kg), median (IQR) | 3.29 (2.90–3.60) |
| Mode of birth (cesarean delivery) | 194 (34) |
| Previous breathing problems before the index hospitalization* | |
| 0 | 469 (82) |
| 1 | 87 (15) |
| 2 | 19 (3) |
| Previous ICU admission | 7 (1) |
| History of eczema | 85 (15) |
| Lifetime antibiotic use | 170 (30) |
| Ever attended daycare | 143 (25) |
| Cigarette smoke exposure at home |
87 (15) |
| Maternal smoking during pregnancy | 87 (15) |
| Parental history of asthma | 186 (32) |
| Parental history of eczema | 110 (19) |
| Clinical presentation | |
| Weight at presentation (kg), median (IQR) | 6.07 (4.70–7.60) |
| Respiratory rate at presentation (per minute), median (IQR) | 48 (40–60) |
| Oxygen saturation at presentation | |
| <90% | 49 (9) |
| 90–93% | 87 (16) |
| ≥94% | 427 (76) |
| Blood eosinophilia (≥4%) | 52 (10) |
| IgE sensitization (%) | 125 (22) |
| Length of hospitalization (day), median (IQR) | 2 (1–3) |
| Corticosteroid use during hospitalization | 71 (12) |
| Respiratory virus | |
| RSV infection | 485 (84) |
| RSV-A | 351 (61) |
| RSV-B | 137 (24) |
| RV infection | 110 (19) |
| RV-A | 50 (9) |
| RV-B | 5 (1) |
| RV-C | 53 (9) |
| Chronic clinical outcome | |
| Childhood asthma† | 156 (27) |
Abbreviations: ICU, intensive care unit; IgE, immunoglobulin E; IQR, interquartile range; RSV, respiratory syncytial virus; RV, rhinovirus.
Data are no. (%) of infants unless otherwise indicated. Percentages may not equal 100, because of rounding and missingness.
Defined as an infant having a cough that wakes him or her at night or causes emesis, or when the child has wheezing or shortness of breath without cough.
Asthma was defined as physician-diagnosis of asthma by age 6 years, plus either asthma medication use (e.g., albuterol inhaler, inhaled corticosteroids, montelukast) or asthma-related symptoms in the preceding year.
Figure 2. Association of nasal airway miRNAs in infants with bronchiolitis with risk of developing asthma .

A. The plot included 1070 miRNAs after quality control and showed the genome-wide distribution of the number of miRNAs in each chromosome. B. The plot shows the association of the 23 DEmiRNAs in the nasal airways with asthma risk. The between-group differences in the expression level were tested by DESeq2 with the negative binomial generalized linear model adjusted for potential confounders, including age, sex, number of previous breathing problems, and IgE sensitization. C. The plot shows the RSV infection-stratified analysis for the association of the 23 DEmiRNAs with the risk of developing asthma. The DESeq2 models adjusted for potential confounders, including age, sex, number of previous breathing problems, and IgE sensitization. D. The plot shows the RV infection-stratified analysis for the association of the 23 DEmiRNAs with the risk of developing asthma. The DESeq2 models adjusted for potential confounders, including age, sex, number of previous breathing problems, and IgE sensitization. Abbreviations: FDR, false discovery rate; RSV, respiratory syncytial virus; RV, rhinovirus.
Nasal miRNAs of Infant Bronchiolitis were Associated with Risk of Developing Asthma
A total of 23 DEmiRNAs were significantly associated with asthma risk (FDR<0.10), with 18 DEmiRNAs being upregulated and 5 DEmiRNAs being downregulated (Figure 2B). Of these, hsa-miR-29a-3p was the most significantly associated with the asthma risk (log2 fold change [FC]=1.44, FDR=5.47×10−15). In the examination of effect modification by virus, 22 of these DEmiRNA had a significant interaction with RSV infection on asthma risk (FDRinteraction<0.05). In the stratified analysis within infants with RSV infection, 15 miRNAs were associated with a significantly higher risk of developing asthma (FDR<0.05; Figure 2C). Additionally, hsa-miR-22-3p had a significant interaction with RV infection on asthma risk (FDRinteraction<0.05). Among infants with RV infection, hsa-miR-22-3p was associated with a significantly higher risk of developing asthma (log2FC=1.98, FDR=5.94×10−6; Figure 2D). However, we did not observe a significant association between miRNAs and asthma development in RV-A or RV-C strata (Figure E2).
DEmiRNAs were Associated with Asthma-related Clinical Features, Tissue Types, and Cell Types
The DEmiRNAs were also associated with major asthma-related clinical features. For example, at the aggregated miRNA level, the DEmiRNAs and non-DEmiRNAs had a difference in the magnitude of association with RSV, RV-A, and RV-B (only 5 children) infection, infant history of eczema and corticosteroids use, serum total IgE level, PPV use, and intensive care use (Figure 3A). At the individual miRNA level, 18 DEmiRNAs were significantly associated with these clinical features (FDR<0.05). For example, hsa-miR-29a-3p was positively associated with infant history of eczema and corticosteroid use, and hsa-let-7b-5p was negatively associated with serum total IgE level (Figure 3B). In the miRNA tissue expression analysis, the DEmiRNAs had differential expression in lung, spleen, and bone marrow (all FDR<0.001) (Figure 4A). In the examination of the miRNA expression in immune cell types, the expression levels of DEmiRNAs were significantly higher in all immune cells (e.g., TH cells, neutrophils) than the non-DEmiRNAs (all FDR<0.001) (Figure 4B). Most of the DEmiRNAs were presented commonly in these immune cells (Figure 4C).
Figure 3. Association of nasal airway DEmiRNAs in infants with bronchiolitis with asthma-related clinical variables.
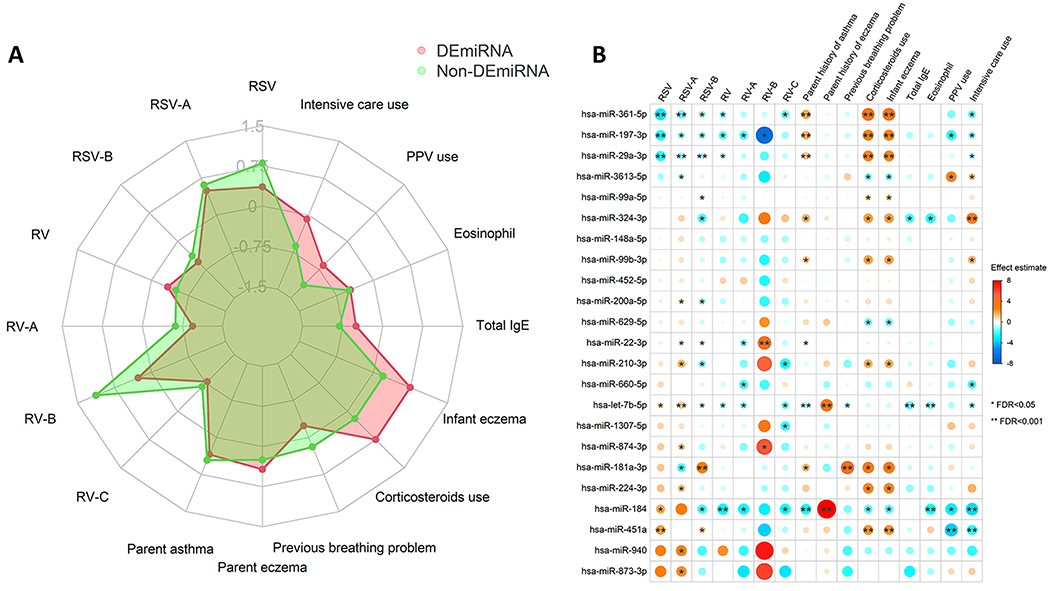
A. Association between the DEmiRNAs or non-DEmiRNAs and that of 16 asthma-related clinical variables in infants with severe bronchiolitis. The red web denotes the average effect estimates of the aggregated DEmiRNAs for the 16 clinical variables. The green web denotes the average effect estimates of the aggregated non-DEmiRNAs for the 16 clinical variables. B. The relationship of the 23 DEmiRNAs with the 16 asthma-related clinical variables was examined by the DESeq2 negative binomial generalized linear model adjusting for age and sex. The blue-to-red gradient in the heatmap denotes the magnitude and direction of the associations. The size of the dot denotes the magnitude of the associations. Abbreviations: FDR, false discovery rate; IgE, immunoglobulin E; PPV, positive pressure ventilation; RSV, respiratory syncytial virus; RSV-A, respiratory syncytial virus A; RSV-B, respiratory syncytial virus B; RV, rhinovirus; RV-A, rhinovirus A; RV-B, rhinovirus B; RV-C, rhinovirus C.
Figure 4. Expression of nasal airway DEmiRNAs in infants with bronchiolitis with tissue and immune cell types.
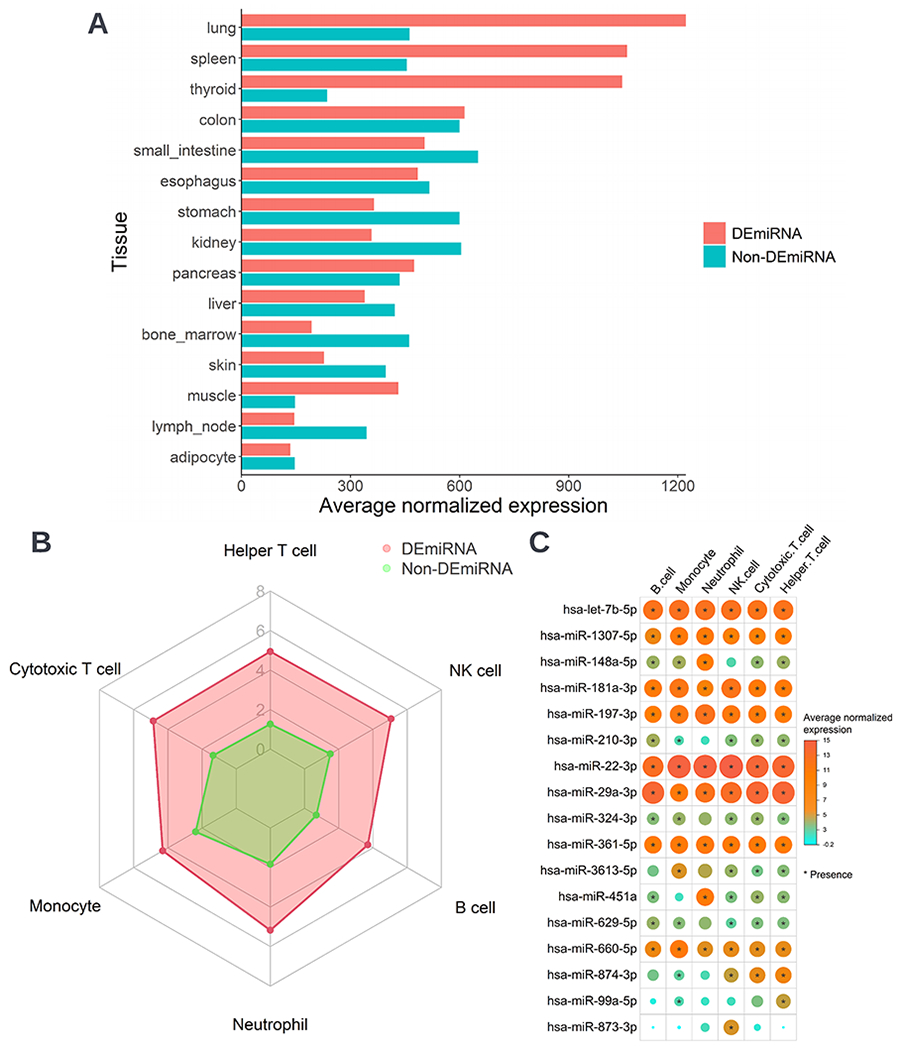
A. Average normalized expression level of the aggregated DEmiRNAs or non-DEmiRNAs in 15 asthma-related tissues. The between-group differences in the expression level were tested by the Wilcoxon rank-sum test. The between-group differences for all tissues were significant (FDR<0.001). B. Average normalized expression level of the aggregated DEmiRNAs or non-DEmiRNAs in six blood immune cell types, including B cell, cytotoxic T cell, helper T cell, monocyte, natural killer cell, and neutrophil cell. The between-group differences in the expression level were tested by the Wilcoxon rank-sum test. The between-group differences for all cell types were significant (FDR<0.001). C. Normalized expression level of the 17 DEmiRNAs in six blood immune cell types. A asterisk denotes the presence of the miRNAs in the cell types. Only the miRNAs which were detected (expression value > 5) in at least 85% of the samples in at least one of the blood cell type were considered as being present. Abbreviation: NK, natural killer.
Identification of Biological Pathways and Network for Risk of Asthma Development
In the gene-set enrichment analysis by using the GO biological process gene set, 108 pathways were differentially enriched (FDR<0.05; Table E4), including asthma-related pathways—e.g., Fc-epsilon receptor (FcɛR), toll-like receptor (TLR), and mitogen-activated protein kinase (MAPK) signaling pathways (Figure 5A). Additionally, in the analysis by using the KEGG gene set, 41 pathways were differentially enriched (FDR<0.05; Table E5), including phosphatidylinositol 3-kinase (PI3K)-protein kinase B (Akt) and MAPK signaling pathways (Figure 5B). The network analysis showed that negatively correlated miRNA-mRNA pairs mapped to the identified immune-related pathways, such as hsa-miR-324-3p/IL13 for FcɛR signaling, hsa-miR-29a-3p/STAT2 for PI3K-Akt signaling, and hsa-let-7b-5p/PTPN22 for TLR signaling (Figure 5C). Finally, the DEmiRNAs were significantly associated with cytokines, such as hsa-miR-29a-3p/IFN-γ (log2FC=−1.05, FDR<0.05), and hsa-miR-324-3p/IL-13 (log2FC=−1.68, FDR<0.05) (Figure 5D).
Figure 5. Nasal airway DEmiRNAs pathways and risk of developing asthma.
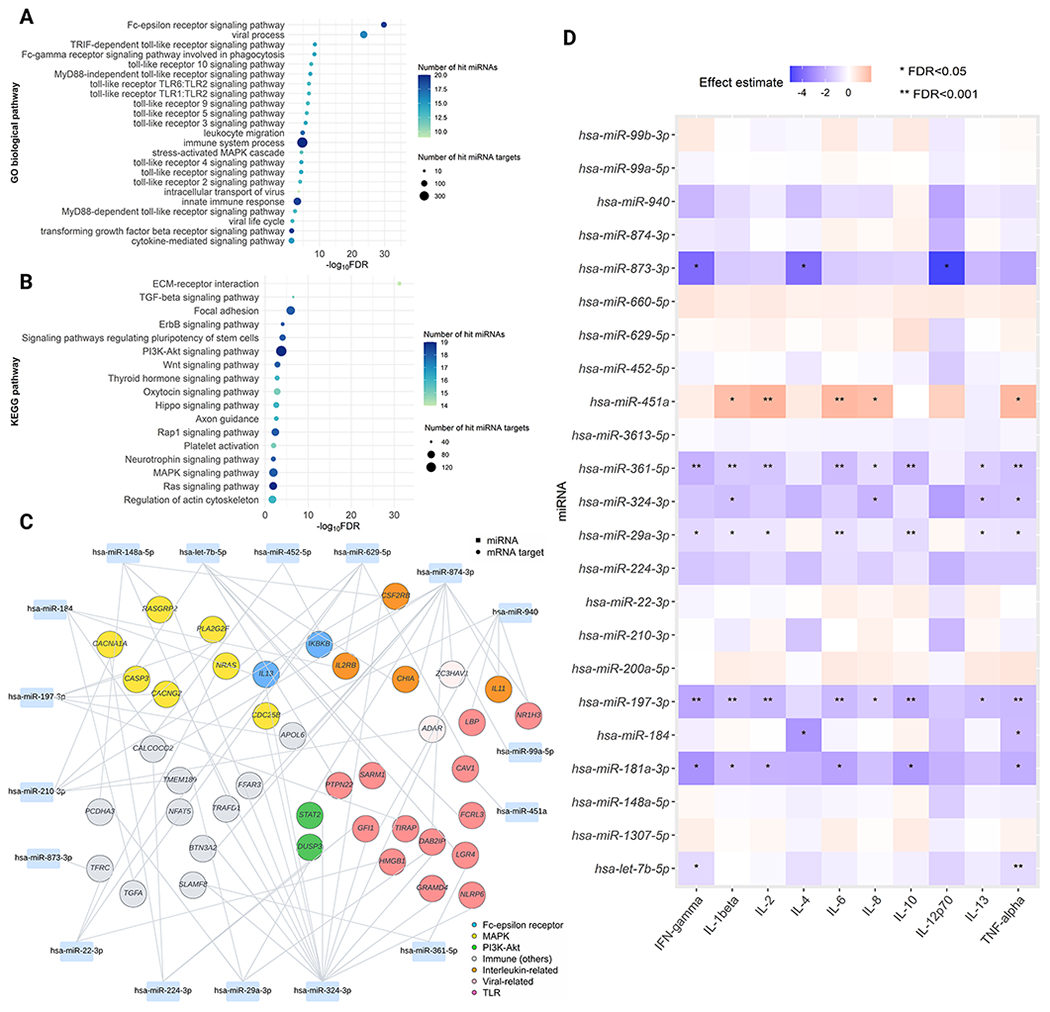
A. Functional enrichment analysis using GO biological process gene sets We identified 108 differentially-enriched pathways (FDR<0.05) associated with the asthma risk. Of these, we selected immune-related pathways to visualize the plot. B. Functional enrichment analysis using KEGG gene sets We identified 41 differentially-enriched pathways (FDR<0.05) associated with the asthma risk. Of these, we selected non cancer-related pathways to visualize the plot. C. DEmiRNA-mRNA target network A rectangle denotes miRNA, a circle denotes mRNA target. mRNA targets that are mapped within major immune-related pathways are highlighted in various colors. In this network, we only included mRNA targets that are negatively correlated (Spearman correlation coefficient<−0.1) with DEmiRNAs and have immune-related functions based on prior knowledge. D. Association of the 23 DEmiRNAs with the ten nasal cytokines. The blue-to-red gradient in the heatmap denotes the magnitude and direction of the associations. Abbreviations: FDR, false discovery rate; IFN, Interferon; IL, interleukin; MAPK, Mitogen-activated protein kinase; PI3K-Akt, phosphatidylinositol 3‑kinase-protein kinase B; TLR, Toll-like receptor; TNF, Tumor necrosis factor.
DISCUSSION
By applying a small RNA-seq technique to a multicenter prospective cohort of infants with severe bronchiolitis, we identified nasal miRNAs, their potential contextual functions, and their longitudinal relationship with incident asthma. More specifically, we found 23 miRNAs significantly associated with the risk of developing asthma—e.g., hsa-miR-22-3p with a higher risk of asthma in infants with RSV or RV infection. Furthermore, we also observed that these DEmiRNAs were associated with asthma-related clinical features, such as infant history of eczema and serum total IgE level. Moreover, by using the integrated miRNA and mRNA data, we found that infants who subsequently developed asthma had differentially-enriched pathways—e.g., TLR, PI3K-Akt and FcɛR signaling pathways. To the best of our knowledge, this is the first study that has demonstrated the potential role for nasal miRNAs in infants with severe bronchiolitis in the pathobiology of developing asthma.
Results in Context
Concordant with our findings, prior studies have suggested that miRNAs are implicated in asthma pathobiology [13, 15–26]. For example, studies have reported that several miRNAs (e.g., hsa-miR-125b) are associated with adult prevalent asthma and its inflammatory features (e.g., the quantity of eosinophils and neutrophils in blood) [13, 17, 19, 24]. Similarly, the literature has also shown that asthma-related miRNAs are involved in immune-related pathways, such as PI3K-Akt [13], MAPK [15, 52, 53], TLRs [20, 24], and TH17 [24]. For example, a recent single-center study of 62 adults with asthma reported that—by applying a microarray approach to sputum specimens—a miRNA network was associated with increased neutrophilic airway inflammation in prevalent asthma, and the miRNAs are enriched for TLR signaling pathway [24]. Additionally, in a single-center case-control study of 35 adults with severe asthma, Rupani et al. have reported that—by applying a PCR approach to bronchoalveolar lavage samples—upregulation of three miRNAs reduces TLR7 expression, which drives impaired innate immune responses to RV [20]. While these studies have collectively suggested the role of the miRNAs in prevalent asthma, to date, no study has evaluated the nasal miRNA in infants—let alone in those with severe bronchiolitis—and their contribution to asthma risk. The current study—with a small RNA-seq approach applied to a large multicenter cohort—builds on previous reports and extends them by identifying the pathobiological role of nasal miRNAs in the development of asthma.
Potential Mechanisms
There are several potential mechanisms linking bronchiolitis—by miRNA post-transcriptional regulation—to the subsequent development of asthma. First, the literature has indicated that miRNAs, as mediators between respiratory virus infections and asthma, modulate airway inflammatory processes [11, 55]. Infants with RV bronchiolitis are more likely to develop asthma as compared to infants with RSV bronchiolitis [56]. Growing evidence has shown that both RSV and RV infections are important triggers for perturbations in miRNA expression, which are actively involved in innate immune response, such as TLRs and NFκB signaling pathways [57–59]. The host innate immune response is the first line of defense against all pathogens (e.g., virus). Thornburg et al. have shown that RSV manipulates host cell gene expression through the regulation of miRNA (e.g., hsa-let-7 families) expression related to the TLRs, NF-κB, or interferon (IFN) signaling pathways in human bronchial epithelial cells and monocyte-derived dendritic cells [60]. In addition, in RSV-induced airways inflammation, a population of IFN-γ secreting TC cells potentially attenuates pathogenic TH2 host response to the RSV G-protein [59]. Of note, our cytokine analysis idenfied that hsa-miR-29a-3p is negatively associated with IFN-γ. This result is consistent with a recent study showing that miRNA-29a-3p upregulation due to pulmonary microbial infection suppresses the immune response by inhibiting IFN-γ expression in T cells, and associated with a higher risk of active and latent pulmonary tuberculosis [61]. Furthermore, studies have reported that miR-22-3p is involved in regulation of asthma-related immune mechanisms (e.g., IFN-γ, NACHT, LRR, and PYD domains-containing protein 3) in both human [15] and mouse [62].
Second, prior research has also showed that miRNAs are involved in the PI3K-Akt signaling pathway, which plays a role in cell proliferation and airway remodelling [63]. For example, Alexandrova et al., by profiling the miRNA expression in bronchial smooth muscle cells from 8 adults with asthma, found that these patients had specific miRNA signatures (e.g., hsa-miR-29a-3p) and that the targeted transcripts were involved in the PI3K-Akt signaling pathway, which plays a role in the airway smooth muscle (ASM) cell growth and proliferation [13]. Consistently, the current study has identified hsa-miR-29a-3p as the top DEmiRNA and had high expression in multiple immune cell types (e.g., TH cells). A recent study has also found a crosstalk between TH cells and ASM in pediatric obesity-related asthma [64]. Furthermore, an in vivo study using an ovalbumin (OVA)-induced mouse model of allergic asthma has found that mmu-miR-221 modulated airway remodelling via PI3K-Akt signaling pathway [65]. Indeed, PI3K is required for growth factor-induced cell migration [66], and activation of PI3K can stimulate DNA synthesis and growth, which all promote airway remodelling [67].
Third, while the research has suggested the roles of miRNAs in airway inflammation and remodelling, the role of miRNAs in IgE-mediated asthma remains unclear. IgE-mediated asthma is characterized by the presence of allergen-specific IgE antibodies, which bind to high-affinity FcɛR [68]. Multiple studies have reported that RSV and RV infection can activate the FcɛR signaling pathway [69, 70], which can trigger allergic airway inflammation [71]. In the current study, among immune-related pathways, we identified FcɛR signaling pathway most significantly associated with the risk of asthma development.
Notwithstanding the complexity of these mechanisms, we believe that the identification of the longitudinal relationship between nasal miRNAs in infancy and childhood asthma is important. Of note, evidence has suggested the role of miRNAs in disease prevention, such as cancer [72]. Thus, our findings, in conjunction with the existent literature, should advance research into the development of miRNA-specific strategies for asthma prevention.
Limitations
Our study has several potential limitations. First, bronchiolitis involves inflammation of the lower airways, in addition to the upper airways. Although the current study used the miRNA data from nasal specimens, research has shown that upper airway specimens offer a reliable representation of inflammatory profiles in the lower airways [73]. Additionally, the use of upper airway specimens is practical because lower airway sampling (e.g., bronchoscopy) would be invasive in young infants. Second, the nasal specimens were obtained at a single time point. While longitudinal molecular data would also be informative, the study objective was to investigate the role of miRNA at the time of bronchiolitis in asthma development. Third, nasal specimens during respiratory infection might have been contaminated with blood immune cells. Accordingly, the miRNA profiles may have partially reflected those of these cells. Accordingly, it is likely that the miRNAs partially reflect the infiltration of blood immune cells. Fourth, our study does not have data on specific RSV genotype variants, while research has suggested different RSV genotype variants may be related with altered functions and/or immunogenicity, potentially leading to an impact on disease severity [74]. Fifth, it is possible that asthma diagnosis (by age 6 years) may have been misclassified and that some children are going to develop asthma at a later age. To address these points, the cohort is currently being followed up to age 9 years. Sixth, the current study does not have mechanistic experiments to validate the identified miRNA functions. Yet, our independent nasal cytokine data has partially validated the miRNA functions, which are consistent with the literature. This study derives well-calibrated hypotheses that facilitate future experiments. Seventh, our data are limited to investigating the effect modification of respiratory infection at infancy on miRNA and asthma development. It is possible that prenatal risk factors (e.g., vertical transmission) [75] may have affected the relationship between miRNA and asthma development. Eighth, we have used miRNA data of publicly available tissue and blood immune cells data from subjects without asthma to investigate the expression of nasal miRNAs from the current study. Although these results elucidate the role of miRNAs in tissue and cell-specific manner, the interpretation of these results requires careful interpretation. Lastly, despite the study sample consisting of racially/ethnically- and geographically-diverse infants, our inferences must be cautiously generalized beyond infants with severe bronchiolitis. Nonetheless, our data remain directly relevant for the 110,000 infants hospitalized yearly in the U.S [2].
Conclusions
In conclusion, by applying small RNA-seq approach to a multicenter cohort of infants with severe bronchiolitis, we found a complex interplay between nasal miRNA, virus, asthma risk factors, and their longitudinal relationship with asthma development. For example, RSV or RV infection modifies the effect of miRNAs on asthma development. Additionally, the data suggest that these miRNAs play key roles in mechanisms relating to asthma, such as innate immunity, airway remodelling, and IgE regulation. For clinicians, our findings provide an evidence base for the early identification of high-risk infants during a critical period of airway development—early infancy. For researchers, these observations should facilitate 1) further understanding of the interplay between virus and host, and their contribution to asthma; 2) further investigations into the development of miRNA-targeted strategies for asthma prevention [11] in infants with severe bronchiolitis—a population with substantial morbidity burden.
Supplementary Material
Take home message:
In infants with severe bronchiolitis, we found a complex interplay of nasal miRNA with virus, asthma risk factors, and their longitudinal relationship with asthma development, facilitating development of miRNA-targeted strategies for asthma prevention.
Acknowledgment
This study was supported by grants from the National Institutes of Health (Bethesda, MD): K01 AI-153558, U01 AI-087881, R01 AI-114552, R01 AI-127507, R01 AI-134940, R01 AI-137091, R01 AI-148338, and UG3/UH3 OD-023253; Massachusetts General Hospital Department of Emergency Medicine Fellowship/Eleanor and Miles Shore Faculty Development Awards Program (Boston, MA); and the Harvard University William F. Milton Fund (Boston, MA). The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We would like to thank the participants and researchers from the Multicenter Airway Research Collaboration (MARC) who significantly contributed or collected data. We also thank Dr. Ignacio Ramos-Tapia for his assistance with statistical analysis.
Funding:
This study was supported by grants from the National Institutes of Health (Bethesda, MD): K01 AI-153558, U01 AI-087881, R01 AI-114552, R01 AI-127507, R01 AI-134940, R01 AI-137091, R01 AI-148338, and UG3/UH3 OD-023253; Massachusetts General Hospital Department of Emergency Medicine Fellowship/Eleanor and Miles Shore Faculty Development Awards Program; and the Harvard University William F. Milton Fund. The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The funding organizations were not involved in the collection, management, or analysis of the data; preparation or approval of the manuscript; or decision to submit the manuscript for publication.
Conflict of interests statement:
Dr. Zhu reports grants from National Institutes of Health during the conduct of the study. Dr. Hahn reports personal fees from Johnson and Johnson, outside the submitted work. Dr. Teach reports grants from National Institutes of Health during the conduct of the study. Dr. Hasegawa reports grants from National Institutes of Health during the conduct of the study; grants from Novartis, outside the submitted work. Dr. Camargo reports grants from National Institutes of Health during the conduct of the study. All other authors have indicated that they have no financial relationships relevant to this article to disclose.
REFERENCES
- 1.Li Y, Wang X, Blau DM, Caballero MT, Feikin DR, Gill CJ, Madhi SA, Omer SB, Simoes EAF, Campbell H, Pariente AB, Bardach D, Bassat Q, Casalegno JS, Chakhunashvili G, Crawford N, Danilenko D, Do LAH, Echavarria M, Gentile A, Gordon A, Heikkinen T, Huang QS, Jullien S, Krishnan A, Lopez EL, Markic J, Mira-Iglesias A, Moore HC, Moyes J, Mwananyanda L, Nokes DJ, Noordeen F, Obodai E, Palani N, Romero C, Salimi V, Satav A, Seo E, Shchomak Z, Singleton R, Stolyarov K, Stoszek SK, von Gottberg A, Wurzel D, Yoshida LM, Yung CF, Zar HJ, Respiratory Virus Global Epidemiology N, Nair H, investigators R. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in children younger than 5 years in 2019: a systematic analysis. Lancet 2022: 399(10340): 2047–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fujiogi M, Goto T, Yasunaga H, Fujishiro J, Mansbach JM, Camargo CA Jr., Hasegawa K Trends in Bronchiolitis Hospitalizations in the United States: 2000-2016. Pediatrics 2019: 144(6): e20192614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carroll KN, Wu P, Gebretsadik T, Griffin MR, Dupont WD, Mitchel EF, Hartert TV. The severity-dependent relationship of infant bronchiolitis on the risk and morbidity of early childhood asthma. J Allergy Clin Immunol 2009: 123(5): 1055–1061.e1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sigurs N, Aljassim F, Kjellman B, Robinson PD, Sigurbergsson F, Bjarnason R, Gustafsson PM. Asthma and allergy patterns over 18 years after severe RSV bronchiolitis in the first year of life. Thorax 2010: 65(12): 1045–1052. [DOI] [PubMed] [Google Scholar]
- 5.Bacharier LB, Cohen R, Schweiger T, Yin-Declue H, Christie C, Zheng J, Schechtman KB, Strunk RC, Castro M. Determinants of asthma after severe respiratory syncytial virus bronchiolitis. J Allergy Clin Immunol 2012: 130(1): 91–100 e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Regnier SA, Huels J. Association between respiratory syncytial virus hospitalizations in infants and respiratory sequelae: systematic review and meta-analysis. Pediatr Infect Dis J 2013: 32(8): 820–826. [DOI] [PubMed] [Google Scholar]
- 7.Hasegawa K, Dumas O, Hartert TV, Camargo CA Jr. Advancing our understanding of infant bronchiolitis through phenotyping and endotyping: clinical and molecular approaches. Expert Rev Respir Med 2016: 10(8): 891–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dumas O, Erkkola R, Bergroth E, Hasegawa K, Mansbach JM, Piedra PA, Jartti T, Camargo CA Jr. Severe bronchiolitis profiles and risk of asthma development in Finnish children. J Allergy Clin Immunol 2022: 149(4): 1281–1285 e1281. [DOI] [PubMed] [Google Scholar]
- 9.Foster PS, Plank M, Collison A, Tay HL, Kaiko GE, Li J, Johnston SL, Hansbro PM, Kumar RK, Yang M, Mattes J. The emerging role of microRNAs in regulating immune and inflammatory responses in the lung. 2013: 253(1): 198–215. [DOI] [PubMed] [Google Scholar]
- 10.Mehta A, Baltimore D. MicroRNAs as regulatory elements in immune system logic. Nat Rev Immunol 2016: 16(5): 279–294. [DOI] [PubMed] [Google Scholar]
- 11.Taka S, Tzani-Tzanopoulou P, Wanstall H, Papadopoulos NG. MicroRNAs in Asthma and Respiratory Infections: Identifying Common Pathways. Allergy Asthma Immunol Res 2020: 12(1): 4–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weidner J, Bartel S, Kilic A, Zissler UM, Renz H, Schwarze J, Schmidt-Weber CB, Maes T, Rebane A, Krauss-Etschmann S, Radinger M. Spotlight on microRNAs in allergy and asthma. Allergy 2021: 76(6): 1661–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alexandrova E, Miglino N, Hashim A, Nassa G, Stellato C, Tamm M, Baty F, Brutsche M, Weisz A, Borger P. Small RNA profiling reveals deregulated phosphatase and tensin homolog (PTEN)/phosphoinositide 3-kinase (PI3K)/Akt pathway in bronchial smooth muscle cells from asthmatic patients. J Allergy Clin Immunol 2016: 137(1): 58–67. [DOI] [PubMed] [Google Scholar]
- 14.Collison A, Herbert C, Siegle JS, Mattes J, Foster PS, Kumar RK. Altered expression of microRNA in the airway wall in chronic asthma: miR-126 as a potential therapeutic target. BMC Pulmonary Medicine 2011: 11(1): 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong X, Xu M, Ren Z, Gu J, Lu M, Lu Q, Zhong N. Regulation of CBL and ESR1 expression by microRNA-223p, 513a-5p and 625-5p may impact the pathogenesis of dust mite-induced pediatric asthma. Int J Mol Med 2016: 38(2): 446–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maes T, Cobos FA, Schleich F, Sorbello V, Henket M, De Preter K, Bracke KR, Conickx G, Mesnil C, Vandesompele J, Lahousse L, Bureau F, Mestdagh P, Joos GF, Ricciardolo FL, Brusselle GG, Louis R. Asthma inflammatory phenotypes show differential microRNA expression in sputum. J Allergy Clin Immunol 2016: 137(5): 1433–1446. [DOI] [PubMed] [Google Scholar]
- 17.Panganiban RP, Wang Y, Howrylak J, Chinchilli VM, Craig TJ, August A, Ishmael FT. Circulating microRNAs as biomarkers in patients with allergic rhinitis and asthma. J Allergy Clin Immunol 2016: 137(5): 1423–1432. [DOI] [PubMed] [Google Scholar]
- 18.Pinkerton M, Chinchilli V, Banta E, Craig T, August A, Bascom R, Cantorna M, Harvill E, Ishmael FT. Differential expression of microRNAs in exhaled breath condensates of patients with asthma, patients with chronic obstructive pulmonary disease, and healthy adults. J Allergy Clin Immunol 2013: 132(1): 217–219. [DOI] [PubMed] [Google Scholar]
- 19.Roffel MP, Boudewijn IM, van Nijnatten JLL, Faiz A, Vermeulen CJ, van Oosterhout AJ, Affleck K, Timens W, Bracke KR, Maes T, Heijink IH, Brandsma CA, van den Berge M. Identification of asthma-associated microRNAs in bronchial biopsies. Eur Respir J 2022: 59(3). [DOI] [PubMed] [Google Scholar]
- 20.Rupani H, Martinez-Nunez RT, Dennison P, Lau LC, Jayasekera N, Havelock T, Francisco-Garcia AS, Grainge C, Howarth PH, Sanchez-Elsner T. Toll-like Receptor 7 Is Reduced in Severe Asthma and Linked to an Altered MicroRNA Profile. Am J Respir Crit Care Med 2016: 194(1): 26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Solberg OD, Ostrin EJ, Love MI, Peng JC, Bhakta NR, Hou L, Nguyen C, Solon M, Nguyen C, Barczak AJ, Zlock LT, Blagev DP, Finkbeiner WE, Ansel KM, Arron JR, Erle DJ, Woodruff PG. Airway epithelial miRNA expression is altered in asthma. Am J Respir Crit Care Med 2012: 186(10): 965–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wardzynska A, Pawelczyk M, Rywaniak J, Makowska J, Jamroz-Brzeska J, Kowalski ML. Circulating miRNA expression in asthmatics is age-related and associated with clinical asthma parameters, respiratory function and systemic inflammation. Respir Res 2021: 22(1): 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kho AT, McGeachie MJ, Moore KG, Sylvia JM, Weiss ST, Tantisira KG. Circulating microRNAs and prediction of asthma exacerbation in childhood asthma. Respir Res 2018: 19(1): 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gomez JL, Chen A, Diaz MP, Zirn N, Gupta A, Britto C, Sauler M, Yan X, Stewart E, Santerian K, Grant N, Liu Q, Fry R, Rager J, Cohn L, Alexis N, Chupp GL. A Network of Sputum MicroRNAs Is Associated with Neutrophilic Airway Inflammation in Asthma. Am J Respir Crit Care Med 2020: 202(1): 51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, Panganiban R, Kho AT, McGeachie MJ, Farnam L, Chase RP, Weiss ST, Lu Q, Tantisira KG. Circulating MicroRNAs and Treatment Response in Childhood Asthma. Am J Respir Crit Care Med 2020: 202(1): 65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boudewijn IM, Roffel MP, Vermeulen CJ, Nawijn MC, Kok K, Terpstra MM, Koppelman GH, Guryev V, van den Berge M. A Novel Role for Bronchial MicroRNAs and Long Noncoding RNAs in Asthma Remission. Am J Respir Crit Care Med 2020: 202(4): 614–618. [DOI] [PubMed] [Google Scholar]
- 27.Hasegawa K, Mansbach JM, Ajami NJ, Espinola JA, Henke DM, Petrosino JF, Piedra PA, Shaw CA, Sullivan AF, Camargo CA Jr., the MARC-35 Investigators. Association of nasopharyngeal microbiota profiles with bronchiolitis severity in infants hospitalised for bronchiolitis. Eur Respir J 2016: 48(5): 1329–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stewart CJ, Mansbach JM, Wong MC, Ajami NJ, Petrosino JF, Camargo CA Jr., Hasegawa K. Associations of nasopharyngeal metabolome and microbiome with severity among infants with bronchiolitis. A multiomic analysis. Am J Respir Crit Care Med 2017: 196(7): 882–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ralston SL, Lieberthal AS, Meissner HC, Alverson BK, Baley JE, Gadomski AM, Johnson DW, Light MJ, Maraqa NF, Mendonca EA, Phelan KJ, Zorc JJ, Stanko-Lopp D, Brown MA, Nathanson I, Rosenblum E, Sayles S 3rd, Hernandez-Cancio S, American Academy of Pediatrics. Clinical practice guideline: the diagnosis, management, and prevention of bronchiolitis. Pediatrics 2014: 134(5): e1474–1502. [DOI] [PubMed] [Google Scholar]
- 30.Hasegawa K, Mansbach JM, Bochkov YA, Gern JE, Piedra PA, Bauer CS, Teach SJ, Wu S, Sullivan AF, Camargo CA Jr. Association of Rhinovirus C Bronchiolitis and Immunoglobulin E Sensitization During Infancy With Development of Recurrent Wheeze. JAMA Pediatr 2019: 173(6): 544–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luna PN, Hasegawa K, Ajami NJ, Espinola JA, Henke DM, Petrosino JF, Piedra PA, Sullivan AF, Camargo CA Jr., Shaw CA, Mansbach JM. The association between anterior nares and nasopharyngeal microbiota in infants hospitalized for bronchiolitis. Microbiome 2018: 6(1): 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mansbach JM, Luna PN, Shaw CA, Hasegawa K, Petrosino JF, Piedra PA, Sullivan AF, Espinola JA, Stewart CJ, Camargo CA Jr. Increased Moraxella and Streptococcus species abundance after severe bronchiolitis is associated with recurrent wheezing. J Allergy Clin Immunol 2020: 145(2): 518–527 e518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mjelle R, Aass KR, Sjursen W, Hofsli E, Saetrom P. sMETASeq: Combined Profiling of Microbiota and Host Small RNAs. iScience 2020: 23(5): 101131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin M Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011 2011: 17(1): 3 % J EMBnet.journal. [Google Scholar]
- 35.Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res 2019: 47(D1): D155–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tarallo S, Ferrero G, De Filippis F, Francavilla A, Pasolli E, Panero V, Cordero F, Segata N, Grioni S, Pensa RG, Pardini B, Ercolini D, Naccarati A. Stool microRNA profiles reflect different dietary and gut microbiome patterns in healthy individuals. Gut 2022: 71(7): 1302–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 2014: 15(12): 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raita Y, Perez-Losada M, Freishtat RJ, Hahn A, Castro-Nallar E, Ramos-Tapia I, Stearrett N, Bochkov YA, Gern JE, Mansbach JM, Zhu Z, Camargo CA, Hasegawa K. Nasopharyngeal metatranscriptome profiles of infants with bronchiolitis and risk of childhood asthma: a multicentre prospective study. Eur Respir J 2022: 60(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu Z, Camargo CA Jr., Raita Y, Freishtat RJ, Fujiogi M, Hahn A, Mansbach JM, Spergel JM, Perez-Losada M, Hasegawa K. Nasopharyngeal airway dual-transcriptome of infants with severe bronchiolitis and risk of childhood asthma: A multicenter prospective study. J Allergy Clin Immunol 2022: 150(4): 806–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujiogi M, Raita Y, Perez-Losada M, Freishtat RJ, Celedon JC, Mansbach JM, Piedra PA, Zhu Z, Camargo CA Jr., Hasegawa K Integrated relationship of nasopharyngeal airway host response and microbiome associates with bronchiolitis severity. Nat Commun 2022: 13(1): 4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 2017: 14(4): 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Camargo CA Jr., Ingham T, Wickens K, Thadhani R, Silvers KM, Epton MJ, Town GI, Pattemore PK, Espinola JA, Crane J, New Zealand A, Allergy Cohort Study G. Cord-blood 25-hydroxyvitamin D levels and risk of respiratory infection, wheezing, and asthma. Pediatrics 2011: 127(1): e180–187. [DOI] [PubMed] [Google Scholar]
- 43.Abreo A, Gebretsadik T, Stone CA, Hartert TV. The impact of modifiable risk factor reduction on childhood asthma development. Clin Transl Med 2018: 7(1): 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu Z, Camargo CA Jr., Raita Y, Fujiogi M, Liang L, Rhee EP, Woodruff PG, Hasegawa K. Metabolome subtyping of severe bronchiolitis in infancy and risk of childhood asthma. J Allergy Clin Immunol 2022: 149(1): 102–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995: 57(1): 289–300. [Google Scholar]
- 46.Zhu Z, Lee PH, Chaffin MD, Chung W, Loh PR, Lu Q, Christiani DC, Liang L. A genome-wide cross-trait analysis from UK Biobank highlights the shared genetic architecture of asthma and allergic diseases. Nat Genet 2018: 50(6): 857–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ludwig N, Leidinger P, Becker K, Backes C, Fehlmann T, Pallasch C, Rheinheimer S, Meder B, Stahler C, Meese E, Keller A. Distribution of miRNA expression across human tissues. Nucleic Acids Res 2016: 44(8): 3865–3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Juzenas S, Venkatesh G, Hubenthal M, Hoeppner MP, Du ZG, Paulsen M, Rosenstiel P, Senger P, Hofmann-Apitius M, Keller A, Kupcinskas L, Franke A, Hemmrich-Stanisak G. A comprehensive, cell specific microRNA catalogue of human peripheral blood. Nucleic Acids Res 2017: 45(16): 9290–9301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reczko M, Maragkakis M, Alexiou P, Grosse I, Hatzigeorgiou AG. Functional microRNA targets in protein coding sequences. Bioinformatics 2012: 28(6): 771–776. [DOI] [PubMed] [Google Scholar]
- 50.Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T, Hatzigeorgiou AG. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res 2015: 43(W1): W460–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Finn PW, Bigby TD. Innate immunity and asthma. Proc Am Thorac Soc 2009: 6(3): 260–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karner J, Wawrzyniak M, Tankov S, Runnel T, Aints A, Kisand K, Altraja A, Kingo K, Akdis CA, Akdis M, Rebane A. Increased microRNA-323-3p in IL-22/IL-17-producing T cells and asthma: a role in the regulation of the TGF-beta pathway and IL-22 production. Allergy 2017: 72(1): 55–65. [DOI] [PubMed] [Google Scholar]
- 53.Zheng R, Du M, Tian M, Zhu Z, Wei C, Chu H, Gan C, Liang J, Xue R, Gao F, Mao Z, Wang M, Zhang Z. Fine Particulate Matter Induces Childhood Asthma Attacks via Extracellular Vesicle-Packaged Let-7i-5p-Mediated Modulation of the MAPK Signaling Pathway. Adv Sci (Weinh) 2022: 9(3): e2102460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu C, Makrinioti H, Saglani S, Bowman M, Lin LL, Camargo CA Jr., Hasegawa K, Zhu Z. Microbial dysbiosis and childhood asthma development: Integrated role of the airway and gut microbiome, environmental exposures, and host metabolic and immune response. Front Immunol 2022: 13: 1028209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Makrinioti H, Camargo CA, Zhu Z, Freishtat RJ, Hasegawa K. Air pollution, bronchiolitis, and asthma: the role of nasal microRNAs. Lancet Respir Med 2022: 10(8): 733–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Makrinioti H, Hasegawa K, Lakoumentas J, Xepapadaki P, Tsolia M, Castro-Rodriguez JA, Feleszko W, Jartti T, Johnston SL, Bush A, Papaevangelou V, Camargo CA Jr., Papadopoulos NG. The role of respiratory syncytial virus- and rhinovirus-induced bronchiolitis in recurrent wheeze and asthma-A systematic review and meta-analysis. Pediatr Allergy Immunol 2022: 33(3): e13741. [DOI] [PubMed] [Google Scholar]
- 57.Hasegawa K, Perez-Losada M, Hoptay CE, Epstein S, Mansbach JM, Teach SJ, Piedra PA, Camargo CA Jr., Freishtat RJ. RSV vs. rhinovirus bronchiolitis: difference in nasal airway microRNA profiles and NFkappaB signaling. Pediatr Res 2018: 83(3): 606–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leon-Icaza SA, Zeng M, Rosas-Taraco AG. microRNAs in viral acute respiratory infections: immune regulation, biomarkers, therapy, and vaccines. ExRNA 2019: 1(1): 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Manti S, Piedimonte G. An overview on the RSV-mediated mechanisms in the onset of non-allergic asthma. Front Pediatr 2022: 10: 998296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thornburg NJ, Hayward SL, Crowe JE Jr. Respiratory syncytial virus regulates human microRNAs by using mechanisms involving beta interferon and NF-kappaB. mBio 2012: 3(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Angria N, Massi MN, Bukhari A, Djaharuddin I, Jumadi O, Ahmad A, Miskad UA, Ladju RB, Santoso A, Halik H. Expression of miRNA-29a-3p and IFN-gamma as biomarkers in active and latent pulmonary tuberculosis. Ann Med Surg (Lond) 2022: 83: 104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guo S, Chen R, Zhang L, Wu M, Wei Y, Dai W, Jiang Y, Kong X. microRNA-22-3p plays a protective role in a murine asthma model through the inhibition of the NLRP3-caspase-1-IL-1beta axis. Exp Physiol 2021: 106(8): 1829–1838. [DOI] [PubMed] [Google Scholar]
- 63.Yoo EJ, Ojiaku CA, Sunder K, Panettieri RA Jr. Phosphoinositide 3-Kinase in Asthma: Novel Roles and Therapeutic Approaches. Am J Respir Cell Mol Biol 2017: 56(6): 700–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yon C, Thompson DA, Jude JA, Panettieri RA Jr., Rastogi D Crosstalk Between CD4+ T Cells and Airway Smooth Muscle in Pediatric Obesity-related Asthma. Am J Respir Crit Care Med 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pan J, Yang Q, Zhou Y, Deng H, Zhu Y, Zhao D, Liu F. MicroRNA-221 Modulates Airway Remodeling via the PI3K/AKT Pathway in OVA-Induced Chronic Murine Asthma. Front Cell Dev Biol 2020: 8: 495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goncharova EA, Ammit AJ, Irani C, Carroll RG, Eszterhas AJ, Panettieri RA, Krymskaya VP. PI3K is required for proliferation and migration of human pulmonary vascular smooth muscle cells. Am J Physiol Lung Cell Mol Physiol 2002: 283(2): L354–363. [DOI] [PubMed] [Google Scholar]
- 67.Krymskaya VP, Ammit AJ, Hoffman RK, Eszterhas AJ, Panettieri RA Jr. Activation of class IA PI3K stimulates DNA synthesis in human airway smooth muscle cells. Am J Physiol Lung Cell Mol Physiol 2001: 280(5): L1009–1018. [DOI] [PubMed] [Google Scholar]
- 68.Humbert M, Bousquet J, Bachert C, Palomares O, Pfister P, Kottakis I, Jaumont X, Thomsen SF, Papadopoulos NG. IgE-Mediated Multimorbidities in Allergic Asthma and the Potential for Omalizumab Therapy. J Allergy Clin Immunol Pract 2019: 7(5): 1418–1429. [DOI] [PubMed] [Google Scholar]
- 69.Dakhama A, Lee YM, Ohnishi H, Jing X, Balhorn A, Takeda K, Gelfand EW. Virus-specific IgE enhances airway responsiveness on reinfection with respiratory syncytial virus in newborn mice. J Allergy Clin Immunol 2009: 123(1): 138–145 e135. [DOI] [PubMed] [Google Scholar]
- 70.Durrani SR, Montville DJ, Pratt AS, Sahu S, DeVries MK, Rajamanickam V, Gangnon RE, Gill MA, Gern JE, Lemanske RF Jr., Jackson DJ. Innate immune responses to rhinovirus are reduced by the high-affinity IgE receptor in allergic asthmatic children. J Allergy Clin Immunol 2012: 130(2): 489–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Crosson T, Wang JC, Doyle B, Merrison H, Balood M, Parrin A, Pascal M, Mindt BC, Seehus CR, Ozcan A, Huang X, Semenara E, Lai NYY, Majdoubi A, Abdulnour RE, Rajchgot T, Rafei M, Foster SL, Thibodeau J, Fritz JH, Levy BD, Woolf CJ, Talbot S. FcepsilonR1-expressing nociceptors trigger allergic airway inflammation. J Allergy Clin Immunol 2021: 147(6): 2330–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ross SA, Davis CD. MicroRNA, nutrition, and cancer prevention. Adv Nutr 2011: 2(6): 472–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Poole A, Urbanek C, Eng C, Schageman J, Jacobson S, O’Connor BP, Galanter JM, Gignoux CR, Roth LA, Kumar R, Lutz S, Liu AH, Fingerlin TE, Setterquist RA, Burchard EG, Rodriguez-Santana J, Seibold MA. Dissecting childhood asthma with nasal transcriptomics distinguishes subphenotypes of disease. J Allergy Clin Immunol 2014: 133(3): 670–678 e612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Midulla F, Di Mattia G, Nenna R, Scagnolari C, Viscido A, Oliveto G, Petrarca L, Frassanito A, Arima S, Antonelli G, Pierangeli A. Novel Variants of Respiratory Syncytial Virus A ON1 Associated With Increased Clinical Severity of Bronchiolitis. J Infect Dis 2020: 222(1): 102–110. [DOI] [PubMed] [Google Scholar]
- 75.Manti S, Esper F, Alejandro-Rodriguez M, Leonardi S, Betta P, Cuppari C, Lanzafame A, Worley S, Salpietro C, Perez MK, Rezaee F, Piedimonte G. Respiratory syncytial virus seropositivity at birth is associated with adverse neonatal respiratory outcomes. Pediatr Pulmonol 2020: 55(11): 3074–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


